Abstract
The response of Escherichia coli central carbon metabolism to genetic and environmental manipulation has been studied by use of a recently developed methodology for metabolic flux ratio (METAFoR) analysis; this methodology can also directly reveal active metabolic pathways. Generation of fluxome data arrays by use of the METAFoR approach is based on two-dimensional 13C-1H correlation nuclear magnetic resonance spectroscopy with fractionally labeled biomass and, in contrast to metabolic flux analysis, does not require measurements of extracellular substrate and metabolite concentrations. METAFoR analyses of E. coli strains that moderately overexpress phosphofructokinase, pyruvate kinase, pyruvate decarboxylase, or alcohol dehydrogenase revealed that only a few flux ratios change in concert with the overexpression of these enzymes. Disruption of both pyruvate kinase isoenzymes resulted in altered flux ratios for reactions connecting the phosphoenolpyruvate (PEP) and pyruvate pools but did not significantly alter central metabolism. These data indicate remarkable robustness and rigidity in central carbon metabolism in the presence of genetic variation. More significant physiological changes and flux ratio differences were seen in response to altered environmental conditions. For example, in ammonia-limited chemostat cultures, compared to glucose-limited chemostat cultures, a reduced fraction of PEP molecules was derived through at least one transketolase reaction, and there was a higher relative contribution of anaplerotic PEP carboxylation than of the tricarboxylic acid (TCA) cycle for oxaloacetate synthesis. These two parameters also showed significant variation between aerobic and anaerobic batch cultures. Finally, two reactions catalyzed by PEP carboxykinase and malic enzyme were identified by METAFoR analysis; these had previously been considered absent in E. coli cells grown in glucose-containing media. Backward flux from the TCA cycle to glycolysis, as indicated by significant activity of PEP carboxykinase, was found only in glucose-limited chemostat culture, demonstrating that control of this futile cycle activity is relaxed under severe glucose limitation.
Access to complete genome sequence information for a number of microorganisms now motivates the development and application of experimental techniques for phenotype characterization (such as transcriptome and proteome analyses), providing arrays of data that can be directly mapped to corresponding arrays of genes (14, 36). The physiological counterpart to such composition arrays is the array of fluxes (reaction rates on a per-unit cell volume or per-unit cell mass basis) for all of the reactions that occur in the organism, for which we use, by analogy, the term fluxome. Approximate fluxome access for certain subsets of metabolism can be attained by methods of metabolic flux analysis, which require data on uptake and efflux rates of certain metabolites outside the cell and which assume a corresponding network of metabolic pathways in the cell (39). Alternatively, by use of more recently introduced methodology based on isotopic imprinting of amino acids by their precursors, the active central carbon pathways and the ratios of their fluxes can be directly determined from two-dimensional (2D) nuclear magnetic resonance (NMR) analysis of hydrolyzed cell protein (30–33). This method, for which we introduce the term METAFoR (metabolic flux ratio) analysis, offers a relatively high throughput access to these key fluxome elements, enabling physiological data arrays to be acquired over a broad range of genetic and environmental conditions.
Specifically, METAFoR analysis quantifies the relative abundance of intact carbon bonds originating from uniformly isotopically labeled source molecules by use of proton-detected 2D 13C-1H correlation NMR spectroscopy (COSY) (30, 34, 42). Such 2D NMR analysis of amino acids obtained from hydrolyzed cell protein permits quantitative analysis of the relative abundance of intact, contiguous fragments in the precursor metabolites of central metabolism, because the carbon backbone of these molecules is conserved in the amino acids. Typically, fractional 13C labeling of amino acids is achieved by growing cells with a mixture of 85 to 90% natural-abundance glucose and 10 to 15% [U-13C6]glucose (22, 27, 30–32, 34). Because alternative pathways leading to common intermediates or products produce different intact fragments originating from a single glucose source molecule (30–32), specific multiplet patterns in the 13C fine structures that reflect the in vivo usage of reactions are generated. Probabilistic equations relate the determined intensities of the multiplet components to the relative abundance of intact carbon fragments (30) and thus allow derivation of intracellular carbon flux ratios (30–33). These data provide not only comprehensive insight into cellular metabolism but also inherent flux indications that can provide critical information for metabolic (net) flux analysis (27, 32).
The active pathways and the flux distribution in central carbon metabolism are critical components of a multidimensional physiological representation of the organism, since this central backbone of metabolism provides energy, cofactor regeneration, and building blocks for biomass synthesis and controls the extent and nature of by-product excretion. A wide array of regulatory responses are embedded in this network on the transcriptional level as well as the protein level. The purpose of this complex regulatory structure is not yet fully elucidated, but the observed insensitivity of growth rates and extracellular fluxes to the overexpression of key enzymes suggests a homeostatic objective of the regulatory system (4, 8, 38).
In this study, we used METAFoR analysis to examine, at the level of flux ratios and operational pathways, how the central carbon physiology of Escherichia coli responds to genetic and environmental manipulations. In addition, we show the extent of variation in these facets of the central carbon network fluxome in several different standard laboratory strains. These METAFoR data show in detail how and under what conditions the E. coli central carbon metabolic network maintains flux ratio homeostasis and when significant alterations arise in both active pathways and flux ratios.
MATERIALS AND METHODS
Strains, plasmids, and media.
Strains and plasmids used in this study are listed in Table 1. All batch cultivations were performed with a minimal medium containing 5 g of glucose per liter, 48 mM Na2HPO4, 22 mM KH2PO4, 10 mM NaCl, and 30 mM (NH4)2SO4. The following components were sterilized separately and then added (per liter of final medium): 1 ml of 1 M MgSO4, 1 ml of 0.1 mM CaCl2, 1 ml of 1 mg of vitamin B1 per liter (filter sterilized), and 10 ml of trace element solution containing (per liter) 0.55 g of CaCl2 · 2H2O, 1 g of FeCl3, 0.1 g of MnCl2 · 4H2O, 0.17 g of ZnCl2, 0.043 g of CuCl2 · 2H2O, 0.06 g of CoCl2 · 6H2O, and 0.06 g of Na2MoO4 · 2H2O. To ensure maintenance of plasmids, ampicillin was added to a final concentration of 25 mg/liter. The medium fed into the glucose-limited chemostat had the same composition as the batch medium, with the following exceptions (per liter): 3.6 g of glucose, 4.7 g of Na2HPO4 · H2O, 3.0 g of KH2PO4, 0.5 g of NaCl, and 1 g of NH4Cl. To enforce nitrogen limitation, the concentration of glucose was increased to 4.5 g/liter, and the concentration of the sole nitrogen source, NH4Cl, was reduced to 0.7 g/liter. Chemostat media were sterilized by passage through a 0.2-μm-pore-size filter, and 10-fold-diluted trace element solution was added after filtration to prevent losses via precipitation.
TABLE 1.
E. coli strains and plasmids used
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| MG1655 | Wild-type K-12 strain (λ− F−rph-1) | 1 |
| JM101 | [F−traD36 lacIq Δ(lacZ)M15 proA+B+ supE thi Δ(lac-proAB)] | 43 |
| PB25 | Pyruvate kinase-deficient JM101 (pykA::kan pykF::cat) | 25 |
| ATCC 11303 | Wild-type B strain; prototroph | American Type Culture Collection |
| KO20 | Ethanol-producing ATCC 11303; chromosomal insertion of the pet operon (pyruvate decarboxylase and alcohol dehydrogenase II) of Z. mobilis into the pyruvate formate-lyase gene | 23 |
| Plasmids | ||
| pTrc99a | E. coli expression vector | Pharmacia |
| pPPec | pTrc99a derivative for expression of the artificial E. coli pykF-pfkA operon | 6 |
| pPYKbs | pTrc99a derivative for expression of the pyruvate kinase from Bacillus stearothermophilus | 6 |
Batch and chemostat cultivations.
All batch cultivations were performed at 30°C. Aerobic batch cultures were grown in 1-liter baffled shake flasks with 150 ml of medium on a gyratory shaker at 200 rpm. Anaerobic batch cultivations were performed with rubber-sealed glass flasks previously flushed with N2 and incubated in a gyratory water bath (G76D; New Brunswick). Chemostats were operated at 37°C in a 1.5-liter bench-top fermentor (Bioengineering) with a working volume of 1.0 liter and a constant dilution rate (D) of 0.2 h−1, meaning that the feed rate was 0.2 liter/h. The working volume was kept constant by removal of effluent from the center of the culture volume by use of a weight-controlled pump. The pH of the culture was maintained at 7.0 by automatic addition of 2.0 M NaOH with a pH controller and was verified periodically by off-line measurements. The airflow was maintained at 1 liter/min with filter-sterilized air by use of a volume flow meter, and the agitation speed was set to 1,200 rpm.
Labeling experiments with chemostats were initiated after the cultures appeared to reach a steady state, inferred from (i) at least five volume changes after adjustment to new conditions and (ii) stable optical density and oxygen and carbon dioxide concentrations in the fermentor effluent gas for at least two volume changes. The feed medium containing 3.6 (or 4.5 in the NH4+-limited experiment) g of unlabeled glucose per liter was then replaced by an identical medium containing 3.24 (4.05) g of glucose labeled by natural abundance per liter and 0.36 (0.45) g of [U-13C6]glucose (13C, >98%; Isotech) per liter. Biomass samples for METAFoR analysis were taken after one volume change, so that 63% of the biomass was fractionally labeled according to the first-order washout kinetics that follow from assuming that the bioreactor contents are well mixed. Batch cultures were grown entirely in media supplemented with 4.5 g of glucose containing 13C at natural abundance per liter and 0.5 g of [U-13C6]glucose per liter. Because the percentage of unlabeled biomass originating from the inoculum was well below 1% in the batch cultures, unlabeled biomass was subsequently neglected in the analysis of the 13C-labeling patterns.
Analytical procedures.
Cell growth during the cultivations was monitored by measuring the optical density at 600 nm (OD600). For cellular dry weight (cdw) determination in selected cases, a known volume of fermentation broth was centrifuged for 10 min in preweighed glass tubes at 4°C and 3,000 × g, washed once with water, and dried at 90°C for 24 h to a constant weight. Samples for extracellular metabolite analysis were centrifuged for 1 min at maximum speed in an Eppendorf tabletop centrifuge to remove the cells. Glucose and ethanol concentrations were determined enzymatically (Synchron CX5CE apparatus; Beckman) with kits supplied by the manufacturer. Acetate (and ethanol, in selected cases) was measured by gas chromatography (5890E chromatograph; Hewlett-Packard) with a Carbowax MD-10 column (Macherey-Nagel) and butyrate as an internal standard. Concentrations of oxygen and carbon dioxide in the feed medium and off gas of bioreactor fermentations were determined with a mass spectrometer (Prima 600; Fisons Instruments).
Determination of physiological parameters.
In batch cultures, the exponential growth phase was identified by log-linear regression of biomass concentration versus time, with growth rate (μ) as the regression coefficient. The biomass yield on the substrate (YX/S) was determined as the coefficient of a linear regression of biomass concentration (X) versus substrate concentration (S) during the exponential growth phase. A predetermined correlation factor (OD600, 0.33) was used to convert the OD600 values into cell concentrations for the calculation of specific conversion rates. The specific consumption rate for a substrate (qS), e.g., glucose and O2—defined as the differential change in S with time (t) normalized to the biomass concentration—was obtained as the coefficient of a linear regression of ΔS (the change in S) versus X divided by μ, on the basis of the relationship St1 − St2 = ΔS = Xt2(qS/μ). The same relationship holds for the specific rate of formation of products (P), e.g., acetate and CO2. This relationship is linear provided that μ and qS are constant. In a steady-state chemostat, μ is constant and equals D. In batch cultures, maximum μ was constant during the exponential growth phase, and all specific rates from batch experiments reported here refer to the exponential phase.
In chemostat cultures, D and thus μ are constant; therefore, the consumption and production rates were determined from the difference between S or P in the feed medium (or air) and S or P in the effluent (or off gas). The relationship qS (or P) = ΔS (or P) (D/X) normalized these rates to the steady-state concentration of biomass, generating the corresponding specific rates.
NMR sample preparation.
For network topology and flux ratio analysis by NMR, a specified amount of culture was harvested and cells were centrifuged at 1,200 × g for 10 min at 4°C. The cell pellet was washed once with 20 mM Tris-HCl (pH 7.6) and centrifuged again. Washed pellets from chemostat cultures were resuspended in the above buffer, and cells were disrupted by sonication on ice three times for 45 s each time at 20% output (XL-2020 sonicator; Hert Systems). Cell debris was removed by centrifugation for 20 min at 9,000 × g. Sonication and centrifugation were repeated until cell lysis was virtually complete, as determined by visual inspection with a microscope. Small debris particles were removed by ultracentrifugation for 30 min at 33,000 × g. Cellular protein in the supernatant was precipitated overnight at −20°C after the addition of 60% (vol/vol) ethanol. The precipitate was resuspended in 6 ml of 6 M HCl and hydrolyzed by incubation in sealed Pyrex glass tubes for 24 h at 110°C. The hydrolysate was filtered through a 0.2-μm-pore-size filter and lyophilized. The dried material was dissolved in 600 μl of 20 mM deuterium chloride (DCl) in D2O, incubated for 2 h at room temperature, centrifuged, and used for the NMR measurements. Washed pellets from batch cultures were directly resuspended in 6 M HCl and hydrolyzed.
NMR spectroscopy and data analysis.
Proton-detected 2D 13C-1H heteronuclear single-quantum COSY was performed with the pulse sequence of Bodenhausen and Ruben (3), which ensures that 1H-13C scalar couplings do not affect the 13C-13C scalar coupling fine structure along the chemical shift ω1(13C) (34). Pulsed-field gradients were used for coherence pathway rejection (2, 40), and a 2-ms spin-lock pulse (24) was used to purge the magnetization arising from 12C-bound protons and the residual 2HOH signal. 13C decoupling during data acquisition was achieved by use of the composite pulse decoupling scheme GARP (28), and quadrature detection in ω1 was accomplished with States-TPPI (18). The spectra were recorded at a 1H resonance frequency of 500 MHz by use of a Bruker DRX500 spectrometer; the sample temperature was 40°C. For each sample, two spectra were measured: one spectrum for the aliphatic resonances, with the 13C carrier set to 42.5 ppm relative to 2,2-dimethyl-2-silapentane-5-sulfonate sodium salt, and one spectrum for the aromatic resonances, with the 13C carrier set to 125.9 ppm. The spectra of the aliphatic resonances were folded along ω1(13C) with a sweep width of 33.8 ppm. The measurement time was 4.5 h per spectrum (1,706 × 256 complex points; t1max, 402 ms; t2max, 102 ms; relaxation delay between scans, 2 s). The spectra of the aromatic resonances were recorded in about 2.5 h (920 × 512 complex points; t1max, 392 ms; t2max, 87 ms; relaxation delay between scans, 2 s). Before Fourier transformation with the program PROSA (12), the time domain data were multiplied in t1 and t2 with sine-bell windows shifted by π/2 (5). The digital resolutions after zero filling were 1.0 Hz/point along ω1 and 2.4 Hz/point along the second frequency axis ω2 for spectra of the aliphatic resonances and 0.6 Hz/point along ω1 and 5.8 Hz/point along ω2 for spectra of the aromatic resonances. One-dimensional 1H NMR spectra (tmax, 1.022 s; 10-s relaxation delay between scans) were recorded to determine the overall degree of 13C labeling in the amino acids from the satellites of isolated proton peaks, which corresponds to P1 in the probabilistic equations of Szyperski (30).
The relative abundance of intact carbon fragments present in the eight principal intermediates that link central carbon metabolism to amino acid biosynthesis was determined from the intensities of the individual multiplet components in the 13C-13C scalar coupling fine structures (27, 30). Flux ratios through several key pathways in central metabolism were then calculated from the abundance of the fragments as described previously (31, 32).
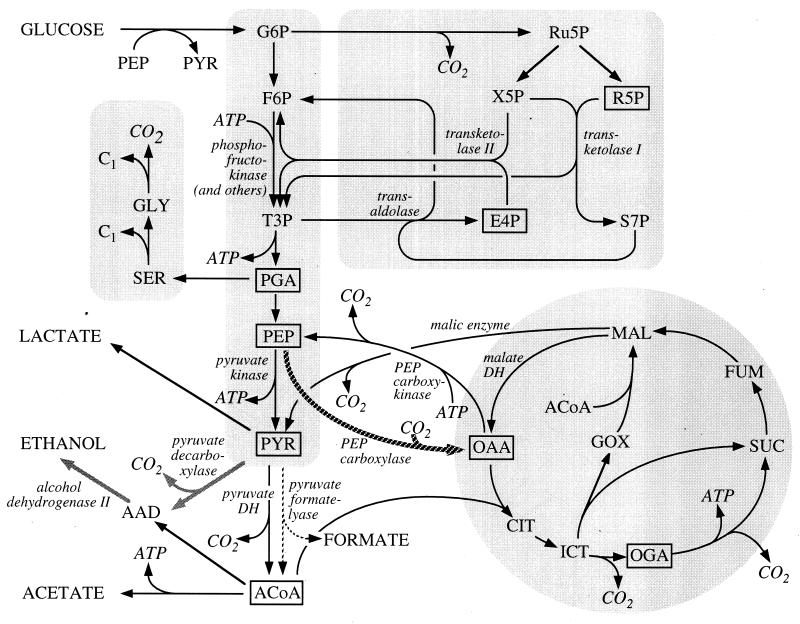
Biochemical reaction master network of E. coli.
As an initial step for METAFoR analysis, a biochemical master network that comprises all currently known reactions of central carbon and amino acid metabolism for the organism under investigation is constructed (30). For E. coli, this network (Fig. 1) was compiled from textbooks (11, 20) and Internet-accessible metabolic databases (16). Inspection of observed intact carbon fragments in the amino acids subsequently allows identification of active biosynthetic pathways (27, 30–32). Such analysis cannot distinguish between trioses that are generated via the methylglyoxal bypass or via the Entner-Doudoroff pathway, since these pathways generate fragment patterns that are indistinguishable from those emerging from the glycolysis and pentose phosphate (PP) pathways, respectively (30). Because the methylglyoxal bypass and the Entner-Doudoroff pathway were reported to be inactive for E. coli cells grown with glucose (11), they were not considered in the presently used network.
FIG. 1.
Biochemical master network with reactions for identified genes or enzyme activities in E. coli. The information was compiled from the EcoCyc database (17) and other sources (11, 20). The reaction sets of glycolysis, the PP pathway, the TCA cycle (including the glyoxylate shunt), and C1 metabolism are shaded, and enzymes catalyzing key reactions are indicated in italics. The hatched arrow highlights the TCA cycle-replenishing anaplerotic reaction, and the broken arrows indicate the anaerobic pyruvate formate-lyase, which is interrupted in strain KO20. The grey arrows indicate the reactions catalyzed by the enzymes encoded on the pet operon of Z. mobilis. The intact carbon fragment patterns of boxed metabolites were directly determined by 13C-1H COSY of proteinogenic amino acids. Abbreviations: F6P, fructose-6-phosphate; Ru5P, ribulose-5-phosphate; X5P, xylulose-5-phosphate; E4P, erythrose-4-phosphate; S7P, seduheptulose-7-phosphate; PGA, 3-phosphoglycerate; SER, serine; GLY, glycine; AAD, acetaldehyde; CIT, citrate; ICT, isocitrate; OGA, oxoglutarate; SUC, succinate; GOX, glyoxylate; and DH, dehydrogenase.
RESULTS
Analysis of glucose- and ammonia-limited chemostat cultures of wild-type E. coli MG1655.
Continuous cultivation was performed with aerobic chemostats at a D (volumetric flow rate/working volume) of 0.2 h−1 under glucose- or ammonia-limited conditions, representing two largely different bioenergetic regimens. Carbon-sufficient (i.e., ammonia-limited) cultures are known to exhibit metabolic behavior that differs from that of carbon-limited cultures with respect to specific substrate consumption rate, maintenance requirements, and by-product secretion (21). This fact is reflected by the physiological data from the cultures described here (Table 2). When the ammonia-limited and the glucose-limited cultures were compared, marked increases were found in the specific glucose consumption rate (qglc) and in the specific rates of production of acetate and pyruvate, while the specific oxygen consumption rate and CO2 evolution rate varied little between the two different conditions; these results indicated that there are only minor changes in respiratory metabolism. The low steady-state biomass concentration in the ammonia-limited culture results from the low ammonia concentration used (13.1 mM). The residual glucose concentration in this culture was slightly above 1 g/liter.
TABLE 2.
Aerobic growth parameters of glucose- or ammonia-limited chemostat cultures of E. coli MG1655 at a D of 0.2 h−1
| Limitation | Biomass
|
Specific rate (mmol g−1 h−1) of:
|
||||||
|---|---|---|---|---|---|---|---|---|
| Concn (g [cdw] liter−1) | Yield on:
|
Glucose consumption | Acetate formation | Pyruvate formation | O2 consumption | CO2 formation | ||
| Glucose (g g of glucose−1) | Nitrogen (g g of N−1) | |||||||
| Glucose | 1.45 | 0.40 | 5.5 | 2.8 | 0 | 0 | 8.3 | 8.7 |
| Ammonia | 0.55 | 0.17 | 3.0 | 6.4 | 4.4 | 0.8 | 10.6 | 9.6 |
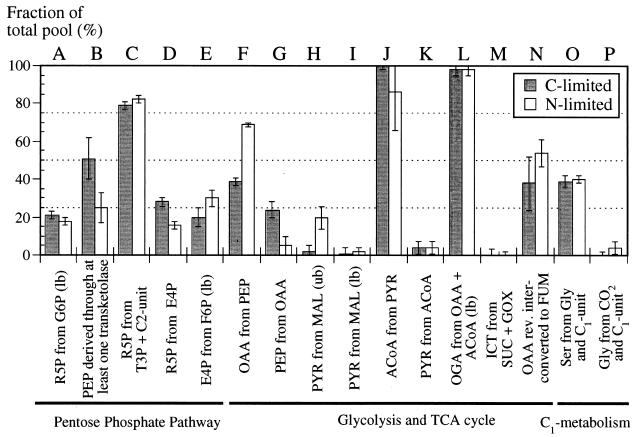
For both cultures, the METAFoR data show evidence of two reactions that are generally considered to be inactive in E. coli grown in glucose-containing media (Fig. 2). These are the gluconeogenic conversion of oxaloacetate (OAA) to phosphoenolpyruvate (PEP) (Fig. 2G), catalyzed by the PEP carboxykinase (10, 11), and the conversion of malate (MAL) to pyruvate (PYR) (Fig. 2H and I) through the malic enzyme. Malic enzyme is normally required for growth on four-carbon compounds (11). Although the detected flux ratio of the latter reaction to all other reactions generating PYR is small, these data illustrate that the in vivo activity of reactions in central metabolism does not necessarily follow straightforward on-off paradigms (11, 20).
FIG. 2.
Origins of metabolic intermediates (A to P) during aerobic growth of E. coli MG1655 in glucose-limited or ammonia-limited chemostats. In certain cases, the NMR data permit the determination only of upper bounds (ub) or lower bounds (lb) on the origin of intermediates. The experimental error (error bars) was estimated from the analysis of redundant 13C scalar coupling fine structures and the signal-to-noise ratio of the 13C-1H COSY spectra by use of the Gaussian law of error propagation. The fraction of the total pool for a particular metabolite quantifies the ratio of this metabolite derived from a specified substrate to the sum of all other substrates that contribute to the pool of this metabolite. In cases where only two reactions contribute to one metabolite, e.g., OAA from PEP and PEP from OAA, the remaining fraction of the total pool can be attributed to the competing reaction. Abbreviations are explained in the text and in the legend to Fig. 1; rev., reversibly.
Cells harvested from the ammonia-limited culture clearly showed a fluxome pattern different from that of cells from the glucose-limited culture (Fig. 2). (i) Nearly double the fraction of OAA molecules was found to be derived from PEP (Fig. 2F), demonstrating an increased contribution from the anaplerotic PEP carboxylase reaction (9) and a corresponding decrease in MAL dehydrogenase activity. This anaplerotic reaction synthesizes the OAA that is required to replenish the pool of tricarboxylic acid (TCA) cycle intermediates, and its relative contribution therefore reflects the extent to which the TCA cycle is used for the biosynthesis of biomass components relative to energy generation (via oxidative phosphorylation) (11). (ii) A decrease in the fraction of PEP molecules originating from OAA was observed (Fig. 2G), providing evidence of reduced fluxes through the gluconeogenic PEP carboxykinase. (iii) An increase in the fraction of PYR derived from MAL was detected (Fig. 2H), indicating increased fluxes through the malic enzyme. (iv) A 50% reduction in the fraction of PEP molecules that were derived through at least one transketolase reaction was registered (Fig. 2B). This decrease would be consistent with an increased contribution from the glycolytic pathway relative to the PP pathway, suggesting that the higher catabolic fluxes in the ammonia-limited culture were mainly supported by glycolysis. (v) Less than 100% of the acetyl coenzyme A (ACoA) molecules were found to originate from PYR (Fig. 2J). This result can be explained by a dilution of the intracellular ACoA pool via exchange with the large, mostly unlabeled extracellular acetate pool that was detected in the ammonia-limited culture but not in the glucose-limited culture (Table 2). This result provides direct evidence for the presence of exchange fluxes between extracellular and intracellular acetate pools, as well as for the reversibility of the reactions connecting ACoA to acetate.
Analysis of wild-type batch cultures harvested at different growth phases.
Chemostat experiments are the most suitable method of analyzing cells under steady-state conditions. On the other hand, a physiological steady state is also attained during the exponential growth phase in batch cultures, which are characterized by unrestricted growth at the maximum specific rate possible under the applied conditions. Because batch cultures enable more efficient parallel analysis of different strains, we adapted batch cultivations for METAFoR analysis of genetic effects on central carbon metabolism.
The underlying principle of METAFoR analysis is the imprinting of central carbon network history into cell protein. This notion implies that, in a transient situation such as a batch cultivation, METAFoR results provide a time average over the interval of isotopic labeling of the biomass. To minimize changes in central carbon metabolism during the labeling period and to undertake the METAFoR analysis over a range of closely related physiological states, batch cultivations were initiated with mid-exponential-phase cultures at a low inoculum density (less than 1% the final culture volume) in medium containing 90% natural-abundance glucose and 10% [U-13C6]glucose. One set of experiments was undertaken to assess the sensitivity of METAFoR results to the sampling time of the labeled culture. These experiments also indicate the extent to which significant changes in central metabolism occur between different sampling times.
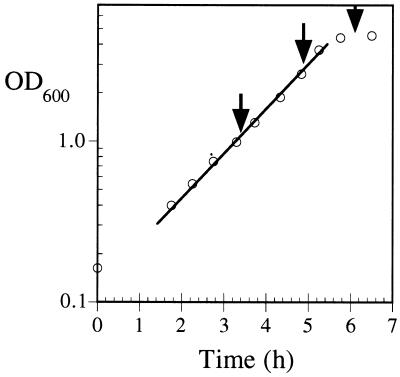
Three batch cultivations of wild-type E. coli B strain ATCC 11303 were initiated in parallel and harvested in the mid-exponential phase, late exponential phase, and stationary phase (OD600s, 1, 2.6, and 4, respectively) (Fig. 3). METAFoR analysis indicates extremely similar flux ratio histories for all three samples, demonstrating that batch cultivations provide consistent central metabolic flux ratios (Table 3) when isotopic labeling of biomass is achieved throughout the exponential growth phase. Slight trends in some of the results, most notably in the fraction of OAA molecules derived from PEP, indicate adjustments in central metabolism that occur as the culture approaches and enters stationary phase. Overall, however, the present data indicate that time of harvest is not a critical parameter in the experiments used for this study. Therefore, in later experiments, all cultures were harvested at OD600s between 0.9 and 1.2.
FIG. 3.
Growth of E. coli ATCC 11303 in aerobic batch cultures. The line represents the best fit to the exponential growth phase data, and the arrows indicate the times of biomass sampling for the METAFoR analysis.
TABLE 3.
Origin of intermediates in E. coli ATCC 11303 harvested during different growth phases from the cultures shown in Fig. 3a
| Metabolite | Fraction of total pool (%) during the following growth phase:
|
||
|---|---|---|---|
| Mid-exponential | Late exponential | Stationary | |
| PP pathway | |||
| R5P from G6P (lower bound) | 29 ± 2 | 25 ± 2 | 24 ± 2 |
| R5P from T3P + S7P | 71 ± 2 | 75 ± 2 | 76 ± 2 |
| Glycolysis and TCA cycle | |||
| OAA from PEP | 44 ± 1 | 40 ± 1 | 38 ± 1 |
| PEP from OAA | 7 ± 4 | <7 | 6 ± 4 |
| PYR from MAL (upper bound) | <5 | <5 | <5 |
| PYR from MAL (lower bound) | <3 | <3 | <3 |
| ACoA from PYR | >98 | >98 | >98 |
| PYR from ACoA | 0 | 0 | 0 |
| OGA from OAA + ACoA | >98 | >98 | >98 |
| ICT from SUC + GOX | <2 | <2 | 0 |
| OAA reversibly interconverted to FUM | 46 ± 11 | 48 ± 10 | 50 ± 10 |
| C1 metabolism | |||
| Ser from Gly + C1 unit | 7 ± 2 | 8 ± 2 | 9 ± 2 |
| Gly from CO2 + C1 unit | 4 ± 3 | 8 ± 3 | 9 ± 3 |
The data were derived from a 13C-1H COSY spectrum centered on the aliphatic region; therefore, some data for the PP pathway that are accessible only through erythrose-4-phosphate incorporated in the aromatic rings of Trp or Phe could not be determined. See the text and the legend to Fig. 1 for abbreviations.
Analysis of aerobic batch cultures of wild-type and mutant E. coli strains during exponential growth.
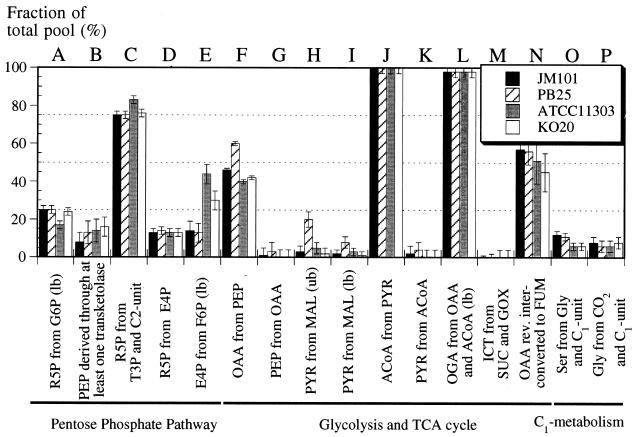
Two commonly used but genetically different E. coli strains, JM101 (a K-12 strain) and ATCC 11303 (a B strain), were grown in shake flask cultures under aerobic conditions for direct comparison of their carbon metabolism. Although ATCC 11303 grew somewhat faster (Table 4), the fluxome data for the two wild-type strains were almost identical (Fig. 4), suggesting that central metabolism is very similar in the two E. coli strains. There were, however, small changes in the origins of metabolites involved in the nonoxidative part of the PP pathway, from ribose-5-phosphate (R5P) to triose-3-phosphate (T3P) (Fig. 4B, C, and E), indicating that ATCC 11303 cells exhibit a higher degree of exchange through transketolase II.
TABLE 4.
Aerobic growth parameters of exponentially growing E. coli strains
| Strain | μ (h−1)a | YX/S (g g−1)b | qglc (g g−1 h−1)c | Concn (mM)d of:
|
|
|---|---|---|---|---|---|
| Acetate | Ethanol | ||||
| PB25 | 0.64 | 0.30 | 2.2 | 2.0 | 0.4 |
| JM101 | 0.60 | 0.30 | 2.0 | 1.5 | 0.4 |
| ATCC 11303 | 0.65 | 0.29 | 2.2 | 1.6 | 1.0 |
| KO20 | 0.70 | 0.28 | 2.5 | 1.6 | 1.5 |
| KO20::pTrc99a | 0.68 | 0.28 | 2.4 | 1.6 | 1.5 |
| KO20::pPPec | 0.65 | 0.30 | 2.2 | 2.2 | 1.5 |
| KO20::pPYKbs | 0.74 | 0.29 | 2.3 | 1.3 | 2.0 |
Standard deviation (SD), ±0.01.
SD, ±0.02.
SD, ±0.2.
At an OD600 of 1.
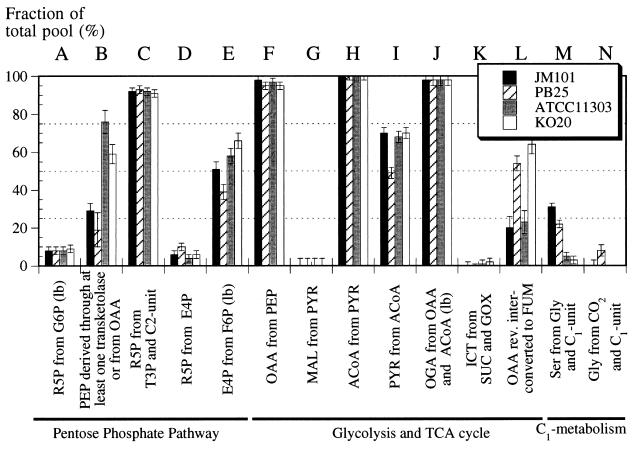
FIG. 4.
Origin of metabolic intermediates (A to P) during aerobic exponential growth of various E. coli strains. For more details, see the legend to Fig. 2.
In additional experiments, we studied the effects arising from knockout mutations which inactivate key central metabolism enzymes for each of the two wild-type strains. Pyruvate kinase-deficient PB25, which is otherwise isogenic with JM101, actually grew faster than its parent but exhibited similar biomass yield and specific glucose consumption rate (Table 4). While the overall METAFoR patterns were rather similar, inactivation of both pyruvate kinase isoenzymes resulted in significant changes at the branch points between glycolysis and the TCA cycle (Fig. 4). Specifically, we found a higher fraction of OAA from PEP (Fig. 4F) and PYR from MAL (Fig. 4H and I) in the mutant. Apparently, the carbon flux from PEP to PYR is redistributed from pyruvate kinase to anaplerotic PEP carboxylase and the malic enzyme, allowing PB25 to generate sufficient pyruvate for fueling the TCA cycle via ACoA to generate energy (Fig. 1). These local changes provide evidence that considerable flexibility in E. coli central carbon metabolism permits the use of alternative pathways to compensate for knockout mutations.
Ethanol-producing E. coli KO20 is derived from ATCC 11303 and is characterized by a single chromosomal insertion of the artificial pet operon, which encodes the Zymomonas mobilis genes for alcohol dehydrogenase II and pyruvate decarboxylase, such that it disrupts the pyruvate formate-lyase gene (23). This strain grew faster and consumed glucose at a higher specific rate than both wild-type strains (Table 4). Although it primarily exhibited oxidative metabolism, it generated 50% more ethanol than did its parent. Despite these genetic and physiological differences, the METAFoR pattern of KO20 was, within experimental error, identical to that of ATCC 11303 (Fig. 4).
Carbon metabolism during exponential growth in aerobic batch cultures of E. coli strains engineered for ethanol production.
We chose ethanol-producing E. coli KO20 (23) as the host for a series of plasmids used for the expression of homologous and heterologous pyruvate kinases and phosphofructokinases (6). The expression of both genes was induced by use of isopropyl-β-d-thiogalactopyranoside (IPTG) (0.01 mM), leading to in vitro activities two- to ninefold above the control level, which was represented by the empty expression vector pTrc99a (Pharmacia). In terms of their growth physiology, all strains showed similar behaviors under aerobic conditions in batch cultures (Table 4). However, by-product formation was altered in the overexpression strains, with KO20::pPPec generating more acetate and KO20::pPYKbs generating more ethanol than KO20. Again, the METAFoR patterns of all strains derived from ATCC 11303 were identical, within experimental error, to those of KO20 in Fig. 4 (data not shown). Thus, expression of the pet operon, medium-copy-number plasmid maintenance, and the overexpression of either a heterologous pyruvate kinase or the homologous phosphofructokinase do not appear to have a pronounced influence on the flux ratios accessible by current METAFoR analysis.
Analysis of anaerobic batch cultures of wild-type and mutant E. coli strains during exponential growth.
The physiological differences among the wild-type and mutant E. coli strains under anaerobic conditions were larger than those observed in aerobiosis (Table 5). The two wild-type strains, JM101 and ATCC 11303, showed significant differences with respect to glucose consumption as well as acetate production rates, both of which were higher in ATCC 11303. Accordingly, the apparent biomass yield was lower in this strain. While pyruvate kinase-deficient PB25 grew slower than its parent, JM101, ethanol-producing KO20 grew fastest of all the strains investigated. Although the specific glucose consumption rates of JM101 and PB25 were identical, the latter was severely impaired in its ability to form the normal anaerobic end products ethanol and acetate. On the other hand, KO20 produced about fivefold more ethanol than any other strain and exhibited a higher glucose consumption rate and a lower biomass yield on glucose than its parent, ATCC 11303.
TABLE 5.
Anaerobic growth parameters of exponentially growing E. coli strains
| Strain | μ (h−1) | YX/S (g g−1) | qglc (g g−1 h−1) | qetoha (mM g−1 h−1) | qacb (mM g−1 h−1) |
|---|---|---|---|---|---|
| PB25 | 0.23 ± 0.01 | 0.078 ± 0.003 | 3.1 ± 0.2 | 17 ± 1 | 7 ± 3 |
| JM101 | 0.30 ± 0.01 | 0.097 ± 0.003 | 3.1 ± 0.2 | 27 ± 5 | 21 ± 3 |
| ATCC 11303 | 0.33 ± 0.01 | 0.075 ± 0.003 | 4.4 ± 0.2 | 27 ± 5 | 54 ± 6 |
| KO20 | 0.36 ± 0.01 | 0.065 ± 0.002 | 5.5 ± 0.2 | 118 ± 10 | 39 ± 6 |
Specific ethanol formation rate.
Specific acetate formation rate.
METAFoR analysis of the anaerobic cultures shown in Table 5 showed that OAA originated almost entirely from PEP via the anaplerotic reaction (Fig. 5F), illustrating the almost total absence of complete TCA cycle operation. This result concurs with the results of a recent study (7) and is consistent with earlier conclusions based on enzyme activity data, which indicated that, under anaerobic conditions, a branched, noncyclic TCA cycle pathway operates mainly to fulfill biosynthetic requirements (35). Because the fragments needed for tracing the activities of malic enzyme and PEP carboxykinase with METAFoR analysis do not appear under anaerobic conditions (27), these reactions are inaccessible to this analysis. Hence, although the experimental data conform to a bioreaction network devoid of malic enzyme (Fig. 5G), its activity cannot be excluded.
FIG. 5.
Origin of metabolic intermediates (A to N) during anaerobic exponential growth of various E. coli strains. For more details, see the legend to Fig. 2.
We cannot independently quantify the relative flux of OAA to PEP via PEP carboxykinase in anaerobically grown cells; therefore, METAFoR analysis cannot distinguish between OAA decarboxylation and PEP synthesis through the PP pathway. When ATCC 11303 (and its derivative, KO20) and JM101 (and its derivative, PB25) are compared, the major difference in the fraction of PEP molecules that are not directly derived from glucose through glycolysis (Fig. 5B) may originate from OAA via decarboxylation, from a higher net flux through the PP pathway, or from increased exchange reactions in the PP pathway. However, the observation that the exchange fluxes in the PP pathway are very similar among all strains (Fig. 5C to E) indicates a major difference in flux through the PEP carboxykinase or PP pathway. The low but significant fraction of R5P found to originate from glucose-6-phosphate (G6P) (Fig. 5A) provides unambiguous evidence for the activity of the oxidative PP pathway during anaerobiosis.
In all cases, we observed a large fraction of PYR molecules that were interconverted at least once to ACoA (Fig. 5I), illustrating the in vivo reversibility of the anaerobic pyruvate formate-lyase reaction, as opposed to the irreversible aerobic pyruvate dehydrogenase reaction (11, 30) (see also Fig. 4K). However, in KO20 the pyruvate formate-lyase gene is disrupted and pyruvate conversion is achieved via the expression of a heterologous pyruvate decarboxylase. Because PYR exchange with ACoA in KO20 is almost identical to that in the reference strain, it appears either that the reversibility of this reaction is similar for pyruvate formate-lyase and pyruvate carboxylase or that an isoenzyme of pyruvate formate-lyase is responsible for this exchange. The uncharacterized product of the E. coli yhaS gene is highly homologous to pyruvate formate-lyase and thus may encode this activity.
Despite their considerably altered by-product formation characteristics, the two engineered strains, PB25 and KO20, exhibit central carbon metabolism surprisingly similar to that of their parent strains. This result can be seen in the fluxome data shown in Fig. 5, where the flux ratios are nearly identical in the parent strains and the modified strains, except for reduced reversibility of the interconversion of OAA to fumarate (FUM) by the parent strains (Fig. 5L). This result provides further evidence for the homeostasis of carbon flux distribution in central metabolism following significant genetic modifications that impinge directly upon this metabolic subsystem.
DISCUSSION
This study provides novel insights into global metabolic network behavior. Variations resulting from different growth conditions and genetic backgrounds in E. coli are monitored by combined physiological and fluxome analyses. A particular strength of the METAFoR analysis used here is the ability to decipher the relative fluxes connecting the lower part of glycolysis with the TCA cycle, namely, the anaplerotic reaction and certain futile cycles which dissipate ATP. On the other hand, flux ratios of the oxidative versus the nonoxidative PP pathways (R5P from G6P; Fig. 2A, 4A, and 5A) and glycolysis versus the PP pathway (PEP molecules derived through at least one transketolase reaction; Fig. 2B, 4B, and 5B) are accessible only as upper and lower bounds, because the pool of pentoses is rapidly and reversibly turned over by transaldolase and transketolase. The key findings are as follows. (i) Intracellular carbon flux ratios in the central metabolism of E. coli are affected only a little by enzyme overexpression and are flexible toward gene disruption. (ii) Of all central carbon fluxes, those in the TCA cycle change most significantly in response to changes in environmental conditions. (iii) Reactions mediated by the malic enzyme and PEP carboxykinase and previously considered to be absent during growth on glucose were identified. (iv) A novel regulation phenomenon in which futile cycling through at least one set of reactions is increased under conditions of a very low extracellular glucose concentration was evident.
Interstrain differences.
The METAFoR pattern of exponentially growing aerobic E. coli revealed surprisingly few interstrain differences (Fig. 4), although major changes in physiological parameters are documented for the various strains used here. One would expect such physiological differences to be reflected in fluxes through central metabolism (Fig. 1), which provides energy, cofactor regeneration, and building blocks for biosynthesis. Strain KO20, for example, which expresses the pet operon of Z. mobilis alcohol dehydrogenase II and pyruvate decarboxylase and has pyruvate formate-lyase knockout mutations, was previously described to exhibit significantly altered by-product formation, with ethanol as the major product (23). Furthermore, overexpression of pyruvate kinase and phosphofructokinase, which are major control enzymes in the glycolytic pathway, were shown to have a profound effect on glucose catabolism in resting E. coli KO20. Specifically, an increased glucose consumption rate was found for KO20::pPYKbs harvested from aerobic precultures (13), and a large shift from ethanol to lactate formation was described for KO20::pPPec harvested from anaerobic precultures (6).
Previously, pyruvate kinase deficiency was reported to alter growth kinetics under aerobic conditions (25). In the present study, JM101 and its pyruvate kinase-deficient derivative, PB25, showed few global changes in the METAFoR pattern. There was a small increase in the fraction of PEP molecules that were derived through at least one transketolase reaction (Fig. 4B), while all other exchange reactions in the PP pathway remained comparable (Fig. 4C to E). These results are consistent with the recent findings of Ponce et al. (26), who observed increased PP pathway activity in strain PB25 compared to strain JM101 by radiorespirometric analysis. Overall similarity in METAFoR patterns in response to genetic modifications seems to be a common feature of exponentially growing cells in aerobic cultures, consistent with the small differences in physiological parameters observed in this situation (Table 4). Under anaerobic conditions, where larger physiological changes were found, METAFoR analysis revealed more pronounced differences (Table 5 and Fig. 5; see also below).
Anaplerosis and the TCA cycle.
METAFoR data provide information about the fraction of OAA molecules that originate from PEP (Fig. 2F, 4F, and 5F); this information quantifies the contribution of the TCA cycle-replenishing anaplerotic reaction to OAA generation, relative to that of MAL dehydrogenase in the TCA cycle (30–32). In the aerobic cultures studied here, the relative flux through anaplerotic PEP carboxylase is about 40%; the value for the ammonia-limited chemostat culture is increased to about 70% (Fig. 2F). Consistent with the extensive overflow metabolism seen in the ammonia-limited culture (Table 2), this increased anaplerosis indicates that, compared with the situation for the glucose-limited culture, a larger portion of the TCA cycle flux is used for biomass formation instead of energy generation. In the anaerobic regimen, the TCA cycle is reduced to a two-branch pathway (30) and OAA is generated by anaplerosis only (Fig. 5F).
Futile cycling.
In the glucose-limited chemostat, a significant fraction of PEP molecules were found to originate from OAA (Fig. 2G). This result indicates in vivo activity of gluconeogenic PEP carboxykinase, an enzyme that is required for growth on carbon sources that are metabolized via the TCA cycle and that has previously been considered to be inactive in cells grown on glucose (11). In principle, PEP molecules could also originate from OAA via the reverse reaction of anaplerotic PEP carboxylase, but based on thermodynamic considerations and the absence of 14CO2 exchange with OAA in enzyme assays (37), this notion is highly unlikely. Hence, we have evidence of an ATP-dissipating futile cycle via PEP carboxylase and ATP-consuming PEP carboxykinase. At the same D under ammonia-limited conditions in the chemostat, this futile cycle appears to be significantly less active (Fig. 2G). Similar activity levels for this cycle were seen with Bacillus subtilis (27) grown in a chemostat under glucose limitation at a D of about 0.1 h−1. In faster growing cells of B. subtilis in this chemostat, however, the contribution of this futile cycle was reduced, and the batch data presented here show it to be absent in exponentially growing E. coli (Fig. 4). On the other hand, for Corynebacterium glutamicum, a similar exchange between the PEP-PYR and OAA-MAL pools was described, not only for glucose-limited chemostat cultures (41) but also for batch cultures (29); however, it is not clear whether or not that pool exchange involved dissipation of ATP via a futile cycle. These data provide evidence for a metabolic regulation phenomenon in E. coli and B. subtilis in which futile cycle activity is less tightly controlled under extreme glucose limitation than under glucose excess, as in slow-growing chemostat cultures. It is tempting to speculate that this reduced control is caused by the extremely low extracellular glucose concentration and a concomitant reduction in catabolite repression. This hypothesis is also supported by the observation that PEP carboxykinase expression in E. coli is repressed by glucose (10).
In the anaerobically grown E. coli B strains ATCC 11303 and KO20, the high upper bound of the fraction of PEP originating from pentoses or OAA indicates a major difference in the metabolism of these strains and strains JM101 and PB25 (Fig. 5B). This difference could result from a higher flux either through the PP pathway or through PEP carboxykinase. From a physiological perspective, however, high fluxes through the oxidative PP pathway appear unreasonable, because anaerobic metabolism cannot reoxidize concomitantly formed NADPH with oxygen and reduced by-products were not detected. Therefore, it is more likely that a futile cycle involving PEP carboxykinase carries higher fluxes in ATCC 11303 and KO20. This scenario would be consistent with the observed higher specific rate of glucose catabolism and the reduced biomass yield compared to those in anaerobic E. coli JM101 cultures.
Exchange reactions.
METAFoR analysis affords a qualitative assessment of several exchange fluxes (30). In the experiments analyzed here, the fraction of R5P molecules originating from T3P and a C2 unit via the transketolase reaction was usually about 70 to 80% (Fig. 2C and 4C) and, under anaerobic conditions, even as high as 90% (Fig. 5C). In contrast, a much lower fraction of R5P molecules originated from erythrose-4-phosphate (Fig. 2D, 4D, and 5D), representing either an exchange via transaldolase or a recycling of PP pathway-generated fructose-6-phosphate to G6P and on to R5P. Similar insights into the PP pathway have previously been reported for batch cultures of E. coli K-12 and B strains by mass spectrometric analysis of 18O-labeling patterns in the ribose moiety-containing nucleotides (15) and by METAFoR analysis (30). Rapid exchange of metabolite pools in the PP pathway was also described for C. glutamicum (19, 41). In B. subtilis, the exchange mediated by transketolase appears to be less significant, since about 50% of the R5P in slow-growing, glucose-limited chemostat cultures was found to contain intact C5 fragments from the source glucose (27).
Three additional exchange fluxes can be assessed by the present methodology (30). First, the reversible interconversion of OAA to FUM was found to be invariant at about 50% in all cases, with the exception of JM101 and ATCC 11303 under anaerobic conditions (Fig. 2N, 4N, and 5L). Second, in C1 metabolism the backward reaction from Gly and a C1 unit to Ser was essentially negligible in aerobic batch cultures but was significant in anaerobic JM101 and PB25 batch cultures as well as in aerobic MG1655 continuous cultures (Fig. 4O and 5M). Third, in apparent contrast to earlier observations with amino acids obtained by hydrolysis of a purified recombinant protein (30, 42) we observed the reverse reaction in the glycine cleavage pathway (Gly from a C1 unit and CO2) under aerobic conditions but not under anaerobic conditions (Fig. 4P and 5N).
ACKNOWLEDGMENTS
This work was supported by the Swiss Priority Program in Biotechnology (SPP BioTech).
We are grateful to L. O. Ingram (University of Florida) and F. Valle (Universidad National Autónoma de México) for providing us with strains used in this study. We thank Nicola Zamboni for technical assistance with the chemostat experiments.
REFERENCES
- 1.Bachmann B J. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 2460–2488. [Google Scholar]
- 2.Bax A, Pochapsky S. Optimized recording of heteronuclear multidimensional NMR spectra using pulsed field gradients. J Magn Reson. 1992;99:638–643. [Google Scholar]
- 3.Bodenhausen G, Ruben D. Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem Phys Lett. 1980;69:185–188. [Google Scholar]
- 4.Brown A J P. Control of metabolic flux in yeasts and fungi. Trends Biotechnol. 1997;15:445–447. doi: 10.1016/S0167-7799(97)01116-5. [DOI] [PubMed] [Google Scholar]
- 5.DeMarco A, Wüthrich K. Digital filtering with a sinusoidal window function: an alternative technique for resolution enhancement in FT NMR. J Magn Reson. 1976;24:201–204. [Google Scholar]
- 6.Emmerling M, Bailey J E, Sauer U. Glucose catabolism of Escherichia coli strains with increased activity and altered regulation of key glycolytic enzymes. Metab Eng. 1999;1:117–127. doi: 10.1006/mben.1998.0109. [DOI] [PubMed] [Google Scholar]
- 7.Fiaux J, Andersson C I J, Holmberg N, Bülow L, Kallio P T, Szyperski T, Bailey J E, Wüthrich K. 13C-NMR flux ratio analysis of Escherichia coli central carbon metabolism in microaerobic bioprocesses. J Am Chem Soc. 1999;121:1407–1408. [Google Scholar]
- 8.Fraenkel D G. Genetics and intermediary metabolism. Annu Rev Genet. 1992;26:159–177. doi: 10.1146/annurev.ge.26.120192.001111. [DOI] [PubMed] [Google Scholar]
- 9.Fraenkel D G. Glycolysis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 189–198. [Google Scholar]
- 10.Goldie H. Regulation of transcription of the Escherichia coli phosphoenolpyruvate carboxykinase locus: studies with pck-lacZ operon fusions. J Bacteriol. 1984;159:832–836. doi: 10.1128/jb.159.3.832-836.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottschalk G. Bacterial metabolism. 2nd ed. New York, N.Y: Springer-Verlag; 1986. [Google Scholar]
- 12.Güntert P, Dötsch V, Wider G, Wüthrich K. Processing of multi-dimensional NMR data with the new software PROSA. J Biomol NMR. 1992;2:619–629. [Google Scholar]
- 13.Hatzimanikatis V, Emmerling M, Sauer U, Bailey J E. Application of mathematical tools for metabolic design of microbial ethanol production. Biotechnol Bioeng. 1998;58:154–161. doi: 10.1002/(sici)1097-0290(19980420)58:2/3<154::aid-bit7>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 14.James P. Protein identification in the post-genome era: the rapid rise of proteomics. Q Rev Biophys. 1997;30:279–331. doi: 10.1017/s0033583597003399. [DOI] [PubMed] [Google Scholar]
- 15.Johnson R, Krasna A I, Rittenberg D. 18O studies on the oxidative and nonoxidative pentose phosphate pathways in wild-type and mutant Escherichia coli cells. Biochemistry. 1973;12:1969–1977. doi: 10.1021/bi00734a021. [DOI] [PubMed] [Google Scholar]
- 16.Karp P. Metabolic databases. Trends Biochem Sci. 1998;23:114–116. doi: 10.1016/s0968-0004(98)01184-0. [DOI] [PubMed] [Google Scholar]
- 17.Karp, P. D., and M. Riley. 11 January 1999, revision date. EcoCyc: encyclopedia of E. coli genes and metabolism. [Online.] http://ecocyc.PangeaSystems.com/ecocyc/ecocyc.html. [10 August 1999, last date accessed.]
- 18.Marion D, Ikura M, Tschudin R, Bax A. Rapid recording of 2D NMR spectra without phase cycling. Application to the study of hydrogen exchange in proteins. J Magn Reson. 1989;85:393–399. [Google Scholar]
- 19.Marx A, de Graaf A A, Wiechert W, Eggeling L, Sahm H. Determination of the fluxes in the central metabolism of Corynebacterium glutamicum by nuclear magnetic resonance spectroscopy combined with metabolite balancing. Biotechnol Bioeng. 1996;49:111–129. doi: 10.1002/(SICI)1097-0290(19960120)49:2<111::AID-BIT1>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 20.Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. [Google Scholar]
- 21.Neijssel O M, Teixeira de Mattos M J, Tempest D W. Growth yield and energy distribution. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1683–1692. [Google Scholar]
- 22.Neri D, Szyperski T, Otting G, Senn H, Wüthrich K. Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434-repressor by biosynthetically directed fractional 13C labeling. Biochemistry. 1989;28:7510–7516. doi: 10.1021/bi00445a003. [DOI] [PubMed] [Google Scholar]
- 23.Ohta K, Beall D S, Mejia J P, Shanmugam K T, Ingram L O. Genetic improvement of Escherichia coli for ethanol production: chromosomal integration of Zymomonas mobilis genes encoding pyruvate decarboxylase and alcohol dehydrogenase II. Appl Environ Microbiol. 1991;57:893–900. doi: 10.1128/aem.57.4.893-900.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otting G, Wüthrich K. Efficient purging scheme for proton-detected heteronuclear two-dimensional NMR. J Magn Reson. 1988;76:569–574. [Google Scholar]
- 25.Ponce E, Flores N, Martinez A, Valle F, Bolivar F. Cloning of the two pyruvate kinase isoenzyme structural genes from Escherichia coli: the relative roles of these enzymes in pyruvate biosynthesis. J Bacteriol. 1995;177:5719–5722. doi: 10.1128/jb.177.19.5719-5722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ponce E, Martinez A, Bolivar F, Valle F. Stimulation of glucose catabolism through the pentose phosphate pathway by the absence of the two pyruvate kinase isoenzymes in Escherichia coli. Biotechnol Bioeng. 1998;58:292–295. doi: 10.1002/(sici)1097-0290(19980420)58:2/3<292::aid-bit25>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 27.Sauer U, Hatzimanikatis V, Bailey J E, Hochuli M, Szyperski T, Wüthrich K. Metabolic fluxes in riboflavin-producing Bacillus subtilis. Nat Biotechnol. 1997;15:448–452. doi: 10.1038/nbt0597-448. [DOI] [PubMed] [Google Scholar]
- 28.Shaka A J, Barker P B, Freemann R. Computer-optimized decoupling scheme for wideband applications and low-level operation. J Magn Reson. 1985;64:547–552. [Google Scholar]
- 29.Sonntag K, Schwinde J, de Graaf A A, Marx A, Eikmanns B J, Wiechert W, Sahm H. 13C NMR studies of the fluxes in the central carbon metabolism of Corynebacterium glutamicum during growth and overproduction of amino acids in batch cultures. Appl Microbiol Biotechnol. 1995;44:489–495. [Google Scholar]
- 30.Szyperski T. Biosynthetically directed fractional 13C-labeling of proteinogenic amino acids: an efficient analytical tool to investigate intermediary metabolism. Eur J Biochem. 1995;232:433–448. doi: 10.1111/j.1432-1033.1995.tb20829.x. [DOI] [PubMed] [Google Scholar]
- 31.Szyperski T. 13C-NMR, MS and metabolic flux balancing in biotechnological research. Q Rev Biophys. 1998;31:41–106. doi: 10.1017/s0033583598003412. [DOI] [PubMed] [Google Scholar]
- 32.Szyperski T, Bailey J E, Wüthrich K. Detecting and dissecting metabolic fluxes using biosynthetic fractional 13C labeling and two-dimensional NMR spectroscopy. Trends Biotechnol. 1996;14:453–459. [Google Scholar]
- 33.Szyperski T, Glaser R W, Hochuli M, Fiaux J, Sauer U, Bailey J E, Wüthrich K. Bioreaction network topology and metabolic flux ratio analysis by biosynthetic fractional 13C-labeling and two-dimensional NMR spectroscopy. Metab Eng. 1999;1:189–197. doi: 10.1006/mben.1999.0116. [DOI] [PubMed] [Google Scholar]
- 34.Szyperski T, Neri D, Leiting B, Otting G, Wüthrich K. Support of 1H-NMR assignments in proteins by biosynthetically directed fractional 13C-labelling. J Biomol NMR. 1992;2:323–334. doi: 10.1007/BF01874811. [DOI] [PubMed] [Google Scholar]
- 35.Thomas A D, Doelle H W, Westwood A W, Gordon G L. Effect of oxygen on several enzymes involved in the aerobic and anaerobic utilization of glucose in Escherichia coli. J Bacteriol. 1972;112:1099–1105. doi: 10.1128/jb.112.3.1099-1105.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tomb J-F. A panoramic view of bacterial transcription. Nat Biotechnol. 1998;16:23. doi: 10.1038/nbt0198-23. [DOI] [PubMed] [Google Scholar]
- 37.Utter M F. Nonoxidative carboxylation and decarboxylation. In: Boyer P D, Lardy H, Myrbäck K, editors. Hydrolytic cleavage (part B) Vol. 5. New York, N.Y: Academic Press, Inc.; 1961. pp. 320–343. [Google Scholar]
- 38.van Dam K, Jansen N. Quantification of control of microbial metabolism by substrates and enzymes. Antonie Leeuwenhoek. 1991;60:209–223. doi: 10.1007/BF00430366. [DOI] [PubMed] [Google Scholar]
- 39.Varma A, Palsson B O. Metabolic flux balancing: basic concepts, scientific, and practical use. Bio/Technology. 1994;12:994–998. [Google Scholar]
- 40.Wider G, Wüthrich K. A simple experimental scheme using pulsed field gradients for coherence pathway rejection and solvent suppression in phase-sensitive heteronuclear correlation spectra. J Magn Reson. 1993;102:239–241. [Google Scholar]
- 41.Wiechert W, Siefke C, de Graaf A A, Marx A. Bidirectional reaction steps in metabolic networks. II. Flux estimation and statistical analysis. Biotechnol Bioeng. 1997;55:118–135. doi: 10.1002/(SICI)1097-0290(19970705)55:1<118::AID-BIT13>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 42.Wüthrich K, Szyperski T, Leiting B, Otting G. Biosynthetic pathways of the common proteinogenic amino acids investigated by fractional 13C labeling and NMR spectroscopy. In: Takai K, editor. Frontiers and new horizons in amino acid research. Amsterdam, The Netherlands: Elsevier Science Publishers; 1992. pp. 41–48. [Google Scholar]
- 43.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]