Abstract
Bacterial glycogen is a polyglucose storage compound that is thought to prolong viability during stationary phase. However, a specific role for glycogen has not been determined. We have characterized SMEG53, a temperature-sensitive mutant of Mycobacterium smegmatis that contains a mutation in glgE, encoding a putative glucanase. This mutation causes exponentially growing SMEG53 cells to stop growing at 42°C in response to high levels of glycogen accumulation. The mutation in glgE is also associated with an altered growth rate and colony morphology at permissive temperatures; the severity of these phenotypes correlates with the amount of glycogen accumulated by the mutant. Suppression of the temperature-sensitive phenotype, via a decrease in glycogen accumulation, is mediated by growth in certain media or multicopy expression of garA. The function of GarA is unknown, but the presence of a forkhead-associated domain suggests that this protein is a member of a serine-threonine kinase signal transduction pathway. Our results suggest that in M. smegmatis glycogen is continuously synthesized and then degraded by GlgE throughout exponential growth. In turn, this constant recycling of glycogen controls the downstream availability of carbon and energy. Thus, in addition to its conventional storage role, glycogen may also serve as a carbon capacitor for glycolysis during the exponential growth of M. smegmatis.
Glycogen is a polysaccharide composed of glucose in an α-1,4-linked linear arrangement with α-1,6 branches. Bacterial glycogen is generally considered a storage compound because it accumulates in stationary phase and under growth-limiting conditions (reviewed in reference 22). Presumably, glycogen serves as a reservoir of carbon and energy during times of starvation. Consistent with this idea, some bacterial mutants unable to produce glycogen have decreased survival under carbon starvation conditions relative to wild-type strains (22). However, the particular physiological role of glycogen has not been resolved. In bacteria such as Bacillus subtilis and Streptomyces coelicolor, glycogen synthesis is associated with sporulation and may provide the resources necessary to drive differentiation (10, 17, 19).
The genetic aspects of glycogen synthesis have been studied intensively in Escherichia coli. Two glycogen-related gene clusters, glgBX and glgCAP, have been characterized (reviewed in reference 23). The genes glgC, glgA, and glgB encode the biosynthetic enzymes ADP-glucose pyrophosphorylase, glycogen synthase, and branching enzyme, respectively, while glgP and glgX encode the catabolic enzymes glycogen phosphorylase and debranching enzyme, respectively (23, 28). A variety of growth-limiting conditions, including low nitrogen, phosphate, or sulfur availability in the presence of excess carbon, promote glycogen synthesis (22). Therefore, the amount of glycogen accumulated by E. coli involves the integration of many physiological signals, and accordingly, glycogen synthesis is a highly regulated process. The global regulatory systems that control glycogen synthesis include catabolite repression, the stringent response, ςs, and csrA (23). Glycogen synthesis is also regulated by the allosteric regulation of ADP-glucose pyrophosphorylase (23).
Earlier work done on mycobacterial glycogen suggested that the features of glycogen accumulation in mycobacteria were similar to those required for other bacteria (2–4, 13). However, no detailed genetic or molecular studies pertaining to glycogen have been reported. Here, we report the characterization of SMEG53, a temperature-sensitive mutant of Mycobacterium smegmatis that inappropriately accumulates glycogen during exponential growth. The growth defect at 42°C is due to a mutation in glgE, a glycogen-associated gene that encodes a putative glucanase. The temperature-sensitive phenotype of SMEG53 can be suppressed by the multicopy expression of garA, a novel effector of glycogen accumulation, or by growth on alternate media. The genetic and phenotypic data for SMEG53 suggest that in M. smegmatis, carbon flows preferentially through a glycogen-recycling system prior to its use in cellular biosynthesis and energy production.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in these studies are given in Table 1. The M. smegmatis mc2155 chromosomal DNA library was a gift from William Jacobs, Jr. (Albert Einstein School of Medicine). The M. tuberculosis H37Rv chromosomal DNA library was a gift from Julia Inamine (Colorado State University). The media used for mycobacterial propagation were Middlebrook 7H9 and 7H10 with ADC supplement (Difco, Detroit, MI). E. coli was grown in Luria broth (Difco, Detroit, Mich.). Antibiotic concentrations were as follows: kanamycin, 10 (mycobacteria) or (E. coli) 25 μg/ml; hygromycin, 100 μg/ml; and carbenicillin, 100 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| M. smegmatis | ||
| mc2155 | ept1 glgE+ | 25 |
| SMEG53 | ept1 glgE1(Ts) | This study |
| E. coli XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| Plasmids | ||
| pMD30 | Extrachromosomal E. coli-mycobacterial shuttle vector | 12 |
| pMH94 | Mycobacterial integrating vector | 18 |
| pAEB225 | M. smegmatis-derived cosmid containing garA | This study |
| pAEB226 | M. smegmatis-derived cosmid containing glgE | This study |
| pAEB234 | 1.7-kb Sau3AI fragment containing garA cloned into pMD30 | This study |
| pAEB235 | 2.4-kb Sau3AI fragment containing glgE cloned into pMD30 | This study |
| pAEB236 | EcoRI/XbaI insert fragment from pAEB234 cloned into pMH94 | This study |
| pAEB237 | M. tuberculosis-derived cosmid containing glgE | This study |
| pAEB238 | M. tuberculosis-derived cosmid containing glgE | This study |
| pAEB239 | KpnI/XbaI insert fragment of pAEB235 cloned into pMH94 | This study |
Mutant generation.
Chemical mutagenesis was performed by using a method developed for Streptomyces (5), with some modifications. A culture of M. smegmatis mc2155 was grown to mid-log phase (optical density at 600 nm [OD600] = 0.8) at 37°C. Following adjustment of the culture pH to 8.5, nitrosoguanidine was added to a final concentration of 100 μg/ml. The cells were exposed to the mutagen for 20 min with shaking at 37°C. Mutagenesis was stopped by centrifuging the cultures at 3,000 × g for 10 min and removing the supernatant. Following resuspension in fresh medium, the bacterial chromosomes were allowed to segregate for 4 h at 30°C. Approximately 300 CFU of mutagenized mycobacteria was spread on 7H10 containing 0.05% Tween 80 and then incubated for 5 to 7 days at 30°C. Individual mutagenized colonies were evaluated for temperature sensitivity at 42°C by replica plating using Accutran filters (Schleicher & Schuell, Keene, N.H.). The colonies that were able to grow at 30°C but not at 42°C were rescreened to confirm the temperature-sensitive phenotype.
Genetic complementation.
SMEG53 was electroporated with an M. smegmatis mc2155 or M. tuberculosis H37Rv extrachromosomal genomic library, using conditions described previously (16). A proportion of the cells were plated at 30°C as an electroporation control, and the remainder were plated at 42°C. The cosmid DNA from colonies able to grow at 42°C was recovered from SMEG53 by electroduction (6). The M. smegmatis genes restoring growth at 42°C to SMEG53 were identified by first constructing sublibraries of pAEB225 and pAEB226. Cosmids were partially digested with Sau3AI, and fragments of 1 to 5 kb were ligated into the BamHI site of pMD30. SMEG53 was reelectroporated with a pool of the resulting constructs followed by selection at 42°C. Subclones able to complement SMEG53 were isolated by electroduction and then sequenced. The proportion of original cosmid constructs containing garA was determined with PCR using the oligonucleotide primers 5′-AAGACAGCAATTTGGGGG-3′ (forward) and 5′-ATGGGTCATCGGCTGTTC-3′ (reverse). The fragment was amplified with Pfu polymerase (Stratagene, La Jolla, Calif.) according to the manufacturer’s specifications. The integrating plasmid containing garA was constructed by removing the insert DNA of pAEB234 with EcoRI and XbaI digestion and then ligating this fragment into similarly digested pMH94. The integrating plasmid containing glgE was constructed by removing the insert DNA of pAEB235 with a KpnI and XbaI digestion, and then ligating this fragment into similarly digested pMH94.
Sequence analysis.
DNA constructs were sequenced with both vector- and insert-specific primers, using an ABI310 automated sequencer with the Taq FS dye-terminator ready reaction mix (Perkin-Elmer, Foster City, Calif.) according to the manufacturer’s specifications. DNA sequences were assembled by using the Sequencher package (GeneCodes, Ann Arbor, Mich.) and analyzed with BLAST (1), PROSITE (15), CLUSTAL W (27), and McBoxshade version 2.11. To determine the sequence of the SMEG53 glgE allele, the gene was amplified by using primers 5′-TTACTGACAAATCCCGCATCC-3′ (forward) and 5′-CTGCTTCTCGTCATCTCGCC-3′ (reverse). A single colony of SMEG53 was suspended in 100 μl of water and then boiled for 5 min. One microliter of the boiled cell mixture was used as the template DNA in the PCR. The DNA was amplified with Pfu polymerase (Stratagene) according to the manufacturer’s specifications in the presence of 2.5% dimethyl sulfoxide. The PCR product was isolated from agarose gels and purified by using a gel extraction kit from Qiagen, Chatsworth, Calif. This purified PCR product was then used in subsequent DNA sequencing reactions.
Glycogen assays.
Glycogen assays were performed on whole cells, using α-amylase and glucose oxidation quantitation according to a previously published protocol (21). Bacterial cells were grown under the appropriate conditions to mid-log phase (OD600 = 0.75 to 0.85) or to saturation, harvested by centrifugation at 3,000 × g for 10 min, and then washed once in an equal volume of water. The resulting cell pellet was stored at −80°C until required. For temperature shift experiments, exponential-phase cells grown at 30°C were diluted to an OD600 of approximately 0.3 in fresh medium and then grown at 42°C to an OD600 of 0.75 to 0.85.
Nucleotide sequence accession numbers.
The DNA sequences of garA and glgE derived from M. smegmatis mc2155 have been deposited in GenBank under accession no. AF173844 and AF172946, respectively.
RESULTS
Phenotypic characterization.
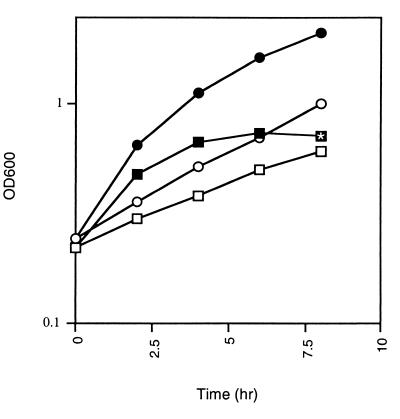
Strain SMEG53 was originally isolated as part of a temperature-sensitive mutant bank of M. smegmatis mc2155 that was generated by nitrosoguanidine mutagenesis (7a). Besides temperature sensitivity, SMEG53 displays several other interesting phenotypic characteristics, including an altered colony morphology (see below) and slightly slower growth at the permissive temperature (Table 2). Another trait of SMEG53 is the suppression of the temperature-sensitive phenotype when it is plated on certain growth media. While SMEG53 is unable to grow at 42°C on 7H10, the medium originally used for the isolation of the temperature-sensitive mutants, it can form colonies when plated on M9 or 7H10 containing an osmolyte such as sucrose or NaCl (data not shown). An interesting phenotype of SMEG53 is evident from the growth curves performed on temperature-shifted cultures. When an exponential-phase culture of SMEG53 is grown at 30°C and then shifted to 42°C, the culture doubles once and then growth ceases. In contrast, the wild-type culture continues to grow exponentially (Fig. 1). The temperature-dependent growth cessation of SMEG53 is reversible, because when the cells are shifted back to 30°C, growth resumes (data not shown). Microscopic examination revealed that SMEG53 cells that had stopped growing for several hours at 42°C were shorter than actively growing mutant cells, which are the same size as wild-type cells (data not shown). We also observed that the growth cessation of SMEG53 at 42°C coincided with a cellular clumping of the culture. These changes in cellular morphology and culture properties are similar to the changes that occur in stationary-phase M. smegmatis cells, particularly those starved for carbon (24). Since the molecular events associated with stationary phase in M. smegmatis have not been characterized, we could not further demonstrate that the mutant enters stationary phase at 42°C.
TABLE 2.
Phenotypic characteristics of M. smegmatis mc2155 and SMEG53
| Strain | Growth temp (°C) | Growth on 7H10 | Glycogen contenta | Doubling time (h)b |
|---|---|---|---|---|
| mc2155 | 30 | + | 7.6 ± 0.4 | 3.4 ± 0.3 (1) |
| 37 | + | 5.8 ± 0.4 | 2.3 ± 0.3 (1) | |
| 42 | + | 10.2 ± 0.2 | 1.3 ± 0.1 (1) | |
| SMEG53 | 30 | + | 13.4 ± 0.7 | 4.6 ± 0.1 (0.7) |
| 37 | + | 117.9 ± 2.8 | 6.0 ± 0.4 (0.4) | |
| 42 | −c | 176.9 ± 5.5 | NDd |
For glycogen determinations, bacterial cells were grown to an OD600 of 0.75 to 0.85 in 7H9 broth. For glycogen quantitation at 42°C, exponential cultures grown at 30°C were diluted to an OD600 of 0.3 and then incubated at 42°C to an OD600 of 0.75 to 0.85. Glycogen was quantitated as nanomoles of free glucose liberated by α-amylase treatment per gram (wet weight) of cells. Each value represents the mean of three assays ± standard deviation.
Calculated from exponential-phase cultures grown in 7H9. Relative growth rates are shown in parentheses.
SMEG53 can grow at 42°C on M9 and 7H9 containing 0.2 M NaCl.
ND, a doubling time could not be accurately determined.
FIG. 1.
Growth curves of SMEG53 and mc2155 cultures at 30°C following a temperature shift from 30 to 42°C. Cells grown exponentially at 30°C were diluted to an OD600 of 0.3 in fresh medium; and then one half was incubated at 30°C, and the other half was incubated at 42°C. Cell densities were determined at OD600 over the time period shown. Samples: SMEG53, 30°C (□); SMEG53, 42°C (■); mc2155, 30°C (○); mc2155, 42°C (●). Shown is a representative example of three independent experiments. The time point where the morphological change is first noted in SMEG53 at 42°C is indicated by the symbol containing the asterisk.
Genetic complementation.
To identify the mutation that causes the temperature-sensitive phenotype in SMEG53, we electroporated the strain with an extrachromosomal M. smegmatis mc2155 genomic DNA library and then selected for cosmids that could restore the ability of the mutant to grow at 42°C. Transformants able to grow at 42°C were recovered at a frequency of 0.2%. Cosmid DNA was isolated from eight of the colonies and then analyzed by restriction enzyme analysis. Based on restriction enzyme patterns, it appeared that two distinct genetic regions restored high-temperature growth to SMEG53 (data not shown). To identify the specific genes involved, sublibraries were constructed from two unrelated cosmids, pAEB225 and pAEB226, and then electroporated into SMEG53. Subclones that restored wild-type growth to SMEG53 at 42°C were isolated from each library. Subsequent DNA sequence analysis of these clones revealed that two different genes restored the ability of SMEG53 to grow at 42°C.
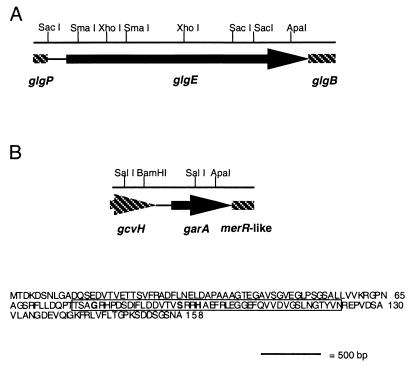
One of the M. smegmatis genes that restores the ability of SMEG53 to grow normally at 42°C is located on plasmid pAEB235. This gene encodes a 78.2-kDa protein that is the apparent homolog of Rv1327c, an M. tuberculosis protein with similarity to glycosyl hydrolases of the α-amylase family (11). The two proteins share an overall similarity of 79%, but the Rv1327c gene product is 40 amino acids larger, the significance of which is unknown (Fig. 2). The M. smegmatis gene product also shares 64% similarity with the Pep1 isoenzymes of S. coelicolor (Fig. 2; reference 10). The pep1 genes are located in a glycogen and trehalose biosynthetic cluster and appear to be involved in polysaccharide catabolism. The genes encoding Rv1327c and the M. smegmatis homolog are also closely linked to genes involved in glycogen metabolism and catabolism. The gene encoding Rv1327c is arranged in an operon with glgB, the gene encoding glycogen branching enzyme, and is immediately adjacent to glgP, the gene encoding glycogen phosphorylase. This gene order has also been conserved in M. smegmatis (Fig. 3A). The similarity of the M. smegmatis gene product to polysaccharide-degrading enzymes and its proximity to other glycogen-associated genes suggest that this protein is involved in glycogen catabolism. For this reason, and in view of other data given below, we have designated this M. smegmatis gene glgE.
FIG. 2.
Amino acid similarities between the M. smegmatis glgE gene product (SMEG), the M. tuberculosis homolog Rv1327c (TB), and one of the S. coelicolor pep1 gene products (PEP1). Alignments were performed with CLUSTAL W, and the amino acid shading was performed with McBoxshade version 2.11. Black shading depicts amino acid residues conserved in all three of the proteins; grey shading depicts residues conserved in two. The histidine mutated in the SMEG53 glgE gene product is indicated by the asterisk.
FIG. 3.
Genetic organization of the SMEG53 complementing genes glgE (A) and garA (B). The region shown depicts the smallest amount of DNA required for the restoration of high-temperature growth to SMEG53 based on subclone insert size or overlapping clone analysis. Shadings denote complete (black) and partial (shaded) open reading frames. Arrows indicate direction of transcription, and lines depict intervening DNA. Also shown is the putative gene product of garA. The FHA domain predicted by PROSITE is boxed, and the three invariant amino acid residues associated with this domain are shown in bold.
The second gene that restores the growth of SMEG53 at 42°C is located on plasmid pAEB234. This gene encodes a 16.6-kDa protein product with 89% overall similarity to the M. tuberculosis protein Rv1827 (11) and 87% overall similarity to the M. leprae protein MLCB1788.36c (accession no. AL008609). The M. smegmatis gene product was predicted to contain a forkhead-associated (FHA) domain (14) at amino acid positions 76 to 125 when used in a search against the PROSITE (15) database. This protein domain mediates the recognition of a phosphorylated partner among members of serine-threonine kinase signal transduction pathways (26). The region corresponding to the FHA domain in the M. smegmatis protein product contains the three invariant glycine, serine, and histidine residues found in all FHA domain-containing proteins (reference 26; Fig. 3B). Database searches using BLAST also revealed that the M. smegmatis gene product has similarity to several of the eukaryotic and bacterial proteins originally used to define the FHA domain such as FraH, CDS1, and KAPP (14) and other mycobacterial proteins that apparently contain this protein motif such as EmbR (7). Another feature of the predicted protein, with unknown significance, is an acidic N-terminal domain (12 of 75 amino acids are aspartate or glutamate). In all three mycobacterial species, the gene encoding the FHA domain-containing protein is the first gene in an operon with another gene of unknown function. The protein product of the second gene has some similarity at the N terminus to the MerR family of transcriptional activators, but computer searches did not reveal a helix-turn-helix motif. Upstream of the gene encoding the FHA domain-containing protein is gcvH, a gene that encodes a protein involved in glycine degradation (Fig. 3B). Given the regulatory role of other proteins containing FHA domains, and our subsequent findings detailed below, we will refer to the second M. smegmatis gene that restores high temperature growth to SMEG53 as garA (glycogen accumulation regulator).
Analysis of the eight original cosmids isolated during genetic complementation studies using PCR and restriction enzyme digestions revealed that two of the constructs contained garA and the remaining six contained glgE (data not shown). Because the majority of the cosmids contained glgE, we looked for the temperature-sensitive mutation in the SMEG53 allele of this gene. Sequence analysis of the SMEG53 glgE allele revealed that the temperature-sensitive mutant has a histidine-to-tyrosine change at amino acid 349 of the protein product. The histidine residue that is mutated in SMEG53 is conserved in the M. tuberculosis Rv1327c protein and the S. coelicolor Pep1 isozymes (Fig. 2). If glgE is the true complementing gene, then the garA gene must act as a multicopy suppressor. To test this hypothesis, garA and glgE were each cloned into an integrating vector to create pAEB236 and pAEB239, respectively. The resulting plasmids were then used to construct partial diploids of SMEG53. Unlike pAEB234, the garA construct made with an extrachromosomal vector, pAEB236 was unable to restore normal growth to SMEG53 at 42°C. In contrast, pAEB239 could restore wild-type growth to SMEG53 as well as pAEB235, the glgE construct made with an extrachromosomal vector (data not shown). Thus, the temperature sensitivity of SMEG53 is most likely attributable to the mutation in the glycogen-associated gene glgE, and the growth defect caused by this mutation can be suppressed by multiple copies of garA.
To determine if any M. tuberculosis genes could restore the growth of SMEG53 at 42°C, the mutant was also electroporated with an extrachromosomal cosmid library of M. tuberculosis H37Rv. Following selection at 42°C, transformants that could grow at the nonpermissive temperature were found to occur at a frequency of 0.5%. The cosmids from 10 of these colonies were isolated and analyzed by restriction enzyme analysis. The restriction patterns obtained for these constructs indicated that all 10 cosmids represented a single genetic region (data not shown). The DNA sequence was determined at either end of the cosmid insert for the constructs pAEB237 and pAEB238. Database searches with this partial sequence information enabled us to map the location of the cosmid inserts within the genome sequence of M. tuberculosis H37Rv. The DNA region common to the cosmids examined was found to contain the Rv1327c gene, the M. tuberculosis glgE homolog. The M. tuberculosis H37Rv garA homolog, Rv1827, is located elsewhere on the chromosome and is not present in these DNA constructs. Thus, the complementation and genetic studies suggest that the M. tuberculosis Rv1327c gene product can functionally substitute for the M. smegmatis glgE gene product.
SMEG53 accumulates glycogen in a temperature-dependent manner.
The identification of the H349Y mutation in the SMEG53 glgE gene product suggested that a perturbation in glycogen levels may be responsible for the temperature-sensitive phenotype of this strain. Therefore, we examined the glycogen content of wild-type and SMEG53 exponential-phase cells grown at various temperatures. At the permissive temperature of 30°C, SMEG53 accumulated approximately twofold more glycogen than the wild-type cells (Table 2). Mutant cells grown at 37°C, a temperature that is permissive for SMEG53 growth, accumulated 20-fold more glycogen than wild-type cells treated similarly (Table 2). Cells that were grown exponentially at 30°C and then shifted to 42°C were also examined for glycogen accumulation. For these studies, the SMEG53 cells were incubated at 42°C until the optical density of the culture no longer increased. Under these conditions, SMEG53 cells accumulated approximately 18-fold more glycogen than the wild-type cells (Table 2). It is noteworthy that both the mutant and the wild type accumulated glycogen in a temperature-dependent manner. Therefore, while the ratio of glycogen accumulation between SMEG53 and the wild-type are similar at 37 and 42°C, the absolute level of glycogen accumulation is highest in the mutant at 42°C (Table 2). We also examined the glycogen content of stationary-phase SMEG53 and wild-type cells grown at 30°C. At this temperature, the overall growth dynamics of both strains are similar, and they reach saturation at the same optical density. In stationary phase, SMEG53 and the wild-type contain similar levels of glycogen (15.6 ± 2.1 and 15.0 ± 0.8, respectively, nmol of free glucose liberated per g of cells). Taken together, these data are consistent with the idea that the H349Y mutation in the glgE gene product of SMEG53 results in a temperature-dependent accumulation of glycogen in this strain during exponential growth. Presumably, with the putative degradative role of GlgE impaired, glycogen synthesis continues unchecked in actively growing SMEG53 cells, and the growth of the mutant ceases. The fact that glycogen can accumulate in SMEG53 implies that there is a continuous synthesis and breakdown of glycogen in actively growing M. smegmatis cells under normal culture conditions.
Growth of SMEG53 is restored by suppression of glycogen accumulation.
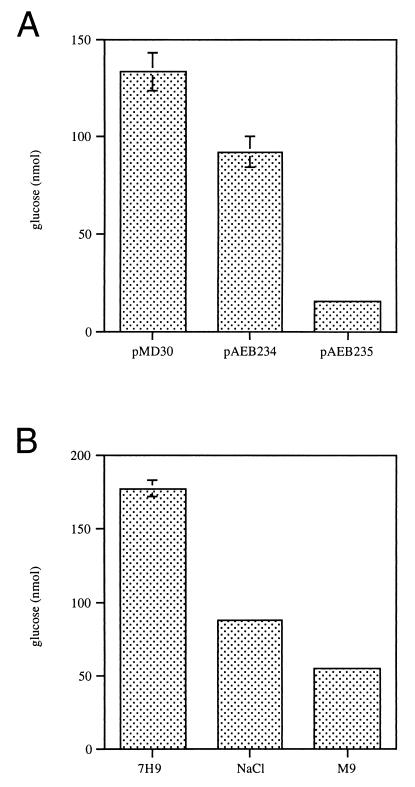
Our data are consistent with the idea that the temperature sensitivity of SMEG53 results from insufficient GlgE-mediated glycogen degradation. If this hypothesis is true, then the restoration of growth to SMEG53 at 42°C by glgE should be accompanied by near-normal levels of glycogen. Indeed, at the nonpermissive temperature the glycogen content of SMEG53 cells containing a plasmid-encoded wild-type copy of glgE was significantly lower than that of mutant cells containing only the cloning vector (Fig. 4A).
FIG. 4.
Glycogen content of SMEG53 grown under conditions suppressing the temperature-sensitive phenotype. Exponentially growing cultures were diluted to an OD600 of 0.3 in fresh medium and then shifted to 42°C. Cells were grown to mid-log phase (OD600 = 0.75 to 0.85), and then glycogen assays performed as before. Each value represents the mean of three assays ± standard deviation. (A) Mutant cells containing pMD30, pAEB234 (garA), or pAEB235 (glgE), grown in 7H9 containing 10 μg of kanamycin per ml. (B) SMEG53 cells grown in 7H9, 7H9 containing 0.2 M NaCl (NaCl), or M9.
Several conditions were found to suppress the temperature sensitivity of SMEG53 but not the differences in growth rate or colony morphology. One of these conditions was the multicopy expression of the garA gene. To better understand why suppression occurs, we compared the glycogen content at 42°C of SMEG53 cells containing an extrachromosomal copy of garA with that of cells carrying the vector alone. The glycogen content of SMEG53 cells overexpressing garA was approximately 1.5-fold lower than that found in cells containing only the cloning vector (Fig. 4A). Although lower, the glycogen content of cells containing garA was still approximately sixfold higher than that of cells containing an extrachromosomal copy of glgE (Fig. 4A). These results indicate that suppression by garA does not arise from tolerance of high glycogen levels. Rather, suppression occurs because the amount of accumulated glycogen falls below a putative threshold level associated with growth cessation. The glycogen content of SMEG53 cells grown at the nonpermissive temperature in 7H9 containing 0.2 M NaCl or M9 was also examined. The glycogen levels of SMEG53 cells grown in M9 or 7H9 containing 0.2 M NaCl at 42°C were two- and threefold lower, respectively, than the glycogen levels found previously in mutant cells grown in 7H9 at 42°C (Fig. 4B). Thus, the mechanism of suppression appears to be the same for SMEG53 cells grown in these alternate media and mutant cells overexpressing garA.
Glycogen accumulation is responsible for altered growth rates and morphology.
As noted above, SMEG53 concomitantly accumulates almost twofold more glycogen and grows at approximately 70% of the rate of the wild-type strain at 30°C. To demonstrate that it is the increase in glycogen accumulation that influences the growth rate of SMEG53, we determined the doubling times of the mutant and wild-type cells grown at 37°C. As previously established, SMEG53 cells accumulate approximately 20-fold more glycogen than the wild-type cells at this temperature. If glycogen does adversely affect growth rate, then the mutant should grow even more slowly than the wild type at 37°C than it does at 30°C. The doubling time of the wild-type strain decreases as the growth temperature increased from 30 to 37°C, but the doubling time of the mutant actually increases at the higher temperature (Table 2). Comparison of the growth rates showed that the SMEG53 cells grow at only 40% of the rate of the wild-type cells at 37°C (Table 2). Thus, as the glycogen levels of SMEG53 increase, the growth rate of the strain decreases.
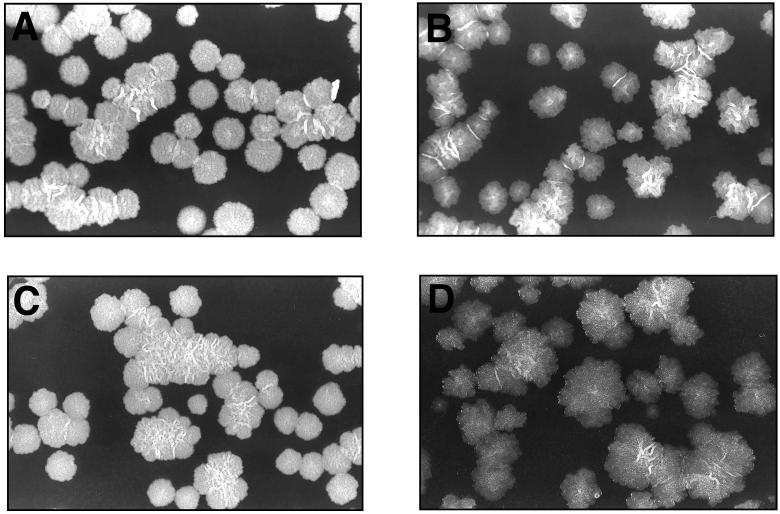
As mentioned earlier, SMEG53 exhibits an altered colony morphology at the permissive temperature. In comparison with the wild-type colonies, SMEG53 colonies grown at 30°C appear slightly transparent with irregular borders (Fig. 5A and B). To demonstrate that glycogen accumulation in exponential phase correlates with colony morphology, we examined SMEG53 cells containing either glgE or garA grown at 42°C. Since mutant cells containing glgE have low levels of glycogen at the nonpermissive temperature, we would expect the colony morphology of these cells to be indistinguishable from the wild-type cell morphology. This was indeed the case (Fig. 5C). Conversely, as we had previously demonstrated, SMEG53 cells transformed with garA and grown at 42°C still contain approximately six times more glycogen than mutant cells containing glgE and grown at 42°C. Therefore, if glycogen accumulation influences colony morphology, we would expect these cells to show a more pronounced change in morphology in comparison with SMEG53 cells containing glgE grown at 42°C. As expected, SMEG53 transformed with garA and grown at 42°C did exhibit a more dramatic morphology change (Fig. 5D). garA did not produce such a morphology change at 42°C when electroporated into the wild-type strain (data not shown). Since mycobacterial colony morphology is influenced by the composition of the cell wall (8, 9, 20), these observations suggest that the accumulation of glycogen in SMEG53 affects cell wall biosynthesis.
FIG. 5.
(A and B) Colonial morphology of wild-type (A) and SMEG53 (B) colonies grown at 30°C on 7H10. (C and D) Colonies of SMEG53 complemented with pAEB235 (glgE; C) or pAEB234 (garA; D) and grown at 42°C on 7H10. The colonies shown in panel A are representative of the morphology found for the wild-type under all conditions tested.
DISCUSSION
The work reported here has revealed several exciting new aspects of glycogen synthesis in M. smegmatis. The finding that the temperature-sensitive mutant SMEG53 accumulates high levels of glycogen suggests that the polysaccharide is being constantly synthesized and then recycled by GlgE throughout exponential growth. Therefore, the absolute levels of glycogen present in exponential-phase M. smegmatis cells represents a net accumulation of polysaccharide that results from coordinated synthesis and degradation. These results contradict the commonly accepted ideas that in bacteria, glycogen levels reflect only the synthetic capabilities of the cell and glycogen degradation occurs exclusively in stationary phase (22, 23). In E. coli, the genes encoding proteins with both glycogen catabolic and metabolic functions may be found located in the same operon (23). In M. smegmatis, glgE, a gene encoding a protein with a predicted role in glycogen catabolism, is in an operon with glgB. This type of genetic arrangement is consistent with the idea that the metabolic and catabolic enzymes associated with glycogen are coordinately expressed and that the gene products work in concert as a single biological process. Though the absolute levels varied, SMEG53 accumulated high levels of glycogen at 42°C under all growth conditions tested. This suggests that glycogen synthesis and recycling constitute a fundamental process in M. smegmatis that occurs during normal growth.
The phenotype of SMEG53 suggests that glycogen accumulation coincides with a limited availability of carbon and energy in the cell. At 30°C, approximately twofold more glycogen accumulates in SMEG53 in comparison to the wild type, and slower growth and an altered colony morphology are observed. At 42°C, glycogen accumulation is 18-fold higher than in the wild type, and the growth of the mutant cannot be sustained. One model that explains these results is that during normal growth, wild-type M. smegmatis preferentially shunts its available carbon into glycogen synthesis prior to using it in metabolism. In SMEG53, the mutation in GlgE interferes with normal glycogen recycling, glucose remains sequestered in glycogen, and the downstream utilization of carbon is compromised. This model is based on the phenotypes observed in SMEG53 during growth in 7H9, a medium that supports the optimal growth of M. smegmatis in vitro. However, a reasonable assumption is that if this type of carbon processing occurs in 7H9, then it may be an intrinsic process that occurs in the cell under all growth conditions. We have observed that under certain growth conditions, including growth in M9 or 7H9 containing 0.2 M NaCl or growth while overexpressing garA, SMEG53 still exhibits high levels of glycogen accumulation, slow growth, and altered colony morphologies but does not stop growing at 42°C. According to our carbon utilization model, glycogen recycling must occur in order for there to be enough carbon for growth. Therefore, one possibility is that the temperature-sensitive phenotype of SMEG53 can be suppressed because the specific growth conditions somehow promote glycogen recycling despite the mutation in GlgE. The growth of M. smegmatis in M9 and 7H9 containing 0.2 M NaCl is slower than it is in 7H9 at all temperatures. While comparisons cannot be made at 42°C, SMEG53 also grows slower in these media than it does in 7H9 at 30°C. It is tempting to speculate that suppression occurs in the mutant because the slow growth is associated with an alternate form of glycogen recycling in M. smegmatis. If so, we would expect that under growth-limiting conditions, M. smegmatis and perhaps other mycobacteria would accumulate low levels of glycogen. However, it has been well established that just the opposite is true: under growth-limiting conditions, mycobacteria accumulate higher levels of glycogen (2–4, 13). In view of this, we favor the hypothesis that slow growth specifically influences glycogen recycling in SMEG53 by lowering the demand for carbon by the cell, thereby creating a condition where the impaired enzymatic activity of the mutated GlgE enzyme can release enough glucose from glycogen to sustain growth. We have observed that SMEG53 cells containing multiple copies of garA grow slower than mutant cells containing either the cloning vector or plasmid-encoded glgE at the permissive temperature. Thus, similar molecular mechanisms of suppression may be operating in SMEG53 cells overexpressing garA and mutant cells grown in alternate media.
The precise role of glgE and garA in the glycogen synthesis and recycling pathway remains to be determined. In E. coli, glgX, the gene encoding glycogen debranching enzyme, is located in an operon with glgB, the gene encoding glycogen branching enzyme (23). While glgE is located in an operon with glgB, GlgE is not the mycobacterial homolog of GlgX. A putative homolog of GlgX is encoded by Rv1564c, a gene located elsewhere in the chromosome (11). Additional studies are needed to determine the role that GlgE fulfills in glycogen catabolism. The garA gene product is a novel effector of glycogen accumulation. The presence of an FHA domain in GarA suggests that this protein is a member of a serine-threonine kinase signal transduction pathway. Analysis of the M. tuberculosis genome sequence indicates that this organism uses serine-threonine kinase signal transduction pathways, but the cellular processes controlled by these regulatory pathways are unknown (11). Further studies with garA and SMEG53 may provide valuable information about the role of serine-threonine kinase signal transduction pathways in mycobacteria.
Why synthesize glycogen and then recycle it during exponential phase? Our data for SMEG53 are best explained by the idea that glycogen acts as a carbon capacitor for glycolysis during the exponential growth of M. smegmatis. In this model, carbon that is not immediately required for glycolysis would be temporarily stored in glycogen and then accessed when needed by glycogen recycling. Such a system would modulate the flow of carbon into glycolysis and prevent a wasteful expenditure of resources. Glycogen is an ideal carbon capacitor for glycolysis because the two processes are linked by their requirement for glucose-6-phosphate. In the case of glycogen synthesis, glucose-6-phosphate is required for the production of glucose-1-phosphate, which is subsequently used in ADP-glucose formation (22, 23). Using the carbon capacitor model, we predict that the conditions favoring a high rate of glycolysis would also favor a high rate of glycogen synthesis. This idea could account for the allosteric activation of ADP-glucose pyrophosphorylase in M. smegmatis by the glycolytic intermediates fructose-1-phosphate and fructose-1,6-bisphosphate (13). The carbon capacitor model also predicts that under conditions where the rate of glycolysis, and therefore the demand for glucose, is low, glycogen recycling should also be low. This idea could explain the accumulation of glycogen under nutrient-limiting conditions and in stationary phase (2–4, 13). Under these conditions, the cell would want to limit the flow of carbon through glycolysis, so more of the carbon would remain stored in glycogen.
The identification of an exponential-phase glycogen synthesis and recycling system in M. smegmatis raises the question of whether other bacteria have such a system. Mutants of E. coli that produce no glycogen or make excessive amounts of the polysaccharide have been isolated, but such mutants grow normally in comparison to the wild type (22, 23). The defects in glycogen accumulation are attributed to decreased or increased biosynthetic capabilities (22, 23). These studies do not preclude the existence of an exponential-phase glycogen synthesis and recycling system since mutants in genes encoding glycogen-degrading enzymes have not been studied. It is possible that in E. coli, a disruption in a gene such as glgX or another amylolytic enzyme-encoding gene, coupled with the appropriate growth conditions, could produce a phenotype similar to that observed in SMEG53. Further studies should determine if exponential-phase glycogen synthesis and recycling is unique to mycobacteria or is a common theme in bacteria.
ACKNOWLEDGMENTS
We thank Michael Ford and Shruti Jain for critical reading of the manuscript and Joelle Porter for excellent technical assistance.
This work was supported by grant AI37848 from the National Institute of Health to G.F.H. and an American Lung Association Fellowship to A.E.B.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antoine A D, Tepper B S. Characterization of glycogens from mycobacteria. Arch Biochem Biophys. 1969;134:207–213. doi: 10.1016/0003-9861(69)90267-7. [DOI] [PubMed] [Google Scholar]
- 3.Antoine A D, Tepper B S. Environmental control of glycogen and lipid content of Mycobacterium tuberculosis. J Bacteriol. 1969;100:538–539. doi: 10.1128/jb.100.1.538-539.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antoine A D, Tepper B S. Environmental control of glycogen and lipid content of Mycobacterium phlei. J Gen Microbiol. 1969;55:217–226. doi: 10.1099/00221287-55-2-217. [DOI] [PubMed] [Google Scholar]
- 5.Baltz R H. Mutagenesis in Streptomyces spp. In: Demain A L, Soloman N A, editors. Manual of industrial microbiology and technology. Washington, D.C.: American Society for Microbiology; 1986. pp. 184–190. [Google Scholar]
- 6.Baulard A, Jourdan C, Mercenier A, Locht C. Rapid mycobacterial plasmid analysis by electroduction between Mycobacterium spp. and Escherichia coli. Nucleic Acids Res. 1992;20:4105. doi: 10.1093/nar/20.15.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belanger A E, Besra G S, Ford M E, Mikus̆ová K, Belisle J T, Brennan P J, Inamine J M. The embAB genes of Mycobacterium avium encode an arabinosyl transferase involved in cell wall arabinan biosynthesis that is the target for the antimycobacterial drug ethambutol. Proc Natl Acad Sci USA. 1996;93:11919–11924. doi: 10.1073/pnas.93.21.11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Belanger, A. E., J. C. Porter, and G. H. Hatfull. Unpublished data.
- 8.Belisle J T, Brennan P J. Chemical basis of smooth and rough variation in mycobacteria. J Bacteriol. 1989;171:3465–3470. doi: 10.1128/jb.171.6.3465-3470.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belisle J T, Klaczkiewicz K, Brennan P J, Jacobs W R, Jr, Inamine J M. Rough morphological variants of Mycobacterium avium: characterization of genomic deletions resulting in the loss of glycopeptidolipid expression. J Biol Chem. 1993;268:10517–10523. [PubMed] [Google Scholar]
- 10.Bruton C J, Plaskitt K A, Chater K F. Tissue-specific glycogen branching isoenzymes in a multicellular prokaryote, Streptomyces coelicolor A3(2) Mol Microbiol. 1995;18:89–99. doi: 10.1111/j.1365-2958.1995.mmi_18010089.x. [DOI] [PubMed] [Google Scholar]
- 11.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Englmeir K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail M A, Rajandream M-A, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 12.Donnelly-Wu M, Jacobs W R, Jr, Hatfull G F. Superinfection immunity of mycobacteriophage L5: applications for genetic transformation of mycobacteria. Mol Microbiol. 1993;7:407–417. doi: 10.1111/j.1365-2958.1993.tb01132.x. [DOI] [PubMed] [Google Scholar]
- 13.Elbein A D, Mitchell M. Levels of glycogen and trehalose in Mycobacterium smegmatis and the purification and properties of the glycogen synthase. J Bacteriol. 1973;113:863–873. doi: 10.1128/jb.113.2.863-873.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann K, Bucher P. The FHA domain: a putative nuclear signalling domain found in protein kinases and transcription factors. Trends Biol Sci. 1995;20:347–349. doi: 10.1016/s0968-0004(00)89072-6. [DOI] [PubMed] [Google Scholar]
- 15.Hoffman K, Bucher P, Falquet L, Bairoch A. The PROSITE database, its status in 1999. Nucleic Acids Res. 1999;27:215–219. doi: 10.1093/nar/27.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacobs W R, Jr, Kalpana G V, Cirillo J D, Pascopella L, Snapper S B, Udani R A, Jones W, Barletta R G, Bloom B R. Genetic systems for mycobacteria. Methods Enzymol. 1991;204:537–555. doi: 10.1016/0076-6879(91)04027-l. [DOI] [PubMed] [Google Scholar]
- 17.Kiel J A K W, Boels J M, Beldmann G, Venema G. Glycogen in Bacillus subtilis: molecular characterization of an operon encoding enzymes involved in glycogen biosynthesis and degradation. Mol Microbiol. 1994;11:203–218. doi: 10.1111/j.1365-2958.1994.tb00301.x. [DOI] [PubMed] [Google Scholar]
- 18.Lee M H, Pascopella L, Jacobs W R, Jr, Hatfull G F. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guerin. Proc Natl Acad Sci USA. 1991;88:3111–3115. doi: 10.1073/pnas.88.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin M C, Schneider D, Bruton C J, Chater K F, Hardisson C. A glgC gene essential only for the first of two spatially distinct phases of glycogen synthesis in Streptomyces coelicolor A3(2) J Bacteriol. 1997;179:7784–7789. doi: 10.1128/jb.179.24.7784-7789.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mdluli K, Slayden R A, Zhu Y, Ramaswamy S, Pan X, Mead D, Crane D D, Musser J M, Barry C E., III Mechanisms involved in the intrinsic isoniazid resistance of Mycobacterium avium. Mol Microbiol. 1998;27:1223–1233. doi: 10.1046/j.1365-2958.1998.00774.x. [DOI] [PubMed] [Google Scholar]
- 21.Parrou J L, François J. A simplified procedure for a rapid and reliable assay of both glycogen and trehalose in whole yeast cells. Anal Biochem. 1997;248:186–188. doi: 10.1006/abio.1997.2138. [DOI] [PubMed] [Google Scholar]
- 22.Preiss J. Bacterial glycogen synthesis and its regulation. Annu Rev Microbiol. 1984;38:419–458. doi: 10.1146/annurev.mi.38.100184.002223. [DOI] [PubMed] [Google Scholar]
- 23.Preiss J. Regulation of glycogen synthesis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: American Society for Microbiology; 1996. pp. 1015–1024. [Google Scholar]
- 24.Smeulders M J, Keer J, Speight R A, Williams H D. Adaptation of Mycobacterium smegmatis to stationary phase. J Bacteriol. 1999;181:270–283. doi: 10.1128/jb.181.1.270-283.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snapper S, Melton R, Keiser T, Jacobs W R., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 26.Sun Z, Hsiao J, Fay D S, Stern D F. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science. 1998;281:272–274. doi: 10.1126/science.281.5374.272. [DOI] [PubMed] [Google Scholar]
- 27.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang H, Liu M Y, Romeo T. Coordinate genetic regulation of glycogen catabolism and biosynthesis in Escherichia coli via the CsrA gene product. J Bacteriol. 1996;178:1012–1017. doi: 10.1128/jb.178.4.1012-1017.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]