Abstract
Lipids are a naturally occurring group of molecules that not only contribute to the structural integrity of the lung preventing alveolar collapse but also play important roles in the anti-inflammatory responses and antiviral protection. Alteration in the type and spatial localization of lipids in the lung plays a crucial role in various diseases, such as respiratory distress syndrome (RDS) in preterm infants and oxidative stress-influenced diseases, such as pneumonia, emphysema, and lung cancer following exposure to environmental stressors. The ability to accurately measure spatial distributions of lipids and metabolites in lung tissues provides important molecular insights related to lung function, development, and disease states. Nanospray desorption electrospray ionization (nano-DESI) and other ambient ionization mass spectrometry techniques enable label-free imaging of complex samples in their native state with minimal to absolutely no sample preparation. However, lipid coverage obtained in nano-DESI mass spectrometry imaging (MSI) experiments has not been previously characterized. In this work, the depth of lipid coverage in nano-DESI MSI of mouse lung tissues was compared to liquid chromatography tandem mass spectrometry (LC-MS/MS) lipidomics analysis of tissue extracts prepared using two different procedures: standard Folch extraction method of the whole lung samples and extraction into a 90% methanol/10% water mixture used in nano-DESI MSI experiments. A combination of positive and negative ionization mode nano-DESI MSI identified 265 unique lipids across 20 lipids subclasses and 19 metabolites (284 in total) in mouse lung tissues. Except for triacylglycerols (TG) species, nano-DESI MSI provided comparable coverage to LC-MS/MS experiments performed using methanol/water tissue extracts and up to 50% coverage in comparison with the Folch extraction-based whole lung lipidomics analysis. These results demonstrate the utility of nano-DESI MSI for comprehensive spatially resolved analysis of lipids in tissue sections. A combination of nano-DESI MSI and LC-MS/MS lipidomics is particularly useful for exploring changes in lipid distributions during lung development, as well as resulting from disease or exposure to environmental toxicants.
Graphical Abstract
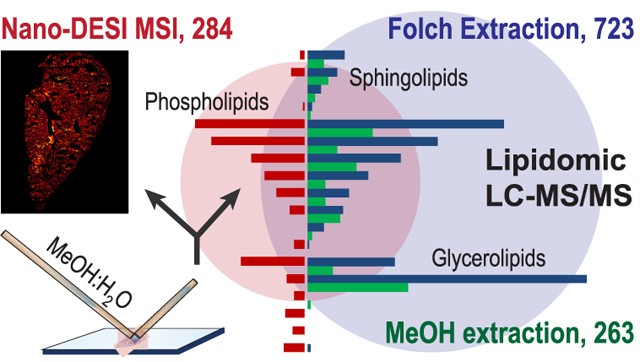
Lipids are naturally occurring biomolecules that play an important role in many cellular processes, including but not limited to membrane dynamics, energy balance, and signal transduction.1 Lipids are the major component of the pulmonary surfactant and, as such, are critical to both developing a healthy lung and preventing lung disease.2 For example, lipids are known to be involved in both inflammation3 and antiviral protection4 of lungs. Many lung diseases, including respiratory distress syndrome (RDS) in preterm infant,5 cystic fibrosis,6 lung cancer,7 pneumonia,8 and asthma,9 are associated with changes both in lipid composition and their spatial localization in the lung tissue.10 Therefore, it is important to thoroughly characterize the spatial distribution of lipids in lungs to further understand lung function, development, and disease states.
Conventional methods, such as high-performance liquid chromatography (HPLC), gas chromatography (GC), and thin-layer chromatography (TLC), followed by mass spectrometry (MS), have been extensively used for the identification of lipids in biological tissues.11−13 However, these techniques do not provide information on their spatial distribution. Mass spectrometry imaging (MSI), on the other hand, enables spatial localization of molecules in biological systems.14−19 Matrix-assisted laser desorption/ionization (MALDI)20 and desorption electrospray ionization (DESI)21 are by far the most widely used ionization techniques in MSI applications. Of particular interest to this study is MALDI imaging of lipids in mouse lung reported by Berry et al.,22 in which spatial distributions of phospholipids containing arachidonic and docosahexaenoic acids were examined. Despite the popularity of MSI techniques, they are often criticized for the relatively low depth of coverage in comparison with LC-MS and LC-MS/MS. Lipidome coverage in MALDI MSI of biological tissues has been examined by Wang et al.23,24 In these studies, best coverage of the lipidome was achieved using hydroxyflavone-based MALDI matrices, which enabled imaging of up to 212 lipids in rat brain23 and 555 unique lipids and metabolites in porcine adrenal gland.24
Alternatively, ambient liquid extraction mass spectrometry techniques such as DESI25 and nanospray desorption electrospray ionization (nano-DESI)26 have been extensively used for imaging of complex biological samples in their native state with minimal sample pretreatment.27−29 Similar to DESI, tissue imaging by nano-DESI MSI is of growing interest for lipidomics analysis since it provides both relative abundances and spatial localization of a large number of lipid species.30 and compensates for matrix effects.31 Furthermore, solvent composition may be adjusted to facilitate extraction of different lipids from tissue samples.32,33 Typical nano-DESI MSI experiments are performed using a mixture of methanol or acetonitrile with water with or without acid. Ionization of selected classes of compounds can be enhanced by adding cationization reagents to the solvent. For example, silver ions have been shown to dramatically enhance the ionization efficiency and facilitate quantitative imaging of prostaglandins, which are difficult to observe using pure solvents.34 In addition, reactive analysis has been used by several groups to enhance lipid coverage in ambient liquid extraction MSI experiments. In particular, charge labeling in reactive DESI using betaine aldehyde as a reagent enabled imaging of cholesterol in rat brain tissues.35 Furthermore, dicationic pairing compounds have been used to generate positive ions of lipids that are typically observed as negative ions thereby enhancing lipid coverage in positive mode.36 Despite the success of these studies, lipid coverage obtained in liquid extraction MSI experiments has not been previously characterized.
In this study, we compare lipid coverage in nano-DESI MSI experiments with the results obtained using a traditional lipidomics workflow. Specifically, nano-DESI MSI of lipids in mouse lung samples was performed using a commonly used extraction solvent composed of 90% methanol (MeOH) and 10% water (H2O). Lipid coverage in nano-DESI MSI was then accessed by comparing accurate m/z values observed in imaging experiments to the mass list generated by LC-MS/MS of identified lipids extracted from an adjacent lung section using both the standard Folch extraction as well as extraction in 90% MeOH and 10% H2O. The results indicate reasonably efficient lipid extraction from tissues sections using a 9:1 MeOH: H2O mixture with about 40% coverage in comparison with the standard LC-MS/MS lipidomics experiment. We demonstrate that a majority of the observed differences in lipid coverage in nano-DESI MSI and LC-MS/MS using MeOH: H2O as an extraction solvent are attributed to the inefficient extraction of triacylglycerols by this solvent system and low ionization efficiency of this lipid class. Our study indicates that nano-DESI MSI experiments provide reasonable coverage of a majority of lipid classes in lung tissues and highlights the need for the development of improved extraction and ionization approaches for obtaining deep lipid coverage in liquid extraction-based MSI experiments.
MATERIALS AND METHODS
Tissue Collection and Handling.
All mice used in this investigation were housed in the Cincinnati Children’s Hospital Medical Center Animal Care Facility according to National Institutes of Health and institutional guidelines for the use of laboratory animals. All experimental protocols were reviewed and approved by Cincinnati Children’s Hospital Research Foundation Institutional Animal Care and Use Committee. C57BL/6 mice from JAX Mice (Jackson Laboratory, Bar Harbor, ME, USA) were sacrificed at postnatal day 28 by CO2 overdose. Lungs were collected, cleaned, embedded in carboxymethyl cellulose (CMC) and stored in a −80 °C freezer. Samples were sectioned into 10 μm thick slices with a Thermo CryoStar NX70 (Thermo Scientific, Waltham, MA) microtome to generate coronal sections of the lungs and were thaw-mounted onto regular glass slides for imaging experiments and stored at −80 °C. Tissue sections were allowed to thaw at room temperature prior to nano-DESI MSI experiments. Adjacent sections were stored in 1.5 mL glass vials at −80 °C for LC-MS/MS analysis.
Methanol Extraction Method.
Methanol extraction of sectioned lung was performed by adding 50 μL of MeOH: H2O (9:1) mixture to ice chilled samples. The samples were well mixed and the debris was removed by centrifugation (10 000g, 10 min, 4 °C). The extract solution was isolated and ready for LC-ESI-MS/MS analyses. A total of 18 lung sections—3 mice (biological replicates) × 3 sections (technical replicates) × 2 ionization modes (positive and negative mode)—were analyzed using this method.
Lipids Analysis by LC-MS/MS.
LC-ESI-MS/MS analyses employed a Waters Aquity UPLC H class system interfaced with a Velos-ETD Orbitrap mass spectrometer (Thermo Fisher Scientific, Waltham, MA). Lipids were separated using a Waters CSH column (3.0 mm × 150 mm x 1.7 μm particle size) over a 34 min gradient (mobile phase A, acetonitrile/ water (40:60) containing 10 mM ammonium acetate; mobile phase B, acetonitrile/isopropanol (10:90) containing 10 mM ammonium acetate) at a flow rate of 250 μL/min.37 An additional section was also analyzed using a Waters HSS T3 column (1.0 mm × 150 mm × 1.8 μm particle size), over a 90 min gradient at a flow rate of 30 μL/min.13 MS analysis of eluting lipids was performed using electrospray ionization in both positive and negative modes with a mass range of 200− 2000 m/z and mass resolving power of 60 000. Tandem mass spectrometry (MS/MS) experiments were performed using HCD (higher-energy collision dissociation) and CID (collision-induced dissociation). In these experiments, a full scan MS event in the Orbitrap was followed by MS/MS of the top 4 ions, alternating between HCD (Orbitrap) and CID (ion trap) using normalized collision energy (NCE) of 35 and 30 arbitrary units, respectively, with an isolation width of 2 and dynamic exclusion of 8 s.
Lipid Data Processing.
LC-MS/MS raw data files were analyzed using LIQUID (Lipid Quantitation and Identification).37 Briefly, fragment ions and associated acyl chain fragment information from tandem mass spectra, the mass measurement error, isotopic profile, extracted ion chromatogram, and retention time of each lipid precursor ion were examined for confident lipid identifications.
Nano-DESI Imaging.
Imaging experiments were performed on an LTQ/Orbitrap XL mass spectrometer (Thermo Fisher Scientific, Waltham, MA) using a custom designed nano-DESI source38 equipped with a shear force microscopy capability.39 Two fused silica capillaries (OD 150 μm × ID 50 μm) were used to assemble the nano-DESI probe. A primary capillary propels the extraction solvent to the sample; a secondary capillary removes the extracted analyte molecules and transfers them to a mass spectrometer inlet. A third capillary (200 × 800 μm, ID × OD) with a 20 μm OD tip serves as a shear force probe, which maintains constant distance between the sample and the nano-DESI probe. This capillary was pulled using a laser-based micropipette puller (P-2000, Sutter Instrument, Novato, CA). Two small piezoelectric plates (3 × 3 × 0.55 mm, Steiner & Martins, Inc. Doral, FL) are attached to the shear force probe. The first plate induces low-amplitude vibration using a sinusoidal waveform supplied by a function generator (Agilent 33220A, Agilent Technologies, Santa Clara, CA). The second plate positioned closer to the sample surface detects the tip oscillation amplitude. The height of the sample is adjusted on the fly based on the amplitude of a resonant oscillation at a selected frequency measured by a lock-in amplifier (Model SR865, Stanford Research Systems, Sunnyvale, CA). The measured amplitude is used to provide positive feedback to the sample positioning XYZ stage controlled by a custom-designed Labview program. The extraction solvent described later in the text was delivered at a flow rate of 500 nL/min. Ionization was achieved by applying a 3.5 kV potential to the syringe needle. The heated capillary inlet was held at 30 V and 250 °C. The capillaries (primary, secondary, and shear-force probe) were positioned using high-resolution micromanipulators (XYZ 500MIM, Quater Research and Development, Bend, OR); two Dino-Lite digital microscopes (AnMo Electronics Corporation, Sanchong, New Taipei, Taiwan) were used to guide the positioning.
Nano-DESI MSI experiments were performed using one of the most commonly used solvent systems, MeOH: H2O 9:1, spiked with internal standards listed in Table S1. The internal standards, which do not interfere with the endogenous lipids and metabolites extracted from the lung tissue, were selected to enable normalization of lipid and metabolite signals as described in previous studies.30,40,41 We confirmed that lipid molecules used as internal standards were not observed in the LC-MS/MS of mouse lung tissue. In positive mode, the constructed images were normalized to the internal standards including a deuterated metabolite (acetylcholine-1,1,2,2-d4) and two odd-chain phospholipids (Lyso-phosphatidylcholine, LPC 19:0, and phosphatidylcholine PC(12:0/13:0)), For negative mode experiments, the images were normalized to standards including deuterated fatty acids (oleic acid-d17 and arachidonic acid-d8) and four odd-chain phospholipids (phosphatidylethanolamine, PE(15:0/15:0), phosphatidylglycerol, PG(15:0/15:0), phosphatidylinositol, PI(16:0/16:0), and phosphatidylserine, PS(17:0/17:0)). Nano-DESI MSI was performed with a spatial resolution of ~100 μm in both positive and negative ionization modes. Representative mass spectra obtained in positive and negative modes are shown in Figure S1. Compounds detected in both ionization modes were combined and identified based on the LC-MS/MS lipidomics data, MS/MS directly from the tissue, accurate mass matching using METLIN42 and LIPID MAPS,43 and comparison with the literature. Of note is that when multiple isoforms or compound identities match one m/z peak, only one of the matches is reported.
Data Collection and Image Processing.
Nano-DESI MSI experiments were performed by scanning line by line the XYZ stage holding the sample under the shear force nano-DESI probe at a constant velocity of 70 μm/s, while acquiring high-resolution mass spectra (m/Δm = 60 000 at m/z 400). The spacing between lines was 100 μm. A contact closure signal was used to synchronize the XYZ stage and Xcalibur acquisition software. Molecules observed in MSI experiments were identified using accurate mass measurement and LC-MS/MS experiments. Mass spectral data were subsequently processed using MSI QuickView,44 a visualization software developed at PNNL. A total of 18 lung sections-3 mice (biological replicates) × 3 sections (technical replicates) × 2 ionization modes (positive and negative mode)-were examined by nano-DESI MSI.
RESULTS AND DISCUSSION
In this study, we examined the spatial localization of lipids and metabolites in mouse lung tissue sections, analyzed common trends in the localization of different lipid classes in the tissue, and assessed lipid coverage by comparing the number of lipid species observed in nano-DESI MSI experiments with the results of LC-MS/MS analysis of tissue homogenates. Although lipid hydrolysis during the analysis cannot be ruled out, previous studies have demonstrated that nano-DESI is more gentle toward chemically labile molecules than ESI.45 This was attributed to the short time the extracted molecules reside in the solvent in comparison with traditional liquid extraction approaches. Furthermore, numerous experiments with both lipid standards and complex lipid mixtures performed in our laboratory indicate that nano-DESI detects intact lipid species in biological samples.
Nano-DESI MSI of Mouse Lung Tissue.
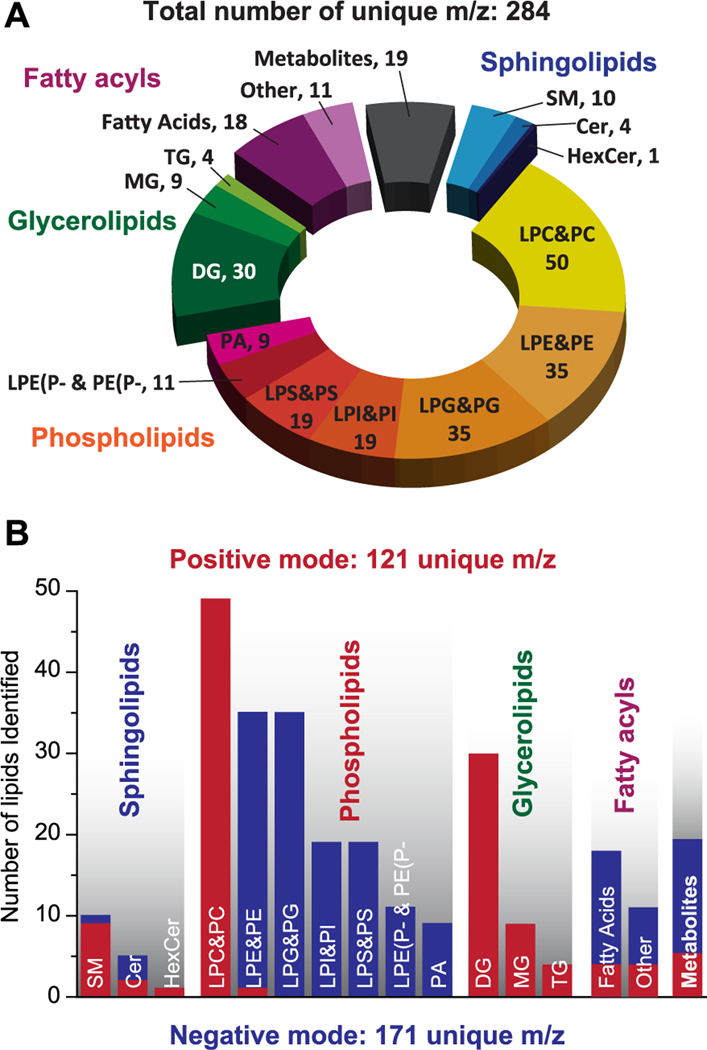
Using nano-DESI MSI, we have successfully detected 263 unique lipids across 20 lipids subclasses and 19 metabolites for a total of 284 identifications (Figure 1A and 1B and Table S2). Of the 284 identified compounds, 121 were observed in positive mode, 171 were observed in negative mode, and 8 were found in both modes. Sample positive and negative mass spectra of lung tissue are shown in Figures S1 and S2.
Figure 1.
All lipids and metabolites detected by the Nano-DESI MSI method on lung tissue sections using extraction solvent system of MeOH: H2O 9:1. (A) Pie chart of 282 unique endogenous compounds identified from both positive and negative ionization mode, which have been divided into different lipid classes and subclasses. (B) Break down of compounds detected in positive (red) and negative ionization mode (blue).
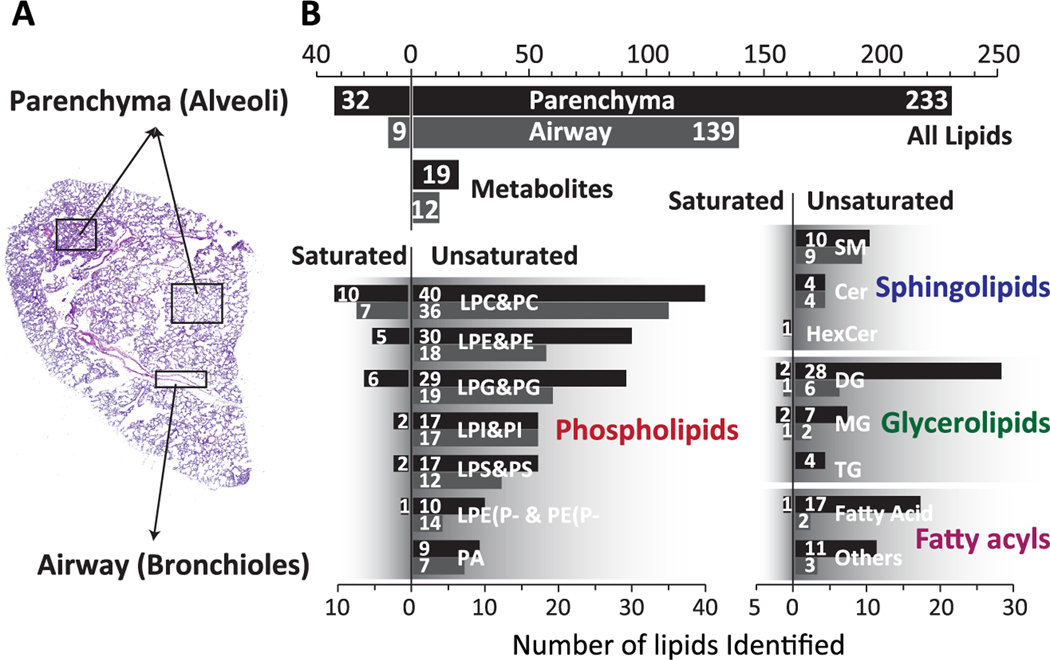
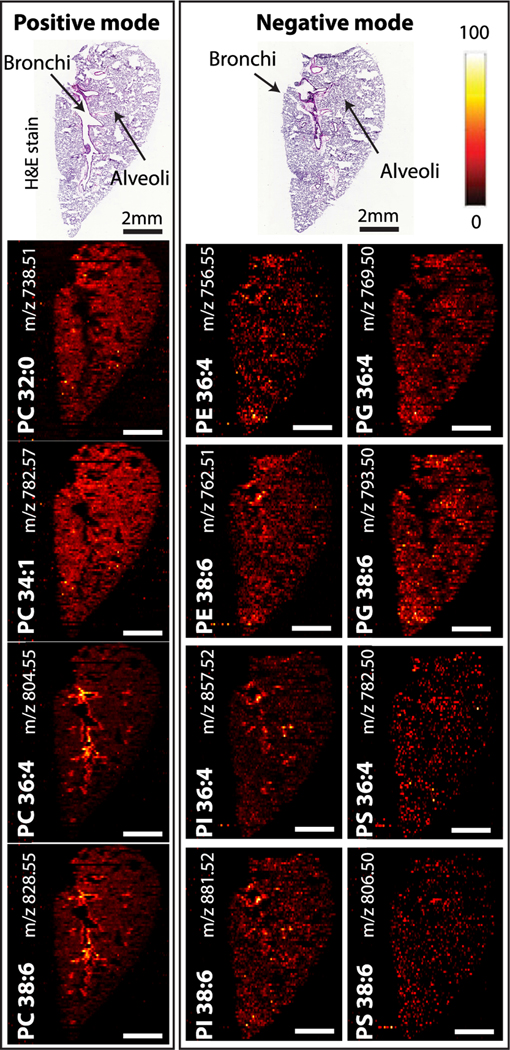
The global distribution of lipids across the lung tissue revealed differences in the lipidome between the alveolar region and airway lining (Figure 2). All of the detected species including 233 unsaturated lipids, 32 saturated lipids, and 19 metabolites show signal in alveolar regions, while approximately only half of them (139 unsaturated lipids, 9 saturated lipids, and 12 other metabolites, total 160) appear enhanced at the airway linings (bronchioles). We found that almost 60% of unsaturated lipids and only 28% of saturated lipids are localized to the bronchioles indicating a pronounced selectivity toward unsaturated lipids in this region of the lung tissue. This selectivity varies among different classes of lipids. For example, glycerolipids (39 in alveoli, 8 in bronchioles) and fatty acyls (28 in alveoli, 5 in bronchioles) are preferentially localized to alveoli. In contrast, phospholipids (152 in alveoli, 113 in bronchioles) and sphingolipids (14 in alveoli, 13 in bronchioles) are more evenly distributed between the airways and alveolar regions. Ion images in Figure 3 indicate that PC, PE, and PI lipid species containing polyunsaturated fatty acids (PC(36:4), PC(38:6), PE(38:6), PI(36:4), and PI(38:6)) are enhanced at the airway while PG and PS containing the same number of fatty acid carbons and double bonds are evenly distributed in the lung parenchyma. A complete list of all the lipid and metabolite ion images observed in the nano-DESI MSI of lung tissue is presented in Table S2.
Figure 2.
Distribution bar graph of imaged lipids (saturated and unsaturated) and metabolites in different anatomical features of mouse lung. (A) All lipids and metabolites detected in airway (bronchioles) and lung parenchyma (alveoli). (B, C) Distributions of different lipid classes in lung.
Figure 3.
Selected ion images acquired over the lung tissue section of phospholipids from both positive and negative ionization modes using shear force microscopy nano-DESI MSI. H&E stained image of a mouse lung tissue section and the ion maps of sodiated PCs detected in the positive ionization mode. The localization of selected PE, PG, PI, and PS containing polyunsaturated fatty acids identified by the negative mode. Scale bars are 2 mm. Intensity scale bar (red hot) ranges from 0% (black) to 100% (white) signal intensity of an individual peak.
These observations are consistent with previous studies.22,46 For example, Berry et al. examined phospholipid distributions in adult mouse lung tissue using MALDI MSI. They discovered that PC, PE, and PI species containing arachidonic (FA 20:4) and docosahexaenoic (FA 22:6) are preferentially localized along the airways. Similarly, our previous study indicated tight localization of polyunsaturated species, including LPC 20:4, LPE 22:6, PC 36:4, and PC 38:6 along the airways.46 The enhanced abundance of polyunsaturated phospholipids and fatty acids along the airways has been attributed to their role as precursors to lipid mediators such as leukotrienes, prostaglandins, and resolvins involved in the lung response to inflammation3,47,48 and pulmonary diseases.49 In contrast, a majority of PC containing more saturated fatty acids (e.g., PC 32:0, PC 34:1) and several PG species (e.g., PG 32:0, PG 32:1) are uniformly distributed across the lung parenchyma indicating alveolar localization. These phospholipids are major components of lung surfactant which is synthesized, stored, and secreted in the alveolar region to prevent the lung from collapsing.
Comparison of Lipid Coverage in Nano-DESI MSI with LC-MS/MS Lipidomics.
One of the goals of this study was to evaluate the depth of lipid coverage in nano-DESI MSI by comparing lipids in mouse lung tissues observed in these experiments with the whole lung lipidome analysis. In particular, LC-MS/MS analyses were performed on lipids extracted from lung tissues using both a standard Folch extraction procedure and extraction using a 9:1 MeOH: H2O mixture employed in nano-DESI MSI experiments. Whole lung lipidome analysis revealed a total of 924 unique lipid species (723 LC-MS/MS peaks) from 3 lipid categories and 21 lipid subclasses with the greatest lipid subclass representation corresponding to triacylglycerol (TG, 253 lipids) followed by PC (128 lipids) and PG (105 lipids) subclasses.13 In comparison, LC-MS/MS analysis of tissue extracts obtained using MeOH: H2O 9:1 as an extraction solvent identified 263 LC-MS peaks including 332 unique lipid species across 19 subclasses. TG (91 lipids), LPC + PC (51 lipids), and LPE + PE (36 lipids) being the most prominent subclasses.
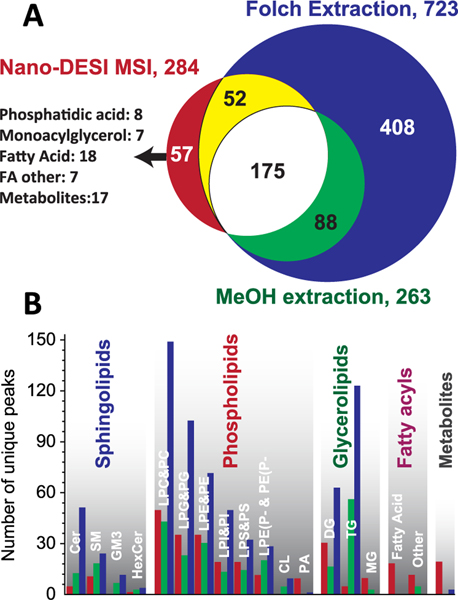
Several factors affect the extraction efficiency of lipids in nano-DESI MSI experiments, including solvent composition, residence time of the probe in a specific location on the sample, and thickness of the tissue section. Furthermore, ionization efficiency and matrix effects known to be particularly important in the absence of separation may dramatically alter ion signals observed experimentally. Collectively, these factors may affect lipid coverage in nano-DESI MSI experiments. For the purpose of comparison between nano-DESI MSI and LC-MS/MS-based lipidomics methods, we have to consider the fact that LC-MS/MS methods identify isomeric and isobaric compounds that cannot be resolved by high resolution mass spectrometry (m/Δm = 60 000 at m/z 400) alone without MS/MS. Assuming that all the isomeric and isobaric compounds identified in LC-MS/MS lipidomics methods contribute to ion signals observed in nano-DESI MSI, we estimate that the observed nano-DESI MSI features may be attributed to 443 unique lipids and 19 nonlipid metabolites (462 in total). This is clearly an upper limit estimate as it is likely that not all of the lipids identified using LC−MS/MS are observed in nano-DESI MSI. We note that our previous MS/MS imaging experiments clearly demonstrated the presence of several isomeric and isobaric species in each isolated m/z window in nano-DESI MSI of mouse uterine sections.50 It is reasonable to assume that the presence of such overlapping species also occurs in imaging of mouse lung tissues. Although it is likely that the number of species observed in nano-DESI MSI experiments is greater than 284 (the number of distinct m/z features observed experimentally) and close to 462 (the estimated number of lipid species by comparison with LC-MS/MS), we cannot unambiguously determine that number. Thus, in order to make a fair comparison between the three methods, we used the number of unique peaks identified by MS rather than the estimated number of lipid species. In comparison to 284 unique peaks observed by nano-DESI MSI, 723 and 263 were identified by LC-MS/MS methods using Folch and MeOH:H2O extraction, respectively. Detailed analysis of the experimental data was performed to obtain a better understanding of the differences in coverage observed in nano-DESI MSI and LC-MS/MS experiments as described below.
As expected, the best lipid coverage was obtained using Folch extraction with the other two methods containing a subset of the total lipid content (Figure 4). Although LC-MS/MS methods identified several small molecules, there is a subset of molecules detected by nano-DESI MSI (57 compounds, including 8 PAs, 7 MGs, 25 FAs, and 17 nonlipid metabolites, colored red) that were absent in the LC-MS/MS lipidomics data. Overall, there are 175 compounds detected by all three methods (white), 52 compounds observed both in nano-DESI MSI and Folch extraction (yellow), and 88 compounds observed in both LC-MS/MS data sets but not in nano-DESI MSI (green). Figure 4B shows a detailed comparison of different lipid classes observed in the three types of experiments. In all three methods, phospholipids were found to be the largest identified class followed by glycerolipids and sphingolipids. In comparison with LC-MS/MS, nano-DESI MSI using MeOH:H2O provides adequate coverage for almost all of the lipid subclasses with the exception of some low-abundance large lipid molecules in negative mode, such as gangliosides (GM3) and cardiolipins (CL). This is likely due to the low extraction efficiency of these lipids in MeOH: H2O. Indeed, although GM3 and CL species have been detected in the conventional lipidomics experiment using Folch extraction (11 GM3s, 14 CLs), they are less abundant in the LC-MS/MS data obtained using MeOH: H2O extraction (6 GM3s, 4 CLs). It is remarkable that lipid coverage in nano-DESI MSI data was comparable to that observed in the LC-MS/MS analysis of the MeOH: H2O tissue extract (284 vs 263 species). Unsurprisingly, both methods using MeOH: H2O as extracting solvents determine only about 40% that of conventional whole lung lipidomics method (284 and 263 vs 723 entities, respectively). It is not only due to the solvents used but also due to the fact that a substantially larger amount of the homogenized lung tissue (a whole lung) was used for the Folch method while only a 10 μm-thick slice was used for the other two methods. The smaller number of phospholipids identified in LC-MS/MS of the MeOH: H2O tissue extract in comparison with nano-DESI MSI using the same solvent may be attributed to greater analyte loses in the LC-MS/MS experiment.
Figure 4.
Comparison of endogenous compounds extraction between nano-DESI MSI (red) and LC-MS/MS lipidomic methods using MeOH: H2O extraction solvents (green) and Folch extraction method (blue). (A) Venn diagram of lipids and metabolites identified by three methods. (B) Side by side comparison of detected compounds in lipid classes and subclasses.
Interestingly, although TG was an abundant subclass in LC-MS/MS data, only few low-abundance TG species were observed in nano-DESI MSI. This result is in agreement with a previous report, which demonstrated lower extraction efficiency of TG species into MeOH: H2O.43 We propose that the ionization efficiency of TG species also plays an important role in the low detection with nano-DESI. More efficient ionization of TG species in LC-MS/MS experiments may be attributed to their complexation with NH4+ in ammonium acetate doped LC solvents in comparison with ionization by sodium or potassium adduct formation in nano-DESI MSI. More efficient detection of TG species in nano-DESI MSI experiments may be achieved by adding ionization enhancers such as silver nitrate,51 ammonium formate,52 or a dicationic reagent53 as described previously. For example, efficient ionization of TG species in canine bladder tissues in DESI experiments has been previously achieved by adding silver nitrate to the spraying solvent,51 while dicationic reagents has been shown to selectively enhance the detection efficiency of TG species in tissue imaging using a single probe.53 Although we did see some improvement in TG signals when using suggested solvent compositions, the overall lipid coverage was actually worse than methanol/water solvent system. A thorough optimization of the solvent composition is necessary to obtain a more comprehensive TG coverage and improve the overall lipid coverage. Collectively, our results indicate that nano-DESI MSI experiments provide reasonable coverage of lipids in tissue sections and identify pathways for further improving the performance of these experiments.
CONCLUSIONS
In this study, we presented a detailed comparison between lipids observed in the whole lung lipidomics LC-MS/MS data and nano-DESI MSI experiments of mouse lung tissue sections. Our results demonstrated that imaging experiments performed with a spatial resolution of 100 μm provide good coverage of lipids while also informing on their spatial localization in tissues. Lipid coverage of close to 50% was obtained in nano-DESI MSI in comparison with the widely used Folch extraction from tissue homogenates followed by LC-MS/MS analysis. These results are very encouraging considering the fact that nano-DESI MSI is a direct sampling method, which relies on extraction of analyte molecules from areas smaller than 100 μm × 100 μm followed by ionization of the extracted molecules without prior separation. The latter is known to result in signal suppression during ionization, which is detrimental to the analysis of compounds with low ionization efficiency. Aside from lipid species, nano-DESI MSI provided insights into the spatial localization of small molecules including FAs and nonlipid metabolites that are not observed in LC-MS/MS lipidomics experiments. Although lipid extraction in nano-DESI MSI experiments using MeOH: H2O as an extraction solvent in fairly efficient for many lipid subclasses, some lipid species were underrepresented in these experiments. Further optimization of the solvent composition is necessary for the efficient detection of TG, GM3, and CL. Finally, high sensitivity, high mass resolving power, and rapid separation prior to analysis using, for example, ion mobility spectrometry54,55 or structural characterization using MS/MS50 could be used to further improve the detection and identification of low abundance lipids along with isomeric and isobaric species.
Supplementary Material
ACKNOWLEDGMENTS
The research described in this paper is supported by Grant U01 HL122703 from the National Heart Lung Blood Institute of NIH (LungMAP). The research was conducted under the Laboratory Directed Research and Development Program at PNNL, a multiprogram national laboratory operated by Battelle for the U.S. Department of Energy (DOE) under Contract DE-AC05-76RL01830. The work was performed in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the U.S. Department of Energy and located at Pacific Northwest National Laboratory (PNNL) in Richland, WA.
Footnotes
ASSOCIATED CONTENTS
Supporting Information
The Supporting Information is available free of charge on the ACSPublicationswebsite at DOI: 10.1021/acs.analchem.9b02045.
Sample positive and negative mass spectra and a complete list of all the lipid and metabolite ion images observed in the nano-DESI MSI of lung tissue (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Hawthorne JN Techniques of Lipidology: Isolation, Analysis and Identification of Lipids, 2nd revised ed., International Congress Series; Elsevier, 2015; Vol. 16. [Google Scholar]
- (2).Ardini-Poleske ME; Clark RF; Ansong C; Carson JP; Corley RA; Deutsch GH; Hagood JS; Kaminski N; Mariani TJ; Potter SS; et al. Am. J. Physiol. Cell. Mol. Physiol. 2017, 313 (5), L733–L740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Stables MJ; Gilroy DW Prog. Lipid Res. 2011, 50 (1), 35–51. [DOI] [PubMed] [Google Scholar]
- (4).Numata M; Chu HW; Dakhama A; Voelker DR Proc. Natl. Acad. Sci. U. S. A. 2010, 107 (1), 320–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Avery ME Arch. Pediatr. Adolesc. Med. 1959, 97 (5_PART_I), 517. [Google Scholar]
- (6).Freedman SD; Katz MH; Parker EM; Laposata M; Urman MY; Alvarez JG Proc. Natl. Acad. Sci. U. S. A. 1999, 96 (24), 13995−14000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Hakomori S. Cancer Res. 1996, 56 (206), 5309–5318. [PubMed] [Google Scholar]
- (8).Ray NB; Durairaj L; Chen BB; McVerry BJ; Ryan AJ; Donahoe M; Waltenbaugh AK; O’Donnell CP; Henderson FC; Etscheidt CA; et al. Nat. Med. 2010, 16 (10), 1120–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Yoder M; Zhuge Y; Yuan Y; Holian O; Kuo S; van Breemen R; Thomas LL; Lum H. Allergy, Asthma Immunol. Res. 2014, 6 (1), 61–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Zemski Berry KA; Murphy RC; Kosmider B; Mason RJ J. Lipid Res. 2017, 58 (5), 926–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Goto-Inoue N; Hayasaka T; Taki T; Gonzalez TV; Setou MJ Chromatogr. A 2009, 1216 (42), 7096–7101. [DOI] [PubMed] [Google Scholar]
- (12).Paglia G; Ifa DR; Wu C; Corso G; Cooks RG Anal. Chem. 2010, 82 (5), 1744–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Dautel SE; Kyle JE; Clair G; Sontag RL; Weitz KK; Shukla AK; Nguyen SN; Kim Y-M; Zink EM; Luders T; et al. Sci. Rep. 2017, 7, 40555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Caprioli RM Proteomics 2016, 16 (11−12), 1607–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Caprioli RM J. Am. Soc. Mass Spectrom. 2015, 26 (6), 850–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Amstalden van Hove ER; Smith DF; Heeren RM A. J. Chromatogr. A 2010, 1217 (25), 3946–3954. [DOI] [PubMed] [Google Scholar]
- (17).Ifa DR; Wu C; Ouyang Z; Cooks RG Analyst 2010, 135 (4), 669–681. [DOI] [PubMed] [Google Scholar]
- (18).Cooks RG; Ouyang Z; Takats Z; Wiseman JM Science (Washington, DC, U. S.) 2006, 311 (5767), 1566–1570. [DOI] [PubMed] [Google Scholar]
- (19).Lietz CB; Gemperline E; Li L. Adv. Drug Delivery Rev. 2013, 65 (8), 1074–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Gessel MM; Norris JL; Caprioli RM J. Proteomics 2014, 107, 71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Takats Z; Wiseman JM; Gologan B; Cooks RG Science (Washington, DC, U. S.) 2004, 306 (5695), 471–473. [DOI] [PubMed] [Google Scholar]
- (22).Berry KAZ; Li B; Reynolds SD; Barkley RM; Gijón MA; Hankin JA; Henson PM; Murphy RC J. Lipid Res. 2011, 52 (8), 1551–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Wang X; Han J; Chou A; Yang J; Pan J; Borchers CH Anal. Chem. 2013, 85 (15), 7566–7573. [DOI] [PubMed] [Google Scholar]
- (24).Wang X; Han J; Pan J; Borchers CH Anal. Chem. 2014, 86 (1), 638–646. [DOI] [PubMed] [Google Scholar]
- (25).Swales JG; Tucker JW; Strittmatter N; Nilsson A; Cobice D; Clench MR; Mackay CL; Andren PE; Takáts Z; Webborn PJH; et al. Anal. Chem. 2014, 86 (16), 8473–8480. [DOI] [PubMed] [Google Scholar]
- (26).Roach PJ; Laskin J; Laskin A. Analyst 2010, 135 (9), 2233–2236. [DOI] [PubMed] [Google Scholar]
- (27).Venter AR; Douglass KA; Shelley JT; Hasman G; Honarvar E. Anal. Chem. 2014, 86 (1), 233–249. [DOI] [PubMed] [Google Scholar]
- (28).Badu-Tawiah AK; Eberlin LS; Ouyang Z; Cooks RG Annu. Rev. Phys. Chem. 2013, 64 (1), 481–505. [DOI] [PubMed] [Google Scholar]
- (29).Espy RD; Teunissen SF; Manicke NE; Ren Y; Ouyang Z; Van Asten A; Cooks RG Anal. Chem. 2014, 86 (15), 7712–7718. [DOI] [PubMed] [Google Scholar]
- (30).Lanekoff I; Thomas M; Laskin J. Anal. Chem. 2014, 86 (3), 1872–1880. [DOI] [PubMed] [Google Scholar]
- (31).Lanekoff I; Stevens SL; Stenzel-Poore MP; Laskin J. Analyst 2014, 139 (14), 3528–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Laskin J; Lanekoff I. Anal. Chem. 2016, 88 (1), 52–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Green FM; Salter TL; Gilmore IS; Stokes P; O’Connor G. Analyst 2010, 135 (4), 731–737. [DOI] [PubMed] [Google Scholar]
- (34).Duncan KD; Fang R; Yuan J; Chu RK; Dey SK; Burnum-Johnson KE; Lanekoff I. Anal. Chem. 2018, 90 (12), 7246–7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Wu C; Ifa DR; Manicke NE; Cooks RG Anal. Chem. 2009, 81 (18), 7618–7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Lostun D; Perez CJ; Licence P; Barrett DA; Ifa DR Anal. Chem. 2015, 87 (6), 3286–3293. [DOI] [PubMed] [Google Scholar]
- (37).Kyle JE; Crowell KL; Casey CP; Fujimoto GM; Kim S; Dautel SE; Smith RD; Payne SH; Metz TO Bioinformatics 2017, 33 (11), 1744–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Lanekoff I; Heath BS; Liyu A; Thomas M; Carson JP; Laskin J. Anal. Chem. 2012, 84 (19), 8351–8356. [DOI] [PubMed] [Google Scholar]
- (39).Nguyen SN; Liyu AV; Chu RK; Anderton CR; Laskin J. Anal. Chem. 2017, 89 (2), 1131–1137. [DOI] [PubMed] [Google Scholar]
- (40).Lanekoff I; Thomas M; Carson JP; Smith JN; Timchalk C; Laskin J. Anal. Chem. 2013, 85 (2), 882–889. [DOI] [PubMed] [Google Scholar]
- (41).Bergman HM; Lundin E; Andersson M; Lanekoff I. Analyst 2016, 141 (12), 3686–3695. [DOI] [PubMed] [Google Scholar]
- (42).Guijas C; Montenegro-Burke JR; Domingo-Almenara X; Palermo A; Warth B; Hermann G; Koellensperger G; Huan T; Uritboonthai W; Aisporna AE; et al. Anal. Chem. 2018, 90 (5), 3156–3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Fahy E; Subramaniam S; Murphy RC; Nishijima M; Raetz CRH; Shimizu T; Spener F; van Meer G; Wakelam MJO; Dennis EA J. Lipid Res. 2009, 50, S9–S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Thomas M; Heath BS; Laskin J; Li D; Liu E; Hui K; Kuprat AP; Kleese Van Dam K; Carson JP Proc. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. EMBS 2012, 2012, 5545–5548. [DOI] [PubMed] [Google Scholar]
- (45).Roach P; Laskin J; Laskin A. Anal. Chem. 2010, 82 (19), 7979–7986. [DOI] [PubMed] [Google Scholar]
- (46).Nguyen SN; Sontag RL; Carson JP; Corley RA; Ansong C; Laskin JJ Am. Soc. Mass Spectrom. 2018, 29 (2), 316–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Samuelsson B; Dahlén SE; Lindgren JÅ; Rouzer CA; Serhan CN Science (Washington, DC, U. S.) 1987, 237 (4819), 1171–1176. [DOI] [PubMed] [Google Scholar]
- (48).Epstein FH; Lewis RA; Austen KF; Soberman RJ N. Engl. J. Med. 1990, 323 (10), 645–655. [DOI] [PubMed] [Google Scholar]
- (49).Riccioni G; Bucciarelli T; Mancini B; Di Ilio C; D’Orazio N. Curr. Med. Chem. 2007, 14 (18), 1966–1977. [DOI] [PubMed] [Google Scholar]
- (50).Lanekoff I; Burnum-Johnson K; Thomas M; Short J; Carson JP; Cha J; Dey SK; Yang P; Prieto Conaway MC; Laskin J. Anal. Chem. 2013, 85 (20), 9596–9603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Jackson AU; Shum T; Sokol E; Dill A; Cooks RG Anal. Bioanal. Chem. 2011, 399 (1), 367–376. [DOI] [PubMed] [Google Scholar]
- (52).Byrdwell WC; Neff WE Rapid Commun. Mass Spectrom. 2002, 16 (4), 300–319. [DOI] [PubMed] [Google Scholar]
- (53).Rao W; Pan N; Tian X; Yang ZJ Am. Soc. Mass Spectrom. 2016, 27 (1), 124–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Bennett RV; Gamage CM; Galhena AS; Fernández FM Anal. Chem. 2014, 86 (8), 3756–3763. [DOI] [PubMed] [Google Scholar]
- (55).Kliman M; May JC; McLean JA Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2011, 1811 (11), 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.