Abstract
The double-stranded DNA bacteriophage PRD1 uses an IncP plasmid-encoded conjugal transfer complex as a receptor. Plasmid functions in the PRD1 life cycle are restricted to phage adsorption and DNA entry. A single phage structural protein, P2, located at the fivefold capsid vertices, is responsible for PRD1 attachment to its host. The purified recombinant adsorption protein was judged to be monomeric by gel filtration, rate zonal centrifugation, analytical ultracentrifugation, and chemical cross-linking. It binds to its receptor with an apparent Kd of 0.20 nM, and this binding prevents phage adsorption. P2-deficient particles are unstable and spontaneously release the DNA with concomitant formation of the tail-like structure originating from the phage membrane. We envisage the DNA to be packaged through one vertex, but the presence of P2 on the other vertices suggests a mechanism whereby the injection vertex is determined by P2 binding to the receptor.
Virus-cell interactions are specific. The first step in virus propagation is the attachment to the cell via recognition of and binding to the receptor structure. Virus receptors consist of cellular surface components that interact with the receptor binding protein, the virus-encoded counterpart of a receptor. This interaction then leads to the delivery of the viral nucleic acid into the cell cytoplasm. A substantial amount of information is available on the initial steps of bacteriophage infection of Escherichia coli T-like and filamentous phages. In tailed phages (like T1, T4, and T5), the tail proteins form a channel spanning either the outer membrane or all envelope layers of the gram-negative cell and this channel is used to inject the genome while the capsid is left outside (for a review, see reference 22). In filamentous phages (fd), the major capsid protein is inserted into the membrane, forming a pore (reviewed in reference 22). Enveloped bacteriophages with virion structure analogous to that of the enveloped viruses of higher organisms exhibit entry mechanisms different from that of the DNA phages, as the entire virion (or a substructure) is internalized by the process of membrane fusion (6, 44). However, the entry mechanism of bacteriophages with a membrane as an internal component of the virion most probably exhibits a novel system for the DNA delivery.
The broad-host-range double-stranded DNA (dsDNA) bacteriophage PRD1, capable of infecting a variety of gram-negative bacteria, is composed of an icosahedral outer protein capsid and an internal membrane (2, 7, 41). The membrane vesicle encloses the 14,925-bp-long linear dsDNA genome (9, 15). The phage genome contains inverted terminal repeats at both ends (48) and a covalently linked protein at each 5′ terminus (4, 5). The DNA is replicated by a phage-encoded polymerase via a protein-priming mechanism (16, 49). The protein coat is composed of the major capsid protein P3 organized on a pseudo T = 25 lattice (15). The fivefold vertices are built up by a spike structure comprised of three proteins, P31, P5, and P2 (45). There are a number of intriguing functional and structural similarities between PRD1 and adenovirus. These include genome organization and replication strategy, capsid organization, and major coat protein fold, as well as the structure and function of the fivefold receptor binding structure (10, 15, 45, 46, 50). In this context, it should be noted that phage PRD1 protein P2 is the structural equivalent of the adenovirus receptor binding spike knob (protein PIV distal domain) (52). For a detailed description of the PRD1 system, see the review by Bamford et al. (3).
PRD1 infects cells harboring a conjugative plasmid, such as IncP plasmid RP4 (43). Plasmid functions are only involved in host cell recognition and DNA penetration (21, 35). The plasmid encodes the phage receptor complex, a cell envelope structure bridging the inner and outer membranes (25). Receptor saturation experiments suggest that there are approximately 25 and 60 receptors per cell in E. coli and Salmonella typhimurium, respectively (20, 30). This IncP-encoded multiprotein structure functions also as a mating pair formation complex-DNA transfer apparatus in bacterial conjugation (32, 42, 55).
Isolation and analysis of phage nonsense mutants revealed that PRD1 adsorption is dependent on one phage-specific structural protein, P2 (36, 37). P2 is a minor protein, the copy number being about 10 per virion (21, 36). In addition, at least three other phage structural proteins (P11, P16, and P18) are involved in the DNA entry (37). In wild-type virus, a tubular tail-like structure, derived from the viral membrane and protruding through one vertex, has been reported (15, 33). Tail formation is dependent on at least protein P18, since a mutation in the corresponding gene prevents tube formation (2). Here we show that purified recombinant P2 is a monomeric protein able to interfere with the binding of the virus to its receptor. It is also a stability factor that secures that the DNA injection process does not start before the virus binds to its receptor. A novel DNA ejection mechanism is proposed involving the formation of the tail tube at the vertex that binds to the receptor.
MATERIALS AND METHODS
Bacteria, plasmids, and phages.
The bacterial strains used in this study were E. coli K-12 HMS174 (F− recA hsdR) (18) and JE2571 (leu thr thi lacY thy pil) (13) and S. typhimurium LT2 DB7154 (supD) (36), DB7156 (supF) (36), DS88 (8), PSA (supE) (36), and SL5676 (ΔH2 H1-i::Tn10 [Tcs] non rev; B. A. D. Stocker, Stanford University, Palo Alto, Calif.). Cells were grown in Luria-Bertani (LB) medium (47). When appropriate, ampicillin (100 μg/ml) and/or chloramphenicol (10 μg/ml) were added. Phage PRD1 (41) was propagated on S. typhimurium DS88, and its mutant derivatives sus1 (36), sus170 (36), and sus539 (45) were propagated on the suppressor strains PSA(pLM2), DB7154(pLM2), and DB7156(pLM2), respectively. For the production of wild-type and mutant phage particles, DS88 cells were infected at a multiplicity of infection of about 5. After lysis, the particles were concentrated and purified by rate zonal sucrose gradient centrifugation, as previously described (8). The virus zones from the sucrose gradients were further purified by ion-exchange chromatography on MemSep cartridges (Millipore) as described by Walin et al. (53). 14C-labeled wild-type and mutant (sus170) PRD1 viruses were produced and purified as described by Kotilainen et al. (30). The phage particles from the lysate were concentrated and purified in sucrose gradients. Two virus zones, containing empty (DNA-less) or filled (DNA-containing) particles, were collected. The specific activities obtained were 2.0 × 105 and 2.4 × 105 cpm/μg of protein for empty and DNA-containing wild-type particles, respectively, and 1 × 105 cpm/μg for the DNA-containing sus170 mutant particles. The plasmids and phages used are listed in Table 1.
TABLE 1.
Plasmids and phages used in this study
| Plasmid or phage | Descriptiona | Relevant property | Selective marker(s)b | Replicon | Reference or source |
|---|---|---|---|---|---|
| pLM2 | Encodes PRD1 receptor | Kmr | IncPα | 38 | |
| pMG59 | pMS470Δ(NdeI-HindIII) Ω(PRD1 3128–4903)c | PRD1 gene II+ | Apr | pMB1 | This study |
| pML123 | pGZ119EHΔ (EcoRI-BamHI) Ω(EcoRI-XmnI adapter, RP4 XmnI-NotI, 18841–30042) | RP4 trbB-trbM+ | Cmr | ColD | 31 |
| pMS470Δ8 | pMS119EHΔ(XbaI-PstI) Ω(pT7-7 XbaI-NdeI, R751 traC AvaI-SphI) | Apr | pMB1 | 1 | |
| pWP471 | pJF119EH Ω(RP4 NspI-HaeII, 45909–46577) | RP4 traF+ | Apr | pMB1 | 54 |
| RP4 | Encodes PRD1 receptor | Apr, Kmr, Tcr | IncPα | 19 | |
| PRD1 | Wild type | 41 | |||
| PRD1sus1 | Amber mutation in gene IX | Defective in DNA packaging | 36 | ||
| PRD1sus170 | Amber mutation in gene II | Defective in adsorption | 36 | ||
| PRD1sus539 | Amber mutation in gene II | Defective in adsorption | 45 |
The cloning vector is indicated, followed by the restriction enzymes used and the origin of the insert in parentheses. PRD1 and RP4 coordinates were taken from the GenBank/EMBL database (accession no. M69077 and L27758, respectively).
Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Kmr, kanamycin resistance; Tcr, tetracycline resistance.
The PCR primers used for amplification of gene II were OP173 (TTATTCATATGGCTAATTTCAACGTGCC) and OP174 (TTTATTAAGCTTACACCTTTGAAATAATTCCGC).
Cloning of gene II.
Standard molecular cloning techniques were performed as described by Sambrook et al. (47). The coding sequence for PRD1 gene II (for nucleotide numbering, see reference 9) was amplified by PCR and inserted into plasmid pMS470Δ8, resulting in plasmid pMG59. For the cloning strategy, see Table 1. The plasmid construct contains the strong controllable Ptac promoter and the ribosome binding site of phage T7 gene 10 (23).
Expression and purification of protein P2.
E. coli HMS174(pMG59) cells were grown in LB medium containing ampicillin at 37°C to a density of about 2 × 108 CFU/ml. Synthesis of protein P2 was induced by the addition of isopropyl-β-d-thiogalactopyranoside to a final concentration of 1 mM, and the cells were incubated for another 3 h at the same temperature. Bacteria were collected (Sorvall GS3 rotor, 5,000 rpm, 15 min, 5°C) and resuspended in 1/50 of the original culture volume of 50 mM Tris-HCl, pH 7.4. After freezing at −20°C and thawing, bacteria were disrupted by three passages through a French pressure cell (15,000 lb/in2). Urea was added to a final concentration of 1 M, and the cell debris was removed by centrifugation (Sorvall SS-34 rotor, 12,000 rpm, 20 min, 5°C). The supernatant was further cleared by centrifugation (Sorvall T865 rotor, 25,000 rpm, 3 h, 10°C). To the resulting supernatant, saturated ammonium sulfate was added to 25% saturation, and the precipitated proteins were collected (SS-34 rotor, 10,000 rpm, 30 min, 5°C). The pellet was resuspended in 50 mM Tris-HCl (pH 8.2)–50 mM NaCl and dialyzed overnight in the cold against the same buffer, with several changes. After clearing of the solution (SS-34, 10,000 rpm, 10 min, 5°C), proteins were loaded at a flow rate of 1 ml/min onto a Source 15Q column (Pharmacia) that had been equilibrated with the same buffer. Bound proteins were eluted by a linear NaCl gradient up to 1 M. Fractions containing P2, as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), were pooled and dialyzed overnight in the cold against 50 mM potassium phosphate, pH 7.2, containing 12% ammonium sulfate. The solution, cleared as described above, was loaded (0.5 ml/min) onto a phenyl-Superose HR 5/5 column (Pharmacia) equilibrated with the same buffer, and bound proteins were eluted with a linear gradient of decreasing ammonium sulfate concentrations. Fractions containing P2 were pooled, and the proteins were precipitated by the addition of ammonium sulfate to 30% saturation. After collection of the proteins by centrifugation as described above, the pellet was resuspended in 50 mM Tris-HCl, pH 7.4, containing 150 mM NaCl, 1 M urea, and 1 mM 2-mercaptoethanol and P2 was further purified by gel filtration on a Hi-Load 16/60 Superdex 200 column (Pharmacia) equilibrated with the same buffer. Fractions containing purified protein P2 were stored on ice.
To obtain 35S-labeled protein P2, cells were grown as described above but in M9 medium supplemented with all of the amino acids except methionine and cysteine. Fifteen minutes after the addition of the inducer, [35S]methionine (>1,000 Ci/mmol; Amersham) was added to the culture (10 μCi/ml) and the incubation was continued for another 4 h. The cells were collected as described above, resuspended in ice-cold water, pelleted again, and frozen at −20°C. One molar urea (in 50 mM Tris-HCl, pH 7.4) was added to the thawed pellet, and the cells were broken by sonication (Ultrasonics Ltd. Sonifier; eight bursts of 30 s with 90-s intervals; the samples were placed on ice-water). The suspension was cleared by centrifugation (SS-34 rotor, 10,000 rpm, 1 h, 5°C), and proteins were precipitated by addition of saturated ammonium sulfate to 25% saturation. Labeled protein P2 was purified as described above, with the exception that the last gel filtration step was omitted.
Determination of P2 monomeric status.
Rate zonal centrifugation was carried out in 5 to 20% sucrose gradients made in 20 mM Tris-HCl (pH 7.4)–150 mM NaCl (Sorvall TH 641 rotor, 35,000 rpm, 24 h, 10°C). Hen egg white lysozyme (14 kDa; Boehringer), bovine serum albumin (67 kDa; Boehringer), and PRD1 P3 trimer (130 kDa) (51) were used as molecular mass markers. Gel filtration experiments were done with a Superdex 200 HR 10/30 column (Pharmacia) equilibrated with 50 mM Tris-HCl (pH 7.4)–150 mM NaCl. The molecular mass standards used were as described above. Cross-linking was performed with increasing concentrations (0.01 to 0.5%) of glutaraldehyde in 50 mM potassium phosphate buffer, pH 7.2. In order to minimize the formation of unspecific intermolecular cross-links, the protein concentration in the samples was adjusted to 0.4 mg/ml. After 2 h of incubation at 22°C, samples were prepared for and analyzed by SDS-PAGE. Sedimentation equilibrium experiments were performed in a Beckman Optima XL-I analytical ultracentrifuge (50Ti analytical rotor, 12,000 rpm, 4°C). P2 was diluted so that the loading concentrations were 0.5, 0.7, and 2 mg/ml. The distribution of the protein was determined by measuring the absorbance at 294, 297, and 301 nm over the cell radius. Data were analyzed by using the Origin computer program (Microcal Inc., Northampton, Mass.). A partial specific volume of 0.7232 ml/g and a monomer molecular mass of 63,821 Da were calculated from the known amino acid sequence of the protein (9).
Phage adsorption test.
Cells were grown to the optimal adsorption phase (approximately 2 × 109 CFU/ml) (30) in LB medium. 14C-labeled PRD1 was adsorbed to the cells for 15 min at 22°C. Cells were then collected by centrifugation and washed twice with 0.5 ml of LB medium. Radioactivity in the supernatant and pellet fractions was measured by liquid scintillation counting.
The reversibility of phage binding was analyzed by using the adsorption test, except that a 30-min equilibration was allowed between the washing steps. In another set of experiments, cells were transferred into 50 mM Tris-HCl (pH 7.2)–0.1% glucose. 14C-labeled phages were added, reversibility was challenged by adding increasing concentrations of NaCl, and the incubation was continued for 15 min. The washing steps were carried out in the used NaCl concentrations.
Inhibition of phage adsorption by protein P2.
Cells were grown to the optimal adsorption phase as described above. A constant number of cells (2 × 108 CFU) and various amounts of P2 (cell lysate or purified protein; see above) were mixed in a volume of 70 μl and incubated for 30 min at 22°C. The ability of P2 to interfere with phage binding was determined either by phage plaque assay or by the phage adsorption test. (i) In the phage plaque assay, approximately 200 PFU was added to the cell-P2 mixture, which was incubated for 15 min at 22°C. Thereafter, cells were collected by centrifugation and the amount of free (nonadsorbed) phage particles was determined by plating the supernatant on S. typhimurium DS88 cells. (ii) In the adsorption test, 14C-labeled PRD1, at a multiplicity of infection of about 3, was added. After 15 min of incubation at 22°C, cells were collected by centrifugation and washed twice with LB medium. Radioactivity in the supernatant and pellet fractions was measured by liquid scintillation counting.
P2 binding assay.
Cells (2 × 108 CFU) and increasing amounts of 35S-labeled protein P2 were mixed and incubated for 45 min at 22°C. Subsequently, cells were collected by centrifugation and the amounts of free and bound P2 were determined by measuring the radioactivity in the supernatant and pellet fractions.
P2 inactivation assays.
Purified protein P2 (150 ng) was incubated for 15 min at different temperatures; this was followed by 10 min of equilibration to 22°C. Cells (2 × 108 CFU) were added, and the adsorption inhibition assay was performed as described above. The effect of denaturing agents on P2 activity was tested by incubating 75 ng of P2 in the presence of 8 M urea or 5 M guanidine hydrochloride (GuHCl) for 15 min at 22°C. After that, the concentration of the denaturing agent was diluted to 1/10 of the original value and the incubation was continued for 30 min. The activity of the preparation was measured by the adsorption inhibition assay.
Polyclonal antiserum.
Polyclonal antibodies against protein P2 were raised by immunizing a rabbit at 20-day intervals. For the primary immunization, the antigen (about 400 μg) was emulsified with complete Freund’s adjuvant. Incomplete adjuvant was used in the subsequent immunizations. The first two immunizations were done with a sample cut out as a single band from a preparative SDS–14% polyacrylamide gel. For the final booster, purified P2 was used as the antigen. The serum was collected 2 weeks after administration of the third booster, and its specificity was determined by immunoblotting as described below. Anti-P2 immunoglobulins G (IgGs) were purified from the serum by chromatography on a GammaBind G Sepharose column (Pharmacia) in accordance with the manufacturer’s instructions.
The antiserum and the purified IgG fraction were tested for neutralization ability. Approximately 150 PFU was treated with antibody dilutions for 1 h at 22°C; this was followed by plating with S. typhimurium DS88 cells. The neutralization titer was the last antibody dilution giving 50% reduction in the plaque count. Radioimmunoprecipitation of 35S-labeled protein P2 was performed by using protein A Sepharose CL-4B (Pharmacia) as previously described (29). The preimmune serum and polyclonal antibodies directed against bacteriophage φ6 protein P4 were used as negative controls.
Virus aggregation tests.
14C-labeled wild-type PRD1 (about 10,000 cpm) or P2-deficient mutant sus170 (about 5,000 cpm) was incubated with serial antibody dilutions for 2 h at 22°C. Virus aggregates were collected by centrifugation, and the radioactivity in the supernatant and pellet fraction was measured by liquid scintillation counting. To test if purified P2 can block virion precipitation, anti-P2 IgGs (1:1,000 dilution) and serial P2 dilutions were incubated for 2 h at 22°C. After that, 14C-labeled wild-type PRD1 (10,000 cpm) was added and the incubation was continued for 1 h at 22°C. Virus aggregates were collected as described above. Virus aggregation was also verified by electron microscopy by using a JEOL 1200 EX electron microscope operating at 60 kV. PRD1 wild-type and mutant (sus1 and sus170) particles were incubated with anti-P2 IgGs (1:500 dilution) for 1 to 2 h at 22°C; this was followed by negative staining (1% potassium phosphotungstate, pH 6.5).
Disruption of phage particles.
Purified wild-type PRD1 particles in 20 mM Tris-HCl (pH 7.2) or in 20 mM potassium phosphate (pH 7.2)–1 mM MgCl2 were treated with 2.5 M GuHCl for 30 min at 37°C (4). Samples were diluted 1:1 with the appropriate buffer and analyzed by rate zonal centrifugation in 10 to 40% sucrose gradients made in the same buffer (Sorvall TH 641 rotor, 30,000 rpm, 45 min, 20°C). Fractions (0.9 ml) were collected starting from the top by using a Piston Gradient Fractionator (BioComp Instruments, Inc.). Fractions were analyzed by SDS-PAGE and Western blotting as described below.
Stability analysis of P2-deficient phage particles.
Affinity-purified DNA-containing wild-type and P2-deficient (sus170 or sus539) phage particles (3 mg of protein/ml) in 20 mM Tris-HCl (pH 7.4) or in 20 mM potassium phosphate (pH 7.2) were stored at 4°C. The viscosity of the preparation was monitored by visual inspection. The particles were also subjected, at appropriate intervals, to rate zonal sedimentation analysis through a linear 5 to 20% sucrose gradient, as well as to electron microscopy (see above). Due to high viscosity, the samples were digested with DNase I (50 μg/ml [final concentration]) for 40 min at 22°C prior to application to carbon-coated grids. The effect of the DNase digestion was also verified by agarose gel electrophoresis.
Analytical methods.
SDS-PAGE was performed as described by Olkkonen and Bamford (39). Western blotting was done essentially as previously described (40), after transfer of the proteins from SDS-polyacrylamide gels onto a polyvinylidene difluoride membrane (Millipore). Protein concentrations were determined with Coomassie brilliant blue by using bovine serum albumin as a standard (12). N-terminal amino acid analysis of the purified protein was performed in a Beckman 890D sequencer after SDS-PAGE and blotting (9). Circular dichroism (CD) spectra were recorded in a Jasco J-720 spectropolarimeter using a 1-mm path-length cell at the temperatures indicated in each case. The protein concentration of the samples was adjusted to approximately 35 μg/ml in 20 mM Tris-HCl buffer, pH 7.4. Each spectrum, recorded at a scan rate of 20 nm/min, was the average of five scans. The background contribution due to the solvent was subtracted in each case. Triton X-114 phase separation was performed essentially as described by Bordier (11), with incubation for 15 min at 30°C. After centrifugation, the radioactivity in the water and Triton X-114 phases was measured by liquid scintillation counting.
RESULTS
Cloning of gene II and purification of the gene product.
The adsorption of phage PRD1 to its receptor on the cell surface is dependent on a single minor phage protein, P2 (21, 37). In order to more closely characterize protein P2, gene II was cloned into plasmid pMS470Δ8. In this plasmid, a high level of protein expression is enabled by the strong Ptac promoter and the ribosome binding sequence of phage T7 gene 10. The biological activity of the gene II construct was verified by genetic complementation using the PRD1 sus170 nonsense mutant, which is defective in P2. The complementation titer of PRD1 sus170 on cells containing the gene II clone was nearly the same as that obtained when the suppressor strain DB7154(pLM2) was used.
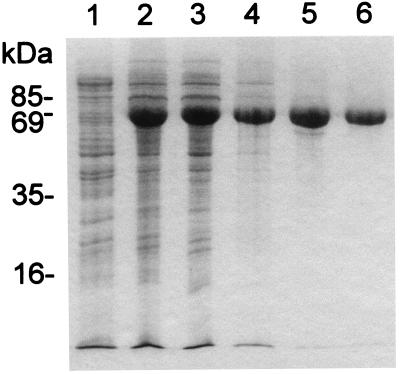
For the purification of P2, cells overproducing the protein were lysed and debris was removed by ultracentrifugation. After precipitation of the proteins with ammonium sulfate, P2 was purified nearly to homogeneity by a combination of ion-exchange, hydrophobic, and gel filtration chromatography steps, with a typical final yield of about 3 mg/liter of culture. The results of the purification steps are summarized in Fig. 1.
FIG. 1.
Purification of protein P2. Samples were taken at different purification stages and analyzed by SDS-PAGE, and the gels were stained with Coomassie blue. Lanes: 1, disrupted HMS174(pMG59) cells prior to induction; 2, disrupted cells after induction; 3, cleared supernatant after ultracentrifugation; 4, pooled fractions after chromatography on Resource Q; 5, P2-containing fractions after phenyl-Superose chromatography; 6, purified P2 after gel filtration. The values on the left indicate the molecular masses of several marker proteins.
In SDS-polyacrylamide gels, the purified protein displayed a molecular mass of approximately 65 kDa, very close to the molecular mass of 63,821 Da calculated from the sequence of gene II (9). The identity of the purified protein was confirmed by N-terminal amino acid sequence analysis. The sequence Ala-Asn-Phe-Asn-Val-Pro-Lys-Leu-Gly-Val was determined, and it was identical to that deduced from the sequence of the gene (9), except for the first Met residue had been removed.
A very similar protocol was used for the purification of 35S-labeled P2. In this case, however, the last gel filtration step was omitted, due to the smaller amount of protein in the samples. The purity of this preparation was at least 90%, as judged by SDS-PAGE and densitometry of the stained gels. P2 appeared to be a soluble protein. The hydrophilicity displayed by the purified protein was tested by Triton X-114 phase separation experiments (11). 35S-labeled P2 was found in the water phase.
Purified protein P2 is a stable asymmetric monomer.
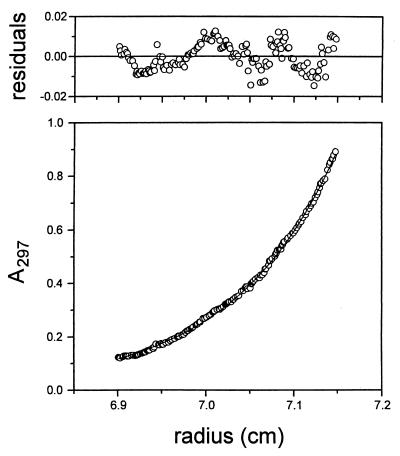
To determine the apparent molecular mass of the isolated protein, P2 was first subjected to rate zonal centrifugation, gel filtration, and cross-linking. In sucrose gradients, P2 sedimented to a position corresponding to a molecular mass of about 40 kDa. Gel filtration experiments indicated a molecular mass of approximately 95 kDa. Incubation of the protein with increasing amounts of glutaraldehyde did not result in detectable cross-linking products (data not shown). Taken together, these results indicate that purified protein P2 is an asymmetric monomer. To confirm this, protein samples at three different concentrations were subjected to analytical ultracentrifugation. Sedimentation equilibrium experiments indicated that the concentration profiles could be fitted by a single-species model with apparent molecular masses of 63,152 ± 189 (0.5 mg/ml), 63,642 ± 276 (0.7 mg/ml), and 58,372 ± 406 (2 mg/ml) Da (Fig. 2).
FIG. 2.
Equilibrium sedimentation of protein P2. The lower graph shows the absorbance distribution obtained at a protein concentration of 0.5 mg/ml and a rotor speed of 12,000 rpm. The thin continuous line represents the single-species model fitted to the data points. The plot of the residuals of the absorbance values is shown in the upper graph.
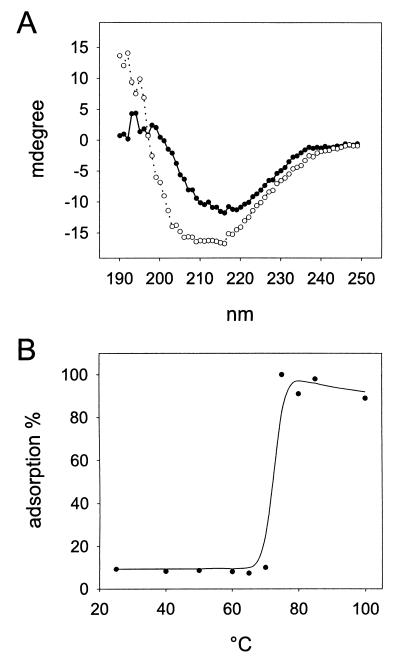
To gain information on the structure and stability of protein P2, the CD spectra of the protein were recorded at different temperatures. Figure 3A shows the typical CD spectrum of the protein recorded at 20°C (closed circles). The minimum observed at 216 nm together with the analysis of the spectrum, done by using the software and model compound libraries provided by the manufacturer of the spectropolarimeter, suggested that the secondary structure of P2 is predominantly (about 50%) β-sheet, with only about 20% α-helices. These values are in good agreement with those obtained by the sequence-based secondary-structure prediction (24).
FIG. 3.
Thermal stability of protein P2. (A) CD spectra of the purified protein recorded at 20°C prior to heating (filled circles) and after heating to 80°C and cooling (open circles). (B) Biological activity of heat-treated P2 measured by its ability to inhibit radioactively labeled phage adsorption.
Protein P2 was stable up to 60°C, as no significant changes in its CD spectrum were seen upon heating. Above this temperature, however, some spectral changes were observed. These changes were shown to be irreversible, since cooling of the sample to 20°C did not restore the original spectral properties of the protein (Fig. 3A). These results suggest that denaturation of the protein occurs at temperatures above 60°C. To confirm this, the effect of heat on the biological activity of P2 was tested by the adsorption inhibition assay. The ability of P2 to inhibit 14C-labeled PRD1 adsorption was drastically diminished at temperatures above 70°C (Fig. 3B), which is in good agreement with the CD data.
Recombinant P2 is biologically active and has high affinity for its receptor.
PRD1 infects various gram-negative bacteria containing a receptor-encoding conjugative plasmid, such as IncP plasmid RP4 (14, 41, 43). The biological activity of recombinant P2 was studied by using bacteria (S. typhimurium DS88) expressing the phage receptor. The ability of P2 to bind to the receptor was analyzed indirectly by determining its ability to interfere with phage binding. If P2 occupies the receptor sites at the cell surface, infective phage particles are not able to attach to the cells. This was measured as increasing radioactivity (14C-labeled PRD1) or plaque count in the supernatant fraction. The activity of P2, both as purified protein and in cell extracts, was tested and compared by using the radioactive phage adsorption method. The amount of protein used was adjusted on the basis of SDS-PAGE analysis. The two preparations were equally active (data not shown), as was the 35S-labeled protein (tested by the phage plaque assay). The stability of P2 was monitored during its storage under different conditions. The protein could be stored on ice for at least 3 months with no detectable loss of activity. The biological activity of P2 was also preserved when it was frozen at −20 or −80°C. Treatment with 8 M urea or 5 M GuHCl, followed by subsequent dilution of the denaturing agent, did not abolish the ability of P2 to interfere with phage adsorption (data not shown).
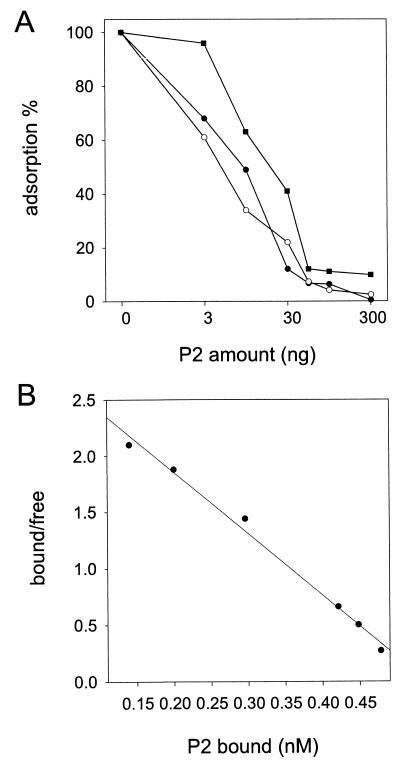
The expression of the PRD1 receptor can be increased from the original level by using recombinant receptor-encoding plasmids (20, 30). We tested the binding of P2 to three different cell types, S. typhimurium DS88 and E. coli JE2571(RP4) and JE2571(pML123, pWP471). The numbers of receptors in these cells are approximately 60, 25, and 80 per cell, respectively (20, 30). In DS88 and JE2571(RP4) cells, the receptor complex is encoded by the original IncP plasmid but expressed in different bacterial backgrounds. In JE2571(pML123, pWP471), genes needed for phage adsorption (the RP4 DNA transfer genes trbB-L and traF; 26, 27, 31, 54) are inserted into multicopy plasmids. Figure 4A shows that the amount of P2 needed to inhibit phage adsorption decreases as the number of receptor complexes decreases.
FIG. 4.
Biological activity of P2. (A) Inhibition of adsorption of labeled PRD1 by purified P2. The relative amount of phage particles associated with cells decreases as the amount of P2 is increased. Adsorption inhibition is shown for three bacterial strains: DS88 (60 receptors/cell; filled circles), JE2571(RP4) (25 receptors/cell; open circles), and JE2571(pML123, pWP471) (80 receptors/cell; filled squares). (B) Binding of 35S-labeled P2 to JE2571(pML123, pWP471) cells. The Scatchard transformation of the data obtained from two independent experiments is shown.
In another set of experiments, the binding of 35S-labeled P2 to the three different bacterial strains was measured directly (data not shown). The Scatchard transformation of the data obtained for the binding of 35S-labeled P2 to JE2571(pML123, pWP471) cells is shown in Fig. 4B. From the slope of the line, a Kd of 0.20 nM, the mean of two independent experiments, was obtained. Duplicate, independent experiments performed with the other two bacterial strains resulted in Kd values of 0.19 and 0.18 nM for DS88 and RP4 cells, respectively. These results indicate a very high affinity of the phage adsorption protein for its receptor.
Stable DNA packaging is secured by protein P2.
Binding of a phage by its adsorption protein to the host must trigger the next step in the infection process. In order to study the role of protein P2 in DNA release from the phage particle, we compared the stability of DNA-containing wild-type and two P2-deficient mutant PRD1 particles (sus170 and sus539) by monitoring the physical properties of highly purified preparations. The wild-type virion preparation did not show any increase in viscosity and displayed a single sedimenting zone in rate zonal centrifugation, even after 3 weeks of storage at 4°C. Increasing viscosity was seen in both of the mutant particle preparations, and it was detectable already a few hours after virus purification. In rate zonal centrifugation experiments, only a barely detectable virus zone was obtained with mutant particles stored at 4°C overnight and practically all of the material was found in the pellet due to aggregation. Electron microscopy of negatively stained P2− particles revealed an increasing number of empty particles with an extended tail structure (Fig. 5). The tails of adjacent particles had a tendency to associate to form larger aggregates. The viscosity of the preparations was abolished by DNase treatment, allowing electron microscopy examination to be made, as well as showing that the cause of viscosity in the preparations was the DNA released from the particles. Electrophoresis of the DNase-treated preparations also confirmed the sensitivity of the mutant particle DNA to DNase (data not shown). These experiments were carried out in 20 mM Tris-HCl (pH 7.4) buffer. It was found that the P2− particles were more stable in potassium phosphate buffer (pH 7.2). They behaved the same as the wild-type particles in rate zonal centrifugation assays, and no considerable increase in viscosity was observed after 3 weeks of storage at 4°C.
FIG. 5.
Stability of P2-deficient particles. Electron micrograph of negatively stained P2− particles (sus539) showing extended tails in empty DNA-less particles. The tail structures have a tendency to associate, forming rosettes. Bar, 200 nm.
Protein P2 is a virion surface component that signals to the phage membrane.
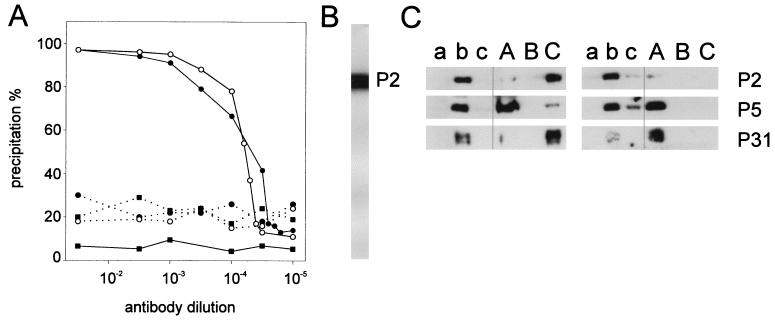
P2 was used as the antigen to produce polyclonal antibodies in a rabbit. Both serum and purified IgGs were tested for the ability to recognize 35S-labeled P2. The recognition of the labeled protein could be detected by a radioimmunoprecipitation assay (0.5 μg of P2/100 μl, 1:50 serum or IgG dilution; data not shown). In Western blotting, the antigen could be detected at antibody dilutions as high as 1:20,000 (Fig. 6B). Both P2 antiserum and the purified IgG fraction recognized and inactivated the intact virus, the highest antibody dilution giving a 50% reduction in plaque count being 1:50 (virus concentration, 1.2 × 103 PFU/ml; data not shown).
FIG. 6.
Determination of P2 location in the virus particle. (A) Antibodies against P2 induce virus aggregation of wild-type PRD1 particles (solid line) but not of P2-deficient particles (dashed line), as shown for anti-P2 serum (filled circles) and purified IgGs (open circles) by the precipitation assay. Preimmune serum (filled squares) was used as a negative control. (B) Specificity of the anti-P2 antibodies as detected by Western blotting. The position of protein P2, determined in an equivalent nitrocellulose strip that had been stained with Coomassie blue, is indicated on the right. (C) Immunological identification of proteins P2, P5, and P31 in sucrose gradient fractions (a, top fraction of the gradient; b, virus zone; c, pellet) of GuHCl-treated (capital letters) and nontreated (lowercase letters) phage particles analyzed by SDS-PAGE. Disruption of purified PRD1 particles was performed in 20 mM potassium phosphate (pH 7.2)–1 mM MgCl2 (left panel) or in 20 mM Tris-HCl (pH 7.4) (right panel). Immunological identification of P2 in a GuHCl-disrupted virus preparation (Tris buffer) was weak. The presence of protein P2 in the gradient top fraction was confirmed by Coomassie staining (10-fold-concentrated sample). The positions of proteins P2, P5, and P31 are indicated on the right.
To determine whether P2 is on the virion surface, we took advantage of the anti-P2 antibodies. 14C-labeled PRD1 wild-type and P2− mutant particles were purified, and the ability of anti-P2 serum to aggregate these particles was tested in a precipitation assay. In the purification of P2− particles, potassium phosphate was used instead of Tris buffer (used for the wild-type virus), as the integrity of the P2− virions was maintained in phosphate but not in the Tris buffer (see above). Anti-P2 serum and IgGs aggregated wild-type virus particles at the high virus concentration (5 × 109 PFU/ml) used, even at high antibody dilutions, as shown in the 14C-labeled PRD1 precipitation assay (Fig. 6A). Under the same conditions, mutant P2-deficient particles were not aggregated (Fig. 6A). Also, purified P2 blocked precipitation, confirming the specificity of the antiserum (data not shown). Virus aggregation was also observed by electron microscopy in samples containing wild-type and mutant (defective in DNA packaging protein P9) PRD1 particles but not with mutant particles devoid of the adsorption protein P2 (data not shown). Together, these results indicate that the P2 molecules are distributed around the virion surface. This also explains the need of high antibody concentrations for inactivation of the virus but low concentrations for its aggregation.
Previous virus disruption studies suggested that protein P2 is associated with the phage membrane while the major capsid protein P3 and a minor protein P5 are released as soluble proteins (2). However, P2 could be dissociated from the virion by heat, high pH, or low SDS concentrations, and thus, a peripheral membrane association was suggested (17). Recent studies revealed that on mutant particles lacking the pentameric vertex protein P31, proteins P2 and P5 were also missing (45). To further characterize the suggested P2 membrane association, we determined the distribution of the fivefold vertex proteins (P31, P5, and P2; 45) in sucrose gradient fractions of disrupted phage particles by using specific antisera. Under the centrifugation conditions (2) used, phage membrane aggregates sedimented to the bottom of the tube whereas released proteins remained at the top of the gradient. Under these conditions, intact virus particles formed a single sedimenting zone. When virus disruption was performed in phosphate buffer, both P2 and P31 were found in the membrane fraction while P3 and P5 remained at the top (Fig. 6C; 4). In Tris buffer, P2 and P31 were also detached from the membrane and behaved like soluble proteins in rate zonal centrifugation (Fig. 6C). Together, these results and studies on the capsid fivefold organization (45) indicate that P2 and P31 are in contact with the phage membrane. Since P2 seems to be a soluble protein, its association with the membrane most probably occurs via interaction with P31.
Both empty and DNA-containing PRD1 particles bind irreversibly to the cell surface.
The phage-receptor interaction is classically considered to occur in two steps: (i) recognition and binding and (ii) transformation of this interaction to the DNA penetration step. The binding of the PRD1 adsorption protein P2 to cells was found to be reversible (see determination of the binding constant). However, PRD1 virions bound to the host cannot be detached from the cells under the conditions studied. In LB medium, after 15 min of adsorption and four subsequent extended washing steps (incubation for 30 min at 22°C), about 65% of both empty and DNA-containing phage particles were associated with cells (data not shown), suggesting that virions are irreversibly bound. Also, if a reversible complex exists between the phage and its receptor, one might expect this complex to be dissociated by a high salt concentration. No significant release of either empty or filled particles adsorbed to cells could be detected when increasing amounts of NaCl (up to a 500 mM final concentration) were added after the adsorption period of 15 min (data not shown). Since empty phage particles are as tightly bound to the cells as are the DNA-containing particles, we conclude that in PRD1 infection, DNA penetration is not a prerequisite for irreversible binding.
DISCUSSION
Recombinant P2 can be produced as a stable, biologically active monomer. By estimating the protein concentration of the 35S-labeled P2 preparation and subsequently measuring the amount of label associated with a defined number of cells, we can calculate the number of P2 molecules bound per cell. Taking into account the number of receptors in RP4-containing cells, a very low number of P2 molecules (close to one) per receptor is obtained. This is consistent with a model in which one P2 molecule mediates the connection between the phage and its receptor.
The anti-P2 serum readily aggregated PRD1 particles. This is in accordance with the idea that protein P2 has a distal location in the spike complex. On the other hand, virus disruption studies revealed that spike complex proteins P31 and P2 are associated with the membrane while the shell composed of major capsid protein P3 is dissociated. It appears that association of the receptor recognition-DNA injection device with the membrane (at the vertices) is independent of the viral protein shell. This is also supported by the previous report that removal of the viral membrane with detergents leaves the protein shell intact except that the fivefold structures are removed with the membrane (15, 34). Recent studies on the organization of the capsid fivefold spike complex indicated that the main protein involved in the DNA packaging, P9, does not interact with the adsorption spike structure (45). These results are consistent with a model in which the DNA packaging machinery occupies a particular vertex of the virion, the remaining 11 vertices being occupied by the spike structure. We cannot, however, exclude the possibility that all of the vertices are identical but with independent DNA packaging and injecting machineries. The latter possibility is less favored based on comparisons to other dsDNA phage packaging systems, in which a multiprotein complex is associated with a specific vertex (28). Both models imply, if P2 is considered as a monomer, either 11 or 12 copies of P2 per virion, values very close to the stoichiometry of 10 determined biochemically by Davis et al. (21).
The two P2-deficient mutants studied, sus170 and sus539, package DNA and do not lack any other structural protein. In Tris buffer (but not in phosphate buffer), these purified mutant particles, unlike the wild-type particles, spontaneously eject DNA and display the tail-like structure presumably involved in DNA ejection (33). Tris buffer destabilizes membranes. This effect was also observed in the virus disruption studies. Removal of P2 apparently allows Tris molecules to gain access to the phage membrane, triggering DNA ejection and tail tube formation.
The above observations can be extended into a model in which protein P2 secures the DNA injection process. Most probably, the association of P2 with the receptor signals, possibly by P2 removal, the activation of the injection process. This P2-receptor association leads to irreversible binding and is independent of the DNA content of the virion, as empty phage particles were found to be as tightly bound to the cells, as were the DNA-containing particles. The tail-like structure is most probably involved in the stabilization of the virus-cell contact. The tail formation is dependent on several phage-encoded proteins, and the involvement of P18 has been previously reported (2). Also, a mutation in gene XXXI prevents tail tube formation and results in loss of the capsid fivefold structure (proteins P2, P5, and P31) and the peripentonal trimers of major capsid protein P3 (45). Since the tail-like structure is readily observed in the P2-deficient mutant, it is likely that P5 and/or P31 contribute to its formation. A noteworthy consequence of this model of DNA injection is that the tail structure would extrude through the particular vertex that makes the contact with the receptor. This mechanism would differ from that of other dsDNA phages, in which the DNA entry and exit vertices are the same, but might be more similar to the one used by adenoviruses, in which all of the fivefold structures are metastable. If there were a specific ejection vertex, the receptor binding should lead to a complex signalling and orientation procedure to ascertain that the tail is ejected toward the cell surface.
ACKNOWLEDGMENTS
We are grateful to Marja-Leena Perälä for her skillful technical assistance. We also thank Nisse Kalkkinen for the N-terminal amino acid sequencing of P2 and Erich Lanka for the RP4 clones. The help of Hans van der Zandt in the analytical ultracentrifugation experiments is gratefully acknowledged.
The electron microscopy was performed at the electron microscopy unit, Institute of Biotechnology, University of Helsinki. This investigation was supported by research grants 37725 and 41400 from the Finnish Academy of Sciences.
REFERENCES
- 1.Balzer D, Ziegelin G, Pansegrau W, Kruft V, Lanka E. KorB protein of promiscuous plasmid RP4 recognizes inverted sequence repetitions in regions essential for conjugative plasmid transfer. Nucleic Acids Res. 1992;20:1851–1858. doi: 10.1093/nar/20.8.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bamford D, Mindich L. Structure of the lipid-containing bacteriophage PRD1: disruption of wild-type and nonsense mutant phage particles with guanidine hydrochloride. J Virol. 1982;44:1031–1038. doi: 10.1128/jvi.44.3.1031-1038.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bamford D H, Caldentey J, Bamford J K H. Bacteriophage PRD1: a broad host range dsDNA Tectivirus with an internal membrane. Adv Virus Res. 1995;45:281–319. doi: 10.1016/s0065-3527(08)60064-0. [DOI] [PubMed] [Google Scholar]
- 4.Bamford D, McGraw T, MacKenzie G, Mindich L. Identification of a protein bound to the termini of bacteriophage PRD1 DNA. J Virol. 1983;47:311–316. doi: 10.1128/jvi.47.2.311-316.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bamford D H, Mindich L. Characterization of the DNA-protein complex at the termini of the bacteriophage PRD1 genome. J Virol. 1984;50:309–315. doi: 10.1128/jvi.50.2.309-315.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bamford D H, Romantschuk M, Somerharju P. Membrane fusion in prokaryotes: bacteriophage φ6 membrane fuses with the Pseudomonas syringae outer membrane. EMBO J. 1987;6:1467–1473. doi: 10.1002/j.1460-2075.1987.tb02388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bamford D H, Rouhiainen L, Takkinen K, Söderlund H. Comparison of the lipid containing bacteriophages PRD1, PR3, PR4, PR5 and L17. J Gen Virol. 1981;57:365–373. doi: 10.1099/0022-1317-57-2-365. [DOI] [PubMed] [Google Scholar]
- 8.Bamford J K H, Bamford D H. Capsomer proteins of bacteriophage PRD1, a bacterial virus with a membrane. Virology. 1990;177:445–451. doi: 10.1016/0042-6822(90)90508-o. [DOI] [PubMed] [Google Scholar]
- 9.Bamford J K H, Hänninen A-L, Pakula T M, Ojala P M, Kalkkinen N, Frilander M, Bamford D H. Genome organization of membrane-containing bacteriophage PRD1. Virology. 1991;183:658–676. doi: 10.1016/0042-6822(91)90995-n. [DOI] [PubMed] [Google Scholar]
- 10.Benson S D, Bamford J K H, Bamford D H, Burnett R. Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell. 1999;98:825–833. doi: 10.1016/s0092-8674(00)81516-0. [DOI] [PubMed] [Google Scholar]
- 11.Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- 12.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 13.Bradley D E. Morphological and serological relationship of conjugative pili. Plasmid. 1980;4:155–169. doi: 10.1016/0147-619x(80)90005-0. [DOI] [PubMed] [Google Scholar]
- 14.Bradley D E, Rutherford E L. Basic characterization of lipid-containing bacteriophage specific for plasmids of the P, N and W compatibility groups. Can J Microbiol. 1975;21:152–163. doi: 10.1139/m75-023. [DOI] [PubMed] [Google Scholar]
- 15.Butcher S J, Bamford D H, Fuller S D. DNA packaging orders the membrane of bacteriophage PRD1. EMBO J. 1995;14:6078–6086. doi: 10.1002/j.1460-2075.1995.tb00298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caldentey J, Blanco L, Bamford D H, Salas M. In vitro replication of bacteriophage PRD1 DNA. Characterization of the protein-primed initiation site. Nucleic Acids Res. 1993;21:3725–3730. doi: 10.1093/nar/21.16.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caldentey J, Luo C, Bamford D H. Dissociation of the lipid-containing bacteriophage PRD1: effects of heat, pH, and sodium dodecyl sulfate. Virology. 1993;194:557–563. doi: 10.1006/viro.1993.1294. [DOI] [PubMed] [Google Scholar]
- 18.Campbell J L, Richardson C C, Studier F W. Genetic recombination and complementation between bacteriophage T7 and cloned fragments of T7 DNA. Proc Natl Acad Sci USA. 1978;75:2276–2280. doi: 10.1073/pnas.75.5.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Datta N, Hedges R W, Shaw E J, Sykes R B, Richmond M H. Properties of an R factor from Pseudomonas aeruginosa. J Bacteriol. 1971;108:1244–1249. doi: 10.1128/jb.108.3.1244-1249.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daugelavičius R, Bamford J K H, Grahn A M, Lanka E, Bamford D H. IncP plasmid encoded cell envelope-associated DNA transfer complex increases the cell permeability. J Bacteriol. 1997;179:5195–5202. doi: 10.1128/jb.179.16.5195-5202.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis T N, Muller E D, Cronan J E., Jr The virion of the lipid-containing bacteriophage PR4. Virology. 1982;120:287–306. doi: 10.1016/0042-6822(82)90031-9. [DOI] [PubMed] [Google Scholar]
- 22.Dreiseikelmann B. Translocation of DNA across bacterial membranes. Microbiol Rev. 1994;58:293–316. doi: 10.1128/mr.58.3.293-316.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dunn J J, Studier F W. Complete nucleotide sequence of bacteriophage T7 genetic elements. J Mol Biol. 1983;166:477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- 24.Garnier J, Osguthorpe D J, Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978;120:97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- 25.Grahn, A. M., J. Haase, D. H. Bamford, and E. Lanka. Components of the RP4 conjugative transfer apparatus form an envelope structure bridging inner and outer membranes of donor cells: implications for related macromolecule transport systems. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 26.Grahn A M, Haase J, Lanka E, Bamford D H. The assembly of a functional phage PRD1 receptor depends on 11 genes of the IncP plasmid mating pair formation complex. J Bacteriol. 1997;179:4733–4740. doi: 10.1128/jb.179.15.4733-4740.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haase J, Lurz R, Grahn A M, Bamford D H, Lanka E. Bacterial conjugation mediated by plasmid RP4: RSF1010 mobilization, donor-specific phage propagation, and pilus production require the same Tra2 core components of a proposed DNA transport complex. J Bacteriol. 1995;177:4779–4791. doi: 10.1128/jb.177.16.4779-4791.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hendrix R. Bacteriophage DNA packaging: RNA gears in a DNA transport machine. Cell. 1998;94:147–150. doi: 10.1016/s0092-8674(00)81413-0. [DOI] [PubMed] [Google Scholar]
- 29.Kenney J M, Hantula J, Fuller S D, Mindich L, Ojala P M, Bamford D H. Bacteriophage φ6 envelope elucidated by chemical crosslinking, immunodetection, and cryoelectron microscopy. Virology. 1992;190:635–644. doi: 10.1016/0042-6822(92)90901-z. [DOI] [PubMed] [Google Scholar]
- 30.Kotilainen M M, Grahn A M, Bamford J K H, Bamford D H. Binding of an Escherichia coli double-stranded DNA virus PRD1 to a receptor coded by an IncP-type plasmid. J Bacteriol. 1993;175:3089–3095. doi: 10.1128/jb.175.10.3089-3095.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lessl M, Balzer D, Weyrauch K, Lanka E. The mating pair formation system of plasmid RP4 defined by RSF1010 mobilization and donor-specific phage propagation. J Bacteriol. 1993;175:6415–6425. doi: 10.1128/jb.175.20.6415-6425.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lessl M, Lanka E. Common mechanisms in bacterial conjugation and Ti-mediated T-DNA transfer to plant cells. Cell. 1994;77:321–324. doi: 10.1016/0092-8674(94)90146-5. [DOI] [PubMed] [Google Scholar]
- 33.Lundström K H, Bamford D H, Palva E T, Lounatmaa K. Lipid-containing bacteriophage PR4: structure and life cycle. J Gen Virol. 1979;43:583–592. doi: 10.1099/0022-1317-43-3-583. [DOI] [PubMed] [Google Scholar]
- 34.Luo C, Butcher S, Bamford D H. Isolation of a phospholipid-free protein shell of bacteriophage PRD1, an Escherichia coli virus with an internal membrane. Virology. 1993;194:564–569. doi: 10.1006/viro.1993.1295. [DOI] [PubMed] [Google Scholar]
- 35.Lyra C, Savilahti H, Bamford D H. High-frequency transfer of linear DNA containing 5′-covalently linked terminal proteins: electroporation of bacteriophage PRD1 genome into Escherichia coli. Mol Gen Genet. 1991;228:65–69. doi: 10.1007/BF00282449. [DOI] [PubMed] [Google Scholar]
- 36.Mindich L, Bamford D, Goldthwaite C, Laverty M, MacKenzie G. Isolation of nonsense mutants of lipid-containing bacteriophage PRD1. J Virol. 1982;44:1013–1020. doi: 10.1128/jvi.44.3.1013-1020.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mindich L, Bamford D, McGraw T, MacKenzie G. Assembly of bacteriophage PRD1: particle formation with wild-type and mutant viruses. J Virol. 1982;44:1021–1030. doi: 10.1128/jvi.44.3.1021-1030.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mindich L, Cohen J, Weisburd M. Isolation of nonsense suppressor mutants in Pseudomonas. J Bacteriol. 1976;126:177–182. doi: 10.1128/jb.126.1.177-182.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olkkonen V M, Bamford D H. Quantitation of the adsorption and penetration stages of bacteriophage φ6 infection. Virology. 1989;171:229–238. doi: 10.1016/0042-6822(89)90530-8. [DOI] [PubMed] [Google Scholar]
- 40.Olkkonen V M, Pekkala P M, Bamford D H. Monoclonal antibodies to the major structural proteins of bacteriophage φ6. Virology. 1988;165:317–320. doi: 10.1016/0042-6822(88)90693-9. [DOI] [PubMed] [Google Scholar]
- 41.Olsen R H, Siak J-S, Gray R H. Characteristics of PRD1, a plasmid-dependent broad host range DNA bacteriophage. J Virol. 1974;14:689–699. doi: 10.1128/jvi.14.3.689-699.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pansegrau W, Lanka E. Enzymology of DNA transfer by conjugative mechanisms. Prog Nucleic Acid Res. 1996;54:197–251. doi: 10.1016/s0079-6603(08)60364-5. [DOI] [PubMed] [Google Scholar]
- 43.Pansegrau W, Lanka E, Barth P T, Figurski D H, Guiney D G, Haas D, Helinski D, Schwab H, Stanisich V A, Thomas C M. Complete nucleotide sequence of Birmingham IncP plasmids: compilation and comparative analysis. J Mol Biol. 1994;239:623–663. doi: 10.1006/jmbi.1994.1404. [DOI] [PubMed] [Google Scholar]
- 44.Poranen, M. M., R. Daugelavičius, P. M. Ojala, and D. H. Bamford. A novel virus-host cell membrane interaction; membrane voltage dependent endocytic-like entry of bacteriophage φ6 nucleocapsid. J. Mol. Biol. 291:575–587. [DOI] [PMC free article] [PubMed]
- 45.Rydman, P. S., J. Caldentey, S. J. Butcher, S. D. Fuller, T. Rutten, and D. H. Bamford. Bacteriophage PRD1 contains a five-fold receptor-binding structure similar to that of adenovirus. Submitted for publication. [DOI] [PubMed]
- 46.Salas M. Protein-priming of DNA replication. Annu Rev Biochem. 1991;60:39–71. doi: 10.1146/annurev.bi.60.070191.000351. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 48.Savilahti H, Bamford D H. Linear DNA replication: inverted terminal repeats of five closely related Escherichia coli bacteriophages. Gene. 1986;49:199–205. doi: 10.1016/0378-1119(86)90280-5. [DOI] [PubMed] [Google Scholar]
- 49.Savilahti H, Caldentey J, Lundström K, Syväoja J E, Bamford D H. Overexpression, purification, and characterization of Escherichia coli bacteriophage PRD1 DNA polymerase. In vitro synthesis of full-length PRD1 DNA with purified proteins. J Biol Chem. 1991;266:18737–18744. [PubMed] [Google Scholar]
- 50.Stewart P L, Fuller S D, Burnett R M. Different imaging of adenovirus: bridging the resolution gap between X-ray crystallography and electron microscopy. EMBO J. 1993;12:2589–2599. doi: 10.1002/j.1460-2075.1993.tb05919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stewart P L, Ghosh S, Bamford D H, Burnett R M. Crystallization of the major coat protein of PRD1, a bacteriophage with an internal membrane. J Mol Biol. 1993;230:349–352. doi: 10.1006/jmbi.1993.1148. [DOI] [PubMed] [Google Scholar]
- 52.van Oostrum J, Burnett R M. Molecular composition of the adenovirus type 2 virion. J Virol. 1985;56:439–448. doi: 10.1128/jvi.56.2.439-448.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walin L, Tuma R, Thomas G J, Jr, Bamford D H. Purification of viruses and macromolecular assemblies for structural investigations using a novel ion exchange method. Virology. 1994;201:1–7. doi: 10.1006/viro.1994.1259. [DOI] [PubMed] [Google Scholar]
- 54.Waters V L, Strack B, Pansegrau W, Lanka E, Guiney D G. Mutational analysis of essential IncP plasmid transfer genes traF and traG and involvement of traF in phage sensitivity. J Bacteriol. 1992;174:6666–6673. doi: 10.1128/jb.174.20.6666-6673.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zechner, E. L., F. de la Cruz, R. Eisenbrandt, A. M. Grahn, G. Koraimann, E. Lanka, G. Muth, W. Pansegrau, C. M. Thomas, C. M. Wilkins, and M. Zatyka. Conjugative DNA transfer processes. In C. M. Thomas (ed.), The horizontal gene pool: bacterial plasmids and gene spread, in press. Harwood Academic Publishers GmbH, Amsterdam, The Netherlands.