Abstract
Chlamydia muridarum (Cm) was detected in 2 colonies of mice with lymphoplasmacytic pulmonary infiltrates by using PCR and immunohistochemistry. This discovery was unexpected, as Cm infection had not been reported in laboratory mice since the 1940s. A Cm specific PCR assay was developed and testing implemented for the resident colonies of 8 vivaria from 3 academic institutions, 58 incoming mouse shipments from 39 academic institutions, and mice received from 55 commercial breeding colonies (4 vendors). To estimate Cm’s global prevalence in research colonies, a database containing 11,387 metagenomic fecal microbiota samples from 120 institutions and a cohort of 900 diagnostic samples from 96 institutions were examined. Results indicate significant prevalence among academic institutions, with Cm detected in 63% of soiled bedding sentinels from 3 institutions; 33% of incoming mouse shipments from 39 academic institutions; 14% of 120 institutions submitting microbiota samples; and 16% of the diagnostic sample cohort. All samples from commercial breeding colonies were negative. In addition, naïve NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice exposed to Cm-shedding mice and/or their soiled bedding developed clinical disease at 21 to 28 d after exposure. These mice had a moderate-to-severe histiocytic and neutrophilic bronchointerstitial pneumonia, with their respiratory epithelium demonstrating inclusions, chlamydial major outer membrane protein immunostaining, and hybridization with a Cm reference sequence (GenBank accession no. U68436). Cm was isolated from lungs, cecum, and feces of a Cm-infected NSG mouse by using HeLa 229 cells. The considerable prevalence of Cm is likely due to widespread global interinstitutional distribution of unique mouse strains and failure to recognize that some of these mice were from enzootically infected colonies. Given that experimental Cm colonization of mice results in a robust immune response and, on occasion, pathology, natural infection may confound experimental results. Therefore, Cm should be excluded and eradicated from enzootically infected mouse colonies.
Abbreviations: Cm, Chlamydia muridarum; EB, elementary body; FFPE, formalin-fixed paraffin embedded; GEM, genetically engineered mouse; IB, inclusion body; IFA, immunofluorescence; IFU, inclusion forming units; IHC, immunohistochemistry; ISH, in-situ hybridization; MOMP, major outer membrane protein; MoPn, mouse pneumonitis virus; MSK, Memorial Sloan Kettering; NSG, NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ; RB, reticulate body; TLR, toll-like receptor
Introduction
Chlamydiae are obligate intracellular bacteria with an extensive host range at the genus level and high host specificity at the species level.15,37 Chlamydia muridarum (Cm) is the only natural chlamydial pathogen of mice identified to date. It has been used extensively to model sexually transmitted Chlamydia trachomatis infection of humans.15,37 Cm was first described in the late 1930s and further investigated in the early 1940s, when scientists identified a putative infectious organism causing respiratory disease and lung pathology in mice used in studies of influenza and “the virus of the common cold.”13,19 The etiologic agent was initially referred to as “mouse pneumonitis virus” (MoPn) in 1944.30 It was subsequently identified as a biovar of C. trachomatis and finally as Cm after phylogenetic analysis, genome sequencing, and comparative genomics.9,16,37,39 Despite widespread use as an experimental model, relatively little is known about the biology of natural infection, as Cm has not been routinely reported in or isolated from research mouse populations since its initial discovery. A closely related strain (denoted as strain SFPD) was isolated from hamsters in the early 1990s.16,43 This strain was shown to share 91% sequence homology of the major outer membrane (MOMP) with the Weiss and Nigg strains of Cm.51 While Cm was likely prevalent in early 20th C mouse colonies, the development and introduction of modern biosecurity practices such as C-section rederivation and barrier husbandry in the middle of the 20th C would have been expected to eliminate the bacterium from most vendors’ colonies. Accordingly, testing of commercial and research colonies for Cm is not routinely performed.
As with other chlamydiae, Cm has a biphasic life cycle comprised of the nonreplicating but infectious ‘elementary body’ (EB) and the replicating but noninfectious ‘reticulate body’ (RB).15,37 EBs enter host mucosal epithelial cells and are incorporated into a membrane-bound compartment (also called an ‘inclusion’ or ‘inclusion body,’ IB) in which they differentiate into RB.15,37 RBs replicate within inclusions before reverting into EBs, which are released and can infect nearby cells.15,37 Natural transmission is now thought to occur primarily via the fecal-oral route, similar to chlamydial infections in domestic animals, with the gastrointestinal tract often being the natural site of colonization.50 Experimental infection supports this idea, as orally inoculated immunocompetent mice remain persistently infected, shedding for up to 260 d, with replication primarily in the cecum and colon. Pulmonary lesions and colonization, as reported in the initial discovery of Cm and in subsequent experimental infections, are presumed to occur through aspiration or inhalation of the organism.30,37,50 Significant differences in host response to Cm infection are dependent on the infected mouse strain, with Cm’s MOMP considered an important antigenic stimulus.28,37
After our unexpected identification of Cm in association with lymphoplasmacytic pulmonary infiltrates in 2 genetically engineered mouse (GEM) strains from distinct colonies, we developed a Cm specific PCR assay and began a multifaceted investigation to determine the prevalence of Cm within our colonies, other institutional mouse colonies, and commercial vendor colonies. We further confirmed the global prevalence of Cm in research mouse colonies by examining fecal microbiome data from a large dataset and testing a large cohort of multi-institutional diagnostic samples and we successfully isolated Cm in cell culture from highly immunocompromised mice used as contact and soiled bedding sentinels.
Materials and Methods
History/index cases.
Cm was initially detected in 2020 in 2 unrelated GEM strains maintained at our institution by 2 laboratories. Histologic examination performed by laboratory A as part of their experimental protocol revealed an unexpected inflammatory lung infiltrate in an immunocompetent, clinically normal, experimentally naïve other than having received a single dose of tamoxifen, GEM strain. Histopathology of the lungs revealed mild to moderate, multifocal, perivascular lymphocytic aggregates in 1 of 3, 6-mo-old Foxp3DTRgCD4CreERT2R26tdTomato, knock-in reporter mice on a C57BL/6 background submitted for evaluation. No histologic evidence of bacterial or fungal pathogens was seen, but organisms known to colonize the respiratory tract and cause inflammation are not always detectable histologically. Accordingly, formalin-fixed paraffin embedded (FFPE) scrolls of lung tissue were assessed for Mycoplasma spp., Pneumocystis spp., and Cm by real time PCR. PCR targeting of the highly conserved chlamydial 23S rRNA gene with duplex fluorescent oligonucleotide probes followed by sequencing of the resulting amplicon surprisingly confirmed the presence of Cm (Molecular Diagnostics, Auburn University, Auburn, AL).12 Commercial PCR assays (IDEXX Bioanalytics, Columbia, MO) did not detect Mycoplasma spp. or Pneumocystis spp. Two additional mice (C57Bl/6-Foxp3tm1Flv/J; C57Bl/6/Foxp3CreERR26tdTomato) were submitted from this colony in 2021 for further analysis including complete blood count, serum biochemistry, histopathology, immunohistochemistry (IHC) for MOMP, and in-situ hybridization (ISH) for Cm mRNA.
Subsequently, complete necropsies were conducted on 2, 4-wk-old, female Cd23-Cre Gen1−/− Mus81fl/fl (conditional knockouts of DNA repair proteins in B cells on a C57BL/6 background) from a rodent colony in Laboratory B. These mice initially presented with runting, mild hunched posture, and alopecia and thinning of hair along the dorsum and bilateral flanks. The skin was characterized by follicular dystrophy compatible with a background lesion described in C57BL/6 mice.46 Both mice also had mild, multifocal, chronic lymphoplasmacytic bronchiolitis and bronchitis. Given these pulmonary lesions and the discoveries detailed for Laboratory A, additional diagnostics were pursued including MOMP IHC, ISH for Cm mRNA in lung tissue, and PCR.
Investigative plan.
After the unexpected detection of Cm in mice from 2 distinct institutional GEM colonies, a multifaceted investigation was undertaken to determine whether this finding was unique and, if Cm was found in other colonies, how widely it was distributed.
After developing a Cm specific PCR assay (described below), the following investigations were initiated: 1) All incoming GEM strains (n = 58) from noncommercial colonies were tested for Cm by fecal PCR; 2) Soiled bedding SW:Tac sentinels employed in our multi-institutional colony health monitoring program (described below) from 97 mouse holding rooms were tested by fecal PCR performed during their annual testing submissions; 3) Two NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were cohoused with 2 cages containing 4 Cm fecal PCR-positive mice each (recently imported from another academic institution) to increase the likelihood of isolating Cm. These mice were reported by their source institution to be an immunocompetent GEM strain with a floxed tumor-suppressor allele. After cohousing as contact sentinels for 1 wk, each of these NSG mice were subsequently cohoused with 4 naïve NSG mice in 2 cages, which continued to receive soiled bedding from the imported mice for an additional week. The mice were observed until morbidity developed, at which point they were euthanized, and a complete necropsy undertaken. IHC for MOMP and ISH were performed on select tissues, and samples of lung, cecum, and feces from a single morbid NSG contact sentinel were collected for subsequent isolation of Cm. The PCR product obtained from FFPE pulmonary scrolls from a single NSG sentinel mouse was sequenced and compared with published Cm sequences; 4) Cm PCR was performed on feces collected on arrival from mice sourced from 55 production rooms from 4 commercial breeding colonies. In addition, several nonresearch mouse populations were surveyed to determine whether Cm was present in mice outside of the research environment. Feces were collected from Mus musculus from 3 pet stores (pet shop A and B in NYC, pet shop C in Michigan), and from a wild population of Mus musculus in New Jersey; 5) Anonymized diagnostic samples (n = 900) were tested for Cm by PCR. Samples included feces, oral or body/fur swabs, tissue (mediastinal lymph node, lung, or spleen), environmental swabs (IVC exhaust air dust, plenum/filter/hose/environment) or filter material); and biologics (cells and/or tissue culture); and, 6) A database containing mouse fecal metagenomic microbiome sequencing data from 11,387 samples from global academic (n = 112) and commercial (n = 8) sources was queried for Cm abundance.
Mice.
Swiss Webster (Tac:SW; Taconic Biosciences, Germantown, NY) and NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG; The Jackson Laboratory, Bar Harbor, ME) were used. All mice were free of mouse hepatitis virus (MHV), Sendai virus, mouse parvovirus (MPV), minute virus of mice (MVM), murine norovirus (MNV), murine astrovirus 2 (MuAstV2), pneumonia virus of mice (PVM), Theiler meningoencephalitis virus (TMEV), epizootic diarrhea of infant mice (mouse rotavirus, EDIM), ectromelia virus, reovirus type 3, lymphocytic choriomeningitis virus (LCMV), K virus, mouse adenovirus 1 and 2 (MAD 1/2), polyoma virus, murine cytomegalovirus (MCMV), mouse thymic virus (MTV), Hantaan virus, mouse kidney parvovirus (MKPV), Mycoplasma pulmonis, CAR bacillus, Chlamydia muridarum, Citrobacter rodentium, Rodentibacter pneumotropicus, Helicobacter spp., segmented filamentous bacterium (SFB), Salmonella spp., Streptococcus pneumoniae, Beta-hemolytic Streptococcus spp., Streptobacillus moniliformis, Filobacterium rodentium, Clostridium piliforme, Corynebacterium bovis, Corynebacterium kutscheri, Staphylococcus aureus, Klebsiella pneumoniae, Klebsiella oxytoca, Pseudomonas aeruginosa, fur mites (Myobia musculi, Myocoptes musculinis, and Radfordia affinis), pinworms (Syphacia spp. and Aspiculuris spp.), Demodex musculi, Pneumocystis spp, Giardia muris, Spironucleus muris, Entamoeba muris, Tritrichomonas muris, and Encephalitozoon cuniculi when studies were initiated.
Husbandry and housing.
Mice were housed in individually ventilated, polysulfone shoebox cages with stainless-steel wire-bar lids and filter tops (experimental and colony; number 19, Thoren Caging Systems, Hazelton, PA) or in solid-bottom polysulfone microisolation cages maintained in a custom-designed quarantine caging system (noncommercial mice imported from other institutions were housed in a dedicated quarantine facility; BioZone Global, Chester, SC). Mice were housed on autoclaved aspen chip bedding (PWI Industries, Quebec, Canada) at a density of no greater than 5 mice per cage. Each cage was provided with a Glatfelter paper bag containing 6 g of crinkled paper strips (EnviroPak, WF Fisher and Son, Branchburg, NJ) for enrichment. Mice were fed either a natural ingredient, closed-source, flash-autoclaved, g-irradiated feed (experimental and colony; LabDiet 5053, PMI, St. Louis, MO) or g-irradiated feed containing 150 PPM fenbendazole and 13 PPM ivermectin (TestDiet, PMI, fed ad libitum during quarantine).47 All mice received ad libitum, autoclaved, reverse osmosis, acidified (pH 2.5 to 2.8 with hydrochloric acid) water in polyphenylsulfone bottles with stainless-steel caps and sipper tubes (Techniplast, West Chester, PA). Cages were changed every 7 d in either a class II, type A2 biologic safety cabinet (LabGard S602-500, Nuaire, Plymouth, MN) or an animal changing station (Nuaire NU-S612-400, Nuaire, Plymouth MN). Rooms were maintained on a 12:12-h light:dark cycle, relative humidity of 30% to 70%, and temperature of 72 ± 2 °F (22.2 ± 1.1 °C). Rooms were classified as receiving mice from all sources, (for example, noncommercial sources [released after testing for specific agents] and commercial breeders), from noncommercial sources (rederived into axenic state and reassociated with defined, vendor-specific flora), and/or from commercial breeders only. The animal care and use program at Memorial Sloan Kettering Cancer Center (MSK) is accredited by AAALAC International, and all animals are maintained in accordance with the recommendations provided in the Guide.23 All animal use described in this investigation was approved by MSK’s IACUC in agreement with AALAS position statements on the Humane Care and Use of Laboratory Animals and Alleviating Pain and Distress in Laboratory Animals.1,2
Mouse importation programs and colony health monitoring.
Mice imported from noncommercial sources like academic institutions and atypical commercial sources (commercial research companies, nonapproved commercial breeders, and approved commercial breeders who perform insufficient testing) are subject to quarantine and testing. The length of quarantine and scope of testing are determined by several factors, including the intended use of the mice and their health status. No imported mice were tested for Cm prior to arrival, as institutions and companies do not routinely test for this agent. Transport crates are received into the quarantine facility, where they are sprayed with chlorine dioxide (Clidox-S 1:18:1 dilution, Pharmacal Laboratories, Naugatuck, CT) and placed in the assigned holding room in a dedicated, physically separated, quarantine facility operated at ABSL-2. Testing is comprised of at least 2 rounds of testing and involves either all mice (when 10 or fewer are received), or a minimum of 10 mice with at least 1 from each cage received. Initial testing (QA) is performed within 72 h of arrival; this testing includes veterinary assessment and PCR on fecal pellet and oral and fur/pelt swabs. Secondary testing (QB) is performed 4 wk after QA and includes serology using 0.25 mL blood collected via the submandibular vein from 1 mouse per cage. PCR and serologic samples are tested by a commercial diagnostic laboratory (Charles River Laboratories, Cambridge, MA) for the above listed agents (see “Mice”). Testing for the presence of Cm was added to the routine institutional quarantine testing panel in October 2021.
The soiled bedding sentinel colony health monitoring program has been previously described.25 In brief, 3- to 5-wk-old female SW:Tac mice are used as soiled bedding sentinels. Each sentinel cage contains 4 mice and surveys a maximum of 280 cages. Every week, each sentinel cage receives approximately 15 mL dirty bedding from each of 40 colony cages, with weekly alternating of selected cages so that all cages are surveyed over a 7-wk period. One sentinel mouse per cage is sampled every 8 wk, with blood collected for serology and feces/pelt swabs for pathogen testing by PCR. Survival blood collection is performed using a sterile 25g needle, with samples taken from either the tail, submandibular or submental vein. One sentinel mouse per cage is euthanized at 6 and 12 mo after placement for additional testing, including blood collection for serology (cardiac puncture after euthanasia by CO2 asphyxiation), pelt and large intestinal content examination for ecto- and endoparasites, and gross necropsy with subsequent histology if lesions are identified. Every 2 mo over a 12 mo period, sentinels are tested for fur mites and pinworms (above), as well as D. musculi, S. muris, MHV, Sendai virus, MPV, MVM, PVM, TMEV, EDIM, MNV, MuAstV2, Reovirus 3, M. pulmonis, R. pneumotropicus, C. bovis, S. aureus, Helicobacter spp., and SFB. Sentinels are also tested every 6 mo for Ectromelia virus, LCMV, K-Virus, MAD1/2, Polyoma virus, MKPV, C. rodentium, Salmonella spp., K. pneumoniae/oxytoca, S. pneumoniae, Beta-hemolytic Strep., P. aeruginosa, and Pneumocystis spp. and every 12 mo for MCMV, MTV, Hantaan virus, Filobacterium rodentium, C. pilliforme, C. kutscheri, S. moniliformis, E. cuniculi, G. muris, E. muris, and T. muris. Testing for Cm was added to the 6- and 12-mo routine soiled bedding colony health surveillance testing panels in December 2021.
Fecal collection.
Fecal pellets for PCR assay or microbiome analysis were collected from the soiled bedding, directly from the mouse, or both. When collected directly, mice were lifted by the base of the tail and allowed to grasp onto the wire-bar lid. Mice were gently restrained in this manner while a sterile 1.5-mL microfuge tube was held below the anus to allow feces to fall directly into the tube. If the mouse did not defecate within a 30-s period, it was returned to the cage and allowed to rest for at least 2 min. Collection was then reattempted until a sample was produced. A total of 2 fecal pellets were collected for each microbiome sample, and 10 fecal pellets were collected for each PCR sample.
Chlamydia muridarum PCR assay and amplicon sequencing.
A proprietary real-time fluorogenic 5¢ nuclease PCR assay specifically targeting a 425 bp region Cm 23S rRNA was used to determine the presence of genomic DNA in samples. Samples that amplified during initial testing were subsequently retested using DNA isolated from a retained lysate sample to confirm the original finding. A positive result was reported when the retested sample was confirmed positive. To monitor for successful DNA recovery after extraction and to assess whether PCR inhibitors were present, an exogenous nucleic acid recovery control assay was added to each sample after the lysis step and prior to magnetic nucleic acid isolation. The concentration of eluted nucleic acid in mock extracted samples (no sample material) was calibrated to approximately 40 copies of exogenous DNA/µL and compared with a 100-copy system suitability control. A second real-time fluorogenic 5¢ nuclease PCR assay was used to target the exogenous template to serve as a sample suitability control and was performed simultaneously with the Cm assay. Nucleic acid recovery control assays for samples that had greater than a log10 loss of template copies compared with control wells were diluted 1:4 and retested, reextracted or both prior to accepting results as valid. A 100-copy/reaction positive control plasmid template containing the Cm target template was co-PCR amplified with the test sample to demonstrate master mix and PCR amplification equipment function. PCR products from select samples were purified and sequenced using the Sanger method (Genomics Core, Tufts University Medical School, Boston, MA). Sequence results were further processed by trimming the sequencing primers and any undetermined nucleotide bases. The clean consensus was analyzed by comparing it to available Cm sequences in GenBank.
Pathology.
After euthanasia by CO2 asphyxiation, a complete necropsy was performed. Gross lesions were recorded, fresh samples of lungs were collected for aerobic culture, and all tissues including heart, thymus, lungs, liver, gallbladder, kidneys, pancreas, stomach, duodenum, jejunum, ileum, cecum, colon, lymph nodes (mandibular, mesenteric), salivary glands, skin (trunk and head), urinary bladder, uterus, cervix, vagina, ovaries, oviducts, adrenal glands, spleen, thyroid gland, esophagus, trachea, spinal cord, vertebrae, sternum, femur, tibia, stifle joint, skeletal muscle, nerves, skull, nasal cavity, oral cavity, teeth, ears, eyes, pituitary gland, and brain were fixed in 10% neutral buffered formalin. After fixation, the skull, spinal column, sternum, femur, and tibia were decalcified in a formic acid and formaldehyde solution (Surgipath Decalcifier I, Leica Biosystems). Tissues were then processed in ethanol and xylene and embedded in paraffin in a tissue processor (Leica ASP6025, Leica Biosystems). Paraffin blocks were sectioned at 5-mm thickness, stained with hematoxylin and eosin (H and E), and examined by a board-certified veterinary pathologist (SC). Scrolls of lung (20 mm) were obtained from paraffin blocks for bacterial genome sequencing (below).
Peripheral blood was collected from a subset of mice by cardiac puncture. For hematology, blood was collected into tubes containing EDTA. Samples were analyzed on an automated hematology analyzer (IDEXX Procyte DX, Columbia, MO) and the following parameters were determined: white blood cell count, red blood cell count, hemoglobin concentration, hematocrit, mean corpuscular volume, mean corpuscular hemoglobin, mean corpuscular hemoglobin concentration, red blood cell distribution width standard deviation and coefficient of variance, reticulocyte relative and absolute counts, platelet count, platelet distribution width, mean platelet volume, and relative and absolute counts of neutrophils, lymphocytes, monocytes, eosinophils, and basophils. For serum chemistry, blood was collected into tubes containing a serum separator and centrifuged. The serum was then eluted for analysis. Serum chemistry was performed using an automated analyzer (Beckman Coulter AU680, Brea, CA). The concentration of the following analytes was determined: alkaline phosphatase, alanine aminotransferase, aspartate aminotransferase, creatine kinase, g-glutamyl transpeptidase, albumin, total protein, globulin, total bilirubin, blood urea nitrogen, creatinine, cholesterol, triglycerides, glucose, calcium, phosphorus, chloride, potassium, and sodium. The Na/K and the albumin/globulin ratios were calculated.
Immunohistochemistry (IHC).
All tissues evaluated by histopathology were screened for chlamydial MOMP using a technique optimized and validated by MSK’s Laboratory of Comparative Pathology. Formalin-fixed paraffin-embedded sections were stained using an automated staining platform (Leica Bond RX, Leica Biosystems). After deparaffinization and heat-induced epitope retrieval in a citrate buffer at pH 6.0, the primary antibody against chlamydial MOMP (NB100-65054, Novus Biologicals, Centennial, CO) was applied at a dilution of 1:500. A rabbit anti-goat secondary antibody (Cat. No. BA-5000, Vector Laboratories, Burlingame, CA) and a polymer detection system (DS9800, Novocastra Bond Polymer Refine Detection, Leica Biosystems) was then applied to the tissues. The 3,3¢-diaminobenzidine tetrachloride (DAB) was used as the chromogen, and the sections were counterstained with hematoxylin and examined by light microscopy. Reproductive tracts from TLR3-deficient mice experimentally infected with Chlamydia muridarum strain Nigg were used as the positive control.7
In situ hybridization (ISH).
Select IHC-positive tissues were included for ISH analysis. The target probe was designed to detect region 581 to 617 of Chlamydia muridarum str. Nigg complete sequence, NCBI Reference Sequence NC_002620.2 (1039538-C1; Advanced Cell Diagnostics, Newark, CA). The target probe was validated on reproductive tracts from mice experimentally inoculated with Chlamydia muridarum strain Nigg.7 Slides were stained on an automated stainer (Leica Bond RX, Leica Biosystems) with RNAscope 2.5 LS Assay Reagent Kit-Red (322150, Advanced Cell Diagnostics) and Bond Polymer Refine Red Detection (DS9390, Leica Biosystems). Control probes detecting a validated positive housekeeping gene (mouse peptidylprolyl isomerase B, Ppib to confirm adequate RNA preservation and detection; 313918, Advanced Cell Diagnostics) and a negative control, Bacillus subtilis dihydrodipicolinate reductase gene (dapB to confirm absence of nonspecific staining; 312038, Advanced Cell Diagnostics) were used. Positive RNA hybridization was identified as discrete punctate chromogenic red dots under bright field microscopy.
Chlamydia muridarum 23S rRNA amplicon sequencing.
DNA was extracted according to the manufacturers’ instructions (MagMAX Total Nucleic Acid Isolation Kit, Life Technologies, Carlsbad, CA and a Qiagen robotic extraction station, Thermo Labsystems, Franklin, MA) from pulmonary tissue scrolls from an NSG mouse that had been used as a contact sentinel. A region of the 23S rRNA gene was amplified using HotStar Taq polymerase (Qiagen) with the primers CHLSE37 (5¢-CTTGGCATTGACAGGCGA-3¢) and CHLSE461 (5¢-GGAGAGTGGTCTCCCCAGATT-3¢), which generated an expected product of 425 bp; primer nomenclature is based on the nucleotide positions of a Cm reference sequence (GenBank accession no., U68436) and was designed to detect within the genus of Chlamydia. Thermal cycling conditions were: denaturation at 95 °C for 15 min, followed by an initial 5 cycles of denaturation at 94 °C for 30 s, primer annealing at 64 °C for 30 s, and extension at 72 °C for 60 s. The annealing temperature was then decreased to 58 °C for 35 cycles; all other parameters remained as described for the initial 5 cycles. PCR products were separated on 2% agarose gels, eluted from the gel (Minelute Kit, Qiagen), and sequenced (ABI 3130XL DNA sequencer, Tufts University Core Facility, Boston, MA).
Isolation and growth of chlamydiae.
Samples (lung, cecum, and feces) from an NSG contact sentinel were macerated, placed in sucrose-phosphate-glutamic acid buffer (pH 7.2), frozen at −80 °C, and shipped on dry ice to Midwestern University where they were again held at −80 °C until isolation was conducted. Methods have been previously described.35 Briefly, all samples were thawed, sonicated, and centrifuged (400 × g for 10 min at 4 °C) to pellet cellular and other debris. The pelleted debris was discarded, and the supernatants were diluted in Dulbecco modified essential medium (DMEM, containing 584 mg/L glutamine, 4.5g/L glucose). Fecal and cecum samples were then filtered through 0.22-mM sterile filter. Samples were then inoculated on confluent monolayers of HeLa 229 in 24-well tissue culture plates at 500 ml per well. Inoculated 24-well plates were then centrifuged at 1,100 g for 1 h at 37 °C. After centrifugation, the plates were incubated for an additional 1 h at 37 °C in 5% CO2. The inocula were then aspirated and replaced with chlamydial growth media (DMEM containing, 10% fetal calf serum, cycloheximide 0.5 mg/mL) and antibiotics (vancomycin 100 µg/mL, gentamicin 50 µg/mL, and amphotericin B 1.25 µg/mL) to inhibit growth of mouse microbial flora.35 Monolayers were then monitored for inclusion formation by scanning twice daily by inverted microscopy. In some cases, initial dilutions were too low and chlamydial cytotoxicity was observed that we believe was due to chlamydial growth, as described elsewhere.4 In other cases, inclusions were observed by 36-, 48-, or 72-h after inoculation; the 24-well plate flask was then fixed in cold methanol for visualization of inclusions by indirect immunofluorescent assay (IFA).10 Duplicate plates were run in parallel for expanding isolates and confirming Chlamydia detection by 16S rRNA PCR, as previously described.49
Fecal microbiome analysis.
Collection kits (Transnetyx Microbiome, Transnetyx, Cordova, TN) containing barcoded sample collection tubes were used. Fecal samples (2 pellets from each mouse) were placed in separate tubes containing DNA stabilization buffer to ensure reproducibility, stability, and traceability, and were shipped for DNA extraction, library preparation, and sequencing (Transnetyx). DNA extraction was performed using the Qiagen DNeasy 96 PowerSoil Pro QIAcube HT extraction kit and protocol for reproducible extraction of inhibitor-free, high molecular weight genomic DNA that captures the true microbial diversity of stool samples. After DNA extraction and quality control, genomic DNA was converted into sequencing libraries using the KAPA HyperPlus library preparation protocol optimized for minimal bias. Unique dual indexed adapters were used to ensure that reads and/or organisms were not mis-assigned. Thereafter, the libraries were sequenced using the NovaSeq instrument (Illumina, San Diego, CA) and protocol via the shotgun sequencing method (a depth of 2 million 2 × 150 bp read pairs), which enables species and strain level taxonomic resolution. Raw data (in the form of FASTQ files) were analyzed using the One Codex analysis software and database. The One Codex Database consists of approximately 114K complete microbial genomes, including 62K distinct bacterial genomes, 48K viral genomes, and thousands of archaeal and eukaryotic genomes.31 Human and mouse genomes are included to screen out host reads. The database was assembled from both public and private sources, with a combination of automated and manual curation steps to remove low quality or mislabeled records. Every individual sequence (NGS read or contig) is compared against the One Codex database by exact alignment using k-mers where k = 31. Based on the relative frequency of unique k-mers in the sample, sequencing artifacts are filtered out of the sample to eliminate false positive results caused by contamination or sequencing artifacts. The relative abundance of each microbial species is estimated based on the depth and coverage of sequencing across every available reference genome.
Results
Index cases – Laboratory A.
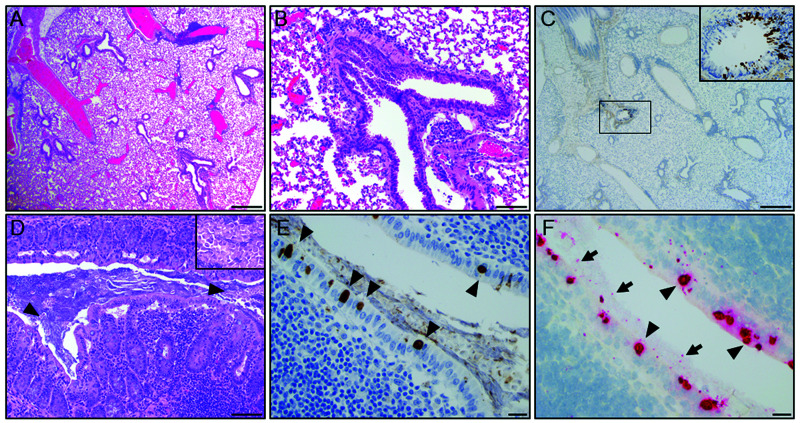
The initial index cases in Laboratory A were 2 experimentally naïve, 1.5-y-old, female, C57Bl/6-Foxp3tm1Flv/J; C57Bl/6/Foxp3CreERR26tdTomato mice that were found to have unexpected pulmonary inflammatory infiltrates on routine experimental histology as described above. One of the 2 mice (C57Bl/6/Foxp3CreERR26tdTomato) had perivascular and peribronchiolar lymphoplasmacytic aggregates (Figure 1A and B). Pulmonary FFPE scrolls from the affected lung were assayed by real time PCR targeting of the chlamydial 23S rRNA gene followed by sequencing of the resulting amplicon. This analysis yielded chlamydial DNA with 98.22% identity to Cm isolate CM001 (GenBank accession: CP027217.1). Subsequently, bronchial epithelium staining for Chlamydia MOMP by IHC was detected in an affected airway (Figure 1C). Additional spontaneous and/or incidental age-related lesions found in these mice included: mild, lymphoplasmacytic enteritis (n = 2); mild cecal trichomoniasis (n = 2); mild multifocal hepatocellular necrosis associated with neutrophilic and histiocytic infiltrates (n = 2); mild, lymphoplasmacytic colitis (n = 1), lymphocytic interstitial nephritis (n = 1); lymphocytic aggregates in the mesentery, mediastinum and the salivary glands (n = 1); and, subcapsular spindle cell hyperplasia in the adrenal gland (n = 1). No microscopic lesions were detected in the cecum or colon of these mice (Figure 1D); however, chlamydial antigen was detected in moderate amounts in surface cecal, and colonic epithelial cells in proximity to gut associated lymphoid tissue (Figure 1E). Chlamydial antigen was not detected in other organs assayed (heart, thymus, kidneys, adrenal glands, liver, gallbladder, spleen, pancreas, stomach, duodenum, jejunum, ileum, salivary glands, lymph nodes, trachea, esophagus, reproductive tract, mammary glands, skin, bones, skeletal muscle, femoral and spinal nerves, teeth, pituitary gland, and brain). Chlamydial 23S rRNA was also detected in select cecal and colonic epithelial cells by ISH (Figure 1F). Red and white blood cell counts and serum chemistry results from these mice were unremarkable.
Figure 1.
Representative histopathology of the lung and cecum from a 1.5-y-old, female, C57Bl/6/Foxp3CreERR26tdTomato mouse. (A) Multifocally, peribronchiolar and perivascular spaces are infiltrated by small to moderate clusters of lymphocytes and histiocytes (scale bar = 500 µm). (B) High magnification field shows lymphocytic and histiocytic infiltrates around a bronchiole (scale bar = 100 µm). (C) IHC of the lung demonstrating focal detection of chlamydial MOMP antigen in bronchiolar epithelial cells (scale bars = 100 µm; 20 µm - brown staining in inset). (D) Representative section of normal mucosa and gut-associated lymphoid tissue (GALT) in the cecum with low numbers of luminal T. muris (arrowheads and inset; scale bars = 100 µm). (E) IHC of the cecum demonstrating detection of intracytoplasmic chlamydial inclusions in surface epithelial cells (arrowhead; scale bar = 20 µm). (F) ISH demonstrates positive staining (red) for Cm mRNA within the cecal epithelium (arrowhead) and lumen (arrow; scale bar = 20 µm).
Laboratory B.
The second set of index cases were 2, 4-wk-old, female Cd23-Cre Gen1−/− Mus81fl/fl mice from Laboratory B. These mice had presented for alopecia and runting. Microscopically, skin lesions were consistent with a spontaneous follicular dystrophy known to occur in mice of this genetic background.46 These mice also demonstrated a mild, multifocal, chronic lymphoplasmacytic nasopharyngitis, tracheitis, bronchiolitis and bronchitis. IHC revealed focal staining of chlamydial MOMP in bronchiolar epithelial cells in normal bronchioles and in bronchioles surrounded by lymphoplasmacytic infiltrates in both mice, as well in surface colonic epithelium in 1 of the 2 mice. Pulmonary FFPE scrolls from 1 of the mice were submitted for genus-specific PCR and amplicon sequencing (as described for the mice from Laboratory A) and yielded similar results. Other microscopic findings seen in these mice included neutrophilic otitis media (n = 2), small intestinal spironucleosis (n = 2), subacute nasopharyngitis (n = 2) and minimal to mild tracheitis and/or hepatitis (n = 1).
Analysis of imported mice for Chlamydia muridarum.
Approximately 33% of the 58 groups of mice imported from academic institutions, pharmaceutical/biotechnology companies, and nonapproved commercial breeders were positive when assessed for Cm infection by fecal PCR (Table 1). Imported mice were classified ‘positive’ if Cm was detected via PCR on samples collected during both QA and QB. In 4 groups, only samples collected during QB were positive. In 3 of 4 of these cases, the laboratory had introduced Cm positive mice from their colony for breeding during quarantine, and therefore these groups were classified as ‘negative.’ The remaining case was not related to the introduction of colony mice. A subsequent additional (3rd) PCR assay was also found to be positive, and the group was therefore classified as ‘positive.’ The initial negative result was presumed to be a false negative. The 19 positive groups had been imported from 16 different institutions. While the preponderance of the groups received had come from other US institutions, a group of mice imported from Germany was also positive. No mice imported during this period were observed to have any clinical abnormalities.
Table 1.
Detection of Chlamydia muridarum in imported mice
| Sample Totals | Institution Totals | |||||
|---|---|---|---|---|---|---|
| # Samples | # Positive | % Positive | # Institutions | # Positive | % Positive | |
| Europe | 8 | 1 | 13% | 7 | 1 | 14% |
| Austria | 1 | 0 | 0% | 1 | 0 | 0% |
| Denmark | 1 | 0 | 0% | 1 | 0 | 0% |
| France | 1 | 0 | 0% | 1 | 0 | 0% |
| Germany | 3 | 1 | 33% | 2 | 1 | 50% |
| UK | 2 | 0 | 0% | 2 | 0 | 0% |
| Asia | 2 | 0 | 0% | 2 | 0 | 0% |
| Japan | 2 | 0 | 0% | 2 | 0 | 0% |
| Northeast US | 28 | 10 | 36% | 15, 1α | 9, 0α | 60%, 0%α |
| Connecticut | 1 | 1 | 100% | 1 | 1 | 100% |
| Distr. Columbia | 1 | 0 | 0% | 1 | 9 | 0% |
| Maryland | 5 | 1 | 20% | 4 | 1 | 25% |
| New York | 16 | 5 | 31% | 7, 1α | 5, 0α | 71%, 0%α |
| Pennsylvania | 5 | 3 | 60% | 3 | 2 | 67% |
| Midwest US | 6 | 3 | 50% | 3 | 2 | 67% |
| Missouri | 5 | 2 | 40% | 2 | 1 | 50% |
| Wisconsin | 1 | 1 | 100% | 1 | 1 | 100% |
| South US | 8 | 2 | 25% | 7 | 2 | 29% |
| Alabama | 1 | 0 | 0% | 1 | 0 | 0% |
| North Carolina | 3 | 0 | 0% | 2 | 0 | 0% |
| Tennessee | 1 | 0 | 0% | 1 | 0 | 0% |
| Texas | 3 | 2 | 67% | 3 | 2 | 67% |
| West US | 6 | 3 | 50% | 3, 1β | 2, 0β | 67%, 0%β |
| California | 6 | 3 | 50% | 3, 1β | 2, 0β | 67, 0%β |
| Total | 58 | 19 | 33% | 37, 1α, 1β | 16,0α, 0β | 43%, 0%α, 0%β |
Results are from academic colonies unless otherwise indicated. α = noncommercial breeder; β = biotechnology company
Colony surveillance (sentinels) for Chlamydia muridarum.
Of the 97 animal holding rooms from 3 institutions that were surveyed by examining soiled bedding sentinel mice for Cm via fecal PCR, 61 (63%) were positive. These positives represented all 8 (100%) of the surveyed vivaria (Table 2). Of the 90 rooms surveyed that received imported mice after testing for excluded agents and release, 61 (68%) were positive for Cm. None of the 7 holding rooms that require receipt from commercial vendors or C-section rederivation of imported mice were Cm-positive.
Table 2.
Detection of Chlamydia muridarum in soiled bedding sentinels
| Facility | Rm. Type | # Rooms | # Positive | % Positive |
|---|---|---|---|---|
| A | 18 | 13 | 72% | |
| STD | 16 | 13 | 81% | |
| R/V | 2 | 0 | 0% | |
| B | 13 | 8 | 62% | |
| STD | 13 | 8 | 62% | |
| C | 3 | 3 | 100% | |
| STD | 3 | 3 | 100% | |
| D | 7 | 4 | 57% | |
| STD | 6 | 4 | 67% | |
| R/V | 1 | 0 | 0% | |
| E | 36 | 19 | 53% | |
| STD | 32 | 19 | 59% | |
| R/V | 4 | 0 | 0% | |
| F | 12 | 8 | 67% | |
| STD | 12 | 8 | 67% | |
| G | 5 | 3 | 60% | |
| STD | 5 | 3 | 60% | |
| H | 3 | 3 | 100% | |
| STD | 3 | 3 | 100% | |
| Total | 97 | 61 | 63% |
STD = Standard sources: animal holding rooms containing mice obtained from commercial breeders and noncommercial sources (released after testing for specific agents). R/V = Rederived/Vendor: animal holding rooms receiving rederived mice from noncommercial sources (rederived into axenic state and reassociated with defined, vendor-specific flora) and/or commercial breeders.
Exposure of NSG mice to Chlamydia muridarum shedding imported mice.
One of the 2 NSG mice that was cohoused with Cm-shedding (confirmed via PCR) imported mice and 4 of the 8 NSG mice that were cohoused with these NSG contacts (which continued to receive soiled bedding from the imported mice) became acutely ill 3 wk after the initial cohoousing with the 2 NSG contact sentinels and were euthanized. The affected mice were lethargic, hunched, had unkempt hair coats, and had lost weight. The remaining 5 NSG mice (including the second contact sentinel and 4 remaining soiled bedding sentinel mice) were euthanized 1 wk later when they developed similar clinical signs.
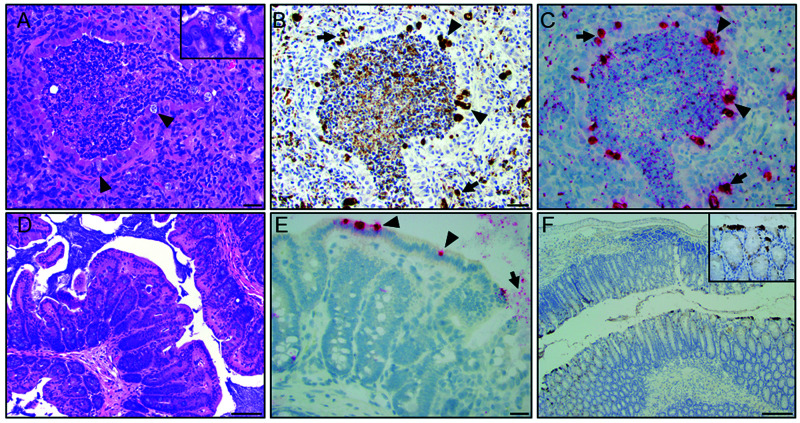
The initial 2 contact sentinels had a moderate to severe, multifocal to coalescing, chronic, histiocytic, and neutrophilic bronchointerstitial pneumonia. Bronchiolar lumens contained degenerate neutrophils, necrotic cells, karyorrhectic debris, fibrin, and proteinaceous eosinophilic fluid (Figure 2A). Bronchiolar epithelial cells often contained clear intracytoplasmic vacuoles with pale-basophilic inclusions compatible with chlamydial inclusions (Figure 2A). Intracytoplasmic inclusions were also seen in macrophages and alveolar epithelial cells within regions of peribronchiolar and alveolar inflammation. Peribronchiolar and alveolar spaces were infiltrated by large numbers of macrophages and neutrophils and were associated with edema or contained pyknotic cells, karyorrhectic debris and fibrin. Additional microscopic findings in these mice included a minimal to mild, multifocal, subacute neutrophilic and histiocytic typhlocolitis with crypt and goblet cell hyperplasia and crypt dilatation. Chlamydial MOMP antigen was detected by IHC in many surface epithelial cells in histologically normal regions of the small and large intestine (Figure 2D and 2F), in cecal segments with the aforementioned inflammation and in histologically normal segments of the trachea, nasopharynx, and lungs of both mice. One mouse had positive staining in the nasal cavity, and another in the spleen. Chlamydial mRNA was also detected in the cecal and colonic epithelium (Figure 2E), and in tracheal and lung epithelial cells of both mice by ISH. The inclusions seen in the lung on H and E correlated well with IHC and ISH staining, detecting chlamydial MOMP antigen and mRNA, respectively, in affected bronchiolar epithelial cells and regions of peribronchiolar and alveolar inflammation (Figure 2B and C). Chlamydial MOMP antigen and mRNA were also detected in the affected bronchiolar lumens admixed with degenerate neutrophils, necrotic debris, and proteinaceous material as described above. Pulmonary FFPE scrolls from the 2 mice were Cm PCR positive. Aerobic pulmonary cultures from both mice were negative.
Figure 2.
Histopathology of the lung and large intestine from a 1-y-old, female, NSG mouse. (A) Representative airway demonstrating a bronchiolar and alveolar inflammation characterized by luminal neutrophilic infiltration mixed with necrotic debris and proteinaceous material. Multifocally, bronchiolar epithelial cells exhibit intracytoplasmic clear vacuoles with pale-basophilic structures compatible with Chlamydial inclusions (arrowhead and inset). Peribronchiolar and alveolar space is infiltrated with moderate numbers of macrophages and neutrophils intermixed with reactive fibroblasts (scale bar = 50 µm). (B) IHC of the lung demonstrating detection of chlamydial MOMP antigen in bronchiolar epithelial cells (arrowhead; brown staining) and areas with peribronchiolar inflammation (arrow, scale bar = 50 µm). (C) ISH demonstrates positive staining (red) for Cm mRNA in bronchiolar epithelial cells (arrowhead) and areas of peribronchiolar inflammation (arrow; scale bar = 50 µm). (D) Representative H and E-stained section of a normal cecal wall (scale bars = 100 µm). (E) High magnification field demonstrates ISH signal (red staining) in the cecal epithelium (arrow) and lumen (arrowhead; scale bar = 50 µm). (F) IHC of descending colon demonstrating detection of intracytoplasmic chlamydial MOMP antigen in surface epithelial cells (inset - brown staining; scale bars = 20 and 200 µm).
A moderate leukocytosis (5.50 to 9.34 × 103/µL, reference range 0.94 to 4.68 × 103/µL) characterized by a moderate to marked neutrophilia (5.18 to 8.77 × 103/µL, reference range 0.54 to 3.16 × 103/µL) was also found in these mice.
Chlamydia muridarum 23S rRNA amplicon sequencing.
Sequence analysis of the amplified DNA 387 bp fragment from a segment of the 23S rRNA gene showed 100% identity with 22 Cm Nigg strain isolates (GenBank accessions: CP027217.1, CP027216.1, CP027215.1, CP027214.1, CP027213.1, CP027212.1, CP027211.1, CP027210.1, CP027209.1, CP027208.1, CP027207.1, CP027206.1, CP007217.1, CP009760.1, CP009609.1, CP009608.1, CP007276.1, CP006975.1, CP006974.1, NR_076163.1, CP063055.1, and AE002160.2) and with a single Cm isolate listed only as MoPn (GenBank accession: U68436.4) and a Cm isolate listed as SFPD, noted as isolated in a hamster (GenBank accession: U68437.2). There was no sequence homology with the single strain Weiss whole genome sequence (GenBank accession: NZ_ACOW00000000.1); however, close examination revealed the sequence was incomplete and lacked the entire amplified DNA 387 bp fragment.
Assessment of commercial breeders, pet store, and wild mice for Chlamydia muridarum.
Chlamydia muridarum was not detected in feces collected from any of the mice received directly from any of the 55 production housing rooms, located in 5 US states and 2 Canadian provinces, of 4 commercial breeders. All 3 samples from Pet Shop A, collected at 2 distinct time points, were positive for Cm. Samples from pet shops B and C were negative. The fecal sample from the single wild mouse population was negative.
Isolation of Chlamydia muridarum.
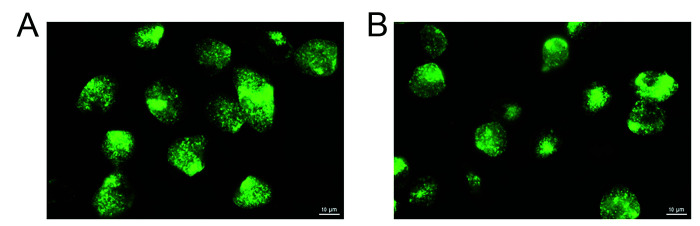
Chlamydia muridarum was isolated from the lung, cecum, and feces of an NSG mouse that was cohoused with imported mice shedding Cm. Cytotoxicity was observed in HeLa 229 cell cultures inoculated with 1:10 dilutions of filtered supernatants from lung, cecum, and feces by 24 h after inoculation. Cytotoxicity is a well-described effect of chlamydial species that carry a cytotoxin orthologous to the clostridial cytotoxin; Cm is known to carry 3 copies.4 Classic chlamydial inclusions were observed 24 h after inoculation when higher dilutions of samples were cultured on HeLa 229 cell monolayers. These inclusions often displayed the typical Brownian movement seen with Cm infection, which was visualized by IFA (Figure 3). Cm was also confirmed in these infected monolayers by PCR for Cm 16S rRNA.
Figure 3.
Fluorescent images of Chlamydia-infected cells. HeLa 229 cells were infected with Chlamydia spp. isolated with an NSG mouse cecum sample. At 30 h after infection, cells were fixed and stained with FITC-labeled anti-Chlamydia spp. antibody (green) and analyzed using an EVOS FL Auto Imaging fluorescent microscope (Life Technologies).
Prevalence of Chlamydia muridarum in diagnostic samples.
The overall prevalence of Cm in diagnostic samples from various institutions and source material was 16% (146 of 900 samples tested). Positive samples were found in 10% (54 of 546) of those collected directly from mice, 30% (85 of 286) of those collected from the environment, and 12% (7 of 58) that were combined animal and environmental collections (Table 3). None of the biologics nor the samples collected from rodent production facilities tested positive. A much greater proportion of samples from academic, governmental, and medical research facilities (133 of 519, 26%) tested positive as compared with samples from pharmaceutical (5 of 249, 2%) or contract research facilities (8 of 76, 10%).
Table 3.
Prevalence of Chlamydia muridarum in diagnostic samples
| Sample Type | # Samples | # Positive | % Positive | |
|---|---|---|---|---|
| Animal | 546 | 54 | 10% | |
| Research1 | 282 | 46 | 16% | |
| Pharmaceutical | 162 | 0 | 0% | |
| Contract Research | 71 | 8 | 11% | |
| Rodent production | 31 | 0 | 0% | |
| Environmental | 286 | 85 | 30% | |
| Research1 | 207 | 80 | 39% | |
| Pharmaceutical | 72 | 5 | 7% | |
| Contract Research | 0 | 0 | 0% | |
| Rodent production | 7 | 0 | 0% | |
| Anim./Enviro. Combination |
58 | 7 | 12% | |
| Research1 | 27 | 7 | 26% | |
| Pharmaceutical | 8 | 0 | 0% | |
| Contract Research | 5 | 0 | 0% | |
| Rodent production | 18 | 0 | 0% | |
| Biologics | 10 | 0 | 0% | |
| Research1 | 3 | 0 | 0% | |
| Pharmaceutical | 7 | 0 | 0% | |
| Contract Research | 0 | 0 | 0% | |
| Rodent production | 0 | 0 | 0% | |
| Total | 900 | 146 | 16% | |
Academic, governmental, and medical research facilities
Fecal microbiome analysis.
The metagenomic microbiome sequencing data results for Cm are provided in Table 4. Overall, institutional prevalence of Cm abundance was 14% (17 of 120 institutions). One institution had abundant Chlamydia at the genus, but not the species level. While the preponderance of the samples was from academic institutions (112 of 120; 93%) located in the United States (91 of 120; 76%), Cm was also detected as a component of the fecal microbiome in European colonies (3 of 23; 13%). None of the 8 commercial institutions, all of which were from North America, had Cm as a component of their intestinal flora. The number of samples assayed from institutions with Cm abundance varied considerably, ranging from as few as 3 to as many as 4,756 samples per institution. The prevalence in samples from these institutions ranged from as low as 1% to as high as 100%. Academic institutions that submitted positive samples were obtained from all regions of the US, with an overrepresentation of samples submitted from the Northeast and the South.
Table 4.
Metagenomic fecal microbiome sequence data for Chlamydia muridarum
| Sample Totals | Institution Totals | ||||||
|---|---|---|---|---|---|---|---|
| Category/ Region | #Samples | #Positive | %Positive | #Institutions | #Positive | %Positive | |
| Academic | 11,106 | 383 | 3% | 112 | 17 | 15% | |
| Australia | 115 | 0 | 0% | 5 | 0 | 0% | |
| Europe | 728 | 47 | 7% | 23 | 3* | 13% | |
| Institution A* | 8 | 2 | 25% | ||||
| Institution B* | 39 | 7 | 18% | ||||
| Institution C* | 52 | 38 | 73% | ||||
| Northeast US | 976 | 223 | 23% | 27 | 5* | 16% | |
| Institution D* | 10 | 8 | 80% | ||||
| Institution E* | 11 | 11 | 100% | ||||
| Institution F* | 36 | 6 | 17% | ||||
| Institution G* | 68 | 32 | 47% | ||||
| Institution H* | 524 | 166 | 32% | ||||
| Midwest US | 1,603 | 15 | 1% | 20 | 1* | 5% | |
| Institution I* | 45 | 15 | 33% | ||||
| Southern US | 7,003 | 46 | 1% | 18 | 5 | 28% | |
| Institution J* | 30 | 3 | 10% | ||||
| Institution K* | 16 | 6 | 38% | ||||
| Institution L* | 40 | 5 | 13% | ||||
| Institution M* | 53 | 4 | 8% | ||||
| Institution N* | 4,756 | 28 | 1% | ||||
| Western US | 681 | 7 | 1% | 19 | 3* | 16% | |
| Institution O* | 3 | 1 | 33% | ||||
| Institution P* | 5 | 5 | 100% | ||||
| Institution Q* | 82 | 1 | 1% | ||||
| Commercial | 429 | 0 | 0% | 8 | 0 | 0% | |
| Canada | 20 | 0 | 0% | 1 | 0 | 0% | |
| Northeast US | 168 | 0 | 0% | 4 | 0 | 0% | |
| Midwest US | 168 | 0 | 0% | 1 | 0 | 0% | |
| Southern US | 73 | 0 | 0% | 2 | 0 | 0% | |
| Total | 11,387 | 383 | 3% | 120 | 17 | 14% | |
Sample details for institutions yielding positive results in each region.
Discussion
We provide compelling evidence that Cm is moderately prevalent and globally distributed in mouse colonies in numerous academic biomedical research institutions. This evidence includes PCR and fecal metagenomic data from 2 large sample sets, testing of imported mice from other academic colonies, and the prevalence in our institutions (MSK, Weill Cornell Medicine, and the Hospital for Special Surgery). The initial identification of Cm occurred in pursuit of the etiology of pulmonary lesions, often reported by comparative pathologists as incidental, in 2 immunocompetent GEM. This was a stunning finding, considering Cm had last been identified in research mice in the 1940s and has not been considered an agent of concern since. Because Cm has not been considered an ‘excluded agent,’ institutions, whether commercial breeders or end users, were not testing for its presence. The noted exception were laboratories using this bacterium in mice as a model for Chlamydia trachomatis infections. If present in mice obtained from commercial breeders, these laboratories would likely have detected its presence in uninfected study controls. Importantly, and consistent with this finding, Cm does not appear to be a component of the gastrointestinal microbiota of commercially bred, globally distributed mice. As such, its moderate prevalence is likely unrelated to distribution of Cm-infected mice by a major commercial breeder(s). This is fortunate, as the prevalence of infection would likely have been considerably higher had this not been the case. The absence of Cm in commercially reared mice is not surprising, as mice bred by commercial breeders have been rederived, or are descendants of mice that have been rederived by C-section or embryo transfer and subsequently fostered onto or transferred into mice derived using a similar method.
This situation begs the question as to the source(s) of Cm and how has it become enzootic in so many colonies around the globe. Cm could have been introduced by Cm-infected wild mice gaining access to research mouse colonies. While no evidence suggests spontaneous or natural infection of research mice since its last reported isolation from laboratory mice in the 1940s, some evidence indicates that Cm is found in wild populations of other rodents (for example, Peromyscus spp).36 In addition, we repeatedly detected Cm in mice obtained from a pet store. These mice are likely sourced from an enzootically infected colony(ies). However, we did not find Cm in the single wild mouse population that we surveyed. Evaluation of additional wild mouse populations is warranted to determine if wild mice pose a high-level risk of exposure to this bacterium. As many research facilities are infested with escaped laboratory mice, some of which likely breed with wild populations, these mice could be Cm-carriers and pose a risk to research colonies. Another source could be escape of a passaged experimental isolate. Numerous scientists have previously used, and continue to use, mice that have been experimentally infected with Cm as a model of human genital chlamydial infections caused by C. trachomatis. Historically, at some institutions, mice experimentally infected with Cm were handled using standard husbandry practices, creating the possibility that contamination of surfaces and equipment could have led to cross-contamination of other institutional mouse colonies.
Regardless of the mechanism or frequency by which Cm was introduced undetected into research mouse colonies, the interinstitutional global distribution of unique GEM strains greatly exacerbated Cm infection and may be the major contributor to its observed prevalence. This statement is supported by the considerable percentage of Cm positive mice imported into our institution. While some institutions rederive mouse strains imported from other institutions, the great majority of academic institutions quarantine imported mice and test for excluded agents, only releasing mice into their colonies if they are negative for these agents. We speculate that institutions using the former method are much less likely to have introduced Cm into their colonies. Institutions that test imported mice may not have had Cm on their exclusion list, making them considerably more likely to have introduced it into their colonies. This suspicion is supported by the finding that, at our institutions, colonies that received mice which were C-section rederived on importation and/or were only obtained from commercial vendors were Cm free. In contrast, colonies that received mice from commercial vendors and from other institutions using a test (for excluded agents) and release system were generally Cm infected. This latter scenario resembles the situation that occurred with Demodex musculi, which has also been widely distributed as a result of the exchange of GEM strains.29
Prior evidence indicates Cm’s presence in European research colonies. In a recent study of next-generation sequencing of the microbiome as a colony health surveillance tool, Cm was detected in fecal pellets from mice housed in conventional facilities.40 Cm was included in a list of pathogenic bacteria but the significance of its detection was not further explored, even though it was described as a non-enteric pathogen.40 Another recent study investigating resistance to Plasmodium yoelii assessed the cecal microbiome of C57BL/6 mice and detected bacteria in the Chlamydia phylum, but without further discussion of this finding.44 The significance of its presence was apparently not recognized in these studies, which is likely a result of the dearth of published data about spontaneous Cm infection since the 1940s.
Perivascular and peribronchiolar lymphoid aggregates (observed in our index cases) are commonly reported as incidental findings in aged mice. In at least some of these cases, Cm infection could have been associated with these lesions.20,41,48 Precedent exists for the presence of a novel agent, or one that is not routinely screened for, to cause lesions that were originally interpreted as spontaneous or incidental findings. For example, pulmonary lesions in rats and interstitial nephritis in mice that had been recognized as incidental findings for a number of years were recently attributed to Pneumocystis carinii and mouse kidney parvovirus respectively.14,26
Once Cm is introduced into a colony, its ease of transmission between cages of mice is uncertain. Previous studies have shown that Cm can be transmitted between mice housed in the same cage, likely by the oral route.10,33 Although no studies have definitively demonstrated aerosol transmission of Cm, other Chlamydiae including C. pneumoniae are transmitted via aerosols.37,39 Given the conservation observed among Chlamydia spp., along with the historically and currently documented respiratory pathology, aerosol transmission cannot be excluded as a potential means of transmission.8,18,37,42 Chlamydial EBs are relatively hardy, having adapted for transient extracellular survival until cell internalization can occur.15,16 This stability is further supported by its cecal and colonic tropism, as this indicates resistance to gastric acidity.50 The relative ease of distribution of Cm through soiled bedding, as reflected by the large number of soiled bedding sentinels that were fecal PCR positive at our institutions, suggests that cross-contamination is both plausible and probable within a vivarium. Furthermore, our data support the value of soiled bedding sentinels, and presumably other methods, such as PCR testing of pooled soiled bedding or exhaust air duct filters, for determining whether Cm is present in a mouse colony.
Although Cm has been used extensively to model C. trachomatis, little is known about the biology of the bacterium after natural infection of its presumed natural host, Mus musculus. Experimental infection with Cm typically involves inoculation of the murine urogenital tract, upper respiratory tract, or on occasion, the gastrointestinal tract. Urogenital inoculation is, by far, the most frequent method of inoculation described in the literature, as Cm is used to model C. trachomatis-induced urogenital disease in humans.3,37 As such, its experimental use fails to model natural or spontaneous Cm infection in several ways. First, the presumed site of natural infection and colonization is the gastrointestinal tract, similar to chlamydiae in other animals.37,50 Second, urogenital inoculation is often performed after administration of progesterone. Finally, the infectious doses used are typically very large (often ranging between 105 and 107 inclusion forming units [IFUs]), which likely exceeds exposures that occur in natural infections. In a recent study, one group orally inoculated C57BL/6 mice with Cm doses ranging from 102 to 106 IFU and found an ID50 of less than 102 IFU.50 The use of inocula that greatly exceed those that mice would be exposed to in an enzootically infected colony may not accurately model natural infection. However, previous studies do provide some information on the biology of Cm infection. For example, although Cm was originally isolated from the respiratory tract, its primary site of infection and continued replication appears to be the gastrointestinal tract, as supported by experimental observations.13,19,30,37,50 Although Cm can acutely infect lung and urogenital tissue in mice, it establishes chronic persistent infections in the gastrointestinal tract when inoculated orally.50 Chlamydia is commonly carried in the gastrointestinal tract in other species, an observation long known to chlamydiologists and veterinarians. Chlamydiae can reside in the gastrointestinal tract for long periods of time without causing clinical disease in virtually all hosts, including birds and mammals. Our current observations are consistent with this finding, as we detected Cm in the intestinal tract epithelium and feces of both immunocompetent and immunocompromised mice.
Experimental Cm infection of immunocompetent mouse strains is generally subclinical while immunodeficient strains often succumb to infection.24,27,50 While immunocompetent mice were historically considered to resolve infection, recent studies suggest immunocompetent animals develop a chronic, subclinical infection without clinical signs and normal leukograms, but remain culture positive and/or have histologic evidence of Cm for 50 to 265 d after infection.38,50 Moreover, immunocompetent strains differ, with BALB/c mice sustaining higher Cm burdens, longer infections and greater respiratory pathology than do C57BL/6 mice.50 Studies indicate that a Th1 immune response is principally required to control Cm infection.6,24,32,34,37,50 Furthermore, the immune response involves at least IgM, IgG, and IgA antibodies, and may be significantly more intense on reinfection.3,37 Immunocompromised strains, including athymic nude, SCID, and other immunodeficient transgenic strains (including IFNg, TLR, STAT1, MHCII/CD4, and Rag1 knockouts) develop more significant clinical signs, greater dissemination of Cm, and more protracted courses of infection.7,10,11,17,27,34,37 Long-term immunologic and other biologic effects are likely after Cm infection, but additional work is needed to more thoroughly elucidate these changes, as they may confound studies that use Cm-infected mice.
As we observed in NSG mice used as sentinels and from which Cm was isolated, mice with a compromised immune system appear unable to control infection, with many developing pneumonia by 30 d after inoculation.10,21,27,34,37 We observed significant clinical disease in severely immunocompromised NSG mice approximately 3 to 4 wk after cohousing with Cm-shedding imported mice. The clinical signs observed were described in the experimentally infected immunodeficient strains noted above. Leukogram changes were consistent with inflammation secondary to infectious processes. In the NSGs, significant pathology was observed in pulmonary sections in association with characteristic chlamydial inclusions in the bronchial epithelium. Inclusions were confirmed to be Chlamydia spp. by IHC MOMP staining and Cm by ISH. Pulmonary lesions noted in these NSG mice share similarities with lesions reported in the lungs from TLR2 deficient mice challenged with Cm intranasally. TLR2 deficient mice exhibited a proinflammatory cytokine profile and an exaggerated neutrophilic infiltration in the airways.21 Cm inclusions were not colocalized with the inflammatory cell infiltrates of the lamina propria of the large intestine. Cm was also seen by both IHC and ISH in histologically normal sections of the small and large intestine and in unaffected regions of the cecum. Thus, these lesions were unlikely to have been triggered by the presence of Cm. They could be a result of enhanced antigenic stimulation from alterations in the mucosal epithelial lining or enhanced trafficking of leukocytes in the lamina propria. While colonization and shedding may be prolonged, especially in gastrointestinal tissues and feces, overt inflammation of the gastrointestinal tract is not associated with chronic experimental infection, and no gastrointestinal pathology was documented in our NSG mice.28 Cm was likely the sole etiology of the noted pulmonary pathology, as no other bacteria were isolated by aerobic culture. Further exploration of Cm-induced disease in highly immunocompromised mouse strains, such as the NSG, could allow modeling of select characteristics of C. trachomatis-induced disease in humans.
Experimental studies with Cm have used serially passaged strains whose kinetics or tissue tropism may differ from that of the ‘field’ strains that are currently circulating in research mouse colonies. The 2 commonly used Cm isolates (Nigg and Weiss) differ significantly from one another in both their in vitro and in vivo characteristics. For example, a recent study demonstrated that the Weiss isolate had greater virulence and a higher replicative capacity than did Nigg after both intranasal and genitourinary inoculations.35 Morphologic differences are also present between the 2 strains, with the Weiss isolate producing smaller IBs than Nigg.35 Finally, 11 mutations differentiate the sequenced Weiss genome and published Nigg sequences.35 These findings demonstrate that even among the well-documented and carefully studied Cm isolates, significant differences in virulence, morphology, and genetics suggest the potential for field isolates to be similarly distinct.
Cm should be included in institutional biosecurity protocols and treated as an excluded agent because it has the potential to affect research and animal welfare. Adding Cm to a list of surveyed agents will allow institutions to understand colony prevalence and make decisions about its eradication. These decisions may include rederivation on importation or treatment. Limited data are available on antimicrobial therapy for Cm in mice. Chlamydial infections in humans and other species are generally treated effectively with doxycycline, which was efficacious in Cm eradication in a study investigating inhibition of chlamydial immunity secondary to antibiotic therapy.45 To date, we have successfully treated, as confirmed by repeated negative fecal PCR assays, several small mouse colonies using commonly available doxycycline (625 PPM) impregnated feed or drinking water dosed (1mg/mL) with the drug. However, additional effective antimicrobial agents will be needed as doxycycline would be contraindicated in studies using murine Tet-on/off systems. Antibiotics would also likely alter the microbiome of treated animals, which may be contraindicated in certain colonies and could further confound the studies in which treated mice are used.5,22
In conclusion, we have demonstrated that Cm has reemerged and is prevalent in academic mouse colonies. We believe this prevalence is at least partially attributable to global collaboration and the sharing of GEM strains among institutions. As Cm infections are likely to result in a host of biologic effects in many mouse strains, it should be treated as an ‘excluded’ agent, with institutional biosecurity protocols implemented to prevent its introduction. As eradication will be necessary for many enzootically infected colonies, additional studies are necessary to identify antimicrobials that can clear affected colonies with minimal undesirable effects that can also confound research. Other important future studies include further investigation of Cm’s natural transmission and biologic effects on its murine host.
Acknowledgments
MSK Core Facilities are supported by MSK’s NCI Cancer Center Support Grant P30 CA008748. The authors would like to thank Sockie Jiao and the staff of the Laboratory of Comparative Pathology for their technical assistance with IHC and ISH, Lisa Eldred and Dr. Michael Palillo for sample collection and animal monitoring, Dr. Adam Michel for his assistance in sample collection and pathology, and Dr. William Shek for his assistance in data analysis and presentation, and Denise Lynch at One Codex for her assistance in analysis of fecal microbiome data.
References
- 1.American Association for Laboratory Animal Services. [Internet]. 2021. Alleviating pain and distress in laboratory animals. [Cited 06 June 2022]. Available at https://www.aalas.org/about-aalas/position-papers/alleviating-pain-and-distress.
- 2.American Association for Laboratory Animal Sciences. [Internet]. 2021. Humane care and use of laboratory animals. [Cited 06 June 2022]. Available at https://www.aalas.org/about-aalas/position-papers/humane-care-and-use.
- 3.Barron AL, Rank RG, Moses EB. 1984. Immune response in mice infected in the genital tract with mouse pneumonitis agent (Chlamydia trachomatis biovar). Infect Immun 44:82–85. 10.1128/iai.44.1.82-85.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belland RJ, Scidmore MA, Crane DD, Hogan DM, Whitmire W, McClarty G, Caldwell HD. 2001. Chlamydia trachomatis cytotoxicity associated with complete and partial cytotoxin genes. Proc Natl Acad Sci USA 98:13984–13989. 10.1073/pnas.241377698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boynton FD, Ericsson AC, Uchihashi M, Dunbar ML, Wilkinson JE. 2017. Doxycycline induces dysbiosis in female C57Bl/6 6NCrl mice. BMC Res Notes 10:644. 10.1186/s13104-017-2960-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunham RC, Rey-Ladino J. 2005. Immunology of Chlamydia infection: implications for a Chlamydia trachomatis vaccine. Nat Rev Immunol 5:149–161. 10.1038/nri1551. [DOI] [PubMed] [Google Scholar]
- 7.Carrasco SE, Hu S, Imai DM, Kumar R, Sandusky GE, Yang XF, Derbigny WA. 2018. Toll-like receptor 3 (TLR3) promotes the resolution of Chlamydia muridarum genital tract infection in congenic C57Bl/6N mice. PLoS One 13:e0195165. 10.1371/journal.pone.0195165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell J, Huang Y, Liu Y, Schenken R, Arulanandam B, Zhong G. 2014. Bioluminescence imaging of Chlamydia muridarum ascending infection in mice. PLoS One 9:e101634. 10.1371/journal.pone.0101634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarke IN. 2011. Evolution of Chlamydia trachomatis. Ann N Y Acad Sci 1230:E11–E18. 10.1111/j.1749-6632.2011.06194.x. [DOI] [PubMed] [Google Scholar]
- 10.Cotter TW, Ramsey KH, Miranpuri GS, Poulsen CE, Byrne GI. 1997. Dissemination of Chlamydia trachomatis chronic genital tract infection in gamma interferon gene knockout mice. Infect Immun 65:2145–2152. 10.1128/iai.65.6.2145-2152.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darville T, O’Neill JM, Andrews CW, Nagarajan UM, Stahl L, Ojcius DM. 2003. Toll-like receptor-2, but not toll-like receptor-4, is essential for development of oviduct pathology in Chlamydial genital tract infection. J Immunol 171:6187–6197. 10.4049/jimmunol.171.11.6187. [DOI] [PubMed] [Google Scholar]
- 12.DeGraves FJ, Gao D, Kaltenboeck B. 2003. High-sensitivity quantitative PCR platform. Biotechniques 34:106–115. 10.2144/03341rr01. [DOI] [PubMed] [Google Scholar]
- 13.Dochez AR, Mills KC, Mulliken B. 1937. A virus disease of Swiss mice transmissible by intranasal inoculation. Proc Soc Exp Biol Med 36:683–686. 10.3181/00379727-36-9357. [DOI] [Google Scholar]
- 14.Edmonson EF, Hsieh WT, Kramer JA, Breed MW, Roelke-Parker ME, Stephens-Devalle J, Pate NM, Bassel LL, Hollingshead MG, Karim BO, Butcher DO, Warner AC, Nagashima K, Gulani J. 2020. Naturally acquired mouse kidney parvovirus infection produces a persistent interstitial nephritis in immunocompetent laboratory mice. Vat Pathol 57:915–925. 10.1177/0300985820953500. [DOI] [PubMed] [Google Scholar]
- 15.Elwell C, Mirrashidi K, Engel J. 2016. Chlamydia cell biology and pathogenesis. Nat Rev Microbiol 14:385–400. 10.1038/nrmicro.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Everett KD, Bush RM, Andersen AA. 1999. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. Nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int J Syst Bacteriol 49:415–440. 10.1099/00207713-49-2-415. [DOI] [PubMed] [Google Scholar]
- 17.Fan Y, Wang S, Yang X. 1999. Chlamydia trachomatis (mouse pneumonitis strain) induces cardiovascular pathology following respiratory tract infection. Infect Immun 67:6145–6151. 10.1128/IAI.67.11.6145-6151.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gogolak FM. 1953. The histopathology of murine pneumonitis infection and the growth of the virus in the mouse lung. J Infect Dis 92:254–272. 10.1093/infdis/92.3.254. [DOI] [PubMed] [Google Scholar]
- 19.Gordon FB, Freeman G, Glampit JM. 1938. A pneumonia-producing filtrable agent from stock mice. Proc Soc Exp Biol Med 39:450–453. 10.3181/00379727-39-10236. [DOI] [Google Scholar]
- 20.Haines DC, Chattopadhyay S, Ward JM. 2001. Pathology of aging B6;129 mice. Toxicol Pathol 29:653–661. 10.1080/019262301753385988. . [DOI] [PubMed] [Google Scholar]
- 21.He X, Nair A, Mekasha S, Alroy J, O’Connell CM, Ingalls RR. 2011. Enhanced virulence of Chlamydia muridarum respiratory infections in the absence of TLR2 activation. PLoS One 6:e20846. 10.1371/journal.pone.0020846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou X, Zhu L, Zhang X, Zhang L, Bao H, Tang M, Wei R, Wang R. 2019. Testosterone disruptor effect and gut microbiome perturbation in mice: early life exposure to doxycycline. Chemosphere 222:722–731. 10.1016/j.chemosphere.2019.01.101. [DOI] [PubMed] [Google Scholar]
- 23.Institute for Laboratory Animal Research. 2011. Guide for the Care and Use of Laboratory Animals: Eighth Edition. Washington, DC: The National Academies Press. [Google Scholar]
- 24.Jupelli M, Guentzel MN, Meier PA, Zhong G, Murthy AK, Arulanandam BP. 2008. Endogenous IFN-gamma production is induced and required for protective immunity against pulmonary chlamydial infection in neonatal mice. J Immunol 180:4148–4155. 10.4049/jimmunol.180.6.4148. [DOI] [PubMed] [Google Scholar]
- 25.Lipman NS, Homberger FR. 2003. Rodent quality assurance testing: use of sentinel animal systems. Lab Anim 32:36–43. 10.1038/laban0503-36. [DOI] [PubMed] [Google Scholar]
- 26.Livingston RS, Besch-Williford CL, Myles MH, Franklin CL, Crim MJ, Riley LK. 2011. Pneumocystis carinii infection causes lung lesions historically attributed to rat respiratory virus. Comp Med 61:45–59. [PMC free article] [PubMed] [Google Scholar]
- 27.Magee DM, Igietseme JU, Smith JG, Bleicker CA, Grubbs BG, Schacter J, Rank RG, Williams DG. 1993. Chlamydia trachomatis pneumonia in the severe combined immunodeficiency (SCID) mouse. Reg Immunol 5:305–311. [PubMed] [Google Scholar]
- 28.Morrison SG, Giebel AM, Toh EC, Spencer HJ, 3rd, Nelson DE, Morrison RP. 2018. Chlamydia muridarum genital and gastrointestinal infection tropism is mediated by distinct chromosomal factors. Infect Immun 86:e00141-18. 10.1128/IAI.00141-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nashat MA, Luchins KR, Lepherd ML, Riedel ER, Izdebska JN, Lipman NS. 2017. Characterization of Demodex musculi infestation, associated comorbidities, and topographic distribution in a mouse strain with defective adaptive immunity. Comp Med 67:315–329. [PMC free article] [PubMed] [Google Scholar]
- 30.Nigg C, Eaton MD. 1944. Isolation from normal mice of a pneumotropic virus which forms elementary bodies. J Exp Med 79:497–510. 10.1084/jem.79.5.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.One Codex. [Internet]. 2022. One Codex reference database. [Cited 06 June 2022]. Available at: https://app.onecodex.com/references.
- 32.Pal S, Tifra DF, de la Maza LM. 2019. Characterization of the horizontal and vertical sexual transmission of Chlamydia genital infections in a new mouse model. Infect Immun 87:e00834-18. 10.1128/IAI.00834-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perry LL, Hughes S. 1999. Chlamydial colonization of multiple mucosae following infection by any mucosal route. Infect Immun 67:3686–3689. 10.1128/IAI.67.7.3686-3689.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poston TB, O’Connell CM, Girardi J, Sullivan JE, Nagarajan UM, Marinov A, Scurlock AM, Darville T. 2018. T cell-independent gamma interferon and B cells cooperate to prevent mortality associated with disseminated Chlamydia muridarum genital tract infection. Infect Immun 86:e00143-18. 10.1128/IAI.00143-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramsey KH, Sigar IM, Schripsema JH, Denman CJ, Bowlin AK, Myers GA, Rank RG. 2009. Strain and virulence diversity in the mouse pathogen Chlamydia muridarum. Infect Immun 77:3284–3293. 10.1128/IAI.00147-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramsey KH, Sigar IM, Schripsema JH, Townsend KE, Barry RJ, Peters J, Platt KB. 2015. Detection of Chlamydia infection in Peromyscus species rodents from sylvatic and laboratory sources. Pathog Dis 74:ftv129. 10.1093/femspd/ftv129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rank R. 2007. Chlamydial Diseases, p 326–344. In: Fox JG, Davisson MT, Quimby FW, Barthold SW, Newcomer CE, Smith AL, editors. The mouse in biomedical research, vol 2. Burlington (MA): Elsevier. [Google Scholar]
- 38.Rank RG, Soderberg LS, Barron AL. 1985. Chronic Chlamydial genital infection in congenitally athymic nude mice. Infect Immun 48:847–849. 10.1128/iai.48.3.847-849.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Read TD, Brunham RC, Shen C, Gill SR, Heidelberg JF, White O, Hickey EK, Peerson J, Utterback T, Berry K, Bass S, Linher K, Weidman J, Khouri H, Craven B, Bowman C, Dodson R, Gwinn M, Nelson W, DeBoy R, Kolonay J, McClarty G, Salzberg SL, Eisen J, Fraser CM. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res 28:1397–1406. 10.1093/nar/28.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scavizzi F, Bassi C, Lupini L, Guerriero P, Raspa M, Sabbioni S. 2021. A comprehensive approach for microbiota and health monitoring in mouse colonies using metagenomic shotgun sequencing. Anim Microbiome 3:53. 10.1186/s42523-021-00113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Snyder JM, Snider TA, Ciol MA, Wilkinson JE, Imai DM, Casey KM, Vilches-Moure JG, Pettan-Brewer C, Pillai SPS, Carrasco SE, Salimi S, Ladiges W. 2019. Validation of a geropathology grading system for aging mouse studies. Geroscience 41:455–465. 10.1007/s11357-019-00088-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Starkey MR, Essilfie AT, Horvat JC, Kim RY, Nguyen DH, Beagley KW, Mattes J, Foster PS, Hansbro PM. 2013. Constitutive production of IL-13 promotes early-life Chlamydia respiratory infection and allergic airway disease. Mucosal Immunol 6:569–579. 10.1038/mi.2012.99. [DOI] [PubMed] [Google Scholar]
- 43.Stills HF, Jr, Fox JG, Paster BJ, Dewhirst FE. 1991. A “new” Chlamydia sp. strain SFPD isolated from transmissible proliferative ileitis in hamsters. Microb Ecol Health Dis 4:S99. [Google Scholar]
- 44.Stough JM, Dearth SP, Denny JE, LeCleir GR, Schmidt NW, Campagna SR, Wilhelm SW. 2016. Functional characteristics of the gut microbiome in C57Bl/6 mice differentially susceptible to Plasmodium yoelii. Front Microbiol 7:1520. 10.3389/fmicb.2016.01520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Su H, Morrison R, Messer R, Whitmire W, Hughes S, Caldwell HD. 1999. The effect of doxycycline treatment on the development of protective immunity in a murine model of chlamydial genital infection. J Infect Dis 180:1252–1258. 10.1086/315046. [DOI] [PubMed] [Google Scholar]
- 46.Sundberg JP, Taylor D, Lorch G, Miller J, Silva KA, Sundberg BA, Roopenian D, Sperling L, Ong D, King LE, Everts H. 2011. Primary follicular dystrophy with scarring dermatitis in C57Bl/6 mouse substrains resembles central centrifugal cicatricial alopecia in humans. Vet Pathol 48:513–524. 10.1177/0300985810379431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thurlow RW, Arriola R, Soll CE, Lipman NS. 2007. Evaluation of a flash disinfection process for surface decontamination of gamma-irradiated feed packaging. J Am Assoc Lab Anim Sci 46:46–49. [PubMed] [Google Scholar]
- 48.Ward JM, Sundberg JP. 2022. The respiratory tract, p 237–258. In: Sundberg JP, Vogel P, Ward JM, editors. Pathology of genetically engineered and other mutant mice. Hoboken (NJ): John Wiley & Sons. [Google Scholar]
- 49.Wooters MA, Kaufhold RM, Field JA, Indrawati L, Heinrichs JH, Smith JG. 2009. A real-time quantitative polymerase chain reaction assay for the detection of Chlamydia in the mouse genital tract model. Diagn Microbiol Infect Dis 63:140–147. 10.1016/j.diagmicrobio.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 50.Yeruva L, Spencer N, Bowlin AK, Wang Y, Rank RG. 2013. Chlamydial infection of the gastrointestinal tract: a reservoir for persistent infection. Pathog Dis 68:88–95. 10.1111/2049-632X.12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang YX, Fox JG, Ho Y, Zhang L, Stills HF, Smith TF. 1993. Comparison of the major outer-membrane protein (MOMP) gene of mouse pneumonitis (MoPn) and hamster SFPD strains of Chlamydia trachomatis with other Chlamydia strains. Mol Biol Evol 10:1327–1342. [DOI] [PubMed] [Google Scholar]