Abstract
Chirality is essential in nearly all biological organizations and chemical reactions but is rarely considered due to technical limitations in identifying L/D isomerization. Using OmpF, a membrane channel from Escherichia coli with an electrostatically asymmetric constriction zone, allows discriminating chiral amino acids in a single peptide. The heterogeneous distribution of charged residues in OmpF causes a strong lateral electrostatic field at the constriction. This laterally asymmetric constriction zone forces the sidechains of the peptides to specific orientations within OmpF, causing distinct ionic current fluctuations. Using statistical analysis of the respective ionic current variations allows distinguishing the presence and position of a single amino acid with different chiralities. To explore potential applications, the disease-related peptide β-Amyloid and its d-Asp1 isoform and a mixture of the icatibant peptide drug (HOE 140) and its d-Ser7 mutant have been discriminated. Both chiral isomers were not applicable to be distinguished by mass spectroscopy approaches. These findings highlight a novel sensing mechanism for identifying single amino acids in single peptides and even for achieving single-molecule protein sequencing.
Introduction
Amino acid isomerization can be induced by post-translational modifications or asymmetric synthesis.1−5 Identifying these isomers in a peptide at a single-molecule level is highly desirable in early disease diagnosis and chiral impurity analysis but challenging due to technical limitations. Mass spectrometry (MS)6 approaches are not easy due to identical mass between chiral isomers. While liquid chromatography-tandem MS is available, only a few chirality-containing peptides are determined based on limited l-/d-type amino acid enzymatic cleavage positional mismatch. Alternatively, circular dichroism is a method based on the differential absorption of circularly polarized light and allows the identification of chiral isomers. This approach could be further improved toward the single-molecule level, based on confining the polarized light into the single-molecule scale.7 However, developing an accessible method for identifying peptides containing chiral amino acids remains cumbersome. The chirality-containing peptides break the sidechain zig-zag alignment,8 which usually lacks the chiral center of the whole molecule. Therefore, the circular dichroism-based approaches are ineffective in identifying such stereo distortion. Thus, no single-molecule technique is available that easily distinguishes chiral amino acids and even positional isomers in single peptides. This lack of an easy method is likely a bottleneck for the discovery of more biological and chemical processes.
Protein nanopores have already been utilized to identify single amino acids9−12 and protein–protein interactions13 by amperometric recognition. Peptides with post-translational modifications are recognized by nanopore-based approaches, to the extent that the sensing principle relies on volume exclusion.9,14 Moreover, the inter-molecule interaction between the target peptides and the nanopore interface can lead to characteristic ionic current patterns for discriminating amino acids with similar volumes.15,16 The nanopore-based single molecule approach has demonstrated the possibility of sensing chiral molecules by designing a specific chiral environment, but the generalization of such a strategy toward peptide-containing chiral amino acid is lacking.17−19 Previous nanopore-based tools remain insufficient in identifying amino acids in peptides with identical volumes. The only difference between chiral isomers is the opposite orientation of the sidechains.
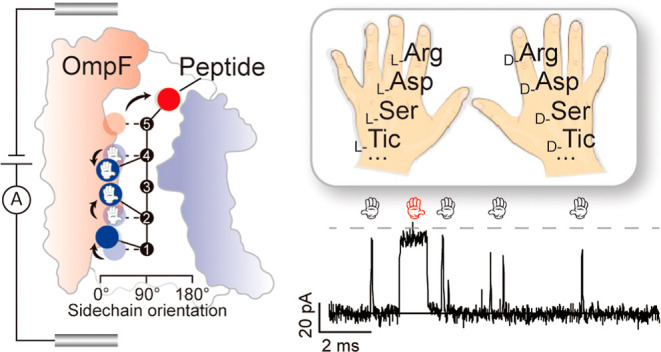
To identify the sidechain orientation, herein, we use OmpF, a bacterial outer-membrane channel, as a chiral biosensor. The crystal structure of OmpF shows that the narrowest size of the constriction zone (CZ) is about 0.7 nm, one of the smallest CZs reported20 among the silent protein channels. The size is close to that of a single amino acid and promises to serve an enhanced stereo confinement.21 Notably, the CZ is composed of a positively charged ladder (K16, R42, R82, and R132) and a negatively charged pocket (D113, E117, and D121) on opposite sides of the CZ (Figure 1a), which creates a lateral electrostatic field (Figures 1b and S1). Due to this electrostatic asymmetry in the CZ, the chirality directs the sidechain orientation during the pore–analyte interaction, which in turn determines the characteristic single-molecule current pattern used for identification purposes. The specific amplitudes of the current blockades and the respective residence time allow us to identify chiral and positional isomers. As examples for potential applications, we identified d-Asp in disease-related β-Amyloid mutants and an icatibant drug (HOE140) mixture with one of the impurities consisting of d-Ser.
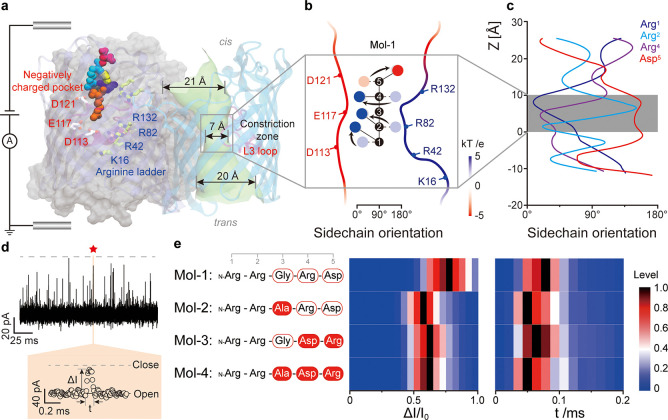
Figure 1.
(a) Depiction of an OmpF trimeric protein sensing a single peptide under the influence of a bias voltage. The asymmetrically distributed charged amino acids across the pore are highlighted in stick presentation and include a prototype peptide n-Arg-Arg-Gly-Arg-Asp (Mol-1) in Van der Waals representation. The monomer on the right highlights the heterogeneous inner tunnel surface with loop L3 highlighted in magenta. (b) Schematic representation of the zig-zag alignment of each amino acid sidechain of Mol-1 in bulk (undertint color), together with arrows representing their reorientation, upon entering OmpF (dark color) during the simulations. For the calculation of the sidechain orientation, the angle was averaged along the channel axis from 0 to 10 Å, accordingly (see Figure S6a). Blue spheres represent arginine sidechains, and red sphere represents the aspartic acid sidechain. The backbone numbering denotes the position of the respective amino acid, starting from the N-terminus. The profile of the electrostatic potential is depicted in Figure S1. (c) Each sidechain orientation of Mol-1, determined quantitatively. An angle of 0° indicates that the sidechain points toward the acidic region, while it is oriented toward the basic ladder at 180°. (d) Typical nanopore-based amperometric readout of Mol-1. The colored panel shows a zoomed-in single-molecule event induced by Mol-1 interaction with OmpF. The current drop ΔI/I0 and residence time t are the two parameters being analyzed. The red star denotes the enlarged events (orange panel) in the original trace. (e) The peptide sequences of Mol-1, 2, 3, and 4 are shown on the left. For the results on the right, a final concentration of 7.6 μM was used for Mol-1 and 3 and 3.7 μM for Mol-2 and 4. The color histograms depict the current drop ΔI/I0 and residence time t measured at a bias voltage of −50 mV in 1.0 M KCl, 10 mM Tris, and 1 mM EDTA at pH 8.0. At least 1000 single-molecule events have been analyzed per peptide. Moreover, the peak values of the distributions have been normalized to one with this maximum value shown in black color. More detailed histograms are delineated in Figure S4.
Results and Discussion
Previous studies revealed that OmpF has a cationic selectivity and showed strong interaction with Arginine at negative bias voltage.23 For the proof-of-concept demonstration, a cationic model peptide N-Arg-Arg-Gly-Arg-Asp (Mol-1) has been designed.22 The Arginine placed at the N-terminus is supposed to head the peptide into OmpF. Further molecular dynamics (MD) simulations support that the positively charged N-terminus is more preferable for the entrance than the negatively charged C-terminus (cf. MD simulations in Figure S2). In the first set of experiments, we added Mol-1 and performed statistical analysis of the current blockades (Figure 1d) at a bias voltage of −50 mV. Note that OmpF rarely produces gating events between −50 to −75 mV (Figure S3). As summarized in Table S1, Gaussian fits to the Mol-1 blockade level (ΔI/I0) gave 65.9 ± 8.7% and a residence time of 47.8 ± 1.2 μs (Figure S4). Both distributions have been plotted in the color map, as shown in Figure 1e. We also conducted steered MD simulations for Mol-1 and analyzed the dynamics of the sidechain orientation of each peptide residue along the pore axis (Figure 1b). The overall salt-bridge network has also been calculated between the peptide and key residues in OmpF (Figure S5). When Mol-1 enters OmpF reaching the CZ, all arginine sidechains are oriented toward the negatively charged pocket, and the aspartic acid sidechain faces toward the positively charged ladder (Figure 1c). Arg2 needs to reorient from the opposite orientation of ca. 135° against the backbone stiffness toward this region. In contrast, Arg4 is more flexible to orient from 100° since the preceding Gly3 has no sidechain. For detailed in silico analysis, see also Supporting Note 1. To this end, we have concluded that each sidechain reorients in response to the lateral electrostatic field in the CZ.
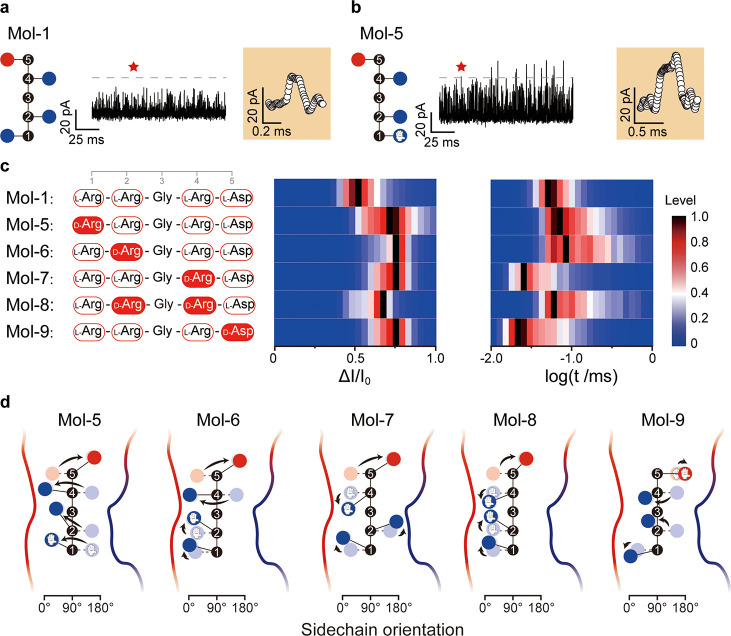
The second series of experiments were devoted to elucidating what degree the backbone stiffness prohibits the sidechain orientation. The peptide N-Arg-Arg-Ala-Arg-Asp (Mol-2) was designed by substituting Gly3 with Ala3, to manipulate the stiffness of the peptide backbone by adding a methyl group. Performing single-molecule analysis, Mol-2 yielded ΔI/I0 decreased to 61.1 ± 0.9% and the residence time stayed almost the same, 46.8 ± 0.7 μs. The salt-bridge created between the C-terminus of Mol-2 and the OmpF vestibule gets stronger than that of Mol-1 (Figure S5). Arg4 orients from approximately 50 to 90°, toward the arginine ladder (Figure S6). To show the effect of the sidechain orientation, the positions Asp5 and Arg4 in the abovementioned peptides have been switched to invert the sidechain orientations, maintaining the zig-zag form. Thus, two peptides have been synthesized, that is, N-Arg-Arg-Gly-Asp-Arg (Mol-3) and N-Arg-Arg-Ala-Asp-Arg (Mol-4). The ΔI/I0 for Mol-3 was 59.4 ± 1.6%, and the t value was 47.2 ± 0.3 μs. However, for Mol-4, we obtained 60.6 ± 0.3% for ΔI/I0 and a t value of 47.6 ± 0.2 μs. Therefore, switching the positions of Arg4 and Asp5 at the C-terminus induces sidechain orientations inversely aligned, compared to Mol-1 and Mol-2, decreasing the ΔI/I0. Overall, by analyzing the current blockades, Mol-1 has the best sidechain, matching the lateral electrostatic field in the CZ of OmpF. The electrostatic force at the CZ is enhanced in lower electrolyte concentration.24 In this work, we decreased the electrolyte concentration from 1.0 to 0.5 M and the t for Mol-1 rose by 38% (Figure 2a). Applying the picture of sidechains reorientation in the asymmetry CZ, we designed chirality-containing peptides and positional isomers (Mol-5 to 9). Placing D-Arg at the 1st position (Mol-5), the residence time t increases to 112.3 ± 33.0 μs (Figures 2b,c and S6). Placing D-Arg at the 2nd position (Mol-6), t increased to 137.9 ± 37.0 μs. The orientation of each sidechain in the CZ is depicted schematically in Figure 2d, in accordance with the dynamics (Figure S8). For both Mol-5 and Mol-6, the calculated salt-bridge interactions got stronger than those for Mol-1, leading to longer t (Figure S9).
Figure 2.
(a,b) Typical nanopore-based amperometric readouts for Mol-1 and Mol-5, using 0.5 M KCl. Red star points denote enlarged events (orange panel) in the original trace. (c) The peptide sequence of Mol-1, together with that of Mol-5, 6, 7, 8, and 9, contain at least one d-amino acid. Final concentrations of 1.5 μM for Mol-1, 7.6 μM for Mol-5, 6, 7, and 8, and 0.8 μM for Mol-9 have been reached in the experiments. The color histograms show the current drop ΔI/I0 and the logarithm of residence time t, measured at a bias voltage of −50 mV in 0.5 M KCl, 10 mM Tris, and 1 mM EDTA at pH 8.0. Moreover, the peak values of the distributions have been normalized to one with this maximum value shown in black. More detailed histograms are delineated in Figure S7. (d) Representation of Mol-5, 6, 7, 8, and 9 containing chiral amino acid reorientation in OmpF. The orientation of each sidechain has been determined from the results of the simulated dynamics delineated in Figure S8, averaged along the channel axis from 0 to 10 Å.
Placing d-Arg at the 4th position (Mol-7) yields a ΔI/I0 of 76.9 ± 3.1% and t of 64.9 ± 0.3 μs. Since the d-Arg4 and l-Asp5 sidechains face toward the same side in bulk, upon entering the CZ of OmpF, the lateral electrostatic field tears them apart toward the opposite channel walls, against the stiffness of the peptide backbone. The analysis of the sidechain orientation dynamics revealed that Arg2 fails to orient toward the negatively charged pocket (Figures 2d and S8). As this conformation is energetically less favorable, Mol-7 resides in the CZ for a shorter time than Mol-1. Considering that Mol-8 contains both D-Arg2 and D-Arg4 (Figure 1d), the measurements resulted in a ΔI/I0 of 73.7 ± 9.1% and t of 104.9 ± 23.0 μs. As expected, the sidechain of D-Arg2 orients from 135 to 10°, while the D-Arg4 and L-Asp5 sidechains remain in the same orientation as in Mol-7 (Figures 2d and S8). For D-Asp5 at the C-terminus (Mol-9), the ionic current ratio ΔI/I0 decreased to 73.7 ± 3.6% and the residence time t to 61.3 ± 0.1 μs. This relatively short t can be attributed to L-Arg4D-Asp5 that requires additional energy to orient them differently against the backbone stiffness, similar to the case of D-Arg4L-Asp5 (Mol-7 and Mol-8).
In short, the measurements of the residence times and reductions in the ionic flux revealed a sensitivity of both parameters for d-amino-acid-containing peptides. Ionic current recordings, using a single OmpF protein, can detect the presence of chiral amino acids including positional isomers. The presence of D-Arg at various positions in the model peptides Mol-5 to Mol-7 caused variations in ionic current blockages and residence times. Note that each monomer of the trimeric OmpF pore is assembled from a single polypeptide, creating a heterogeneous sensing environment. The lateral electrostatic field at the CZ enforces the reorientation of the peptide sidechains. As a control, we used an OmpF triple mutant (R42E, R82E, and R132E) which eliminates the strong asymmetric lateral charge distribution at the CZ. This OmpF triple mutant did not allow for chiral discrimination, which further proves the importance of the lateral electrostatic field (Figure S10). This is markedly different from the commonly used pore-forming toxins which are self-assembled from repeats of monomers. Wild type aerolysin, as an example, is not capable of detecting if a peptide contains chiral isomers (Figure S11).
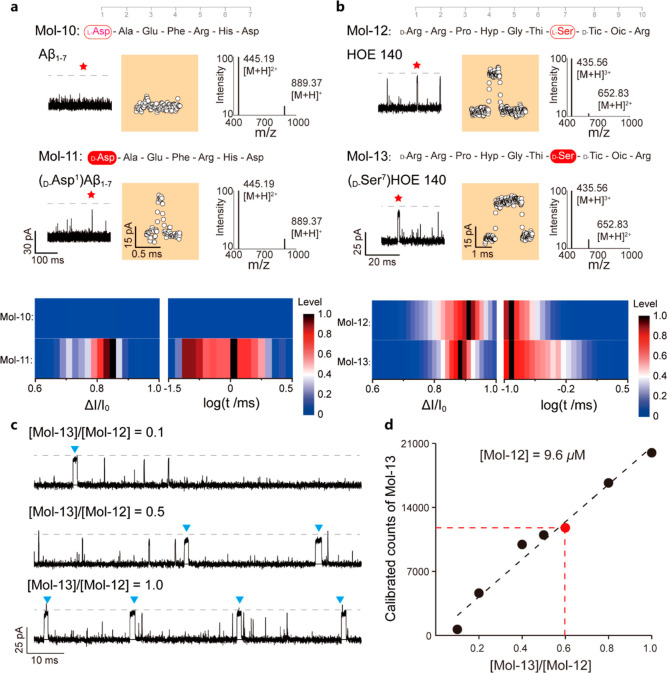
As a possible application, we characterized disease- and drug-relevant peptide chirality-containing isomers. The Amyloid β peptide (Aβ) is a crucial indicator for the progression of Alzheimer’s disease. Its D-Asp1 mutation causes unusual aggregates in vivo.25,26 To measure this critical segment of β-Amyloid, N-Asp-Ala-Glu-Phe-Arg-His-Asp (Mol-10) and its isomer D-Asp1 (Mol-11), OmpF-based nanopore assay was performed. Contrary to Mol-10 hardly interacting with OmpF, Mol-11 interacts with the OmpF nanopore, with typical current blockades being counted (Figure 3a). The ratio ΔI/I0 from Mol-11 has a value of 82 ± 9%, and t is 219 ± 89 μs. We ascribe this finding to the fact that the sidechain of Arg5 faces toward the negatively charged pocket in the presence of D-Asp1, altering the orientation of the subsequent latter sidechains (Figure S12). A varying concentration of Mol-11 in the fixed high interference concentration of Mol-10 of 56.2 μM shows that event counts of the D-Asp1 mutant Mol-11 can be linearly fitted to the increasing concentration ratio of [Mol-11]/[Mol-10] (Figure S13a). Therefore, OmpF could achieve identification of the rare species of the chirality mutant from the mixture. Moreover, our results demonstrate that the OmpF report of Mol-11 from the mixture is independent of the Mol-10 concentration (Figure S13b), and the events from Mol-11 are distinguishable from Mol-10. The OmpF identification of the D-Asp1 isoform of β-Amyloid (Mol-11) from the WT β-Amyloid (Mol-10) would facilitate the qualitative evaluation of the isomers in pathogenesis and disease progression.
Figure 3.
(a) Current tracing of the β-Amyloid (1–7) (Mol-10) and its D-Asp1 mutant (Mol-11) monitored using the OmpF-based nanopore assay at a bias voltage of −75 mV. The colored panel depicts a zoomed-in single-molecule event. MS characterization of both Mol-10 and Mol-11 is plotted aside, with molecular ion peaks overlapped. Bottom panel: electrochemical characterization of the Mol-10 and 11interaction with OmpF, showing the current drop and logarithm of the residence time. No interaction of Mol-10 with OmpF has been observed. A final concentration of 56.2 μM has been reached for Mol-10 and 11. (b) Current traces of HOE140 (Mol-12) and the D-Ser7 impurity (Mol-13) were measured using a single OmpF nanopore with a voltage of −50 mV. Zoomed-in color panels represent star-highlighted single molecule event. MS characterization of both Mol-12 and Mol-13 is plotted at the right side. Identical molecular ion peaks were measured. Bottom panel: Electrochemical characterizations of Mol-12 and Mol-13 with unique signature pattern interactions with OmpF. Among the experiments, a final concentration of 19.2 μM for Mol-12 and 9.6 μM for Mol-13 was reached in 0.5 M KCl, 10 mM Tris, and 1 mM EDTA, at pH 8.0. At least 1000 single-molecule events have been analyzed. The red star denotes enlarged events (orange panel) in the original trace. (c) Current traces of the mixture of HOE140 and its D-Ser7 impurity at different molar ratios, whose scattering is plotted in Figure S15. The blue triangles indicate the current pattern for detection of a translocation event. (d) Calibrated counts of Mol-13 against the molar ratio to Mol-12 in the mixture. The concentration of Mol-12 is 9.6 μM. For the detailed calibration process, please refer to Figure S16 and Supporting Note 5.
A second peptide, the so-called icatibant peptide drug HOE140, is a highly selective antagonist at the bradykinin B2 receptor.27 The sequence of HOE140 is N-D-Arg-Arg-Pro-Hyp-Gly-Thi-Ser-D-Tic-Oic-Arg (Mol-12), containing chiral amino acids. Early studies hinted at the fact that the peptide containing D-Ser7 isomer (Mol-13) is the most difficult to be probed among all the impurities, when using GC chromatography.27 Both the drug and its isomer were measured using the present OmpF protocol, and generated specific current distribution patterns of current blockades, as shown in Figure 3b–d. Characterizing the blockade events for Mol-12, we got 89 ± 17% for ΔI/I0 and 89 ± 19 μs for t. The presence of Mol-13 yielded a ΔI/I0 of 80 ± 11% and t value of 62 ± 28 μs. Comparing Mol-12 and Mol-13, the D-Asp1L-Asp2 combination of Mol-12 at the N-terminus of the sequence nicely matches the electrostatic distribution in the CZ, similar to Mol-5, while the inverted orientation at D-Ser7 in Mol-13 leads to a decrease of the residence time of about 50%, compared to Mol-12. Additionally, an all L-type variant of HOE140 (Mol-14, Figure S14) was synthesized and sensed with OmpF to further prove the mechanism of the sidechain orientation in the CZ.
Finally, a mixture of Mol-12 and Mol-13 has been measured using OmpF with various molar ratios (Figure 3c,d). The distinct single molecule distribution pattern could be used for quantitatively and qualitatively determining the impurity of Mol-13 (Figures 3d and S15). The linear relationship between calibrated counts of Mol-13 and the concentration ratio of Mol-13 to Mol-12 is independent of the concentration of Mol-12 (Figures 3d and S17). It is noteworthy that by performing MS characterization (cf. Supporting Note 4), neither Mol-10 and Mol-11 nor Mol-12 and Mol-13 were identifiable (Figure 3a,b), suggesting the limitation in identifying the stereochemistry of amino acids, while the nanopore-based approach presented here is more applicable. Therefore, OmpF allows discrimination of chiral impurities from the icatibant drug, providing a single-molecule method for quality control of the peptide drugs.
Conclusions
In summary, the lateral electrostatic field in the CZ of OmpF leads to the sidechain reorientation during peptide translocation. The resulting changes in the current drop and residence times allow identification of peptides containing chiral amino acids and positional isomer at the single-molecule level. As for potential detection examples, we measured disease-related β-Amyloid and HOE140 impurities containing chiral amino acids. All peptides have been probed using OmpF based on electric recognition. One of the major concerns of OmpF-sensing peptides is the short residence time. Many speed-controlled approaches can potentially be utilized to boost the peptide sensing capability through nanopores, for example, forming peptide-DNA conjugation for enzyme-directed amino acid stepwise scanning.10,28,29 Moreover, we found strong indication that the chirality of the polar sidechain from serine (Mol-13) creates additional interactions with the residues in the CZ of OmpF (e.g., Thr112,122 and Asn101), thus from serine (Mol-13) creates additional interactions with the residues in the CZ of OmpF (e.g., Thr112,122 and Asn101), thus increasing the residence time to hundreds of milliseconds. This finding might further explain the possible discrimination of the presence of D-Ser and L-Tic containing polar and hydrophobic sidechains, respectively. In conclusion, using nanopores with the lateral electrostatic field can control the sidechain orientation. This can be employed to detect properties that cannot be distinguished using nanopores to preserve the symmetry. Similarly, a lateral polar or hydrophobic environment in the CZ might allow for identification of sidechain polarities or hydrophobicity properties. According to the structure and functions of each residue at the CZ,30,31 the enhanced laterally asymmetric effect can be achieved by introducing critical sensing residues (e.g., containing polar sidechains: Ser and Gln and hydrophobic sidechains: Phe and Trp) on the CZ. Lastly, an asymmetric CZ allows for high sensitivity to the sidechain orientation, bringing a new mechanism into proteomic studies. Along with the small confining size of the CZ in OmpF, which is compatible with the volume of 1–2 amino acids, asymmetries in the CZ provide additional opportunities for nanopore sensing in single-molecule proteomic studies which have not been explored so far.
Acknowledgments
Dr. Xin Hua from Southeast University is acknowledged for fruitful discussion on mass spectrometry experiments. Junge Li from Nanjing University is thanked for mutant protein preparation.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.2c03923.
Experimental section, simulation details, characterizations, and nanopore data analysis results (PDF)
Author Present Address
⊥ Currently employed at Los Alamos National Laboratory, Los Alamos, NM 87544, United States
Author Contributions
# J.W., J.D.P., and F.G. contributed equally.
This research was supported by the National Natural Science Foundation of China (21922405, 22027806, 61901171, and 22090050), Sino-German Mobility Programme (M-0143), and programs for high-level entrepreneurial and innovative talents introduction of Jiangsu Province. Y-L. Y is sponsored by National Ten Thousand Talent Program for young top-notch talent.
The authors declare no competing financial interest.
Supplementary Material
References
- Kuzyk A.; Schreiber R.; Fan Z.; Pardatscher G.; Roller E.-M.; Högele A.; Simmel F. C.; Govorov A. O.; Liedl T. DNA-based self-assembly of chiral plasmonic nanostructures with tailored optical response. Nature 2012, 483, 311–314. 10.1038/nature10889. [DOI] [PubMed] [Google Scholar]
- Mukherjee S.; Yang J. W.; Hoffmann S.; List B. Asymmetric enamine catalysis. Chem. Rev. 2007, 107, 5471–5569. 10.1021/cr0684016. [DOI] [PubMed] [Google Scholar]
- Tang W. J.; Zhang X. M. New chiral phosphorus ligands for enantioselective hydrogenation. Chem. Rev. 2003, 103, 3029–3070. 10.1021/cr020049i. [DOI] [PubMed] [Google Scholar]
- Martínez-Rodríguez S.; Martínez-Gómez A. I.; Rodríguez-Vico F.; Clemente-Jiménez J. M.; Las Heras-Vázquez F. J. Natural Occurrence and industrial applications of d-amino acids: an overview. Chem. Biodiversity 2010, 7, 1531–1548. 10.1002/cbdv.200900245. [DOI] [PubMed] [Google Scholar]
- Zhong C.; Zhu N.; Zhu Y.; Liu T.; Gou S.; Xie J.; Yao J.; Ni J. Antimicrobial peptides conjugated with fatty acids on the side chain of D-amino acid promises antimicrobial potency against multidrug-resistant bacteria. Eur. J. Pharm. Sci. 2020, 141, 105123. 10.1016/j.ejps.2019.105123. [DOI] [PubMed] [Google Scholar]
- Domingos S. R.; Pérez C.; Schnell M. Sensing chirality with rotational spectroscopy. Annu. Rev. Phys. Chem. 2018, 69, 499–519. 10.1146/annurev-physchem-052516-050629. [DOI] [PubMed] [Google Scholar]
- Solomon M. L.; Saleh A. A. E.; Poulikakos L. V.; Abendroth J. M.; Tadesse L. F.; Dionne J. A. Nanophotonic platforms for chiral sensing and separation. Acc. Chem. Res. 2020, 53, 588–598. 10.1021/acs.accounts.9b00460. [DOI] [PubMed] [Google Scholar]
- Berkholz D. S.; Driggers C. M.; Shapovalov M. V.; Dunbrack R. L. Jr.; Karplus P. A. Nonplanar peptide bonds in proteins are common and conserved but not biased toward active sites. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 449–453. 10.1073/pnas.1107115108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouldali H.; Sarthak K.; Ensslen T.; Piguet F.; Manivet P.; Pelta J.; Behrends J. C.; Aksimentiev A.; Oukhaled A. Electrical recognition of the twenty proteinogenic amino acids using an aerolysin nanopore. Nat. Biotechnol. 2020, 38, 176–181. 10.1038/s41587-019-0345-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkerhoff H.; Kang A. S. W.; Liu J.; Aksimentiev A.; Dekker C. Multiple rereads of single proteins at single-amino acid resolution using nanopores. Science 2021, 374, 1509–1513. 10.1126/science.abl4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z.-L.; Huo M.-Z.; Ying Y.-L.; Long Y.-T. Biological nanopore approach for single-molecule protein sequencing. Angew. Chem., Int. Ed. 2021, 60, 14738–14749. 10.1002/anie.202013462. [DOI] [PubMed] [Google Scholar]
- Chen X.; Zhang Y.; Arora P.; Guan X. Nanopore Stochastic Sensing Based on Non-covalent Interactions. Anal. Chem. 2021, 93, 10974–10981. 10.1021/acs.analchem.1c02102. [DOI] [PubMed] [Google Scholar]
- Thakur A. K.; Movileanu L. Real-time measurement of protein-protein interactions at single-molecule resolution using a biological nanopore. Nat. Biotechnol. 2019, 37, 96–101. 10.1038/nbt.4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas F. L. R.; Sarthak K.; Lenting E. M.; Coltan D.; van der Heide N. J.; Versloot R. C. A.; Aksimentiev A.; Maglia G. The manipulation of the internal hydrophobicity of FraC nanopores augments peptide capture and recognition. ACS Nano 2021, 15, 9600–9613. 10.1021/acsnano.0c09958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.-Y.; Ying Y.-L.; Yu J.; Liu S.-C.; Wang Y.-Q.; Li S.; Long Y.-T. Revisiting the origin of nanopore current blockage for volume difference sensing at the atomic level. JACS Au 2021, 1, 967–976. 10.1021/jacsau.1c00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cressiot B.; Bacri L.; Pelta J. The Promise of Nanopore Technology: Advances in the Discrimination of Protein Sequences and Chemical Modifications. Small Methods 2020, 4, 2000090. 10.1002/smtd.202000090. [DOI] [Google Scholar]
- Boersma A. J.; Bayley H. Continuous Stochastic Detection of Amino Acid Enantiomers with a Protein Nanopore. Angew. Chem., Int. Ed. 2012, 51, 9606–9609. 10.1002/anie.201205687. [DOI] [PubMed] [Google Scholar]
- Kang X.-F.; Cheley S.; Guan X.; Bayley H. Stochastic Detection of Enantiomers. J. Am. Chem. Soc. 2006, 128, 10684–10685. 10.1021/ja063485l. [DOI] [PubMed] [Google Scholar]
- Cooper J. A.; Borsley S.; Lusby P. J.; Cockroft S. L. Discrimination of supramolecular chirality using a protein nanopore. Chem. Sci. 2017, 8, 5005–5009. 10.1039/c7sc01940h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan S. W.; Schirmer T.; Rummel G.; Steiert M.; Ghosh R.; Pauptit R. A.; Jansonius J. N.; Rosenbusch J. P. Crystal structures explain functional-properties of 2 Escherichia coli porins. Nature 1992, 358, 727–733. 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- Mitscha-Baude G.; Stadlbauer B.; Howorka S.; Heitzinger C. Protein transport through nanopores illuminated by long-time-scale simulations. ACS Nano 2021, 15, 9900–9912. 10.1021/acsnano.1c01078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Prajapati J. D.; Kleinekathöfer U.; Winterhalter M. Dynamic interaction of fluoroquinolones with magnesium ions monitored using bacterial outer membrane nanopores. Chem. Sci. 2020, 11, 10344–10353. 10.1039/d0sc03486j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Bafna J.; Bhamidimarri S.-P.; Winterhalter M. Small molecule permeation across membrane channels: Chemical modification to quantify transport across OmpF. Angew. Chem., Int. Ed. 2019, 58, 4737–4741. 10.1002/anie.201814489. [DOI] [PubMed] [Google Scholar]
- Qiu Y.; Lin C. Y.; Hinkle P.; Plett T. S.; Yang C.; Chacko J. V.; Digman M. A.; Yeh L. H.; Hsu J. P.; Siwy Z. S. Highly charged particles cause a larger current blockage in micropores compared to neutral particles. ACS Nano 2016, 10, 8413–8422. 10.1021/acsnano.6b03280. [DOI] [PubMed] [Google Scholar]
- Sakai-Kato K.; Naito M.; Utsunomiya-Tate N. Racemization of the amyloidal beta Asp1 residue blocks the acceleration of fibril formation caused by racemization of the Asp23 residue. Biochem. Biophys. Res. Commun. 2007, 364, 464–469. 10.1016/j.bbrc.2007.10.014. [DOI] [PubMed] [Google Scholar]
- Roher A. E.; Lowenson J. D.; Clarke S.; Wolkow C.; Wang R.; Cotter R. J.; Reardon I. M.; Zürcher-Neely H. A.; Heinrikson R. L.; Ball M. J.; et al. Structural alterations in the peptide backbone of beta-amyloid core protein may account for its deposition and stability in Alzheimer’s disease. J. Biol. Chem. 1993, 268, 3072–3083. 10.1016/s0021-9258(18)53661-9. [DOI] [PubMed] [Google Scholar]
- Ermer J.; Gerhardt J.; Siewert M. Quality control of peptide drugs. Chiral amino acid analysis versus standard for icatibant acetate. Arch. Pharm. 1995, 328, 635–639. 10.1002/ardp.19953280902. [DOI] [PubMed] [Google Scholar]
- Chen Z.; Wang Z.; Xu Y.; Zhang X.; Tian B.; Bai J. Controlled movement of ssDNA conjugated peptide through Mycobacterium smegmatis porin A (MspA) nanopore by a helicase motor for peptide sequencing application. Chem. Sci. 2021, 12, 15750–15756. 10.1039/d1sc04342k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S.; Zhang J.; Wang Y.; Guo W.; Zhang S.; Liu Y.; Cao J.; Wang Y.; Wang L.; Ma F.; Zhang P.; Chen H.-Y.; Huang S. Single molecule ratcheting motion of peptides in a Mycobacterium smegmatis porin A (MspA) Nanopore. Nano Lett. 2021, 21, 6703–6710. 10.1021/acs.nanolett.1c02371. [DOI] [PubMed] [Google Scholar]
- Karshikoff A.; Spassov V.; Cowan S. W.; Ladenstein R.; Schirmer T. Electrostatic properties of two porin channels from Escherichia coli. J. Mol. Biol. 1994, 240, 372–384. 10.1006/jmbi.1994.1451. [DOI] [PubMed] [Google Scholar]
- Lou K. L.; Saint N.; Prilipov A.; Rummel G.; Benson S. A.; Rosenbusch J. P.; Schirmer T. Structural and functional characterization of OmpF porin mutants selected for larger pore size. I. Crystallographic analysis. J. Biol. Chem. 1996, 271, 20669–20675. 10.1074/jbc.271.34.20669. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.