Abstract
A novel cellulosomal scaffoldin gene, termed cipV, was identified and sequenced from the mesophilic cellulolytic anaerobe Acetivibrio cellulolyticus. Initial identification of the protein was based on a combination of properties, including its high molecular weight, cellulose-binding activity, glycoprotein nature, and immuno-cross-reactivity with the cellulosomal scaffoldin of Clostridium thermocellum. The cipV gene is 5,748 bp in length and encodes a 1,915-residue polypeptide with a calculated molecular weight of 199,496. CipV contains an N-terminal signal peptide, seven type I cohesin domains, an internal family III cellulose-binding domain (CBD), and an X2 module of unknown function in tandem with a type II dockerin domain at the C terminus. Surprisingly, CipV also possesses at its N terminus a catalytic module that belongs to the family 9 glycosyl hydrolases. Sequence analysis indicated the following. (i) The repeating cohesin domains are very similar to each other, ranging between 70 and 90% identity, and they also have about 30 to 40% homology with each of the other known type I scaffoldin cohesins. (ii) The internal CBD belongs to family III but differs from other known scaffoldin CBDs by the omission of a 9-residue stretch that constitutes a characteristic loop previously associated with the scaffoldins. (iii) The C-terminal type II dockerin domain is only the second such domain to have been discovered; its predicted “recognition codes” differ from those proposed for the other known dockerins. The putative calcium-binding loop includes an unusual insert, lacking in all the known type I and type II dockerins. (iv) The X2 module has about 60% sequence homology with that of C. thermocellum and appears at the same position in the scaffoldin. (v) Unlike the other known family 9 catalytic modules of bacterial origin, the CipV catalytic module is not accompanied by a flanking helper module, e.g., an adjacent family IIIc CBD or an immunoglobulin-like domain. Comparative sequence analysis of the CipV functional modules with those of the previously sequenced scaffoldins provides new insight into the structural arrangement and phylogeny of this intriguing family of microbial proteins. The modular organization of CipV is reminiscent of that of the CipA scaffoldin from C. thermocellum as opposed to the known scaffoldins from the mesophilic clostridia. The phylogenetic relationship of the different functional modules appears to indicate that the evolution of the scaffoldins reflects a collection of independent events and mechanisms whereby individual modules and other constituents are incorporated into the scaffoldin gene from different microbial sources.
The cellulosome is a multiprotein complex consisting of cellulolytic and hemicellulolytic enzymes which has been described mainly in anaerobic clostridia (5, 7, 13, 25). The cellulosomal enzymes are attached to a large, multimodular, noncatalytic subunit called scaffoldin. Four scaffoldin genes have been sequenced from the following clostridial species: Clostridium thermocellum (cipA) (15), Clostridium cellulovorans (cbpA) (46), Clostridium cellulolyticum (cipC) (38), and Clostridium josui (cipJ) (for clarity, the CipA scaffoldin from C. josui is renamed CipJ in this communication) (24). All four contain multiple type I cohesin domains which integrate type I dockerin-tagged enzymes into the cellulosome complex. In addition, a family IIIa cellulose-binding domain (CBD) in the scaffoldin is responsible for the binding of the complex to its substrate, cellulose (39). Another class of domain, a unique C-terminal dockerin domain (categorized as a type II dockerin), has also been identified—thus far, only in the scaffoldin of C. thermocellum. The type II dockerin is involved in anchoring this scaffoldin to the bacterial cell wall by interacting selectively with a type II cohesin, borne by a series of cell surface proteins (44). Only three such anchoring proteins have been described, all in C. thermocellum (7). Finally, X2 modules of unknown function (11a) have been found in all four scaffoldin genes.
Acetivibrio cellulolyticus was first isolated from sewage sludge and proved to be a highly efficient cellulolytic bacterium (26, 41, 42). The strain was classified in a new genus of cellulolytic, gram-negative, non-spore-forming, anaerobic mesophilic bacteria. Nevertheless, recent 16S ribosomal DNA analysis has suggested that A. cellulolyticus is closely related to the clostridia (32).
In an earlier work (30), A. cellulolyticus was found to resemble C. thermocellum in a variety of cellulosome-related biochemical, immunochemical, and ultrastructural properties. Notably, the cell surface topology of A. cellulolyticus exhibited perhaps the most dramatic display of exocellular protuberance structures yet observed (27). The critical question that remained, however, was whether such organisms produced cellulosomes.
In recent years, the detection of cellulosome-related “signature sequences” (such as cohesin or dockerin domains) in a protein has become a clear indication that a given bacterium produces a cellulosome (1). We therefore decided to try to supplement the previous biochemical evidence, obtained for A. cellulolyticus, with genetic information. In doing so, we discovered a 10-kb EcoRI fragment which contained an intact scaffoldin gene. The gene, termed cipV, was sequenced in its entirety, and its modular arrangement and sequence similarities with other known scaffoldins were analyzed. The structural organization of the CipV scaffoldin was found to resemble that of CipA from C. thermocellum. Nevertheless, it differs from that and other scaffoldins thus far described by the inclusion of a sequence consistent with a family 9 glycosyl hydrolase as an integral part of its polypeptide chain. The entry of CipV into the family of scaffoldins warrants phylogenetic treatment of its functional modules vis-à-vis those of the four other known family members.
MATERIALS AND METHODS
Organism and growth conditions.
A. cellulolyticus ATCC 33288 was purchased from the German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany). The cells were grown anaerobically at 37°C in serum bottles containing an American Type Culture Collection recommended medium (1207 BC medium for A. cellulolyticus) which included either cellobiose (Sigma Chemical Co., St. Louis, Mo.) or cellulose (Avicel; E. Merck AG, Darmstadt, Germany) as a carbon source. The cells were grown to mid-exponential phase (36 to 48 h), the culture was centrifuged (10,000 × g; 10 min), and both the supernatant fluids and the cells were stored at −20°C for further use.
Isolation and identification of candidate scaffoldin band(s) from A. cellulolyticus.
Cell-free culture fluids of A. cellulolyticus were mixed with a 1% volume of a 10-mg/ml suspension of amorphous cellulose (29). The suspension was shaken at low speed at room temperature for 30 min and centrifuged at 10,000 × g for 30 min, and the supernatant fluids were discarded. The pellet was resuspended in sodium dodecyl sulfate (SDS) sample buffer (34), and the cellulose-adsorbed proteins were separated by SDS–6% polyacrylamide gel electrophoresis (PAGE). The gel was cut into three parts. One was stained with Coomassie brilliant blue R250, destained, and photographed. The remaining two parts were transferred electrophoretically onto nitrocellulose sheets, one of which was stained with rabbit antibodies specific for the cellulosome from C. thermocellum while the other was stained with Griffonia simplicifolia GS-I lectin, as described previously (30). The primary antibody was visualized with a second (goat anti-rabbit) antibody-peroxidase conjugate, and the glycosylated bands were visualized by peroxidase-conjugated lectin (both obtained from Sigma). Prestained, low-range calibrated molecular weight standards were obtained from Bio-Rad Laboratories (Hercules, Calif.). The cellulosome from C. thermocellum was prepared by the affinity digestion procedure (35).
Peptide sequencing.
Candidate protein bands, which were recognized both by anticellulosomal antibodies and by GS-I lectin, were extracted from the SDS-PAGE gel. The extracted proteins were subjected to proteolysis with Lys-C, and the resultant peptides were resolved by reverse-phase high-performance liquid chromatography and collected manually (33). The purified peptide peaks were analyzed and sequenced by Edman degradation (Protein Center, Technion, Haifa, Israel).
PCR cloning.
A. cellulolyticus genomic DNA was isolated as described by Murray and Thompson (36). Oligonucleotide primers were generated from the peptide sequence. PCR was carried out at various annealing temperatures (40 to 60°C) in order to obtain specific amplified products with the genomic DNA as a template. The purified PCR products were cloned into the pGEM-T Easy vector system (Promega, Madison, Wis.) and sequenced. The sequences were compared with GenBank and known cellulosome-related proteins.
Southern blotting.
Genomic DNA (1 μg) was cleaved with various restriction enzymes, e.g., BamHI, HindIII, EcoRI, NcoI, SacI, and PstI, and the fragments were separated in 1% agarose gels. Relevant DNA fragments were labeled by the digoxigenin system (Boehringer Mannheim, Mannheim, Germany), and Southern blotting was performed according to the manufacturer’s instructions.
Construction and screening of genomic library.
A. cellulolyticus genomic DNA (10 μg) was cleaved by EcoRI and ligated with EcoRI-predigested and alkaline phosphatase-treated pUC19. The ligation was transferred into E. coli XL-1 Blue (Stratagene, La Jolla, Calif.) and plated onto Luria broth-ampicillin-X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)-IPTG (isopropyl-β-d-thiogalactopyranoside) plates. The above-described labeled probe was used for screening the genomic library. Hybridization was carried out according to the manufacturer’s instructions (Boehringer Mannheim). Putative positive clones were verified by dot hybridization.
Nucleotide sequence accession number.
The DNA sequence for the cipV gene reported here was deposited in the GenBank database under accession no. AF155197.
RESULTS
Identification of scaffoldin and sequencing of relevant peptide segment.
Cell culture extracts of cellobiose-grown A. cellulolyticus were treated with amorphous cellulose, and the adsorbed fraction was subjected to SDS-PAGE. The separated proteins were blotted onto nitrocellulose membranes and examined with cellulosome-specific antibodies and GS-I lectin from G. simplicifolia. The antibodies were elicited in rabbits with intact C. thermocellum cells (31). The whole-cell preparation was adsorbed onto mutant cells and the residual antibody species proved relatively selective for the cellulosomal scaffoldin subunit. The Griffonia lectin is selective for α-galactosyl moieties, which appear to be characteristic determinants of cellulolytic bacteria in various species (30). The lectin was previously found to recognize cellulosomal oligosaccharides from C. thermocellum and Bacteroides cellulosolvens (16, 17).
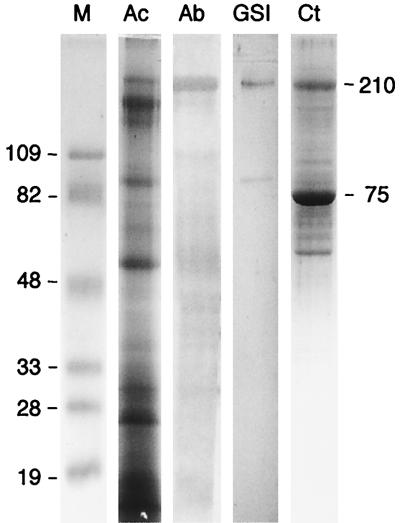
With these probes, a 210-kDa protein band was identified by both the antibodies and the lectin (Fig. 1). It is noteworthy that this band stained relatively poorly with Coomassie blue compared to other bands (e.g., the ∼180-kDa band). On the basis of the observed immuno-cross-reactivity and lectin-specific staining pattern, the high-molecular-weight (210,000) band was considered a primary candidate for the scaffoldin subunit of the A. cellulolyticus cellulosome. This rationale provided a focused approach for identification of the putative scaffoldin, thereby saving us time and unproductive efforts on unsuitable prospects.
FIG. 1.
Identification of putative cellulosome-related proteins in A. cellulolyticus. Ac, SDS-PAGE crude cell-free culture fluids adsorbed to amorphous cellulose; Ab, Western blot analysis with antibodies specific for the scaffoldin subunit from C. thermocellum; GSI, blotted protein bands cross-reacting with GS-I lectin from G. simplicifolia; M, prestained, low-range molecular mass markers; Ct, C. thermocellum cellulosome standard. The relative molecular masses of designated bands are indicated. Note that both the antibody and lectin probes labeled a relatively faint 210-kDa Coomassie-stained polypeptide band.
The candidate Coomassie blue-stained protein band was extracted from the SDS-PAGE gel and subjected to proteolysis with Lys-C protease. Several peptides were purified and sequenced. The sequences obtained were compared with those of known cellulosome-related proteins from other species, and a 17-residue peptide (N′-VEFFNAGTQAQSNSIYP-C′) appeared to be remarkably conserved, compared to a known segment of the family III CBDs.
Amplification of an internal CBD fragment.
A degenerate forward primer, pAC1 (5′-GTK GAA TTY TTY AAY GCN GG-3′), was designed from the N terminus (N′-VEFFNA-C′) of the above 17-residue peptide sequence, which was homologous with family IIIa CBDs. A degenerate reverse primer, pCBD (5′-TGW KYR WAR TTW SWC CAG TC-3′), was then designed from a particularly conserved region of known family IIIa CBDs. (Abbreviations for degenerate nucleotides are as follows: K, G or T; Y, C or T; W, A or T; R, A or G; S, C or G; N, A, C, G, or T.) With these two primers, a 350-bp fragment (AC3) was amplified specifically from the A. cellulolyticus genomic DNA by primers pAC1 and pCBD. Cloning and sequencing showed the deduced polypeptide from AC3 had significantly high homology (approximate 50% identity) with known family III CBDs. As expected, the sequence of the AC3 N terminus was identical with the initial sequenced peptide (i.e., N′-GTQAQSNSIYP-C′), which indicated that AC3 was indeed part of the gene encoding a putative scaffoldin subunit.
Cloning and DNA sequencing of the cipV scaffoldin gene.
The mass of the putative scaffoldin subunit from the A. cellulolyticus cellulosome is very similar to that of the 210,000-Da S1 band, which corresponds to the scaffoldin subunit from C. thermocellum (Fig. 1). Consequently, the gene encoding the putative scaffoldin from A. cellulolyticus would be expected to exhibit a size (∼6 kb) similar to that of the scaffoldin gene (cipA) from C. thermocellum.
A. cellulolyticus chromosomal DNA was cleaved with various restriction enzymes, and Southern blotting analysis, with AC3 as a probe, detected a ∼10-kb EcoRI band. The size of this fragment was particularly appealing, since, if a prospective ∼6-kb gene was correctly situated there, it might contain the entire gene in a single fragment. To clone the 10-kb fragment, an A. cellulolyticus genomic library was constructed in pUC19 with fully EcoRI-digested chromosomal DNA. The plasmid library was screened by colony hybridization with the AC3 probe, and a colony containing the vector with the ∼10-kb EcoRI insert (pACE) was detected. The authenticity of the insert was confirmed by Southern blotting of EcoRI- and PstI-digested A. cellulolyticus chromosomal DNA. By using a combined PstI and HindIII digest, fragments from the insert were subcloned into pUC19 and then sequenced. Sequencing analysis indicated that the 10-kb insert contained two open reading frames (ORFs) apparently related to cellulosomal proteins. One ORF represented the entire gene encoding the scaffoldin subunit of A. cellulolyticus, termed CipV. The second ORF represented an incomplete gene containing at least two repeating type II cohesin domains at its N terminus (unpublished data).
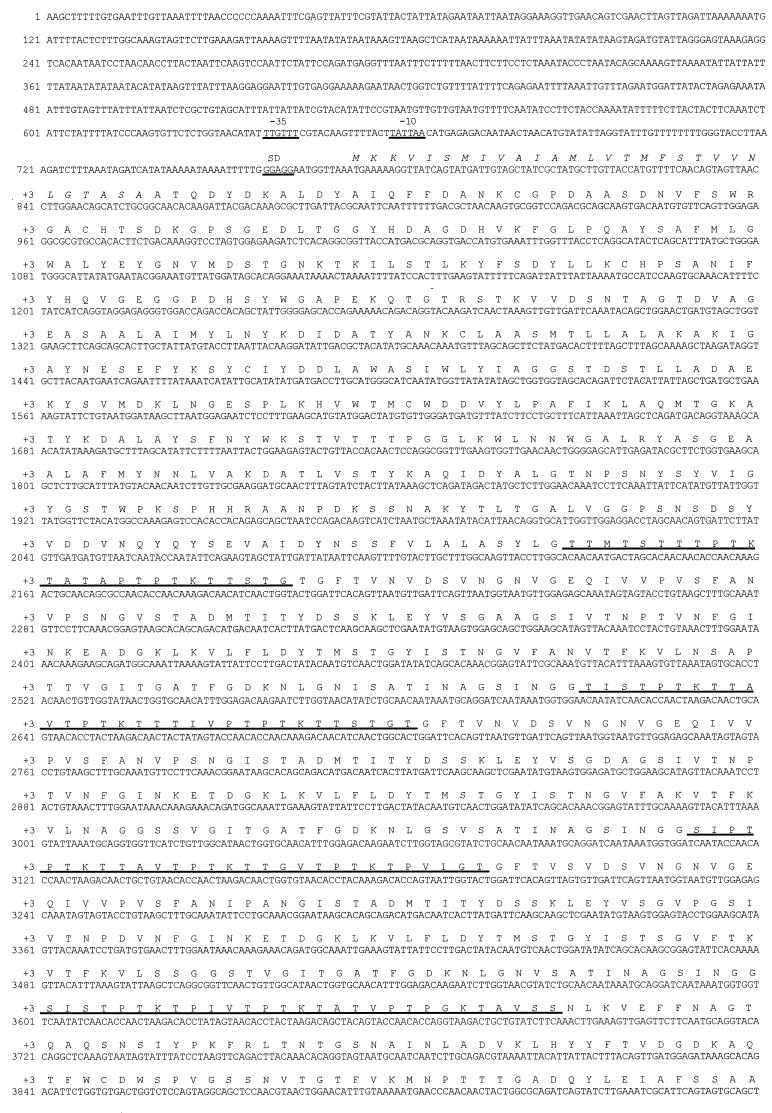
The complete sequence of the intact cipV gene is shown in Fig. 2. The A. cellulolyticus scaffoldin gene consists of 5,748 nucleotides encoding a 1,915-residue polypeptide—the largest scaffoldin described to date. The deduced polypeptide exhibited a calculated molecular mass of 199,496 Da, which is consistent with that (excluding saccharide components) of the identified candidate scaffoldin subunit (Fig. 1). The authenticity of the reading frame encoding CipV was also confirmed by an internal amino acid sequence of the 17-residue segment located in the resident CBD which is identical with that of the peptide originally identified from the putative scaffoldin band.
FIG. 2.
Nucleotide and deduced amino acid sequences of the A. cellulolyticus scaffoldin subunit (CipV). The proposed Shine-Dalgarno sequence (SD) and the −10 and −35 regions of the putative promoter are indicated. The signal sequence is shown in italics, the intermodular linker sequences are underlined, and the C-terminal dockerin domain is designated by a wavy underline. See Table 2 for the positions of the linkers relative to the adjoining modular components, i.e., the N-terminal catalytic module, the CBD, the seven cohesins, and domain X. The overall modular arrangement of CipV is presented in Fig. 3.
The start codon (ATG) is preceded 8 bp upstream by a typical Shine-Dalgarno sequence (GGAGG), homologous to other potential ribosome-binding sites found in various C. thermocellum genes and completely complementary to 5 bases (boldface) in the 3′ end of the 16S rRNA of Bacillus subtilis (3′-UCUUUCCUCCACUAC-5′). A potential promoter is located 110 bp from the ATG start codon with conserved −10 (5′-TATTAA-3′) and −35 (5′-TTGTTT-3′) regions (boldface).
The N terminus of CipV commences with a putative 29-residue signal peptide sequence, with a typical positively charged N-terminal end, a hydrophobic core, and a more polar carboxylic end with alanines at positions −1 and −3 (50).
DISCUSSION
The cellulosome is a massive complex that contains multiple types of enzymes that work synergistically to degrade crystalline cellulose and other plant cell wall polysaccharides. The enzymes are incorporated into the complex by virtue of a noncatalytic scaffoldin subunit. Continued insight into how such a complex is constructed can provide a model for other multicomponent protein systems.
Only a few complete scaffoldin sequences are currently available. Each new sequence contributes new and vital information to our understanding of the structural organization of cellulosomes and the interaction of their manifold functional modules with each other and with the substrate.
Previously, biochemical and immunochemical methods were used to detect cellulosome-like entities in various cellulolytic bacteria. In recent years, however, conclusive establishment of the presence of cellulosomes in a given bacterium has involved the identification of cellulosome signature sequences, which typically include cohesin and dockerin domains (1). In the present study, the entire A. cellulolyticus scaffoldin subunit, CipV, was sequenced. Comparative sequence analysis of its functional modules with those of the previously sequenced scaffoldins provides new insight into the structural arrangement and phylogeny of this intriguing family of microbial proteins.
The modular organization of CipV.
CipV includes all of the major domains found so far in cellulosomal scaffoldin proteins, with the surprising and unprecedented addition of a family 9 glycosyl hydrolase sequence as an integral part of the deduced polypeptide chain. Until this work, catalytic modules have been found only in free enzymes or in nonscaffoldin cellulosomal subunits.
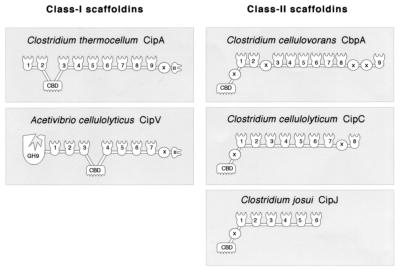
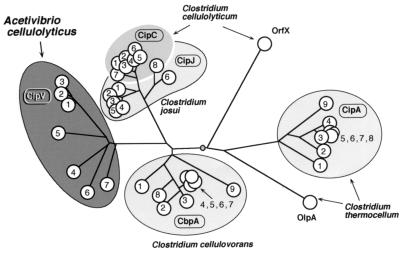
Comparison of the domain organization of the CipV sequence with those of the other clostridial species (Fig. 3) reveals the highest similarity with the C. thermocellum scaffoldin (15). Both scaffoldins contain an internal CBD, flanked by multiple copies of cohesin domains, with a single X2 module of unknown function and with a type II dockerin at the C terminus. Nevertheless, the precise position of the CBD and the number of cohesins (seven versus nine) are clearly different in the two strains. In contrast, scaffoldins from C. cellulovorans, C. cellulolyticum, and C. josui exhibit N-terminal CBDs, with multiple cohesin domains interspersed with one or more copies of internal X2 modules.
FIG. 3.
Structural organization of the known scaffoldins from different cellulosome species. The cohesin domains are designated by numbers according to their sequential positions relative to the amino terminus. Also indicated are the CBDs, the X2 domains (X), the type II dockerins (II), and the CipV GH9. The modular organization is based on the sequences of the respective scaffoldins, available from GenBank with the following accession numbers: CipA from C. thermocellum, L08665; CbpA from C. cellulovorans, M73817; CipC from C. cellulolyticum, U40345; and CipJ from C. josui, AB004845.
Despite the intrinsic family 9 catalytic module of the A. cellulolyticus CipV, it can be catalogued together with that of C. thermocellum to compose the class I scaffoldins, on the basis of its internal CBD and C-terminal dockerin domain. These features distinguish the class from the other currently known scaffoldins of class II (Fig. 3).
The signal peptide.
The sequence of the CipV signal peptide is particularly homologous to that of C. thermocellum and also has similarity to those of the other known cellulosomal scaffoldins (Table 1). In fact, the interspecies homology among the scaffoldin signal peptides is much more pronounced than that between the genes for the scaffoldins and the other cellulosomal components even within the same species. The observed homology among the scaffoldin signal peptides might indicate a specialized functional role, perhaps related to the assembly of the cellulosome, such that attachment of the enzymatic subunits to scaffoldin occurs while the latter is still associated to the cell surface. In this context, it has been observed early on (4) that the cellulosomal subunits, and the scaffoldin subunit in particular, from C. thermocellum do not appear in the free, uncomplexed form.
TABLE 1.
Sequences and alignment of signal peptides from known scaffoldin genes
| Gene | Bacterium | Signal sequence alignmenta | GenBank accession no. |
|---|---|---|---|
| cipV | A. cellulolyticus | MKKVISMIVAIAMLVTMFSTVVNLGTASA | AF155197 |
| cipA | C. thermocellum | MRKVISMLLVVAMLTTIFAAMIPQ-TVSA | L08665 |
| cbpA | C. cellulovorans | MQKKKSLNLLLA-LMMVFALVLPSIPALA | M73817 |
| cipC | C. cellulolyticum | MRKK-SLAFLLA-LTMLVTLLGAQLTAFA | U40345 |
| cipJ | C. josui | MRKK-SLAFLLA-LTMLVTLLGAQLTAFA | AB004845 |
Dashes represent gaps.
The catalytic module.
Sequence analysis indicates that the proposed catalytic module of CipV belongs to family 9 of the glycosyl hydrolases (GH9). GH9 is a common component of cellulolytic enzymes in bacteria and plants (11, 11a, 12).
Microbial family 9 cellulases commonly conform to one of four thematic modular arrangements (5, 6). The simplest GH9 theme is typical of many plant cellulases and consists of a solitary catalytic module. The others contain different adjoining accessory or helper modules. For example, endoglucanase E4 from Thermomonospora fusca (43) includes a family IIIc CBD immediately downstream from its GH9 catalytic module. A third theme is exemplified by endoglucanase CelD from the C. thermocellum cellulosome (23), which bears an immunoglobulin (Ig)-like domain upstream of the catalytic module (20). A fourth thematic type also contains an Ig-like domain in the same position, but in addition, it includes an N-terminal family IV CBD. In this respect, the CipV catalytic module of the A. cellulolyticus scaffoldin subunit is similar to that of the plant GH9 cellulases in that it lacks an adjoining helper module.
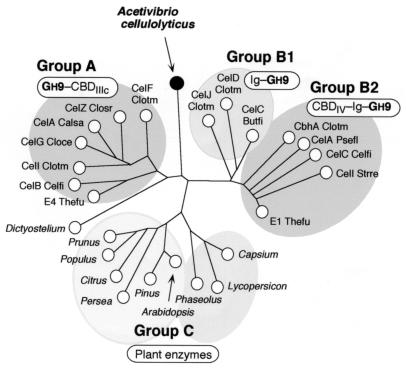
This thematic arrangement of the GH9 cellulases is mirrored in the sequences of their catalytic modules alone, and the divergent sequences are reflected in the phylogenetic relationship of the parent cellulases (Fig. 4). Thus, the catalytic modules from cellulases, which include a fused family IIIc CBD (group A), all map within the same branch. On the other hand, the catalytic modules that bear an adjacent Ig-like domain fall into a cluster on the opposite side of the tree. Cellulases which have the Ig-like domain only (group B1) occupy a small separate branch, and those that also include a family IV CBD (group B2) develop distally to form a separate subcluster. Another large cluster (group C) represents plant enzymes that generally lack adjoining helper modules. The catalytic module of CipV is distinct from the groups depicted in Fig. 4, occupying a position adjacent to the plant enzymes.
FIG. 4.
Phylogenetic analysis of the N-terminal family 9 catalytic module of CipV and its relationship with other family 9 members. The analysis of the following catalytic modules was performed with GenBee based on the GenBank sequences (accession numbers in parentheses): solid circle, CipV Acece, CipV scaffoldin from A. cellulolyticus (this communication), and Dictyostelium, endoglucanase from Dictyostelium discoideum (M33861). Group A enzymes: CelF Clotm, endoglucanase F from C. thermocellum (X60545); CelZ Closr, exoglucanase Z from Clostridium stercorarium (X55299); CelA Calsa, cellulase A from Caldocellum saccharolyticum (L32742); CelG Cloce, endoglucanase G from C. cellulolyticum (M87018); CelI Clotm, endoglucanase I from C. thermocellum (L04735); CelB Celfi, endoglucanase B from Cellulomonas fimi (M64644); E4 Thefu, endo- or exoglucanase E4 from T. fusca (M73322). Group B1 enzymes: CelJ Clotm, cellulase J from C. thermocellum (D83704); CelD Clotm, endoglucanase D from C. thermocellum (X04584); CelC Butfi, endoglucanase C from Butyrivibrio fibrisolvens (X55732). Group B2 enzymes: CbhA Clotm, cellobiohydrolase A from C. thermocellum (X80993); CelA Psefl, endoglucanase A from Pseudomonas fluorescens (X12570); CelC Celfi, endoglucanase C from C. fimi (X57858); CelI Strre, endoglucanase I from Streptomyces reticuli (X65616); E1 Thefu, endoglucanase E1 from T. fusca (L20094). Plant enzymes: cellulases from Prunus persica (X96853), Populus alba (D32166), Citrus sinensis (AF000135), Persea americana (M17634), Pinus radiata (X96853), Arabidopsis thaliana (X98543), Phaseolus vulgaris (M57400), Capsium annuum (X97189), and Lycopersicon esculentum (U20590).
The discrete position of the CipV catalytic module in the phylogenetic tree underscores the fact that it lacks an adjoining helper module and represents a new class of scaffoldin-associated family 9 glycosyl hydrolase.
Interdomain linker segments.
The sequences of the linkers, which connect adjacent cellulosomal domains of A. cellulolyticus, are shown in Table 2. With the exception of linkers 6, 8, and 9, the relatively long linker segments of the A. cellulolyticus scaffoldin are rich in prolines and threonines, and extended stretches of their sequences are remarkably similar. The long linkers are reminiscent of those of the C. thermocellum scaffoldin subunit but not those of C. cellulovorans, C. cellulolyticum, or C. josui (for a comparison, see Table 1 of reference 3). The high incidence of prolines suggests that the linkers form extended configurations, such as the plant cell wall extensins (52), that physically separate the various domains. In addition, proline-rich regions of proteins have been suggested to cause rapid and nonspecific binding (51), which in the case of scaffoldins may promote intermodular and/or intersubunit protein-protein interactions. The numerous threonines would be suitable glycosylation sites, as demonstrated for the C. thermocellum scaffoldin (18).
TABLE 2.
Sequences of intermodular linker segments in the CipV scaffoldin from A. cellulolyticus
| Linker | Flanking modules | Sequence |
|---|---|---|
| 1 | GH9-Coh1 | TTMTSTTTPTKTATAPTPTKTTSTGT |
| 2 | Coh1-Coh2 | TISTPTKTTAVTPTKTTTIVPTPTKTTSTGT |
| 3 | Coh2-Coh3 | SIPTPTKTTAVTPTKTTGVTPTKTPVIGT |
| 4 | Coh3-CBD | SISTPTKTPIVTPTKTATVPTPGKTAVSS |
| 5 | CBD-Coh4 | NGTVVTPTTVVTPTKTATATPTKTATATPTKTSTPNPN |
| 6 | Coh4-Coh5 | DGK |
| 7 | Coh5-Coh6 | NVTPTKTATATPTVTPTKTSTATATPTGTQT |
| 8 | Coh6-Coh7 | KKDDPN |
| 9 | Coh7-X2 | IVSEGTTVSG |
The first linker sequence that separates the CipV GH9 from cohesin 1 is indicative of the scaffoldins and dissimilar to those that usually flank GH9 modules, notably the characteristic linker segment that joins a GH9 module to a family IIIc CBD (43). The lack of such a linker and its replacement by a Pro- Thr-rich linker underscores the special nature of the CipV GH9 module.
The cohesin domains.
Multiple alignment of the seven repeating cohesin domains of CipV reveals sequence homology of between 59 and 92%. The most diversity occurs between CohV-1 and CohV-6. In contrast, the sequences of the first three N-terminal cohesins are very similar (about 90% identity).
Multiple-sequence alignment among the type I cohesins showed a close interspecies relationship, which would suggest that all of these cohesin domains would assume the same general structural fold as that of the recently determined cohesin structures (45, 47). The cohesin sequences can be compared by phylogenetic analysis (8). An unrooted phylogenetic tree of the known type I cohesins (Fig. 5) indicates that the cellulosomal cohesins from each species generally form a tight cluster. Interestingly, the tree places the CipV cohesins on a separate branch between those of the other mesophilic bacteria (i.e., C. cellulovorans, C. cellulolyticum, and C. josui), while the cellulosomal cohesins of thermophilic C. thermocellum occupy an opposing branch on the tree. The two known noncellulosomal type I cohesins (OlpA from C. thermocellum and OrfX from C. cellulolyticum) maintain discrete positions on the tree which radiate away from the other cohesin clusters. It should be mentioned that, when analyzed together, the type I and type II cohesins form two distinct clusters on opposite sides of the resultant phylogenetic tree (2).
FIG. 5.
Phylogenetic relationship of the CipV cohesin domains with other type I cohesins. The sequences of the scaffoldin-borne cohesins were obtained from the GenBank accession numbers shown in the legend to Fig. 3. The nonscaffoldin, type I cohesins were OlpA from C. thermocellum (X67506) and OrfX from C. cellulolyticum (AF081458). The darkly shaded oval represents the weighted centroid of the tree.
The CBD.
The internal CipV CBD belongs to the family III CBDs, classified according to sequence alignment (48). The CBDs of this family are separated into two functionally different types. One type (comprising the family IIIa and IIIb CBDs) binds strongly to crystalline cellulose, and another (the aforementioned family IIIc CBDs fused to a family 9 glycosyl hydrolase) fails to bind crystalline cellulose but reportedly serves in a helper role in the hydrolysis of a single cellulose chain (5, 43).
The CBDs of families IIIa and IIIb were previously proposed to be distinguished by the nature of the parent protein, the family IIIa CBDs being component parts of cellulosomal scaffoldin subunits and the family IIIb CBDs being the targeting agent for free noncellulosomal enzymes (3). In this context, the family IIIa CBDs contain a 9-residue segment called the scaffoldin loop which includes a proposed cellulose-binding tyrosine residue (Y67, numbered according to the system of Tormo et al. [49]). This loop, together with the distinctive tyrosine, is missing in all of the family IIIb CBDs. In addition, another putative binding residue (position 57) is different in the two subfamilies: a histidine appears in this position in the family IIIa CBDs, and in those of family IIIb the residue is a tryptophan.
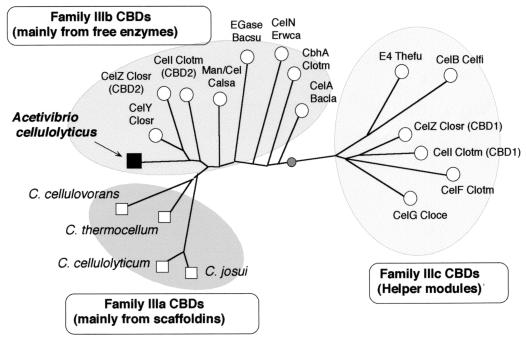
The phylogenetic relationship of the family III CBDs follows the general pattern of their functions (Fig. 6). Thus, all of the family IIIc CBDs form a distinct cluster on one side of the tree. On the opposite side of the weighted centroid are scattered the family IIIb CBDs. The family IIIa CBDs from the clostridial scaffoldins occupy a single branch, which emanates from an intermediate position among the family IIIb CBDs.
FIG. 6.
Phylogenetic analysis of the CipV CBD and its relationship to other scaffoldin and nonscaffoldin family III CBDs. Scaffoldin CBDs are shown as squares, and enzyme-borne CBDs are shown as circles. The sequences were obtained from the respective GenBank accession numbers (in parentheses): CelY Closr, endoglucanase Y from Clostridium stercorarium (Z69359); Man/Cel Calsa, mannanase or cellulase from Caldocellum saccharolyticum (L01257); EGase Bacsu, endoglucanase from B. subtilis (M16185); CelN Erwca, cellulase N from Erwinia carotovora subsp. atroseptica (L39788); CelA Bacla, cellulase A from Bacillus lautus (M76588). The sources of the sequences of other family III CBDs are shown in the legends to Fig. 3 and 4. The darkly shaded oval represents the weighted centroid of the tree.
According to the phylogenetic tree, the CipV CBD clearly belongs to the family IIIb CBDs. It forms a separate subbranch with several other CBDs of this family, derived from clostridial cellulases. In addition, the CipV CBD lacks the scaffoldin loop (and its intrinsic tyrosine residue) and has a tryptophan rather than a histidine at position 57. These observations immediately suggest that the scaffoldin loop is not strictly definitive of the scaffoldins, and the relationship among the family IIIa and IIIb CBDs is not necessarily a precise function of its parent protein (i.e., a free noncellulosomal enzyme versus a cellulosomal scaffoldin subunit). In any case, further attempts at categorizing this particular family of CBDs should await a larger collection of relevant sequences.
The X2 module.
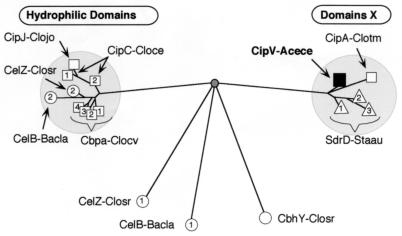
The function of this particular type of module (currently designated the X2 module by Coutinho and Henrissat [11a]) remains unknown. Originally, the X2 module was alternatively referred to as the hydrophilic domain in C. cellulovorans (as opposed to the hydrophobic cohesins [46]) and as domain X in C. thermocellum (28), and the latter distinction is relevant to the current discussion. Interestingly, the phylogenetic relationship of the homologous domains (Fig. 7) appears to reflect the overall modular organization of the parent scaffoldin subunit (Fig. 3). Thus, the sequences of the domains X from the group I scaffoldins (i.e., A. cellulolyticus CipV and C. thermocellum CipA) form a small cluster on the phylogenetic tree that maps on the opposite pole of the weighted centroid from the hydrophilic domains of the group II scaffoldins.
FIG. 7.
Phylogenetic distribution of X2 modules. The scaffoldin-derived modules are shown as squares, the enzyme-derived modules are shown as circles, and cell wall-associated proteins (Sdr) from S. aureus (GenBank accession no. AJ005646) are shown as triangles. The numbers indicate repeated modules, according to their sequential arrangement with respect to the N terminus of the given protein. The accession numbers of the other proteins are provided in the legends to Fig. 3, 4, and 6.
It is interesting to note that related X2 domains occur in a few free cellulases as well, which either occupy divergent branches or cluster together with the group II hydrophilic domains. Another set of newly described cell wall-associated proteins from Staphylococcus aureus (21, 22) has also been shown to contain repeated domains (called B motifs), which appear to be closely related to the domains X in the group I scaffoldins.
Until we know the precise function(s) of the X2 module, it will be hard to assess the biological and/or structural consequences of the observed sequence-based clustering.
Type II dockerin domain.
Like that of the CipA scaffoldin from C. thermocellum, the C terminus of CipV exhibits a type II dockerin domain, which presumably interacts with a type II cohesin of a putative anchoring protein. This is only the second such dockerin domain to be discovered. Moreover, the type II cohesins of the incomplete ORF that appears immediately downstream from the scaffoldin gene signify a potential anchoring protein in A. cellulolyticus, suggestive of the anchoring protein that occurs in a similar position on the C. thermocellum genome (14). Both of the known type II dockerins appear in tandem with a domain X, and this modular dyad may represent a functional theme that implies interaction with a type II cohesin and consequent anchoring to the cell surface.
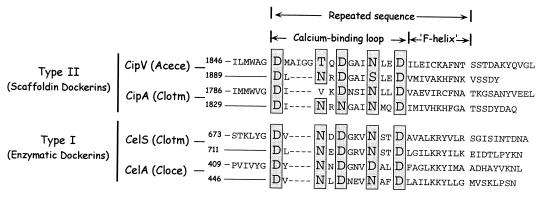
The A. cellulolyticus CipV dockerin sequence is organized in a seven-division arrangement, as described previously for both the type I and type II dockerins (37). The dockerins include a 22-residue duplicated sequence that contains a 12-residue calcium-binding loop (Fig. 8) (10). The designated calcium-binding residues usually involve conserved aspartic acids, asparagines, and sometimes hydroxyamino acids (serines or threonines) (9), and such residues indeed appear at appropriate positions within the type II CipV dockerin. However, the most striking feature of this dockerin is a 4-residue insert in the first duplicated calcium-binding motif. Digressions from the canonical EF-hand calcium-binding motif are infrequent but have been observed in some type I dockerins as well as in the only other known type II dockerin from C. thermocellum, in which one of the usual calcium-binding residues is replaced by a valine. Such replacements or inserts could result in reduced binding affinity for calcium, or alternative components (e.g., backbone atoms) may compensate for the replaced side chain (19).
FIG. 8.
Deduced amino acid sequence alignment of the C-terminal type II dockerin domain from CipV with that of C. thermocellum and their relationship to selected type I dockerins from various cellulosomal enzyme subunits. The GenBank accession numbers for CelA (family 5 glycosyl hydrolase from C. cellulolyticum) and CelS (family 48 glycosyl hydrolase from C. thermocellum) are M93096 and L06942, respectively.
Molecular evolution of cellulosome-related proteins.
Comparison of the structural arrangement of the CipV scaffoldin subunit with the phylogenetic relationship among the different modular types is revealing. Although ancestral information is lacking from the unrooted phylogenetic trees, the branching points indicate the relationship between the neighboring intra- or interspecies homologues.
The phylogeny of the various cellulosomal components from the cellulosome-producing bacteria does not necessarily reflect the phylogenetic relationships of the bacteria themselves (40). Moreover, the phylogenetic relationships among the individual types of functional modules of the A. cellulolyticus scaffoldin are, in some instances, quite different. For example, the CipV cohesins are more similar to other mesophilic cohesins than to those of C. thermocellum, whereas the X2 module, the type II dockerin, and the linkers are clearly more similar to those of C. thermocellum. Furthermore, the CipV CBD is unlike those of the other known scaffoldins and is classified in family IIIb on the basis of sequence homology.
The phylogenetic clustering of the cohesins of the A. cellulolyticus scaffoldin indicates that their evolutionary acquisition may have involved initial lateral gene transfer of a single cohesin, followed by domain insertion, multiplication and/or shuffling, and then divergence by conventional mutagenesis (i.e., accumulation of point mutations leading to compositional assimilation). At some point in the process, the genetic material that encoded the cohesin(s) joined that of the other types of functional modules and linkers to form the scaffoldin gene. It is not clear when speciation of A. cellulolyticus occurred, with respect to the development of the cellulosomal genes. In this context, it is interesting to note that A. cellulolyticus has been proposed to be a member of the greater clostridial assemblage on the basis of 16S ribosomal DNA sequences (32).
Taken together, the phylogenetic information appears to indicate that the evolution of the scaffoldins reflects a collection of independent events and mechanisms. The individual functional modules and other constituents appear to have been obtained from different microbial sources, incorporated into the scaffoldin gene, and modified to meet the needs of the overall system. Future sequence data from additional scaffoldins and other cellulosome-related components will undoubtedly refine our views of the phylogenetics of cellulosome structure.
The most extraordinary finding, however, was the discovery that the family 9 glycosyl hydrolase sequence is an integral part of CipV. This may indicate the central importance of such an enzyme to cellulosome action and may portend a trend in as-yet-undescribed scaffoldins from other microorganisms.
ACKNOWLEDGMENTS
This work was supported by a contract from the European Commission (Biotechnology Programme; BIO4-97-2303) and by grants from the Israel Science Foundation (administered by the Israel Academy of Sciences and Humanities, Jerusalem, Israel). Additional support was provided by the Otto Meyerhof Center for Biotechnology, established by the Minerva Foundation, Munich, Germany.
REFERENCES
- 1.Bayer E A, Chanzy H, Lamed R, Shoham Y. Cellulose, cellulases and cellulosomes. Curr Opin Struct Biol. 1998;8:548–557. doi: 10.1016/s0959-440x(98)80143-7. [DOI] [PubMed] [Google Scholar]
- 2.Bayer, E. A., S.-Y. Ding, A. Mechaly, Y. Shoham, and R. Lamed. Emerging phylogenetics of cellulosome structure. In H. Gilbert (ed.), Advances in carbohydrate bioengineering, in press. The Royal Society of Chemistry, London, United Kingdom.
- 3.Bayer E A, Morag E, Lamed R, Yaron S, Shoham Y. Cellulosome structure: four-pronged attack using biochemistry, molecular biology, crystallography and bioinformatics. In: Claeyssens M, Nerinckx W, Piens K, editors. Carbohydrases from Trichoderma reesei and other microorganisms. London, United Kingdom: The Royal Society of Chemistry; 1998. pp. 39–67. [Google Scholar]
- 4.Bayer E A, Setter E, Lamed R. Organization and distribution of the cellulosome in Clostridium thermocellum. J Bacteriol. 1985;163:552–559. doi: 10.1128/jb.163.2.552-559.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayer E A, Shimon L J W, Lamed R, Shoham Y. Cellulosomes: structure and ultrastructure. J Struct Biol. 1998;124:221–234. doi: 10.1006/jsbi.1998.4065. [DOI] [PubMed] [Google Scholar]
- 6.Bayer, E. A., Y. Shoham, and R. Lamed. The cellulosome: an exocellular organelle for degrading plant cell wall polysaccharides. In R. J. Doyle (ed.), Glycomicrobiology, in press. Plenum, New York, N.Y.
- 7.Béguin P, Lemaire M. The cellulosome: an exocellular, multiprotein complex specialized in cellulose degradation. Crit Rev Biochem Mol Biol. 1996;31:201–236. doi: 10.3109/10409239609106584. [DOI] [PubMed] [Google Scholar]
- 8.Belaich J-P, Tardif C, Belaich A, Gaudin C. The cellulolytic system of Clostridium cellulolyticum. J Biotechnol. 1997;57:3–14. doi: 10.1016/s0168-1656(97)00085-0. [DOI] [PubMed] [Google Scholar]
- 9.Bränden C I, Tooze J. Introduction to protein structure. New York, N.Y: Garland Publishers, Inc.; 1991. p. 22. [Google Scholar]
- 10.Chauvaux S, Béguin P, Aubert J-P, Bhat K M, Gow L A, Wood T M, Bairoch A. Calcium-binding affinity and calcium-enhanced activity of Clostridium thermocellum endoglucanase D. Biochem J. 1990;265:261–265. doi: 10.1042/bj2650261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coutinho, P. M., and B. Henrissat. 27 August 1999, revision date. CAZy website, Carbohydrate-active enzymes server. [Online.] http://afmb.cnrs-mrs.fr/∼pedro/CAZY/db.html. [22 September 1999, last date accessed.]
- 11a.Coutinho, P. M., and B. Henrissat. 9 September 1999, revision date. CAZyModO website, Carbohydrate-active enzymes server. [Online.] http://afmb.cnrs-mrs.fr/∼pedro/DB/db.html. [22 September 1999, last date accessed.]
- 12.Coutinho P M, Henrissat B. The modular structure of cellulases and other carbohydrate-active enzymes: an integrated database approach. In: Ohmiya K, Hayashi K, Sakka K, Kobayashi Y, Karita S, Kimura T, editors. Genetics, biochemistry and ecology of cellulose degradation. Tokyo, Japan: Uni Publishers Co., Ltd.; 1999. pp. 15–23. [Google Scholar]
- 13.Felix C R, Ljungdahl L G. The cellulosome—the exocellular organelle of Clostridium. Annu Rev Microbiol. 1993;47:791–819. doi: 10.1146/annurev.mi.47.100193.004043. [DOI] [PubMed] [Google Scholar]
- 14.Fujino T, Béguin P, Aubert J-P. Organization of a Clostridium thermocellum gene cluster encoding the cellulosomal scaffolding protein CipA and a protein possibly involved in attachment of the cellulosome to the cell surface. J Bacteriol. 1993;175:1891–1899. doi: 10.1128/jb.175.7.1891-1899.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerngross U T, Romaniec M P M, Kobayashi T, Huskisson N S, Demain A L. Sequencing of a Clostridium thermocellum gene (cipA) encoding the cellulosomal SL-protein reveals an unusual degree of internal homology. Mol Microbiol. 1993;8:325–334. doi: 10.1111/j.1365-2958.1993.tb01576.x. [DOI] [PubMed] [Google Scholar]
- 16.Gerwig G, de Waard P, Kamerling J P, Vliegenthart J F G, Morgenstern E, Lamed R, Bayer E A. Novel O-linked carbohydrate chains in the cellulase complex (cellulosome) of Clostridium thermocellum. J Biol Chem. 1989;264:1027–1035. [PubMed] [Google Scholar]
- 17.Gerwig G, Kamerling J P, Vliegenthart J F G, Morag (Morgenstern) E, Lamed R, Bayer E A. Novel oligosaccharide constituents of the cellulase complex of Bacteroides cellulosolvens. Eur J Biochem. 1992;205:799–808. doi: 10.1111/j.1432-1033.1992.tb16844.x. [DOI] [PubMed] [Google Scholar]
- 18.Gerwig G, Kamerling J P, Vliegenthart J F G, Morag E, Lamed R, Bayer E A. The nature of the carbohydrate-peptide linkage region in glycoproteins from the cellulosomes of Clostridium thermocellum and Bacteroides cellulosolvens. J Biol Chem. 1993;268:26956–26960. [PubMed] [Google Scholar]
- 19.Hohenester H, Maurer P, Hohenadl C, Timpl R, Jansonius J N, Engel J. Structure of a novel extracellular Ca2+-binding module in BM-40. Nat Struct Biol. 1996;3:67–73. doi: 10.1038/nsb0196-67. [DOI] [PubMed] [Google Scholar]
- 20.Joliff G, Béguin P, Aubert J-P. Nucleotide sequence of the cellulase gene celD encoding endoglucanase D of Clostridium thermocellum. Nucleic Acids Res. 1986;14:8605–8613. doi: 10.1093/nar/14.21.8605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Josefsson E, McCrea K W, Ni Eidhin D, O’Connell D, Cox J, Hook M, Foster T J. Three new members of the serine-aspartate repeat protein multigene family of Staphylococcus aureus. Microbiology. 1998;144:3387–3395. doi: 10.1099/00221287-144-12-3387. [DOI] [PubMed] [Google Scholar]
- 22.Josefsson E, O’Connell D, Foster T J, Durussel I, Cox J. The binding of calcium to the B-repeat segment of SdrD, a cell surface protein of Staphylococcus aureus. J Biol Chem. 1998;273:31145–31152. doi: 10.1074/jbc.273.47.31145. [DOI] [PubMed] [Google Scholar]
- 23.Juy M, Amit A G, Alzari P M, Poljak R J, Claeyssens M, Béguin P, Aubert J-P. Crystal structure of a thermostable bacterial cellulose-degrading enzyme. Nature. 1992;357:39–41. [Google Scholar]
- 24.Kakiuchi M, Isui A, Suzuki K, Fujino T, Fujino E, Kimura T, Karita S, Sakka K, Ohmiya K. Cloning and DNA sequencing of the genes encoding Clostridium josui scaffolding protein CipA and cellulase CelD and identification of their gene products as major components of the cellulosome. J Bacteriol. 1998;180:4303–4308. doi: 10.1128/jb.180.16.4303-4308.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karita S, Sakka K, Ohmiya K. Cellulosomes, cellulase complexes, of anaerobic microbes: their structure models and functions. In: Onodera R, Itabashi H, Ushida K, Yano H, Sasaki Y, editors. Rumen microbes and digestive physiology in ruminants. Tokyo, Japan: Japan Scientific Society Press; 1997. pp. 47–57. [Google Scholar]
- 26.Khan A W. Cellulolytic enzyme system of Acetivibrio cellulolyticus, a newly isolated anaerobe. J Gen Microbiol. 1980;121:499–502. [Google Scholar]
- 27.Lamed E, Naimark J, Morgenstern E, Bayer E A. Scanning electron microscopic delineation of bacterial surface topology using cationized ferritin. J Microbiol Methods. 1987;7:233–240. [Google Scholar]
- 28.Lamed R, Bayer E A. The cellulosome concept—a decade later! In: Shimada K, Hoshino S, Ohmiya K, Sakka K, Kobayashi Y, Karita S, editors. Genetics, biochemistry and ecology of lignocellulose degradation. Tokyo, Japan: Uni Publishers Co., Ltd.; 1993. pp. 1–12. [Google Scholar]
- 29.Lamed R, Kenig R, Setter E, Bayer E A. Major characteristics of the cellulolytic system of Clostridium thermocellum coincide with those of the purified cellulosome. Enzyme Microb Technol. 1985;7:37–41. [Google Scholar]
- 30.Lamed R, Naimark J, Morgenstern E, Bayer E A. Specialized cell surface structures in cellulolytic bacteria. J Bacteriol. 1987;169:3792–3800. doi: 10.1128/jb.169.8.3792-3800.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamed R, Setter E, Bayer E A. Characterization of a cellulose-binding, cellulase-containing complex in Clostridium thermocellum. J Bacteriol. 1983;156:828–836. doi: 10.1128/jb.156.2.828-836.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin C, Urbance J W, Stahl D A. Acetivibrio cellulolyticus and Bacteroides cellulosolvens are members of the greater clostridial assemblage. FEMS Microbiol Lett. 1994;124:151–155. doi: 10.1111/j.1574-6968.1994.tb07277.x. [DOI] [PubMed] [Google Scholar]
- 33.Matsudaira P. A practical guide to protein and peptide purification for microsequencing. 2nd ed. New York, N.Y: Academic Press; 1993. [Google Scholar]
- 34.Morag E, Bayer E A, Lamed R. Relationship of cellulosomal and noncellulosomal xylanases of Clostridium thermocellum to cellulose-degrading enzymes. J Bacteriol. 1990;172:6098–6105. doi: 10.1128/jb.172.10.6098-6105.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morag E, Bayer E A, Lamed R. Affinity digestion for the near-total recovery of purified cellulosome from Clostridium thermocellum. Enzyme Microb Technol. 1992;14:289–292. [Google Scholar]
- 36.Murray M G, Thompson W F. Rapid isolation of high-molecular-weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pagès S, Belaich A, Belaich J-P, Morag E, Lamed R, Shoham Y, Bayer E A. Species-specificity of the cohesin-dockerin interaction between Clostridium thermocellum and Clostridium cellulolyticum: prediction of specificity determinants of the dockerin domain. Proteins. 1997;29:517–527. [PubMed] [Google Scholar]
- 38.Pagès S, Belaich A, Fierobe H-P, Tardif C, Gaudin C, Belaich J-P. Sequence analysis of scaffolding protein CipC and ORFXp, a new cohesin-containing protein in Clostridium cellulolyticum: comparison of various cohesin domains and subcellular localization of ORFXp. J Bacteriol. 1999;181:1801–1810. doi: 10.1128/jb.181.6.1801-1810.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poole D M, Morag E, Lamed R, Bayer E A, Hazlewood G P, Gilbert H J. Identification of the cellulose binding domain of the cellulosome subunit S1 from Clostridium thermocellum. FEMS Microbiol Lett. 1992;99:181–186. doi: 10.1016/0378-1097(92)90022-g. [DOI] [PubMed] [Google Scholar]
- 40.Rainey F A, Stackebrandt E. 16S rDNA analysis reveals phylogenetic diversity among the polysaccharolytic clostridia. FEMS Microbiol Lett. 1993;113:125–128. doi: 10.1111/j.1574-6968.1993.tb06501.x. [DOI] [PubMed] [Google Scholar]
- 41.Saddler J N, Khan A W. Cellulase production by Acetivibrio cellulolyticus. Can J Microbiol. 1980;26:760–765. [Google Scholar]
- 42.Saddler J N, Khan A W. Cellulolytic enzyme system of Acetivibrio cellulolyticus. Can J Microbiol. 1981;27:288–294. doi: 10.1139/m81-045. [DOI] [PubMed] [Google Scholar]
- 43.Sakon J, Irwin D, Wilson D B, Karplus P A. Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca. Nat Struct Biol. 1997;4:810–818. doi: 10.1038/nsb1097-810. [DOI] [PubMed] [Google Scholar]
- 44.Salamitou S, Raynaud O, Lemaire M, Coughlan M, Béguin P, Aubert J-P. Recognition specificity of the duplicated segments present in Clostridium thermocellum endoglucanase CelD and in the cellulosome-integrating protein CipA. J Bacteriol. 1994;176:2822–2827. doi: 10.1128/jb.176.10.2822-2827.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimon L J W, Bayer E A, Morag E, Lamed R, Yaron S, Shoham Y, Frolow F. The crystal structure at 2.15 Å resolution of a cohesin domain of the cellulosome from Clostridium thermocellum. Structure. 1997;5:381–390. doi: 10.1016/s0969-2126(97)00195-0. [DOI] [PubMed] [Google Scholar]
- 46.Shoseyov O, Takagi M, Goldstein M A, Doi R H. Primary sequence analysis of Clostridium cellulovorans cellulose binding protein A. Proc Natl Acad Sci USA. 1992;89:3483–3487. doi: 10.1073/pnas.89.8.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tavares G A, Béguin P, Alzari P M. The crystal structure of a type I cohesin domain at 1.7 Å resolution. J Mol Biol. 1997;273:701–713. doi: 10.1006/jmbi.1997.1326. [DOI] [PubMed] [Google Scholar]
- 48.Tomme P, Warren R A J, Miller R C, Kilburn D G, Gilkes N R. Cellulose-binding domains—classification and properties. In: Saddler J M, Penner M H, editors. Enzymatic degradation of insoluble polysaccharides. Washington, D.C.: American Chemical Society; 1995. pp. 142–161. [Google Scholar]
- 49.Tormo J, Lamed R, Chirino A J, Morag E, Bayer E A, Shoham Y, Steitz T A. Crystal structure of a bacterial family-III cellulose-binding domain: a general mechanism for attachment to cellulose. EMBO J. 1996;15:5739–5751. [PMC free article] [PubMed] [Google Scholar]
- 50.Von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williamson M P. The structure and function of proline-rich regions in proteins. Biochem J. 1994;297:249–260. doi: 10.1042/bj2970249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woessner J P, Goodenough U W. Zygote and vegetative cell wall proteins in Clamydomonas reinhardtii share a common epitope (SerPro)x. Plant Sci. 1992;83:65–76. [Google Scholar]