Abstract
Background
Compared with the precise targeting of drug-resistant mutant cancer cells, strategies for eliminating non-genetic adaptation-mediated resistance are limited. The pros and cons of the existence of inflammasomes in cancer have been reported. Nevertheless, the dynamic response of inflammasomes to therapies should be addressed.
Methods
Tumor-derived exosomes were purified by differential ultracentrifugation and validated by nanoparticle tracking analysis and transmission electron microscopy. A proximity ligation assay and interleukin-1β (IL-1β) level were used for detecting activation of NLRP3 inflammasomes. RNA sequencing was used to analyze the exosomal RNAs. MIR21 knocked out human monocytic THP cells and mir21 knocked out murine oral cancer MTCQ1 cells were generated for confirming the exosomal delivery of microRNA (miR)-21. Syngeneic murine models for head and neck cancer (C57BLJ/6J), breast cancer (BALB/C) and lung cancer (C57BL/6J) were applied for examining the impact of Snail-miR21 axis on inflammasome activation in vivo. Single-cell RNA sequencing was used for analyzing the tumor-infiltrated immune cells. Head and neck patient samples were used for validating the findings in clinical samples.
Results
We demonstrated that in cancer cells undergoing Snail-induced epithelial-mesenchymal transition (EMT), tumor cells suppress NLRP3 inflammasome activities of tumor-associated macrophages (TAMs) in response to chemotherapy through the delivery of exosomal miR-21. Mechanistically, miR-21 represses PTEN and BRCC3 to facilitate NLRP3 phosphorylation and lysine-63 ubiquitination, inhibiting NLRP3 inflammasome assembly. Furthermore, the Snail-miR-21 axis shapes the post-chemotherapy tumor microenvironment (TME) by repopulating TAMs and by activating CD8+ T cells. In patients with head and neck cancer, the Snail-high cases lacked post-chemotherapy IL-1β surge and were correlated with a worse response.
Conclusions
This finding reveals the mechanism of EMT-mediated resistance beyond cancer stemness through modulation of post-treatment inflammasome activity. It also highlights the dynamic remodeling of the TME throughout metastatic evolution.
Keywords: head and neck neoplasms; immunity, innate; macrophages; tumor microenvironment
WHAT IS ALREADY KNOWN ON THIS TOPIC
Therapeutic resistance emerges during late-stage progression of cancers.
WHAT THIS STUDY ADDS
The epithelial-mesenchymal transition-undergoing cancer cells deliver microRNA-21-abundant exosomes to suppress inflammasome activities of macrophages for enhancing resistance.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
A dynamic interplay between cancer and microenvironments indicates the importance of real-time adjustment of therapeutic strategies alongside cancer progression.
Introduction
The emergence of therapeutic resistance during cancer progression is a major obstacle in combating advanced cancers. The reasons for the acquired resistance of malignant tumors include the genetic evolution of cancer cells and non-genetic adaptation to therapies.1 The non-genetic plasticity of cancer cells enables lineage switching to adapt to the environment and facilitates the evasion of anticancer immunosurveillance. Compared with the rapid advances in sequencing techniques and precise targeting of resistant mutants, strategies for eliminating non-genetic adaptation-mediated resistance are relatively limited.
The dynamic switch between epithelial and mesenchymal states is crucial for cancer metastasis and therapeutic resistance.2 In addition to the well-established impact of epithelial-mesenchymal transition (EMT) on cancer stemness and therapeutic resistance,3 accumulated evidence, including findings reported by our previous studies, supports the bidirectional interaction between EMT-undergoing cancer cells and host immune cells.4–7 EMT is well known for both engendering immunosuppression8–10 and for promoting the creation of proinflammatory tumor microenvironments (TMEs).5 11 12 Compared with the well-established role of EMT reported in tumor immunosuppression, the impact of EMT on the modulation of the response of host innate immune cells in response to stimuli/treatments is less clear.
Inflammasomes, the multimolecular complexes that consist of an NOD-like receptor scaffold, caspase activation, adapter proteins such as ASC (also known as Pycard), and caspase-1, are considered major sensors of innate immunity which function in response to danger signals in mammals.13 Although chronic inflammation has been noted as a critical event in tumorigenesis and cancer progression, the contradictory roles of inflammasomes in the TME have been demonstrated in different contexts. The pro-cancer and anticancer effects of inflammasomes have both been reported.14–17 Diverse stimuli such as ATP, bacterial components, or nucleic acids induce the assembly of inflammasomes, leading to caspase-1 activation and secretion of interleukin-1β (IL-1β) and IL-18.13 Post-treatment dying cancer cells release danger signals, such as nucleic acids, high-mobility group box 1 (HMGB1) protein, and ATP, which trigger innate immunity and inflammation.18–20 However, knowledge about the impact of the post-therapeutic activation of inflammasomes on treatment response is relatively limited.
The present study demonstrated that in cancers undergoing Snail-induced EMT, tumor cells suppressed the activity of inflammasomes of tumor-associated macrophages (TAMs) in response to chemotherapy through the delivery of exosomal microRNA (miR)-21. The miR-21 modulates NLRP3 phosphorylation and lysine 63-ubiquitination to inhibit the assembly of NLRP3 inflammasomes, thereby shaping the post-chemotherapy TME and reducing the chemotherapy response. This finding reveals the mechanism of EMT-mediated therapeutic resistance beyond cancer stemness through modulation of post-treatment inflammasome activities. It also highlights the dynamic remodeling of the TME throughout metastatic evolution.
Materials and methods
Cell lines and plasmids
Human head and neck squamous cell carcinoma (HNSCC) cell line FaDu, human monocytic leukemia cell line THP-1, human embryonic kidney cell line 293T, BALB/c mouse breast carcinoma cell line 4T1, and C57BL/6J mouse lung carcinoma cell line LLC1 were originally sourced from the American Type Culture Collection. The HNSCC cell line OECM-1 and the C57BL/6J murine oral cancer cell line MTCQ1 were kindly provided by Dr Kuo-Wei Chang (National Yang Ming Chiao Tung University of Taiwan). The pCDH-Snail, pCDH-Zeb1 and pCDH-MIR21 plasmids were generated via insertion of full-length cDNA (SNAI1: NM_005985; ZEB1: NM_030751.4, MIR21: NR_029493.1) into the pCDH-CMV-MCS-EF1-puro vector. The pcDNA3-Flag-NLRP3 plasmid was provided by Dr Szu-Ting Chen (National Yang Ming Chiao Tung University of Taiwan). The 3’-untranslated region (UTR) of BRCC3 (NM_001018055) was cloned into pMIR-REPORTER to generate pMIR-BRCC3-wt, and site-directed mutagenesis was performed to generate the miR-21 binding-site-mutated pMIR-BRCC3-mut.
Patient with HNSCC samples
Five independent sets of samples were used for the experiments. The informed consent was obtained from all patients in this study. The information of the patient samples is detailed in online supplemental methods and online supplemental tables 1-5.
jitc-2022-004832supp001.pdf (3.3MB, pdf)
jitc-2022-004832supp002.xlsx (10.5KB, xlsx)
jitc-2022-004832supp003.xlsx (11.2KB, xlsx)
jitc-2022-004832supp004.xlsx (10.7KB, xlsx)
jitc-2022-004832supp005.xlsx (11KB, xlsx)
jitc-2022-004832supp006.xlsx (10.7KB, xlsx)
Single-cell RNA-sequencing
To obtain cells for single-cell RNA-sequencing (scRNA-seq) analysis, 1×106 MTCQ1-WT/MTCQ1mir21–/– cells were inoculated into the subcutaneous region of C57BL/6 mice. Cisplatin (5 mg/kg) was administered intraperitoneally on the 14th day, and tumors were harvested on the 17th day. Tumors were dissociated using a tumor dissociation kit (Miltenyi Biotec), and dead cells were removed using a dead cell removal kit (MACS). CD45+ tumor-infiltrating immune cells were isolated using microbeads (Miltenyi Biotec). Cell viability detected via trypan blue staining was over 85% for subsequent sequencing. For library construction and sequencing, we used the droplet-based scRNA-seq (10x Genomics Chromium Single Cell 3’ Reagent Kit V.3.1, no. 1000121) for single-cell library preparation. After the conduction of reverse transcription, complementary DNA sequencing was performed using the Illumina NovaSeq 6000 (Illumina). The quality control (QC) and filtering steps were performed using a loupe browser. Briefly, data on cells with low total counts and high mitochondrial gene expression were filtered. The scRNA-seq reads were processed using a 10x Genomics Cell Ranger pipeline and analyzed using the Partek Flow software (Partek, St. Louis, Missouri, USA). Clustering of cells in our dataset was performed using a t-distributed stochastic neighbor embedding (t-SNE) algorithm in Partek Flow. To identify the t-SNE subclusters present in different types of immune cells, the average gene expression of each cluster was identified using cluster identity predictor (CIPR).21 Gene Ontology (GO) analysis of the differentially expressed genes between clusters was performed using DAVID22 and GSEA.23 Partek Flow was used for gene-specific analysis, macrophage re-clustering, macrophage trajectory analysis, and volcano plot generation.
Data and code availability
The accession numbers for the data reported in this paper are GEO: GSE99474, GSE172326, GSE178537, and GSE 181300.
Statistical analysis
Statistical analyses were performed using GraphPad Prism V.8 (GraphPad Software). Two-sided independent Student’s t-test (normal distribution) Pearson’s correlation test was used to analyze the correlation between the two continuous factors. All statistical data were derived from at least three independent biological replicates, and each experiment contained at least two technical replicates. P value ≤0.05 was considered statistically significant (*: ≤0.05, **: ≤0.01, ***: ≤0.001).
RNA-seq analysis of patient with HNSCC samples, RNA-seq for exosomal small RNA, preparation of conditional media and macrophages, purification and characterization of tumor-derived exosomes, tissue processing and data generation for Visium spatial gene expression, generation of the MIR21/mir21 knockout cell lines, quantitative real-time PCR, immunoblotting and immunoprecipitation, immunohistochemistry, multiplex immunofluorescence staining of HNSCC samples, proximity ligation assay, luciferase reporter assay, SYTOX green assay, and animal experiments: The information of the experiments is detailed in online supplemental methods.
Results
Supernatants derived from Snail-expressing cancer cells suppress the NLRP3 inflammasome activity
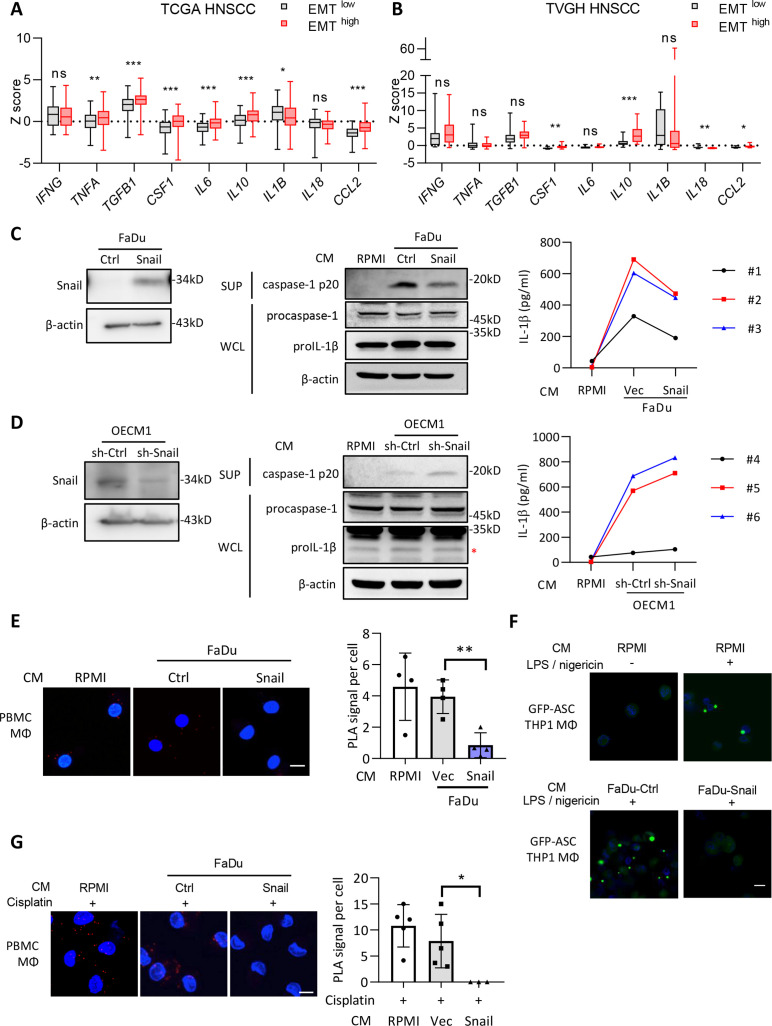
In the present study, we considered HNSCC as the major model for experiments based on the crucial role of EMT reported in HNSCC progression and on our previous findings reported in HNSCC EMT studies.24 25 Since Snail has been identified as the major EMT regulator affecting microenvironments, we performed multiplex immunofluorescence staining to examine the immune cells in HNSCC samples with different expression levels of Snail. A noticeable increase in the infiltration of CD163+ TAMs was observed in samples with higher Snail expression (online supplemental figure 1A and online supplemental table 1). Next, we analyzed the correlation between EMT and inflammatory cytokines in the RNA-seq data of HNSCC samples retrieved from The Cancer Genome Atlas (TCGA) and the cohort of Taipei Veterans General Hospital (TVGH) (figure 1A–B and online supplemental table 2). The samples were categorized as EMThigh or EMTlow according to the calculated EMT score,26 and a higher expression of transforming growth factor ß (TGF-β) validated the feasibility of the categorization. Increased expression of cytokines related to cancer progression and immunosuppression, such as IL-6, tumor necrosis factor (TNF-α) (in the TCGA cohort), and IL-10 (in both cohorts), was observed in the EMThigh group, which was consistent with previous reports.4 27 28 Intriguingly, a trend of reduced levels of IL-1ß (in the TCGA cohort) or IL-18 (in the TVGH cohort) was noted in the EMThigh group, indicating the potential influence of EMT on inflammasome activity. We further applied spatial transcriptomic technology in a HNSCC sample to validate the relationship between EMT and inflammasome activation. A trend of a reversed relationship between the gene expression profile of EMT and inflammasome-related genes was shown (online supplemental figure 1C), (online supplemental table 6), indicating the potential of EMT in suppression of inflammasome activity in HNSCC.
Figure 1.
The supernatants of Snail-expressing cancer cells inhibit activation of NLRP3 inflammasomes. (A–B) Analysis of the expression of the inflammation-related genes in the TCGA HNSCC database (A) or Taipei Veterans General Hospital (TVGH) database. The samples were categorized as the EMThigh and EMTlow group according to the expression of the EMT-related genes (VIM, FN1, CDH2, ITBG6, FOXC2, MMP2, MMP3, MMP9, SOX10, SNAI1, SNAI2, TWIST1, GSC, CDH1, DSP, TJP1) (see Chae et al).26 n=521 for TCGA, n=65 for TVGH. The boxplots show the minimum, first quartile, medium, third quartile, and maximum. *p<0.05, **p<0.01, ***p<0.001, ns=no significance by Student’s t-test. (C) Left, western blot of Snail in FaDu-Ctrl/Snail. β-actin was a loading control. Middle, immunoblots of cleaved caspase-1 p20 in supernatants (SUP), pro-caspase-1 and pro-IL-1β in whole cell lysates (WCL) of PBMC-derived macrophages cultivated in conditioned media (CM) from FaDu cells stably transfected with Snail or a control vector (Ctrl) for 48 hours. RPMI was a control for CM. β-actin was a loading control for immunoblots of WCL. Right, ELISA for analyzing the level of secreted IL-1β from PBMC-derived macrophages cultivated with the conditional media from FaDu-Snail/FaDu-Vec. Peripheral blood mononuclear cells were isolated from three different healthy donors (case #1, #2, and #3). (D) Left, western blot of Snail in OECM1-sh-Ctrl/sh-Snail. β-actin was a loading control. Middle, immunoblots of cleaved caspase-1 p20 in SUP, pro-caspase-1 and pro-IL-1β in WCL of PBMC-derived macrophages cultivated in CM from OECM1 cells receiving short hairpin RNA against Snail (sh-Snail) or a control sequence (sh-Ctrl) for 48 hours. RPMI was a control for CM. β-actin was a loading control for immunoblots of WCL. Right, ELISA for analyzing the level of secreted IL-1β from macrophages cultivated with the conditional media from FaDu-Snail/FaDu-Vec. Peripheral blood mononuclear cells were isolated from three different healthy donors (case #4, #5, and #6). (E) Left, representative images of proximity ligation assay (PLA) for detecting NLRP3 and ASC interaction in PBMC-derived macrophages cultivated in CM from FaDu-Vec or FaDu-Snail. The red dots indicate the PLA signals. Scale bar, 10 μm. Right, quantification of number of PLA signals per cell. For each group, at least a total of 18 cells from four randomly selected fields were used for PLA quantification. Data represent means±SD. **p<0.01 by Student’s t-test. (F) Representative immunofluorescent images of THP1-derived macrophages transfected with GFP-tagged ASC, stimulated with nigericin and cultivated with the CM from FaDu-Ctrl, FaDu-Snail, or RPMI for 24 hours. Scale bar, 10 µm. (G) PLA for detecting NLRP3 and ASC interaction in cisplatin-activated PBMC-derived macrophages incubated with CM from FaDu-Vec/FaDu-Snail. Left, representative images. Red dots indicate PLA signals. Scale bar, 10 µm. Right, quantification of PLA signals per cell. For each group, at least total 16 cells from five random selections (three for FaDu-Snail) were used for PLA quantification. Data represent means±SD. *p<0.05 by Student’s t-test. Scale bar, 10 µm. EMT, epithelial-mesenchymal transition; HNSCC, head and neck squamous cell carcinoma; IL, interleukin; PBMC, peripheral blood mononuclear cells; TCGA, The Cancer Genome Atlas.
jitc-2022-004832supp007.xlsx (12.7KB, xlsx)
We have previously shown that Snail-expressing cancer cells recruit TAMs through the secretion of CCL2, CCL5, and TNF-α to facilitate cancer progression.4 Snail-expressing cancer cells secrete exosomes to promote M2 polarization of TAMs.6 We examined the influence of Snail on inflammasome activity. Snail was overexpressed in FaDu, a human HNSCC cell line harboring low-endogenous Snail (left panel of figure 1C). Snail knockdown was performed in the high-endogenous Snail human HNSCC cell line OECM14 (left panel of figure 1D). The supernatant derived from FaDu-Snail inhibited the release of active caspase-1 and IL-1β from activated peripheral blood monoclear cells (PBMC)-derived macrophages compared with the control cells (figure 1C). Knockdown of Snail in OECM1 potentiated the release of active caspase-1 and IL-1β (figure 1D). Consistent results were observed in the human monocytic cell line THP1-derived activated macrophages. Supernatants from FaDu-Snail repressed IL-1β secretion from THP1-derived activated macrophages. Supernatants from OECM1-Snail knockdown cells did not potentiate the secretion of IL-1β to a significant level (online supplemental figure 2A). In contrast, the supernatant derived from FaDu cells expressing another major EMT regulator, Twist1, did not affect the secretion of active caspase-1 and IL-1β from THP1-derived activated macrophages (online supplemental figure 2B).
We further confirmed the effect of Snail-expressing cancer cells on NLRP3 inflammasome assembly. When the NLRP3 inflammasome is activated, NLRP3 establishes interactions with ASC to recruit and activate caspase-1. Concurrently, the complexes are assembled into a giant pyroptosome. Proximity ligation assay (PLA) was performed to examine the interaction between NLRP3 and ASC in PBMC-derived and THP1-derived macrophages. The supernatant derived from FaDu-Snail repressed the interaction between NLRP3 and ASC in activated macrophages compared with the control cells (figure 1E and online supplemental figure 2C). The FaDu-Snail supernatant also reduced pyroptosome formation in activated macrophages (figure 1F). As chemotherapy reportedly activates the NLRP3 inflammasome,14 20 we examined whether Snail-overexpressing cancer cells suppressed chemotherapy-activated inflammasomes in macrophages. Cisplatin treatment activated inflammasomes in macrophages as expected, and FaDu-Snail supernatant suppressed the activity of inflammasomes, as evidenced by the reduction of active caspase-1 and IL-1β production (online supplemental figure 2D). The FaDu-Snail supernatant also reduced the NLRP3-ASC interaction in cisplatin-activated macrophages (figure 1G). Altogether, the above-mentioned results indicate that the supernatant derived from FaDu-expressing HNSCC suppresses NLRP3 inflammasome activity.
Snail-expressing cancer cells suppress NLRP3 inflammasome activity through the secretion of miR-21-abundant exosomes
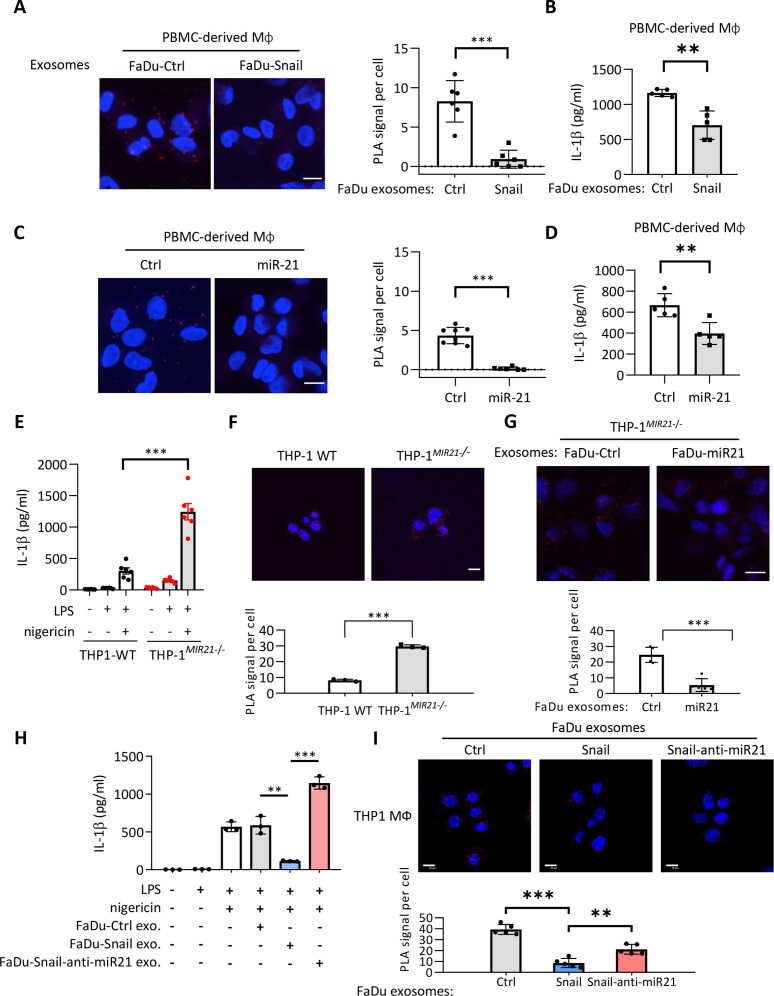
Next, we determined the major player(s) in the suppression of inflammasome activity in the supernatant of Snail-expressing cancer cells. We found that IL-6, IL-8, CCL2, and CCL5 were the major cytokines regulated by Snail.4 29 However, neutralization of these cytokines did not affect NLRP3-ASC interaction or pyroptosome formation in activated macrophages subjected to treatment with the supernatant derived from either FaDu-Snail or control cells (online supplemental figure 3A-B). As tumor-secreted exosomes (TEXs) have been critically involved in remodeling microenvironments,30 31 we investigated whether Snail-expressing cancer cell-secreted exosomes modulated the inflammasome activity in macrophages. The exosomes were confirmed via direct visualization by performing transmission electron microscopy and nanoparticle tracking analysis, and results showed the presence of 50–150 nm nanovesicles with bilayer membranes (online supplemental figure 3C). The markers of exosomes, including CD9, CD63, CD81, and Alix, were examined to confirm the successful purification of cancer-secreted exosomes (online supplemental figure 3D). We investigated the effect of TEXs on the inflammasome activity of macrophages. A significant reduction in the NLRP3-ASC interaction was noted when activated macrophages were subjected to treatment with FaDu-Snail-secreted exosomes compared with those derived from FaDu-control cells (figure 2A). Consistently, a decrease in IL-1β secretion was also noted when macrophages were incubated with FaDu Snail-secreted exosomes (figure 2B). These data indicate that Snail-expressing cell-secreted exosomes suppress the activity of the NLRP3 inflammasome in activated macrophages.
Figure 2.
Exosomes from Snail-miR-21 axis activated cancer cells suppress NLRP3 inflammasome activity of macrophages. (A) Representative images of PLA for detecting NLRP3 and ASC interaction in PBMC-derived macrophages incubated with the exosomes from FaDu-Vec/FaDu-Snail. The red dots indicate the PLA signals. Scale bar, 10 µm. Right, quantification of number of PLA signals per cell. For each group, at least a total of 50 cells from six randomly-selected fields were used for PLA quantification. Data represent means±SD. ***p<0.001 by Student’s t-test. (B) ELISA for analyzing the level of secreted IL-1β by PBMC-derived macrophages incubated with exosomes from FaDu-Vec/FaDu-Snail. n=5 independent experiments (each experiment contains two technical replicates). Data represent means±SD. **p<0.01 by Student’s t-test. (C) Left, representative images of PLA for detecting NLRP3 and ASC interaction in PBMC-derived macrophages transduced with miR-21 or a control agomir. The red dots indicate the PLA signals. Scale bar, 10 µm. Right, quantification of number of PLA signals per cell. For each group, at least a total of 60 cells from randomly-selected fields (six for miR-21 group and eight for ctrl group) were used for PLA quantification. Data represent means±SD. ***p<0.001 by Student’s t-test. (D) IL-1β ELISA of PBMC-derived macrophage. miR-21 agomir or a control sequence (Ctrl) was transduced to macrophage. n=5 independent experiments (each experiment contains two technical replicates). Data represent means±SD. **p<0.01 by Student’s t-test. (E) ELISA for analyzing the level of secreted IL-1β by macrophages derived from wild-type THP1 (THP1-WT) or MIR21-knockout THP1 (THP1MIR21–/–). The macrophages were treated with/without LPS (1 µg/mL) and nigericin (5 µM). n=6 (each contains two technical replicates) for each group. Data represent means±SD. ***p<0.001 by Student’s t-test. (F) Representative images of PLA for detecting NLRP3 and ASC interaction in THP1-WT/THP1MIR21–/–-derived activated macrophages. The red dots indicate the PLA signals. Scale bar, 10 µm. Right, quantification of number of PLA signals per cell. For each group, at least a total of 400 cells from five randomly selected fields were used for PLA quantification. Data represent means±SD. ***p<0.001 by Student’s t-test. (G) Representative images of PLA for detecting NLRP3 and ASC interaction in THP1MIR21–/–-derived macrophages incubated with exosomes from FaDu cells transfected with miR-21-expressing vector (FaDu-miR21) or a control vector (FaDu-Ctrl). The red dots indicate the PLA signals. Scale bar, 10 µm. For each group, at least a total of 400 cells from five randomly selected fields were used for PLA quantification. Data represent means±SD. ***p<0.001 by Student’s t-test. (H) ELISA for analyzing the level of secreted IL-1β by THP1-derived macrophages. The macrophages were treated with/without LPS (1 µg/mL) and nigericin (5 µM) and the exosomes from FaDu cells transfected with a control vector (FaDu-ctrl), Snail-expressing vector (FaDu-Snail), Snail and anti-miR-21 (FaDu-Snail-anti-miR-21). n=3 (each contains two technical replicates) for each group. Data represent means±SD. **p<0.01, ***p<0.001 by Student’s t-test. (I) Representative images of PLA for detecting NLRP3 and ASC interaction in THP1MIR21–/–-derived activated macrophages incubated with exosomes from FaDu cells transfected with a control vector, a miR-21-expressing vector, or Snail together with an antagomir for miR-21 (Snail-anti-miR21). The red dots indicate the PLA signals. Scale bar, 10 µm. For each group, at least a total of 400 cells from five randomly selected fields were used for PLA quantification. Data represent means±SD. **p<0.01, ***p<0.001 by Student’s t-test. IL, interleukin; miR, micro RNA; PBMC, peripheral blood monoclear cells; PLA, proximity ligation assay.
MicroRNAs (miRNAs) are important components of regulatory networks in innate immunity.32 We have previously shown that MIR21 transcription was regulated by Snail, and miR-21-abundant exosomes secreted from Snail-overexpressing HNSCC promote M2-like polarization of TAMs.6 We hypothesized that exosomal miRNAs might provide a link between cancer cells and innate immune signaling pathways. To this end, we performed exosomal miRNA sequencing in the human HNSCC cell line FaDu versus immortalized human gingival mucosa cells SG. We investigated the miRNAs that were especially enriched in TEXs, the tumor-specific exosomal miRNAs, by focusing on the top 10 miRNAs in FaDu-derived exosomes. Among them, miR-21 was found to be the most significantly enriched in the TEXs (online supplemental table 7). As EMT is closely linked to miRNA dysregulation,33 and as Snail dominates the expression of several miRNAs in cancer cell,34 35 we explored the major Snail-regulated exosomal miRNA(s) by examining the expression of the top-ranked tumor exosomal miRNAs in FaDu-Snail versus FaDu-control and OECM1 that received a short hairpin RNA (shRNA) against Snail or a control sequence. Among these, miR-21, miR-10a, and miR-191 showed consistency in upregulated levels of FaDu-Snail-secreted exosomes and downregulated levels of exosomes derived from the Snail-knockdown OECM1 cells (online supplemental figure 4A). We further examined the impact of the three miRNAs on NLRP3 inflammasome activation. Transfection of miR-21, but not miR-10a/miR-191, into THP1-derived macrophages significantly attenuated the interaction between NLRP3 and ASC (online supplemental figure 4B). Consistently, miR-21 reduced the assembly of NLRP3 and ASC (figure 2C) and IL-1β secretion in PBMC-derived macrophages (figure 2D). Together with our previous finding which highlighted that Snail upregulated exosomal miR-21 levels through direct or indirect (via Zeb1) activation of MIR21 transcription,6 the results indicated the potential involvement of Snail-regulated miR-21 in the suppression of NLRP3 inflammasome activity.
jitc-2022-004832supp008.xlsx (19.3KB, xlsx)
To further elucidate the role of exosomal miR-21 in the regulation of NLRP3 inflammasome activity in macrophages, we generated MIR21 knockout THP1 cells using the CRISPR-Cas9 technology (online supplemental figure 4C). A significant increase in IL-1β secretion was noted in the supernatant of THP1MIR21–/– -derived macrophages subjected to treatment with LPS and nigericin compared with the wild-type THP1-derived macrophages (figure 2E). Knockout of miR-21 in THP1 increased NLRP3-ASC interaction in THP1-dervied macrophages (figure 2F). Treatment of THP1MIR21–/–-derived macrophages with exosomes harvested from miR-21-overexpressed FaDu cells reduced NLRP3-ASC interaction (figure 2G). To further confirm the role of Snail-regulated miR-21 in the suppression of inflammasome activity, we knocked down miR-21 in Snail-overexpressing FaDu cells (FaDu-Snail) and examined the impact of the TEXs on IL-1β secretion of macrophages. The efficacy of the miRZip anti-miR-21 was validated by the reporter assay (online supplemental figure 4D). Downregulation of miR-21 reporter activity was shown in Snail-overexpressing FaDu cells, and the successful knockdown of miR-21 was confirmed by the restoration of reporter activity in FaDu-Snail transfected with anti-miR-21 (online supplemental figure 4E). We next examined the impact of the Snail-miR-21 axis in cancer cells on the inflammasome activity of macrophages through delivering exosomes. The macrophages were treated with TEXs derived from FaDu-control, FaDu-Snail, and FaDu-Snail-anti-miR21. Overexpression of Snail in FaDu cells suppressed IL-1β secretion of macrophages, and knockdown of miR-21 in FaDu-Snail restored the IL-1β level (figure 2G). Consistently, overexpression of Snail in FaDu cells reduced NLRP3-ASC interaction (detected by PLA) and knockdown of miR-21 in FaDu-Snail restored the interaction (figure 2H). In summary, these results indicate that exosomal miR-21 from Snail-overexpressing cancer cells attenuates NLRP3 inflammasome activation of macrophages.
MiR-21 modulates NLRP3 phosphorylation and polyubiquitination to suppress inflammasome activity
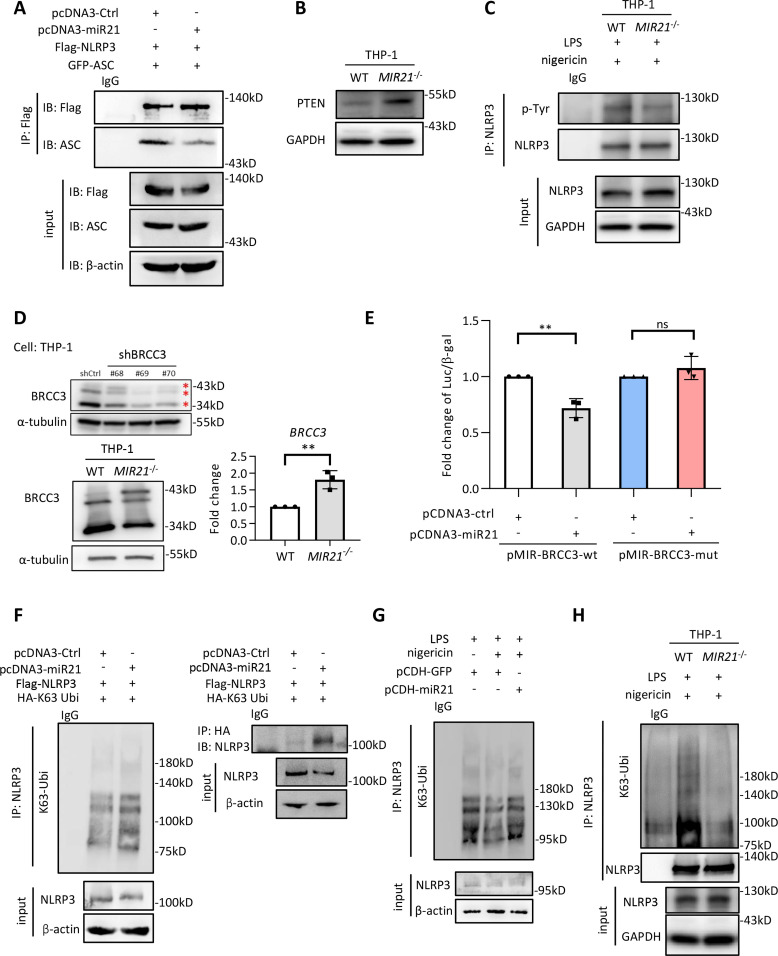
We next investigated the mechanism of miR-21-mediated suppression of NLRP3 inflammasome activity. Neither miR-21 agomir nor the miR-21-abundant exosomes (harvested from Snail-expressing cancer cells) affected the messenger RNA (mRNA) levels of NLRP3/ASC (online supplemental figure 5A) or the protein stability of NLRP3/ASC (online supplemental figure 5B). Knockout of miR-21 did not downregulate the expression of A20, a negative regulator of nuclear factor kappa B (NF-κB),36 in LPS/nigericin-stimulated THP1 cells (online supplemental figure 5C). Therefore, we considered that miR-21 repressed inflammasome activity by reducing the interaction between NLRP3 and ASC. Transfection with miR-21 reduced the interaction between NLRP3 and ASC (figure 3A; see also figure 2C). Post-translational modifications of NLRP3, including phosphorylation and K63-ubiquitination, are reportedly crucial for the assembly of NLRP3 inflammasomes for subsequent activation.20 37 38 Next, we investigated the mechanisms responsible for the miR-21-regulated NLRP3 inflammasome activation. Regarding phosphorylation-regulated NLRP3 inflammasome activities, PTEN has been shown to dephosphorylate tyrosine 32 of NLRP3 to facilitate NLRP3 inflammasome activation.20 PTEN has been observed to be a direct target suppressed by miR-21.39 Here, we showed that knockout of MIR21 in THP-1 cells upregulated PTEN (figure 3B) and reduced NLRP3 phosphorylation (figure 3C). In addition to phosphorylation, polyubiquitination of NLRP3 is critical in the regulation of inflammasome activity. Furthermore, NLRP3 is the substrate of BRCA1/BRCA2-containing complex subunit 3 (BRCC3), a deubiquitinase that specifically cleaves lysine 63-linked polyubiquitin chains.38 Intriguingly, BRCC3 contains the seed regions of miR-21 predicted by using TargetScan, and the sequences of the seed regions are preserved across species (online supplemental figure 5D), indicating the potential for miR-21-mediated inflammasome inactivation by targeting BRCC3. Ectopic expression of miR-21 in PBMC-derived macrophages downregulated BRCC3 expression (online supplemental figure 5E). Increased BRCC3 protein and mRNA expression levels were noted in the MIR21 knockout subline of THP1 cells (figure 3D). Co-incubation of activated macrophages with the supernatants derived from FaDu-Snail versus FaDu-control cells showed that the FaDu-Snail supernatant upregulated miR-21 expression and downregulated the expression levels of PDCD4 (a known target of miR-21) and BRCC3 in macrophages (online supplemental figure 5F). A consistent result was observed in OECM1 cells that received shRNA against Snail versus a control sequence. The supernatants derived from Snail-knockdown OECM1 cells reduced miR-21 and increased BRCC3 and PDCD4 expression (online supplemental figure 5G). The reporter containing the wild-type or mutated miR-21 binding sites of BRCC3 3’-UTR was generated to investigate the regulation of BRCC3 by miR-21 (online supplemental figure 5H). Ectopic miR-21 suppressed the wild-type BRCC3 reporter, whereas mutation of the miR-21 binding motifs abrogated this effect (figure 3E).
Figure 3.
The miR-21-containing exosomes represses BRCC and PTEN to reduce NLRP3 inflammasome activity. (A) Immunoprecipitation (IP)-western blots to show the interaction of NLRP3 and ASC in HEK293T cells transfected with Flag-NLRP3, GFP-ASC and miR-21/control vector. IgG is a control for IP. (B) Immunoblots for examining the expression of PTEN in macrophages derived from THP-WT or THP1MIR21–/–. (C) IP-western blots to show the tyrosine phosphorylation of THP-WT/THP1MIR21–/–-derived macrophages primed by LPS (1 µg/mL) and activated by nigericin (5 µM). IgG is a control for IP. (D) Left upper, western blot of BRCC3 in THP1 cells receiving short hairpin RNA against BRCC3. Left lower, western blots of BRCC3 in THP-WT/THP1MIR21–-derived macrophages. α-tubulin was a loading control. Right, quantitative real-time PCR for examining the relative expression of BRCC3 in THP-WT/THP1MIR21–/–-derived macrophages. n=3 (each contains two technical replicates). Data shows mean±SD. **p<0.01 by Student’s t-test. (E) BRCC3 3’-UTR reporter assay. The wild-type or miR-21 binding site mutated 3’-UTR reporter constructs of BRCC3 (pMIR-BRCC3-wt and pMIR-BRCC3-mut), pcDNA3-miR21/control vector and β-galactosidase were co-transfected to HEK293T cells. Data represent means±SD. **p<0.01 by Student’s t-test. n=3 independent experiments (each contains two technical replicates). (F) IP-western blot to show the K63-ubiquitylated NLRP3 in HEK293T cells transfected with miR-21 or a control vector. NLRP3 (left panel) or HA-tagged K63 ubiquitin (right panel) was immunoprecipitated for immunoblotting. IgG is a control for IP. (G) IP-western blot to show the K63-ubiquitylated NLRP3 in THP1-derived activated macrophages primed by LPS (1 µg/mL) and transfected with miR-21 or a control vector. Nigericin (5 µM) was used to activate inflammasome. IgG is a control for IP. (H) IP-western blot to show the K63-ubiquitylated NLRP3 in THP1-WT/THP1MIR21–/–derived activated macrophages. IgG is a control for IP. miR, micro RNA; UTR, untranslated region.
We examined whether miR-21 inhibited NLRP3 inflammasome activation by suppressing NLRP3 deubiquitination. NLRP3 was immunoprecipitated from HEK293T cells co-transfected with NLRP3 and K63 ubiquitin plus miR-21/control vector. Overexpression of miR-21 increased the level of K63 ubiquitination of NLRP3 (figure 3F). Stable expression of miR-21 inhibited the deubiquitination of NLRP3 in THP-1 cells stimulated with nigericin (figure 3G). Reduced K63 ubiquitination was observed in macrophages derived from THP-1MIR21–/– cells compared with wild-type THP1 cells (figure 3H). Together, these results indicate that miR-21 or miR-21-abundant exosomes from Snail-expressing cells suppress NLRP3 inflammasome activity by targeting PTEN to enrich tyrosine phosphorylation of NLRP3, and by targeting BRCC3 to enhance K63 ubiquitination of NLRP3.
Snail and miR-21 inhibit chemotherapy-induced NLRP3 inflammasome activation and attenuate chemotherapy responses in vivo
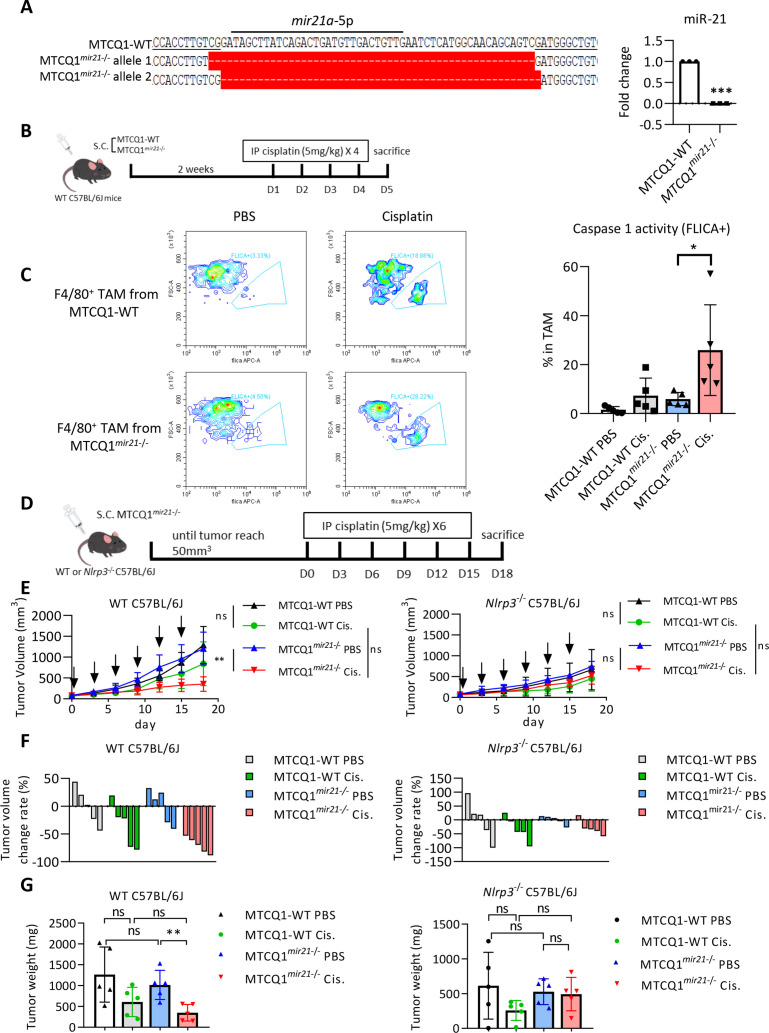
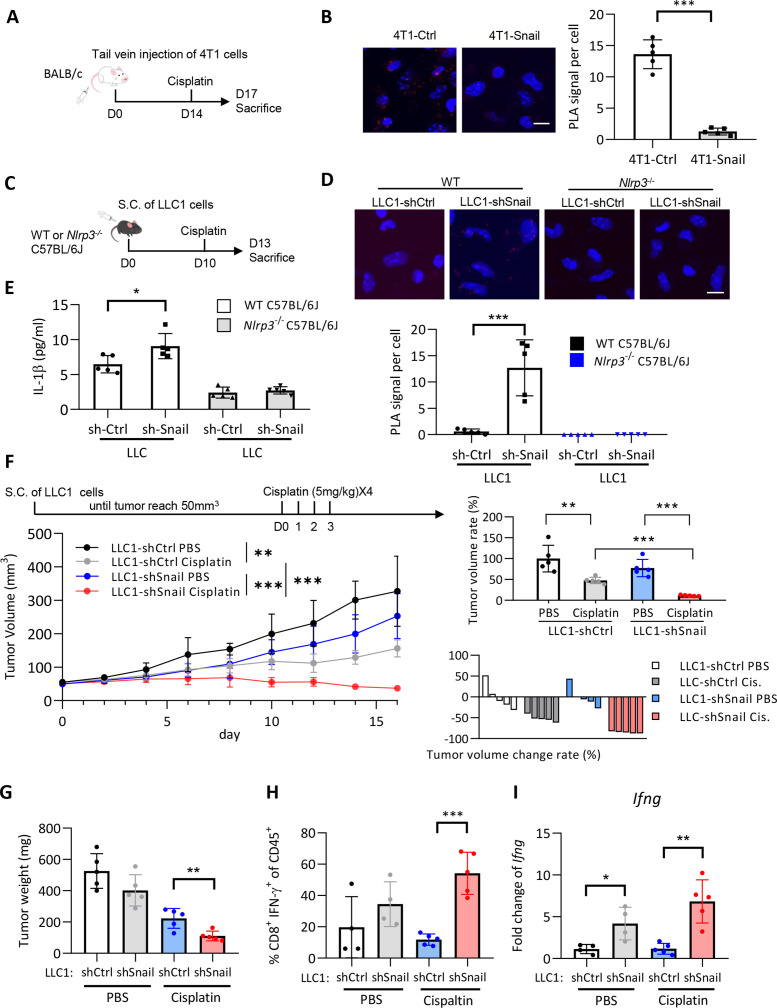
NLRP3 inflammasome-produced active caspase-1 and IL-1β are necessary for chemotherapy-induced antitumor immunity.14 We hypothesized that Snail-regulated miR-21 reduced activation of the NLRP3 inflammasome of macrophages which attenuates chemotherapy-induced antitumor immunity of cancer cells, which contributed to the development of chemotherapy resistance. To this end, we used three syngeneic murine tumor models for the experiments. We first investigated the role of tumorous mir-21 in chemotherapy response in a syngeneic murine HNSCC model40 (the oral squamous cell carcinoma cell line MTCQ1 derived from C57B6/J mice) and generated the mir21 knockout murine HNSCC cell line MTCQ1mir21–/– (figure 4A). We inoculated wild-type and mir21 knockout MTCQ1 cells into the subcutaneous region of wild-type C57B6/J mice to enable the formation of syngeneic HNSCC tumors, following which treatments were conducted with cisplatin. Tumors were harvested 4 days after treatment to assess caspase-1 activity to indicate inflammasome activity (figure 4B). Cisplatin treatment increased caspase-1 activity of F4/80+ TAMs from wild-type MTCQ1 tumors, whereas knockout of mir21 significantly enhanced caspase-1 activation in TAMs (figure 4C; the gating strategy of caspase 1-positive cells is illustrated in online supplemental figure 6A). To investigate the impact of mir21-regulated Nlrp3 inflammasomes on chemotherapy responses, we inoculated the wild-type murine MTCQ1 cells or MTCQ1mir21–/– cells to the wild-type or Nlrp3–/– C57B6/J mice. Cisplatin was intraperitoneally injected to the tumor-bearing mice, and the mice were sacrificed on the 18th day after the commencement of cisplatin treatment (figure 4D). In wild-type mice, knockout of mir-21 in MTCQ1 cells potentiated the tumor suppressive effect of cisplatin, whereas the mir-21 knockout-potentiated chemotherapy response was abrogated in Nlrp3–/– mice (figure 4E–F). We examined whether cisplatin influence the viability of TAMs from murine MTCQ-WT and MTCQmir21–/– -formed tumors by 7-Aminoactinomycin D (7-AAD). The result showed that cisplatin treatment had a slight trend to reduce the viability of F4/80+ macrophages in MTCQ-WT-formed tumors, and the effect was also noted in MTCQmir21–/–-formed tumors (online supplemental figure 6B). We next investigated the impact of inflammasome activation on pyroptosis of human THP1 cells-derived macrophages. Knockout of miR-21 increased LPS/nigericin-induced cleavage of gasdermin D and IL-1β. A SYTOX green assay also validated the increased pyroptosis in macrophages derived from THP1MIR21–/– cells. Re-expression of miR-21 in THP1MIR21–/– cells reduced SYTOX green-stained cells (online supplemental figure 6C-D).
Figure 4.
Depletion of mir-21 potentiates response to chemotherapy in oral cancer cells. (A) Left, genomic sequence of mir21a-5p of wild-type murine oral cancer cell line MTCQ1 (MTCQ-WT) and mir-21 knockout subline MTCQ1mir21–/–. Right, quantitative real-time PCR for analyzing the expression of mir-21 of MTCQ-WT and MTCQ1mir21–/–. Data represent means±SD. ***p<0.001 by Student’s t-test. n=3 independent experiments (each experiment contains two technical replicates). (B) Schema of the experiment for assaying inflammasome activities of murine tumors. 1×106 of MTCQ1-WT or MTCQ1mir21–/– cells were inoculated to the subcutaneous region of the wild-type C57BL/6J mice for 2 weeks. Cisplatin 5 mg/kg was given intraperitoneally for 4 consecutive days. The mice were sacrificed on the fifth day after the start of cisplatin injection. F4/80+ tumor associated macrophages (TAMs) were harvested and caspase 1 activity (FLICA+) was analyzed by flow cytometry. (C) Left, representative data of the flow cytometry analysis for the caspase 1 activity of the TAMs (F4/80+FLICA+) after treated with cisplatin or the control PBS. n=5 for each group. Data shows mean±SD. *p<0.05 by Student’s t-test. (D) Schema of the experiment of (E) and (F). 1×106 of wild-type MTCQ1 cells (MTCQ1-WT) or MTCQ1mir21−/− cells were inoculated to the subcutaneous region of the wild-type or Nlrp3−/− C57BL/6J mice. After the tumor size reached 50 mm3 (day 0), intraperitoneal injection of cisplatin (50 mg/kg) or PBS was given every 3 days for a total of six doses, and the tumor size were measured every 3 days. Mice were sacrificed at the 18th day and tumor weight were measured. (E) The volume the MTCQ1-WT/MTCQ1mir21−/−-formed tumors in wild-type or Nlrp3−/− mice treated with cisplatin or PBS. n=5 for each group. Data shows mean±SD. **p<0.01, ns=no significance by Student’s t-test. (F) The waterfall plots to indicate the tumor volume change of each mouse. (G) The weight of the MTCQ1-WT/MTCQ1mir21−/−-formed tumors in wild-type or Nlrp3−/− mice treated with cisplatin or PBS. n=5 for each group. Data shows mean±SD. **p<0.01 ns=no significance by Student’s t-test. miR, micro RNA; PBS, phosphate buffered saline.
We next investigated the effect of Snail on chemotherapy-induced inflammasome activation in vivo. The murine mammary cancer cell line 4T1 transfected with Snail or a control vector (4T1-Snail/4T1-control; online supplemental figure 7A) was intravenously injected into BALB/c mice to establish pulmonary colonization of tumors. Cisplatin was administered 14 days after the tumor cell injection. Mice were euthanized 3 days later, and the F4/80+ macrophages were isolated from the lungs of mice to investigate NLRP3 inflammasome activation by PLA (figure 5A). Overexpression of Snail in murine 4T1 cells significantly reduced the interaction between NLRP3 and ASC in F4/80+ macrophages (figure 5B). A reduced level of serum IL-1β was noted in mice that received orthotopic implantation of 4T1-Snail and were subsequently treated with cisplatin compared with the 4T1-control group (online supplemental figure 7B). We used another syngeneic mouse model to confirm the effect of Snail on the suppression of inflammasome activation. We delivered shRNA against Snail or a control sequence into the murine Lewis lung carcinoma cell line LLC1 (LLC1-shSnail or LLC1-control, respectively; online supplemental figure 7C). LLC1-shSnail/LLC1-control cells were inoculated into the subcutaneous area of wild-type or Nlrp3–/– C57BL/6J mice. Cisplatin was injected 10 days after tumor cell inoculation, and the mice were euthanized on the 13th day (figure 5C). In wild-type mice, knockdown of Snail in cancer cells increased the interaction between NLRP3 and ASC in TAMs and the serum level of IL-1β. NLRP3-ASC interaction in TAMs was not observed in Nlrp3–/– mice, as expected (figure 5D). The elevation of serum IL-1β in the LLC1-shSnail group was also not noted in Nlrp3–/– mice (figure 5E).
Figure 5.
Snail limits chemotherapy-induced NLRP3 inflammasome activation in vivo. (A) Schema of the experiment. 2.5×105 4T1 cells expressing Snail or a control vector were intravenously injected into the tail veins of BALB/c mice. 4T1 tumor-bearing mice received a single injection of cisplatin (5 mg/kg) at the 14th day after tumor cell injection. The mice were sacrificed at the 17th day and the F4/80+ macrophages isolated from the lungs of the mice were subjected to PLA analysis. (B) PLA signal of NLRP3 and ASC interaction in pulmonary macrophages of 4T1-Snail/4T1-Ctrl. Left, the representative immunofluorescent images for showing the interaction of ASC-NLRP3 in F4/80+ macrophages isolated from lungs of mice. The red dots indicate the PLA signals. Right, quantification of PLA signals per cell. For each mouse, four high power fields from random selection were used for PLA quantification. Scale bar, 10 µm. n=5 for each group. ***p<0.001 by Student’s t-test. (C) Schema for animal experiment. 5×105 LLC1 cells were inoculated into the subcutaneous area of the WT or Nlrp3–/– C57BL/6J mice. A single dose of cisplatin (5 mg/kg) was given at the 10th day after tumor cell injection. The mice were sacrificed at the 13th day after tumor injection. The F4/80+ TAMs were harvested for PLA for detecting the NLRP3-ASC interaction. (D) PLA signal of NLRP3 and ASC interaction. Upper, the representative immunofluorescent images for showing the NLRP3-ASC interaction in F4/80+ TAMs isolated from tumors. For each mouse, at least a total of 30 cells from eight randomly-selected fields were used for PLA quantification. Lower, quantification of PLA signals per cell. Scale bar, 10 µm. ***p<0.001 by Student’s t-test. (E) Serum IL-1β level of mice 3 days after cisplatin treatment (n=5). Data shows means±SD. *p<0.05 by Student’s t-test. (F) Left upper, schema of the experiment. LLC1 cells (2.5×105) were inoculated to the subcutaneous region of C57BL/6 mice. Cisplatin (5 mg/kg) was given at 0, 1, 2, 3 day after tumor cell injection. The mice were sacrificed at the 16th day after cisplatin/PBS treatment. Left lower, tumor volume curve. Right upper, tumor volume change presented in %. Right lower: a waterfall plot to show the volume change of each tumor. n=5 for each group. Tumor volume is shown in mean±SD, and tumor volume reduction rate shows mean±SD. **p<0.01, ***p<0.001 by Student’s t-test. (G) The tumor weight of LLC1-formed tumors in (F). Data shows mean±SD. **p<0.01 by Student’s t-test. (H) Quantification of the percentages of CD8+IFNγ+ tumor-infiltrating lymphocytes among CD45+ tumor-infiltrating leukocytes in different groups of mice as panel (F). The harvested tumors were dissociated and the tumor-infiltrating leukocytes were analyzed by flow cytometry. For PBS groups, n=4; for cisplatin groups, n=5. ***p<0.001 by Student’s t-test. (I) Quantitative real-time PCR for analyzing the expression level of Ifng in different groups of LLC1-formed tumors as panel (F) and (G). For PBS groups, n=4; for cisplatin groups, n=5. *p<0.05, **p<0.01 by Student’s t-test. IFN, interferon; IL, interleukin; PBS, phosphate buffered saline; PLA, proximity ligation assay; TAM, tumor-associated macrophages.
We next confirmed the role of the Snail-regulated NLRP3 inflammasome in chemotherapy responses. The effect of Snail knockdown exerted on chemotherapy-mediated tumor suppression, the recruitment of interferon (IFN)-γ-expressing CD8+ tumor-infiltrating lymphocytes (TILs), and the expression of Ifng were examined in the LLC1 syngeneic mouse model (the gating strategy of CD45+CD8+IFN-γ+ TILs is illustrated in online supplemental figure 7D). When mice were not treated with cisplatin, knockdown of Snail caused a borderline decrease in tumor volume/weight. In contrast, the effect of Snail-knockdown-induced tumor suppression was augmented when mice received cisplatin treatment (figure 5F–G). IL-1β affects CD8+ T cells to enhance their effector function in response to antigen stimulation.14 41 The present study found that the knockdown of Snail increased the proportion of IFN-γ-expressing CD8+ TILs after cisplatin treatment (figure 5H). Suppression of Snail also enhanced the expression of Ifng in the harvested tumors (figure 5I). To further validate the Snail-miR-21 axis in regulating tumor response to cisplatin, we generated the murine LLC1-sh-ctrl, LLC1-sh-Snail, and LLC1-sh-Snail-mir21 sublines for in vivo experiments. The expression of Snail and mir-21 was validated in the corresponding cell lines (online supplemental figure 7E). Snail knockdown potentiated the effect of cisplatin treatment, and the expression of mir-21 in Snail-knockdown LLC1 cells abrogated this effect (online supplemental figure 7F-G). Taken together, these results suggest that Snail attenuates cisplatin-induced NLRP3 inflammasome activation of TAMs, which contributes to the chemoresistance of cancer cells.
Tumorous miR-21 repopulates the infiltrated immune cells in syngeneic tumors
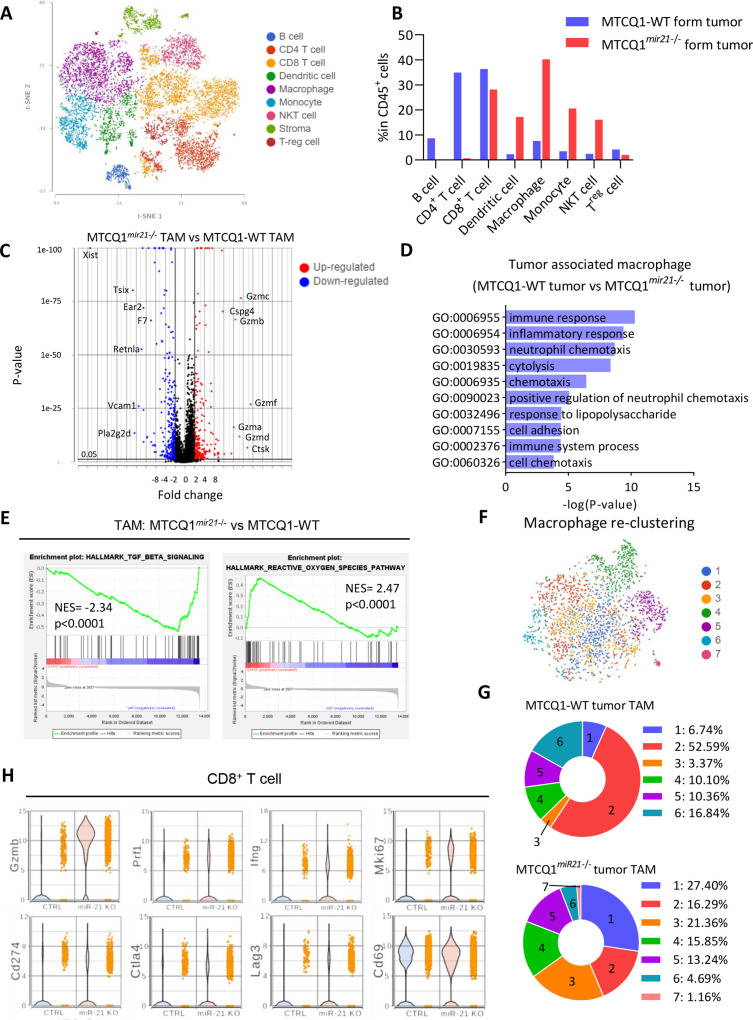
To understand the influence of mir-21 expression in chemotherapy-treated oral cancers on infiltrated immune cells, scRNA-seq was performed using the CD45+ cells sorted from the wild-type/mir21 knockout murine oral cancer MTCQ1 cell (MTCQ-WT/MTCQmir21–/–)-derived tumors 72 hours after chemotherapy (online supplemental figure 8A; online supplemental table 8). We used CIPR to annotate the cell clusters of the CD45+ cells,21 and different types of immune cells were identified accordingly (figure 6A and online supplemental figure 8B). The following pattern of infiltrated immune cells was distinct between the wild-type and mir-21 KO cancer cell-derived tumors: a significant increase in the proportion of macrophages, dendritic cells, monocytes, and natural killer cells and a decrease in the proportion of CD4+ T cells and regulatory T cells were noted in murine MTCQmir21–/–-derived tumors (figure 6B).
Figure 6.
Tumorous mir-21 shapes the infiltrated immune cells of syngeneic murine oral cancers. (A) The t-distributed stochastic neighbor embedding and immune cell type clustering (see Liu et al)28 of CD45+ cells in MTCQ1-WT and MTCQ1mir21–/–-formed tumors 3 days after cisplatin (5 mg/kg) injection. (B) Different immune cell type distribution between MTCQ1-WT and MTCQ1mir21–/–-formed tumor. (C) Volcano plots of the differential expressed genes of the TAMs from MTCQmir21–/– versus MTCQ1-WT-formed tumors. Red, upregulated genes; blue, downregulated genes. (D) GO enrichment analysis of the biological pathways of the TAMs from MTCQ1 mir21–/– versus MTCQ1-WT-formed tumors. (E) GSEA of the TGF-β signaling gene set (M5896) and reactive oxygen species pathway gene set (M5938) in MTCQ1mir21–/– TAMs versus MTCQ1-WT TAMs. (F) Re-clustering of the TAMs from MTCQ1-WT and MTCQ1mir21–/– -formed tumors. (G) Distribution of the TAM clusters of MTCQ1-WT or MTCQ1mir21–/– TAM. (H) Violin plots for showing the expression of the T cell activation genes (Gzmb, Pfr1, Ifng, Mki67) and immune checkpoint genes (Cd274, Ctla4, Lag3, Cd69) expression in CD8+ T cells of the MTCQ1-WT and MTCQ1mir21–/– tumors. GSEA, Gene Set Enrichment Analysis; GO, Gene Ontology; miR, micro RNA; TAM, tumor-associated macrophages; TGF-ß: transforming growth factor ß.
jitc-2022-004832supp009.xlsx (10.6KB, xlsx)
Next, we analyzed the TAMs in MTCQ-WT- and MTCQmir21–/–-derived tumors. The results showed that TAMs from MTCQmir21–/–-derived tumors harbored an immunoactive gene signature compared with that from MTCQ-WT-derived tumors (figure 6C, online supplemental table 9). GO analysis revealed that the pathways related to immune responses and inflammation were significantly enriched in TAMs of MTCQ1mir21–/–-derived tumors (figure 6D, online supplemental table 10). GSEA demonstrated that compared with the TAMs of MTCQ1-WT-derived tumors, TAMs of MTCQmir21–/–-derived tumors were associated with a more significant reactive oxygen species-related signature and downregulated expression of the TGF-β-related signature (figure 6E). Since the polarity of macrophages affects the therapeutic efficacy of tumors significantly,42 we further analyzed the subclusters of TAMs in these two groups (figure 6F; online supplemental table 11). Re-clustering of macrophages demonstrated a distinct distribution of macrophage subgroups among TAMs from MTCQmir21–/–- and MTCQ-WT-derived tumors. A significantly higher proportion of cluster 1 was noted in the MTCQ1mir21–/– group, whereas a dominant cluster 2 was observed in the MTCQ1-WT group (figure 6G). Pathway analysis showed that TAMs other than cluster 2 TAMs were associated with an immunoactive signature (online supplemental figure 8C), (online supplemental table 12). However, there is no significant trend of M1/M2 polarity among these seven clusters. Cluster 1 macrophages tended to coexpress both M1 and M2 genes compared with the other clusters (online supplemental figure 8D). Single-cell trajectory analysis of the macrophages/monocytes from tumors revealed that the order of macrophage polarization was cluster 6 to clusters 2 and 3, followed by separation into clusters 1, 4, and 5 (online supplemental figure 9A). In addition to the expression signature in macrophages, the gene expression signature was also distinct between the dendritic cells obtained from the MTCQ1mir21–/– and MTCQ1-WT groups (online supplemental figure 9B), (online supplemental table 13).
jitc-2022-004832supp010.xlsx (1.6MB, xlsx)
jitc-2022-004832supp011.xlsx (21.4KB, xlsx)
jitc-2022-004832supp012.xlsx (10.1KB, xlsx)
jitc-2022-004832supp013.xlsx (38.4KB, xlsx)
jitc-2022-004832supp014.xlsx (1.5MB, xlsx)
Next, we investigated the influence of tumorous mir-21 expression on the infiltrated T cells. The violin plots showed that knockout of mir-21 in tumor cells increased the expression of the cytotoxic genes (Grmzb, Prf1), activated T-cell genes (Ifng, Mki67) in T cells, suggesting that the activity and cytotoxicity of T cell was enhanced in mir-21 knockout tumors. A mild increase of the immune checkpoints (Cd274, Ctla4, Lag3, and Cd69) was noted in TILs of mir-21 knockout tumors (figure 6H). Regarding the expression of the genes related to T cell exhaustion, a reduced Tcf1 and an increased Tbet was noted. The level of Tim3 and Pd1 was not changed significantly (online supplemental figure 9C). In summary, these data suggest that an increased antitumor activity of both macrophages and CD8+ T cells was observed in the infiltrated immune cells of mir21–/– tumors after chemotherapy.
The Snail-miR-21 axis correlates with the suppression of NLRP3 inflammasome activity and a worse response to chemotherapy in patients with head and neck cancer
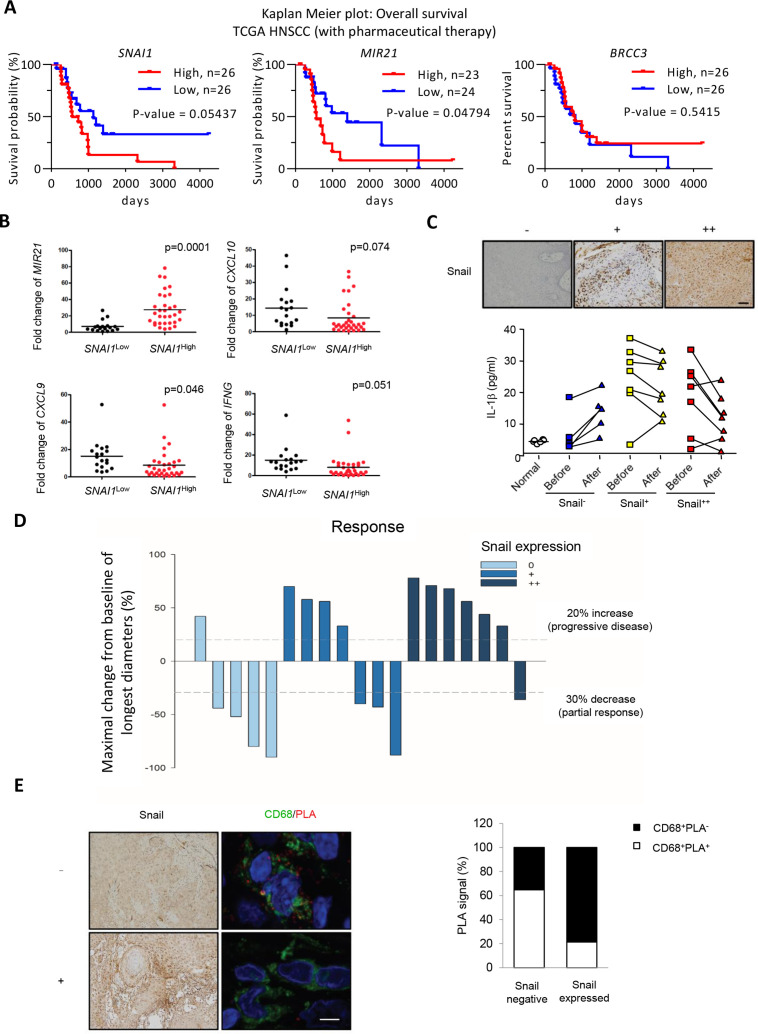
Finally, we validated the clinical significance of the Snail-miR-21 axis in the regulation of inflammasome activity and chemosensitivity in patients with HNSCC. We first examine the prognostic impact of Snail, miR-21, and BRCC3 in the HNSCC TCGA database. The result showed that in the total population of TCGA-HNSCC, a higher expression of SNAI1 was correlated with a worse overall survival (p=0.03858). The expression of BRCC3 and MIR21 did not have significant impact on the overall survival of HNSCC. We further analyzed the subgroup of patients with HNSCC receiving pharmaceutical treatment (n=53). SNAI1 or MIR21 upregulation correlated with worse an overall survival, whereas BRCC3 expression did not have a significant prognostic impact (figure 7A). This result indicates that the Snail-miR-21 axis has a negative impact on patient with HNSCC survival especially in patients receiving pharmaceutical therapies, which is consistent with our hypothesis that the Snail-miR-21 axis influences the treatment responses of patients with HNSCC.
Figure 7.
Snail limits chemotherapy-induced NLRP3 inflammasome activation in patients with HNSCC. (A) Kaplan-Meier plots for analyzing the influence of differential expression of SNAI1, MIR21, and BRCC3 on overall survival of patients with HNSCC with pharmaceutical therapy from TCGA database. The log-rank p value is shown in each panel. (B) Quantitative real-time PCR for analyzing the expression level of MIR21, CXCL10, IFNG and SNAI1 in HNSCC samples. n=50. The SNAI1High (n=32) is defined as the level higher than mean value, and the SNAI1Low (n=18) is defined as the level lower than mean value. (C) Upper, IHC staining of Snail with differential expressions in representative HNSCC samples. Scale bar, 50 µm. Lower, serum IL-1β level in patient with HNSCC before and 1 day after chemotherapy. (D) A waterfall plot for illustrating the response to chemotherapy of patients with HNSCC in (C). (E) Left, IHC staining of Snail (left) and immunofluorescent staining (right) of CD68 (green)/NLRP3-ASC PLA (red) in representative HNSCC samples. Right, quantification of the results. Three Snail-negative and two Snail-positive HNSCC samples were used in the experiment. For each sample, at least five CD68+ TAMs were quantified for PLA signals. The result is shown as the percentage of PLA-positive among CD68+ TAMs. Scale bar, 5 µm. HNSCC, head and neck squamous cell carcinoma; IHC, immunohistochemistry; PLA, proximity ligation assay; TAM, tumor-associated macrophages; TCGA, The Cancer Genome Atlas.
We next confirmed the findings in three independent sets of samples which were used in this study. In the first group, we retrospectively analyzed the expression of SNAI1 and IFN-γ gene expression signatures in 50 HNSCC samples. Increased expression of SNAI1 correlated with higher miR-21 and downregulated expression of IFN-γ signature genes IFN-γ, CXCL9, and CXCL10 (figure 7B). In the second group, we prospectively collected serum samples from 19 patients with HNSCC who received chemotherapy. The samples were obtained 1 day before and 1 day after chemotherapy to examine the level of IL-1β to indicate the activity of inflammasomes. The expression of Snail in the corresponding tumors was also examined. In patients with low Snail expression, upregulated serum IL-1β levels were observed after chemotherapy. When tumors expressed a higher level of Snail (Snail + ~ ++), chemotherapy could not induce a post-treatment surge in IL-1β levels (figure 7C). The treatment response was concordant with the expression of Snail, that is, a low percentage of tumor response to chemotherapy was noted in patients with Snail +~++ (figure 7D). In the third group, we examined the correlation between Snail and activated NLRP3 inflammasome in TAMs in five tumor samples. PLA could detect activation of the NLRP3 inflammasome in tumor samples probed with anti-NLRP3 and anti-ASC antibodies. Immunofluorescence staining of CD68 was performed to indicate the activation of the NLRP3 inflammasome in TAMs. Reduced abundance of CD68+PLA+ TAMs was observed in Snail-overexpressing HNSCC cells (figure 7E). In summary, the clinical sample data support that the Snail-miR21 axis regulates the activity of the NLRP3 inflammasome to attenuate the response to chemotherapy in HNSCC.
We summarize our finding in online supplemental figure 10. In cancer cells with high expression of Snail, the tumor-derived exosomes are enriched with miR-21. The miR-21 suppresses the expression of PTEN and BRCC3 in macrophages, which results in the phosphorylation and K63 polyubiquitination of NLRP3, leading to the disassembly and inactivation of NLRP3 inflammasomes. NLRP3 inflammasome inactivation lowers IL-1β secretion, thereby the chemotherapy-induced inflammation and immunogenic cell death is reduced.
Discussion
Despite the arguments put forth regarding inflammasome activation in cancer progression and therapeutic responses, recent studies support the role of inflammasome activation in augmentation of the chemotherapy response via enhancement of antitumor immunity. Chemotherapy-induced damage-associated molecular patterns include extracellular ATP, a potent activator of the NLRP3 inflammasome,43 which activates the inflammasome of dendritic cells to produce IL-1β and IL-18 to expand immune signals and to recruit CD8+ T-cells for antitumor immune responses.14 A recent study has shown that phosphorylation of NLRP3 in myeloid cells reduces NLRP3-ASC interaction and inhibits inflammasome activity, resulting in the development of chemoresistance.20 Intriguingly, our clinical data showed that in patients with advanced HNSCC harboring a higher Snail expression, the basal level of serum IL-1β was relatively high compared with the Snail-low cases. However, the post-treatment surge in IL-1β levels was not observed in the Snail-high HNSCC and was associated with a worse chemotherapy response (figure 7C–D). The high basal level and lack of response of IL-1β in Snail-high advanced HNSCC cases implicate pro-tumorous inflammatory microenvironments of aggressive tumors with blunted chemotherapy-triggered inflammasome activation. Together with these findings, our study clarifies the context-dependent role of the inflammasome of innate immune cells in cancer, in which inflammasome-induced proinflammatory signals favor tumor growth. In contrast, therapy-induced inflammasome activation facilitates chemotherapy-induced antitumor immunity and treatment responses.
Our data showed that miR-21 was the most abundant exosomal miRNA in HNSCC, consistent with reports regarding the significant oncogenic role and abundance of miR-21 in different types of cancers.44 The miR-21 exerts its oncogenic effects by targeting different tumor suppressors, such as PTEN,39 PDCD4,45 46 and IGFBP347 to promote tumor growth. However, the role of miR-21 in immune cells is more complicated than its prominent pro-tumorous effects in cancer cells. It is well established that miR-21 is predominantly expressed in M2 macrophages.6 48 49 In this study, we showed that miR-21 suppresses NLRP3 inflammasome activation in TAMs. Nevertheless, miR-21 has been reported to activate NLRP3 inflammasome in septic shock patients by repressing A20 to activate NF-κB for increasing the transcription of inflammasome components.36 The possible explanation for the differential role of miR-21 in regulating NLRP3 inflammasome in different disease context is as follows. NLRP3 inflammasome activation can be regulated at both the transcription level of the inflammasome components for regulating the amount of inflammasome subunits and post-translational modification of NLRP3 for regulating inflammasome assembly. In patients with cancer, the primary TMEs contain abundant inflammatory cytokines,50 and macrophages in primary tumors expressed both the M1 and M2 phenotype.5 We consider that the basal level of the components of NLRP3 inflammasome is relatively high in patients with cancer, the assembly of inflammasome is therefore crucial for regulating its activities. In patients with septic shock, activation of inflammasomes is the triggering event for the subsequent immune response, therefore induction of components of the expression of NLRP3 inflammasome is important.
Compared with the prominent oncogenic role in cancer cells, contradictory findings were noted regarding the antitumor immunity of miR-21. Knockout of mir-21 in mice slowed the proliferation of both CD4+ and CD8+ cells and accelerated the growth of engrafted tumors.51 However, tumorous miR-21 is inversely associated with the densities of CD3+ and CD45RO+ cells in colorectal cancers.52 Furthermore, miR-21 promotes M2 polarization,49 and our previous work has shown that miR-21-containing TEXs are potent regulators of TAMs through the promotion of M2-like activation.6 Here, we further investigated the role of Snail-miR-21 axis in antitumor immunity. Snail-driven miR-21-abundant TEXs inhibited NLRP3 inflammasome activity of macrophages. In HNSCC, an increased infiltration of TAMs was observed in samples with a higher expression of Snail (online supplemental figure 1A). Knockout of mir-21 in the murine oral cancer cell line MTCQ (MTCQmir21–/–) upregulated caspase-1 activity of TAMs when treated with cisplatin (figure 4C). In the scRNA-seq analysis, knockout of mir-21 reduced the proportion of CD4+ TILs but activated CD8+ TILs. Regarding the innate immune cells, depletion of mir-21 prominently enriched and shaped the gene expression signature of innate immune cells such as macrophages and dendritic cells to an antitumor profile (figure 6). Taken together, we consider that the malignant behaviors of miR-21-abundant cancers are attributed to both the oncogenic effect of intracancerous miR-21 and the TME-modulating effect of exosomal miR-21. In tumor-infiltrated immune cells, miR-21 may majorly affect innate immune cells in response to stimuli, highlighting the importance of exosomal transmission of miR-21.
In conclusion, we demonstrate the development of an immune adaptation-related chemoresistance driven by Snail during metastatic evolution. The plasticity of cancer cells dynamically shapes the tumor-infiltrated immune cells in response to therapeutic stimuli, indicating the necessity of real-time adjustment of treatment strategies alongside tumor progression.
jitc-2022-004832supp015.pdf (170.3KB, pdf)
Acknowledgments
We thank Professor Shie-Liang Hsieh (Genomics Research Center of Academia Sinica, Taiwan) and Nien-Jung Chen (Institute of Microbiology and Immunology, National Yang Ming Chiao Tung University, Taiwan) for providing the Nlrp3-/- C57BL/6 mice and technique supports; Dr Szu-Ting Chen (Institute of Clinical Medicine, National Yang Ming Chiao Tung University, Taiwan) for the pcDNA3-Flag-NLRP3 plasmids and critical comments; Tri-Biotech (Taiwan) for technical support related to the exosomal micro RNA sequencing; Dr Tsai-Yu Tzeng (Cancer Progression Research Center, National Yang Ming Chiao Tung University, Taiwan) for providing the sgRNA/Cas9 expression vector and assistance in the CRISPR/Cas9 technology; and Man-Ting Yu (National Yang Ming Chiao Tung University University) for the excellent technical support. We thank Division of Experimental Surgery, Department of Surgery of Taipei Veterans General Hospital for the assistance in processing clinical samples.
Footnotes
Contributors: M-HY is the guarantor of the study and supervised the whole study. M-HY, H-YC, and C-HH conceived and designed the experiments. H-YC and C-HH performed most of the experiments with the help of P-HL, Y-TC, and DS-SH; and they analyzed the data. M-HY, S-KT and P-YC provided clinical samples and patient care. M-HY collected all demographic data, interpreted the immunohistochemistry results, analyzed the relevant clinical parameters, and performed the statistical comparisons. H-YC, C-HH and M-HY wrote the paper.
Funding: This work was supported by grants from the Ministry of Science and Technology (109-2320-B-010-042 and 108-2314-B-010-020-MY3 to M-HY); the National Health Research Institutes (NHRI-EX109-10919BI to M-HY); Taipei Veterans General Hospital (V110C-139 and V109E-007-01 to M-HY); Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (to M-HY), and the Ministry of Health and Welfare, Center of Excellence for Cancer Research (MOHW110-TDU-B-211-144019 to M-HY).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available upon reasonable request.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study involves human participants and was approved by Institutional Review Board of Taipei Veterans General Hospital (TVGH-IRB certificate No. 2014-03-004AC; No. 2017-05-013AC; No. 2018-06-001BC). Participants gave informed consent to participate in the study before taking part.
References
- 1.Marine J-C, Dawson S-J, Dawson MA. Non-Genetic mechanisms of therapeutic resistance in cancer. Nat Rev Cancer 2020;20:743–56. 10.1038/s41568-020-00302-4 [DOI] [PubMed] [Google Scholar]
- 2.Ye X, Weinberg RA. Epithelial-Mesenchymal plasticity: a central regulator of cancer progression. Trends Cell Biol 2015;25:675–86. 10.1016/j.tcb.2015.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shibue T, Weinberg RA. Emt, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol 2017;14:611–29. 10.1038/nrclinonc.2017.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsu DS-S, Wang H-J, Tai S-K, et al. Acetylation of snail modulates the cytokinome of cancer cells to enhance the recruitment of macrophages. Cancer Cell 2014;26:534–48. 10.1016/j.ccell.2014.09.002 [DOI] [PubMed] [Google Scholar]
- 5.Lee C-C, Lin J-C, Hwang W-L, et al. Macrophage-Secreted interleukin-35 regulates cancer cell plasticity to facilitate metastatic colonization. Nat Commun 2018;9:3763. 10.1038/s41467-018-06268-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsieh C-H, Tai S-K, Yang M-H. Snail-overexpressing cancer cells promote M2-like polarization of tumor-associated macrophages by delivering miR-21-abundant exosomes. Neoplasia 2018;20:775–88. 10.1016/j.neo.2018.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol 2019;20:69–84. 10.1038/s41580-018-0080-4 [DOI] [PubMed] [Google Scholar]
- 8.Kudo-Saito C, Shirako H, Takeuchi T, et al. Cancer metastasis is accelerated through immunosuppression during snail-induced EMT of cancer cells. Cancer Cell 2009;15:195–206. 10.1016/j.ccr.2009.01.023 [DOI] [PubMed] [Google Scholar]
- 9.Dongre A, Rashidian M, Reinhardt F, et al. Epithelial-To-Mesenchymal transition contributes to immunosuppression in breast carcinomas. Cancer Res 2017;77:3982–9. 10.1158/0008-5472.CAN-16-3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taki M, Abiko K, Ukita M, et al. Tumor immune microenvironment during epithelial-mesenchymal transition. Clin Cancer Res 2021;27:4669–79. 10.1158/1078-0432.CCR-20-4459 [DOI] [PubMed] [Google Scholar]
- 11.Li C-W, Xia W, Huo L, et al. Epithelial-Mesenchymal transition induced by TNF-α requires NF-κB-mediated transcriptional upregulation of Twist1. Cancer Res 2012;72:1290–300. 10.1158/0008-5472.CAN-11-3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katsura A, Tamura Y, Hokari S, et al. ZEB1-regulated inflammatory phenotype in breast cancer cells. Mol Oncol 2017;11:1241–62. 10.1002/1878-0261.12098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo H, Callaway JB, Ting JP-Y. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat Med 2015;21:677–87. 10.1038/nm.3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghiringhelli F, Apetoh L, Tesniere A, et al. Activation of the NLRP3 inflammasome in dendritic cells induces IL-1beta-dependent adaptive immunity against tumors. Nat Med 2009;15:1170–8. 10.1038/nm.2028 [DOI] [PubMed] [Google Scholar]
- 15.Allen IC, TeKippe EM, Woodford R-MT, et al. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J Exp Med 2010;207:1045–56. 10.1084/jem.20100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huber S, Gagliani N, Zenewicz LA, et al. IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature 2012;491:259–63. 10.1038/nature11535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ershaid N, Sharon Y, Doron H, et al. Nlrp3 inflammasome in fibroblasts links tissue damage with inflammation in breast cancer progression and metastasis. Nat Commun 2019;10:4375. 10.1038/s41467-019-12370-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kroemer G, Galluzzi L, Kepp O, et al. Immunogenic cell death in cancer therapy. Annu Rev Immunol 2013;31:51–72. 10.1146/annurev-immunol-032712-100008 [DOI] [PubMed] [Google Scholar]
- 19.Son S, Shim D-W, Hwang I, et al. Chemotherapeutic agent paclitaxel mediates priming of NLRP3 inflammasome activation. Front Immunol 2019;10:1108. 10.3389/fimmu.2019.01108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Y, Wang H, Hao Y, et al. Myeloid PTEN promotes chemotherapy-induced NLRP3-inflammasome activation and antitumour immunity. Nat Cell Biol 2020;22:716–27. 10.1038/s41556-020-0510-3 [DOI] [PubMed] [Google Scholar]
- 21.Ekiz HA, Conley CJ, Stephens WZ, et al. CIPR: a web-based R/shiny APP and R package to annotate cell clusters in single cell RNA sequencing experiments. BMC Bioinformatics 2020;21:191. 10.1186/s12859-020-3538-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene Lists using David bioinformatics resources. Nat Protoc 2009;4:44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 23.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang M-H, Hsu DS-S, Wang H-W, et al. Bmi1 is essential in Twist1-induced epithelial-mesenchymal transition. Nat Cell Biol 2010;12:982–92. 10.1038/ncb2099 [DOI] [PubMed] [Google Scholar]
- 25.Yang W-H, Lan H-Y, Huang C-H, et al. Rac1 activation mediates Twist1-induced cancer cell migration. Nat Cell Biol 2012;14:366–74. 10.1038/ncb2455 [DOI] [PubMed] [Google Scholar]
- 26.Chae YK, Chang S, Ko T, et al. Epithelial-Mesenchymal transition (EMT) signature is inversely associated with T-cell infiltration in non-small cell lung cancer (NSCLC). Sci Rep 2018;8:2918. 10.1038/s41598-018-21061-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yadav A, Kumar B, Datta J, et al. Il-6 promotes head and neck tumor metastasis by inducing epithelial-mesenchymal transition via the JAK-STAT3-SNAIL signaling pathway. Mol Cancer Res 2011;9:1658–67. 10.1158/1541-7786.MCR-11-0271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu C-Y, Xu J-Y, Shi X-Y, et al. M2-Polarized tumor-associated macrophages promoted epithelial-mesenchymal transition in pancreatic cancer cells, partially through TLR4/IL-10 signaling pathway. Lab Invest 2013;93:844–54. 10.1038/labinvest.2013.69 [DOI] [PubMed] [Google Scholar]
- 29.Hwang W-L, Yang M-H, Tsai M-L, et al. Snail regulates interleukin-8 expression, stem cell-like activity, and tumorigenicity of human colorectal carcinoma cells. Gastroenterology 2011;141:279–91. 91 e1-5. 10.1053/j.gastro.2011.04.008 [DOI] [PubMed] [Google Scholar]
- 30.Peinado H, Alečković M, Lavotshkin S, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through Met. Nat Med 2012;18:883–91. 10.1038/nm.2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou W, Fong MY, Min Y, et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 2014;25:501–15. 10.1016/j.ccr.2014.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baltimore D, Boldin MP, O'Connell RM, et al. Micrornas: new regulators of immune cell development and function. Nat Immunol 2008;9:839–45. 10.1038/ni.f.209 [DOI] [PubMed] [Google Scholar]
- 33.Iorio MV, Croce CM. Microrna dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med 2012;4:143–59. 10.1002/emmm.201100209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hwang W-L, Jiang J-K, Yang S-H, et al. Microrna-146A directs the symmetric division of Snail-dominant colorectal cancer stem cells. Nat Cell Biol 2014;16:268–80. 10.1038/ncb2910 [DOI] [PubMed] [Google Scholar]
- 35.Vetter G, Saumet A, Moes M, et al. miR-661 expression in SNAI1-induced epithelial to mesenchymal transition contributes to breast cancer cell invasion by targeting nectin-1 and StarD10 messengers. Oncogene 2010;29:4436–48. 10.1038/onc.2010.181 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Xue Z, Xi Q, Liu H, et al. miR-21 promotes NLRP3 inflammasome activation to mediate pyroptosis and endotoxic shock. Cell Death Dis 2019;10:461. 10.1038/s41419-019-1713-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Juliana C, Fernandes-Alnemri T, Kang S, et al. Non-Transcriptional priming and deubiquitination regulate NLRP3 inflammasome activation. J Biol Chem 2012;287:36617–22. 10.1074/jbc.M112.407130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Py BF, Kim M-S, Vakifahmetoglu-Norberg H, et al. Deubiquitination of NLRP3 by BRCC3 critically regulates inflammasome activity. Mol Cell 2013;49:331–8. 10.1016/j.molcel.2012.11.009 [DOI] [PubMed] [Google Scholar]
- 39.Meng F, Henson R, Wehbe-Janek H, et al. Microrna-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 2007;133:647–58. 10.1053/j.gastro.2007.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y-F, Chang K-W, Yang I-T, et al. Establishment of syngeneic murine model for oral cancer therapy. Oral Oncol 2019;95:194–201. 10.1016/j.oraloncology.2019.06.026 [DOI] [PubMed] [Google Scholar]
- 41.Ben-Sasson SZ, Wang K, Cohen J, et al. IL-1β strikingly enhances antigen-driven CD4 and CD8 T-cell responses. Cold Spring Harb Symp Quant Biol 2013;78:117–24. 10.1101/sqb.2013.78.021246 [DOI] [PubMed] [Google Scholar]
- 42.DeNardo DG, Ruffell B. Macrophages as regulators of tumour immunity and immunotherapy. Nat Rev Immunol 2019;19:369–82. 10.1038/s41577-019-0127-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mariathasan S, Weiss DS, Newton K, et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature 2006;440:228–32. 10.1038/nature04515 [DOI] [PubMed] [Google Scholar]
- 44.Bautista-Sánchez D, Arriaga-Canon C, Pedroza-Torres A, et al. The promising role of miR-21 as a cancer biomarker and its importance in RNA-based therapeutics. Mol Ther Nucleic Acids 2020;20:409–20. 10.1016/j.omtn.2020.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frankel LB, Christoffersen NR, Jacobsen A, et al. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem 2008;283:1026–33. 10.1074/jbc.M707224200 [DOI] [PubMed] [Google Scholar]
- 46.Asangani IA, Rasheed SAK, Nikolova DA, et al. Microrna-21 (miR-21) post-transcriptionally downregulates tumor suppressor PDCD4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 2008;27:2128–36. 10.1038/sj.onc.1210856 [DOI] [PubMed] [Google Scholar]
- 47.Yang CH, Yue J, Pfeffer SR, et al. Microrna-21 promotes glioblastoma tumorigenesis by down-regulating insulin-like growth factor-binding protein-3 (IGFBP3). J Biol Chem 2014;289:25079–87. 10.1074/jbc.M114.593863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caescu CI, Guo X, Tesfa L, et al. Colony stimulating factor-1 receptor signaling networks inhibit mouse macrophage inflammatory responses by induction of microRNA-21. Blood 2015;125:e1–13. 10.1182/blood-2014-10-608000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xue J, Xiao T, Wei S, et al. miR-21-regulated M2 polarization of macrophage is involved in arsenicosis-induced hepatic fibrosis through the activation of hepatic stellate cells. J Cell Physiol 2021;236:6025–41. 10.1002/jcp.30288 [DOI] [PubMed] [Google Scholar]
- 50.Greten FR, Grivennikov SI. Inflammation and cancer: triggers, mechanisms, and consequences. Immunity 2019;51:27–41. 10.1016/j.immuni.2019.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He W, Wang C, Mu R, et al. Mir-21 is required for anti-tumor immune response in mice: an implication for its bi-directional roles. Oncogene 2017;36:4212–23. 10.1038/onc.2017.62 [DOI] [PubMed] [Google Scholar]
- 52.Mima K, Nishihara R, Nowak JA, et al. Microrna MIR21 and T cells in colorectal cancer. Cancer Immunol Res 2016;4:33–40. 10.1158/2326-6066.CIR-15-0084 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2022-004832supp001.pdf (3.3MB, pdf)
jitc-2022-004832supp002.xlsx (10.5KB, xlsx)
jitc-2022-004832supp003.xlsx (11.2KB, xlsx)
jitc-2022-004832supp004.xlsx (10.7KB, xlsx)
jitc-2022-004832supp005.xlsx (11KB, xlsx)
jitc-2022-004832supp006.xlsx (10.7KB, xlsx)
jitc-2022-004832supp007.xlsx (12.7KB, xlsx)
jitc-2022-004832supp008.xlsx (19.3KB, xlsx)
jitc-2022-004832supp009.xlsx (10.6KB, xlsx)
jitc-2022-004832supp010.xlsx (1.6MB, xlsx)
jitc-2022-004832supp011.xlsx (21.4KB, xlsx)
jitc-2022-004832supp012.xlsx (10.1KB, xlsx)
jitc-2022-004832supp013.xlsx (38.4KB, xlsx)
jitc-2022-004832supp014.xlsx (1.5MB, xlsx)
jitc-2022-004832supp015.pdf (170.3KB, pdf)
Data Availability Statement
The accession numbers for the data reported in this paper are GEO: GSE99474, GSE172326, GSE178537, and GSE 181300.
Data are available upon reasonable request.