Abstract
Unique cytoplasmic filaments are found in the treponeme genus of spirochete bacteria. Their function is unknown, but their location underneath the periplasmic flagellar filaments (PFF) suggests a role in motility and/or cell structure. To better understand these unique structures, the gene coding for the cytoplasmic filaments, cfpA, was identified in various treponemal species. Treponema phagedenis cfpA was 2,037 nucleotides long, and the encoded polypeptide showed 78 to 100% amino acid sequence identity with the partial sequence of CfpA from T. denticola, T. vincentii, and T. pallidum subsp. pertenue. Wild-type T. phagedenis and a PFF-deficient isolate were analyzed by electron microscopy to assess the structural relationship of the cytoplasmic filaments and the PFF. The number of cytoplasmic filaments per cell of T. phagedenis (mean, 5.7) was compared with the number of PFF at each end of the cell (mean, 4.7); the results suggest that there is no direct one-to-one correlation at the cell end. Moreover, a structural link between these structures could not be demonstrated. The cytoplasmic filaments were also analyzed by electron microscopy at different stages of cell growth; this analysis revealed that they are cleaved before or during septum formation and before the nascent formation of PFF. A PFF-deficient mutant of T. phagedenis possessed cytoplasmic filaments similar to those of the wild type, suggesting that intact PFF are not required for their assembly and regulation. The extensive conservation of CfpA among pathogenic spirochetes suggests an important function, and structural analysis suggests that it is unlikely that the cytoplasmic filaments and the flagellar apparatus are physically linked.
The treponemes are a group of spirochetes associated with a wide range of diseases. Treponema pallidum subsp. pallidum is the causative agent of syphilis, a sexually transmitted disease (33); T. pallidum subsp. pertenue is associated with yaws, a disease transmitted by skin contact (3); and T. vincentii and T. denticola are associated with gingivitis and periodontitis (8, 41, 42). Other treponemes, including T. phagedenis, are host associated, and their role in disease is unclear.
Treponemes are helical or flat wave-shaped bacteria that are motile, and their polar flagellar filaments are located in the periplasmic space. In addition, all treponemes have an unusual structure in the cytoplasm: a bundle of filaments located just underneath the cytoplasmic membrane (15–17). These cytoplasmic filaments are apparently located below the bundle of periplasmic flagellar filaments (PFF) (15–18). The individual cytoplasmic filaments maintain a ribbon-like configuration (9), and the periodicity of the helix of the bundle is equivalent to the helical periodicity of the cell (44). The cytoplasmic filaments are not transitory and are found in cells at all growth stages (9). The function of the cytoplasmic filament structures is unknown.
Masuda and Kawata (31) purified the cytoplasmic filaments of different strains of treponemes, including T. denticola TD2 and T. phagedenis biotypes Kazan and Reiter. In each case, the major constituent of the cytoplasmic filaments was an 82-kDa protein. You et al. (43) cloned and sequenced the cytoplasmic filament gene, cfpA, from T. pallidum subsp. pallidum. However, analysis of the deduced amino acid sequence did not provide sufficient clues to determine the function of these distinctive structures. The location of the filaments and their helical shape suggest involvement in cell motility, maintenance of cell structure, or cell division (9).
The aims of this work were to determine the sequence conservation of the cytoplasmic filament polypeptide among pathogenic and nonpathogenic spirochetes, to elucidate the structural relationship of the cytoplasmic filament ribbon and the flagellar apparatus, and to determine the structural characteristics of the cytoplasmic filaments during cell division. We found that the amino acid sequence of CfpA is well conserved among different species of cultivable and noncultivable treponemes, whether or not they are associated with disease. Electron microscopy studies suggest that the cytoplasmic filaments and the flagellar apparatus are not physically linked on a one-to-one basis, although an interaction of the two types of structures cannot be excluded. Finally, the fate of the cytoplasmic filaments during cell division was elucidated. This analysis provides an initial genetic and structural characterization of the cytoplasmic filaments as a basis for future work to elucidate the function of these unique structures.
MATERIALS AND METHODS
Strains, reagents, and culture and molecular methods.
T. phagedenis Kazan 5, flagellar filament-deficient mutant T55 of T. phagedenis Kazan 5 (26), and T. vincentii ATCC 35580 were grown in Spirolate broth (Becton Dickinson Microbiology Systems, Cockeysville, Md.) with 10% heat-inactivated rabbit serum in an anaerobic chamber. T. denticola ATCC 33520 was grown in New Oral Spirochete medium (4) with 10% heat-inactivated rabbit serum and 10 μg of cocarboxylase per ml at 36°C in an anaerobic chamber (Coy Laboratory Products Inc., Grass Lake, Mich.) with an atmosphere of 85% nitrogen, 10% hydrogen, and 5% carbon dioxide. T. pallidum subsp. pertenue Haiti B cells were kindly provided by Konrad Wicher (Wadsworth Center, Albany, N.Y.).
Oligonucleotides were synthesized at the Molecular Genetics Core Facility of the Wadsworth Center with a PerSeptive Biosystems 8909 (PE Biosystems, Foster City, Calif.). Automated DNA sequencing was done with a DyeDeoxy terminator cycle sequencing kit (PE Biosystems), a PE 9600 ThermalCycler, and an ABI Prism 377 sequencer (Perkin-Elmer, Foster City, Calif.).
Chromosomal DNA from the different strains was isolated by standard methods (30). Plasmid miniprep DNA was isolated by standard methods (30). Midi columns (Qiagen Corp., Chatsworth, Calif.) were used for purification prior to DNA sequencing.
The PCR was performed with Taq polymerase, reagents, and thermal cyclers available from Perkin-Elmer. DNA probes were prepared by amplification and labeled with digoxigenin by use of the Genius System (Boehringer Mannheim Corp., Indianapolis, Ind.).
Restriction endonucleases were purchased from New England BioLabs, Inc. (Beverly, Mass.). Ligations were performed either with the Rapid DNA Ligation Kit (Boehringer) or directly with the TOPO TA Cloning Kit Dual Promoter (Invitrogen, Carlsbad, Calif.). Escherichia coli Top 10 One Shot was used as the recipient for cloning experiments with vector pZErO-2 or pCRII-TOPO (Invitrogen).
α-35S-deoxynucleoside triphosphate for DNA sequencing and [γ-32P]ATP for the primer extension reaction were purchased from Amersham Life Science, Inc. (Arlington Heights, Ill.).
Identification of cfpA. (i) T. phagedenis.
To identify T. phagedenis cfpA, the T. pallidum subsp. pallidum cfpA sequence (43) and the N-terminal amino acid sequence of T. phagedenis Kazan 5 CfpA (43) were used to design two degenerate oligonucleotide primers: NTPHDE1N (5′-AA[C,T]GT[A,C,G,T]TT[C,T]CC[A,C,G,T]GA[A,G]AA[A,G]CC-3′) and CTPHDE1R (5′-[A,C,G,T]GT[C,T]TTCAT[A,C,G,T]CC[A,C,G,T]AC[A,G]TC-3′); corresponding regions in T. pallidum subsp. pallidum CfpA are amino acids 11 to 18 and 651 to 656. After amplification of T. phagedenis DNA, the PCR product obtained was digested with HindIII to generate a 317-bp DNA fragment, ligated in pZErO-2 digested with HindIII, and transformed into competent E. coli Top 10 One Shot cells. On the basis of the DNA sequence of this PCR product, synthetic oligonucleotides PHAGHIN5 (5′-AAATACCATGTAGGAATTCG-3′) and PHAGHIN3 (5′-ATTATCAGTATCCGATTGAG-3′) were used to produce a digoxigenin-labeled probe by amplification. A T. phagedenis genomic library (28) in Lambda ZAP Express (Stratagene, La Jolla, Calif.) was screened by use of a plaque hybridization assay with the digoxigenin-labeled probe (40), positive plaques were purified, and the insert was sequenced with synthetic oligonucleotides.
To obtain the 5′ end of the T. phagedenis cfpA sequence, which was not present in the clones sequenced, the One Strand PCR technique (36) was used. The amplification reaction for the first step was carried out at 93°C for 1 min, 55°C for 1 min, and 72°C for 1 min for a total of 30 cycles with primer CFPAPH4R (5′-GGAGCTTTGTGTTCAAGTG-3′) and purified chromosomal DNA of T. phagedenis as a template. For the second step, 1 μl from the first reaction was used as a template, together with CFPAPH4R and a short, nonspecific primer, OS1290 (5′-GTGGAATGCGA-3′) (1). The amplification reaction was carried out at 93°C for 1 min, 60°C for 1 min, and 72°C for 1 min for a total of 30 cycles. The resulting product was cloned in EcoRV-digested vector pZErO-2 and sequenced. From this sequence, a specific primer, OSPCRFO2 (5′-GAGTTTGTAAGAATTGTTAGCAGC-3′), was synthesized. The product of amplification of T. phagedenis chromosomal DNA with primers OSPCRFO2 and CFPAPH4R, which contains the promoter, was cloned and sequenced to obtain the promoter region.
(ii) T. denticola.
A degenerate set of primers was synthesized on the basis of the similarity between the cfpA sequences of T. phagedenis and T. pallidum subsp. pallidum. NTPHDE1N (5′-AA[C,T]GT[A,C,G,T]TT[C,T]CC[A,C,G,T]GA[A,G]AA[A,G]CC-3′) and DENTCF2R (5′-AG[ACGT]AC[TG]TT[ACGT]GCCAT[ACGT]CC[AG]TC-3′) (corresponding regions in T. pallidum subsp. pallidum CfpA are amino acids 11 to 18 and 570 to 577) were used to amplify T. denticola chromosomal DNA, and the product (1,675 bp) was cloned in EcoRV-digested pZErO-2 and sequenced. The 3′ end of the sequence was obtained by the One Strand PCR technique (36) with purified chromosomal DNA of T. denticola as a template. The first primer was DENTCF9 (5′-GACTTTCGTTCATAGAAGCAG-3′), and the second primer was OS1283 (5′-GCGATCCCCA-3′) (1). The amplification reaction was the same as that for T. phagedenis cfpA. The resulting product was cloned with the TOPO TA Cloning Kit Dual Promoter and sequenced. From this sequence, a specific primer, DENTCF10R (5′-ACTTTCGTTCATAGAAGCAGCC-3′), was synthesized. The product of amplification of T. denticola chromosomal DNA with primers DENTCF9 and DENTCF10R was cloned in vector pCRII-TOPO.
(iii) T. pallidum subsp. pertenue.
Two oligonucleotide primers were synthesized on the basis of the T. pallidum subsp. pallidum cfpA sequence (43); MCFPAL2 (5′-CGCGGATCCATGGCAAGTTTAGATCTACC-3′) and CCFPPAL2 (5′-GATCAGATCTCGCTAGAGTTCGCGAATG-3′). The product of amplification of T. pallidum subsp. pertenue chromosomal DNA was cloned with the TOPO TA Cloning Kit Dual Promoter and sequenced with primers based on the T. pallidum subsp. pallidum cfpA sequence (GenBank accession no. U32683).
(iv) T. vincentii.
The DNA sequence of T. vincentii cfpA was cloned by amplification of chromosomal DNA with primers CFPAPH14 (5′-CGCGGATCCAAGGAGATAGCAAATGGC-3′) and CFPAPH15 (5′-CTAGTCTAGAGCAATTACTAAAGTTCACG-3′) and sequenced with primers based on the T. phagedenis cfpA sequence.
Primer extension.
To identify the start site of transcription of the T. phagedenis cfpA gene, primer extension was performed. T. phagedenis RNA was isolated and purified by use of an Rnaid Plus Kit with SPIN (Bio 101, Inc., Vista, Calif.). Primer labeling with 32P by use of T4 polynucleotide kinase and the primer extension reaction were carried out with a Primer Extension System (Promega Corp., Madison, Wis.) and 1 pmol of T. phagedenis primer OSPCRA2 (5′-CTTGATCCGACCGCACTGGG-3′). The annealing step was carried out at 65°C for 3 min, followed by 52°C for 20 min. For determining the lengths of the primer extension products, a DNA ladder was generated by PCR with the control sequence from an AmpliCycle sequencing kit (Perkin-Elmer) and the forward M13 primer. Burst-Pak Sequencing Gels (Owl Scientific, Inc., Woburn, Mass.) were used to prepare 6% polyacrylamide gels for electrophoresis. Autoradiography was performed with XAR 2 film (Eastman Kodak Co., New Haven, Conn.) at −70°C with an intensifying screen.
Isolation and purification of cytoplasmic filaments.
The cytoplasmic filaments were prepared according to a modification of the protocol of Masuda and Kawata (31). After sonication of the cells, 50 mM EDTA and 2% Triton X-100 (final concentrations) were added to the samples, which were then stored at 4°C overnight. The samples were layered on top of a gradient of 14, 48, 80, and 100% glycerol in 10 mM Tris-HCl (pH 7.4)–4 mM EDTA–1 M urea. The samples were centrifuged in an SW28 rotor at 20,000 × g for 20 min in an XL-90 ultracentrifuge (Beckman Intruments, Inc., Palo Alto, Calif.). The different fractions were dialyzed against 10 mM Tris-HCl (pH 7.4)–1 mM EDTA.
Electron microscopy.
To prepare cells for visualization of the cytoplasmic filaments, 2 ml of an exponential-phase culture of wild-type T. phagedenis or flagellar filament-deficient mutant T55 was centrifuged for 1 min at 10,000 × g. The cells were resuspended in 2 ml of sterile distilled water, stored at 4°C overnight, centrifuged, and resuspended in 100 μl of sterile distilled water prior to use.
Negative staining was used for the visualization of cytoplasmic filaments in the cells or of purified cytoplasmic filaments. Drops (40 μl) of samples were placed on dental wax. Formvar-coated copper grids were floated on the drops for 2 to 4 min. Excess liquid was removed by wicking with filter paper, and the grids were immediately washed by floatation on 2 drops of double-distilled water. After the grids were washed, excess water was removed, the grids were briefly floated on 2% sodium phosphotungstate (pH 7.0), liquid was removed by wicking, and the samples were viewed with a Zeiss (LEO) 910 transmission electron microscope operating at 80 keV. The negatives were enlarged photographically.
Sequence analysis.
Sequences were analyzed with programs available through Wisconsin Package version 9.1 (Genetics Computer Group, Madison, Wis.).
Nucleotide sequence accession numbers.
The nucleotide sequences of cfpA from T. phagedenis, T. pallidum subsp. pertenue, T. denticola ATCC 33520, and T. vincentii ATCC 35580 have been deposited in GenBank under accession no. AF037069, AF062736, AF062737, and AF080570, respectively.
RESULTS
Identification and conservation of cfpA.
The entire T. phagedenis cfpA open reading frame was sequenced, whereas the T. denticola, T. vincentii, and T. pallidum subsp. pertenue cfpA sequences represent 98, 99, and 98%, respectively, of the T. phagedenis cfpA open reading frame. The nucleotide and amino acid sequences of cytoplasmic filaments were extensively conserved among these different species of treponemes. The percentage of sequence identity was calculated for each pair by use of the GAP program and is shown in Table 1. The alignment of the nucleotide sequences from the five different treponemes revealed several gaps from 3 to 15 nucleotides long at the 3′ end of the sequences, with no frameshifts (data not shown). The alignment of the T. vincentii and T. phagedenis cfpA sequences showed that the sequences were identical. Notably, the CfpA sequence of T. pallidum subsp. pallidum differs from that of T. pallidum subsp. pertenue by only 3 amino acids in the NH2-terminal region (C31→Y, H51→R, and E110→G).
TABLE 1.
Identity between cytoplasmic filament sequences from different treponemes
| Treponeme | % Nucleotide (amino acid) sequence identity of:
|
|||
|---|---|---|---|---|
| T. phagedenis | T. denticola | T. vincentii | T. pallidum subsp. pallidum | |
| T. denticola | 76.5 (82.7) | |||
| T. vincentii | 100 (100) | 76.8 (82.9) | ||
| T. pallidum subsp. pallidum | 72.4 (84.9) | 69.4 (77.6) | 73.2 (84.9) | |
| T. pallidum subsp. pertenue | 73.0 (84.8) | 69.4 (77.8) | 73.0 (84.8) | 99.8 (99.6) |
Analysis of the cytoplasmic filament sequences by use of the BLAST (2) or FASTA (37) protocols revealed no sequences in the GenBank database having significant similarity to cfpA.
In T. phagedenis, the two partial predicted open reading frames preceding and following cfpA are similar to those found at the same relative positions in T. pallidum subsp. pallidum (11, 43). The products of both T. pallidum genes (GenBank accession no. TP0750 and TP0747) are of unknown function and are not similar to any known prokaryotic proteins.
Identification of the cfpA promoter by primer extension.
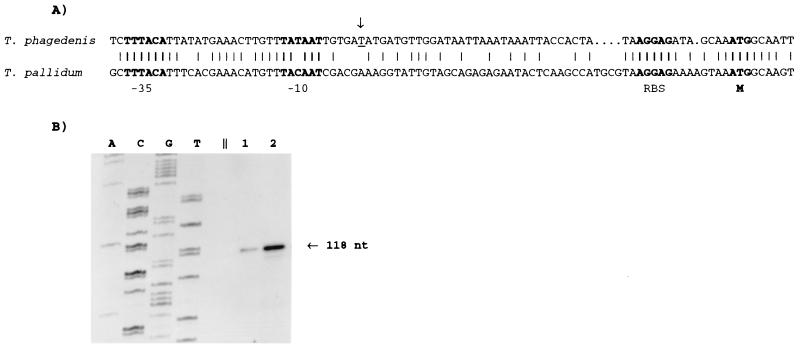
Primer extension analysis revealed the transcription start site of T. phagedenis cfpA, and a putative promoter was identified upstream of the start site (Fig. 1A and B). This promoter differed by only 1 nucleotide from the consensus sigma 70 promoter sequence found in E. coli (12). A similar promoter sequence was previously noted in T. pallidum subsp. pallidum (43).
FIG. 1.
Identification of the cfpA promoter. (A) Nucleotide sequence identity of promoter regions, including proposed −10 and −35 regions of PcfpA. The arrow indicates the start site of transcription, determined by a primer extension assay with T. phagedenis. RBS, putative ribosome binding site; M, first amino acid of the CfpA polypeptide. Identical nucleotides in the T. phagedenis and T. pallidum subsp. pallidum sequences are indicated by vertical lines. (B) Primer extension assay done to determine the start site of transcription of T. phagedenis cfpA. A, C, G, and T indicate nucleotides used to generate a size ladder with unrelated DNA. Lanes 1 and 2 contain the primer extension reaction products (1 and 5 μl, respectively). nt, nucleotides.
The likely strength of the promoter as well as the presumed toxicity of the gene precluded cloning of T. phagedenis cfpA gene with its native promoter in E. coli. This situation is consistent with previous data concerning cfpA from T. pallidum subsp. pallidum and flaA, another gene producing abundant protein in treponemes; both are preceded by a sigma 70-like promoter (20, 21, 43). We were able to clone T. phagedenis cfpA under the control of the T7 promoter and found that the expression of T. phagedenis CfpA was toxic to E. coli (data not shown).
T. phagedenis cytoplasmic filaments span the length of the cell.
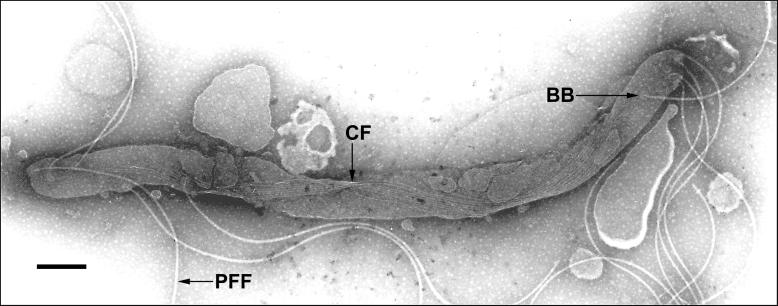
The cytoplasmic filaments are associated in a ribbon-like structure in the cytoplasm of the treponemes (15, 17, 44) and originate or terminate at each of the cell ends (15, 16, 18). A similar structure was found in T. phagedenis Reiter (31). We wanted to clarify the location of the filaments throughout the length of the cell. However, the filaments are difficult to visualize along the entire length of the cell, due to the low contrast of the filaments in the cell when observed by electron microscopy with negative staining. The complete ribbon of filaments was observed in 11 cells. In each case, a single ribbon of filaments spanned the cell from one end to the other (Fig. 2). In 10 of the 11 cells, the numbers of cytoplasmic filaments differed at both ends, suggesting ongoing filament synthesis (Table 2). The protein units apparently are assembled to the nascent filament at one end and progress along the ribbon, using it as a template (Fig. 3).
FIG. 2.
The cytoplasmic filament ribbon spans the entire cytoplasmic compartment. After removal of the outer membrane, the flagellar filaments are liberated and two helical turns of the cytoplasmic filament (CF) ribbon can be seen. This cell corresponds to cell no. 8 in Table 2. T. phagedenis cells were prepared and stained as described in Materials and Methods. BB, flagellar basal body. Bar, 250 nm.
TABLE 2.
Distribution of the numbers of cytoplasmic and flagellar filaments at both ends in 11 T. phagedenis cells
| Cell no. | No. ofa:
|
|||
|---|---|---|---|---|
| PFF1 | CF1 | CF2 | PFF2 | |
| 1 | 3 | 5 | 4 | 4 |
| 2 | 4 | 5 | 6 | 4 |
| 3 | 4 | 5 | 6 | 4 |
| 4 | 4 | 5 | 6 | 5 |
| 5 | 5 | 5 | 6 | 5 |
| 6 | 5 | 5 | 7 | 5 |
| 7 | 5 | 6 | 6 | 5 |
| 8 | 5 | 7 | 8 | 5 |
| 9 | 5 | 5 | 4 | 6 |
| 10 | 5 | 5 | 6 | 6 |
| 11 | 5 | 8 | 7 | 6 |
PFF1, PFF at end one; CF1, cytoplasmic filaments at end one; CF2, cytoplasmic filaments at end two; PFF2, PFF at end two.
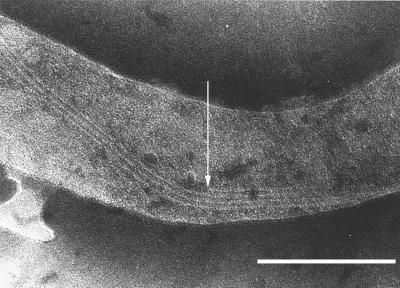
FIG. 3.
Assembly of cytoplasmic filaments in vivo. The arrow shows one end of a cytoplasmic filament likely in construction and adjacent to a ribbon of cytoplasmic filaments. T. phagedenis cells were prepared and stained as described in Materials and Methods. Bar, 250 nm.
Relationship of cytoplasmic filaments and PFF.
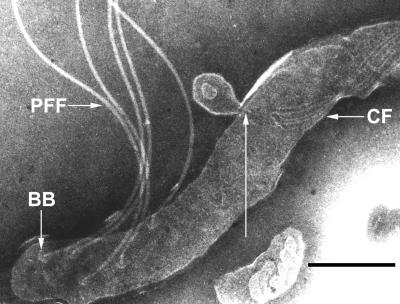
The cytoplasmic filaments and the flagellar filaments are the only two filamentous structures visible inside a treponemal cell by electron microscopy with negative staining. A potential physical link between these two types of structures, through the flagellar basal body, has been discussed by Hovind-Hougen and Birch-Andersen (18), and reports of copurification of cytoplasmic filaments and flagellar basal bodies (43) support this hypothesis. To investigate this structural feature, T. phagedenis cells were extensively analyzed to determine the termini of the cytoplasmic filaments. In most cells, the ends of the cytoplasmic filament bundle were found to disperse near the last turn of the ribbon and terminate close, but not linked, to the insertion point of the flagella. However, on rare occasions the ends of an easily discernible group of cytoplasmic filaments were not dispersed and terminated at a unique point (Fig. 4). On the basis of extensive electron microscopy analysis, we believe that the ends of the cytoplasmic filament ribbon are not dispersed and terminate at a unique anchor in the cell membrane. The sample treatment required to visualize the cytoplasmic filaments probably destabilizes some protein structures present in the cell, possibly explaining why the cytoplasmic filaments often appear dispersed at the last turn of the ribbon. In addition to the structural evidence, a modified version of the purification protocol of Masuda and Kawata (31) enabled the purification of cytoplasmic filaments of T. phagedenis without flagellar filaments or basal bodies, showing that there is no tight link between these two membrane-associated structures (22).
FIG. 4.
The cytoplasmic filament ribbon ends at a unique point connected to the membrane. The long arrow indicates a potential cytoplasmic filament (CF) ribbon attachment site associated with a membrane. This region has undergone evagination and would not be found in normal cells. T. phagedenis cells were prepared and stained as described in Materials and Methods. BB, flagellar basal body. Bar, 250 nm.
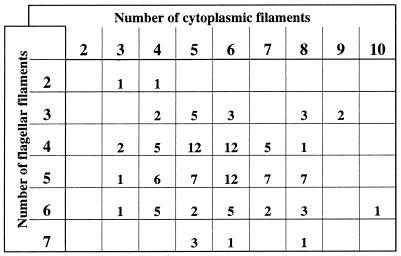
A direct count of the numbers of flagellar filaments versus cytoplasmic filaments at each cell end was done to determine whether there was a one-to-one relationship between the two types of structures in T. phagedenis, which would suggest a direct interaction with the basal body. Overall, 3 to 10 cytoplasmic filaments were observed (mean, 5.7; 127 cell ends) at the ends of the cell. Two to eight flagellar filaments per cell end were observed (mean, 4.7; 202 cell ends). In most cases, there was a difference of one flagellar filament at both ends of the cell (73%; 49 cells), consistent with previous observations (14). A detailed analysis of the numbers of both structures at 118 cell ends is presented in Fig. 5. A direct comparison of the flagellar filament number versus the number of cytoplasmic filaments at one end of the cell did not show any correlation. In fact, the number of cytoplasmic filaments was often larger than the number of flagellar filaments (68% of the cell ends observed) (Fig. 5).
FIG. 5.
Distribution of the numbers of cytoplasmic and flagellar filaments in T. phagedenis cell ends. The number of each filament type in 118 cell ends was evaluated. The number of cells with seven or more cytoplasmic filaments is probably slightly underestimated due to the low contrast of the filamentous structure. Blank spaces indicate that no cells having this defined number of filaments were observed.
Analysis of the numbers of both structures visible in the same whole cell was done with 11 well-preserved cells (Table 2). The noncongruent numbers of flagellar and cytoplasmic filaments at one end or at both ends of a cell ruled out a one-to-one interaction between these types of structures.
Cytoplasmic filaments in a flagellar filament-deficient mutant.
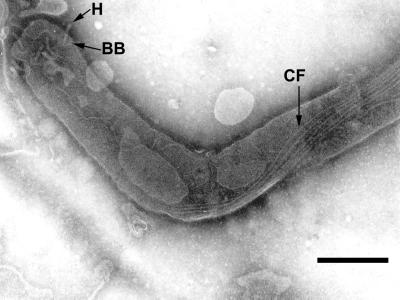
Cells of the T. phagedenis flagellar filament-deficient mutant T55 have the same general shape as wild-type cells, except for the shape of the bent ends (7, 26). No flagellar filaments were produced by this mutant, but the basal body and the hook were visible, as were the cytoplasmic filaments (Fig. 6). Electron microscopy showed that the cytoplasmic filament ribbon had the same helical shape and position in mutant cells as in wild-type cells. In addition, the absence of flagellar filaments did not appear to adversely influence the synthesis or number of cytoplasmic filaments per cell (data not shown).
FIG. 6.
The cell end of a T. phagedenis flagellar filament-deficient mutant has a wild-type cytoplasmic filament structure. The hook (H) and the basal body (BB) of the flagellar apparatus as well as the cytoplasmic filaments (CF) can be seen, showing that flagellar filaments are not needed for cytoplasmic filament assembly. Bar, 250 nm.
Cytoplasmic filaments and cell division.
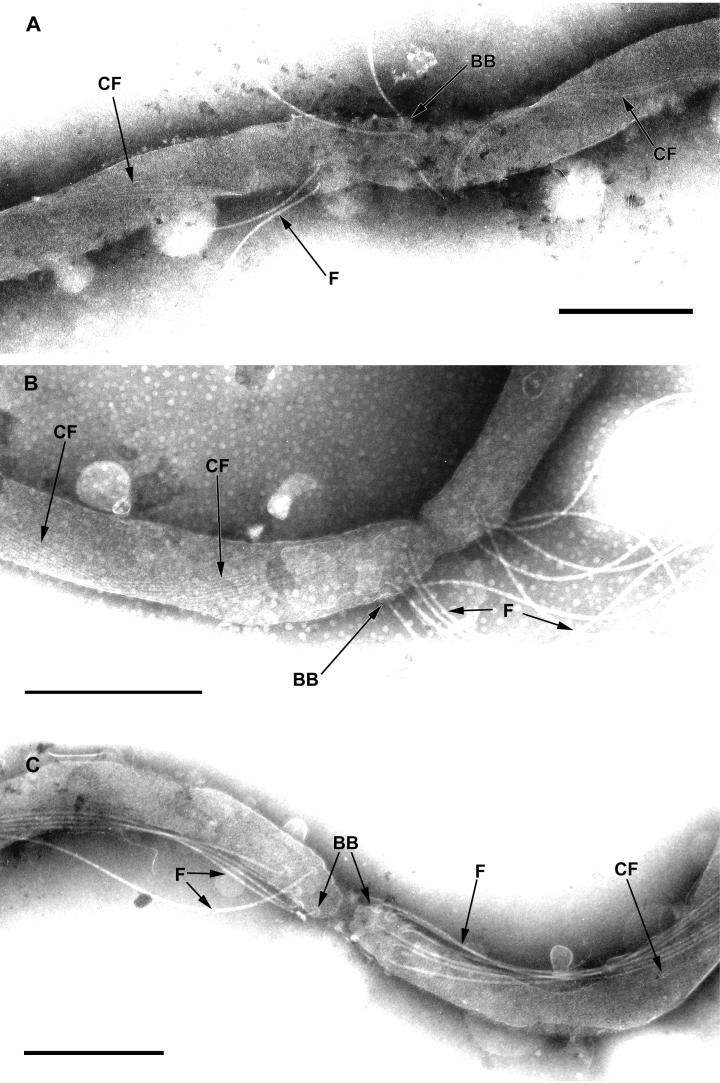
To investigate the fate of the cytoplasmic filaments during cell division, we analyzed cells during various stages of growth by electron microscopy. The flagellar filaments are just forming (as indicated by their short length) when the septum at the cell division site is visible (Fig. 7A), and the cytoplasmic filaments are already visible as two independent ribbons in the future cells. When the two cells are further along in the division process, flagellar filament synthesis appears to be completed, as indicated by the mature length (Fig. 7B). Later, the separate cytoplasmic cylinders of both cells are visible, yet the cells are still attached by the outer membrane (Fig. 7C). In rare cases when we were able to count the cytoplasmic filaments on both sides of the division site, the same numbers were observed. It appears that the ribbon of cytoplasmic filaments is cut at the division site before or during the early stages of the formation of the flagellar apparatus.
FIG. 7.
Cytoplasmic filament and flagellar filament formation during cell division. (A) Early cell division step. Four cytoplasmic filaments (CF) in individual bundles can be seen on both sides of the cell division septum. Flagellar basal bodies (BB) and hooks linked to forming flagellar filaments (F) are visible on both sides of the cell division constriction site. Bar, 500 nm. (B) Later in cell division, the flagellar filaments have completed formation on both sides of the cell division septum. Bar, 250 nm. (C) Two individual cytoplasmic cylinders are visible, but the two daughter cells have not completely separated. Bar, 250 nm.
DISCUSSION
The cytoplasmic filament protein CfpA is the major constituent of the cytoplasmic filament, a unique structure present in the cytoplasm of Treponema cells (31, 43). The gene encoding the cytoplasmic filament has nucleotide and deduced amino acid sequences that are well conserved among different species of treponemes. Although some gaps were noted in the alignment of several treponemal CfpA amino acid sequences, the coding region remained intact, with no frameshifts. The sequence similarity is independent of the habitat, mode of transmission of the bacteria, or the ability to cultivate the organism in the laboratory. These results extend and are consistent with the results of Masuda and Kawata (31), which indicated that the cytoplasmic filament protein shows serologic cross-reactivity among various treponemes. Moreover, CfpA, previously identified as TpN83 in T. pallidum subsp. pallidum (35), is an antigenic protein (5) and likely plays a role in the immune response. In this report, a limited number of amino acid changes were found between T. pallidum subsp. pallidum CfpA and T. pallidum subsp. pertenue CfpA. A small number of sequence variations within antigenic proteins of these subspecies has previously been noted (6, 34). Our data show that the observed sequence variations between the two subspecies are not limited to membrane proteins.
How and where the cytoplasmic filaments terminate at the cell end are major questions related to the function of these structures. We attempted to resolve these questions by using electron microscopy, comparison of numbers of cytoplasmic filaments to numbers of flagellar filaments, and analysis of a flagellar filament-deficient mutant. The data reported here show that the cytoplasmic filaments span the length of the cell and therefore differ from the flagellar filaments, which originate at each end of the cell and terminate in the central region. Previous reports presented the hypothesis of a direct physical relationship between flagellar basal body and the cytoplasmic filament ends (18, 43). No evidence of such association was revealed by electron microscopy of the samples observed in this study. The proximal ends of the cytoplasmic filament bundle were found close but not connected to the flagellar basal body, as previously described (14), and most of the time the observed cytoplasmic filament ribbon was dissociated into individual filaments at the cell ends. In rare instances, grouped cytoplasmic filaments were observed associated with a membranous structure. We suggest that the cytoplasmic filament ribbon terminates at a unique anchor associated with the membrane at the cell end, as shown in Fig. 4. Biochemical study of this structure, designed to determine the protein-protein interaction partners, should resolve the nature of the components involved.
An alternative approach to resolving whether the cytoplasmic filaments are attached to the flagellar basal body in a one-to-one relationship is to compare the number of cytoplasmic filaments with the number of flagellar filaments. Extensive analysis and counting of filaments at cell ends revealed that there was no one-to-one correlation between the number of cytoplasmic filaments and the number of flagellar filaments or flagellar basal bodies. Interestingly, the number of cytoplasmic filaments differed at the ends of the same cells in most of the samples observed, a result which may be attributed to ongoing cytoplasmic filament synthesis. The discrepancy between the number of cytoplasmic filaments and the number of flagellar filaments may also be an indication of noncoordinate synthesis of the cytoplasmic and periplasmic filaments.
The electron microscopic analysis of a flagellar filament-deficient mutant of T. phagedenis allowed us to observe the cytoplasmic filament structure in the absence of the flagellar filament structure. The flagellar basal body and hook were still intact in this mutant. The wild type and the flagellar filament-deficient mutant had morphologically indistinguishable cytoplasmic filament structures. Moreover, wild-type T. denticola and a well-defined FliK flagellar filament-deficient mutant (27) have morphologically indistinguishable cytoplasmic filament structures (23). The absence of the flagellar filaments in both cases did not interfere with cytoplasmic filament formation and regulation in a flagellar filament-deficient mutant, indicating that intact flagellar filaments are not required for cytoplasmic filament formation. The use of a flagellar filament-deficient mutant also permitted the isolation and purification of cytoplasmic filaments and confirmed that they were composed of one major polypeptide band (data not shown).
The fate of the cytoplasmic filaments was examined during various stages of cell division. The severing of the cytoplasmic filaments and the transverse binary fission during cell division appear to be synchronized together with the formation of the flagellar apparatus. This analysis documents the severing of cytoplasmic filaments before or during the early stages of cell division and before flagellar filament synthesis in treponemal cells. When the constriction ring of the cell division site was visible, basal bodies, hooks, and nascent flagellar filaments were observed. Although visualization of the bundle of cytoplasmic filaments on both sides of the constriction site rarely occurred, in each observed sample the cytoplasmic filaments were already severed and the numbers of cytoplasmic filaments were the same on both sides. We hypothesize that the ribbon of cytoplasmic filaments is cut at or near the cell division septum site. This hypothesis implies that a specific mechanism cuts the filament bundle and creates a new anchor in the inner membrane. After the formation of the flagellar filaments, the constriction of the septum continues until the two cytoplasmic cylinders are separated underneath a unique outer membrane. As a result, very long cells consisting of one outer membrane and two or more cytoplasmic cylinders resulting from cell division are often observed in young cultures of T. phagedenis (data not shown) (14, 39). The final step would be the complete separation of the two newly formed daughter cells. The cell division steps are similar to previous data obtained with spirochetes (13, 19, 29, 32, 38, 39), and it is now shown that the cytoplasmic filament ribbon is severed during the cell division process, before the establishment of the flagellar apparatus.
The identification of the gene and the promoter for cfpA in T. denticola and structural analysis are the first steps leading to a better understanding of cytoplasmic filament function. Further insights could be obtained by specific inactivation of cfpA. Gene inactivation in T. denticola is feasible (10, 24, 25, 27) and enables phenotypic study of engineered mutants. Further detailed structural analysis together with the study of a knockout mutation of the cytoplasmic filament gene will provide critical information on the function and regulation of this unique structure. The location of the filaments and their helical shape suggest involvement in cell motility, maintenance of cell structure, or cell division (9); each of these proposed functions is a good potential target for the development of new antimicrobial drugs.
ACKNOWLEDGMENTS
We acknowledge the Wadsworth Center Molecular Genetics and Electron Microscopy Core Facilities and the Wadsworth Center Photography Unit for technical assistance. We thank Konrad Wicher for providing T. pallidum subsp. pertenue cells and Nyles Charon for providing flagellar filament-deficient mutant T55 of T. phagedenis Kazan 5.
J.I. was supported in part by a basic research grant from Health Research Incorporated. This work was supported by Public Health Research Service grant AI34354 from the National Institutes of Health.
REFERENCES
- 1.Akopyanz N, Bukanov N O, Westblom T U, Kresovich S, Berg D E. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 1992;20:5137–5142. doi: 10.1093/nar/20.19.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angulo J J, Watson J J, Wedderburn C C, Leon-Blanco F, Varella G. Electronmicrography of Treponemas from cases of yaws, pinta, and so-called Cuban form of pinta. Am J Trop Med. 1951;31:458–478. doi: 10.4269/ajtmh.1951.s1-31.458. [DOI] [PubMed] [Google Scholar]
- 4.Atlas R M. Handbook of microbiological media. 2nd ed. New York, N.Y: CRC Press, Inc.; 1997. [Google Scholar]
- 5.Baughn R E, Musher D M. Evidence that autologous idiotypic regulation of anti-arginine-glycine-aspartic acid autoantibodies may influence development and progression of syphilitic lesions in infected rabbits. Infect Immun. 1992;60:3861–3871. doi: 10.1128/iai.60.9.3861-3871.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centurion-Lara A, Arroll T, Castillo R, Shaffer J M, Castro C, Van Voorhis W C, Lukehart S A. Conservation of the 15-kilodalton lipoprotein among Treponema pallidum subspecies and strains and other pathogenic treponemes: genetic and antigenic analyses. Infect Immun. 1997;65:1440–1444. doi: 10.1128/iai.65.4.1440-1444.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charon N C, Goldstein S F, Curci K, Limberger R J. The bent-end morphology of Treponema phagedenis is associated with short, left-handed, periplasmic flagella. J Bacteriol. 1991;173:4820–4826. doi: 10.1128/jb.173.15.4820-4826.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi B K, Paster B J, Dewhirst F E, Gobel U B. Diversity of cultivable and uncultivable oral spirochetes from a patient with severe destructive periodontitis. Infect Immun. 1994;62:1889–1895. doi: 10.1128/iai.62.5.1889-1895.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eipert S R, Black S H. Characterization of the cytoplasmic fibrils of Treponema refringens (Nichols) Arch Microbiol. 1979;120:205–214. doi: 10.1007/BF00423067. [DOI] [PubMed] [Google Scholar]
- 10.Fenno J C, Wong G W, Hannam P M, McBride B C. Mutagenesis of outer membrane virulence determinants of the oral spirochete Treponema denticola. FEMS Microbiol Lett. 1998;163:209–215. doi: 10.1111/j.1574-6968.1998.tb13047.x. [DOI] [PubMed] [Google Scholar]
- 11.Fraser C M, Norris S J, Weinstock G M, White O, Sutton G G, Dodson R, Gwinn M, Hickey E K, Clayton R, Ketchum K A, Sodergren E, Hardham J M, McLeod M P, Salzberg S, Peterson J, Khalak H, Richardson D, Howell J K, Chidambaram M, Utterback T, McDonald L, Artiach P, Bowman C, Cotton M D, Fujii C, Garland S, Hatch B, Horst K, Roberts K, Sandusky M, Weidman J, Smith H O, Venter J C. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 12.Hawley D K, McClure W R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holt S C. Anatomy and chemistry of spirochetes. Microbiol Rev. 1978;42:114–160. doi: 10.1128/mr.42.1.114-160.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hovind-Hougen K. Determination by means of electron microscopy of morphological criteria of value for classification of some spirochetes, in particular treponemes. Acta Pathol Microbiol Scand Sect B. 1976;255(Suppl. 255):1–41. [PubMed] [Google Scholar]
- 15.Hovind-Hougen K. Further observations on the ultrastructure of Treponema pallidum Nichols. Acta Pathol Microbiol Scand Sect B. 1972;80:297–304. doi: 10.1111/j.1699-0463.1972.tb00163.x. [DOI] [PubMed] [Google Scholar]
- 16.Hovind-Hougen K. The ultrastructure of cultivable treponemes. I. Treponema phagedenis, Treponema vincentii, and Treponema refringens. Acta Pathol Microbiol Scand Sect B. 1974;82:329–344. [PubMed] [Google Scholar]
- 17.Hovind-Hougen K, Birch-Andersen A, Jensen H J. Ultrastructure of cells of Treponema pertenue obtained from experimentally infected hamsters. Acta Pathol Microbiol Scand Sect B. 1976;84:101–108. doi: 10.1111/j.1699-0463.1976.tb01909.x. [DOI] [PubMed] [Google Scholar]
- 18.Hovind-Hougen K, Birch-Andersen A. Electron microscopy of the endoflagella and microtubules in Treponema Reiter. Acta Pathol Microbiol Scand Sect B. 1971;79:37–50. doi: 10.1111/j.1699-0463.1971.tb00031.x. [DOI] [PubMed] [Google Scholar]
- 19.Hughes R, Olander H J, Williams C B. Swine dysentery: pathogenicity of Treponema hyodysenteriae. Am J Vet Res. 1975;36:971–977. [PubMed] [Google Scholar]
- 20.Isaacs R D, Hanke J H, Guzman-Verduzco L M, Newport G, Agabian N, Norgard M V, Lukehart S A, Radolf J D. Molecular cloning and DNA sequence analysis of the 37-kilodalton endoflagellar sheath protein gene of Treponema pallidum. Infect Immun. 1989;56:3403–3411. doi: 10.1128/iai.57.11.3403-3411.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isaacs R D, Radolf J D. Expression in Escherichia coli of the 37-kilodalton endoflagellar sheath protein of Treponema pallidum by use of the polymerase chain reaction and a T7 expression system. Infect Immun. 1990;58:2025–2034. doi: 10.1128/iai.58.7.2025-2034.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Izard, J. Unpublished data.
- 23.Izard, J., and W. A. Samsonoff. Unpublished data.
- 24.Li H, Arakawa S, Deng Q D, Kuramitsu H. Characterization of a novel methyl-accepting chemotaxis gene, dmcB, from the oral spirochete Treponema denticola. Infect Immun. 1999;67:694–699. doi: 10.1128/iai.67.2.694-699.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Ruby J, Charon N, Kuramitsu H. Gene inactivation in the oral spirochete Treponema denticola: construction of an flgE mutant. J Bacteriol. 1996;178:3664–3667. doi: 10.1128/jb.178.12.3664-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Limberger R J, Charon N W. Treponema phagedenis has at least two proteins residing together on its periplasmic flagella. J Bacteriol. 1986;166:105–112. doi: 10.1128/jb.166.1.105-112.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Limberger R J, Slivienski L L, Izard J, Samsonoff W A. Insertional inactivation of Treponema denticola tap1 results in a nonmotile mutant with elongated flagellar hooks. J Bacteriol. 1999;181:3743–3750. doi: 10.1128/jb.181.12.3743-3750.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Limberger R J, Slivienski L L, Samsonoff W A. Genetic and biochemical analysis of the flagellar hook of Treponema phagedenis. J Bacteriol. 1994;176:3631–3637. doi: 10.1128/jb.176.12.3631-3637.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Listgarten M A, Socransky S S. Electron microscopy of axial fibrils, outer envelope, and cell division of certain oral spirochetes. J Bacteriol. 1964;88:1087–1103. doi: 10.1128/jb.88.4.1087-1103.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 31.Masuda K, Kawata T. Isolation and characterization of cytoplasmic fibrils from treponemes. Microbiol Immunol. 1989;33:619–630. doi: 10.1111/j.1348-0421.1989.tb02012.x. [DOI] [PubMed] [Google Scholar]
- 32.Nauman R K, Holt S C, Cox C D. Purification, ultrastructure, and composition of axial filaments from Leptospira. J Bacteriol. 1969;98:264–280. doi: 10.1128/jb.98.1.264-280.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nichols H J, Hough W H. Demonstration of Spirochaeta pallida in the cerebrospinal fluid. JAMA. 1913;60:108–110. [Google Scholar]
- 34.Noordhoek G T, Hermans P W, Paul A N, Schouls L M, van der Sluis J J, van Embden J D. Treponema pallidum subspecies pallidum (Nichols) and Treponema pallidum subspecies pertenue (CDC 2575) differ in at least one nucleotide: comparison of two homologous antigens. Microb Pathog. 1989;6:29–42. doi: 10.1016/0882-4010(89)90005-3. [DOI] [PubMed] [Google Scholar]
- 35.Norris S J. Polypeptides of Treponema pallidum: progress toward understanding their structural, functional, and immunologic roles. Microbiol Rev. 1993;57:750–779. doi: 10.1128/mr.57.3.750-779.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park B C, Kim J S, Lee D S, Byun S M. One-strand PCR: a comparably efficient method for chromosomal walking. BioTechniques. 1996;21:592–594. [PubMed] [Google Scholar]
- 37.Pearson W R. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 38.Ritchie A E, Ellinghausen H C. Electron microscopy of leptospires. I. Anatomical features of Leptospira pomona. J Bacteriol. 1965;89:223–233. doi: 10.1128/jb.89.1.223-233.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ryter A, Pillot J. Etude au microscope electronique de la structure interne du Treponeme reiter. Ann Inst Pasteur. 1963;104:496–501. [PubMed] [Google Scholar]
- 40.Schleicher & Schuell, Inc. Transfer and immobilization of nucleic acids to S&S solid supports. Keene, N.H: Schleicher & Schuell, Inc.; 1987. [Google Scholar]
- 41.Simonson L G, Goodman C H, Bial J J, Morton H E. Quantitative relationship of Treponema denticola to severity of periodontal disease. Infect Immun. 1988;56:726–728. doi: 10.1128/iai.56.4.726-728.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smibert R M. Genus III. Treponema Schaudin 1905, 1728AL. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. pp. 49–57. [Google Scholar]
- 43.You Y, Elmore S, Colton L L, Mackenzie C, Stoops J K, Weinstock G M, Norris S J. Characterization of the cytoplasmic filament protein gene (cfpA) of Treponema pallidum subsp. pallidum. J Bacteriol. 1996;178:3177–3187. doi: 10.1128/jb.178.11.3177-3187.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zemper E D, Black S H. Morphology of freeze-etched Treponema refringens (Nichols) Arch Microbiol. 1978;117:227–238. doi: 10.1007/BF00738540. [DOI] [PubMed] [Google Scholar]