Abstract
The antimicrobial resistance (AMR) crisis from bacterial pathogens is frequently emerging and rapidly disseminated during the sustained antimicrobial exposure in human-dominated communities, posing a compelling threat as one of the biggest challenges in humans. The frequent incidences of some common but untreatable infections unfold the public health catastrophe that antimicrobial-resistant pathogens have outpaced the available countermeasures, now explicitly amplified during the COVID-19 pandemic. Nowadays, biotechnology and machine learning advancements help create more fundamental knowledge of distinct spatiotemporal dynamics in AMR bacterial adaptation and evolutionary processes. Integrated with reliable diagnostic tools and powerful analytic approaches, a collaborative and systematic surveillance platform with high accuracy and predictability should be established and implemented, which is not just for an effective controlling strategy on AMR but also for protecting the longevity of valuable antimicrobials currently and in the future.
Keywords: antimicrobial resistance (AMR), AMR bacterial adaptation and evolutionary processes, advancements in biotechnology and computer science, reliable diagnostic tools, surveillance platform
Introduction
The increasing prevalence of antimicrobial resistance (AMR) among bacterial pathogens poses one of the greatest threats to public health all over the world (Yue et al., 2021; Resistance, 2016), mainly because of the overuse of broad-spectrum antibiotics and potential antimicrobial-bacterium mismatch in empirical selections (Pan Hang et al., 2018; Jiang et al., 2019; Wang et al., 2019). Scientific reports indicate that death due to ineffective treatment of multidrug-resistant bacteria reaches over 100 cases daily in the European Union and the United States of America. Meanwhile, the increasing resistance to the “last-resort” antibiotics continuously concerns public health worldwide (Biswas et al., 2019; Elbediwi et al., 2019; Elbediwi et al., 2020).
To identify common AMR and guide diagnostics and effective treatment for bacterial infections, a gold-standard laboratory test used to determine the antimicrobial sensitivity of AMR organisms, termed antimicrobial susceptibility tests (AST), has been adopted in clinics for over 50 years. Traditional AST involves broth microdilution and disc diffusion on agar to determine the minimum inhibitory concentration (MIC). The MIC value indicates the lowest concentration of antibiotics where bacterial growth stops, and the growth level is usually measured by absorbance or light scattering. Before and during testing, multiple turnarounds of culturing the bacteria in the sample and isolating colonies and growth needed to be accomplished before AST. The whole pipeline takes at least 48 h, which usually provides the results after using an empirical prescription of antibiotics that might be misused. It has been estimated that around one-fifth of prescribed antibiotics from 2013 to 2015 were inappropriate in the United Kingdom (Smieszek et al., 2018). Also, currently, with over 500 million infected cases, COVID-19 has increased the setbacks in fighting against AMR (Knight et al., 2021; Ma et al., 2021). As many patients suffering from COVID-19 are being prescribed inappropriate antibiotics, this scenario might surge drug-resistant infections during the pandemic (Kariyawasam et al., 2022), particularly in developing countries where people lack antimicrobial stewardship programs and reliable diagnostics. Numerous AMR-associated outbreaks have been reported during the COVID-19 pandemic, such as carbapenemase-producing Enterobacteriaceae (Farfour et al., 2020; Tiri et al., 2020), highlighting the urgent demand for an efficient AMR surveillance system.
With decreasing sequencing costs and advancements in bioinformatics toolkits, whole-genome sequencing (WGS) for AMR diagnosing has been considered an efficient avenue for combating AMR (Schurch and van Schaik, 2017). The genotype-based alternative promises a quicker AMR detection than phenotypic AST by bypassing routine culturing for early detection and providing genomic insights into the mechanisms of AMR action, as well as critical clinical information, including pathogen species and virulence factors (Li et al., 2022). Utilizing the increasing WGS data and robust computerized analysis, machine learning has emerged to address the global AMR increase (Ren et al., 2021). It is a subfield of artificial intelligence focusing on developing algorithms to provide accurate and reliable predictions for clinical decisions. Conceptually, the genotype-based machine learning pipeline primarily requires the following steps: data selection (e.g., single-nucleotide polymorphisms and MIC value); encoding to be interpreted by algorithms; data training for model construction; evaluation and optimization by other dataset (Ren et al., 2021). The key challenge of these genotypic strategies that rely on the detection of AMR genes or SNPs to predict AMR phenotypes is the prevalent discrepancy between genotype and phenotype, which may produce detrimental effects on patients by prescribing inappropriate or unnecessary broad-spectrum antimicrobials (Yee et al., 2021). The phenotype of an organism is the observed expression of its genotype but is affected by multiple environmental variables (Davis et al., 2011; Rapun Mas et al., 2019; Urmi et al., 2020). The significance of the genotype–phenotype relationship emphasizes both their inherent correlation and the intricate uncertainty that should not be ignored but evaluated especially upon fighting AMR. Furthermore, AMR strains like Pseudomonas aeruginosa exhibit high plasticity of resistance phenotype driven by environmental changes (Dotsch et al., 2015). Overall, these phenomena suggest that bacteria may prefer different survival strategies under pressure circumstances. Various environmental signals, including antimicrobial and non-antimicrobial compounds, could significantly affect various susceptible phenotypes (Paudyal and Yue, (2019); Berendonk et al., 2015). AMR bacteria is an essential evolutionary outcome under selection pressure, and numerous unknown factors are involved, which implies limitations of single-dimension surveillance and diagnostic approaches. Therefore, more information on specific bacteria-host or -environment dynamics should also be considered to predict AMR.
The COVID-19 pandemic has seriously affected global efforts against AMR (Tomczyk et al., 2021), magnifying the insufficiency of available AMR surveillance capabilities. Accurate and rapid identification of antimicrobial resistance is still highly demanded, and a fine-tuned surveillance system remains the top priority of global public health. Since the culture-dependent traditional approaches cannot cope with the new challenge, various emerging novel methods have shown potential to solve the dilemma. Herein, we summarize these methods and propose an informed database/platform of resistant strains coordinated with reliable rapid molecular assays, multi-omics analysis, and machine learning to improve the performance of the established AMR surveillance system.
Traditional phenotypical diagnostics
Traditional phenotypical diagnostics require isolating the initial bacterial colony from a clinical sample after incubation on a solid medium overnight. For the following broth microdilution assay, the monomicrobial cell is suspended under serial 2-fold dilutions with testing antibiotics in micro-wells. The lowest concentration of antibiotics in which bacteria cannot grow is recorded as the MIC value. For disc diffusion assay, the bacteria is spread onto an agar plate containing a fixed antibiotic concentration. After culturing overnight, the diameter of the circle zone of inhibition is measured. These two results are usually the basis for selecting effective antibiotics for bacterial infections. These traditional diagnoses of antimicrobial susceptibility of AMR strains have long relied on bacteria culturing, which is a time-consuming and labor-intensive process (Burnham et al., 2017). Moreover, the time lag of bacteria growth required for the conventional standard test may change the susceptibility to antimicrobials, particularly for important pathogens such as Yersinia pestis (Inglesby et al., 2000) and Mycobacterium tuberculosis (Votintseva et al., 2017). Although the culture-based antimicrobial susceptibility test used in clinical laboratories produces reproducible results, these assays in vitro cannot approximate the complicated interaction between microorganisms and the host in the tissue (Burnham et al., 2017), which may ultimately direct wrong drug selection (Wang et al., 2019; Elbediwi et al., 2021; Xu et al., 2021; Chen et al., 2022).
To tackle the dilemma of traditional phenotypical diagnostics, microbiologists have made novel efforts on establishing effective AST methodologies. Thore et al. (1977) found that ampicillin treatment induces a significant decrease of intracellular ATP, and an increase in inhibitory zone diameters, according to which they developed the luciferase assay-aided AST by measuring intracellular ATP of bacteria. For antifungal drug testing on Candida albicans, Pore established a flow cytofluorometric susceptibility test (FCST) by determining the cellular fluorescence intensity owing to antibiotic-caused membrane damage and the resulting uptake of propidium iodide or rose Bengal (Pore, 1990). To rapidly detect antibiotic resistance of M. tuberculosis, Riska et al. (1999) used phage with the luciferase reporter to infect the bacteria, and the causal detectable light indicates the infection quantitatively. Fredborg et al. (2013) introduced a real-time optical detection system to image bacterial growth and antimicrobial susceptibility. The automatically generated graphs supported by imaging material showed the antibiotic effects on bacteria, and it was able to screen 96 bacteria–antibiotic combinations at once. Idelevich et al. (2017) developed a methodology based on the BacterioScanTM216R laser scattering technology and following statistical analysis to rapidly discriminate the resistant and susceptible phenotypes of Gram-positive bacteria. Kang et al. (2019) built a droplet microfluidic device consisting of four integrated microdroplet arrays with each holding over 8,000 docking sites, which was capable of screening four antibiotics/pathogens simultaneously and assessing antibiotic sensitivity in 15–30 min. Greater flexibility can be achieved by operating microdroplet arrays individually or jointly. Combined with different aptamers, it has been divided into droplet microfluidics active microfluidics, paper microfluidics, and capillary-driven microfluidics (Han et al., 2021). But it has not been fully applied in clinical settings because of fabrication and operational complexity and less portability (Needs et al., 2020). More phenotypical diagnostics methods are summarized in Table 1.
TABLE 1.
Brief summary of different phenotypic detection technologies.
| Technology | Short description | Advantage | Disadvantage | References |
|---|---|---|---|---|
| Disk diffusion method with short incubation | Measure the inhibition zones after short-time incubation | Relatively rapid | A poorly controlled, unstandardized technic | Barry et al. (1973) |
| The firefly luciferase ATP assay | Growth of microorganisms is paralleled by an increase in ATP levels, and the level of ATP can be determined by the light produced by luciferase assay | Simple and highly sensitive | In many bacterial strains, accumulation of extracellular ATP may be prevented by the presence of ATP-ase activity | Thore et al. (1977) |
| Flow cytometry | Use flow cytometry to detect the membrane damage caused by drugs through increased cellular fluorescence | New and rapid; Improved sensitivity and standardization of the susceptibility test for the relatively large-celled fungi |
PI and RB might be toxic to fungi upon binding to internal cell contents | Pore, (1990) |
| Luciferase Reporter Phage | When infected with mycobacteria, it will produce quantifiable light, the Bronx Box can detect the light | Rapid, reliable, inexpensive, simple, and low-tech manner | Lack of further validation | Riska et al. (1999) |
| Digital time-lapse microscopy | System introduces real-time tracking bacterial growth and antimicrobial susceptibility and generated graphs | The oCelloScope system is faster, portable, and requires low sample volumes to perform high-throughput bacterial susceptibility testing | The system is only suited for the imaging of fluid samples | Fredborg et al. (2013) |
| Microfluidic agarose channel system | Immobilize bacteria in microfluidic culture chamber, track single-cell growth by microscopy, and analyze the time lapse images of single bacterial cells to determine MICs | Fast and accurate | Low throughput and not friendly to use | Choi et al. (2013) |
| Forward Laser Scattering | Use narrow angle forward laser scattering to measure the light scattered from bacteria suspended in a liquid sample | The device is easy-to-use and has compact design and greatly shortens the time of AST | Unable to detect multiple resistance phenotypes | Idelevich et al. (2017) |
| MALDI-TOF MS direct-on-target microdroplet growth assay | Incubate the microorganisms on MALDI-TOF MS target and then detect the microorganisms grown by MALDI-TOF MS. | Rapid and accurate | This study uses only two different species | Idelevich et al. (2018) |
| Accelerate Pheno™ system | Transfer the BCB supernatant to a vial that is introduced into the device, then test automatically | Easy-to-use and fast | The number and characteristics of included samples in the study are limited | Descours et al. (2018) |
| Highly parallelized droplet microfluidic platform | Load water-in-oil droplets into four parallel arrays and then monitor the bacterial growth through the time-lapse imaging function | Fast and consumes less; screen four bacteria/drug combinations simultaneously | Only allow to detect the presence of a small proportion of resistant phenotypes; operation complexity | Kang et al. (2019) |
DNA-based detection
Offering speed and accuracy advantages compared with the traditional gold-standard phenotypic assay, molecular detection on the genetic determinants of resistance phenotype has been widely used in clinical settings. These DNA-based molecular assays can be primarily classified into three major platforms according to their underlying mechanics: polymerase chain reaction (PCR) or RT-PCR (Waseem et al., 2019), microfluidics-supported (Bai et al., 2022), and clustered regularly interspaced short palindromic repeats (CRISPR)-based methodologies (Kaminski et al., 2021). PCR has been extensively used today as a laboratory routine to identify bacteria from multiple environments and resistance genes (Rohde et al., 2017). Multiplex PCR, an optimized PCR by adding several primers, was described to detect nine clinically antibiotic resistance genes of Staphylococcus aureus in a single run including mecA (encoding methicillin resistance), aacA-aphD (aminoglycoside resistance), tetK, tetM (tetracycline resistance), erm(A), erm(C) (macrolide-lincosamide-streptogramin B resistance), vat(A), vat(B), and vat(C) (streptogramin A resistance) (Strommenger et al., 2003). With the high-throughput trait of microfluidic technology and rapid amplification at low temperature (37–42°C) of recombinase polymerase amplification (RPA), RPA-based microfluidics has been reported to identify M. tuberculosis by targeting 16 S rRNA with a sensitivity of 11 CFU/ml in 25 min (Tsaloglou et al., 2018); 10 copies of methicillin-resistant S. aureus DNA mixed with human whole blood were detected in 30 min by probing related SNP (Yeh et al., 2017); the major pathogenic bacteria in urinary tract infections Escherichia coli, Proteus mirabilis, P. aeruginosa, and S. aureus were successfully detected with detection limits of 100 CFU/ml from urine samples within 40 min by similar assay (Chen et al., 2018). The two leading platforms CRISPR-Cas13 based SHERLOCK (specific high-sensitivity enzymatic reporter unlocking) and CRISPR-Cas12 based DETECTR (DNA endonuclease-targeted CRISPR trans reporter) have realized that pathogen detections such as Zika and dengue viruses, bacterial isolates (E. coli; Klebsiella pneumoniae; P. aeruginosa; M. tuberculosis; S. aureus), AMR genes (K. pneumoniae carbapenemase and New Delhi metallo-beta-lactamase 1 (NDM-1), and even could detect cancer mutations with attomolar sensitivity and single-base mismatch specificity (Gootenberg et al., 2017; Chen et al., 2018). This CRISPR-based diagnostics pipeline involves pre-amplification of the target sequence, target recognizing, and cleaving by Cas nuclease, and result visualization. Once the target sequence is base-paired with the guide RNA of the CRISPR complex, the Cas protein cleaves the surrounding reporters (described as “collateral cleavage”), resulting in a detectable signal as a positive detection. CRISPR-Cas12 has already been used to diagnose M. tuberculosis with a sensitivity of two copies (Ai et al., 2019) and identify subspecies of the bacterium by targeting rpoB and erm genes (Xiao et al., 2020). Curti et al. (2020) successfully applied CRISPR-Cas12a to identify carbapenem-resistant genes such as bla KPC, bla NDM, and bla OXA of K. pneumoniae at a pM level within 30 min. Shen et al. (2020) proposed a Cas13a-based system termed as APC-Cas by integrating an allosteric probe, and it was able to detect Salmonella in milk samples with single-cell sensitivity. A brief introduction of other different molecular assays is summarized in Table 2.
TABLE 2.
A brief introduction of different molecular detection technologies.
| Technology | Target | Description | Performance | References |
|---|---|---|---|---|
| Multicomponent nucleic acid enzyme-gold nanoparticle (MNAzyme-GNP) platform | Methicillin-resistant Staphylococcus aureus | Amplified target gene is chemically denatured and blocked to prevent rehybridization. When activated by blocked amplicons, MNAzyme cleaves the linker DNA, rendering GNPs monodispersed. In the absence of the target gene, the linker DNA remains intact owing to inactive MNAzyme and causes GNPs to aggregate | 100 DNA copies/μL | Abdou Mohamed et al. (2021) |
| CRISPR-Cas9 triggered two-Step Isothermal amplification method | Escherichia coli O 157:H7 | The target virulence gene sequences are recognized and cleaved by the CRISPR-Cas9 sy (Sun et al., 2020) stem and trigger strand displacement amplification and rolling circle amplification | 4 CFU/ml | Sun et al. (2020) |
| A clustered regularly interspaced short palindromic repeat (CRISPR)-mediated surface-enhanced Raman scattering (SERS) assay | S. aureus, Acinetobacter baumannii, and Klebsiella pneumoniae with multidrug-resistance | The Au MNP-dCas9/gRNA probe and genomic DNA mixed in a single reaction tube. Next, methylene blue (MB) is added to the tube. Finally, the MDR bacterial gene-bound Au MNP-dCas9/gRNA probe is collected with the assistance of an external magnet, and the SERS measurement is accomplished | fM level | Kim et al. (2020) |
| FLASH (Finding Low Abundance Sequences by Hybridization) | S. aureus with antimicrobial resistance | Combines CRISPR/Cas9 and multiplex PCR | 35 copies | Quan et al. (2019) |
| A paper-based chip integrated with loop-mediated isothermal amplification (LAMP) and the “light switch” molecule [Ru (phen)2dppz]2+ | Methicillin resistant S. aureus, E. coli, Listeria Monocytogenes, and Salmonella | The amplification reagents can be embedded into test spots of the chip in advance, thus simplifying the detection procedure. [Ru (phen)2dppz]2+ was applied to intercalate into amplicons for product analysis, enabling this assay to be operated in a wash-free format | 100 copies/μL | Li et al. (2018) |
| Droplet Digital PCR | Clarithromycin-resistant Helicobacter pylori | A method to simultaneously quantify H. pylori clarithromycin-resistant (mutant) and -susceptible (wild-type) 23 S rRNA gene alleles in both stomach and stool samples using droplet digital PCR. | Discriminate the clarithromycin resistance strain DNAs (A2143G, A2142G, and A2142C) mixed with the wild-type strain at ratio of 0:1, 1:100, 1:10, 1:1, 10:1, 100:1, and 1:0 | Sun et al. (2018) |
| Digital real-time loop-mediated isothermal amplification (dLAMP) assay | E. coli | AST results can be obtained by using digital nucleic acid quantification to measure the phenotypic response of samples exposed to an antibiotic for 15 min | Ultrafast (7 min) | Schoepp et al. (2017) |
WGS emerges as a powerful tool for understanding the genetic makeup of bacteria AMR. As the high-throughput sequencing technology developed in the mid-2000 s, it can illustrate a landscape of the whole resistome in a couple of days with a sophisticated downstream bioinformatic pipeline (Chan, 2016; Goodwin et al., 2016). The new advancement in sequencing technology shortens the time of the whole procedure from days to a few hours (Shendure et al., 2017). According to these known genetic determinants, mass software tools have been designed to detect and predict drug resistance (Henry et al., 2014; Sallet et al., 2019). An immense amount of sequencing data generated annually is used to build a global genotype–phenotype database contributing to a worldwide surveillance system on the AMR strains (Gardy and Loman, 2017). Once the public-health-threatening “criminal” is identified by fingerprinting with a genetic test, its AMR profile will be provided for treatment by the database in time. The Antibiotic Resistance Monitoring, Analysis, and Diagnostics Alliance (https://joinarmada.org/), a nonprofit global organization combatting superbugs with a database of bacterial genomes, has been established to create a specific “criminal database” of AMR pathogens, collecting an unprecedented amount of bacterial strains and details of their antibiotic resistance profiles, genetic identity and epidemiology from a global network of hospitals, veterinarians, scientists, and other advocates. Alcock et al. (2020) developed the Comprehensive Antibiotic Resistance Database providing data of curated reference sequences and SNPs, models, and algorithms, helping users with genotype analysis and phenotype prediction of AMR. The notable AMR databases also include ResFinder (Zankari et al., 2012), ARG-ANNOT (Gupta et al., 2014), the and National Center for Biotechnology Information Pathogen Detection Reference Gene catalog (Sayers et al., 2022). These primary AMR databases curate information from the scientific research into the collection for further sequence analysis and knowledge integration. WGS has also been applied for the risk assessment of probiotic lactic acid bacteria (Peng et al., 2022). However, to detect the genetic determinants of AMR, knowledge or prior research regarding which gene is responsible for the resistance is required for the method before diagnostics. Since complex features and mechanisms of AMR remain obscure, the genotype–phenotype method seems less valuable than expected for AMR identification (Wu et al., 2021; Hu et al., 2022).
Machine learning facilitates the underappreciated antimicrobial resistance prediction
Learning essential characteristics directly from the data of genomic sequences, machine learning obviates the need for prior knowledge of resistant strains caused by unknown mechanisms. It has been proven valuable for predicting AMR with an unbiased method (Martens et al., 2016; Her and Wu, 2018; Wheeler et al., 2018). Unlike direct detection of AMR genes, machine learning often takes more extensive features indiscriminately into AMR identification, such as SNP, K-mer, and pan-genome (Her and Wu, 2018; Nguyen et al., 2019; Ren et al., 2021), and even some parameters affecting the performance and reliability of machine learning-based antibiotic susceptibility tests need to be evaluated (Hicks et al., 2019). Different from the traditional DNA alignment approach, machine learning is mathematically oriented by algorithms including logistic regression (LR), support vector machine (SVM), random forest (RF), and convolutional neural network (CNN) (Her and Wu, 2018). In order to confirm the WGS data to the valid format of existing classification algorithms, these data are usually encoded before analysis by the algorithms as mentioned above for model construction, and standard encoding methods include Label, One-Hot, and frequency matrix chaos game representation (FCGR), etc., (Her and Wu, 2018). The final prediction model will be tested and evaluated by other datasets, and the model established might vary for different coding methods and algorithms/classifiers involved. Leveraged with big data from WGS or other NGS technologies, machine learning facilitates potential AMR prediction and helps direct informed drug decisions (Moradigaravand et al., 2018; Ren et al., 2021). Recent literature in the field is presented in Table 3.
TABLE 3.
Recent progresses on ML for AMR prediction.
| Strain | Extracted feature | Algorithm | Predicting target | Result | References |
|---|---|---|---|---|---|
| E. coli | Pan-genome | Support Vector Machine; Naïve Bayes (NB); Adaboost; Random Forest | Meropenem; gentamicin; ciprofloxacin; trimethoprim/sulfa; ethoxazole ampicillin; cefazolin; ampicillin/sulbactam; ceftazidime; cefepime; piperacillin/tazobactam; tobramycin; ceftriaxone | Support Vector Machinen for 12 drugs’ AUC: 0.67–0.82 | Her and Wu, (2018) |
| Naïve Bayes (NB) for 12 drugs’ AUC:0.69–0.85 | |||||
| Adaboost for 12 drugs’ AUC: 0.54–0.86 | |||||
| Random Forest for 12 drugs’ AUC: 0.51–0.82 | |||||
| Nontyphoidal Salmonella | k-mer | XGBoost | Ampicillin; amoxicillin-clavulanic acid; ceftriaxone; azithromycin; chloramphenicol; ciprofloxacin; trimethoprim-sulfamethoxazole; sulfisoxazole; cefoxitin; gentamicin; kanamycin; nalidixic acid; streptomycin; tetracycline; ceftiofur | XGBoost for 12 drugs’ Accuracy: 0.33–0.91 | Nguyen et al. (2019) |
| E. coli | SNP | Support Vector Machine; Logistic Regression; Random Forest; Convolutional Neural Network | Ciprofloxacin; cefotaxime; ceftazidime; gentamicin | Support Vector Machine for 4 drugs’ Accuracy: 0.75–0.88 | Ren et al. (2021) |
| Logistic Regression for four drugs’ Accuracy: 0.77–0.85 | |||||
| Random Forest for 4 drugs’ Accuracy: 0.77–0.92 | |||||
| Convolutional Neural Network for four drugs’ Accuracy: 0.71–0.84 | |||||
| K. pneumoniae; A. baumannii | k-mer | Machine Classification; Random Forest; Random Forest Regression | Ciprofloxacin; azithromycin | Three model’s Accuracy for CIP: ≥0.93; Three model’s Accuracy for AZI: 0.57–0.94 | Hicks et al. (2019) |
| Bacitracin Vancomycin | Protein sequences | Support Vector Machine | AMR or non-AMR | Classification accuracies 87%–90% | Chowdhury et al. (2020) |
| Neisseria gonorrhoeae | pan-genome | Logistic Regression; Random Forest; Gradient Boosting Decision Tree; Support Vector Machine | Penicillin; tetracycline; cefixime; ciprofloxacin; azithromycin | AUC and Recall values of the training and testing datasets were >0.80 in at least one machine learning model for all antibiotics | Li et al. (2020) |
Clinical application of machine learning for AMR diagnosing, preventing, and understanding still stands in the infancy stage because of the complexity of AMR itself and the deployment of machine learning for clinical practice. That genomic-based machine learning aiming to predict the MIC value is the foremost strategy for AMR prediction (Coelho et al., 2013; Pesesky et al., 2016; Nguyen et al., 2019). But challenges/limitations remain to be overcome. Training data, (e.g., MIC value) might not be available for all laboratories and sections, which would affect the clinical effectiveness of the model, which suggests unified standardization of operating protocols or guidelines is highly demanded. For algorithms used in the model, different ones reach different outcomes, indicating a comparative study of algorithms should be conducted before applying for better accuracy. In addition, the prediction result should be understandable to intended users. For the breadth of machine learning prediction, studies have primarily predicted the most common antibiotics used on a few common bacterial pathogens, but the clinical encountered pathogens far outrange the prediction covering. Special effort should also be taken on the highly clinically significant organisms, including vancomycin-non-susceptible S. aureus (Shariati et al., 2020) and the ones with AMR phenotypic plasticity, including P. aeruginosa (Khaledi et al., 2020). Furthermore, data on AMR to new antibiotics (e.g., eravacycline, ceftazidime-avibactam, and cefiderocol) are also required to examine in clinical settings. With the exponential increase of available biological data, massive investments in computational power, critical advancements in algorithm performance, and increasing global involvement worldwide, machine learning will accelerate the clinical diagnosis of AMR to a new level.
Transcriptomic and proteomic approaches
The DNA-based method only detects the existence of these potential resistance DNA traits without providing information regarding efficient transcription, expression, and antimicrobial susceptibility. The feedback in bacterial transcriptome and proteome to antibiotics exposure offers real-time dynamic information of relevant genes. Their expression change in mRNA and protein levels is endowed with key diagnostic values for AMR detection. Some efforts have also been made in transcriptome and proteome, but remain limited. Steinberger-Levy et al. (2016) established a research model by exposing 53 strains of Y. pestis to the growth-inhibiting concentrations of ciprofloxacin for different time periods and investigates the expression changes of specific marker genes by transcriptomic DNA microarray analysis. Eleven transcripts with significant change were identified as the potential biomarkers, of which four mRNAs (recA, pla2, recN, and dinI) were selected to determine the susceptibility by performing quantitative RT-PCR, and the results are consistent with MIC values. Rohde et al. (2016) used fluorescence in situ hybridization and immunofluorescence tests to locate and quantify the mRNA and protein existence of the TEM β-lactamase, conferring ampicillin resistance in the E. coli. Chen et al. (2020) performed a shotgun proteomics assay and WGS on four isolates of Campylobacter jejuni and analyzed the data in the Comprehensive Antibiotic Resistance Database for AMR detection. It was found that both genomic and proteomic approaches can identify molecular determinants responsible for resistance to tetracycline and ciprofloxacin, in line with their phenotypes. These methods require several steps and analyses, which are more suitable for basic research, but not feasible for the field application. For rapid RNA detection, the RNA-targeting CRISPR-associated enzyme Cas13a directly binds to the target RNA and releases the positive signal (Shinoda et al., 2021). Combined with RT-RPA that transforms the RNA to cDNA and amplifies DNA with primers, the DNA-targeting enzyme Cas12a can also be applied to the RNA detections (Malci et al., 2022). In clinical practice, rapid molecular diagnostics help distinguish viral infections from bacterial infections, preventing unnecessary treatment of antibiotics. Only molecular-based methods cannot differentiate which bacteria contain the AMR element (plasmids containing AMR gene) in the mixed bacterial infection of clinical use. If the carrying AMR genes among various bacterial populations can be identified specifically, we might use a more targeting or effective strategy against AMR increase, suggesting this field might need further investigation.
Current challenge
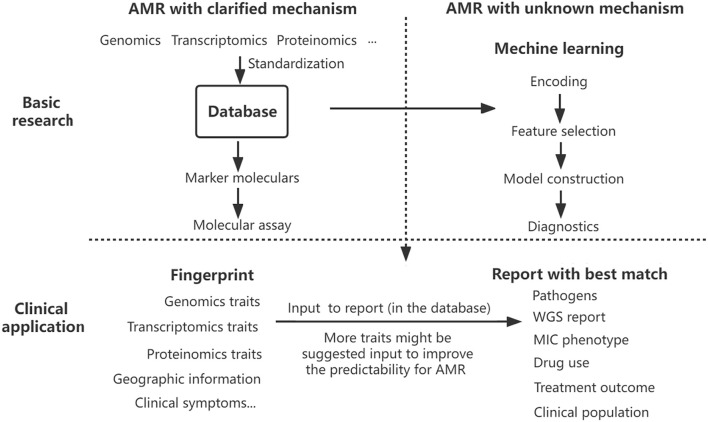
A comprehensive analysis has suggested more factors to make more precision diagnostics and informed preclinical decisions (Sommer et al., 2017). To deal with this compiling problem, a full-scale assessment system should be set up to predict AMR precisely. Establishing a database on genotype–phenotype data is a good start, but a more comprehensive platform of AMR strain is highly needed for the precision diagnostics of AMR. Utilizing the multi-dimension data collected from the genome, transcriptome, and proteome profiles of AMR bacteria isolated from hospitals, doctor offices, clinical laboratories, communities, and veterinary sources, a ‘criminal database’ of AMR pathogens can be geographically and phylogenetically established. After fingerprinting those known genetic makers by rapid molecular test and sampling information, the database will report with the best match containing current and historical essential details, including WGS report, pathogen, MIC phenotype, drug use and treatment outcomes, clinical population, and other accessory information. With the data collected from multiple sources, bacterial AMR patterns will be well analyzed by machine learning, enabling reliable digital models of a particular drug, pathogen, and clinical population. Machine learning will act as an alternative to pinpoint that unknown resistance. Surveillance and diagnostic interaction significantly improve antimicrobial selection and epidemiological monitoring in the pipeline. Once database standardization is well-established, this constantly updated global detection platform combining diagnostics, surveillance, and prediction of AMR would improve proactive identification and mitigate the emerging crisis of AMR (Figure 1).
FIGURE 1.
Proposed AMR surveillance model. Various data on antibiotic resistance have been collected into the database, which provides marker molecules to detect AMR with clarified mechanism and fuels the model training of machine learning to predict the AMR with an unknown mechanism. The results of rapid molecular detection on multi-traits and other information help fingerprint the pathogens, and the database interface will report the best matched or advised treatment decision directly. Taking advantage of the rapidity of molecular assays and the precision of machine learning fueled by a constant flow of multi-dimensional data, the AMR surveillance platform optimizes drug selection, antimicrobial stewardship, and epidemiological monitoring.
From the clinical perspective, the priority is confirming whether antimicrobial therapies are needed in a particular case; the second is which drug is suitable for an optimized treatment if a bacterial infection is already present. Apparently, the most clinical urgency for patients and physicians is not knowing which bacteria is resistant to what antimicrobial but determining which narrow-spectrum drug can eliminate the pathogen. Most diagnostic methods aim to identify drug-resistant determinants of superbugs, but few studies explored if there are any drug-susceptible determinants or other multi-omics traits in AMR strains. These drug-susceptible feature data can be used for model training in machine learning, and molecular-based assays can detect drug-susceptible markers directly. More innovative efforts are needed in this area in the future.
Future direction
In the coming decade, advancements in technical and computational tools for multi-omics approaches are continually improving knowledge regarding the diverse AMR mechanisms and integrating a deep mechanistic understanding of AMR determinants with a broader systematic analysis of microorganisms. The ultimate goal will lead to a revolutionary change in AMR diagnostics, significantly guiding the surveillance of AMR threats and finally slowing down the rising crisis.
Author contributions
HW, CJ, HL, and RY wrote the manuscript; JC, YL, and MY revised and confirmed the manuscript.
Funding
This study was supported by the National Program on Key Research Project of China (2019YFE0103900 and 2017YFC1600103) as well as the European Union’s Horizon 2020 Research and Innovation Programme under Grant Agreement No. 861917—SAFFI, National Natural Science Foundation of China (31872837 & 32150410374), Zhejiang Provincial Natural Science Foundation of China (LR19C180001), and Zhejiang Provincial Key R&D Program of China (2022C02024, 2021C02008, and 2020C02032).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Abdou Mohamed M. A., Kozlowski H. N., Kim J., Zagorovsky K., Kantor M., Feld J. J., et al. (2021). Diagnosing antibiotic resistance using nucleic acid enzymes and gold nanoparticles. ACS Nano 15 (6), 9379–9390. 10.1021/acsnano.0c09902 PubMed Abstract | 10.1021/acsnano.0c09902 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Ai J. W., Zhou X., Xu T., Yang M., Chen Y., He G. Q., et al. (2019). CRISPR-based rapid and ultra-sensitive diagnostic test for Mycobacterium tuberculosis . Emerg. Microbes Infect. 8 (1), 1361–1369. 10.1080/22221751.2019.1664939 PubMed Abstract | 10.1080/22221751.2019.1664939 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcock B. P., Raphenya A. R., Lau T. T. Y., Tsang K. K., Bouchard M., Edalatmand A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48 (D1), D517–D525. 10.1093/nar/gkz935 PubMed Abstract | 10.1093/nar/gkz935 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Ji J., Ji F., Wu S., Tian Y., Jin B., et al. (2022). Recombinase polymerase amplification integrated with microfluidics for nucleic acid testing at point of care. Talanta 240, 123209. 10.1016/j.talanta.2022.123209 PubMed Abstract | 10.1016/j.talanta.2022.123209 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Barry A. L., Joyce L. J., Adams A. P., Benner E. J. (1973). Rapid determination of antimicrobial susceptibility for urgent clinical situations. Am. J. Clin. Pathol. 59 (5), 693–699. 10.1093/ajcp/59.5.693 PubMed Abstract | 10.1093/ajcp/59.5.693 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Berendonk T. U., Manaia C. M., Christophe M., Despo F. K., Eddie C., Fiona W., et al. (2015). Tackling antibiotic resistance: the environmental framework. Nat. Rev. Microbiol. 13 (5), 310–317. 10.1038/nrmicro3439 PubMed Abstract | 10.1038/nrmicro3439 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Biswas S., Li Y., Elbediwi M., Yue M. (2019). Emergence and dissemination of mcr-carrying clinically relevant Salmonella typhimurium monophasic clone ST34. Microorganisms 7 (9), E298. 10.3390/microorganisms7090298 PubMed Abstract | 10.3390/microorganisms7090298 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham C. A. D., Leeds J., Nordmann P., O'Grady J., Patel J. (2017). Diagnosing antimicrobial resistance. Nat. Rev. Microbiol. 15 (11), 697–703. 10.1038/nrmicro.2017.103 PubMed Abstract | 10.1038/nrmicro.2017.103 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Chan K. G. (2016). Whole-genome sequencing in the prediction of antimicrobial resistance. Expert Rev. anti. Infect. Ther. 14 (7), 617–619. 10.1080/14787210.2016.1193005 PubMed Abstract | 10.1080/14787210.2016.1193005 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Chen C. Y., Clark C. G., Langner S., Boyd D. A., Bharat A., McCorrister S. J., et al. (2020). Detection of antimicrobial resistance using proteomics and the comprehensive antibiotic resistance database: A case study. Proteomics. Clin. Appl. 14 (4), e1800182. 10.1002/prca.201800182 PubMed Abstract | 10.1002/prca.201800182 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. G., Xu Y. C., Yan H., Zhu Y. Z., Wang L., Zhang Y., et al. (2018a). Sensitive and rapid detection of pathogenic bacteria from urine samples using multiplex recombinase polymerase amplification. Lab. Chip 18 (16), 2441–2452. 10.1039/c8lc00399h PubMed Abstract | 10.1039/c8lc00399h | Google Scholar [DOI] [PubMed] [Google Scholar]
- Chen J. S., Ma E., Harrington L. B., Da Costa M., Tian X., Palefsky J. M., et al. (2018b). CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360 (6387), 436–439. 10.1126/science.aar6245 PubMed Abstract | 10.1126/science.aar6245 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Zhao L., Liu H., Ding Q., Jia C., Liao S., et al. (2022). Nanoporous silver nanorods as surface-enhanced Raman scattering substrates. Biosens. Bioelectron. 202, 114004. 10.1016/j.bios.2022.114004 PubMed Abstract | 10.1016/j.bios.2022.114004 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Choi J., Jung Y. G., Kim J., Kim S., Jung Y., Na H., et al. (2013). Rapid antibiotic susceptibility testing by tracking single cell growth in a microfluidic agarose channel system. Lab. Chip 13 (2), 280–287. 10.1039/c2lc41055a PubMed Abstract | 10.1039/c2lc41055a | Google Scholar [DOI] [PubMed] [Google Scholar]
- Chowdhury A. S., Call D. R., Broschat S. L. (2020). PARGT: a software tool for predicting antimicrobial resistance in bacteria. Sci. Rep. 10 (1), 11033. 10.1038/s41598-020-67949-9 PubMed Abstract | 10.1038/s41598-020-67949-9 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho J. R., Carrico J. A., Knight D., Martinez J. L., Morrissey I., Oggioni M. R., et al. (2013). The use of machine learning methodologies to analyse antibiotic and biocide susceptibility in Staphylococcus aureus . PLoS One 8 (2), e55582. 10.1371/journal.pone.0055582 PubMed Abstract | 10.1371/journal.pone.0055582 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curti L. A., Pereyra-Bonnet F., Repizo G. D., Fay J. V., Salvatierra K., Blariza M. J., et al. (2020). CRISPR-based platform for carbapenemases and emerging viruses detection using Cas12a (Cpf1) effector nuclease. Emerg. Microbes Infect. 9 (1), 1140–1148. 10.1080/22221751.2020.1763857 PubMed Abstract | 10.1080/22221751.2020.1763857 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. A., Besser T. E., Orfe L. H., Baker K. N., Lanier A. S., Broschat S. L., et al. (2011). Genotypic-phenotypic discrepancies between antibiotic resistance characteristics of Escherichia coli isolates from calves in management settings with high and low antibiotic use. Appl. Environ. Microbiol. 77 (10), 3293–3299. 10.1128/AEM.02588-10 PubMed Abstract | 10.1128/AEM.02588-10 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descours G., Desmurs L., Hoang T. L. T., Ibranosyan M., Baume M., Ranc A. G., et al. (2018). Evaluation of the Accelerate Pheno system for rapid identification and antimicrobial susceptibility testing of Gram-negative bacteria in bloodstream infections. Eur. J. Clin. Microbiol. Infect. Dis. 37 (8), 1573–1583. 10.1007/s10096-018-3287-6 PubMed Abstract | 10.1007/s10096-018-3287-6 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Dotsch A., Schniederjans M., Khaledi A., Hornischer K., Schulz S., Bielecka A., et al. (2015). The Pseudomonas aeruginosa transcriptional landscape is shaped by environmental heterogeneity and genetic variation. mBio 6 (4), e00749. 10.1128/mBio.00749-15 PubMed Abstract | 10.1128/mBio.00749-15 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbediwi M., Beibei W., Pan H., Jiang Z., Biswas S., Li Y., et al. (2020). Genomic characterization of mcr-1-carrying Salmonella enterica serovar 4, [5], 12:i:- ST 34 clone isolated from pigs in China. Front. Bioeng. Biotechnol. 8, 663. 10.3389/fbioe.2020.00663 PubMed Abstract | 10.3389/fbioe.2020.00663 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbediwi M., Li Y., Paudyal N., Pan H., Li X., Xie S., et al. (2019). Global burden of colistin-resistant bacteria: Mobilized colistin resistance genes study (1980-2018). Microorganisms 7 (10), E461. 10.3390/microorganisms7100461 PubMed Abstract | 10.3390/microorganisms7100461 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbediwi M., Shi D., Biswas S., Xu X., Yue M. (2021). Changing patterns of Salmonella enterica serovar rissen from humans, food animals, and animal-derived foods in China, 1995-2019. Front. Microbiol. 12, 702909. 10.3389/fmicb.2021.702909 PubMed Abstract | 10.3389/fmicb.2021.702909 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farfour E., Lecuru M., Dortet L., Le Guen M., Cerf C., Karnycheff F., et al. (2020). Carbapenemase-producing Enterobacterales outbreak: Another dark side of COVID-19. Am. J. Infect. Control 48 (12), 1533–1536. 10.1016/j.ajic.2020.09.015 PubMed Abstract | 10.1016/j.ajic.2020.09.015 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredborg M., Andersen K. R., Jorgensen E., Droce A., Olesen T., Jensen B. B., et al. (2013). Real-time optical antimicrobial susceptibility testing. J. Clin. Microbiol. 51 (7), 2047–2053. 10.1128/JCM.00440-13 PubMed Abstract | 10.1128/JCM.00440-13 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardy J. L., Loman N. J. (2017). Towards a genomics-informed, real-time, global pathogen surveillance system. Nat. Rev. Genet. 19 (1), 9–20. 10.1038/nrg.2017.88 PubMed Abstract | 10.1038/nrg.2017.88 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin S., McPherson J. D., McCombie W. R. (2016). Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 17 (6), 333–351. 10.1038/nrg.2016.49 PubMed Abstract | 10.1038/nrg.2016.49 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg J. S., Abudayyeh O. O., Lee J. W., Essletzbichler P., Dy A. J., Joung J., et al. (2017). Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356 (6336), 438–442. 10.1126/science.aam9321 PubMed Abstract | 10.1126/science.aam9321 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S. K., Padmanabhan B. R., Diene S. M., Lopez-Rojas R., Kempf M., Landraud L., et al. (2014). ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 58 (1), 212–220. 10.1128/Aac.01310-13 PubMed Abstract | 10.1128/Aac.01310-13 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Liu Y., Yin J., Yue M., Mu Y. (2021). Microfluidic devices for multiplexed detection of foodborne pathogens. Food Res. Int. 143, 110246. 10.1016/j.foodres.2021.110246 PubMed Abstract | 10.1016/j.foodres.2021.110246 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Henry V. J., Bandrowski A. E., Pepin A. S., Gonzalez B. J., Desfeux A. (2014). OMICtools: an informative directory for multi-omic data analysis. Oxford: Database. 10.1093/database/bau069 10.1093/database/bau069 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Her H. L., Wu Y. W. (2018). A pan-genome-based machine learning approach for predicting antimicrobial resistance activities of the Escherichia coli strains. Bioinformatics 34 (13), i89–i95. 10.1093/bioinformatics/bty276 PubMed Abstract | 10.1093/bioinformatics/bty276 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks A. L., Wheeler N., Sanchez-Buso L., Rakeman J. L., Harris S. R., Grad Y. H. (2019). Evaluation of parameters affecting performance and reliability of machine learning-based antibiotic susceptibility testing from whole genome sequencing data. PLoS Comput. Biol. 15 (9), e1007349. 10.1371/journal.pcbi.1007349 PubMed Abstract | 10.1371/journal.pcbi.1007349 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Hou P., Teng L., Miao S., Zhao L., Ji S., et al. (2022). Genomic investigation reveals a community typhoid outbreak caused by contaminated drinking water in China, 2016. Front. Med. 9, 753085. 10.3389/fmed.2022.753085 10.3389/fmed.2022.753085 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idelevich E. A., Hoy M., Gorlich D., Knaack D., Grunastel B., Peters G., et al. (2017). Rapid phenotypic detection of microbial resistance in gram-positive bacteria by a real-time laser scattering method. Front. Microbiol. 8, 1064. 10.3389/fmicb.2017.01064 PubMed Abstract | 10.3389/fmicb.2017.01064 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idelevich E. A., Sparbier K., Kostrzewa M., Becker K. (2018). Rapid detection of antibiotic resistance by MALDI-TOF mass spectrometry using a novel direct-on-target microdroplet growth assay. Clin. Microbiol. Infect. 24 (7), 738–743. 10.1016/j.cmi.2017.10.016 PubMed Abstract | 10.1016/j.cmi.2017.10.016 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Inglesby T. V., Dennis D. T., Henderson D. A., Bartlett J. G., Ascher M. S., Eitzen E., et al. (2000). Plague as a biological weapon: Medical and public health management. Working group on civilian biodefense. JAMA 283 (17), 2281–2290. 10.1001/jama.283.17.2281 PubMed Abstract | 10.1001/jama.283.17.2281 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Jiang Z., Paudyal N., Xu Y., Deng T., Li F., Pan H., et al. (2019). Antibiotic resistance profiles of Salmonella recovered from finishing pigs and slaughter facilities in henan, China. Front. Microbiol. 10, 1513. 10.3389/fmicb.2019.01513 PubMed Abstract | 10.3389/fmicb.2019.01513 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski M. M., Abudayyeh O. O., Gootenberg J. S., Zhang F., Collins J. J. (2021). CRISPR-based diagnostics. Nat. Biomed. Eng. 5 (7), 643–656. 10.1038/s41551-021-00760-7 PubMed Abstract | 10.1038/s41551-021-00760-7 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Kang W., Sarkar S., Lin Z. S., McKenney S., Konry T. (2019). Ultrafast parallelized microfluidic platform for antimicrobial susceptibility testing of gram positive and negative bacteria. Anal. Chem. 91 (9), 6242–6249. 10.1021/acs.analchem.9b00939 PubMed Abstract | 10.1021/acs.analchem.9b00939 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Kariyawasam R. M., Julien D. A., Jelinski D. C., Larose S. L., Rennert-May E., Conly J. M., et al. (2022). Antimicrobial resistance (AMR) in COVID-19 patients: a systematic review and meta-analysis (november 2019-june 2021). Antimicrob. Resist Infect. Control 11 (1), 45. 10.1186/s13756-022-01085-z PubMed Abstract | 10.1186/s13756-022-01085-z | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaledi A., Weimann A., Schniederjans M., Asgari E., Kuo T. H., Oliver A., et al. (2020). Predicting antimicrobial resistance in Pseudomonas aeruginosa with machine learning-enabled molecular diagnostics. EMBO Mol. Med. 12 (3), e10264. 10.15252/emmm.201910264 PubMed Abstract | 10.15252/emmm.201910264 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Lee S., Seo H. W., Kang B., Moon J., Lee K. G., et al. (2020). Clustered regularly interspaced short palindromic repeats-mediated surface-enhanced Raman scattering assay for multidrug-resistant bacteria. ACS Nano 14, 17241–17253. 10.1021/acsnano.0c07264 10.1021/acsnano.0c07264 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Knight G. M., Glover R. E., McQuaid C. F., Olaru I. D., Gallandat K., Leclerc Q. J., et al. (2021). Antimicrobial resistance and COVID-19: Intersections and implications. Elife 10, e64139. 10.7554/eLife.64139 PubMed Abstract | 10.7554/eLife.64139 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Zhou X., Liu H., Deng H., Huang R., Xing D. (2018). Simultaneous detection of antibiotic resistance genes on paper-based chip using [Ru(phen)2dppz](2+) turn-on fluorescence probe. ACS Appl. Mat. Interfaces 10 (5), 4494–4501. 10.1021/acsami.7b17653 10.1021/acsami.7b17653 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Li Y., Kang X., Ed-Dra A., Zhou X., Jia C., Müller A., et al. (2022) Genome-based assessment of antimicrobial resistance and virulence potential for non-Pullorum/Gallinarum Salmonella serovars recovered from dead poultry in China. Microbiol. Spectr., e0096522. 10.1128/spectrum.00965-22 10.1128/spectrum.00965-22 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Lin J., Hu Y., Zhou J. (2020). PARMAP: A pan-genome-based computational framework for predicting antimicrobial resistance. Front. Microbiol. 11, 578795. 10.3389/fmicb.2020.578795 PubMed Abstract | 10.3389/fmicb.2020.578795 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma E. S. K., Kung K. H., Chen H. (2021). Combating antimicrobial resistance during the COVID-19 pandemic. Hong Kong Med. J. 27 (6), 396–398. 10.12809/hkmj215124 PubMed Abstract | 10.12809/hkmj215124 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Malci K., Walls L. E., Rios-Solis L. (2022). Rational design of CRISPR/Cas12a-RPA based one-pot COVID-19 detection with design of experiments. ACS Synth. Biol. 11 (4), 1555–1567. 10.1021/acssynbio.1c00617 PubMed Abstract | 10.1021/acssynbio.1c00617 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens K., Hallin J., Warringer J., Liti G., Parts L. (2016). Predicting quantitative traits from genome and phenome with near perfect accuracy. Nat. Commun. 7, 11512. 10.1038/ncomms11512 PubMed Abstract | 10.1038/ncomms11512 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradigaravand D., Palm M., Farewell A., Mustonen V., Warringer J., Parts L. (2018). Prediction of antibiotic resistance in Escherichia coli from large-scale pan-genome data. PLoS Comput. Biol. 14 (12), e1006258. 10.1371/journal.pcbi.1006258 PubMed Abstract | 10.1371/journal.pcbi.1006258 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needs S. H., Donmez S. I., Bull S. P., McQuaid C., Osborn H. M. I., Edwards A. D. (2020). Challenges in microfluidic and point-of-care phenotypic antimicrobial resistance tests. Front. Mech. Eng. 6. 10.3389/fmech.2020.00073 10.3389/fmech.2020.00073 | Google Scholar [DOI] [Google Scholar]
- Nguyen M., Long S. W., McDermott P. F., Olsen R. J., Olson R., Stevens R. L., et al. (2019). Using machine learning to predict antimicrobial MICs and associated genomic features for nontyphoidal Salmonella. J. Clin. Microbiol. 57 (2), e01260–18. 10.1128/JCM.01260-18 PubMed Abstract | 10.1128/JCM.01260-18 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H., Li X., Fang W., Yue M. (2018). Analysis of major human and foodborne pathogens and their resistance to antimicrobials in the USA in the past two decades: Implications for surveillance and control of antimicrobial resistance in China. J. Zhejiang Univ. Agric. Life Sci. 44 (2), 237–246. 10.3785/j.issn.1008-9209.2018.03.124 10.3785/j.issn.1008-9209.2018.03.124 | Google Scholar [DOI] [Google Scholar]
- Paudyal N., Yue M. (2019) Antibiotic resistance in the “dark matter”. Clin. Infect. Dis. 69 (2), 379–380. 10.1093/cid/ciz007 PubMed Abstract | 10.1093/cid/ciz007 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Peng X., Ed-Dra A., Yue M. (2022). Whole genome sequencing for the risk assessment of probiotic lactic acid bacteria. Crit. Rev. Food Sci. Nutr., 1–19. 10.1080/10408398.2022.2087174, 10.1080/10408398.2022.2087174 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Pesesky M. W., Hussain T., Wallace M., Patel S., Andleeb S., Burnham C. D., et al. (2016). Evaluation of machine learning and rules-based approaches for predicting antimicrobial resistance profiles in gram-negative bacilli from whole genome sequence data. Front. Microbiol. 7, 1887. 10.3389/fmicb.2016.01887 PubMed Abstract | 10.3389/fmicb.2016.01887 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pore R. S. (1990). Antibiotic susceptibility testing of Candida albicans by flow cytometry. Curr. Microbiol. 20 (5), 323–328. 10.1007/BF02091913 10.1007/BF02091913 | Google Scholar [DOI] [Google Scholar]
- Quan J., Langelier C., Kuchta A., Batson J., Teyssier N., Lyden A., et al. (2019). FLASH: a next-generation CRISPR diagnostic for multiplexed detection of antimicrobial resistance sequences. Nucleic Acids Res. 47 (14), e83. 10.1093/nar/gkz418 PubMed Abstract | 10.1093/nar/gkz418 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapun Mas L., Angulo Lopez I., Cecilio Irazola A., Betran Escartin A. I. (2019). Discrepancy in the genotypic versus phenotypic testing for resistance to rifampicin in Mycobacterium tuberculosis. A case report. Enferm. Infecc. Microbiol. Clin. 37 (3), 212–213. 10.1016/j.eimc.2018.03.015 10.1016/j.eimc.2018.03.015 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Ren Y., Chakraborty T., Doijad S., Falgenhauer L., Falgenhauer J., Goesmann A., et al. (2021). Prediction of antimicrobial resistance based on whole-genome sequencing and machine learning. Bioinformatics 38, 325–334. 10.1093/bioinformatics/btab681 10.1093/bioinformatics/btab681 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resistance R. o. A. (2016). “Tackling drug-resistant infections globally: final report and recommendations,” in Review on antimicrobial resistance. Google Scholar [Google Scholar]
- Riska P. F., Su Y., Bardarov S., Freundlich L., Sarkis G., Hatfull G., et al. (1999). Rapid film-based determination of antibiotic susceptibilities of Mycobacterium tuberculosis strains by using a luciferase reporter phage and the Bronx Box. J. Clin. Microbiol. 37 (4), 1144–1149. 10.1128/JCM.37.4.1144-1149.1999 PubMed Abstract | 10.1128/JCM.37.4.1144-1149.1999 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde A., Hammerl J. A., Boone I., Jansen W., Fohler S., Klein G., et al. (2017). Overview of validated alternative methods for the detection of foodborne bacterial pathogens. Trends Food Sci. Technol. 62, 113–118. 10.1016/j.tifs.2017.02.006 10.1016/j.tifs.2017.02.006 | Google Scholar [DOI] [Google Scholar]
- Rohde A., Hammerl J. A., Dahouk S. A. (2016). Rapid screening for antibiotic resistance elements on the RNA transcript, protein and enzymatic activity level. Ann. Clin. Microbiol. Antimicrob. 15 (1), 55. 10.1186/s12941-016-0167-8 PubMed Abstract | 10.1186/s12941-016-0167-8 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallet E., Gouzy J., Schiex T. (2019). EuGene: An automated integrative gene finder for eukaryotes and prokaryotes. Methods Mol. Biol. 1962, 97–120. 10.1007/978-1-4939-9173-0_6 PubMed Abstract | 10.1007/978-1-4939-9173-0_6 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Sayers E. W., Bolton E. E., Brister J. R., Canese K., Chan J., Comeau D. C., et al. (2022). Database resources of the national center for biotechnology information. Nucleic Acids Res. 50 (D1), D20–D26. 10.1093/nar/gkab1112 PubMed Abstract | 10.1093/nar/gkab1112 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoepp N. G., Schlappi T. S., Curtis M. S., Butkovich S. S., Miller S., Humphries R. M., et al. (2017). Rapid pathogen-specific phenotypic antibiotic susceptibility testing using digital LAMP quantification in clinical samples. Sci. Transl. Med. 9 (410), eaal3693. 10.1126/scitranslmed.aal3693 PubMed Abstract | 10.1126/scitranslmed.aal3693 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurch A. C., van Schaik W. (2017). Challenges and opportunities for whole-genome sequencing-based surveillance of antibiotic resistance. Ann. N. Y. Acad. Sci. 1388 (1), 108–120. 10.1111/nyas.13310 PubMed Abstract | 10.1111/nyas.13310 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Shariati A., Dadashi M., Moghadam M. T., van Belkum A., Yaslianifard S., Darban-Sarokhalil D. (2020). Global prevalence and distribution of vancomycin resistant, vancomycin intermediate and heterogeneously vancomycin intermediate Staphylococcus aureus clinical isolates: a systematic review and meta-analysis. Sci. Rep. 10 (1), 12689. 10.1038/s41598-020-69058-z PubMed Abstract | 10.1038/s41598-020-69058-z | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J., Zhou X., Shan Y., Yue H., Huang R., Hu J., et al. (2020). Sensitive detection of a bacterial pathogen using allosteric probe-initiated catalysis and CRISPR-Cas13a amplification reaction. Nat. Commun. 11 (1), 267. 10.1038/s41467-019-14135-9 PubMed Abstract | 10.1038/s41467-019-14135-9 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J., Balasubramanian S., Church G. M., Gilbert W., Rogers J., Schloss J. A., et al. (2017). DNA sequencing at 40: past, present and future. Nature 550 (7676), 345–353. 10.1038/nature24286 PubMed Abstract | 10.1038/nature24286 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Shinoda H., Taguchi Y., Nakagawa R., Makino A., Okazaki S., Nakano M., et al. (2021). Amplification-free RNA detection with CRISPR-Cas13. Commun. Biol. 4 (1), 476. 10.1038/s42003-021-02001-8 PubMed Abstract | 10.1038/s42003-021-02001-8 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smieszek T., Pouwels K. B., Dolk F. C. K., Smith D. R. M., Hopkins S., Sharland M., et al. (2018). Potential for reducing inappropriate antibiotic prescribing in English primary care. J. Antimicrob. Chemother. 73 (2), ii36–ii43. 10.1093/jac/dkx500 PubMed Abstract | 10.1093/jac/dkx500 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer M. O. A., Munck C., Toftkehler R. V., Dan I. A. (2017). Prediction of antibiotic resistance: time for a new preclinical paradigm? Nat. Rev. Microbiol. 15 (11), 689–696. 10.1038/nrmicro.2017.75 PubMed Abstract | 10.1038/nrmicro.2017.75 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Steinberger-Levy I., Shifman O., Zvi A., Ariel N., Beth-Din A., Israeli O., et al. (2016). A rapid molecular test for determining Yersinia pestis susceptibility to ciprofloxacin by the quantification of differentially expressed marker genes. Front. Microbiol. 7, 763. 10.3389/fmicb.2016.00763 PubMed Abstract | 10.3389/fmicb.2016.00763 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strommenger B., Kettlitz C., Werner G., Witte W. (2003). Multiplex PCR assay for simultaneous detection of nine clinically relevant antibiotic resistance genes in Staphylococcus aureus . J. Clin. Microbiol. 41 (9), 4089–4094. 10.1128/Jcm.41.9.4089-4094.2003 PubMed Abstract | 10.1128/Jcm.41.9.4089-4094.2003 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Talarico S., Yao L., He L., Self S., You Y., et al. (2018). Droplet digital PCR-based detection of clarithromycin resistance in Helicobacter pylori isolates reveals frequent heteroresistance. J. Clin. Microbiol. 56 (9), e00019–18. 10.1128/JCM.00019-18 PubMed Abstract | 10.1128/JCM.00019-18 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Wang Y., Zhang L., Liu S., Zhang M., Wang J., et al. (2020). CRISPR-Cas9 triggered two-step isothermal amplification method for E. coli O157:H7 detection based on a metal-organic framework platform. Anal. Chem. 92 (4), 3032–3041. 10.1021/acs.analchem.9b04162 PubMed Abstract | 10.1021/acs.analchem.9b04162 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Thore A., Nilsson L., Hojer H., Ansehn S., Brote L. (1977). Effects of ampicillin on intracellular levels of adenosine triphosphate in bacterial cultures related to antibiotic susceptibility. Acta Pathol. Microbiol. Scand. B 85 (2), 161–166. 10.1111/j.1699-0463.1977.tb01690.x PubMed Abstract | 10.1111/j.1699-0463.1977.tb01690.x | Google Scholar [DOI] [PubMed] [Google Scholar]
- Tiri B., Sensi E., Marsiliani V., Cantarini M., Priante G., Vernelli C., et al. (2020). Antimicrobial stewardship program, COVID-19, and infection control: Spread of carbapenem-resistant Klebsiella pneumoniae colonization in ICU COVID-19 patients. What did not work? J. Clin. Med. 9 (9), E2744. 10.3390/jcm9092744 PubMed Abstract | 10.3390/jcm9092744 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomczyk S., Taylor A., Brown A., de Kraker M. E. A., El-Saed A., Alshamrani M., et al. (2021). Impact of the COVID-19 pandemic on the surveillance, prevention and control of antimicrobial resistance: a global survey. J. Antimicrob. Chemother. 76 (11), 3045–3058. 10.1093/jac/dkab300 PubMed Abstract | 10.1093/jac/dkab300 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaloglou M. N., Nemiroski A., Camci-Unal G., Christodouleas D. C., Murray L. P., Connelly J. T., et al. (2018). Handheld isothermal amplification and electrochemical detection of DNA in resource-limited settings. Anal. Biochem. 543, 116–121. 10.1016/j.ab.2017.11.025 PubMed Abstract | 10.1016/j.ab.2017.11.025 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Urmi U. L., Nahar S., Rana M., Sultana F., Jahan N., Hossain B., et al. (2020). Genotypic to phenotypic resistance discrepancies identified involving beta-lactamase genes, blaKPC, blaIMP, blaNDM-1, and blaVIM in uropathogenic Klebsiella pneumoniae . Infect. Drug Resist. 13, 2863–2875. 10.2147/IDR.S262493 PubMed Abstract | 10.2147/IDR.S262493 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Votintseva A. A., Bradley P., Pankhurst L., Del Ojo Elias C., Loose M., Nilgiriwala K., et al. (2017). Same-day diagnostic and surveillance data for tuberculosis via whole-genome sequencing of direct respiratory samples. J. Clin. Microbiol. 55 (5), 1285–1298. 10.1128/JCM.02483-16 PubMed Abstract | 10.1128/JCM.02483-16 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Biswas S., Paudyal N., Pan H., Li X., Fang W., et al. (2019). Antibiotic resistance in Salmonella typhimurium isolates recovered from the food chain through national antimicrobial resistance monitoring system between 1996 and 2016. Front. Microbiol. 10, 985. 10.3389/fmicb.2019.00985 PubMed Abstract | 10.3389/fmicb.2019.00985 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waseem H., Jameel S., Ali J., Saleem Ur Rehman H., Tauseef I., Farooq U., et al. (2019). Contributions and challenges of high throughput qPCR for determining antimicrobial resistance in the environment: A critical review. Molecules 24 (1), E163. 10.3390/molecules24010163 PubMed Abstract | 10.3390/molecules24010163 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler N. E., Gardner P. P., Barquist L. (2018). Machine learning identifies signatures of host adaptation in the bacterial pathogen Salmonella enterica . PLoS Genet. 14 (5), e1007333. 10.1371/journal.pgen.1007333 PubMed Abstract | 10.1371/journal.pgen.1007333 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B., Ed-Dra A., Pan H., Dong C., Jia C., Yue M. (2021). Genomic investigation of Salmonella isolates recovered from a pig slaughtering process in hangzhou, China. Front. Microbiol. 12, 704636. 10.3389/fmicb.2021.704636 PubMed Abstract | 10.3389/fmicb.2021.704636 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao G., Zhang S., Liang Z., Li G., Fang M., Liu Y., et al. (2020). Identification of Mycobacterium abscessus species and subspecies using the Cas12a/sgRNA-based nucleic acid detection platform. Eur. J. Clin. Microbiol. Infect. Dis. 39 (3), 551–558. 10.1007/s10096-019-03757-y PubMed Abstract | 10.1007/s10096-019-03757-y | Google Scholar [DOI] [PubMed] [Google Scholar]
- Xu Y., Zhou X., Jiang Z., Qi Y., Ed-Dra A., Yue M. (2021). Antimicrobial resistance profiles and genetic typing of Salmonella serovars from chicken embryos in China. Antibiot. (Basel) 10 (10), 1156. 10.3390/antibiotics10101156 10.3390/antibiotics10101156 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee R., Dien Bard J., Simner P. J. (2021). The genotype-to-phenotype dilemma: How should laboratories approach discordant susceptibility results? J. Clin. Microbiol. 59 (6), e00138–20. 10.1128/JCM.00138-20 PubMed Abstract | 10.1128/JCM.00138-20 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E. C., Fu C. C., Hu L., Thakur R., Feng J., Lee L. P. (2017). Self-powered integrated microfluidic point-of-care low-cost enabling (SIMPLE) chip. Sci. Adv. 3 (3), e1501645. ARTN e150164510. 10.1126/sciadv.1501645 PubMed Abstract | 10.1126/sciadv.1501645 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue M., Bai L., Song H., Fang W.(2021) Impacts of microbial food safety in China and beyond. Foodborne Pathog. Dis. 18 (8), 508–509. 10.1089/fpd.2021.29015.int PubMed Abstract | 10.1089/fpd.2021.29015.int | Google Scholar [DOI] [PubMed] [Google Scholar]
- Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67 (11), 2640–2644. 10.1093/jac/dks261 PubMed Abstract | 10.1093/jac/dks261 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]