Abstract
Escherichia coli strains that are deficient in the Ada and Ogt DNA repair methyltransferases display an elevated spontaneous G:C-to-A:T transition mutation rate, and this increase has been attributed to mutagenic O6-alkylguanine lesions being formed via the alkylation of DNA by endogenous metabolites. Here we test the frequently cited hypothesis that S-adenosylmethionine (SAM) can act as a weak alkylating agent in vivo and that it contributes to endogenous DNA alkylation. By regulating the expression of the rat liver SAM synthetase and the bacteriophage T3 SAM hydrolase proteins in E. coli, a 100-fold range of SAM levels could be achieved. However, neither increasing nor decreasing SAM levels significantly affected spontaneous mutation rates, leading us to conclude that SAM is not a major contributor to the endogenous formation of O6-methylguanine lesions in E. coli.
S-Adenosylmethionine (SAM) is a ubiquitous intracellular methyl donor. As a methyl donor, SAM normally donates its methyl group in enzyme-catalyzed reactions, such as Dam-catalyzed methylation of Escherichia coli DNA for strand discrimination during mismatch repair. However, in vitro studies in which 3H-labeled SAM was incubated with DNA suggest that SAM can also act as a chemical alkylating agent, spontaneously transferring its methyl group to DNA via non-enzyme-catalyzed reactions (2, 29, 37). Chemical alkylating agents can produce a number of deleterious lesions in DNA, including the mutagenic O6-methylguanine (O6MeG) lesion and the lethal, replication-blocking lesion 3-methyladenine (6). E. coli strains deficient in the DNA methyltransferases that repair O6MeG lesions (namely, Ada and Ogt) are extremely sensitive to mutation induction by alkylating agents (6, 36). ada ogt cells also display an elevated rate of spontaneous mutation, particularly at G:C base pairs (20, 36, 45, 47), suggesting that E. coli is chronically exposed to an endogenous source of alkylating agents that induce O6-alkylguanine DNA lesions (20, 36, 45). Endogenously formed N-nitroso compounds were recently implicated as one source of mutagenic alkylation damage in ada ogt E. coli (40, 45). However, SAM has also been repeatedly cited as a potential endogenous alkylating agent, because of its in vitro alkylating activity and its ubiquitous presence in cells (18, 24, 37). Our strategy for testing the hypothesis that SAM behaves as an endogenous mutagenic alkylating agent was to determine whether changing SAM levels has any influence on spontaneous mutation rates in E. coli.
SAM is synthesized from methionine and ATP by SAM synthetase, encoded in E. coli by the metK gene (38). Because SAM acts as a strong feedback inhibitor of MetK (1, 31), as well as a transcriptional corepressor of the metK gene and other genes in the pathway of SAM and methionine biosynthesis (38), increasing SAM levels by modulating MetK activity is problematic. Further, SAM levels cannot be increased by simply adding SAM to growing cells, because E. coli does not take up SAM from medium and standard techniques for permeabilizing E. coli to SAM cannot be used with living cells (32). Decreasing SAM levels in E. coli has also been problematic; studies of E. coli and Salmonella typhimurium metK mutants suggest that they tend to retain relatively high SAM levels despite marked deficiencies in SAM synthetase activity (11, 13, 14, 39). Although certain metK temperature-sensitive mutants were reported to have a decrease in SAM levels of 200-fold or more, these mutants later proved to be genetically unstable (23, 39).
In this study, we used the regulated expression of two foreign enzymes to evade E. coli’s stringent regulation of SAM levels, namely, expression of rat liver SAM synthetase (RLSS) and bacteriophage T3 SAM hydrolase (T3SH). RLSS is a highly conserved homolog of the bacterial MetK SAM synthetase, except that RLSS appears to be less susceptible to feedback inhibition than MetK (25, 31). Expression of the RLSS cDNA in E. coli (for protein structure studies) was found to increase intracellular SAM levels more than 100-fold (1). The T3SH protein depletes intracellular SAM levels by catalyzing the hydrolysis of SAM into methylthioadenosine and homoserine. The expression of SAM hydrolase in E. coli, either during T3 infection (7) or from a cloned gene (4, 15), has been shown to inhibit SAM-dependent processes, such as RNA and DNA methylation (4, 7, 15). Thus, expression of the T3SH gene has been exploited to lower SAM pools in E. coli (4, 15, 46) and plants (10).
Here, we test the popular hypothesis that SAM is an endogenous DNA-alkylating agent that can contribute to the mutator phenotype seen in O6MeG repair-deficient cells, by monitoring spontaneous mutation in E. coli cultures with very large differences in intracellular SAM levels. Our results suggest that SAM is not an important contributor to the formation of endogenous mutagenic O6MeG lesions.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The genotypes of the E. coli strains used in these experiments are listed in Table 1. FC215 contains an episomal lacZ allele that reverts to lacZ+ only by G:C-to-A:T transition mutations, allowing use of this strain to monitor G:C-to-A:T base substitutions (5, 20, 26). FC218 is a DNA repair methyltransferase (ada ogt)-deficient derivative of FC215 (20). To monitor mutations to rifampin resistance, we used the rifampin-sensitive strain CSH106, which is virtually isogenic to FC215, as FC215 was derived by introducing the lacZ episome into a rifampin-resistant isolate of the parental strain of CSH106 (20). CSH106 also contains a revertible episomal lacZ allele (Table 1) (5, 26), but the lacZ marker was not monitored in this strain. The dam-13::Tn9 allele was moved from strain GM7365 into FC215 and CSH106 by standard P1 transduction techniques (26). Putative dam-13::Tn9 transductants were screened for sensitivity to 2-aminopurine.
TABLE 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| FC215 | ara Δ(gpt-lac)5 Rifr F′ lac proA+B+; from CSH102 G:C to A:T | 20 |
| FC218 | ogt-1::Kanr Δada-25::Camr FC215 | 20 |
| E7014 | ΔlacIZ | 20 |
| CSH106 | ara Δ(gpt-lac)5 F′ lac proA+B+; A:T to G:C | 26 |
| GM7365 | dam-13::Tn9 | M. Marinus (21) |
| Plasmids | ||
| pBAD24 | Ampr, arabinose inducible | J. Beckwith (12) |
| pBAD-RLSS | RLSS cDNA in pBAD24 | This work; L. Alvarez (1) |
| pBAD-T3SH | T3SH gene in pBAD24 | This work; R. Bestwick (10) |
The T3SH coding sequence was obtained on plasmid pAG-930 from R. Bestwick at Agritope, Inc., Beaverton, Oreg. This clone contains an A-to-G substitution at the fourth base of the coding sequence that converts an isoleucine into a valine, creating an NcoI site at the initial ATG codon (10). The T3 gene was subcloned from pAG-930 as an NcoI/PstI fragment into pBAD24 (12) (kindly provided by L.-M. Guzman and J. Beckwith, Harvard Medical School, Boston, Mass.) to create the plasmid pBAD-T3SH. The RLSS cDNA (1) was obtained from L. Alvarez (Instituto de Investigaciones Biomedicas, Consejo Superior de Investigaciones Científìcas, Madrid, Spain) in the plasmid pSSRL-4a, amplified by PCR with an upstream primer that starts at the second RLSS codon (AAT) and a downstream primer that incorporates a KpnI site three bases after the RLSS stop codon, and subcloned into NcoI-Klenow fragment-KpnI-treated pBAD24 (12) to create the plasmid pBAD-RLSS. The coding sequences in pBAD-T3SH and pBAD-RLSS are under the control of the arabinose-inducible PBAD promoter.
Preparation of cell extracts.
Cell extracts for high-performance liquid chromatography (HPLC) and genomic DNA for restriction digestion were prepared from cultures of FC218 grown under conditions comparable to those used for Lac+ mutation assays (see below). Overnight cultures were diluted 106-fold into Luria-Bertani (LB) medium containing ampicillin (100 μg/ml) and 0.2% arabinose and grown for 18 h at 37°C. In some experiments, samples were removed after 8 h, when the culture density was approximately 2 × 108 CFU/ml, to measure SAM levels during exponential growth. Both the exponential samples and the 18-h cultures had their titers determined on LB-ampicillin plates to determine CFU per milliliter and were harvested by centrifugation (15 min at 5,000 × g) at 4°C. Pelleted cultures were resuspended on ice in chilled minimal A salts (26), and aliquots were microcentrifuged; the resulting pellets were frozen in liquid nitrogen and stored at −80°C until they were used for preparing HPLC extracts or genomic DNA.
Preparation of HPLC extracts.
SAM levels were determined by HPLC. To prepare extracts for HPLC, frozen pellets were resuspended in chilled 0.01 N hydrochloric acid (HCl) and sonicated on ice. After centrifugation, supernatants were deproteinized by filtering them through Centricon 10 ultrafiltration columns (Amicon) at 4°C.
HPLC analysis.
SAM levels were measured by reversed-phase ion-pair HPLC by a modification of previous isocratic elution protocols (50). The mobile phase consisted of 87% 5 mM heptanesulfonic acid (Fisher), the pH being adjusted to 3.2 with HCl, and 13% acetonitrile (Fisher). The flow rate was 0.8 ml/min through a Waters Nova-Pak C18 column (3.9 by 300 mm), and absorbance was monitored at 254 nm. The number of picomoles of SAM in the ultrafiltered samples was determined by comparison with a standard solution of 10 μM SAM (p-toluenesulfonate salt; Sigma) in 0.01 N HCl, after correcting for impurities observed during HPLC runs. All runs, including standard runs, were done two or more times. SAM levels were expressed as picomoles per 109 CFU and compared by a two-tailed t test (Student or Welch) of the difference between means. Micromolar SAM values were calculated by using the number of CFU and an intracellular water volume of 0.924 × 10−15 liters (28). The ultrafiltration process did not affect the concentration of SAM in a standard solution as determined by HPLC.
DNA digestion and analysis.
Genomic DNA was isolated from frozen stationary-phase cell pellets. DNA concentration was quantified and gel images were captured by using the Bio-Rad Gel Doc 1000 with Molecular Analyst software. Digestions were for 30 min (DpnI) or 15 min (DpnII and undigested). DpnI was used at 1 U/μg of DNA, and DpnII was used at 4 U/μg of DNA.
Cell morphology and growth curves.
Microscopic images were taken from stationary-phase cultures used to prepare HPLC extracts (see above). The microscope system consisted of a Nikon Eclipse E800 microscope (Nikon, Tokyo, Japan), an Optronics Engineering DEI-750 camera (Optronics Engineering, Goleta, Calif.), and Scion Image 1.60 software (Scion, Frederick, Md.). All images were taken at an ×600 magnification with an oil-immersion objective. For growth curves, overnight cultures of FC215 and FC218 pBAD-T3SH were diluted 105-fold into LB-ampicillin with or without 0.2% arabinose, and their titers were determined on LB-ampicillin plates at various times after inoculation.
Lac+ reversion assays.
Overnight cultures were diluted 106-fold into LB-ampicillin containing 0.2% arabinose to induce expression from the PBAD promoter. Twenty-five 1- or 1.2-ml cultures were dispensed and grown with aeration for approximately 18 h at 37°C. To determine CFU per milliliter, 7 of the 25 cultures had their titers determined on glucose-M9 (26)-ampicillin plates. To determine the number of Lac+ revertants, cultures were plated on lactose-M9-ampicillin plates, and Lac+ colonies were counted after 2 days at 37°C. For wild-type strains, the entire culture was concentrated, resuspended in M9 salts, and plated. For the ada ogt strains, which have a higher mutation rate, only 100 μl of culture was plated. To prevent growth on lactose at the lower plating density, the ada ogt cells were plated with 50 μl of concentrated E7014 “scavenger” cells (20). Scavenger cells were prepared by growing 100-ml LB-ampicillin cultures of E7014 to stationary phase, washing the cells in M9 salts, and resuspending them in 5 ml of M9 salts to form a concentrated cell suspension.
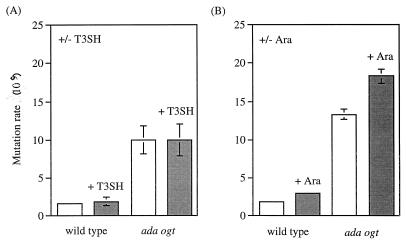
The effect of SAM hydrolase expression on the G:C-to-A:T mutation rate was examined in two ways. In three experiments (see Fig. 3B), wild-type and ada ogt cells containing the SAM hydrolase plasmid (pBAD-T3SH) were grown in the presence or absence of the inducer arabinose. Cultures were prepared and plated essentially as described above except that overnight cultures were diluted 105-fold into LB-ampicillin plus or minus 0.2% arabinose. In two additional experiments (Fig. 3A), strains containing pBAD-T3SH or the control plasmid pBAD24 were compared after growth in 0.2% arabinose. For these experiments, the entire culture was plated without scavenger cells.
FIG. 3.
The effect of decreased SAM levels on the G:C-to-A:T mutation rate in wild-type and ada ogt E. coli. The mutation rate was determined in the lacZ allele as described in Materials and Methods. (A) Strains containing T3SH were compared to strains containing a control plasmid under inducing conditions. The mutation rates for the ada ogt control strain (ada ogt plus pBAD24) and the ada ogt strain expressing SAM hydrolase (ada ogt plus pBAD-T3SH) were not significantly different by a two-sided t test of the difference between means (P > 0.9). (B) Strains containing T3SH were compared under inducing and noninducing conditions. The mutation rates for induced and noninduced ada ogt-plus-pBAD-T3SH cells were not significantly different by a two-sided t test of the difference between means (P > 0.5).
Mutation rates were calculated according to the method of Lea and Coulson (17), and mutation rates were compared by a two-tailed t test of the difference between means.
Antibiotic resistance mutation assays.
Overnight cultures of wild-type or dam FC215 or CSH106 cells containing a control plasmid or the SAM hydrolase plasmid were diluted 106-fold into LB-ampicillin plus 0.2% arabinose, and 15 1-ml cultures were dispensed and grown for 14 to 18 h at 37°C. The cultures were concentrated and plated on LB-nalidixic acid (40 μg/ml)-ampicillin plates (strain FC215) or LB-rifampin (100 μg/ml)-ampicillin plates (strain CSH106). Prior to concentration, aliquots from 4 of the 15 cultures had their titers determined on LB-ampicillin plates to determine CFU per milliliter. Nalidixic acid-resistant (Nalr) or rifampin-resistant (Rifr) colonies were counted 48 h after plating. Mutant frequencies were calculated by dividing the median number of mutants from the 15 cultures by the mean number of CFU plated.
RESULTS
Construction of plasmids and strains for modulating SAM levels.
We set out to modulate SAM levels in E. coli in order to test the hypothesis that SAM behaves as an endogenous alkylating agent that can produce mutagenic O6MeG DNA lesions. To modulate SAM levels, we cloned the coding regions of the RLSS and bacteriophage T3SH genes under the control of the arabinose promoter in the vector pBAD24. The resulting constructs, pBAD-RLSS and pBAD-T3SH, were transformed into a wild-type E. coli strain used for monitoring G:C-to-A:T transition mutations in an episomal lacZ gene (FC215) and into its ada ogt DNA repair methyltransferase-deficient derivative (FC218 [Table 1]).
Modulation of SAM levels.
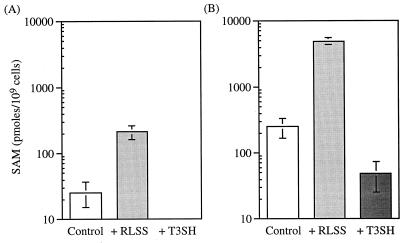
To determine whether SAM levels changed upon induction of the SAM synthetase and SAM hydrolase constructs, we used HPLC to measure intracellular SAM pools in ada ogt E. coli grown in the presence of 0.2% arabinose. Although SAM levels were higher in stationary-phase cells than in exponential-phase cells (compare control bars in Fig. 1A and 1B), we found that SAM levels were consistently elevated in cells expressing SAM synthetase and decreased in cells expressing T3SH, regardless of growth phase. Thus, SAM levels were increased more than eightfold in exponentially growing cells expressing RLSS (Fig. 1A) and 20-fold in stationary-phase cells harvested 10 h later (Fig. 1B). Likewise, T3SH expression decreased the SAM pool in exponentially growing cells (Fig. 1B) to undetectable levels (at least a sevenfold reduction under the conditions used) and decreased SAM levels in stationary-phase cells more than fivefold (Fig. 1B). We therefore achieved an overall range of SAM levels of at least 56-fold in exponentially growing cells and 100-fold in stationary-phase cells. Assuming an intracellular water volume of 0.924 × 10−15 liters for exponential cells (28), our SAM values for exponential control and exponential RLSS cells are the equivalent of 28 and 228 μM, respectively. By comparison, reported SAM values for E. coli or S. typhimurium in different medium and growth conditions range from 1 μM (41) to 100 μM (3) to as high as 10 mM (again assuming a cell volume of 10−15 liters) (39).
FIG. 1.
SAM synthetase (RLSS) and SAM hydrolase (T3SH) constructs modulate SAM levels in E. coli. (A) SAM levels in exponentially growing cells (n = 3). SAM levels in T3SH-containing cells were below the level of detection. (B) SAM levels in stationary-phase cells (n = 5 for control and RLSS, and n = 4 for T3SH). SAM levels in RLSS cells are significantly different from those in control cells by a t test (for exponential cells, P < 0.01, and for stationary cells, P < 0.001). SAM levels in stationary T3SH cells are not significantly different from those in exponential cells (P < 0.1). Error bars show standard errors of the means.
The effect of increased SAM levels on spontaneous G:C-to-A:T mutations.
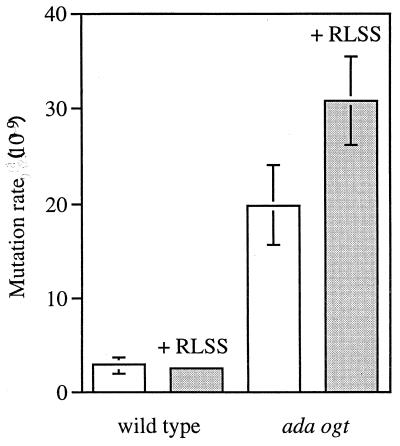
If SAM is an endogenous alkylating agent that contributes to G:C-to-A:T mutations in ada ogt E. coli, increasing intracellular SAM levels would be expected to elevate the G:C-to-A:T mutation rate. We therefore measured spontaneous G:C-to-A:T mutation rates in ada ogt and wild-type strains expressing the SAM synthetase vector or a control vector. Mutation rates were determined by a Lac+ reversion assay, with a lacZ strain that reverts to lacZ+ solely by G:C-to-A:T transitions (5, 20). MacKay et al. (20) reported that ada ogt E. coli had an elevated rate of spontaneous G:C-to-A:T mutations. We likewise observed a roughly sevenfold increase in G:C-to-A:T mutations in ada ogt cells over those in wild-type cells (see Fig. 2). When SAM levels were increased by inducing SAM synthetase, there was no increase in mutation rate in the wild-type strain (Fig. 2). However, there was a slight increase (1.6-fold) in mutation rate in the ada ogt cells upon SAM overproduction. Although this slight increase was observed consistently, it did not reach statistical significance (P > 0.1). We infer that SAM is not a strong endogenous mutagen in E. coli.
FIG. 2.
The effect of increased SAM levels on the G:C-to-A:T mutation rate in wild-type and ada ogt E. coli. The mutation rate was determined in the lacZ allele as described in Materials and Methods. In agreement with previous results, the mutation rate for the ada ogt control strain (ada ogt plus pBAD24) is significantly higher than the mutation rate for the wild-type control strain (wild type plus pBAD24) by a two-sided t test of the difference between means (P < 0.02). However, the mutation rate for the ada ogt strain expressing SAM synthetase (ada ogt plus pBAD-RLSS) is not significantly different (P > 0.1) from the mutation rate for the ada ogt control strain (ada ogt plus pBAD24).
The effect of decreased SAM levels on spontaneous G:C-to-A:T mutations.
To further test the hypothesis that SAM contributes to the normal burden of spontaneous G:C-to-A:T mutations in ada ogt E. coli, we compared mutation rates in cells expressing or not expressing the SAM hydrolase protein. We tested the influence of SAM hydrolase in two ways: (i) in cells grown under arabinose-inducing conditions with the SAM hydrolase vector or the same vector without the T3SH gene and (ii) in cells containing the SAM hydrolase vector, grown under inducing or noninducing conditions. Figure 3 shows that, in both types of experiment, induction of SAM hydrolase did not significantly affect ada ogt-dependent G:C-to-A:T mutations. Thus, normal SAM levels do not appear to be an important contributor to mutagenic alkylation damage in ada ogt E. coli.
SAM hydrolase induction decreases DNA methylation by Dam methyltransferase.
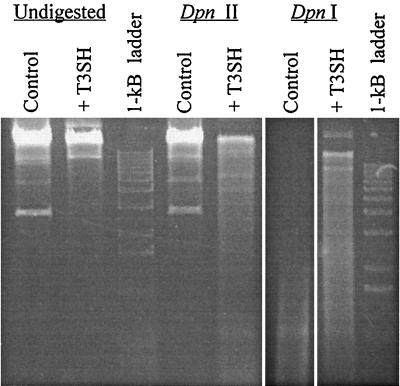
Unlike bacterial DNA, newly replicated T3 DNA is completely unmethylated by E. coli methylases, in part because the viral SAM hydrolase depletes cellular SAM pools during the infection process (7). Similarly, expression of the cloned SAM hydrolase gene has been shown to decrease Dam and Dcm methylation of M13 DNA (15) and M-BamHII methylation of plasmid DNA (4). We tested whether induction of our T3SH construct decreased Dam methylation of chromosomal DNA by digestion with restriction enzymes that cut at GATC (dam) sequences in a methylation-dependent fashion (4, 15, 49). Figure 4 shows DpnI and DpnII digestion of genomic DNA isolated from ada ogt E. coli cells that were grown to stationary phase in the presence of 0.2% arabinose. DpnII, which cuts only at unmethylated GATC sites, was able to partially digest DNA from SAM hydrolase-expressing cultures but had no apparent activity on DNA isolated from cells containing the control vector. This result implies that the genomes of cells grown with a fraction of the normal SAM pool contain a significant number of fully unmethylated GATC sites. Digestion with DpnI, which cuts rapidly at fully methylated GATC sites and approximately 60 times more slowly at hemimethylated GATC sites (49), cleaved DNA from control cells more completely than DNA from cells expressing SAM hydrolase. We thus infer that a reduction in SAM pool size results in increased hemimethylated or unmethylated GATC sites in the E. coli genome. Together, these results demonstrate that expression of the cloned SAM hydrolase interferes with Dam methylation of chromosomal DNA. SAM hydrolase expression also decreased Dam methylation of plasmid DNA (data not shown).
FIG. 4.
SAM hydrolase (T3SH) induction decreases methylation of DNA by the Dam methyltransferase. The enzyme DpnII cleaves only unmethylated DNA; note the increased cleavage of DNA from SAM hydrolase-induced cells relative to that of DNA from cells containing the control vector. The enzyme DpnI cleaves methylated DNA rapidly and hemimethylated DNA very slowly and does not cleave unmethylated DNA. Note the decreased cleavage by DpnI of DNA from SAM hydrolase-induced cells relative to that of DNA from control cells.
Does lowering SAM pools induce a dam mutator phenotype?
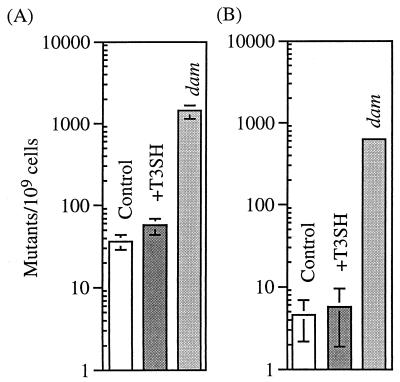
E. coli dam mutants have a strong mutator phenotype (9, 21), suggesting that cells with a SAM hydrolase-induced reduction in Dam methylation (Fig. 4) might also be mutators. To investigate whether depletion of SAM pools induces a dam mutator phenotype, we looked at the effect of SAM hydrolase expression on two mutational markers that have been shown to increase in dam strains, Nalr and Rifr (9, 21). Figure 5 shows that, while a dam mutation strongly increased the Nalr and Rifr mutation frequency in two wild-type strains, induction of the SAM hydrolase construct had no effect or only a slight effect on mutation frequency. Thus, the extent of the decrease in SAM pools and Dam methylation seen in this study was not sufficient to provoke a dam mutator phenotype.
FIG. 5.
Depletion of SAM pools by SAM hydrolase (T3SH) does not increase mutation at alleles that show an increase in mutation frequency in dam mutator cells. Rifr (A) was assayed for CSH106, and Nalr (B) was assayed for FC215, the wild-type strain used for Lac+ reversion assays. Mutation frequency is the median number of mutant colonies from 15 cultures divided by the mean number of cells plated (for the control strains, n = 5; for the T3SH strains, n = 3; for the dam strains, n = 2).
The influence of SAM levels on cell growth and morphology.
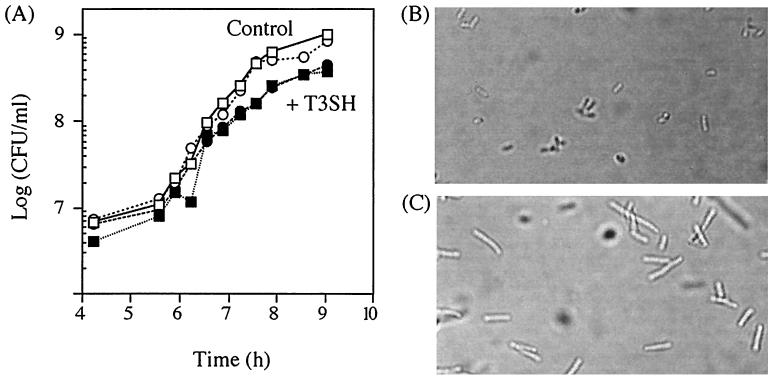
Induction of SAM hydrolase had only a moderate effect on cell growth and appearance under the conditions used in this study. When wild-type (FC215) or ada ogt (FC218) cells containing the SAM hydrolase vector were grown from about 104 CFU/ml to saturation in the presence or absence of inducer, induced and control cells grew at a similar rate until early exponential phase. Cells expressing SAM hydrolase subsequently were saturated at a cell density (CFU per milliliter) that was two- to threefold lower than that of the control cells (Fig. 6A). Cells expressing SAM hydrolase were also more elongated at saturation than control cells (Fig. 6B and C). When we used a construct in which the SAM hydrolase gene was under the control of the strong trc promoter or when we grew cells in minimal medium, we observed more extensive filamentation. These observations are consistent with previous observations that E. coli cells infected with M13 bacteriophage expressing the cloned SAM hydrolase gene tend to elongate and occasionally undergo filamentation during growth (15) and that SAM synthetase-deficient metK84 mutants undergo filamentation under certain growth conditions (30).
FIG. 6.
The effects of SAM hydrolase (T3SH) induction on cell growth and morphology. (A) SAM hydrolase induction decreases growth rate and final cell number at saturation in both wild-type FC215 cells (■, with T3SH; □, control) and ada ogt FC218 cells (●, with T3SH; ○, control). (B and C) SAM hydrolase-induced cells (C) are more elongated than control cells (B) after growth to saturation in LB-ampicillin. Both images are at an ×600 magnification.
Induction of SAM synthetase had no obvious effects during growth in LB medium. However, SAM synthetase overexpression may cause a mild auxotrophy for methionine, as induced cells (but not control cells) had a lower growth rate in methionine-free minimal medium than in methionine-supplemented medium (data not shown). Given that methionine is the precursor to SAM and that SAM is a corepressor of the methionine biosynthesis genes, it is not surprising that overproduction of SAM may induce a partial requirement for methionine. A more complete methionine auxotrophy was previously reported for Bacillus subtilis cells overexpressing the B. subtilis metE SAM synthetase gene (54).
DISCUSSION
Several lines of evidence point to the existence of endogenous cellular compounds that can alkylate DNA and cause spontaneous, alkylation-induced mutation. First, the alkyl lesions O6MeG and 7MeG have been detected in DNA from humans, mice, and rats that were not patently exposed to alkylating agents (16, 24, 33, 44). The 3MeA free base can also be found in human urine, and some of this 3MeA burden is believed to result from release of endogenously alkylated DNA bases (24, 34). Second, bacteria and yeast that are deficient in the DNA methyltransferases that repair mutagenic O6MeG lesions display elevated spontaneous mutation rates (20, 36, 45, 47, 51–53). In E. coli, the majority of the ada ogt-dependent spontaneous mutations have been shown to be G:C-to-A:T transitions (20, 47), consistent with their arising from replication past endogenous O6MeG residues (20).
In 1996, Taverna and Sedgwick (45) reported that ada ogt E. coli defective in the moa gene, which encodes a cofactor involved in nitrosation reactions, had a reduction in the accumulation of G:C-to-A:T Lac+ mutants in stationary phase. The moa effects strongly implicate the nitrosation of amines and amides in the production of endogenous mutagenic alkylating agents. However, the fact that the moa deficiency does not completely abolish ada ogt-dependent spontaneous mutation suggests that there are additional sources of mutagenic alkylating agents in E. coli besides moa-catalyzed nitrosation products.
One compound that has been repeatedly cited as a potential endogenous alkylating agent is the natural cellular compound and universal methyl donor SAM (18, 20, 24, 33, 37, 52). In fact, SAM has been shown to spontaneously alkylate DNA in vitro (2, 29, 37, 42). To test the hypothesis that SAM acts as an endogenous alkylating agent in vivo, we modulated SAM pools in E. coli by expressing the RLSS and T3SH genes. We found that expression of these genes produced an up-to-100-fold overall change in intracellular SAM levels. Furthermore, we found that the molar concentration of SAM during exponential growth was 28 μM in control cells and 228 μM in cells expressing SAM synthetase. By comparison, incubation of DNA in vitro with 7 to 12 μM 3H-labeled SAM (2, 37) produced detectable levels of 7MeG, and possibly O6MeG (2), in DNA. Thus, the basal and, in particular, the elevated SAM levels that we observed are theoretically high enough to alkylate DNA in vivo.
When we expressed the SAM synthetase gene in ada ogt E. coli, we found that SAM overproduction increased the G:C-to-A:T mutation rate by a modest 1.6-fold. This difference does not achieve statistical significance but was nevertheless consistent. Increasing SAM pool size in the wild-type (ada+ ogt+) background had no effect on mutation rate. The consistency of the SAM synthetase-induced increase in mutation rate and its specificity to the ada ogt strain suggest that SAM may be able to produce a modest number of mutagenic O6MeG lesions when present at elevated levels in the cell. Under the growth conditions used for the mutation assays, induction of SAM synthetase elevated SAM levels from 8- to 20-fold. These increases are disproportionately large compared with the modest 1.6-fold increase that SAM synthetase induction caused in the mutation rate. The observation that a substantial increase in SAM levels is needed to provoke a modest increase in mutation argues that SAM is not a major contributor to ada ogt-dependent G:C-to-A:T mutations in E. coli.
The expression of SAM hydrolase decreased SAM levels from fivefold to more than sevenfold but, in agreement with the SAM synthetase experiments, had no effect on spontaneous mutation rates. However, the SAM decrease clearly did have biological consequences, as DNA restriction digestion studies show that Dam-catalyzed DNA methylation was reduced in the presence of SAM hydrolase. In fact, considering both the SAM hydrolase and the SAM synthetase vectors, we were able to examine SAM’s potential mutagenic effects over a 100-fold range in SAM levels. However, spontaneous mutation rates barely changed over this range.
MacKay et al. (20) reported that the ada ogt-dependent increase in G:C-to-A:T mutations occurs both during exponential growth in culture and in nondividing cells during incubation on plates. Others (47) have suggested that methyltransferase deficiency increases mutations only in nondividing cells on plates, not in exponentially growing cells. We found that RLSS and T3SH expression modulated SAM levels both during exponential phase and in stationary phase, immediately prior to plating. We therefore would expect the altered SAM pool to be able to affect both exponential-phase mutations and mutations occurring in nondividing cells on plates, if SAM was, in fact, acting as an endogenous alkylating agent.
Although SAM has been shown to act as an alkylating agent in vitro (2, 29, 37, 42), some have argued that SAM’s alkylation profile may be typical of SN2 alkylating agents that produce primarily nonmutagenic nitrogen lesions and relatively few mutagenic oxygen lesions like O6MeG (29, 37). Such a profile would be consistent with our finding that SAM does not significantly affect O6MeG-driven, ada ogt-dependent spontaneous mutation. The observation that a substantial increase in SAM levels causes only a very modest increase in O6MeG-driven G:C-to-A:T mutations suggests that, if SAM does alkylate DNA in vivo, it may act as an SN2 agent, producing primarily 3MeA and 7MeG (18, 37), rather than producing mutagenic levels of O6MeG.
SAM is the methyl donor for Dam-catalyzed methylation of DNA in E. coli, and we and others have demonstrated that DNA is hypomethylated at dam sites in cells expressing SAM hydrolase (15). The observation that dam cells are partially SOS induced led Hughes et al. (15) to suggest that hypomethylated, hydrolase-expressing cells may also be SOS induced (15), which could itself increase spontaneous mutation. SAM depletion might also be expected to produce a dam-like mutator phenotype by preventing discrimination between old and new strands during methyl-directed mismatch repair. However, we found that the level of SAM hydrolase expression used in these studies did not increase the frequency of two mutational events known to increase in dam mutator or constitutively SOS-induced strains (9, 27). In fact, the effects of SAM depletion on a dam-related mutator phenotype might be hard to predict; if depleted SAM levels delayed rather than prevented Dam methylation, this might have an antimutator effect by extending the time window during which methyl-directed mismatch repair can act (8, 35).
SAM overproduction might also have been predicted to cause a dam-related mutator phenotype. Dam overexpression in E. coli increases spontaneous mutation by allowing more rapid GATC methylation after replication and thereby shortening the time window for methyl-directed mismatch repair (22, 43). If SAM overproduction allowed more rapid Dam methylation, it would also increase spontaneous mutation. Thus, the small increase in G:C-to-A:T mutations that we saw in ada ogt E. coli might be attributable to Dam-induced methylation, rather than O6MeG formation. However, this explanation is unlikely for two reasons. First, the increase in mutation rate was specific to the ada ogt strain; no increase was seen in the ada+ ogt+ background. Second, normal E. coli SAM levels do not appear to be limiting, since increasing Dam methylase levels readily increases methylation (43) and Dam-induced mutation (22).
We found that cells expressing SAM hydrolase were generally more elongated than control cells. This phenotype may relate to DNA hypomethylation (15), as dam cells have also been reported to be elongated in appearance (48). Elongation in dam cells may be linked to partial SOS induction, although such elongation does not require the sfiA gene, whose induction normally leads to filamentation during the SOS response (48). Interestingly, we also saw filamentation when SAM hydrolase was expressed in an sfiA background (data not shown). Alternatively, the appearance of elongated cells in dam cultures (and possibly in SAM hydrolase-expressing cultures) may be a consequence of abnormal replication patterns resulting from lack of methylation at replication origins (19, 48). Another explanation for elongation was recently suggested by Newman et al. (30). They reported that metK84 mutants deficient in SAM synthetase activity undergo extensive filamention under certain growth conditions and suggested that this phenotype results from deficiency in methylation of a cell division protein leading to inhibition of cell division.
In summary, we have successfully modulated SAM pools in E. coli over a wide (100-fold) range by using two non-E. coli genes, RLSS and T3SH, and found that decreases in SAM pools led to hypomethylation at dam sites and changes in cell morphology and growth rates. Our initial goal was to test the popular belief that SAM produces endogenous DNA damage that could contribute to spontaneous mutations. Our data suggest that SAM is not a major contributor to the elevation in spontaneous mutations seen with ada ogt E. coli. Nitrosation reactions triggered by the product of the moa gene appear to account for a significant fraction of spontaneous mutations in stationary-phase ada ogt cells (45). However, the fact that a moa deficiency does not completely abolish the increased spontaneous mutation in ada ogt cells suggests that there are still other sources of endogenous alkylation yet to be uncovered.
ACKNOWLEDGMENTS
We thank L. Alvarez for the SSRL (RLSS) cDNA, R. Bestwick for the SAM hydrolase gene, and L.-M. Guzman and J. Beckwith for pBAD24.
This work was supported by grants to L.D.S. from the National Institute of Environmental Health Sciences (P01-E03926) and the National Cancer Institute (R01-55042). L.D.S. is a Burroughs Wellcome Toxicology Scholar. L.M.P. was supported by a fellowship from the Pharmaceutical Research and Manufacturers of America Foundation and by a Training Program in Environmental Health Sciences grant from the National Institute of Environmental Health Sciences (5 T32 ES07155).
REFERENCES
- 1.Alvarez L, Mingorance J, Pajares M A, Mato J M. Expression of rat liver S-adenosylmethionine synthetase in Escherichia coli results in two active oligomeric forms. Biochem J. 1994;301:557–561. doi: 10.1042/bj3010557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrows L R, Magee P N. Nonenzymatic methylation of DNA by S-adenosylmethionine in vitro. Carcinogenesis. 1982;3:349–351. doi: 10.1093/carcin/3.3.349. [DOI] [PubMed] [Google Scholar]
- 3.Borczuk A, Stock A, Stock J. S-Adenosylmethionine may not be essential for signal transduction during bacterial chemotaxis. J Bacteriol. 1987;169:3295–3300. doi: 10.1128/jb.169.7.3295-3300.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collier G B, Mattson T L, Connaughton J F, Chirikjian J G. A novel Tn10 tetracycline regulon system controlling expression of the bacteriophage T3 gene encoding S-adenosyl-L-methionine hydrolase. Gene. 1994;148:75–80. doi: 10.1016/0378-1119(94)90236-4. [DOI] [PubMed] [Google Scholar]
- 5.Cupples C G, Miller J H. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc Natl Acad Sci USA. 1989;86:5345–5349. doi: 10.1073/pnas.86.14.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C.: American Society for Microbiology; 1995. [Google Scholar]
- 7.Gefter M, Hausmann R, Gold M, Hurwitz J. The enzymatic methylation of ribonucleic acid and deoxyribonucleic acid. X. Bacteriophage T3-induced S-adenosylmethionine cleavage. J Biol Chem. 1966;241:1995–2006. [PubMed] [Google Scholar]
- 8.Geiger J R, Speyer J F. A conditional antimutator in E. coli. Mol Gen Genet. 1977;153:87–97. doi: 10.1007/BF01036000. [DOI] [PubMed] [Google Scholar]
- 9.Glickman B W, Radman M. Escherichia coli mutator mutants deficient in methylation-instructed DNA mismatch correction. Proc Natl Acad Sci USA. 1980;77:1063–1067. doi: 10.1073/pnas.77.2.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Good X, Kellogg J A, Wagoner W, Langhoff D, Matsumura W, Bestwick R K. Reduced ethylene synthesis by transgenic tomatoes expressing S-adenosylmethionine hydrolase. Plant Mol Biol. 1994;26:781–790. doi: 10.1007/BF00028848. [DOI] [PubMed] [Google Scholar]
- 11.Greene R C, Hunter J S, Coch E H. Properties of metK mutants of Escherichia coli K-12. J Bacteriol. 1973;115:57–67. doi: 10.1128/jb.115.1.57-67.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guzman L-M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hafner E W, Tabor C W, Tabor H. Isolation of a metK mutant with a temperature-sensitive S-adenosylmethionine synthetase. J Bacteriol. 1977;132:832–840. doi: 10.1128/jb.132.3.832-840.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hobson A C, Smith D A. S-Adenosylmethionine synthetase in methionine regulatory mutants of Salmonella typhimurium. Mol Gen Genet. 1973;126:7–18. doi: 10.1007/BF00333477. [DOI] [PubMed] [Google Scholar]
- 15.Hughes J A, Brown L R, Ferro A J. Expression of the cloned coliphage T3 S-adenosylmethionine hydrolase gene inhibits DNA methylation and polyamine biosynthesis in Escherichia coli. J Bacteriol. 1987;169:3625–3632. doi: 10.1128/jb.169.8.3625-3632.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang H, Konishi C, Kuroki T, Huh N. A highly sensitive and specific method for quantitation of O-alkylated DNA adducts and its application to the analysis of human tissue DNA. Environ Health Perspect. 1993;99:269–271. doi: 10.1289/ehp.9399269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lea D E, Coulson C A. The distribution of the number of mutants in bacterial populations. J Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 18.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 19.Lobner-Olesen A, Hansen F G, Rasmussen K V, Martin B, Kuempel P L. The initiation cascade for chromosome replication in wild-type and Dam methyltransferase deficient Escherichia coli cells. EMBO J. 1994;13:1856–1862. doi: 10.1002/j.1460-2075.1994.tb06454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacKay W J, Han S, Samson L D. DNA alkylation repair limits spontaneous base substitution mutations in Escherichia coli. J Bacteriol. 1994;176:3224–3230. doi: 10.1128/jb.176.11.3224-3230.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marinus M G, Carraway M, Frey A Z, Brown L, Arraj J A. Insertion mutations in the dam gene of Escherichia coli K-12. Mol Gen Genet. 1983;192:288–289. doi: 10.1007/BF00327681. [DOI] [PubMed] [Google Scholar]
- 22.Marinus M G, Poteete A, Arraj J A. Correlation of DNA adenine methylase activity with spontaneous mutability in Escherichia coli K-12. Gene. 1984;28:123–125. doi: 10.1016/0378-1119(84)90095-7. [DOI] [PubMed] [Google Scholar]
- 23.Markham, G. D. 1995. Personal communication.
- 24.Marnett L J, Burcham P C. Endogenous DNA adducts: potential and paradox. Chem Res Toxicol. 1993;6:771–785. doi: 10.1021/tx00036a005. [DOI] [PubMed] [Google Scholar]
- 25.Mato J M, Alvarez L, Ortiz P, Pajares M A. S-Adenosylmethionine synthesis: molecular mechanisms and clinical implications. Pharmacol Ther. 1997;73:265–280. doi: 10.1016/s0163-7258(96)00197-0. [DOI] [PubMed] [Google Scholar]
- 26.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 27.Miller J H, Low K B. Specificity of mutagenesis resulting from the induction of the SOS system in the absence of mutagenic treatment. Cell. 1984;37:675–682. doi: 10.1016/0092-8674(84)90400-8. [DOI] [PubMed] [Google Scholar]
- 28.Moat A G, Foster J W. Microbial physiology. 3rd ed. New York, N.Y: Wiley-Liss; 1995. [Google Scholar]
- 29.Naslund M, Segerback D, Kolman A. S-Adenosylmethionine, an endogenous alkylating agent. Mutat Res. 1983;119:229–232. doi: 10.1016/0165-7992(83)90165-3. [DOI] [PubMed] [Google Scholar]
- 30.Newman E B, Budman L I, Chan E C, Greene R C, Lin R T, Woldringh C L, D’Ari R. Lack of S-adenosylmethionine results in a cell division defect in Escherichia coli. J Bacteriol. 1998;180:3614–3619. doi: 10.1128/jb.180.14.3614-3619.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oden K L, Clarke S. S-Adenosyl-L-methionine synthetase from human erythrocytes: role in the regulation of cellular S-adenosylmethionine levels. Biochemistry. 1983;22:2978–2986. doi: 10.1021/bi00281a030. [DOI] [PubMed] [Google Scholar]
- 32.Paoni N F, Koshland D E., Jr Permeabilization of cells for studies on the biochemistry of bacterial chemotaxis. Proc Natl Acad Sci USA. 1979;76:3693–3697. doi: 10.1073/pnas.76.8.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park J W, Ames B N. 7-Methylguanine adducts in DNA are normally present at high levels and increase on aging: analysis by HPLC with electrochemical detection. Proc Natl Acad Sci USA. 1988;85:7467–7470. doi: 10.1073/pnas.85.20.7467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prevost V, Shuker D E. Cigarette smoking and urinary 3-alkyladenine excretion in man. Chem Res Toxicol. 1996;9:439–444. doi: 10.1021/tx9501041. [DOI] [PubMed] [Google Scholar]
- 35.Radman M, Wagner R. Effects of DNA methylation on mismatch repair, mutagenesis, and recombination in Escherichia coli. Curr Top Microbiol Immunol. 1984;108:23–28. doi: 10.1007/978-3-642-69370-0_3. [DOI] [PubMed] [Google Scholar]
- 36.Rebeck G W, Samson L. Increased spontaneous mutation and alkylation sensitivity of Escherichia coli strains lacking the ogt O6-methylguanine DNA repair methyltransferase. J Bacteriol. 1991;173:2068–2076. doi: 10.1128/jb.173.6.2068-2076.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rydberg B, Lindahl T. Nonenzymatic methylation of DNA by the intracellular methyl group donor S-adenosyl-L-methionine is a potentially mutagenic reaction. EMBO J. 1982;1:211–216. doi: 10.1002/j.1460-2075.1982.tb01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saint-Girons I, Parsot C, Zakin M M, Barzu O, Cohen G N. Methionine biosynthesis in Enterobacteriaceae: biochemical, regulatory, and evolutionary aspects. Crit Rev Biochem. 1988;23(Suppl. 1):S1–S42. doi: 10.3109/10409238809083374. [DOI] [PubMed] [Google Scholar]
- 39.Satishchandran C, Taylor J C, Markham G D. Novel Escherichia coli K-12 mutants impaired in S-adenosylmethionine synthesis. J Bacteriol. 1990;172:4489–4496. doi: 10.1128/jb.172.8.4489-4496.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sedgwick B. Nitrosated peptides and polyamines as endogenous mutagens in O6-alkylguanine-DNA alkyltransferase deficient cells. Carcinogenesis. 1997;18:1561–1567. doi: 10.1093/carcin/18.8.1561. [DOI] [PubMed] [Google Scholar]
- 41.Spoerel N, Herrlich P, Bickle T A. A novel bacteriophage defence mechanism: the anti-restriction protein. Nature. 1979;278:30–34. doi: 10.1038/278030a0. [DOI] [PubMed] [Google Scholar]
- 42.Swann P F, Waters T R, Moulton D C, Xu Y Z, Zheng Q, Edwards M, Mace R. Role of postreplicative DNA mismatch repair in the cytotoxic action of thioguanine. Science. 1996;273:1109–1111. doi: 10.1126/science.273.5278.1109. [DOI] [PubMed] [Google Scholar]
- 43.Szyf M, Avraham-Haetzni K, Reifman A, Shlomai J, Kaplan F, Oppenheim A, Razin A. DNA methylation pattern is determined by the intracellular level of the methylase. Proc Natl Acad Sci USA. 1984;81:3278–3282. doi: 10.1073/pnas.81.11.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan B H, Bencsath F A, Gaubatz J W. Steady-state levels of 7-methylguanine increase in nuclear DNA of postmitotic mouse tissues during aging. Mutat Res. 1990;237:229–238. doi: 10.1016/0921-8734(90)90004-b. [DOI] [PubMed] [Google Scholar]
- 45.Taverna P, Sedgwick B. Generation of an endogenous DNA-methylating agent by nitrosation in Escherichia coli. J Bacteriol. 1996;178:5105–5111. doi: 10.1128/jb.178.17.5105-5111.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Val D L, Cronan J E., Jr In vivo evidence that S-adenosylmethionine and fatty acid synthesis intermediates are the substrates for the LuxI family of autoinducer synthases. J Bacteriol. 1998;180:2644–2651. doi: 10.1128/jb.180.10.2644-2651.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vidal A, Abril N, Pueyo C. DNA sequence analysis of spontaneous lacI−d mutations in O6-alkylguanine-DNA alkyltransferase-proficient and -deficient Escherichia coli. Mutagenesis. 1998;13:367–373. doi: 10.1093/mutage/13.4.367. [DOI] [PubMed] [Google Scholar]
- 48.Vinella D, Jaffe A, D’Ari R, Kohiyama M, Hughes P. Chromosome partitioning in Escherichia coli in the absence of Dam-directed methylation. J Bacteriol. 1992;174:2388–2390. doi: 10.1128/jb.174.7.2388-2390.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walsh, P. R. 1997. Personal communication.
- 50.Wilson M J, Shivapurkar N, Poirier L A. Hypomethylation of hepatic nuclear DNA in rats fed with a carcinogenic methyl-deficient diet. Biochem J. 1984;218:987–990. doi: 10.1042/bj2180987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiao W, Samson L. In vivo evidence for endogenous DNA alkylation damage as a source of spontaneous mutation in eukaryotic cells. Proc Natl Acad Sci USA. 1993;90:2117–2121. doi: 10.1073/pnas.90.6.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiao W, Samson L. The Saccharomyces cerevisiae MGT1 DNA repair methyltransferase gene: its promoter and entire coding sequence, regulation and in vivo biological functions. Nucleic Acids Res. 1992;20:3599–3606. doi: 10.1093/nar/20.14.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamada M, Sedgwick B, Sofuni T, Nohmi T. Construction and characterization of mutants of Salmonella typhimurium deficient in DNA repair of O6-methylguanine. J Bacteriol. 1995;177:1511–1519. doi: 10.1128/jb.177.6.1511-1519.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yocum R R, Perkins J B, Howitt C L, Pero J. Cloning and characterization of the metE gene encoding S-adenosylmethionine synthetase from Bacillus subtilis. J Bacteriol. 1996;178:4604–4610. doi: 10.1128/jb.178.15.4604-4610.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]