Abstract
Klebsiella pneumoniae (Kp) is a gram-negative opportunistic pathogen that causes severe pneumonia, pyelonephritis, and sepsis in immunocompromised hosts. During a 4-mo interval, several NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) breeders and pups in our facilities were diagnosed with Kp infections. An initial 6 adult and 1 juvenile NSG mice were submitted for necropsy and histologic examination because of acute onset of diarrhea and death. The evaluation revealed typhlocolitis in 2 of the mice and tritrichomoniasis in all 7. Escherichia coli positive for polyketide synthase (pks+) and Kp were isolated from the intestines. Given a history of sepsis due to pks+ E. coli in NSG mice in our facilities and determination of its antimicrobial susceptibility, trimethoprim–sulfamethoxazole (TMP–SMX) was administered to the colony in the drinking water for 4 wk. After this intervention, an additional 21 mice became ill or died; 11 of these mice had suppurative pneumonia, meningoencephalitis, hepatitis, metritis, pyelonephritis, or sepsis. Kp was cultured from pulmonary abscesses or blood of 10 of the mice. Whole-genome sequencing (WGS) indicated that the Kp isolates contained genes associated with phenotypes found in pore-forming Kp isolates cultured from humans with ulcerative colitis and primary sclerosing cholangitis. None of the Kp isolates exhibited a hyperviscous phenotype, but 13 of 14 were resistant to TMP–SMX. Antimicrobial susceptibility testing indicated sensitivity of the Kp to enrofloxacin, which was administered in the drinking water. Antibiotic sensitivity profiles were confirmed by WGS of the Kp strains; key virulence and resistance genes to quaternary ammonia compounds were also identified. Enrofloxacin treatment resulted in a marked reduction in mortality, and the study using the NSG mice was completed successfully. Our findings implicate intestinal translocation of Kp as the cause of pneumonia and systemic infections in NSG mice and highlight the importance of identification of enteric microbial pathogens and targeted antibiotic selection when treating bacterial infections in immunocompromised mice.
Abbreviations: Kp, Klebsiella pneumoniae; MKPV, mouse kidney parvovirus; NSG, NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ; pks, polyketide synthase; TMP–SMX, trimethoprim–sulfamethoxazole; WGS, whole-genome sequencing
Severely immunocompromised NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) mice typically are used for oncology, stem cell biology, diabetes, and infectious disease research. The combination of mutations in these mice includes a unique MHC haplotype, a deletion in the C5 gene, a defect in the double-stranded repair mechanisms necessary for the gene rearrangement of antigen-specific receptors, and a knockout of the IL2 receptor γ chain.21,37 These deficits result in mice with no mature B or T cells, dysregulation of antigen presenting-cells, no NK cells, no hemolytic complement, and impaired cytokine signaling.41 These mice are routinely used for xenotransplantation, including implantation of human cell lines. Appropriate husbandry practices are critical because these mice are highly susceptible to infection with opportunistic agents such as Escherichia coli strains positive for polyketide synthase (pks+), Klebsiella oxytoca, Enterococcus spp., and Candida albicans, which have been associated with meningitis or meningoencephalitis, myocarditis, pneumonia, pyelonephritis, cystitis, metritis, and septicemia in NSG and other immunocompromised mouse strains.4,6,17,43
Klebsiella pneumoniae (Kp) is a gram-negative encapsulated bacterium typically found in the environment and gastrointestinal tract of mammals.22 In both human and veterinary medicine, Kp is considered an emergent and common opportunistic pathogen associated with an array of diseases.33 This organism can acquire resistance to multiple antibiotics and is an important cause of nosocomial infections in humans that include urinary tract infections, pneumonia, septicemia, and soft-tissue infections and particularly occur in hospitalized immunocompromised patients.30,33,34 Classic and hypervirulent strains of Kp have been implicated in cases of pneumonia, urogenital infections, and sepsis in laboratory, companion, and domestic animals.15,35 In laboratory rodents, Kp commonly colonizes the gastrointestinal tract of healthy animals and rarely causes disease in immunocompetent hosts; however, Klebsiella spp. infections in immunocompromised mice have been associated with spontaneous disease, including sepsis, lymphadenitis, pneumonia, empyema, hepatic and renal abscesses, endocarditis and myocarditis, and thrombosis.6,35
This report documents an epidemiologic study of an outbreak of systemic Kp infection in a restricted-access colony of NSG mice that occurred despite the administration of trimethoprim sulfamethoxazole (TMP–SMX) in the drinking water. We hypothesized that a virulent phenotype was responsible for the invasive TMP–SMX-resistant Kp identified in this outbreak and that the systemic Kp originated from translocation of the organism from the intestine due to intestinal barrier compromise.28,30,33,40,42
Materials and Methods
Case presentation.
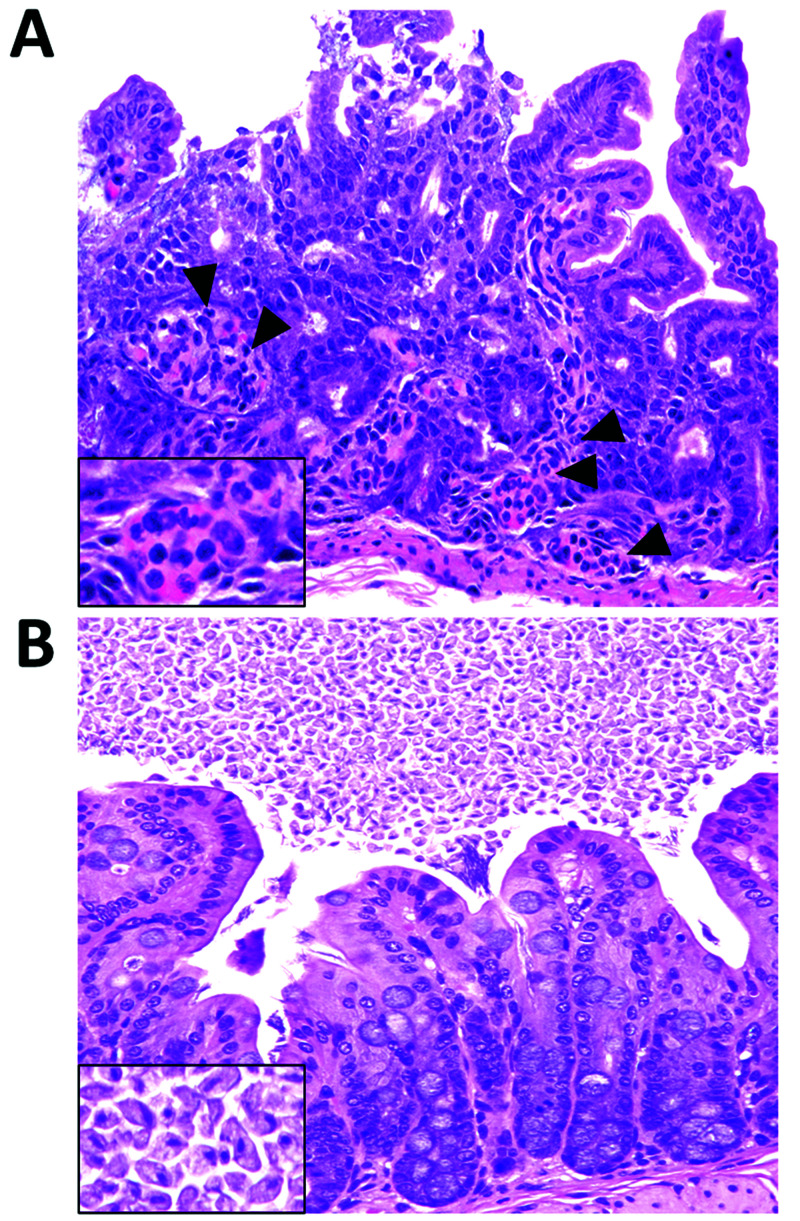
An increase in diarrhea and mortality was noted primarily among NSG dams and their litters in 2 rooms (later combined into one). Some neonatal mice had undergone intracranial injection with human glial cells in a biologic hood located in a procedure room that was also used for procedures in immunocompetent mice. Over 3 wk, 7 mice (6 adults, 1 juvenile) were presented for clinical evaluation, with 4 animals having evidence of diarrhea and 2 of these showing acute neutrophilic enterotyphlocolitis (Figure 1A).
Figure 1.
Representative histology of the gastrointestinal tract of NSG mice prior to antibiotic therapy. (A) Mild multifocal acute enteritis in an NSG mouse infected with pks+ E. coli. The lamina propria of the small intestine is multifocally infiltrated by low numbers of neutrophils (arrowheads, inset). (B) Tritrichomoniasis in the large intestine of an NSG mouse. The cecum is colonized by numerous Tritrichomonas organisms (inset). Magnification, 40×.
E. coli was isolated from the blood of the 2 mice with enterotyphlocolitis and the cecum of 4, and Kp was isolated from one blood sample and 4 cecal samples. The E. coli isolates were further characterized for the presence of genotoxic virulence factors. Three isolates contained the pks gene cluster encoding the genotoxin colibactin, previously shown to contribute to systemic disease in immunocompromised mice (Il2rg−/−Rag2−/−, c-KitW-sh/W-sh).4 In addition, E. coli was identified from culture of rectal swabs from 5 mice in the room. In addition, a single sentinel CD1 mouse from the room had shown a positive nasal wash for Kp in the preceding year but had no gross lesions. Cecal colonization with Tritrichomonas spp. was noted commonly on fecal exams of immunocompetent CD1 sentinel mice and was also detected in all 7 NSG mice submitted for histopathological evaluation (Figure 1B). This protozoal infestation was considered to be an incidental finding, given that the large intestines of NSG mice were intermittently colonized with this protozoan without evidence of inflammation or clinical disease. Given these findings, pks+ E. coli was presumed to be the causative agent of morbidity and mortality in these mice. Antimicrobial sensitivity testing revealed that all isolated E. coli strains were sensitive to enrofloxacin and trimethoprim sulfonamide (TMP-SMX); therefore, TMP–SMX was provided in the drinking water at 1.5 mg/mL (approximately 300 mg/kg) for 4 wk. During and after treatment, 21 additional mice were submitted and evaluated, as described below.
Mice.
All original mice had been purchased from Jackson Laboratories (Bar Harbor, ME). The breeding colony had been maintained at an AAALAC-accredited facility for approximately 3 years, in polycarbonate microisolation caging with autoclaved hardwood bedding (Sani-Chips, PJ Murphy, Jewett, PA) and pelleted food (Prolab RMH 3000, Purina Mills, St. Louis, MO) and deionized water. Mice were housed in clean autoclaved cages with food and bedding twice weekly. The mice were transferred by grasping the base of the tail with rubber-tipped forceps that had been submerged in quaternary ammonia disinfectant solution (Quatricide PV, Pharmacal Research Laboratories, Waterbury, CT) prepared as a 1:64 dilution. The solution was refreshed monthly based on manufacturer’s recommendations. The forceps were disinfected before use and between cages during cage changing. At the time of outbreak, cages containing bedding, feed pellets, and water bottle were autoclaved before housing mice; cages were changed on a bench top and maintained as static cages.
All affected mice were on IACUC-approved protocols. Mice were either found dead or euthanized by using CO2 at a 10% to 30% chamber replacement rate followed by intracardiac exsanguination. Health monitoring was performed via dirty-bedding sentinels (CD1 Elite, Charles River Laboratories, Wilmington, MA) that had been bred and maintained in a separate barrier facility. All sentinel mice during the previous and subsequent years had been negative on serology for mouse hepatitis virus, enzootic diarrhea of infant mice virus, minute virus of mice, Sendai virus, norovirus, pneumonia virus of mice, reovirus 3, Theiler encephalomyelitis virus, lymphocytic choriomeningitis virus, K virus, Mycoplasma pulmonis, and CAR bacillus and on ELISA for mouse parvovirus, Ectromelia, polyomavirus, and mouse adenoviruses. Rectal swabs were all negative for Salmonella and Citrobacter rodentium. Anal tape tests, skin scrapings, and sodium nitrate solution fecal flotation analyses were all negative for parasites. Direct duodenal smears were negative for protozoa, but cecal smears were intermittently positive for Tritrichomonas and Entomoeba spp., which are not excluded pathogens in mice maintained in this facility.
Postmortem examination.
Full necropsies were performed at the Comparative Pathology Laboratory (Division of Comparative Medicine, Massachusetts Institute of Technology). Representative tissues from all organ systems were fixed in 10% neutral-buffered formalin, and paraffin-embedded sections (thickness, 5 μm) were stained with hematoxylin and eosin. To verify the presence of bacteria in tissue sections, Brown–Brenn modified Gram stains were performed in a subset of cases. To demonstrate the presence of neutrophils in lesions, formalin-fixed sections from a subset of cases were incubated for 1 h with rat antimouse Ly6G (dilution, 1:1000; clone 1A8, BioXCell, West Lebanon, NH). The immunohistochemical procedure was performed by using the antirat IgG horseradish peroxidase detection kit (BD Biosciences, San Jose, CA) and developed (ARK K3954, Dako, Agilent, Santa Clara, CA) according to manufacturer instructions.
Microbiology.
In light of gross necropsy findings, culture samples were obtained from blood (n = 10), cecum (n = 17), lung (n = 6), liver (n = 1), and small intestine (n = 1). The samples were placed into Brucella broth containing 10% glycerol and homogenized. The homogenates were streaked onto chocolate agar and blood agar–MacConkey agar split plates (Becton Dickinson, Franklin Lakes, NJ), inoculated into trypticase soy broth (Becton Dickinson) for enrichment, and incubated for 18 to 24 h at 37 °C in 5% CO2. Pure cultures were obtained by restreaking single morphologically different colonies onto another blood agar plate and speciated by using API 20E identification strips (bioMerieux, Marcy l’Etoile, France). To assess the elimination of Kp after antibiotic treatment, fecal samples saved in bacterial freeze media were plated on MacConkey, xylose–lysine–deoxycholate, and Hektoen enteric selective and differential agar or medium and on blood agar, a general-purpose enrichment medium. The bacterial plates were incubated for 18 to 24 h at 37 °C in 5% CO2.
K. pneumoniae fluorescent in situ hybridization.
Fluorescent in situ hybridization was performed on formalin-fixed, paraffin-embedded tissues by using the probe 5′-CCT ACA CAC CAG CGT GCC-3′ (Integrated DNA Technologies, Coralville, IA), as previously reported.4 Slides were deparaffinized in xylene and rehydrated in a descending ethanol series. After air drying, the slides were covered in hybridization buffer containing 10 ng/μL of probe at 74.5 °C and maintained overnight at 48 °C in a dark, humidified chamber. Slides were rinsed with water and incubated in 2 wash buffers for 15 min each at 48 °C. The slides were then rinsed, air dried, mounted in antifade reagent with DAPI (ProLong Gold, Thermo Fisher, Rockford, IL), and examined under fluorescence (Axioskop 2 plus, Zeiss, Oberkochen, Germany). Slides coated with Kp and Enterococcus faecalis colonies were used as positive and negative controls, respectively. Two negative controls were used for this experiment, one without a probe and one with a nontarget E. coli-specific probe.
Antimicrobial sensitivity.
Antimicrobial susceptibility testing of strains of Kp and E. coli was performed by using the disc diffusion method recommended by the Clinical and Laboratory Standards Institute.10,11 Antimicrobials tested included ampicillin, ampicillin–clavulanic acid, ampicillin–sulbactam, cephalothin, enrofloxacin, gentamicin, and TMP–SMX. E. coli ATCC 29322 was included as the reference control. Minimum inhibitory concentrations for enrofloxacin and a quaternary ammonium compound (Quatricide PV) were determined by using the broth microdilution method as recommended by the Clinical and Laboratory Standards Institute and others.10,11 Minimum bacteriocidal concentration values for microbiocides were obtained by using a modification of a previously published protocol.23
String test.
The hyperviscosity phenotype of Kp isolates was initially evaluated by using the string test.11 Briefly, a sterile plastic inoculation loop was placed on top of and slowly removed from a colony grown on an agar plate. The string test was positive when a viscous string measuring 5 mm or longer was produced by the colony clinging to the loop.30,33,40
Whole-genome and molecular characterization.
Representative mouse Kp isolates (n = 15), comprising 13 isolates previously described in brief27 and 2 additional isolates, were submitted for whole-genome sequencing (WGS). Genomic DNA was purified from Kp isolates that had been grown overnight in LB broth. Barcoded libraries were created by using QIAseq FX DNA library kit (Qiagen, Germantown, MD), sequenced with MiSeq (Illumina, San Diego, CA), and trimmed by using BBDuk (Geneious, San Diego, CA); contigs were assembled and annotated by using SPAdes and RAST hosted by PATRIC.27 For comparative genomic analysis, genomes of representative Klebsiella spp. were acquired from PATRIC.12 In addition, the Similar Genome Finder Tool hosted by PATRIC was used to identify the 10 genomes most closely related to the mouse Kp genomes. Whole-genome phylogenetic analysis was performed by using OrthoFinder (version 2.3.1).13 Average nucleotide identity values were calculated by using PYANI (version 0.2.10).31 All Klebsiella spp. genomes were analyzed by using Kleborate to predict multilocus sequencing type, K and O antigen types, virulence genes, and antibiotic-resistance gene profiles.25 In addition, mouse Kp genomes were analyzed against the Virulence Factor Database by using BLAST to identify other potential virulence factor genes as well as homologs to 97 gene sequences associated with the pore-forming Kp strains isolated from human patients with ulcerative colitis and primary sclerosing cholangitis.26,28
Plasmid DNA was extracted by using the QIAprep Spin Miniprep Kit (Qiagen) according to the manufacture’s protocol. Antibiotic-resistance gene cassettes were detected from purified plasmid DNA through PCR analysis of intl1 (forward primer, 5′-GAC TTG CGC TGC CCT ACC TCT CAC-3′; reverse primer, 5′-TTG GCG GCC TTG CTG TTC TAC-3′) and sul1 (forward primer, 5′-AGG CGG ACT GCA GGC TGG TTA-3′; reverse primer: 5′-GGC CGG AAG GTG AAT GCT A-3′) by using the following thermocycling program: initial denaturation at 95 °C for 3 min, followed by 35 cycles consisting of denaturation at 95 °C for 1 min, annealing at 63.7 °C for 1 min, and elongation at 72 °C for 2 min, with a final incubation at 72 °C for 10 min.
E. coli PCR amplification.
E. coli samples were boiled and centrifuged at 12,000 × g. The resulting supernatant was used for PCR analysis to identify genotoxins. Colibactin is a genotoxic peptide encoded by clb genes that lie within the polyketide synthase (pks) pathogenicity island. These genes were identified by using 2 sets of primers: clbA (forward, 5′-CAG ATA CAC AGA TAC CAT TCA-3′; reverse, 5′-CTA GAT TAT CCG TGG CGA TTC-3′) and clbQ (forward, 5′-TTA TCC TGT TAG CTT TCG TTC-3′; reverse, 5′-CTT GTA TAG TTA CAC AAC TAT TTC-3′).16
Tritrichomonas muris PCR analysis.
The histologic diagnosis of T. muris was confirmed and speciated through PCR analysis of frozen samples of cecal tissue or contents. DNA was extracted by using QIAamp Fast DNA Stool Mini Kit (Qiagen); PCR amplification was performed in a 25-µL reaction volume by using the Expand High Fidelity PCR System according to manufacturer instructions (Sigma-Aldrich, St Louis, MO). Amplification of tritrichomonas DNA was performed by using the forward primer 5′-AGC GGA AAA GAA ACT AAC T-3′ and reverse primer 5′-CAT CGG TCT CAC AGC AT-3′, which targeted the 28S ribosomal RNA (expected amplicon, 260 bp). The cycling parameters consisted of an initial denaturing step of 3 min at 95 °C, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 61 °C for 30 s, elongation at 72 °C for 30 s, and a final incubation at 72 °C for 4 min.
PCR amplification and quantitative PCR analyses for mouse kidney parvovirus (MKPV).
Total DNA from frozen kidneys and formalin-fixed, paraffin-embedded kidneys of NSG mice was prepared as previously described.19 PCR amplification was performed in a 25-µL reaction volume by using the Expand High Fidelity PCR System (Sigma-Aldrich) according to the manufacturer’s instructions. MKPV-specific primers were designed to target NS1 region of the MKPV genome as described previously.32 The double-stranded nucleotide sequences of selected PCR products were determined by using Sanger sequencing (Quintara Biosciences, Cambridge, MA).
Statistics.
Fisher exact tests were used to evaluate the associations between Kp infection, age, sex, tritrichomonas status, systemic lesions, and MKPV infection. A P value of 0.05 or less was considered significant (JMP, version 14, SAS Institute, Cary, NC).
Data availability.
Genomes of Kp have been deposited in GenBank under the following accession numbers: SULO00000000, SULP00000000, SULQ00000000, SULR00000000, SULS00000000, SULT00000000, SULU00000000, SULV00000000, SULW00000000, SULX00000000, SULY00000000, SULZ00000000, SUMA00000000, JAEAGK000000000, and JAEAGL000000000.
Results
During and after TMP–SMX treatment, 21 NSG mice (13 females, 8 males; 14 adults, 7 pups) were submitted for diagnostic evaluation. Kp was isolated from cecal contents, blood, or extraintestinal lesions in 17 of the 21 mice examined. Kp was isolated from the cecum of adult mice (P = 0.0254) and from extraintestinal tissues, including lungs, blood, and liver, in 57% (12 of 21) of the mice (Table 1). Kp was exclusively isolated from the cecum in 4 additional mice. Cecal cultures from 12 mice recovered Kp and other bacterial organisms, including Enterococcus faecalis (n = 10), E. coli (n = 4), Enterococcus cloacae (n = 3), and one each of Enterococcus amnigenus, Enterococcus faecium, Streptococcus uberis, S. xylosus, and Aeromonas hydrophilia (Table 1). Two of the E. coli isolates from the ceca of 2 different mice were pks+.
Table 1.
Culture results of NSG mice after TMP–SMX treatment
| Site cultured (number of samples) | Culture resultsa |
|---|---|
| Lungs (n = 6) | |
| With pneumonia (n = 5) |
K. pneumoniae (n = 5) E. coli (n = 1) |
| Without pneumonia (n = 1) | K. pneumoniae (n = 1) |
| Blood (n = 10) | |
| With other systemic lesionsb (2) |
K. pneumoniae (n = 1) No growth (n = 1) |
| No other systemic lesions (7) |
K. pneumoniae (n = 5) E. coli (n = 1) No growth (n = 1) |
| Cecum (n = 18) |
K. pneumoniae (n = 16) Enterococcus faecalis (n = 10) E. coli (n = 4) Enterococcus cloacae (n = 3) Aeromonas hydrophilia (n = 1) Enterococcus amnigenus (n = 1) Enterococcus faecium (n = 1) Streptococcus uberis (n = 1) Streptococcus xylosus (n = 1) |
In cases that grew Kp (n = 16), 12 yielded pure cultures of Kp from lungs, blood, or other organs
Other systemic lesions include meningitis, epicarditis, metritis, splenitis, hepatitis, peritonitis, pyelonephritis, and cystitis
Histologic findings.
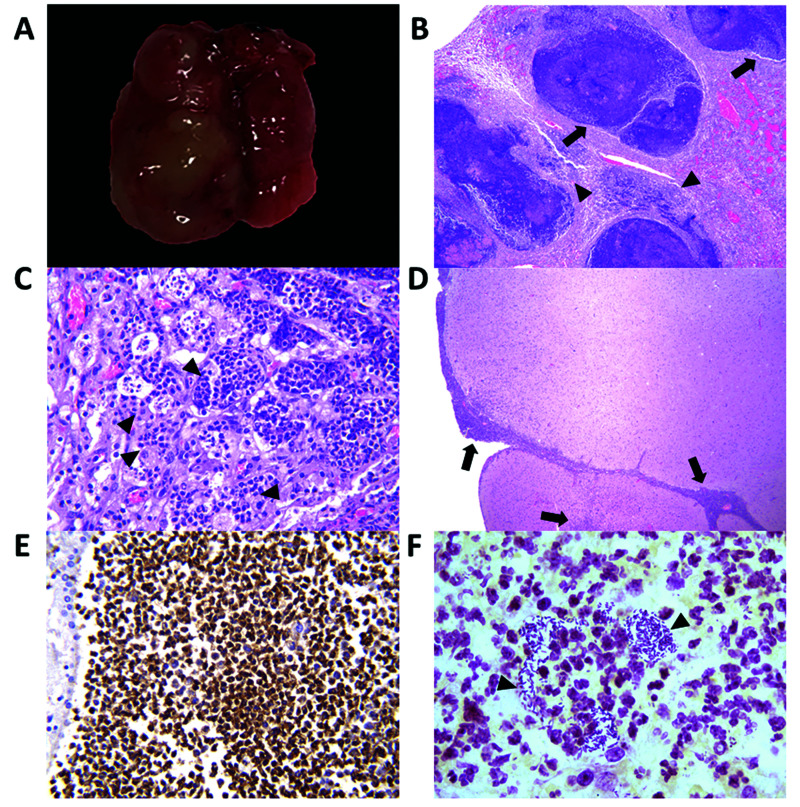
Histologically, the lungs of 5 NSG mice contained multifocal to coalescing abscesses that completely effaced the architecture of the bronchi, bronchioles, and alveolar septa (Figure 2A through C). These abscesses contained large numbers of degenerate neutrophils intermixed with karyorrhectic debris, necrotic cells, mineralization, fibrin, edema, and coccobacilli forming microcolonies. At the periphery, these abscesses were surrounded by fibrous connective tissue, large to moderate numbers of foamy macrophages, and fewer neutrophils. Kp was recovered from bacterial cultures of the pulmonary abscesses and cecum (Table 1). Fluorescent in situ hybridization specific for Kp highlighted the presence of rod-shaped bacteria associated with leukocytes along the edges of pulmonary abscesses (not shown). The meninges, lateral and fourth ventricles, and Virchow–Robin spaces of 2 additional mice were filled with large numbers of degenerate neutrophils admixed with microcolonies of rod-shaped bacteria (Figure 2D through F), fewer macrophages, congestion, and polymerized fibrin. The adjacent cerebral cortex was infiltrated by multifocal clusters of neutrophils intermixed with areas of malacia, gliosis, and edema (not shown). Two other mice had areas of multifocal random neutrophilic hepatitis, one of which also had concurrent suppurative neutrophilic pyelonephritis with papillary necrosis. The uterine walls of 2 breeders were multifocally infiltrated by low to moderate numbers of degenerate neutrophils admixed with fibrinous exudate and few hemosiderin-laden macrophages. These breeders exhibited suppurative inflammation in the peritoneum and/or mediastinum. One mouse had suppurative pyelonephritis and chronic tubulointerstitial nephropathy.
Figure 2.
Suppurative pneumonia and meningoencephalitis in NSG mice infected with K. pneumoniae. (A) Representative gross image of an affected lung from an NSG mouse shows multifocal to coalescing abscesses in the left pulmonary lobe. (B) Bronchioles (arrows) and alveoli (arrowheads) are effaced by multiple abscesses and areas of necrosis. Magnification, 4×. (C) Alveoli are infiltrated by multiple dense clusters of degenerate neutrophils (arrowheads) admixed with few foamy macrophages, karyorrhectic debris, fibrin, and congestion. Magnification, 40×. (D) An affected brain from an NSG mouse. The meninges, cortex, and Virchow–Robins spaces are infiltrated by multifocal to coalescing regions of suppurative inflammation (arrows). Hematoxylin and eosin stain; magnification, 4×. (E) Immunohistochemistry for Ly6G in the cerebrum demonstrates the presence of neutrophils in areas of meningitis (brown staining of neutrophils). Magnification, 40× (F) Gram staining of the meninges shows dense clusters of extracellular, gram-negative, rod-shaped bacteria (arrowheads) associated with neutrophilic inflammation. Magnification, 100×.
Whole-genomic and molecular characterization.
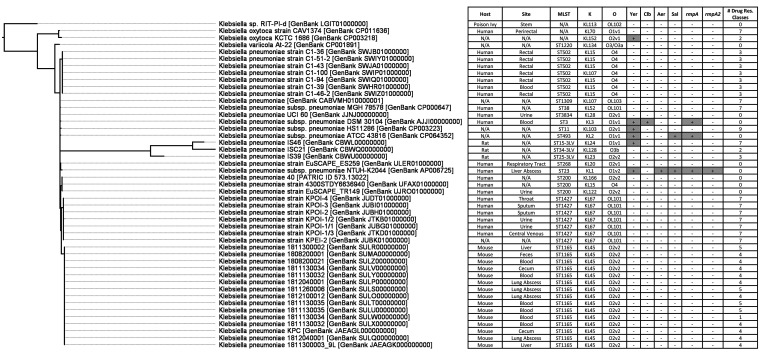
WGS was performed on 15 representative Kp isolates to further study their pathogenic potential and antibiotic resistance profiles. Whole-genome phylogenetic and average nucleotide analyses determined that the mouse Kp isolates were most similar to each other (range, 99.96% to 100.00%). Using the Similar Genome Finder Tool hosted by PATRIC, the 10 genomes most closely related to the mouse Kp isolates were human strains cultured from human blood, rectal swabs, sputum, throat, or urine. Overall, the mouse Kp isolates were 98.93% to 99.30% similar to these genomes and to representative Kp strains from humans and rats (Table S1). In general, average nucleotide identity values for mouse Kp strains were more similar to nonmouse strains not encoding rpmA (99.21% ± 0.00%) than to genomes harboring this hypervirulence gene (99.13% ± 0.00%). Unlike the representative Kp genomes from humans and rats, all 15 mouse Kp genomes were ST1166 and had the predicted K capsular and O antigens of K45:O2.v2 (Figure 3).
Figure 3.
Whole-genome phylogenetic tree of mouse Kp isolates compared with representative Klebsiella spp. genomes. The Kp mouse genomes were most similar to classic Kp strains with multidrug antibiotic resistance profiles that had been isolated from humans. # Drug Res. Classes, number of antibiotic resistant gene classes; Aer, aerobactin; Clb, colibactin; K, K capsule antigen type; MLST, multilocus sequence type; O, O antigen type; rpmA, regulator of mucoid phenotype A; rpmA2, regulator of mucoid phenotype A2; Sal, salmochelin; Yer, yersiniabactin.
None of the Kp mouse genomes harbored rmpA, rmpA2, or magA genes responsible for the hyperviscomucoid phenotype. In agreement with this finding, none of the 23 mouse Kp isolates evaluated had the hyperviscomucoid phenotype, as indicated by a negative string test. All 15 mouse Kp genomes contained multiple genes related to capsular production (wzi, wza, wzb, wzc, gnd, wcaA, wcaJ, and galF) but lacked other capsular genes traditionally found in this gene cluster (cpsB, cpsG). In addition, these 15 genomes contained the virulence factors for lipopolysaccharide synthesis, including the genes uge and wabG. Gene clusters responsible for type 1 fimbriae (fimA–fimI, fimK), type 3 fimbriae (mrkA–mrkD, mrkF–mrkI), and enterobactin siderophore biosynthesis were present in all 15 genomes, but none of these genomes encoded genes for the synthesis of aerobactin, salmochelin, yersiniabactin, or colibactin. In addition, the 15 mouse Kp genomes contained the khe gene, which encodes for hemolysin, but hemolytic activity was not observed phenotypically. The Kp mouse genomes also encoded 28 homologous gene sequences with a putative type VI secretion system, reactive oxygen species degradation, and carbohydrate update or metabolism functions, and have previously been associated with the pore-forming cytotoxicity caused by Kp strains isolated form human patients with primary sclerosing cholangitis.28
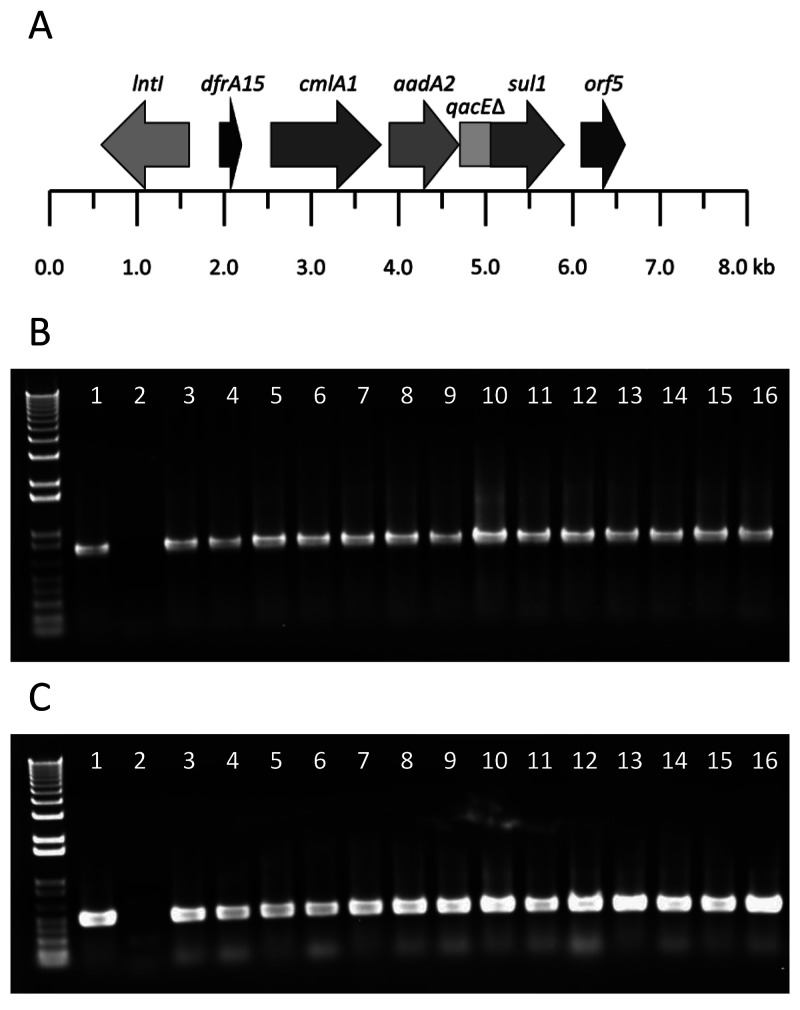
Fourteen of the 15 mouse Kp isolates encoded identical antibiotic-resistance gene cassettes containing aadA2, cmlA1, sul1, dfrA15, and qacEΔ1 for aminoglycoside, phenicol, sulfonamide, trimethoprim, and quaternary ammonium compound resistance, respectively (Figure 4A). BLAST analysis against the nr/nt database revealed that this cassette was most homologous to class 1 integron plasmid sequences from Aeromonas caviae, Kp, E. coli, Enterobacter cloacae, Salmonella enterica, and Enterobacter hormaechei (coverage, 57.0% to 98.0%; percentage identity, 97.7% to 100.0%; Table S2). PCR assays using purified plasmid DNA and targeting the intI and sul1 genes of this antibiotic-resistance gene cassette confirmed its presence in the 14 mouse Kp isolates. In total, plasmid DNA from 20 of 23 mouse Kp isolates were positive for both antibiotic resistance gene cassettes (Figure 4B and C). Furthermore, all isolates also encoded the SHV61 extended spectrum β-lactamase gene. None of the genomes contained fluoroquinolone resistance genes, but qacEΔ1 and cepA, cation efflux pumps associated with chlorhexidine resistance, were present in the genomes of all 15 mouse Kp isolates and were located on the chromosome.
Figure 4.
(A) Gene structure of plasmid-encoded class 1 integron antibiotic resistance gene cassettes present in mouse Kp genomes. (B) PCR analysis for intI sequence from purified plasmid DNA of representative mouse Kp genomes. (C) PCR for sul sequence from purified plasmid DNA of representative mouse Kp genomes. Lanes indicate 16 different representative isolates.
Antimicrobial sensitivity profile of Kp.
Antimicrobial sensitivity testing by disc diffusion demonstrated that 13 of 14 Kp isolates were resistant to TMP–SMX, but all were susceptible to enrofloxacin and gentamicin, as were the original E. coli strains (Table 2). All isolates (n = 14) were resistant to ampicillin; 13 of 14 were sensitive to ampicillin–clavulanic acid, ampicillin–sulbactam, and cephalothin. For confirmation, minimal inhibitory concentration determinations by broth dilution were performed for enrofloxacin; all 14 Kp isolates were susceptible at 0.25 µg/mL or less. Enrofloxacin was provided in the water at 0.25 mg/mL continuously thereafter. The treatment eliminated deaths associated with Kp infection in this breeding colony of NSG mice. Furthermore, to assess treatment efficacy and elimination of Kp, fecal samples saved in bacterial freeze media from several cages were cultured in general-purpose, selective, and differential media. Whereas bacterial growth occurred on general-purpose blood agar plates, no growth was observed in selective and differential media for gram-negative bacteria. These findings suggest that Kp was successfully eliminated from the colony after treatment with enrofloxacin. With regard to the quaternary ammonium product, the minimum inhibitory concentration for the Kp isolates ranged from 0.7 to 5.4 µg/mL and the minimum bacteriocidal concentration ranged from 0.7 to 21.4 µg/mL.
Table 2.
Antimicrobial disc diffusion data (millimeters) from K. pneumoniae isolates
| Sample ID | Ampicillin | Amoxicillin–Clavulanic acid | Ampicillin–Sulbactam | Cephalothin | Enrofloxacin | Gentamicin | Trimethoprim–Sulfamethoxazole |
|---|---|---|---|---|---|---|---|
| 1808210021 Bood | 0 * | 25 | 20 | 20 | 27 | 18 | 0 * |
| 1808210001 Feces | 0 * | 0 * | 0 * | 0 * | 29 | 19 | 0 * |
| 1811130032 Blood | 0 * | 25 | 19 | 20 | 26 | 19 | 0 * |
| 1811130032 Blood | 0 * | 20 | 18 | 20 | 27 | 21 | 0 * |
| 1811130034 Blood | 0 * | 21 | 19 | 19 | 25 | 18 | 0 * |
| 1811130034 Cecum | 0 * | 20 | 21 | 20 | 26 | 20 | 0 * |
| 1811130035 Blood | 0 * | 24 | 17 | 20 | 26 | 21 | 20 |
| 1811130035 Blood | 0 * | 25 | 18 | 19 | 27 | 18 | 0 * |
| 1811260006 Lung abscess | 0 * | 23 | 19 | 19 | 26 | 21 | 0 * |
| 1811300002 Liver | 0 * | 22 | 20 | 19 | 27 | 21 | 0 * |
| 1812040001 Lung abscess | 0 * | 23 | 19 | 20 | 26 | 19 | 0 * |
| 1812100012 Lung abscess | 0 * | 22 | 19 | 20 | 27 | 21 | 0 * |
| 1811300003 Cecum | 0 * | 21 | 19 | 19 | 27 | 20 | 0 * |
| 1811300003 Liver | 0 * | 25 | 21 | 19 | 26 | 19 | 0 * |
| Resistant | ≤ 13 | ≤ 13 | ≤ 11 | ≤ 14 | ≤ 16 | ≤ 12 | ≤ 10 |
| Intermediate | 14–16 | 14–17 | 12–14 | 15–17 | 17–20 | 13–14 | 11–15 |
| Sensitive | ≥ 17 | ≥ 18 | ≥ 15 | ≥ 18 | ≥21 | ≥ 15 | ≥ 16 |
E. coli ATCC 25922, Reference control
Bolded values indicate antibiotic resistance.
Coinfection with T. muris and MKPV.
The ceca of NSG mice (19 of 22) frequently contained a heavy burden of protozoal organisms that exhibited morphologic features consistent with T. muris. These mice did not have typhlocolitis but did show goblet cell hyperplasia and epithelial cell degeneration and tufting, with numerous exfoliated epithelial cells in the intestinal lumina. Tritrichomonas DNA was detected in 4 of 6 cecal samples submitted for evaluation. Sequence analysis of 28s rRNA amplicons revealed 100% identity with T. muris (accession no., Z18255). Kp was significantly (P = 0.0254) detected in ceca cocolonized with T. muris versus those without T. muris.
Because MKPV has been detected in NSG breeders housed in the same room where the outbreaks occured,19 we prospectively collected kidneys from NSG mice, including the 7 early diarrhea cases and the 21 subsequent cases. MKPV was detected in the kidneys of 15 of the 28 NSG mice submitted for evaluation; of these 15 MKPV cases, 2 were from the initial 7 mice with diarrhea, and 13 were from the subsequent cohort. Five mice were either bacteremic or had lesions consistent with Kp infection; however, MKPV status was not significantly associated with Kp infection or the presence of systemic inflammatory lesions. Of the 15 MKPV cases, 2 had chronic tubulointerstitial lesions consistent with MKPV-induced inclusion body nephropathy, whereas the remaining 13 kidneys did not. The mice with inclusion body nephropathy had MKPV levels that were quantified as over 10,000 times higher than in MKPV-positive mice without histologic lesions. The majority of the latter MKPV cases had fewer than 10 copies of MKPV per μg of mouse DNA. MKPV in situ hybridization signals (RNAscope) were detected in the nucleus and cytoplasm of affected tubular epithelial cells in the 2 mice with chronic inclusion body nephropathy (data not shown). MKPV infection was not associated with sex, age, or tritrichomonas status. A subset of these mice (n = 10) was included in a separate report of MKPV infection from our group.19
Discussion
In the current epidemiologic study, we determined that the disease outbreak in our NSG mice was a classic presentation of opportunistic Kp infection, which typically results in pulmonary abscesses, bacteremia, or systemic suppurative inflammation in infected immunocompromised mice.5,35 Contrary to what we initially suspected, the Kp strains isolated were neither phenotypically hyperviscous nor genetically characterized as hypervirulent, but they still contributed to significant morbidity and mortality. Kp strains vary greatly in their pathogenicity, which stems from a variety of virulence factors. Although Kp hypervirulent strains are increasing in medical importance, even classic Kp strains contain multiple virulence factors and can have antibiotic resistance.24,33,36 For example, classic Kp strains produce a thick capsule, lipopolysaccharide, types 1 and 3 fimbriae, and siderophores,30 all of which were present in these isolates. Virulence factors specifically associated with hypervirulent strains, such as the genes rmpA and magA that mediate capsule production,30 were not present in our isolates. Two isolates derived from liver and cecum of the same bacteremic mouse were originally identified as positive on the string test. When WGS did not support this finding due to the lack of rmpA and magA, these strains were retested and found to be string test negative. The viscosity of colonies can be affected by growing conditions, resulting in borderline viscosity that can result in a false positive.40 In contrast, truly hyperviscous colonies often produce strings that extend multiple centimeters, well beyond the 5-mm threshold we used. If the string test is not definitive, confirmatory tests such as the sedimentation test, or PCR analysis of rmpA or magA can be used. In one study, we used WGS to confirm our results of negative string tests.40
These apparently classic Kp strains harbored antibiotic resistance genes to a wide range of antibiotic classes, as confirmed via antimicrobial sensitivity testing and WGS. In the initial 7 cases, culture results had mixed growth of commensals and potentially pathogenic bacteria, such as pks+ E. coli and Kp. These polymicrobial results were difficult to interpret and required ancillary phenotypic and molecular assays to better characterize these organisms. Our PCR testing and clinical history lead us to believe pks+ E. coli was the major contributor to morbidity, and the choice to use of TMP–SMX was based on antimicrobial testing of these isolates. All Kp cases of pulmonary abscesses, bacteremia, or sepsis developed after the administration of TMP–SMX. In addition, our laboratory records indicated that Kp was rarely isolated from the nares of sentinel mice or lesions from sick mice. We suspect that Kp circulating in asymptomatic breeders was transmitted via the fecal–oral route to pups and other breeders in the colony. Kp strains isolated from humans and animals are well known to acquire resistance to various antibiotics, including TMP–SMX.24,34 Treatment with TMP–SMX likely enriched the large intestines of NSG mice with antibiotic-resistant Kp that was then shed in the feces and spread to naïve mice. In addition, continuous antibiotic treatment in mice can create a dysbiosis accompanied by a superspreader phenotype of intestinal pathogens, in this case Kp.42 Although we did not quantify Kp in the feces, the high percentage of Kp in cecal contents implies that Kp was high in feces. Our WGS analysis of Kp isolates and their antimicrobial susceptibility patterns confirmed that most Kp isolates were resistant to TMP–SMX, perhaps explaining the sudden increase in Kp-associated deaths in this colony of NSG mice. The fact that Kp was commonly found as a pure culture in blood and pulmonary abscesses further supports the source of the disease outbreak as invasive Kp and not one of the other commensal organisms originally isolated from the fecal samples of mice with diarrhea.
Previously, 97 orthologous genes were identified that correlated with the pore-forming cytotoxicity caused by Kp strains isolated from human patients with primary sclerosing cholangitis.28 A BLASTP analysis revealed that 28 of these orthologous sequences were also present in our 15 mouse Kp strains. These genes included putative functions associated with the type IV secretion system, reactive oxygen species degradation, and carbohydrate update or metabolism. Although the genes and mechanisms responsible for pore-forming cytotoxicity are unknown, the presence of these 28 genes indicates that the mouse Kp strains are genetically similar to extraintestinal Kp strains associated with intestinal barrier disruption and liver pathology. Further in vitro experiments are required to ascertain whether the presence of these 28 orthologous genes in the rodent Kp strains can exhibit pore-forming cytotoxicity that allows translocation of the bacteria and ensuing sepsis.28
In addition to traditional antibiotics, these Kp genomes were also resistant to quaternary ammonium compounds. The responsible genes, such as qac family, encode for efflux pumps that eliminate both drug residues and quaternary ammonium compounds from the bacterium.7 Decreased susceptibility to these commonly used antiseptic and disinfectant agents makes elimination from the environment difficult and heightens the risk reinfection.39
Quaternary ammonium disinfectants are commonly used in vivaria as surface-acting detergents with antibacterial properties. The product used in our facilities, Quatricide (Pharmacal Waterbury, CT), combines 2 of these compounds, didacyldimethyl ammonia chloride and n-alkyl dimethyl benzyl ammonia chloride. The use of this detergent as a disinfectant of forceps used to transfer mice into clean cages raised concern due to our recognition that the Kp isolate recovered in this outbreak encoded an efflux-based system, the qac-mediated resistance genes qacEΔ1 and cepA. The qacE gene and its attenuated variant qacEΔ1 is commonly found in gram-negative bacteria, including the Enterobacteriaceae, to which Kp belongs.39 Select studies conducted in the United Kingdom noted the presence of qacEΔ1 and cepA genes and a reduced susceptibility to biocides.1,2 In contrast, other reports indicated the presence of qacEΔ1 and cepA genes had no significant effect on Kp biocide resistance.3,29
In this outbreak, we speculated that the presence of the qacEΔ1 and cepA genes in the Kp isolates increased resistance to the quaternary ammonium product we used. The practical relevance of this possibility may be discounted given that the recommended concentration of the disinfectant is usually higher than what the targeted bacteria can tolerate.38 To address this possibility, we determined the minimal inhibitory concentration and minimum biocidal concentration of the disinfectant.11 The values obtained were well within the sensitivity ranges as compared with the concentration of the working solution (686 µg/mL). Nevertheless, the qacEΔ1 and cepA genes, together with other mechanisms such as various multidrug efflux genes or modifications in the cell wall of Kp, may have contributed to the development of Kp resistance to the disinfectant and facilitated the spread of Kp during the current outbreak.8 We recently changed our sanitation program; we have replaced the disinfectant used with Peroxigard (Virox Technologies, Ontario, Canada).
We viewed the association of cecal Tritrichomonas spp. with Kp cocolonization as incidental, given that typhlitis was not observed in these cases. Tritrichomonas spp. in the intestines of laboratory mice are considered commensal organisms.5 Recent studies have shown that experimental infections of laboratory mice with Tritrichomonas spp. alters the epithelial and immune cell homeostasis in the intestinal mucosa, making them more susceptible to inflammation in the large intestine.9,14,20 Such effects may have contributed to facilitating translocation of Kp from the intestines into the bloodstream in cases of enteric Kp infection and sepsis.
MKPV is a recently identified murine pathogen responsible for inclusion body nephropathy in immunocompromised mice, including NSG.32 We recently detected MKPV from both archived and fresh kidney samples from immunocompromised mice with or without inclusion body nephropathy.19 Although inclusion body nephropathy was rare in the current cohort, we found low levels of MKPV in 54% of mice analyzed. Whether these mice would have progressed to IBN had they reached advanced age is unknown. In the current cases, MKPV status was not associated with Kp infection; however, we have previously observed MKPV-positive immunocompromised mice with inclusion body nephropathy and concurrent urosepsis and meningitis from pks+ E. coli.19 Similar to Kp, pks+ E. coli is a common gastrointestinal inhabitant of mice that can sometimes cause systemic disease.18,19,41 The similarities between these 2 outbreaks of systemic infections prompts additional questions; future studies are needed to determine whether MKPV infections in immunocompromised mice enhance their susceptibility to systemic bacterial infections and other comorbidities.
Once Kp was identified as the causative agent of this outbreak, we made appropriate changes to treatment and husbandry protocols. Mice were transitioned from autoclaved static cages to IVC and cages were changed in a biosafety cabinet. Autoclaved food, water, and bedding continued to be used. Enrofloxacin was provided in the water indefinitely. Although we originally had considered a shorter course of treatment, the investigators’ prolonged experimental timeline and continued use of the colony required this long-term therapeutic approach. Feces collected from the colony after 2 y of enrofloxacin treatment did not grow Kp, suggesting that the organism had not developed resistance against this drug class. In addition, the colony continued to do well clinically, with no subsequent cases of systemic bacterial infection reported.
In summary, this study revealed that classic (nonhyperviscous) Kp strains have the potential to induce systemic disease in severely immunocompromised mice. The alteration of the intestinal microflora after TMP–SMX therapy may have enhanced the susceptibility of these NSG breeders and pups to Kp infections. Although we were unable to determine when and how Kp was introduced into this colony of NSG mice housed under SPF conditions, retrospective analysis of culture results from sentinel mice revealed that Kp was detected rarely in sentinel CD1 mice from the room in question. Treatment of the colony with oral antibiotics clearly contributed to increased colonization density of a Kp superspreader phenotype, promoting rapid spread of Kp in the colony. Our findings suggest that translocation of enteric Kp may have resulted in the systemic inflammatory lesions we observed. In view of the enhanced susceptibility of NSG mice to opportunistic pathogens, these mice should be housed under optimal and rigorous management practices. Importantly, the change of antibiotic treatment from TMP–SMX to enrofloxacin successfully controlled this outbreak of Kp.
Supplementary Materials
Average nucleotide identity values
Integron BLAST results
Acknowledgments
We thank and acknowledge Joanna Richards and Caroline Atkinson for their expertise in histology. We also acknowledge Kara Procter and Carolyn Madden for their assistance in our diagnostic laboratory. The efforts of all were critical for the completion of this project and the continued diagnostic activities of DCM. We thank Parisa Zarringhalam for manuscript preparation. This research was supported by grant NIH P30 – ES02109 (JGF).
References
- 1.Abuzaid A, Hamouda A, Amyes SG. 2012. Klebsiella pneumoniae susceptibility to biocides and its association with cepA, qacΔE and qacE efflux pump genes and antibiotic resistance. J Hosp Infect 81:87–91. 10.1016/j.jhin.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 2.Abuzaid AA, Amyes SG. 2015. The genetic environment of the antiseptic resistance genes qacEΔ1 and cepA in Klebsiella pneumoniae. J Chemother 27:139–144. 10.1179/1973947814Y.0000000181. [DOI] [PubMed] [Google Scholar]
- 3.Azadpour M, Nowroozi J, Goudarzi GR, Mahmoudvand H. 2015. Presence of qacEΔ1 and cepA genes and susceptibility to a hospital biocide in clinical isolates of Klebsiella pneumoniae in Iran. Trop Biomed 32:109–115. [PubMed] [Google Scholar]
- 4.Bakthavatchalu V, Wert KJ, Feng Y, Mannion A, Ge Z, Garcia A, Scott KE, Caron TJ, Madden CM, Jacobsen JT, Victora G, Jaenisch R, Fox JG. 2018. Cytotoxic Escherichia coli strains encoding colibactin isolated from immunocompromised mice with urosepsis and meningitis. PLoS One 13:e0194443. 10.1371/journal.pone.0194443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barthold SW, Percy DH, Griffey SM, editors. 2016. Pathology of Laboratory Rodents and Rabbits, Fourth edition. Ames, (IA): John Wiley & Sons, Inc. 10.1002/9781118924051 [DOI] [Google Scholar]
- 6.Bleich A, Kirsch P, Sahly H, Fahey J, Smoczek A, Hedrich HJ, Sundberg JP. 2008. Klebsiella oxytoca: opportunistic infections in laboratory rodents. Lab Anim 42:369–375. 10.1258/la.2007.06026e. [DOI] [PubMed] [Google Scholar]
- 7.Buffet-Bataillon S, Tattevin P, Bonnaure-Mallet M, Jolivet-Gougeon A. 2012. Emergence of resistance to antibacterial agents: the role of quaternary ammonium compounds–a critical review. Int J Antimicrob Agents 39:381–389. 10.1016/j.ijantimicag.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 8.Cervinkova D, Babak V, Marosevic D, Kubikova I, Jaglic Z. 2013. The role of the qacA gene in mediating resistance to quaternary ammonium compounds. Microb Drug Resist 19:160–167. 10.1089/mdr.2012.0154. [DOI] [PubMed] [Google Scholar]
- 9.Chudnovskiy A, Mortha A, Kana V, Kennard A, Ramirez JD, Rahman A, Remark R, Mogno I, Ng R, Gnjatic S, Amir ED, Solovyov A, Greenbaum B, Clemente J, Faith J, Belkaid Y, Grigg ME, Merad M. 2016. Host-Protozoan Interactions Protect from Mucosal Infections through Activation of the Inflammasome. Cell 167:444–456.e414. 10.1016/j.cell.2016.08.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute. [Internet]. 2018. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. CLSI standard M07. Wayne, PA: Clinical and Laboratory Standards Institute. Available at: https://www.researchgate.net/file.PostFileLoader.html?id=564ceedf5e9d97daf08b45a2&assetKey=AS%3A297254750572544%401447882463055 [Google Scholar]
- 11.Clinical and Laboratory Standards Institute. [Internet]. 2022. Performance Standards for Antimicrobial Susceptibilty Testing. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute. Available at: https://clsi.org/standards/products/microbiology/documents/m100/ [Google Scholar]
- 12.Davis JJ, Wattam AR, Aziz RK, Brettin T, Butler R, Butler RM, Chlenski P, Conrad N, Dickerman A, Dietrich EM, Gabbard JL, Gerdes S, Guard A, Kenyon RW, Machi D, Mao C, Murphy-Olson D, Nguyen M, Nordberg EK, Olsen GJ, Olson RD, Overbeek JC, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomas C, VanOeffelen M, Vonstein V, Warren AS, Xia F, Xie D, Yoo H, Stevens R. 2020. The PATRIC Bioinformatics Resource Center: expanding data and analysis capabilities. Nucleic Acids Res 48:D606–D612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emms DM, Kelly S. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol 20:238. 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Escalante NK, Lemire P, Cruz Tleugabulova M, Prescott D, Mortha A, Streutker CJ, Girardin SE, Philpott DJ, Mallevaey T. 2016. The common mouse protozoa Tritrichomonas muris alters mucosal T cell homeostasis and colitis susceptibility. J Exp Med 213:2841–2850. 10.1084/jem.20161776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ewers C, Stamm I, Pfeifer Y, Wieler LH, Kopp PA, Schonning K, Prenger-Berninghoff E, Scheufen S, Stolle I, Gunther S, Bethe A. 2014. Clonal spread of highly successful ST15-CTX-M-15 Klebsiella pneumoniae in companion animals and horses. J Antimicrob Chemother 69:2676–2680. 10.1093/jac/dku217. [DOI] [PubMed] [Google Scholar]
- 16.Feng Y, Mannion A, Madden CM, Swennes AG, Townes C, Byrd C, Marini RP, Fox JG. 2017. Cytotoxic Escherichia coli strains encoding colibactin and cytotoxic necrotizing factor (CNF) colonize laboratory macaques. Gut Pathog 9:71. 10.1186/s13099-017-0220-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foreman O, Kavirayani AM, Griffey SM, Reader R, Shultz LD. 2011. Opportunistic bacterial infections in breeding colonies of the NSG mouse strain. Vet Pathol 48:495–499. 10.1177/0300985810378282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.García A, Mannion A, Feng Y, Madden CM, Bakthavatchalu V, Shen Z, Ge Z, Fox JG. 2016. Cytotoxic Escherichia coli strains encoding colibactin colonize laboratory mice. Microbes Infect 18:777–786. 10.1016/j.micinf.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ge Z, Carrasco SE, Feng Y, Bakthavatchalu V, Annamalai D, Kramer R, Muthupalani S, Fox JG. 2020. Identification of a new strain of mouse kidney parvovirus associated with inclusion body nephropathy in immunocompromised laboratory mice. Emerg Microbes Infect 9:1814–1823. 10.1080/22221751.2020.1798288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howitt MR, Lavoie S, Michaud M, Blum AM, Tran SV, Weinstock JV, Gallini CA, Redding K, Margolskee RF, Osborne LC, Artis D, Garrett WS. 2016. Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science 351:1329–1333. 10.1126/science.aaf1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito M, Hiramatsu H, Kobayashi K, Suzue K, Kawahata M, Hioki K, Ueyama Y, Koyanagi Y, Sugamura K, Tsuji K, Heike T, Nakahata T. 2002. NOD/SCID/gamma(c)(null) mouse: an excellent recipient mouse model for engraftment of human cells. Blood 100:3175–3182. 10.1182/blood-2001-12-0207. [DOI] [PubMed] [Google Scholar]
- 22.Kaur CP, Vadivelu J, Chandramathi S. 2018. Impact of Klebsiella pneumoniae in lower gastrointestinal tract diseases. J Dig Dis 19:262–271. 10.1111/1751-2980.12595. [DOI] [PubMed] [Google Scholar]
- 23.Knapp L, Amezquita A, McClure P, Stewart S, Maillard JY. 2015. Development of a protocol for predicting bacterial resistance to microbicides. Appl Environ Microbiol 81:2652–2659. 10.1128/AEM.03843-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar V, Sun P, Vamathevan J, Li Y, Ingraham K, Palmer L, Huang J, Brown JR. 2011. Comparative genomics of Klebsiella pneumoniae strains with different antibiotic resistance profiles. Antimicrob Agents Chemother 55:4267–4276. 10.1128/AAC.00052-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lam MMC, Wick RR, Watts SC, Cerdeira LT, Wyres KL, Holt KE. 2021. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun 12:4188. 10.1038/s41467-021-24448-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu B, Zheng D, Jin Q, Chen L, Yang J. 2019. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res 47 D1:D687–D692. 10.1093/nar/gky1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mannion A, Fabian N, Stair M, Dzink-Fox J, Carrasco SE, Buckley-Jordan E, Annamalai D, Fox JG. 2019. Draft Genome Sequences of Klebsiella pneumoniae Strains Isolated from Immunocompromised NOD-scid Gamma Research Mice. Microbiol Resour Announc 8:e00869-19. 10.1128/MRA.00869-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamoto N, Sasaki N, Aoki R, Miyamoto K, Suda W, Teratani T, Suzuki T, Koda Y, Chu PS, Taniki N, Yamaguchi A, Kanamori M, Kamada N, Hattori M, Ashida H, Sakamoto M, Atarashi K, Narushima S, Yoshimura A, Honda K, Sato T, Kanai T. 2019. Gut pathobionts underlie intestinal barrier dysfunction and liver T helper 17 cell immune response in primary sclerosing cholangitis. Nat Microbiol 4:492–503. 10.1038/s41564-018-0333-1. [DOI] [PubMed] [Google Scholar]
- 29.Naparstek L, Carmeli Y, Chmelnitsky I, Banin E, Navon-Venezia S. 2012. Reduced susceptibility to chlorhexidine among extremely-drug-resistant strains of Klebsiella pneumoniae. J Hosp Infect 81:15–19. 10.1016/j.jhin.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 30.Paczosa MK, Mecsas J. 2016. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol Mol Biol Rev 80:629–661. 10.1128/MMBR.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pritchard L, Glover RH, Humphris S, Elphinstone JG, Toth IK. 2016. Genomics and taxonomy in diagnostics for food security: Soft-rotting enterobacterial plant pathogens. Anal Methods 8:12–24. 10.1039/C5AY02550H. [DOI] [Google Scholar]
- 32.Roediger B, Lee Q, Tikoo S, Cobbin JCA, Henderson JM, Jormakka M, O’Rourke MB, Padula MP, Pinello N, Henry M, Wynne M, Santagostino SF, Brayton CF, Rasmussen L, Lisowski L, Tay SS, Harris DC, Bertram JF, Dowling JP, Bertolino P, Lai JH, Wu W, Bachovchin WW, Wong JJ, Gorrell MD, Shaban B, Holmes EC, Jolly CJ, Monette S, Weninger W. 2018. An Atypical Parvovirus Drives Chronic Tubulointerstitial Nephropathy and Kidney Fibrosis. Cell 175:530–543.e524. 10.1016/j.cell.2018.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russo TA, Marr CM. 2019. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev 32:e00001-19. 10.1128/cmr.00001-19. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanchez GV, Master RN, Clark RB, Fyyaz M, Duvvuri P, Ekta G, Bordon J. 2013. Klebsiella pneumoniae antimicrobial drug resistance, United States, 1998-2010. Emerg Infect Dis 19:133–136. 10.3201/eid1901.120310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneemilch HD. 1976. A naturally acquired infection of laboratory mice with Klebsiella capsule type 6. Lab Anim 10:305–310. 10.1258/002367776781035224. [DOI] [PubMed] [Google Scholar]
- 36.Shon AS, Bajwa RP, Russo TA. 2013. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence 4:107–118. 10.4161/viru.22718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shultz LD, Schweitzer PA, Christianson SW, Gott B, Schweitzer IB, Tennent B, McKenna S, Mobraaten L, Rajan TV, Greiner DL. 1995. Multiple defects in innate and adaptive immunologic function in NOD/LtSz-scid mice. J Immunol 154:180–191. [PubMed] [Google Scholar]
- 38.Smith K, Hunter IS. 2008. Efficacy of common hospital biocides with biofilms of multi-drug resistant clinical isolates. J Med Microbiol 57:966–973. 10.1099/jmm.0.47668-0. [DOI] [PubMed] [Google Scholar]
- 39.Vijayakumar R, Sandle T. 2019. A review on biocide reduced susceptibility due to plasmid-borne antiseptic-resistant genes-special notes on pharmaceutical environmental isolates. J Appl Microbiol 126:1011–1022. 10.1111/jam.14118. [DOI] [PubMed] [Google Scholar]
- 40.Walker KA, Miller VL. 2020. The intersection of capsule gene expression, hypermucoviscosity and hypervirulence in Klebsiella pneumoniae. Curr Opin Microbiol 54:95–102. 10.1016/j.mib.2020.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whary MT, Baumgarth N, Fox JG, Barthold S. Biology and Diseases of Mice, p 43–137. In: Fox JG, Anderson L, Otto G, Pritchett-Corning K, Whary MT, editors. Laboratory Animal Medicine. London: Academic Press. [Google Scholar]
- 42.Young TM, Bray AS, Nagpal RK, Caudell DL, Yadav H, Zafar MA. 2020. Animal model to study Klebsiella pneumoniae gastrointestinal colonization and host-to-host transmission. Infect Immun 88:e00071-20. 10.1128/IAI.00071-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zadrozny LM, Brinster LR, Rosenzweig BA, Howard KE. 2018. Outbreak of Opportunistic Ascending Pyelonephritis with Numerous Yeast after Experimental Humanization Surgery in NOD.Cg-Prkdc(scid) Il2rg(tm1Wjl)/SzJ and NOD.Cg-Rag1(tm1Mom) Il2rg(tm1Wjl)/SzJ Immunodeficient Mice. Comp Med 68:353–359. 10.30802/AALAS-CM-17-000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Average nucleotide identity values
Integron BLAST results
Data Availability Statement
Genomes of Kp have been deposited in GenBank under the following accession numbers: SULO00000000, SULP00000000, SULQ00000000, SULR00000000, SULS00000000, SULT00000000, SULU00000000, SULV00000000, SULW00000000, SULX00000000, SULY00000000, SULZ00000000, SUMA00000000, JAEAGK000000000, and JAEAGL000000000.