Abstract
The majority of Kenya’s > 3 million camels have antibodies against Middle East respiratory syndrome coronavirus (MERS-CoV), although human infection in Africa is rare. We enrolled 243 camels aged 0–24 months from 33 homesteads in Northern Kenya and followed them between April 2018 to March 2020. We collected and tested camel nasal swabs for MERS-CoV RNA by RT-PCR followed by virus isolation and whole genome sequencing of positive samples. We also documented illnesses (respiratory or other) among the camels. Human camel handlers were also swabbed, screened for respiratory signs, and samples were tested for MERS-CoV by RT-PCR. We recorded 68 illnesses among 58 camels, of which 76.5% (52/68) were respiratory signs and the majority of illnesses (73.5% or 50/68) were recorded in 2019. Overall, 124/4692 (2.6%) camel swabs collected from 83 (34.2%) calves in 15 (45.5%) homesteads between April–September 2019 screened positive, while 22 calves (26.5%) recorded reinfections (second positive swab following ≥ 2 consecutive negative tests). Sequencing revealed a distinct Clade C2 virus that lacked the signature ORF4b deletions of other Clade C viruses. Three previously reported human PCR positive cases clustered with the camel infections in time and place, strongly suggesting sporadic transmission to humans during intense camel outbreaks in Northern Kenya.
Keywords: Middle East respiratory syndrome coronavirus, zoonosis, spillover events, MERS-CoV epidemiology, Horn of Africa
1. Introduction
Middle East respiratory syndrome coronavirus (MERS-CoV) is a betacoronavirus similar to two other recently emergent coronaviruses, severe acute respiratory syndrome coronavirus (SARS-CoV), and SARS-CoV-2, but different because it has a domestic animal reservoir that serves as the primary source of human infections [1,2]. First reported in humans in 2012, MERS-CoV infection causes mild to severe respiratory disease in humans with a case fatality rate of 35%, whereas in camels, the documented infections are mostly asymptomatic with rare cases of rhinorrhea, conjunctivitis, coughing/honking, or diarrhea [3,4,5,6]. As of March 2022, the World Health Organization (WHO) had confirmed more than 2600 human cases and 900 deaths across 27 countries globally, most of them (>80%) in the Middle East and Asia, and none in Africa [7,8]. The global MERS-CoV human case count has been declining since 2016. However, two MERS-CoV cases and one death have been reported in Qatar this year, with both cases reporting contact with camels [9].
Zoonotic transmission of MERS-CoV from camels to humans is often followed by limited human-to-human transmission, occasionally leading to larger chains of transmission in humans and resulting in large clusters of human cases, especially within health care settings [6,10]. Clinical MERS-CoV disease has not been reported in Sub Saharan Africa, where >80% of the world’s dromedary camel population are reared by pastoral communities living in arid regions. Apart from a report of three asymptomatic infections in Kenya and evidence of virus exposure (antibodies) among camel handlers, autochthonous transmission in humans has been rare, despite extensive human–camel contact and >80% MERS-CoV seroprevalence among adult camels [11,12,13,14,15]. This absence of human MERS-CoV disease in Africa may be due to immunological factors, or presence of a genetically different local virus that exhibits high intrinsic adaptation to camels but poor transmissibility or virulence in humans, or both [16,17].
Several groups have conducted community and hospital-based studies in the Horn of Africa (HOA) region, confirming high virus prevalence in camels, but not detecting synchronous infection or clinical disease in humans [11,12,18,19]. Three clades of MERS-CoV circulating in the Middle East, Asia, and Africa have been described, all antigenically similar with capacity to cross-neutralize but with discernable genetic variations in the spike, ORF3, and ORF4b genes [20,21]. Clade A and B viruses circulate predominantly in the Middle East and Asia, while clade C appears to be found primarily in Africa [20,21,22]. A recent study using ex vivo and in vivo inoculations suggested that the three clades exhibit geographical dominance and that clade C viruses may be less pathogenic than clades A and B [20,21]. To track the circulating variant of MERS-CoV, its transmission dynamics in camels, and investigate zoonotic transmission, we established and followed a prospective cohort of camel calves and their handlers in Northern Kenya.
2. Materials and Methods
2.1. Study Site, Design, and Enrollment
The study was conducted in Marsabit county in Northern Kenya among communities bordering Ethiopia—an arid region where nomadic pastoral livestock production systems are the main economic activity [23,24]. Inhabitants keep camels, goats, cattle, and sheep, and >80% of the homesteads, often comprising several households, own or manage camels in a herd and share communal resources such as grazing grounds and watering points [25]. The camel population outnumbers humans by 18 to 1 among camel-keeping households [26]. Camel herds are owned or managed by one household or homestead and are usually reared in the ‘home’ herd (consisting of young, pregnant, or nursing camels kept close to the homestead) and the ‘fora’ herd (consisting of the rest of the adult camels herded far away from the homestead) [25].
We identified camel-keeping homesteads within a 50-km radius from Marsabit town through key informant interviews. A homestead was defined as a grouping of dwellings that jointly owned and reared camels in a single herd. Recruitment of camel calves aged 0–24 months was conducted on a rolling basis over the study period. Newborns and calves ≤6 months were preferentially enrolled to increase chances of detecting primary infections before development of immunity from natural infection, as has been shown in other camel studies [27]. Concurrently, we enrolled compound residents who reported any contact with enrolled camels through activities such as milking, herding, feeding, cleaning barns, slaughtering, or grooming camels (this group of people is referred to here as “camel handlers”) within the enrolled homesteads.
2.2. Sample Size
To calculate the sample size of the camel cohort, we assumed a MERS-CoV exposure of 77% among infant camels, an annual attack rate of 15%, with a power of 80% at the 95% confidence interval [25,28]. We also accounted for an estimated 40% loss to follow-up during the study. No power calculation was done for the camel handlers and individuals >1 year of age who had contact with enrolled camels [15].
2.3. Enrollment, Follow-Up of Camel and Human Cohort and Data Collection
Trained animal health assistants or nurses conducted enrollment and follow-up visits. We used pretested questionnaires on Android® tablets running RedCap® application to collect camel and human data during enrollment and follow-up visits. At enrollment the herd-owner and animal handler were consented, and a baseline questionnaire administered to collect social demographic characteristics of the homestead. We collected data on camel herd structure, herding practices (nomadic or sedentary), and herd mixing while watering or grazing. For each enrolled camel, we captured details such as age, sex, any current illness, if the camel belonged to the ‘home’ herd or the ‘fora’ herd, and we then ear-tagged the calves for ease of follow-up. Camel illness data, collected without knowledge of any infection status, was classified as either a respiratory illness sign (if the camel had nasal discharge, sneezing, cough/honking, or hyper-lacrimation) or other illness sign (if the camel had tick infestation, skin and mouth lesions, lethargy, weight loss, anorexia, or diarrhea). Physical injuries in camels were not documented.
Subsequently, a homestead was visited twice monthly and data on clinical illness in camels was collected in a structured questionnaire, followed by collection of deep nasal swabs from each camel. A follow-up was considered successful when a camel was swabbed, and clinical signs were documented by study staff. Complete follow-up was defined as calves that were available for all their study visits while intermittent follow-up was defined as instances where calves missed some study visits after enrollment but were available for final sample collection at the end of the study period. Loss to follow-up (LTFU) included camels that were not traced for the final sample collection at the end of the follow-up period.
Trained study nurses also collected nasopharyngeal and oropharyngeal swabs from enrolled camel handlers monthly, and when participants called the study toll-free-line to report signs of respiratory illness such as cough, fever, running nose, nasal stuffiness, sore throat, or chest pain. Detailed enrollment and follow-up procedures of camel handlers were as previously described [15].
Data was stored temporarily in RedCap® which was backed up daily in cloud servers. Periodic reports were compiled and shared with the study team and the Ministry of Health.
2.4. Sample Collection, Shipping, and Storage
A Dacron swab (Puritan®) was used to swab both nostrils of each camel and was immediately placed in cryovials containing viral transport medium. For the camel handlers, nasopharyngeal (NP) and oropharyngeal (OP) swabs were similarly collected using polyester tipped swabs at enrollment and monthly thereafter during the follow-up visits.
All specimen tubes were barcoded and transported to the field laboratory within four hours using cool boxes maintained between 4–8 °C. At the field laboratory, samples were stored temporarily at −160 °C in dry shippers in a field laboratory in Marsabit town for up to 72 h. All specimens were later shipped to the U.S. Centers for Disease Control and Prevention (CDC) supported Kenya Medical Research Institute (KEMRI) laboratories in Nairobi for long-term storage and testing.
2.5. Real Time Reverse Transcription PCR Testing
MERS-CoV RNA in camel swabs was detected by real time RT-PCR at the KEMRI-CDC laboratories as described previously [29]. Briefly, total nucleic acids were extracted from 200 µL of the sample, followed by a standard RT-PCR test targeting the envelope (E) and two distinct regions of the nucleocapsid (N) genes. A sample was considered positive if all three PCR targets were positive (defined by a Cycle Threshold/CT value < 40).
2.6. MERS-CoV Isolation and Genotypic Analysis
Virus isolation was attempted for all RT-PCR positive camel nasal swabs (CT values below 20) using Vero cells at the biosafety level 3 (BSL-3) KEMRI-CDC laboratory in Kisumu, Kenya. Inoculated cells were incubated at 37 °C for five days and observed daily for cytopathic effects (CPEs). The culture supernatant and cells were examined for the presence of virus by the real time RT-PCR N2 assay targeting the MERS-CoV N gene and immunofluorescence assay using a rabbit antibody against the MERS-CoV N protein [30].
MERS-CoV culture isolates and aliquots of PCR positive samples were selected on the basis of high virus titers (low CT values below 30) for full-genome sequencing, which was performed at the CDC Genomics and Discovery laboratory in Atlanta, USA, following methods described previously [31,32]. Overlapping primer pairs that span the entire MERS-CoV genome were designed and developed against the MERS-CoV reference genome (JX869059). Briefly, RNA was first reverse transcribed by random hexamers to generate MERS-CoV-complementary deoxyribonucleic acid (cDNA) and then amplified using multiple multiplexing PCRs. The resulting multiple PCR amplicon pools from each sample were subject to fragmentation followed by barcoding libraries using the NEBNext Ultra II DNA library prep kit (New England Biolabs, Ipswich, MA, USA) or using the PCR Barcoding Expansion Pack 1–96 (EXP-PBC096) (Oxford Nanopore Technologies, Oxford, United Kingdom). Sequencing was performed using an Illumina MiSeq instrument (San Diego, CA, USA) and the Oxford nanopore GridION. Sanger sequencing was used to fill gaps if initial Illumina sequencing failed to recover the complete genome sequences.
All full sequences were deposited in GenBank (accession numbers OK094446-OK094454). The full and partial genome sequences were aligned in MAFFT v7.013 [33] including sequences from this study and a collection of representative MERS-CoV sequences retrieved from GenBank.
2.7. Statistical Analysis
R statistical software version 4.2.1 (R Core Team, Vienna, Austria, 2022) was used to detail the epidemiologic data. Q-GIS version 2.18.15 was used to generate maps. Attack rates were estimated as the proportion of camels with a sample positive for MERS-CoV out of the total number of camels under observation that were tested during the study period. The point estimates and 95% confidence interval for the attack rates estimates were reported. A repeat infection was defined as a camel with a second or third PCR positive sample following at least two successive PCR negative samples. Chi-square test was performed to evaluate for differences among the MERS-CoV infected and re-infected camels compared to non-infected camels, and odds ratios and the corresponding 95% confidence intervals (95% CIs) were reported. Phylogenetic analysis was inferred using the maximum likelihood (ML) method available in PhyML version 3.0 [34], assuming a general time-reversible (GTR) model with a discrete gamma distributed rate variation among sites (Gamma4) and an SPR tree swapping algorithm, and visualized in Mega version 6 [35].
3. Results
3.1. Description of Homesteads, Camel Herds, and the Enrolled Calves
From April 2018 to March 2020, we enrolled and followed 243 camel calves (18.5%) from a total of 1311 camels in 33 homesteads. About half of the camel calves (51.0%) were females, and the median age of the cohort at enrollment was four months (interquartile range, IQR = 1–9 months). Most of the enrolled camels (88.1%) were maintained close to the homestead in the home herd.
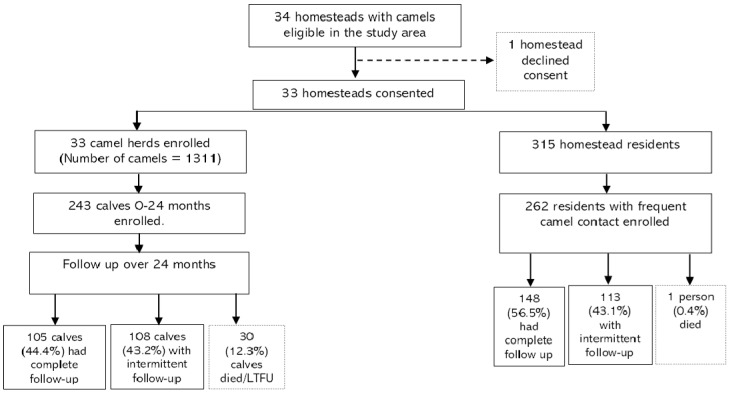
Almost all camel herds (>90%) practiced communal grazing with several other herds and shared watering points. Seven of the 33 homesteads (21.9%) acquired camels through purchase into their herds during the two-year study period, although the new camels were not enrolled in the cohort. The number of follow-up visits after enrollment ranged between 0–45 visits per camel, with a median of 17 follow-up visits (IQR = 7–31). In total, 105 calves (44.4%) had a complete follow-up, 108 calves (43.2%) were available intermittently over the study period, 30 calves died, and 16 were lost to follow-up immediately after enrollment (Figure 1). Overall, the cohort was followed up for a total of 270.6 camel-years equivalent to a median of 16 follow-up visits per camel (interquartile range, IQR = 6–30). A total of 30 calves died during the two-year follow-up, almost all (90.0%) due to predators and injuries.
Figure 1.
Study flow chart showing screening, enrollment, and follow-up of the linked camel–human cohort.
3.2. Clinical Disease in Camels
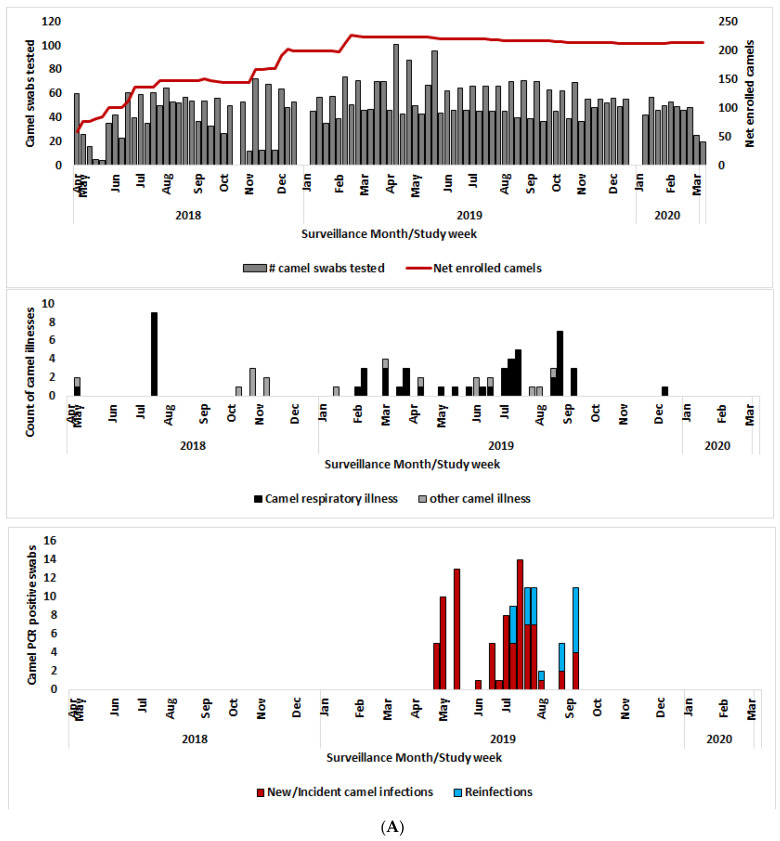
During the 2-year follow-up period, 68 illness episodes were recorded in 58 (23.9%) of the 243 camels, 70% of them between April and September 2019 (Figure 2A). A total of 52 illness episodes (76.5%) were respiratory illness signs, comprised of increased nasal discharge (28/52 or 53.8%), hyper-lacrimation (48.1%), coughing/honking (15.4%), and sneezing (3.8%). The respiratory illness incidence rate was 192.2 episodes per 1000 camel years. The remaining 16 non-respiratory illnesses comprised of anorexia (8/68 or 11.8%), lethargy (8.8%), enlarged lymph nodes (8.8%), weight loss (8.8%), diarrhea (7.4%), and skin and oral lesions (5.9%).
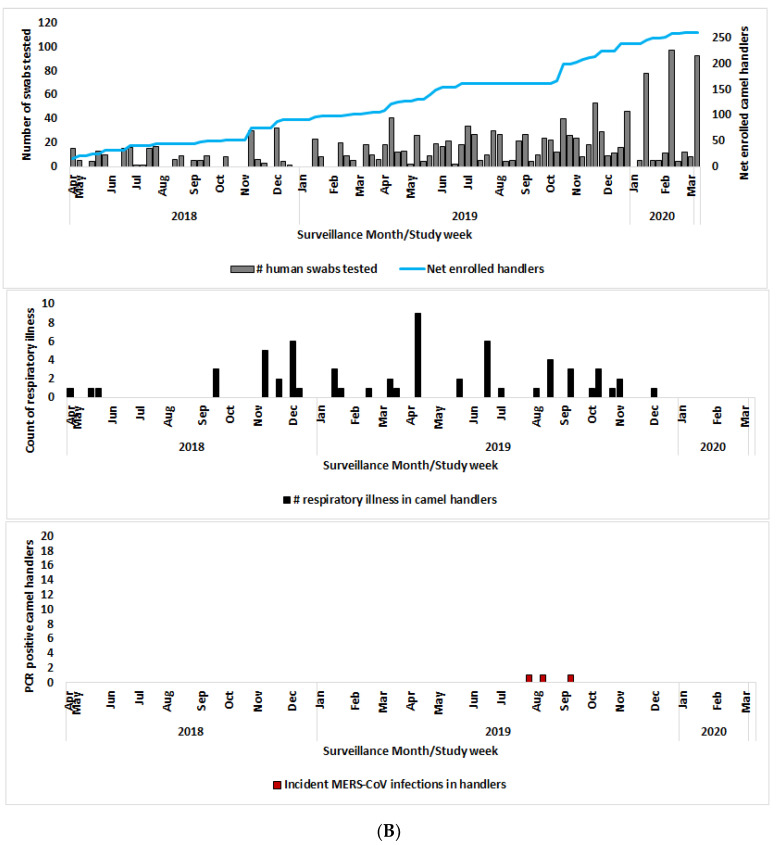
Figure 2.
(A) Net enrollment, number of swabs tested, respiratory illness signs, and PCR positive swabs enrolled camels; (B) Net enrollment, number of swabs tested, respiratory illness signs, and PCR positive swabs enrolled camel handlers.
3.3. Confirmation of MERS-CoV Outbreak in Camels
Of the 4692 nasal swabs collected and tested between April 2018 and March 2020, 124 swabs (2.6%) collected from 83 calves in 15 homesteads were positive for the MERS-CoV virus by RT-PCR. All the positive samples were collected between April and September 2019, the same period when >70% of the camel clinical disease was documented, henceforth referred to as the outbreak period (Figure 2A). During the outbreak period, 162 calves (66.7% of enrolled cohort) from 20 homesteads (60.6% of enrolled homesteads) were sampled and a total of 1853 swabs (39.5% of total swabs) were collected, or a median of 14 samples per camel (IQR 5–16). During this outbreak period, completeness of sampling did not vary by age or sex (Table 1).
Table 1.
Attack rates by age and sex among enrolled calves tested at least once during the MERS-CoV outbreak period in Northern Kenya, 2019 (n = 162).
| Variable | Camels on Follow-Up during Outbreak Period ϕ (n = 162) | Swabs Collected (April–September 2019) (n = 1853) | Median Swabs per Calf (IQR) | PCR Positive (1st PCR Positive) n = 83 | Attack Rates ¶ % (95% CI) | p-Value |
|---|---|---|---|---|---|---|
| By Sex | ||||||
| Male | 86 (53.1%) | 963 (52.0%) | 14 (5–16) | 41 (49.4%) | 47.7% (36.8, 58.7) | Ref |
| Female | 76 (46.9%) | 890 (48.0%) | 14 (5–16) | 42 (50.6%) | 55.3% (43.4, 66.7) | 0.93 |
| By Age (in months) ƛ | ||||||
| 0–4 | 57 (35.2%) | 595 (32.1%) | 15 (5–16) | 24 (28.9%) | 42.1% (29.1, 55.9) | Ref |
| 5–12 | 20 (12.3%) | 285 (15.4%) | 16 (15–16) | 14 (16.9%) | 70.0% (45.7, 88.1) | 0.04 µ |
| 13–18 | 45 (27.8%) | 481 (26.0%) | 13 (5–15) | 22 (26.5%) | 48.9% (33.7, 64.2) | 0.49 |
| 19+ | 40 (24.7%) | 492 (26.6%) | 14 (11–15) | 23 (27.7%) | 57.5% (40.9, 72.9) | 0.14 |
| All calves | 162 (100.0%) | 1853 (100.0%) | 14 (5–16) | 83(100.0%) | 51.2% (43.3, 59.2) | NA |
ϕ 162 calves included in the analysis were sampled during the outbreak period (April–September 2019); ¶ Attack rates were computed for the first infections; ƛ Calf age as at end of March 2019; µ Fisher’s exact test (FET).
The 83 calves involved in the outbreak were drawn from 15 of the 20 homesteads on follow-up at that time, giving a camel attack rate of 51.2% (95% CI 43.3–59.2%) and herd attack rate of 75.0% (95% CI 50.9–91.3%). The duration of viral RNA detection in a camel during the outbreak ranged from 2–6 weeks (Supplementary Figure S1). PCR detection in camels did not vary by sex, however, camels aged between 5–12 months were more likely to be infected when compared to camels from birth–4 months (odds ratio, OR = 3.21, 95% CI = 1.08–9.56 and p = 0.04).
Of the 83 PCR-positive calves, 20 (24.1%) had clinical signs of illness within 2 weeks of the sampling date, and of these, 19 calves had respiratory signs, including nasal discharge, coughing, and increased lacrimation.
3.4. Reinfections
Of the 83 PCR positive calves, 61 (73.5%) were infected once, 22 (26.5%) were reinfected, while one camel had three separate infections, giving a total of 106 separate infections (Table 2 and Supplementary Table S2). The median duration between re-infections was 59 days (IQR 49.5–78.5). The median age of the calves at first infection was 16 months (IQR = 7–22 months). The distribution of reinfected camels is shown in Table 2. Compared to females, male calves were more likely to be reinfected (OR = 2.88, 95% CI 1.03–8.09) and the difference was statistically significant. By age, calves 13–18 months were more likely to be re-infected compared to calves 0–4 months (OR = 5.83, 95% CI 1.15–37.93).
Table 2.
Repeat infections among camel calves during an outbreak of MERS-CoV in camels in Northern Kenya, n = 22.
| Variable | Camels with Repeat Infections * n (%) | OR (95% CI) | p-Value |
|---|---|---|---|
| Sex | |||
| Female | 7 (31.8%) | Ref | |
| Male | 15 (68.2%) | 2.88 (1.03–8.09) | 0.04 |
| By Age (months) | |||
| 0–4 | 3 (13.6%) | Ref | |
| 5–12 | 1 (4.5%) | 0.54 (0.01–7.67) | 1.00 |
| 13–18 | 10 (45.5%) | 5.83 (1.15–37.93) | 0.02 |
| 19+ | 8 (36.4%) | 3.73 (0.72–24.80) | 0.09 |
| All calves | 22 (100.0%) |
* Repeat infection was defined as a camel with a second or third PCR positive sample following at least two successive negative samples.
3.5. Human MERS-CoV Infections and the Spatial and Temporal Correlation with the Camel Outbreak
We previously reported three asymptomatic human MERS-CoV infections by RT-PCR from the cohort of 262 camel handlers [15]. The rest of the humans, sampled monthly, were negative for MERS-CoV by PCR (Figure 2B). Here, we build on this by correlating the spatial and temporal occurrence of human clinical illness within the handler’s cohort (n = 262) with the MERS-CoV outbreak in their camels.
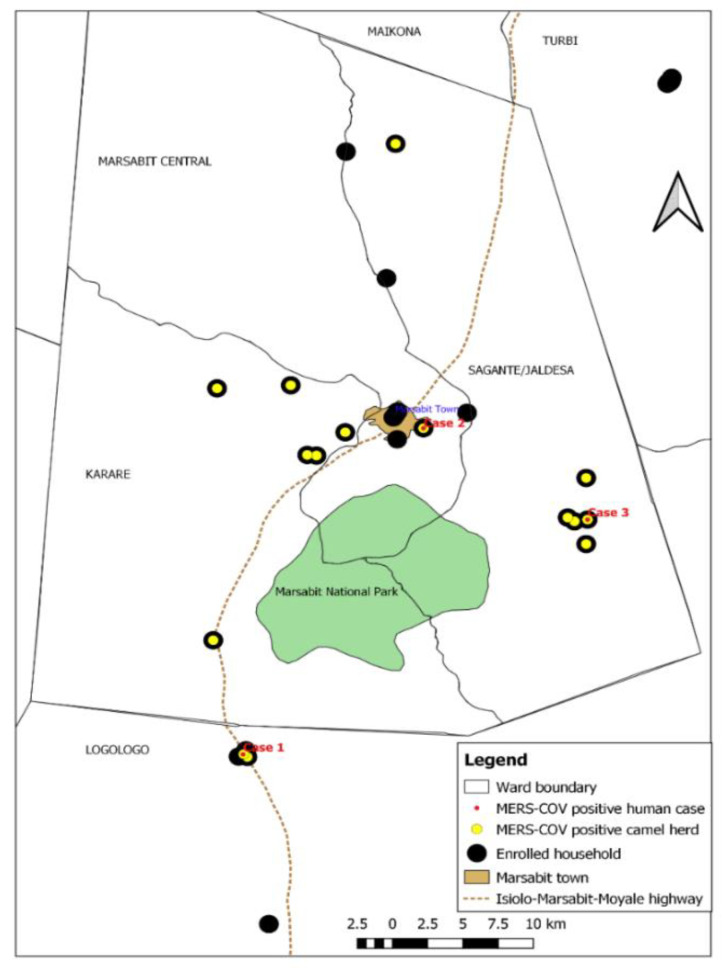
When analyzed alongside the camel data, the first human case (Case 1) tested positive in the last week of July 2019 and was a 20-year-old female spouse of a camel handler with two camels, one of which was enrolled in the cohort study, but this camel did not test PCR positive. However, two neighboring compounds (<2.5 Km away) with 14 enrolled calves had positive camels, while Case 1 tested positive. Case 2 was a 50-year-old male camel herder, diagnosed PCR positive in mid-August 2019, two weeks after one of the enrolled camels in his homestead was also tested positive. Case 3 was a 24-year-old camel herder who tested positive on 12th September 2019, the same day when three out of four camels sampled in his homestead also tested positive (Figure 3, Supplementary Table S3 and Supplementary Figure S1).
Figure 3.
Spot-map of the study area showing the location of enrolled camel homesteads (black spots), herds involved in the MERS-CoV outbreak (yellow spots), and human infections (red dots). The three human MERS-CoV infections clustered with camel infections.
3.6. Whole Genome Sequencing of MERS-CoV Isolates from Camels
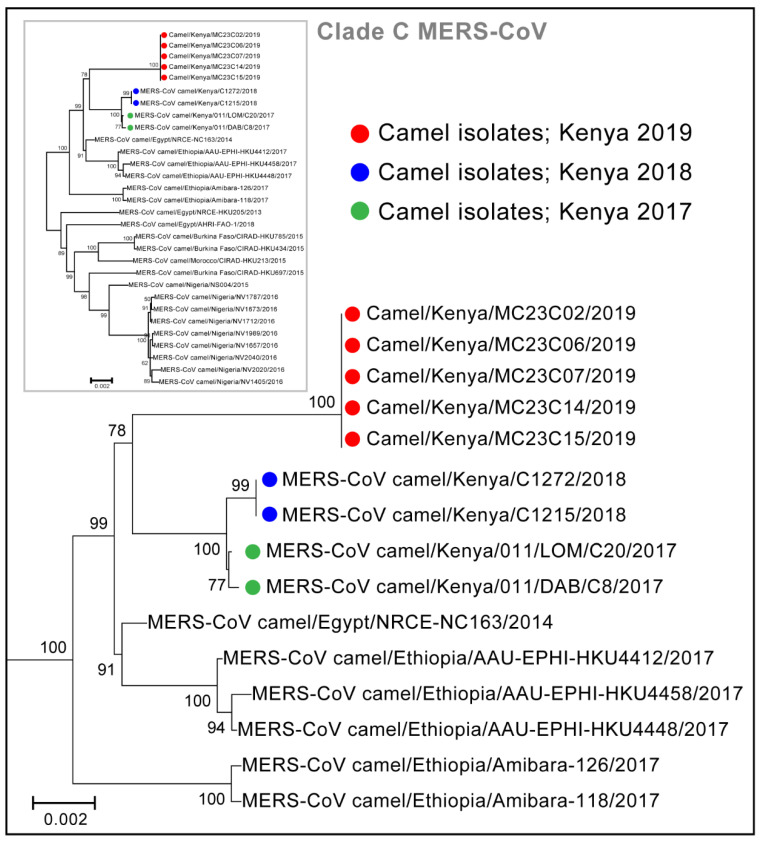
From six nasal swab samples, taken from six different camels, that had CT values <20, we obtained four virus isolates. The virus isolates and eight additional PCR positive swabs from seven camels (Ct < 30) were subjected to whole genome sequencing. A total of nine whole-genome sequences were generated (sequence measuring 30,112 kilobases) representing sequences from five unique camels in a herd that was involved in the May 2019 outbreak peak. Four camels had two sequences generated from isolates derived from nasal swabs collected at two successive sampling points. One camel had a sequence determined directly from a nasal swab. All nine genome sequences in this study were identical and were clustered with the clade C, alongside other African MERS-CoV (Figure 4 and Supplementary Figure S4). Within Clade C, the new Kenyan strains clustered with sub-clade C2 alongside other strains from Kenya and Ethiopia collected in 2017 and 2018. All the nine genomes from this study had identical 12-nucleotide deletions near the 3′ end of OFR3, resulting in nucleic acid deletion between 25,792–25,803 and causing 4 amino acid deletion (ORF3: 87T-, 88E-, 89H-, 90V-) compared to the GenBank sequence JX869059). The 12 nucleotide deletions started slightly (6 nucleotides) upstream of a 42-nucleotide deletion identified in Clade C1.1 strains from camels in Nigeria in 2016 (Supplementary Figure S2). These ORF3 deletions did not affect the stop codon and were not detected in Clade A and B viruses. We did not detect previously reported deletions in ORF4b of Clade C viruses. Our strains also had one nucleotide insertion at the 5′ non-coding region, which was also detected in viruses collected in Ethiopia in 2017 but not in the older Kenya viruses nor in other Clade C viruses.
Figure 4.
Phylogenetic tree of camel-derived MERS-CoV sequences from Clade C. The sequences indicated by the green and blue colored dots were isolated from camels in Kenya in earlier studies. Sequences denoted by the red dots were isolated from our study and were based on whole genome sequences (sequences measuring 30,112 kilobases).
4. Discussion
In this two-year cohort study, we detected an outbreak of MERS-CoV infection among 243 camels in Northern Kenya, affecting up to three-quarters of homesteads under surveillance. This camel outbreak was accompanied by increased respiratory illness signs and possibly repeat infections in camels, and likely resulted in acute camel-to-human MERS-CoV transmission events in three camel handlers as previously reported [15]. Sequencing of the camel isolates revealed genetically distinct clade C sub clade C2 MERS-CoV virus closely related to other sequences from the region. Spatial and temporal clustering of camel and human MERS-CoV cases strongly supports sporadic zoonotic spillover of Clade C MERS-CoV virus, as suggested in other studies [14,15,36].
Despite intense camel infection, and regular swabbing of handlers, only three positive but asymptomatic camel handlers were detected, suggesting that camel-to-human transmission is uncommon and not severe in this setting. This result is in agreement with a number of studies in the Horn of Africa region showing high anti MERS-CoV seroprevalence in camels but minimal detection of the virus or antibodies in humans [15,25]. A study of camels, other livestock, and wildlife in Sudan and Qatar found widespread camel exposure but no human infection [19]. Another study conducted in 2013 by our group found 18 out of 760 (2.4%) people positive for MERS-CoV antibodies by ELISA but negative by neutralization test, suggesting low level immune responses consistent with the possibility of asymptomatic infections [12]. In a past study, the three camel handlers were reported to be either young or middle-aged and were all asymptomatic [15]. We have not been able to explore risk factors for human infection due to the small number of infected camel handlers who all reported frequent camel contact.
The camel MERS-CoV outbreak was accompanied by increased respiratory illness signs in camels across the three outbreak peaks, signaling potential increased shedding events from camels that could contribute to transmission to camels and humans. A few studies have shown that camels infected with MERS-CoV may show a mild clinical illness. In this study, we were able to pick an increased intensity of reporting of camel illness, mostly mild respiratory signs which may underscore a possible role of syndromic surveillance in camels in this region [27,37,38,39]. The onset of the outbreak in April coincided with the long rains and peak calving season in northern Kenya. This season is usually characterized by frequent herd mixing during the Sorio traditional ceremony, celebrated in March-April when all livestock are brought back to the homestead for communal celebrations, creating a possible trigger of disease outbreaks [25]. However, even without cultural events such as Sorio, camel herds are reared in a nomadic pastoralist system with frequent mixing at watering points and grazing fields, providing ample opportunities for inter-herd transmission.
Within our camel cohort, the attack rate was high across all age groups in this study. However, the attack rate was highest among the 5–12-month-old camels, possibly reflecting a first infection in a young susceptible cohort following the waning of maternally derived antibodies. At 6–12 months, camel calves reared in the range production system are weaned and leave the home heard to join adult camels in the fora herd, possibly exposing them to infections once protection from the maternally derived antibodies wanes [25]. Past studies in the Middle East have shown widespread infections of young camels during outbreaks and suggested the possibility of reinfections [27,38,40].
We also observed a likely < 2-week to 6-week period of virus shedding for most camels, although repeat infections were common (a quarter of infected camels had repeat infections). The fact that even second or third MERS-CoV detections were clustered in time suggests that these were more likely repeat infections than prolonged or intermittently shedding. Additionally, CT values for the positive results also showed high viral shedding (heavy viral load), which is consistent with primary shedding as opposed to contamination. Lately, in the case of SARS-CoV-2, it has become apparent that coronaviruses can indeed cause multiple repeat infections [41]. The repeat infections and high attack rates could explain the protracted outbreak that lasted 5 months and could underlie the mechanism of virus maintenance in camels.
Our study found the clade C sub-clade C2 MERS-CoV to be responsible for this outbreak. Few recent studies have associated the African MERS-CoV strains with deletions in accessory genes, particularly ORF3 and ORF4b genes, that are likely to affect viral replication and pathogenicity [18,20,21,22]. Clade C1 viruses from West and North Africa, which have deletions in ORF4b, exhibited lower replication in human cells and in ex vivo human bronchus and lung tissues [20]. This study and others show deletions near the 3′ end of ORF3, however, the importance of this on virus replication and pathogenicity has not been determined [21,22]. The likely explanations behind presence of only clade C viruses in the Horn of Africa remain elusive. First, although MERS-CoV viruses may have originated from the Horn of Africa, with serologic evidence of the virus in camels detected >30 years before confirmation of the first human clinical cases of MERS-CoV [42] studies including this one have not detected the entire spectrum of the viruses in the region [13,21]. Second, clade C African viruses have not been detected in the Middle East, despite the fact that >60% of camels consumed in that region are imported from the Horn of Africa [22].
This study had some limitations. The biweekly follow-up schedule limited our ability to accurately characterize the natural history of MERS-CoV infection in camels. This was further compounded by the intermittent follow-up of about half of the enrolled camels. Being a nomadic pastoralist community, animals and people move over large distances for water and livestock feeding grounds, and this contributed to significant loss to follow-up, especially among adult camels that are also more difficult to restrain and sample. It is possible that some infections were missed between the biweekly sampling dates, especially if viral shedding was for a short period, as shown in this and other past studies [27,39]. The lack of serological data for camels and humans in this paper limited our ability to verify missed infections by checking for seroconversion, and therefore our attack rates may be underestimated. As we did not enroll all camels in herds, some infections would also have been missed in adult camels, even if the calves in our cohort were not infected.
5. Conclusions
In conclusion, our study describes the infection and transmission dynamics of MERS-CoV among camels and closely associated humans in the Horn of Africa. These findings shed light on the natural maintenance and transmission cycle of the virus in the region, perhaps driven by pathogen fidelity, geography, and host factors. Our study showed high levels of clade C MERS-CoV virus transmission among East African camels, with sporadic spillover to persons with camel contact. The possibility of virus evolution in the face of high transmission and the possible introduction of other clades through camel and human movements across the Arabian Peninsula underscores the need for continued virologic and serologic surveillance in camels and humans to understand the public health implications of MERS-CoV at the human–camel interface in Africa [1,21,43].
Acknowledgments
We thank the following staff for their contribution to data and specimen collection; Arithi Mutembei, Jack Omolo, Diba Denge, Boru Dub Wato, Millicent Minayo, Moshe Alando and Doris Marwanga. We appreciate the camel herders in Marsabit who participated in this study. We also thank the administrative team at the Washington State University Global Health Program for facilitating all study logistics.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/v14081743/s1, Figure S1: Sequence index plots of MERS-CoV infection in camels and camel handlers in 15 homesteads; Figure S2: The genetic sequences of the Kenya MERS-CoV virus sequences (bottom of the list) showing 12 nucleotide deletions at the 3′ region of the ORF3 (arrow). Available sequences from African camels and the Middle East have also been included for comparison purposes; Table S1: Distribution of PCR Positivity by enrolled herd among 33 camel herds in Marsabit; Table S2: Dates of PCR positive results and cycle threshold values for calves involved in multiple outbreak peaks, n = 22. Reinfections are shown in brown cell shade; Table S3: Demographic characteristics, clinical and contact history of the 3 human MERS-CoV cases detected in Northern, Kenya. This updated table is adopted from Munyua et al., 2021 [15]; Table S4: Comparison of MERS-CoV genetic sequences from nasal swabs and culture isolates of camel samples from Marsabit, Kenya.
Author Contributions
Conceptualization, I.N., E.A.H., M.M., J.G., A.M., M.K.N., M.-A.W. and P.M.M.; methodology, I.N., E.A.H., S.T., J.O., H.O., M.M., J.G., A.M., B.B., M.K.N., M.-A.W. and P.M.M.; validation, E.A.H., S.T., W.J., J.L.H., N.J.T., E.M.O., L.M., C.O. (Caroline Ochieng), C.O. (Cynthia Ombok), D.L. and A.H.-R.; formal analysis, I.N., E.A.H., J.L.H., N.J.T., C.O. (Caroline Ochieng), Y.T., J.Z. and P.M.M.; investigation, I.N., E.H, J.O, H.O., M.M, J.G., A.M., B.B., M.K.N., M.-A.W. and P.M.M.; resources, M.K.N., M.-A.W. and P.M.M.; data curation, I.N., E.M.O. and C.O. (Cynthia Ombok); writing—original draft preparation, I.N., E.A.H., S.T. and J.L.H.; writing—review and editing, W.J., H.O., M.M., E.M.O., J.G., J.D., A.M., D.L., A.H.-R., M.K.N., M.-A.W. and P.M.M.; visualization, I.N. and C.O. (Cynthia Ombok); supervision, M.M., A.M., M.K.N., M.-A.W. and P.M.M.; project administration, I.N.; funding acquisition, M.K.N., M.-A.W. and P.M.M. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committees of the Kenya Medical Research Institute (KEMRI) Scientific and Ethical Review Unit (SERU) and Animal Care and Use Committee (ACUC), approval number SSC3472, and by the Institutional Review Board of Washington State University human and animal ethical committees (approval numbers 16245 and 6124) and a reliance approval by US CDC (approval number 7065). Administrative approvals were also obtained from the Kenya Ministries of Health, and Agriculture, Livestock and Fisheries, and Marsabit County government prior to commencement of the studies.
Informed Consent Statement
Written informed consent was obtained from all camel owners and individual handlers prior to the questionnaire interview or sampling.
Data Availability Statement
All data underlying this article are available in the article.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results. The findings and conclusions in this manuscript are those of the authors and do not necessarily represent the official position of the US Centers for Disease Control and Prevention or the Government of Kenya.
Funding Statement
Funding for the project was provided by U.S. Centers for Disease Control and Prevention under the research co-operative agreement with Washington State University (CDC 1U01GH002143). Additional financial support was provided by the US National Institute of Allergy and Infectious Disease/National Institutes of Health (NIAID/NIH), grants number U01AI151799 through the Centre for Research in Emerging Infectious Diseases—East and Central Africa (CREID-ECA).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Da Costa V.G., Moreli M.L., Saivish M.V. The Emergence of SARS, MERS and Novel SARS-2 Coronaviruses in the 21st Century. Arch. Virol. 2020;165:1517–1526. doi: 10.1007/s00705-020-04628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mohd H.A., Al-Tawfiq J.A., Memish Z.A. Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Origin and Animal Reservoir. Virol. J. 2016;13:87. doi: 10.1186/s12985-016-0544-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Abdallat M.M., Payne D.C., Alqasrawi S., Rha B., Tohme R.A., Abedi G.R., Nsour M.A., Iblan I., Jarour N., Farag N.H., et al. Hospital-Associated Outbreak of Middle East Respiratory Syndrome Coronavirus: A Serologic, Epidemiologic, and Clinical Description. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2014;59:1225–1233. doi: 10.1093/cid/ciu359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adney D.R., van Doremalen N., Brown V.R., Bushmaker T., Scott D., de Wit E., Bowen R.A., Munster V.J. Replication and Shedding of MERS-CoV in Upper Respiratory Tract of Inoculated Dromedary Camels. Emerg. Infect. Dis. 2014;20:1999–2005. doi: 10.3201/eid2012.141280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khalafalla A.I., Lu X., Al-Mubarak A.I.A., Dalab A.H.S., Al-Busadah K.A.S., Erdman D.D. MERS-CoV in Upper Respiratory Tract and Lungs of Dromedary Camels, Saudi Arabia, 2013–2014. Emerg. Infect. Dis. 2015;21:1153–1158. doi: 10.3201/eid2107.150070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haagmans B.L., Dhahiry S.H.S.A., Reusken C.B.E.M., Raj V.S., Galiano M., Myers R., Godeke G.-J., Jonges M., Farag E., Diab A., et al. Middle East Respiratory Syndrome Coronavirus in Dromedary Camels: An Outbreak Investigation. Lancet Infect. Dis. 2014;14:140–145. doi: 10.1016/S1473-3099(13)70690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.WHO Middle East Respiratory Syndrome Coronavirus (MERS-CoV) [(accessed on 26 January 2022)]. Available online: http://www.who.int/emergencies/mers-cov/en/
- 8.ECDC Geographical Distribution of Confirmed MERS-CoV Cases by Reporting Country from April 2012 to 3 May 2022. [(accessed on 6 July 2022)]. Available online: https://www.ecdc.europa.eu/en/publications-data/geographical-distribution-confirmed-mers-cov-cases-reporting-country-april-2012-1.
- 9.ECDC MERS-CoV Worldwide Overview. [(accessed on 6 July 2022)]. Available online: https://www.ecdc.europa.eu/en/middle-east-respiratory-syndrome-coronavirus-mers-cov-situation-update.
- 10.Azhar E.I., El-Kafrawy S.A., Farraj S.A., Hassan A.M., Al-Saeed M.S., Hashem A.M., Madani T.A. Evidence for Camel-to-Human Transmission of MERS Coronavirus. [(accessed on 10 June 2021)]. Available online: https://www.nejm.org/doi/10.1056/NEJMoa1401505. [DOI] [PubMed]
- 11.Kiambi S., Corman V.M., Sitawa R., Githinji J., Ngoci J., Ozomata A.S., Gardner E., von Dobschuetz S., Morzaria S., Kimutai J., et al. Detection of Distinct MERS-Coronavirus Strains in Dromedary Camels from Kenya, 2017. Emerg. Microbes Infect. 2018;7:195. doi: 10.1038/s41426-018-0193-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Munyua P., Corman V.M., Bitek A., Osoro E., Meyer B., Müller M.A., Lattwein E., Thumbi S.M., Murithi R., Widdowson M.-A., et al. No Serologic Evidence of Middle East Respiratory Syndrome Coronavirus Infection among Camel Farmers Exposed to Highly Seropositive Camel Herds: A Household Linked Study, Kenya, 2013. Am. J. Trop. Med. Hyg. 2017;96:1318–1324. doi: 10.4269/ajtmh.16-0880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liljander A., Meyer B., Jores J., Müller M.A., Lattwein E., Njeru I., Bett B., Drosten C., Corman V.M. MERS-CoV Antibodies in Humans, Africa, 2013–2014. Emerg. Infect. Dis. 2016;22:1086–1089. doi: 10.3201/eid2206.160064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiyong’a A.N., Cook E.A.J., Okba N.M.A., Kivali V., Reusken C., Haagmans B.L., Fèvre E.M. Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Seropositive Camel Handlers in Kenya. Viruses. 2020;12:396. doi: 10.3390/v12040396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munyua P.M., Ngere I., Hunsperger E., Kochi A., Amoth P., Mwasi L., Tong S., Mwatondo A., Thornburg N., Widdowson M.A., et al. Low-Level Middle East Respiratory Coronavirus Syndrome among Camel Handlers, Kenya, 2019. Emerg. Infect. Dis. 2021;27:1201–1205. doi: 10.3201/eid2704.204458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hemida M.G., Al-Naeem A., Perera R.A.P.M., Chin A.W.H., Poon L.L.M., Peiris M. Lack of Middle East Respiratory Syndrome Coronavirus Transmission from Infected Camels. Emerg. Infect. Dis. 2015;21:699–701. doi: 10.3201/eid2104.141949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zumla A., Hui D.S., Perlman S. Middle East Respiratory Syndrome. Lancet Lond. Engl. 2015;386:995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ommeh S., Zhang W., Zohaib A., Chen J., Zhang H., Hu B., Ge X.-Y., Yang X.-L., Masika M., Obanda V., et al. Genetic Evidence of Middle East Respiratory Syndrome Coronavirus (MERS-Cov) and Widespread Seroprevalence among Camels in Kenya. Virol. Sin. 2018;33:484–492. doi: 10.1007/s12250-018-0076-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farag E., Sikkema R.S., Mohamedani A.A., de Bruin E., Munnink B.B.O., Chandler F., Kohl R., van der Linden A., Okba N.M.A., Haagmans B.L., et al. MERS-CoV in Camels but Not Camel Handlers, Sudan, 2015 and 2017. Emerg. Infect. Dis. 2019;25:2333–2335. doi: 10.3201/eid2512.190882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chu D.K.W., Hui K.P.Y., Perera R.A.P.M., Miguel E., Niemeyer D., Zhao J., Channappanavar R., Dudas G., Oladipo J.O., Traoré A., et al. MERS Coronaviruses from Camels in Africa Exhibit Region-Dependent Genetic Diversity. Proc. Natl. Acad. Sci. USA. 2018;115:3144–3149. doi: 10.1073/pnas.1718769115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Z., Hui K.P.Y., So R.T.Y., Lv H., Perera R.A.P.M., Chu D.K.W., Gelaye E., Oyas H., Njagi O., Abayneh T., et al. Phenotypic and Genetic Characterization of MERS Coronaviruses from Africa to Understand Their Zoonotic Potential. Proc. Natl. Acad. Sci. USA. 2021;118:e2103984118. doi: 10.1073/pnas.2103984118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El-Kafrawy S.A., Corman V.M., Tolah A.M., Masaudi S.B.A., Hassan A.M., Müller M.A., Bleicker T., Harakeh S.M., Alzahrani A.A., Alsaaidi G.A., et al. Enzootic Patterns of Middle East Respiratory Syndrome Coronavirus in Imported African and Local Arabian Dromedary Camels: A Prospective Genomic Study. Lancet Planet. Health. 2019;3:e521–e528. doi: 10.1016/S2542-5196(19)30243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Notenbaert A.M.O., Herrero M.T., Kruska R.L., You L., Wood S., Thornton P.K., Omolo A. Classifying Livestock Production Systems for Targeting Agricultural Research and Development in a Rapidly Changing World. International Livestock Research Institute; Nairobi, Kenya: 2009. [(accessed on 10 June 2021)]. 41p. Discussion Paper No. 19. Available online: https://www.researchgate.net/publication/258207462_Classifying_livestock_production_systems_for_targeting_agricultural_research_and_development_in_a_rapidly_changing_world. [Google Scholar]
- 24.Robinson T.P., Food and Agriculture Organization of the United Nations, editor. Global Livestock Production Systems. Food and Agriculture Organization of the United Nations; Rome, Italy: 2011. [Google Scholar]
- 25.Ngere I., Munyua P., Harcourt J., Hunsperger E., Thornburg N., Muturi M., Osoro E., Gachohi J., Bodha B., Okotu B., et al. High MERS-CoV Seropositivity Associated with Camel Herd Profile, Husbandry Practices and Household Socio-Demographic Characteristics in Northern Kenya. Epidemiol. Infect. 2020;148:1–31. doi: 10.1017/S0950268820002939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marsabit County Marsabit County Integrated Development Plan 2013–2017. [(accessed on 8 August 2019)]. Available online: https://www.undp.org/content/dam/kenya/docs/Democratic%20Governance/Marsabit%20County%20%20Revised%20CIDP.pdf.
- 27.Ali M.A., Shehata M.M., Gomaa M.R., Kandeil A., El-Shesheny R., Kayed A.S., El-Taweel A.N., Atea M., Hassan N., Bagato O., et al. Systematic, Active Surveillance for Middle East Respiratory Syndrome Coronavirus in Camels in Egypt. Emerg. Microbes Infect. 2017;6:e1. doi: 10.1038/emi.2016.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clin Calc Sample Size Calculator. [(accessed on 30 December 2019)]. Available online: https://clincalc.com/Stats/SampleSize.aspx.
- 29.Corman V.M., Ölschläger S., Wendtner C.-M., Drexler J.F., Hess M., Drosten C. Performance and Clinical Validation of the RealStar® MERS-CoV Kit for Detection of Middle East Respiratory Syndrome Coronavirus RNA. J. Clin. Virol. 2014;60:168. doi: 10.1016/j.jcv.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corman V., Eckerle I., Bleicker T., Zaki A., Landt O., Eschbach-Bludau M., van Boheemen S., Gopal R., Ballhause M., Bestebroer T. Detection of a Novel Human Coronavirus by Real-Time Reverse-Transcription Polymerase Chain Reaction. Eurosurveillance. 2012;17:20285. doi: 10.2807/ese.17.39.20285-en. [DOI] [PubMed] [Google Scholar]
- 31.Paden C.R., Tao Y., Queen K., Zhang J., Li Y., Uehara A., Tong S. Rapid, Sensitive, Full-Genome Sequencing of Severe Acute Respiratory Syndrome Coronavirus 2. Emerg. Infect. Dis. 2020;26:2401–2405. doi: 10.3201/eid2610.201800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yusof M.F., Queen K., Eltahir Y.M., Paden C.R., Al Hammadi Z.M.A.H., Tao Y., Li Y., Khalafalla A.I., Shi M., Zhang J., et al. Diversity of Middle East Respiratory Syndrome Coronaviruses in 109 Dromedary Camels Based on Full-Genome Sequencing, Abu Dhabi, United Arab Emirates. Emerg. Microbes Infect. 2017;6:1–10. doi: 10.1038/emi.2017.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katoh K., Standley D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 35.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mok C.K.P., Zhu A., Zhao J., Lau E.H.Y., Wang J., Chen Z., Zhuang Z., Wang Y., Alshukairi A.N., Baharoon S.A., et al. T-Cell Responses to MERS Coronavirus Infection in People with Occupational Exposure to Dromedary Camels in Nigeria: An Observational Cohort Study. Lancet Infect. Dis. 2021;21:385–395. doi: 10.1016/S1473-3099(20)30599-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Njenga K., Kemunto N., Kahariri S., Holmstrom L., Oyas H., Biggers K., Riddle A., Gachohi J., Muturi M., Mwatondo A., et al. High Real-Time Reporting of Domestic and Wild Animal Diseases Following Rollout of Mobile Phone Reporting System in Kenya. PLoS ONE. 2021;16:e024411948. doi: 10.1371/journal.pone.0244119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madani T.A., Abuelzein E.-T.M.E., Hussien H.S., Bashri M.A., Hassan A.M., Azhar E.I. Monitoring of the Middle East Respiratory Syndrome Coronavirus Activity in a Secluded Herd of Camels Kept Under Field Conditions. Vector-Borne Zoonotic Dis. 2021;21:994–1002. doi: 10.1089/vbz.2021.0020. [DOI] [PubMed] [Google Scholar]
- 39.Meyer B., Juhasz J., Barua R., Das Gupta A., Hakimuddin F., Corman V.M., Müller M.A., Wernery U., Drosten C., Nagy P. Time Course of MERS-CoV Infection and Immunity in Dromedary Camels. Emerg. Infect. Dis. 2016;22:2171–2173. doi: 10.3201/eid2212.160382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hemida M.G., Alnaeem A., Chu D.K., Perera R.A., Chan S.M., Almathen F., Yau E., Ng B.C., Webby R.J., Poon L.L., et al. Longitudinal Study of Middle East Respiratory Syndrome Coronavirus Infection in Dromedary Camel Herds in Saudi Arabia, 2014–2015. Emerg. Microbes Infect. 2017;6:e56. doi: 10.1038/emi.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cohen J.I., Burbelo P.D. Reinfection With SARS-CoV-2: Implications for Vaccines. Clin. Infect. Dis. 2021;73:e4223–e4228. doi: 10.1093/cid/ciaa1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Corman V.M., Jores J., Meyer B., Younan M., Liljander A., Said M.Y., Gluecks I., Lattwein E., Bosch B.-J., Drexler J.F., et al. Antibodies against MERS Coronavirus in Dromedary Camels, Kenya, 1992–2013. Emerg. Infect. Dis. 2014;20:1319–1322. doi: 10.3201/eid2008.140596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galloway S.E., Paul P., MacCannell D.R., Johansson M.A., Brooks J.T., MacNeil A., Slayton R.B., Tong S., Silk B.J., Armstrong G.L., et al. Emergence of SARS-CoV-2 B.1.1.7 Lineage—United States, December 29, 2020–January 12, 2021. Morb. Mortal. Wkly. Rep. 2021;70:95–99. doi: 10.15585/mmwr.mm7003e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data underlying this article are available in the article.