Abstract
Natriuretic peptides regulate multiple physiologic systems by activating transmembrane receptors containing intracellular guanylyl cyclase domains, such as GC-A and GC-B, also known as Npr1 and Npr2, respectively. Both enzymes contain an intracellular, phosphorylated pseudokinase domain (PKD) critical for activation of the C-terminal cGMP-synthesizing guanylyl cyclase domain. Because ATP allosterically activates GC-A and GC-B, we investigated how ATP binding to the PKD influenced guanylyl cyclase activity. Molecular modeling indicated that all the residues of the ATP-binding site of the prototypical kinase PKA, except the catalytic aspartate, are conserved in the PKDs of GC-A and GC-B. Kinase-inactivating alanine substitutions for the invariant lysine in subdomain II or the aspartate in the DYG-loop of GC-A and GC-B failed to decrease enzyme phosphate content, consistent with the PKDs lacking kinase activity. In contrast, both mutations reduced enzyme activation by blocking the ability of ATP to decrease the Michaelis constant without affecting peptide-dependent activation. The analogous lysine-to-alanine substitution in a glutamate-substituted phosphomimetic mutant form of GC-B also reduced enzyme activity, consistent with ATP stimulating guanylyl cyclase activity through an allosteric, phosphorylation-independent mechanism. Mutations designed to rigidify the conserved regulatory or catalytic spines within the PKDs increased guanylyl cyclase activity, increased sensitivity to natriuretic peptide, or reduced the Michaelis constant in the absence of ATP, consistent with ATP binding stabilizing the PKD in a conformation analogous to that of catalytically active kinases. We conclude that allosteric mechanisms evolutionarily conserved in the PKDs promote the catalytic activation of transmembrane guanylyl cyclases.
Introduction
There are three natriuretic peptides (NPs) in mammals. Two homodimeric single membrane-spanning guanylyl cyclase (GC) receptors mediate the majority of the physiological effects of these NPs by increasing intracellular cGMP concentrations (1–4). However, cGMP-independent functions of NPs have also been described that may be mediated by the NP clearance receptor (5). Atrial NP (ANP) and B-type NP (BNP) activate guanylyl cyclase-A (GC-A, also known as Npr1), which stimulates natriuresis and inhibits cardiac hypertrophy. C-type NP (CNP) activates guanylyl cyclase-B (GC-B, also known as Npr2), which stimulates long bone growth, meiotic arrest in oocytes, and neuronal bifurcation (1, 6). GC-A and GC-B are homologous, with the intracellular portions of the proteins being 78% identical (7). They also share five conserved phosphorylation sites (8).
NP binding causes a 24º counterclockwise rotation in the extracellular domain of one GC-A monomer with respect to the other monomer (9). As this conformational change propagates through the receptor to the C-terminal catalytic domain, the binding signal travels through an intracellular pseudokinase domain (PKD) that is constitutively phosphorylated on at least six residues (8, 10, 11). The chemically determined phosphorylation sites identified in rat and human GC-A are Ser487, Ser497, Thr500, Ser502, Ser506, Ser510, and Thr513 according to rat numbering (Fig. 1C) (8, 12). Also using rat numbering, the chemically identified phosphorylation sites in rat and human GC-B are Ser513, Thr516, Ser518, Ser523, Ser526, and Thr529 (8). Additionally, a functional screen identified a single serine residue in the juxtamembrane region conserved in both GC-A (Ser473) and GC-B (Ser489) (13). The identities of the kinases responsible for phosphorylating these sites are unknown. Phosphorylation of the PKD is absolutely required for propagation of the NP binding signal to the GC catalytic domain of both receptors (10, 11, 14, 15). Data supporting the essential role of phosphorylation are extensive and are derived from experiments as diverse as enzymatic dephosphorylation GC-A and GC-B in membranes to the expression of phosphomimetic forms of GC-A and GC-B that are resistant to inactivation by dephosphorylation in mice (13–21).
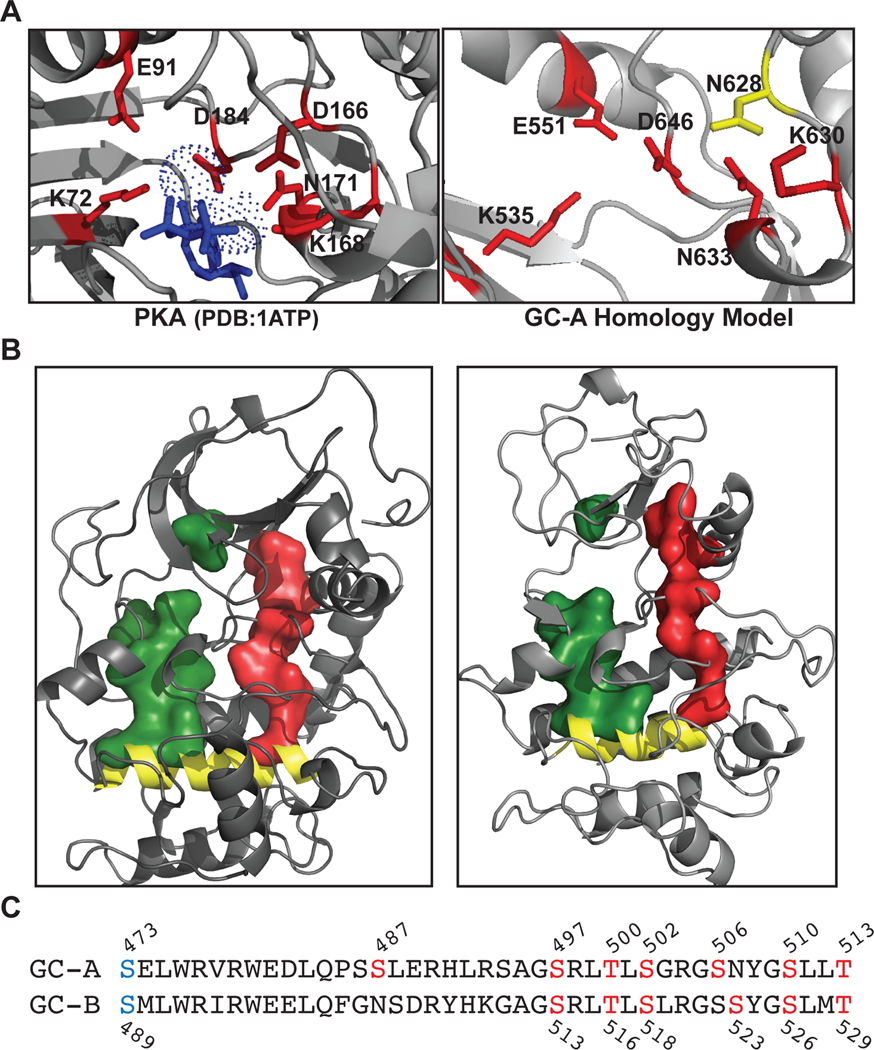
Fig. 1. The R- and C-spines and residues that directly bind and transmit the allosteric ATP-binding signal in PKA are conserved in the PKD of GC-A.
(A) Structure of the active site of the kinase domain of PKA and the LCK-based homology model of the corresponding site in the PKD of GC-A showing conservation of the residues (red) that interact with ATP (blue) in the active site. The catalytic base (Asp166) in PKA is not present in GC-A, where this residue is replaced with Asn628 (yellow). (B) Structures showing the R- (red) and C- (green) spines, and the F-helix (yellow) in the kinase domain of PKA and the homology model of the PKD of GC-A. (C) Serine and threonine residues mutated in non-phosphorylated or phosphomimetic constructs are indicated with their position numbers. Residues in red are phosphorylation sites that were chemically determined (8), and those in blue were identified by a functional screen (13).
In addition to NPs, adenosine triphosphate (ATP) is required for the activation of GC-A and GC-B (22). However, understanding how ATP activates GC-A and GC-B is complicated by the ability of ATP to serve as a substrate for receptor phosphorylation as well as bind both an allosteric site in the PKD and an allosteric site in the catalytic domain (23, 24). In substrate-velocity experiments, ATP serves as an allosteric activator that causes a shift from positive cooperative kinetics in the absence of ATP to linear kinetics in the presence of ATP, which ultimately reduces the Michaelis constant of the guanylyl cyclase catalytic domain, a measure of affinity for its GTP substrate, an order of magnitude in an NP-dependent manner (24). This 10-fold decrease in the Michaelis constant is necessary for the enzyme to function at cellular concentrations of GTP. To identify the ATP binding site mediating these effects, two separate groups covalently cross-linked ATP analogs to the PKD of GC-A and suggested that this process is required for the ability of ATP to increase GC-A activity (25, 26). However, the exact ATP binding site in the PKD was not determined. Because protein kinase A (PKA) is the most widely studied mammalian protein kinase, here we used knowledge of allosteric activation of PKA and other kinases and pseudokinases to better understand how the intrinsic PKDs of GC-A and GC-B regulate the guanylyl cyclase activities of these receptors.
In PKA the transition from the inactive to the active state occurs through phosphorylation of the activation loop. The phosphorylated activation loop then forms a salt-bridge with a histidine residue in the αC-helix located in the N-terminal lobe of the kinase. The regulatory (R)-spine, a network of internal hydrophobic amino acids, then assembles into a rigid structure that properly orients all the components necessary for catalysis in the active site. Upon ATP binding to the kinase active site, the adenine ring of ATP docks between the N-terminal lobe and the C-terminal lobe of the kinase, thus fusing another network of hydrophobic amino acids called the catalytic (C)-spine (27). The triphosphate tail of ATP and the associated Mg2+cations form interactions with multiple polar and charged amino acids lining the active site. The most notable of these are the invariant lysine in subdomain II and the Mg2+-chelating aspartate in the Asp-Phe-Gly (DFG) loop. Both the lysine and aspartate residues are conserved in GC-A and GC-B, and alanine substitutions of these residues provided critical data supporting the conclusions in this study. In PKA, both spines are docked on the αF-helix in the C-terminal lobe, and the assembly of both spines rigidifies the entire kinase domain with coordinated motion in the most catalytically competent state (28). In pseudokinases, phosphotransferase activity is markedly diminished or absent due to the loss of residues required for catalysis, but ATP-dependent allostery is often maintained (29–32).
Here, we created a homology model that revealed that the PKDs of GC-A and GC-B contain the conserved hydrophobic regulatory and catalytic spine elements common to known protein kinases and pseudokinases, which implies that the allosteric activation mechanisms of protein kinases and pseudokinases may be conserved in receptor GCs as well. In addition, the PKDs in GC-A and GC-B contain all of the ATP-binding residues that are conserved in most protein kinases exception for the catalytic aspartate, which suggests that these domains in GC-A and GC-B lack intrinsic phosphotransferase activity (33). However, low kinase activity was observed for the putative pseudokinase ErbB3 (also known as HER3) and for the JH2 pseudokinase domain of JAK2, both of which also lack the conserved catalytic aspartate, suggesting that the PKDs of GC-A and GC-B may also contain muted, but regulatory, amounts of phosphotransferase activity (34, 35).
Here, we investigated why the PKD is conserved in receptor GCs, first by addressing the possibility that the PKD of GC-A may have intrinsic autophosphorylating activity by making single amino acid inactivating mutations that disrupt the activity of most protein kinases. However, these mutations failed to reduce the phosphate content of GC-A or GC-B. In contrast, these same mutations markedly decreased the ability of NPs to decrease the Michaelis constant of GC-A and GC-B guanylyl cyclase activity in an ATP-dependent manner. Finally, we developed a dynamic hydrophobic core model of allostery common to kinases and pseudokinases and used it to evaluate how these PKDs stimulate receptor guanylyl cyclase activity.
Results
Canonical ATP-interacting and hydrophobic core residues in GC-A and GC-B are conserved
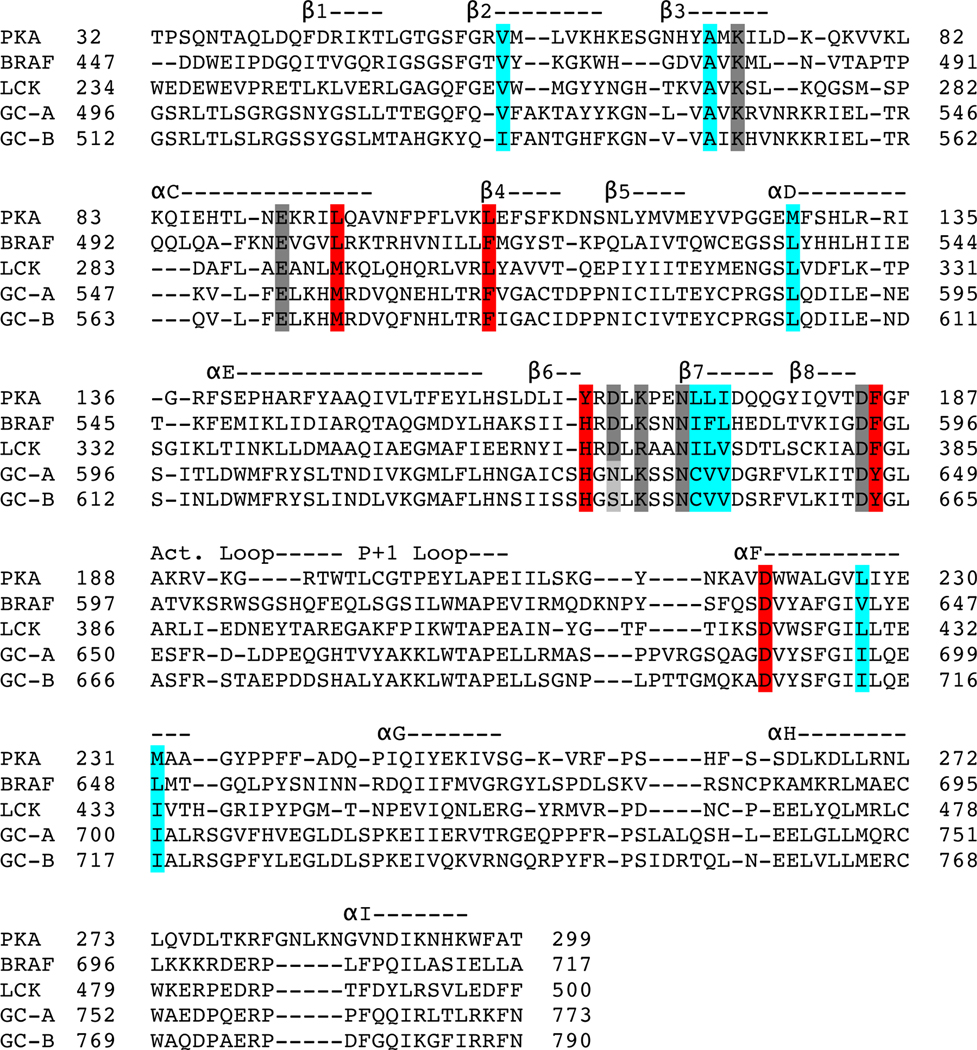
We generated a homology model for the PKD of rat GC-A based on the structure of the kinase domain of human LCK, which has the greatest amino acid identity (32%) to the PKD of GC-A of all known protein kinases (Fig. 1). We use rat GC-A and rat GC-B sequence and numbering throughout our paper. We compared the ATP binding pocket of the GC-A homology model to that of PKA, focusing on the residues required for binding of ATP and catalysis in PKA (Lys72, Glu91, Asp166, Lys168, Asn171, and Asp184) (Fig. 1A). Except for Asp166, the catalytic base in PKA, all residues that directly interact with ATP in PKA are conserved in the PKDs in GC-A and GC-B (Fig 1A and Table 1). We also generated space-filling models that demonstrate that residues composing the R-spine and C-spine in PKA are conserved in GC-A and GC-B (Fig 1B and Table 1). Thus, the structural framework required for ATP binding and allosteric transmission of the ATP binding signal in PKA is conserved in NP-stimulated GCs. We also generated nonphosphorylatable and phosphomimetic versions of GC-A and GC-B by mutating the serine and threonine phosphorylation sites in the juxtamembrane region immediately upstream of the PKD to alanine or glutamate, respectively (Fig. 1C), to use as controls for subsequent guanylyl cyclase activity assays and phosphoprotein analyses. To more thoroughly compare our homology model of the PKD of GC-A, we performed a structural alignment in PyMol using our GC-A homology model and the structures of reference kinases (PKA, PDB: 1ATP; BRAF, PDB: 1UWH; LCK, PDB: 3LCK) (Fig. 2). Previously, many researchers had assumed the Gly-x-Gly-x-x-x-Gly motif in the PKDs of GC-A and GC-B (Gly503-Gly509 in GC-A) fulfilled the role of the glycine-rich (G)-loop of protein kinases found between the β1 and β2 strands. However, when these glycines were mutated to alanine to prevent the backbone flexibility necessary for their function in protein kinases there was no effect on guanylyl cyclase activity (36). In our model of GC-A, we observed that the glycine to alanine substitutions actually preceded or were within the β1 strand, not in the β1-β2 loop as previously predicted. In the β1-β2 loop of our model of GC-A, there is a degraded G-loop motif, which is expected to decrease ATP affinity and may explain why high concentrations of ATP are necessary to fully activate GC-A and GC-B (24).
Table 1.
Amino acids that are important for ATP binding and allosteric transmission of the ATP binding signal are conserved between PKA and GC-A and GC-B.PKA is a representative example of serine-threonine kinases. BRAF is a representative example of Tyr-like serine-threonine kinases, and LCK is a representative example of Tyr kinases and was the source of the homology model.
| PKA | BRAF | LCK | GC-A | GC-B |
|---|---|---|---|---|
| ATP-Interacting Amino Acids | ||||
| K72 | K483 | K273 | K535 | K551 |
| E91 | E501 | E288 | E551 | E567 |
| D166 | D576 | D364 | N628 | S644 |
| K168 | K578 | R366 | K630 | K646 |
| N171 | N581 | N369 | N633 | N649 |
| D184 | D594 | D382 | D646 | D662 |
| Regulatory (R)-Spine | ||||
| L106 (RS4) | F516 | L303 | F566 | F582 |
| L95 (RS3) | L505 | M292 | M555 | M571 |
| F185 (RS2) | F595 | F383 | Y647 | Y663 |
| Y164 (RS1) | H574 | H362 | H626 | H642 |
| D220 (RS0) | D638 | D422 | D689 | D706 |
| Catalytic (C)-Spine | ||||
| V57 | V471 | V259 | L511 | L527 |
| A70 | A481 | A271 | A533 | A549 |
| M128 | L537 | L324 | L588 | L604 |
| L172 | I582 | I370 | C634 | C650 |
| L173 | F583 | L371 | V635 | V651 |
| I174 | L584 | V372 | V636 | V652 |
| L227 | V601 | L429 | I696 | I713 |
| M231 | L605 | I433 | I700 | I717 |
Fig. 2. Structural alignment of the PKD of GC-A with crystal structures from other kinases.
The homology model of the PKD of GC-A and the structures of PKA, BRAF, and LCK were aligned in PyMol. A linear representation of this structural alignment is shown with secondary structure features annotated above the alignment. α, alpha helix; β, beta sheet; Act Loop, activation loop. Residues comprising the R-spine (red) and C-spine (cyan) and those that interact with ATP (gray) are highlighted. See also Table 1.
Alanine substitutions for the conserved Lys in subdomain II or Asp in the DYG-loop inactivate GC-A and GC-B
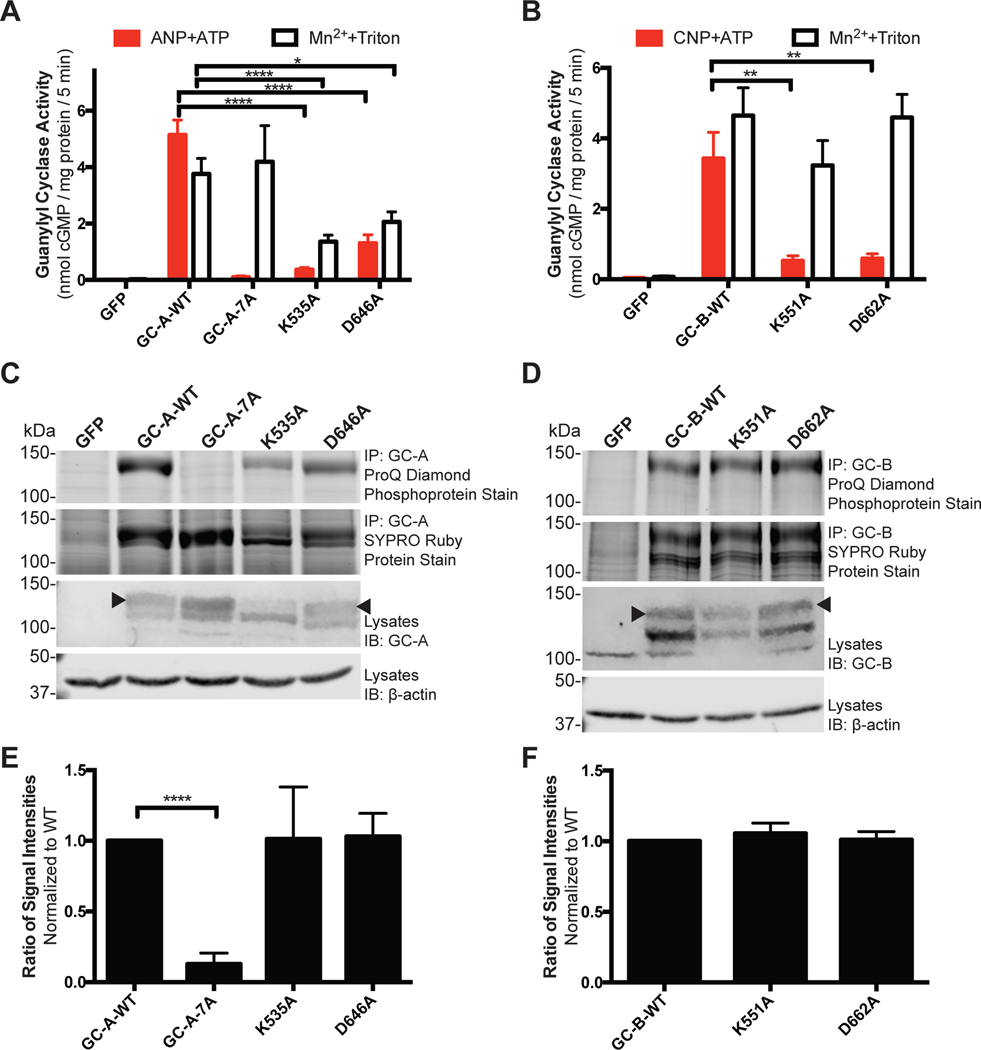
Because the most commonly used kinase-inactivating mutations (corresponding to Lys72 and Asp184 in PKA) are conserved in GC-A (Lys535 and Asp646) and GC-B (Lys551 and Asp662), we used alanine substitutions at these sites to investigate the effects of presumed decreased ATP binding to the PKD on guanylyl cyclase activity. We prepared crude membranes from HEK293T cells transfected with plasmids encoding wild-type GC-A (GC-A-WT), GC-A-7A, a non-phosphorylated mutant containing alanine substitutions at the known phosphorylation sites S473A, S497A, T500A, S502A, S506A, S510A, and T513A, and the single alanine substitution mutants GC-A-K535A and GC-A-D646A (Fig. 3A). We then assayed the membranes for guanylyl cyclase activity under maximal physiologic activation conditions (5mM MgCl2, 1mM ATP, and 1μM ANP) or under synthetic conditions that yield near maximal activity independently of enzyme phosphorylation or ANP binding (5mM MnCl2 and 1% Triton X-100) and are used as a measure of properly folded and active catalytic domain. Activation by manganese and detergent is thought to result from disruption of hydrophobic autoinhibitory interactions (37). We observed strong reductions in ANP-dependent guanylyl cyclase activity for the GC-A-K535A and GC-A-D646A mutants compared to the activity of the wild-type enzyme. However, we also observed substantial reductions in detergent-dependent activity for each mutant (Fig. 3A). To confirm the observed effects were due to changes in conserved mechanisms in the receptor GC family, we performed guanylyl cyclase activity assays on the homologous receptor, GC-B, and the analogous mutants GC-B-K551A and GC-B-D662A. Again, we observed reductions in CNP-dependent guanylyl cyclase activity for GC-B-K551A and GC-B-D662A (Fig. 3B). Unlike the GC-A mutants, we did not detect significant reductions in detergent-dependent activity of the GC-B mutants, which allowed us to rule out reductions in GC-B protein as a possible explanation for the reductions in CNP-dependent enzymatic activity for these mutant enzymes.
Fig. 3. Decreased NP-dependent guanylyl cyclase activity in Lys and Asp mutants is not explained by changes in protein abundance or phosphorylation.
(A) Guanylyl cyclase activity of membranes from HEK293T cells expressing the indicated WT and mutant forms of GC-A in the presence of GTP, ANP, ATP, and MgCl2 (ANP+ATP) or Triton X-100 and MnCl2 (Mn2++Triton). n = 8 independent experiments. (B) Guanylyl cyclase activity of membranes from HEK293T cells expressing the indicated WT and mutant forms of GC-B in the presence of GTP, CNP, ATP, and MgCl2 (CNP+ATP) or Triton X-100 and MnCl2 (Mn2++Triton). n = 3 independent experiments. (C) ProQ Diamond Phosphopreotein Gel Stain and SYPRO Ruby Protein Gel Stain of immunoprecipitated (IP) WT and mutant GC-A and immunoblot (IB) of the same samples used for guanylyl cyclase activity assays in (A). The arrowhead indicates the fully processed form of GC-A. Data are representative of n = 7 independent experiments. (D) ProQ Diamond Phosphopreotein Gel Stain and SYPRO Ruby Protein Gel Stain of immunoprecipitated WT and mutant GC-B and immunoblot of the same samples used for guanylyl cyclase activity assays in (B). The arrowhead indicates the fully processed form of GC-B. Data are representative of n = 3 independent experiments. (E) Quantification of experiments in (C). n = 7 independent experiments. (F) Quantification of experiments in (D). n = 3 independent experiments. Error bars represent the SEM.
Reduced ligand-dependent guanylyl cyclase activity of the GC-A K535A and D646A and GC-B K551A and D662A mutants does not result from reduced protein processing or phosphorylation
Theoretically, the alanine substitution mutations could reduce the activity of GC-A and GC-B by decreasing the amount of fully processed enzyme (38), reducing receptor phosphorylation (14, 15), or interfering with allosteric activation. To determine why the K535A and D646A mutations in GC-A and the K551A and D662A mutations in GC-B reduce guanylyl cyclase activity, we measured the amounts of protein and phosphate in the same samples. Protein abundance was determined by western blotting of lysates or by SYPRO Ruby staining of immunopurified proteins (Fig. 3C, D). We determined the stoichiometry of phosphorylation by first measuring phosphate abundance by ProQ Diamond staining followed by SYPRO Ruby staining of the same gel to determine protein concentrations (Fig. 3C, D) (39, 40).
We detected phosphorylated wild-type GC-A and GC-B as dark, diffuse bands when stained with ProQ Diamond, consistent with migration of multiple phosphorylated species that differ in molecular weight due to differing amounts of glycosylation and phosphorylation (39). Very low or no staining of GC-A-7A with alanine substitutions for known phosphorylation sites demonstrated that ProQ Diamond dye is specific for phosphate. SYPRO Ruby staining of the same gel used for phosphate detection indicated similar abundances of wild-type and mutant proteins. Comparable amounts of ProQ Diamond and SYPRO Ruby staining for each protein indicated that the stoichiometry of phosphorylation was similar between wild-type receptors and the two alanine mutants. Again, these data are consistent with the PKDs of GC-A and GC-B lacking intrinsic autophosphorylating activity. To quantify the relative stoichiometry of phosphorylation of GC-A and GC-B, the ratio of ProQ Diamond signal divided by SYPRO Ruby signal for the upper band of GC-A and GC-B was calculated and plotted (Fig. 3E, F). The relative stoichiometry of phosphorylation of the Lys and Asp substitution mutants was not different from each other or their wild-type counterparts. These data indicate that the decrease in guanylyl cyclase activity observed for both ATP binding site mutations cannot be explained by decreased receptor phosphorylation.
Although we observed reductions in the detergent-dependent guanylyl cyclase activity and the amount of fully glycosylated and phosphorylated enzyme for the GC-A-K535A and GC-A-D646A mutants, none of these decreases could account for the reduction in ANP- and ATP-dependent guanylyl cyclase activity. In contrast, both the detergent-dependent guanylyl cyclase activity and the species migrating as the upper band were not different between GC-B-WT, GC-B-K551A, and GC-B-D662A, but CNP and ATP-dependent guanylyl cyclase activity was markedly reduced by the mutations. Together, these data indicate that both the K535A and D646A mutations in GC-A and the K551A and D662A mutations in GC-B inactivate these enzymes by a process that cannot be explained by reductions in the fully glycosylated, phosphorylated forms of these enzymes.
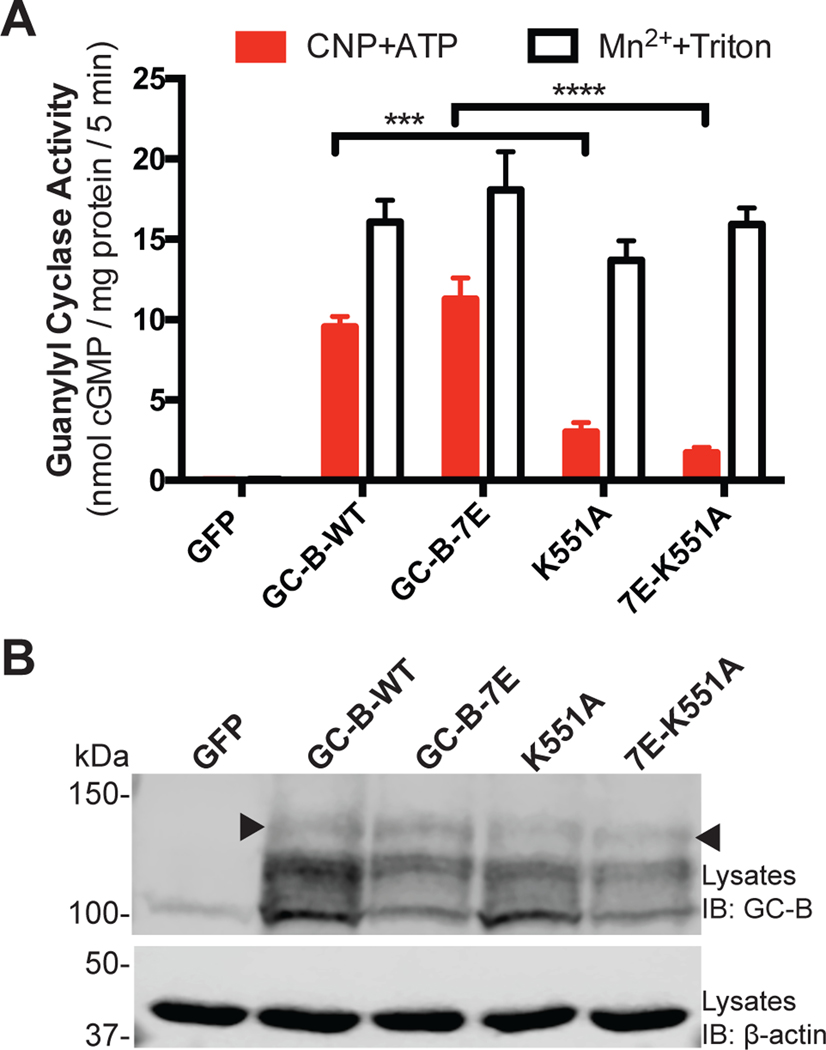
To further rule out confounding effects of changes in phosphorylated residues, we performed guanylyl cyclase activity assays on the glutamate-substituted, phosphomimetic mutant of GC-B called GC-B-7E, which behaves kinetically identical to GC-B-WT (13), with or without the the K551A mutation (7E-K551A and GC-B-7E, respectively). We observed similar decreases in CNP-dependent guanylyl cyclase activity as a result of the K551A mutation in both GC-B-WT and GC-B-7E (K551A and 7E-K551A respectively) with no change in detergent-dependent GC activity (Fig. 4A) and maintained presence of the species migrating as the upper band (Fig. 4B). These data are consistent with Lys551 participating in the ATP dependent activation of GC-B in a manner that does not require changes in GC-B protein concentrations or known phosphorylation sites.
Fig. 4. The GC-B-K551A mutant has reduced CNP-dependent enzymatic activity that is not explained by changes in phosphorylation.
(A) Guanylyl cyclase activity of membranes from HEK293T cells expressing the indicated WT and mutant forms of GC-B in the presence of GTP, CNP, ATP, and MgCl2 (CNP+ATP) or Triton X-100 and MnCl2 (Mn2++Triton). n = 5 independent experiments. (B) Immunoblot (IB) showing GC-B in samples from (A). The arrowhead indicates the fully processed form of GC-B with guanylyl cyclase activity (38). Data are representative of n = 3 experiments. Error bars represent the SEM.
The Lys and Asp substitutions in the PKD specifically reduce activation of GC-A and GC-B by ATP but not by NPs
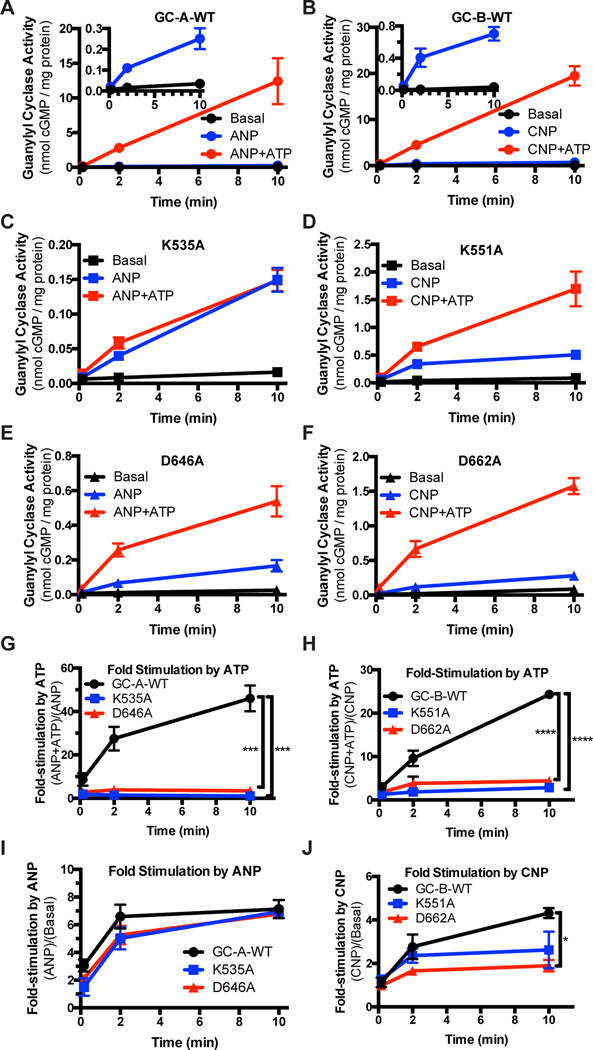
We also investigated the ability of mutations in the PKDs of GC-A and GC-B to inhibit activation by ATP and NPs. Crude membranes from HEK293T cells transfected with plasmids expressing the indicated receptors were assayed for various amounts of time with physiologic concentrations of GTP in the absence of both NP and ATP (basal), with only NP (ANP for GC-A and CNP for GC-B), or with both NP and ATP, conditions that mimic physiologic activation (Fig. 5). We observed higher activity for wild-type versions of GC-A (Fig. 5A) and GC-B (Fig. 5B) when assayed in the presence of both NP and ATP at all time points compared to activities measured for any mutant (Fig. 5C–F). The activities of GC-A-WT and GC-B-WT were substantially increased by NP in the absence of ATP (insets in Fig. 5A, B). Fold activation observed with ATP as determined by activity measured in the presence of ATP and either ANP (for GC-A) or CNP (for GC-B) divided by activity measured in the presence of only ANP or CNP increased with time for GC-A-WT and GC-B-WT, respectively, but not for any of the mutants. The maximal fold activation by ATP was 46-fold for GC-A-WT but was only 3-fold for the GC-A-D646A mutant and less than 2-fold for the GC-A-K535A mutant (Fig. 5G) with similar patterns observed for GC-B-WT and the analogous GC-B-K551A and GC-B-D662A mutants (Fig. 5H).
Fig. 5. Conserved Lys and Asp residues in PKDs are required for activation of GC-A and GC-B by ATP but not by NP.
(A) Guanylyl cyclase activity of membranes from HEK293T cells expressing GC-A-WT at the indicated time points after adding GTP in the presence of MgCl2 (Basal), ANP plus MgCl2 (ANP), or ANP, ATP, and MgCl2 (ANP+ATP). Basal and ANP activity are shown in the inset. n = 3 independent experiments. (B) Guanylyl cyclase activity of membranes from HEK293T cells expressing GC-B-WT at the indicated time points after adding GTP in the presence of MgCl2 (Basal), CNP and MgCl2 (CNP), or CNP, ATP, and MgCl2 (CNP+ATP). Basal and ANP activity are shown in the inset. n = 3 independent experiments. (C-F) Guanylyl cyclase activity measurements as in (A) and (B) for membranes from cells expressing GC-A-K535A (C), GC-B-K551A (D), GC-A-D646A (E), or GC-B-D662A (F). n = 3 independent experiments. (G) Fold stimulation of WT and mutant GC-A guanylyl cyclase activity by ATP calculated from (A), (C), and (E) and plotted as a function of time. n = 3 independent experiments. (H) Fold stimulation of WT and mutant GC-B guanylyl cyclase activity by ATP calculated from (B), (D), and (F) and plotted as a function of time. n = 3 independent experiments. (I) Fold stimulation of WT and mutant GC-A guanylyl cyclase activity by ANP calculated from (A), (C), and (E) and plotted as a function of time. n = 3 independent experiments. (J) Fold stimulation of WT and mutant GC-B guanylyl cyclase activity by CNP calculated from (B), (D), and (F) and plotted as a function of time. n = 3 independent experiments. Error bars represent the SEM.
We also determined fold-activation by NP alone by measuring activity in the presence of only NP divided by activity determined in the absence of NP (basal). We hypothesized that this ratio would decrease if the single alanine mutations were not specific for ATP binding and instead globally affected the structural integrity of the PKD (Fig. 5I, J). We observed no difference in fold activation of wild-type or mutant GC-A by ANP and only small differences in of in wild-type or mutant GC-B by CNP. Fold activation of GC-B-D662A by CNP was significantly different from GC-B-WT but was not on the same order as the differences observed in fold activation by ATP. These data indicate that canonical mutations in protein kinases or pseudokinases that diminish ATP-mediated activation also diminish the ability of ATP to activate GC-A and GC-B with little or no effect on the stability of the PKD.
Loss of cooperativity occurs through ATP binding to an allosteric site in the catalytic domain
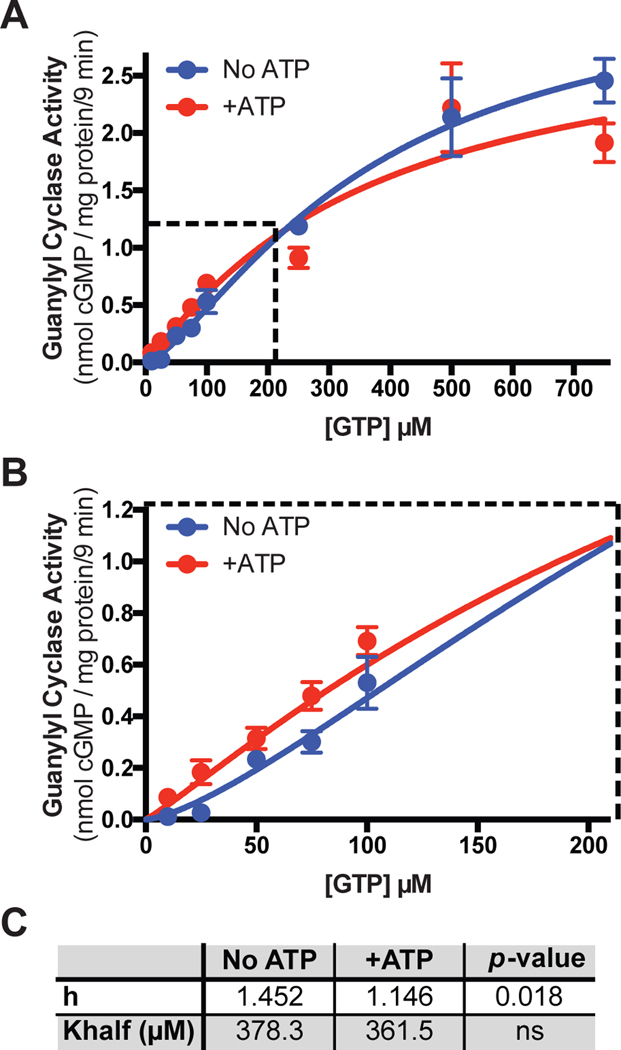
Ill-defined allosteric regulatory sites have been reported for the PKD (25, 26) as well as in the catalytic (guanylyl cyclase) domain of GC-A (23, 24, 41). To determine which site is responsible for the effects of ATP on cooperativity in enzyme activity assays, we performed substrate-velocity assays on membranes prepared from Cos7 cells expressing a form of GC-A containing all domains except the PKD (Fig. 6A, B) (42). Using this construct, we observed a Hill slope of 1.4 in the absence of ATP that was reduced to 1.1 when ATP was added to the reaction mixture (Fig. 6C). However, we failed to observe a substantial decrease in Km upon the addition of ATP in the presence of 1μM ANP as was observed for the full length wild-type version of GC-A (Fig. 6C) (24). These data indicate that ATP binding to the allosteric site in the catalytic domain mediates the positive cooperative effects but is not sufficient to decrease the Km, which requires NP-binding to the extracellular domain (24) and may require ATP binding to the canonical ATP binding site in the PKD.
Fig. 6. A GC-A mutant lacking the pseudokinase domain contains a cooperative, ATP-binding, allosteric site.
(A-B) Guanylyl cyclase activity in membranes from Cos7 cells expressing a form of GC-A in which the PKD domain was deleted and in the presence of ANP, MgCl2, and increasing concentrations of GTP with or without the addition of ATP. n = 4 independent experiments. The dashed area in (A) is enlarged in (B). n = 4 independent experiments. (C) A table showing the measured Hill slope (h) and Khalf of the plots in (A) and (B) in the absence and presence of ATP and their respective p-values. Error bars represent the SEM.
Lys and Asp substitutions in the PKD prevent the ATP-dependent reduction in the Michaelis constants of GC-A and GC-B
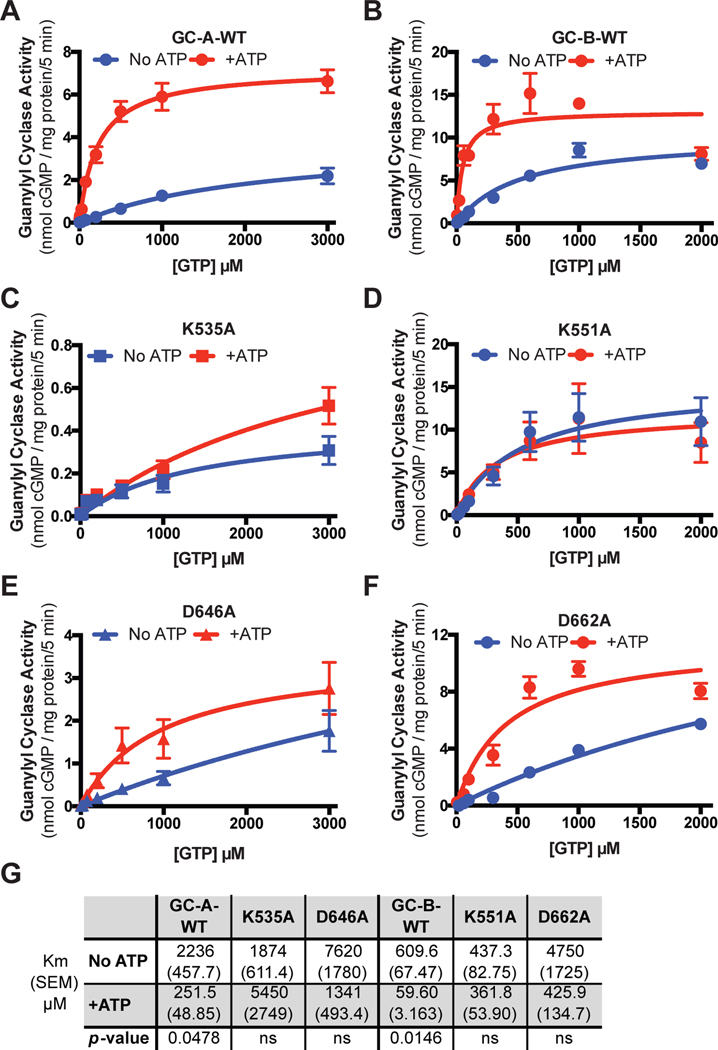
To determine if the conserved ATP binding site in the PKD of GC-A and GC-B is required for the ATP-dependent reduction in the Km, we assayed crude membranes from HEK293T cells transiently transfected with plasmids expressing GC-A-WT, GC-A-K535A, GC-A-D646A, GC-B-WT, GC-B-K551A, or GC-B-D662A with saturating concentrations of the appropriate NP and increasing substrate concentrations in the presence or absence of ATP (Fig. 7A–F). ATP reduced the Michaelis constant of GC-A-WT 10.1-fold and reduced the Michaelis constant of GC-B-WT 10.6-fold (Fig. 7G), consistent with previous reports (36). In contrast, ATP failed to reduce the Km of GC-A-K535A and GC-A-D646A (Fig. 7C, E) or GC-B-K551A and GC-B-D662A (Fig. 7D, F). These data indicate that ATP binding to the conserved binding site in the PKD of GC-A and GC-B is required for allosteric activation of these enzymes.
Fig. 7. Lys and Asp mutations increase the Michaelis constants for GC-A and GC-B.
(A) Guanylyl cyclase activity of membranes from HEK293T cells expressing GC-A-WT in the presence of MgCl2, ANP, and increasing concentrations of GTP with or without ATP. n = 4 independent experiments. (B) Guanylyl cyclase activity of membranes from HEK293T cells expressing GC-B-WT in the presence of MgCl2, CNP, and increasing concentrations of GTP with or without ATP. n = 3 independent experiments. (C–F) Guanylyl cyclase activity as in (A) and (B) for membranes from cells expressing GC-A-K535A (C), GC-B-K551A (D), GC-A-D646A (E), or GC-B-D662A (F). n = 3 or 4 independent experiments. (G) A table showing the measured Michaelis constants of the indicated WT and mutant forms of GC-A and GC-B in the absence and presence of ATP and their associated p-values. Error bars represent the SEM.
To directly determine the effect of the ATP binding site mutations on ATP binding, we removed the catalytic domain from N-terminally FLAG-tagged versions of the wild-type and mutant versions of GC-B and tested each for the ability to bind 8-azido-2’, 3’-biotinyl-ATP, as originally described for ATP binding to the PKD of GC-A by De Lean and colleagues (26). Unfortunately, non-specific binding to each construct was too high to determine any changes in ATP binding (Fig. S1). We also employed the method used by Jaleel et. al using soluble GC-A intracellular domain constructs bound to ATP-agarose (43). Intracellular domains were present in the soluble fraction (Fig. S2A) and were retained during spin filtration to remove endogenous ATP (Fig. S2B). Unfortunately, control wild-type constructs did not demonstrate substantial binding to the ATP-agarose resin using effective ATP concentrations of 250μM. Furthermore, the weak binding observed was not reduced by the addition of 1mM ATP (Fig. S2C). The lack of ATP binding in our assay is not surprising considering that the EC50 for activation of GC-A and GC-B by ATP is approximately 0.1mM (36). If this EC50 is indicative of the dissociation constant, then these results are not unexpected and suggest that conventional non-equilibrium ATP-binding assays used for kinases are not likely to work for these enzymes with low affinity for ATP. Also, because mutations at the invariant lysine (Lys72 in PKA) have failed to inhibit ATP binding in other kinases (44, 45), two explanations for the loss of ATP-dependent functions are possible. The first is that mutations reduce binding to ATP, whereas the second possibility is that the mutation does not affect ATP binding directly, but rather inhibits the ability of ATP to transfer the allosteric effects of ATP binding.
Mutations designed to rigidify the R- and C-spines in the PKD increase allosteric activation
Because large protein segments can contribute to enzyme dynamics, loss-of-function mutations can be misleading if changes in amino acids that do not normally directly contribute to allostery nevertheless inhibit transfer of the allosteric signal. Therefore, we used gain-of-function mutations that are predicted to increase the transmission of hypothesized allosteric signals through the PKD to test our argument that conserved allosteric mechanisms of the PKD participate in the regulation of these enzymes. In kinases and pseudokinases, the assembly and disassembly of the R-spine and C-spine are important for the conformational dynamics that transmit the allosteric signal to the effector module (27, 46, 47). Therefore, we generated three individual mutants in these motifs, which are described below.
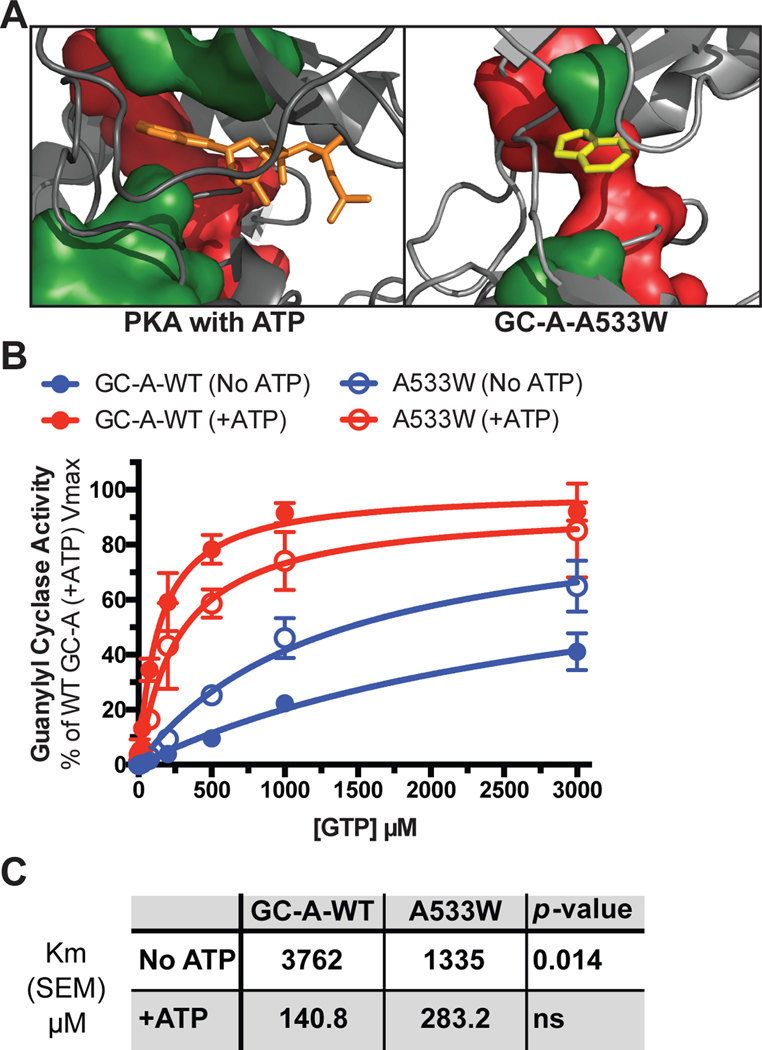
First, GC-A-A533W contains a tryptophan in place of a C-spine alanine in the N-terminal lobe of the PKD (green sidechain in Fig. 8A). The increased bulk of the tryptophan side chain is hypothesized to partially fill the adenine binding pocket of the ATP binding site, which is envisioned to partially mimic an ATP-bound state and rigidify the C-spine. Others used a similar Ala-to-aromatic mutation at this site to engineer a kinase that is catalytically dead but capable of allosterically activating binding partners (48). As predicted, GC-A-A533W reduced the Km in the absence of ATP compared to the Km for GC-A-WT (Fig. 8B, C). These data are consistent with activation of GC-A and GC-B by transmission of the allosteric signal through the C-spine as described for other kinases and pseudokinases.
Fig. 8. The GC-A-A533W mutation partially mimics the ATP-bound state of GC-A.
(A) A model demonstrating how ATP (orange) rigidifies the C-spine (green) in PKA and how the Trp in the GC-A-A533W mutant (yellow) docks into the same pocket. The R-spine is shown in red. (B) Substrate-velocity guanylyl cyclase assays in membranes from HEK293T cells expressing WT GC-A or GC-A-A533W in the presence of MgCl2, ANP, and increasing concentrations of GTP with or without ATP. n = 4 independent experiments. (C) A table showing the measured Michaelis constants and their associated p-values. Error bars represent the SEM.
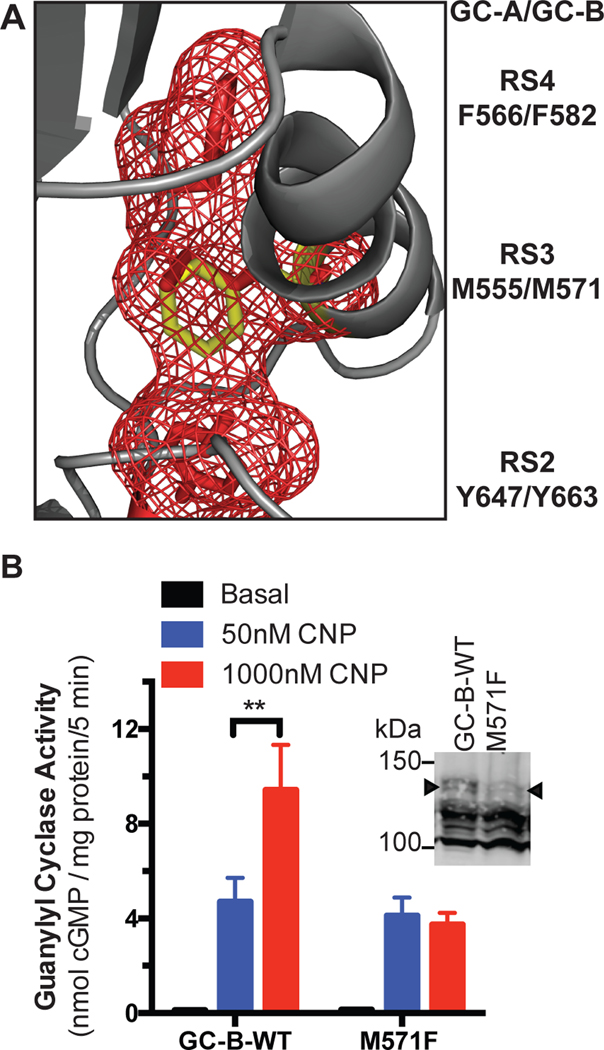
The second unique mutation, GC-B-M571F, lies at the R-spine 3 (RS3) position (Fig. 9A), which is the residue in both the αC-helix and the R-spine that, upon assembly of the R-spine, stabilizes the αC-helix in an active conformation (49, 50). GC-B-M571F is analogous to the BRAF L505F activating mutation (Table 1) (49). The R-spine residues in the N-lobe in GC-A and GC-B are similar to those found in the RAF kinases, which are located close to GC-A and GC-B in the phylogenetic tree of the human kinome (51). As predicted, we observed increased guanylyl cyclase activity at sub-saturating CNP concentrations for GC-B-M571F (Fig. 9B). The activity of the GC-B-M571F mutant was reduced but this was explained by reduced abundance of the completely processed form of the receptor (Fig. 9B, inset). These data suggest that the EC50 for CNP activation of GC-B is modulated by the conformation of the αC-helix and that the R-spine is involved in transmitting the NP binding signal to the catalytic domains of GC-A and GC-B.
Fig. 9. The GC-B-M571F mutation increases guanylyl cyclase activity at sub-saturating concentrations of CNP.
(A) A homology model showing the location of M571F in the C-helix. Met571 is located in the C-helix (grey) and contributes to the R-spine in position 3 (RS3). The R-spine residues RS2, RS3, and RS4 are shown in red. The M571F mutant is shown in yellow. (B) Guanylyl cyclase activity in membranes from HEK293T cells expressing GC-B-WT and GC-B-M571F in the presence of GTP, MgCl2, and ATP with the indicated concentations of CNP. Basal, no CNP. n = 3 independent experiments. Inset: Immunoblot for GC-B in the samples used for guanylyl cyclase assays. The arrowhead indicates the fully processed form of GC-B indicative of guanylyl cyclase activity (38). n = 3 independent experiments. Error bars represent the SEM.
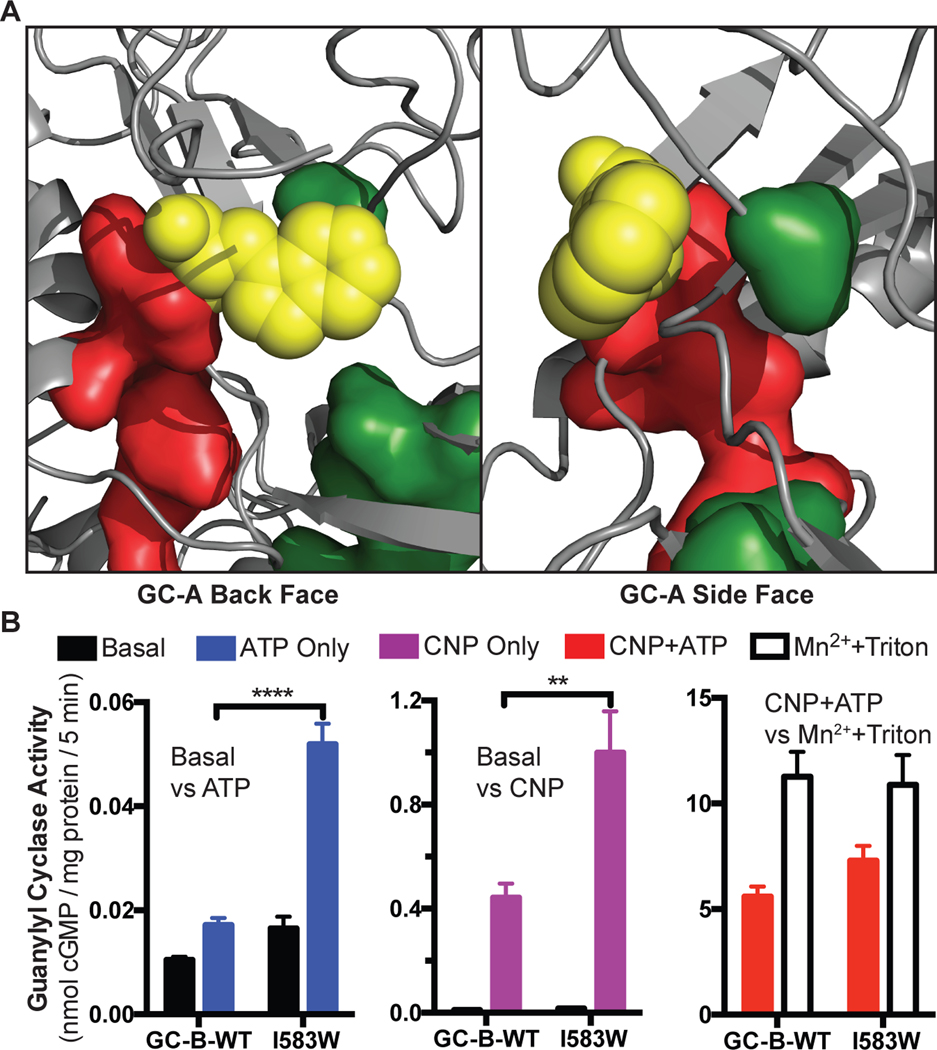
The third mutation is GC-B-I583W, which is located immediately C-terminal to the final R-spine residue (RS4) and is positioned to strengthen interactions between the R and C spines (Fig. 10A). We hypothesize that this residue rigidifies and increases the transmission of both the ATP and the NP binding signals to the catalytic domain. Consistent with this notion, we observed that the guanylyl cyclase activity of GC-B-I583W increased by more than 300% in the presence of ATP alone compared to GC-B-WT. Saturating concentrations of CNP in the absence of ATP increased the activity of the GC-B-I583W mutant almost 250% compared to GC-B-WT. However, activity measured for GC-B-I583W in the presence of saturating concentrations of ATP and CNP only mildly increased to 130% of GC-B-WT activity (Fig. 10B). Together, these data suggest that R-spines and the C-spines are present in GC-A and GC-B and function as allosteric regulatory modules as observed for bona fide kinases and pseudokinases.
Fig. 10. The GC-B-I583W mutation increases stimulation by either ATP or ANP alone.
(A) A homology model of GC-A shows the location of the GC-A-V567W substitution that is analogous to the GC-B-I583W substitution. Yellow spheres represent the substituted Trp. This model shows how the Trp bridges the R-spine (red) and C-spine (green) to increases the activity of GC-B in the absence of either CNP or ATP. (B) Guanylyl cyclase activity of membranes from HEK293T cells expressing GC-B-WT or GC-B-I583W in the presence of GTP and MgCl2, under the indicated conditions. For Mn2++Triton conditions, MnCl2 was substituted for MgCl2. n = 3 independent experiments. Error bars represent the SEM.
Discussion
PKDs are highly conserved from nematodes to humans in the receptor GC family (52, 53), and phylogenetic data indicate that the PKD and GC domains coevolved (54). Despite being the first PKDs to be identified (31, 55), the exact function of these domains in GCs has yet to be determined (54). Initial studies by Chinkers and Garbers reported that removal of the PKD from GC-A led to maximal activity of the enzyme in the absence ANP or ATP. Hence, they suggested that the PKD represses the GC domain (56). Subsequently, groups led by Sharma and DeLean reported that ATP binds to the PKD, but neither group determined exactly where ATP binds and if it binds to the highly conserved ATP binding site that the PKDs share with catalytically kinase domains (25, 26). The Van Den Akker group working on purified hinge-catalytic domain of GC-A (23), and our group working on full-length GC-A and GC-B (24), reported evidence of an additional allosteric site in the guanylyl cyclase catalytic domain, which results in positive cooperativity or linear kinetics in the absence or presence of ATP, respectively. Thus, with multiple ATP-binding sites it has been particularly challenging to determine which effect of ATP is due to engagement of which ATP-binding site. Here, we used strategic loss-of-function coupled to gain-of-function mutations in the PKDs of GC-A and GC-B to demonstrate that these domains are conserved to allosterically stimulate catalytic activity in a manner consistent with the known ability of pseudokinase domains to allosterically affect other enzymatic activities (57).
ATP in broken cell preparations stimulates GC-linked receptors by serving as a source of phosphate for the receptor phosphorylation by an unknown kinase as well as by binding to an allosteric site that decreases the Michaelis constant (Km) of the receptor (36, 58, 59). The PKDs of GC-A and GC-B lack the catalytic aspartate that is responsible for deprotonation of the substrate hydroxyl group, but other kinase domains lacking the same residue retain small amounts of phosphotransferase activity (34, 35). Thus, we investigated whether the PKDs in GC-A and GC-B contain phosphotransferase activity based on conservation of the canonical protein kinase domain. However, inactivating mutations in functionally critical residues failed to reduce the stoichiometry of phosphorylation of GC-A. Hence, we were unable to test the hypothesis suggesting that the PKDs of GC-A and GC-B possess intrinsic autophosphorylating protein kinase activity. Nonetheless, from these experiments, we can conclude that if GC-A and GC-B do contain phosphotransferase activity, they do not adhere to canonical kinase mechanisms because they show no requirement for three conserved residues (corresponding to Lys72, Asp166 and Asp184 in PKA) that are critical for phosphate transfer in most protein kinases.
Results from our GC assays were consistent with a critical role for ATP binding to the PKD in the allosteric stimulation of GC activity. It is unknown if the PKD retains its repressive function on the catalytic domain or if any repressive function can be relieved in the absence of the extracellular and transmembrane domains, so the intracellular constructs of GC-A used in our ATP-agarose assays may exist in a form that has even weaker affinity for ATP than the already weak EC50 of approximately 0.1 mM for ATP on the full-length enzymes. However, with intracellular concentrations of ATP in the 1–10 millimolar range, weak affinity is sufficient to elicit full allosteric activity of the PKDs of GC-A and GC-B. Because we observed little or no difference in NP-induced activation of wild-type or mutant enzymes but observed large decreases in the activation of the mutant enzymes by ATP, we conclude that the reduced activity of the mutant enzymes was due to reduced interactions with ATP, not due to destabilization of the domain structure. Regarding the activating mutations, we observed that GC-A-A533W did not completely mimic the ATP-bound state. This is not surprising because the full effect of ATP binding to kinases includes electrostatic interactions involving the triphosphate tail that would not be accounted for by this tryptophan substitution. Furthermore, these proteins are known to be highly dynamic, so we would not expect the tryptophan substitution to completely fill the adenine binding pocket, thus leaving the possibility for ATP binding despite a partially occluded active site. Because intracellular concentrations of ATP are generally high, we do not believe ATP concentrations are likely to be regulatory in this process. In contrast, we suggest that these ATP-dependent allosteric processes are required to reduce the Km of the enzymes to physiologic GTP concentrations of 0.1mM as a result of NP binding to the extracellular domain.
ATP also stimulates the heat-stable enterotoxin receptor, GC-C, and the retinal guanylyl cyclase, GC-E. Both of these receptors contain PKDs that lack the catalytic base but retain the invariant lysine (Lys535 in GC-A) and the Mg2+-chelating aspartate (Asp646 in GC-A) and are known to be activated by ATP in guanylyl cyclase assays mimicking physiologic conditions (32, 43, 60, 61). Furthermore, both the entire R-spine and the C-lobe portion of the C-spine are conserved in GC-C and GC-E. Substrate-velocity assays for GC-C revealed that the Vmax of the enzyme increased under both basal and peptide-stimulated conditions with the addition of ATP-γ-S with no change in Km as observed with GC-A and GC-B (62). This subtle difference in allosteric regulation is not surprising because the PKD of GC-C is not phosphorylated and cannot substitute for the PKD of GC-A in chimeric GC constructs whereas the PKDs of GC-A and GC-B can substitute for each other (63). Additionally, the PKDs coevolved with the corresponding GC domains (54); therefore, the PKD of GC-C evolved specific interactions to activate its own GC domain and not the GC domain in GC-A and GC-B. This suggests that the mechanisms by which kinases and pseudokinases allosterically activate their partner proteins are used to transfer activation signals from the PKDs to the GC domains in the entire mammalian membrane GC receptor family, not only for GC-A and GC-B, although the exact mechanisms of allosteric signal transfer may differ between proteins.
Allostery in protein kinases occurs through changes in the dynamics of the protein that begin with large-scale motions involving key motifs like the R- and C-spines that are assembled and ultimately transition to more ordered, small-scale elements within the protein (27, 46, 47). Because many amino acids are involved in stabilizing the overall structure of a given protein, mutations in far-reaching segments of the protein may have negative effects on allosteric regulation through their effects on the overall stability and dynamics of the protein. As such, we emphasized mutations that are unlikely to impair the overall stability of the PKD and would promote allosteric functions in a manner consistent with what is known about other kinase domains and PKDs. We designed activating mutations in GC-A and GC-B that mimic allosteric functions in known kinases and pseudokinases. This led to a model wherein the PKDs of GC-A and GC-B integrate allosteric inputs from ATP and NPs and provide a single, unified output to the catalytic domain based on the allosteric dynamics of the PKD. We hypothesize that in the basal state the PKD is dynamic, nonsynchronous, and has large-scale motions that preclude the catalytic domain from adopting the most active conformation. In the presence of both NP and ATP, the PKD transitions to more small-scale motions, thus increasing the probability that the catalytic domain will adopt the most active conformation. Future mechanistic studies describing how the R-spine and C-spine interact with each other and are assembled in GC-A and GC-B may explain how known mutations activate or inactivate membrane GCs (32, 38, 64–66).
Materials and Methods
Reagents
The cyclic GMP radioimmunoassay kit was from Perkin Elmer, and the cGMP Direct ELISA kit was from Enzo. NPs were from Sigma Aldrich. Protease inhibitors and microcystin were from Roche and Cayman Chemical Company, respectfully. 8-azido-ATP was from Jena Biosciences. Innova Biosciences γ-linked ATP-agarose was purchased from Novus Biologicals. The pSVL-GC-A-Δkin plasmid expressing the PKD deleted version of rat GC-A and the pSVL-FLAG-GC-A-INT plasmid expressing a soluble rat GC-A intracellular domain were kind gifts from Dr. Michael Chinkers at the University of South Alabama (42).
Homology modeling of the PKD of GC-A and alignments of GC-A and GC-B
A BLAST search identified the Src-family kinases as possessing some of the highest sequence similarity with the PKD sequence among human protein kinases. We therefore used the X-ray structure of the Src-family kinase LCK (PDB ID: 3LCK) to create a homology model of the PKD, using the SWISS-MODEL server. The initial homology model spanned residues Gly516 in the putative G-loop of the PKD, through Phe772 in the terminal alpha helix of the domain (helix αI). The homology model is missing what would be the first beta strand of the kinase domain (preceding the G-loop), due to poor sequence homology in this region. This strand is conserved in eukaryotic protein kinases, and we noted that the absence of this segment in the PKD model leaves a hydrophobic surface exposed on the top of the N-terminal lobe of the PKD, comprised of residues Phe521, Val534, Tyr527, Cys577, and Leu579, which are conserved in GC-A homologs. To create a putative model for this N-terminal segment, which includes the known phosphorylation sites, we manually docked the corresponding segment from LCK (residues 239–253) into the homology model of the PKD using PyMOL and mutated the sequence to match that of the PKD. This speculative model of the N-terminal segment positions the regulatory phosphorylation sites immediately N-terminal to the PKD, on the back surface in the vicinity of theαC-helix, which is an allosteric hotspot in protein kinases. To generate the alignment (Fig. 2) we aligned the structures of PKA, BRAF, and LCK (PDBs: 1ATP, 1UWH, 3LCK, respectively) with the homology model of GC-A’s PKD using the align function in PyMol. The sequence for the PKD of GC-B was aligned to GC-A using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/).
Cell culture and transfections
HEK293T cells were maintained and transfected using the calcium and phosphate method as previously described (67). Cos7 cells were maintained as previously described (56) and transfected using Lipofectamine 2000 from Invitrogen.
Guanylyl cyclase assays
Cos7 cells were used when pSVL expression plasmids were used because proteins encoded by pSVL plasmids express better in Cos7 cells than in HEK293T cells. HEK293T cells were used for all other experiments. Ten-cm plates of cells were washed twice with cold PBS and scraped into 600μl of phosphatase inhibitor buffer (25mM HEPES pH 7.4, 20% glycerol, 50mM NaCl, 50mM NaF, 2mM EDTA, 0.5μM microcystin, 1X Roche cOmplete EDTA-free Protease Inhibitors). Cells were lysed by sonication at 40% power (Misonix Sonicator XL) for 5 seconds, then the insoluble membrane fraction was pelleted by centrifugation at 25,000 x g for 15 minutes. The supernatant was aspirated and membranes washed with 400μl phosphatase inhibitor buffer before being resuspended in phosphatase inhibitor buffer for the GC assay. Activity assays were performed as described previously (38) using 20μl of prepared crude membranes, 20μl of a 5X activation mix containing the divalent metal (5mM MgCl2 or MnCl2 at reaction volume) as well as any other activators used (1mM ATP, 1μM ANP, 1μM CNP, or 1% v/v Triton X-100 at reaction volume) and adding 60μl of prewarmed reaction cocktail to final reaction conditions of 25mM HEPES pH 7.4, 50mM NaCl, 500mM isobutyl-methyl-xanthine, 0.5μM microcystin, 1mM EDTA, 0.1% BSA, and 5mM creatine phosphate and 0.1μg/ml creatine kinase as a nucleotide triphosphate regeneration system. If not otherwise indicated, GTP concentrations were 0.1 mM. Substrate velocity assays used GTP concentrations ranging from 0 – 3mM. Time course assays used timepoints at 10 seconds, 2 minutes, and 10 minutes. Cyclic GMP content in single-substrate concentration assays was determined by radioimmunoassay (56) or by ELISA by diluting NaOAc-buffered guanylyl cyclase assay samples in 0.1N HCl and following the manufacturer’s instructions. Cyclic GMP content in substrate-velocity assays was determined by radioimmunoassay as previously described (24) or by purification followed by ELISA estimation of cGMP concentrations. Substrate Velocity assays tested by ELISA were terminated in 110mM ZnOAc and 110 mM Na2CO3 and purified on acidified alumina as previously described (68) with the exception of eluting in 3ml 200mM ammonium formate to separate ATP and GTP from cGMP. Eluent was used undiluted in ELISA measurements according to manufacturer’s instructions with the exception of being performed in 200mM ammonium formate rather than 0.1N HCl. Data were normalized to a control value [Vmax for WT GC-A (+ATP)] in Fig 8 due to variations in transfection efficiency. To remove this confounding error associated with transfection efficiency and preserve all differences due to the treatments or mutations we expressed all activity data in Fig 8B as a percentage of the Vmax for GC-A-WT (+ATP).
Immunoprecipitations, SDS-PAGE, and gel staining
Transfected cells from the indicated number of 10-cm plates were lysed in 1ml per 10-cm plate of immunoprecipitation buffer (58) containing protease and phosphatase inhibitors (50mM HEPES pH 7.4, 100mM NaCl, 50mM NaF, 10mM NaH2PO4, 2mM EDTA, 1% NP-40, 0.5% deoxycholic acid, 0.1% SDS, 0.5μM microcystin, 1X Roche cOmplete EDTA-free Protease Inhibitors), aliquots of pre-cleared lysates were taken for loading controls, and the indicated proteins were immunoprecipitated with 2μl of rabbit 6325 antiserum (10) per 10-cm plate for GC-A proteins or 2μl of rabbit 6327 antiserum (11) per 10-cm plate for GC-B proteins and 25μl of a 50% slurry of Protein-A-agarose beads (Repligen) per 10-cm plate overnight. The beads were washed three times with immunoprecipitation buffer lacking NaCl. After boiling for 5 minutes in 2X reducing sample buffer, samples were fractionated on 8% resolving gels. The gel was first stained with ProQ-Diamond (Molecular Probes) and then stained with SYPRO Ruby protein stain (Molecular Probes) as previously described (38). Lysates used for full length GC-A or GC-B western blots were fractionated on 8% resolving gels, and lysates used for β-actin or soluble GC-A intracellular domains were fractionated on 10% resolving gels.
Western blotting
Resolving gels were blotted to an Immobilon-FL PVDF membrane (Millipore). Membranes were blocked with Odyssey Blocking Buffer (LiCor Biosciences) diluted 1:1 with PBS and probed with either a 1:5000 dilution of rabbit 6325 antiserum for GC-A western blots or rabbit 6327 antiserum for GC-B western blots followed by 1:15,000 goat anti-rabbit IRDye 680-conjugated secondary antibody (LiCor Biosciences). FLAG western blots were probed using a 1:5000 dilution of FLAG-M2 monoclonal antibody (Sigma-Aldrich) followed by 1:15000 goat anti-mouse IRDye 800-conjugated secondary antibody (LiCor Biosciences). Actin western blots were probed using a 1:2000 dilution of a monoclonal mouse β-actin antibody (Sigma-Aldrich) followed by 1:15000 goat anti-mouse IRDye 800-conjugated secondary antibody. All western blots were visualized on a LiCor instrument as described previously (69).
Densitometry and ratio calculations
Images from gel staining and western blotting were quantified in LiCor ImageStudio. Ratios of arbitrary units were calculated by dividing the ProQ-Diamond signal by the SYPRO Ruby signal from the same band on the same gel.
8-Azido-2’/3’-Biotinyl-ATP synthesis and binding
Biotin was esterified to the ribose ring of 8-azido-ATP similar to the method used by Schafer and de Lean with the following modifications (26, 70): A 2.5-fold molar excess of carbonyldiimidazole over biotin was used to activate the biotin carboxyl group, and 8-azido-ATP was added as a 10mM stock in buffer as supplied by Jena Biosciences with a 22.5-fold molar excess of biotin over 8-azido-ATP. The reaction was allowed to incubate until precipitates were clarified into solution. No further reaction was observed after this point. The completed reaction was purified by HPLC using a 1.5×15cm column of AGMP-1 (anion exchange resin, Bio-Rad) and a concave-upward gradient of trifluoroacetic acid from 3–300mM at 3ml/minute over 60 minutes. The eluted nucleotides were detected using a Beckman 166 UV detector set at 280nm. The biotinylated product eluted at 56.1 minutes and was collected and dialyzed against ultrapure water in 500 MWCO dialysis tubing at 40C to remove trifluoroacetic acid. After dialysis, samples were lyophilized and stored at −20ºC until use. Unreacted 8-azido ATP was collected (elution time 48.1 minutes), dialyzed as above, and used for further rounds of synthesis and purification of the biotinylated product. By analysis of HPLC peak intensities, the reactions yielded 40% product. Membranes were prepared from transfected HEK293T cells as above and resuspended in a binding buffer consisting of 50mM HEPES pH 7.4, 100mM NaCl, 50mM NaF, 0.5μM CNP, 20% glycerol, 2mM MgCl2, 1μM Microcystin-LR, 1x Roche Complete Protease Inhibitors, and with or without 1mM ATP. Samples were incubated at room temperature 1 hour to allow hormone binding and competitive ATP binding, then 100μM 8-azido-2’/3’-biotinyl-ATP was added. Samples were vortexed briefly and incubated on ice for 10 minutes before crosslinking in a Stratalinker 2400 for 3 minutes. The samples were centrifuged at 25K x g for 15 minutes at 4ºC to pellet the membranes, supernatants were aspirated, and pellets were solubilized in 1ml immunoprecipitation buffer. Solubilized samples were immunoprecipitated overnight using 20μl of FLAG-M2-agarose beads, fractionated by SDS-PAGE, and samples were split to run on two identical 8% resolving gels. Both gels were transferred to Immobilon-FL PVDF membrane and blocked with Odyssey Blocking Buffer diluted 1:1 with PBS. One blot detected total protein abundance by blotting against the N-terminal FLAG epitope as described above, and one-blot detected biotin using a 1:2000 LiCor IRDye-800 Streptavidin in blocking buffer plus 0.15% Tween-20 and 0.08% SDS.
ATP-agarose binding assay
Soluble FLAG-tagged intracellular domain samples of wild-type GC-A were prepared by detaching transiently transfected Cos7 cells from four 10-cm plates per construct using PBS supplemented with 5mM EDTA for 10 minutes at 37ºC. Total lysate controls were prepared by solubilizing one 10-cm plate in RIPA buffer as with the immunoprecipitations. Intact cells in PBS + EDTA were pelleted by centrifugation at 300xg for 3 minutes. Supernatants were aspirated and the cell pellet resuspended in ATP-agarose interaction buffer (50mM HEPES pH 7.4, 150mM NaCl, 10mM MgCl2). Cells were lysed by sonication for 10 seconds at 40% power. The insoluble fraction was pelleted by centrifugation at 25K x g, and the soluble fraction used for further analysis. Endogenous ATP from cells was removed using Pall Nanosep 10K MWCO spin filters to retain our protein of interest but elute ATP in the flow through. Successive rounds of concentration and dilution in the same ATP-agarose interaction buffer were used until 99% of the endogenous ATP had been removed. Buffer exchanged soluble fraction was incubated with ATP-agarose resin (effective ATP concentration = 250μM) with or without the addition of 1mM ATP for 90 minutes at 4ºC with rotation. Agarose beads were briefly washed with ATP-agarose interaction buffer prior to boiling in 2x reducing sample buffer for fractionation by SDS page. The appropriate band was verified by western blotting against the GC-A C-terminus in whole cell lysates and soluble fraction of GC-A transfected cells where it is expected to be present, and in the insoluble fraction of GC-A transfected cells or lysates from GFP-transfected cells where it is not expected to be present. Buffer exchange controls to demonstrate the retention of the GC-A intracellular constructs were further verified by western blotting against the N-terminal FLAG epitope. The band was detected at 60 kDa in agreement with its published molecular weight (42).
Statistical analysis
All statistical analyses were performed in Graphpad Prism. Differences among measured values in groups of two were analyzed by unpaired t-test. Differences among measured values in groups of three or more were analyzed first by ANOVA or two-way ANOVA (if multiple variables were present in the experiment) to determine if any significant differences are present in the data. We then computed specific p-values of any significant differences using Tukey’s test to correct for multiple comparisons. p < 0.05 were considered significant. *, **, and *** indicate statistical significance at p<0.05, p<0.01, and p<0.005 respectively.
Supplementary Material
Fig. S1. 8-azido-2’/3’-biotinyl-ATP binding.
Fig. S2. ATP-agarose interaction assay.
Acknowledgements:
We thank Dr. Gianluigi Veglia (University of Minnesota) and Dr. Laurinda Jaffe (University of Connecticut Health Center) for expert discussions and for constructive comments on this manuscript.
Funding:
This work was funded by NIH grants T32AR050938 to ABE and NIHR01GM098309 to LRP. Grants from the University of Minnesota of Academic Health Center for Faculty Research Development, Office of the Vice President for Research Grants, and the Minnesota Partnership for Biotechnology and Medical Genomics Grant, Fund for Science and Hormone Receptor Fund to LRP also support this work.
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability:
All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References and Notes
- 1.Kuhn M.2016. Molecular Physiology of Membrane Guanylyl Cyclase Receptors. Physiol Rev 96: 751–804. [DOI] [PubMed] [Google Scholar]
- 2.Potter LR 2011. Natriuretic Peptide Metabolism, Clearance and Degradation. The FEBS Journal 278: 1808–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Potter LR 2011. Guanylyl cyclase structure, function and regulation. Cellular signalling 23: 1921–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duda T, Yadav P, and Sharma RK. 2011. Allosteric modification, the primary ATP activation mechanism of atrial natriuretic factor receptor guanylate cyclase. Biochemistry 50: 1213–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moyes AJ, Khambata RS, Villar I, Bubb KJ, Baliga RS, Lumsden NG, Xiao F, Gane PJ, Rebstock AS, Worthington RJ, Simone MI, Mota F, Rivilla F, Vallejo S, Peiró C, Sánchez Ferrer CF, Djordjevic S, Caulfield MJ, MacAllister RJ, Selwood DL, Ahluwalia A, and Hobbs AJ. 2014. Endothelial C-type natriuretic peptide maintains vascular homeostasis. J Clin Invest 124: 4039–4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Potter LR, Abbey-Hosch S, and Dickey DM. 2006. Natriuretic peptides, their receptors, and cyclic guanosine monophosphate-dependent signaling functions. Endocrine reviews 27: 47–72. [DOI] [PubMed] [Google Scholar]
- 7.Schulz S, Green CK, Yuen PS, and Garbers DL. 1990. Guanylyl cyclase is a heat-stable enterotoxin receptor. Cell 63: 941–948. [DOI] [PubMed] [Google Scholar]
- 8.Yoder AR, Stone MD, Griffin TJ, and Potter LR. 2010. Mass spectrometric identification of phosphorylation sites in guanylyl cyclase a and B. Biochemistry 49: 10137–10145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogawa H, Qiu Y, Ogata CM, and Misono KS. 2004. Crystal structure of hormone-bound atrial natriuretic peptide receptor extracellular domain: rotation mechanism for transmembrane signal transduction. J Biol Chem 279: 28625–28631. [DOI] [PubMed] [Google Scholar]
- 10.Potter LR, and Hunter T. 1998. Phosphorylation of the kinase homology domain is essential for activation of the A-type natriuretic peptide receptor. Molecular and cellular biology 18: 2164–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Potter LR, and Hunter T. 1998. Identification and characterization of the major phosphorylation sites of the B-type natriuretic peptide receptor. The Journal of biological chemistry 273: 15533–15539. [DOI] [PubMed] [Google Scholar]
- 12.Schroter J, Zahedi RP, Hartmann M, Gassner B, Gazinski A, Waschke J, Sickmann A, and Kuhn M. 2010. Homologous desensitization of guanylyl cyclase A, the receptor for atrial natriuretic peptide, is associated with a complex phosphorylation pattern. FEBS J 277: 2440–2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoder AR, Robinson JW, Dickey DM, Andersland J, Rose BA, Stone MD, Griffin TJ, and Potter LR. 2012. A Functional Screen Provides Evidence for a Conserved, Regulatory, Juxtamembrane Phosphorylation Site in Guanylyl Cyclase A and B. PLoS ONE 7: e36747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Potter LR, and Garbers DL. 1992. Dephosphorylation of the guanylyl cyclase-A receptor causes desensitization. The Journal of biological chemistry 267: 14531–14534. [PubMed] [Google Scholar]
- 15.Potter LR 1998. Phosphorylation-dependent regulation of the guanylyl cyclase-linked natriuretic peptide receptor B: dephosphorylation is a mechanism of desensitization. Biochemistry 37: 2422–2429. [DOI] [PubMed] [Google Scholar]
- 16.Shuhaibar LC, Egbert JR, Edmund AB, Uliasz TF, Dickey DM, Yee SP, Potter LR, and Jaffe LA. 2016. Dephosphorylation of juxtamembrane serines and threonines of the NPR2 guanylyl cyclase is required for rapid resumption of oocyte meiosis in response to luteinizing hormone. Dev Biol 409: 194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson JW, Zhang M, Shuhaibar LC, Norris RP, Geerts A, Wunder F, Eppig JJ, Potter LR, and Jaffe LA. 2012. Luteinizing hormone reduces the activity of the NPR2 guanylyl cyclase in mouse ovarian follicles, contributing to the cyclic GMP decrease that promotes resumption of meiosis in oocytes. Developmental biology 366: 308–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Egbert JR, Shuhaibar LC, Edmund AB, Van Helden DA, Robinson JW, Uliasz TF, Baena V, Geerts A, Wunder F, Potter LR, and Jaffe LA. 2014. Dephosphorylation and inactivation of NPR2 guanylyl cyclase in granulosa cells contributes to the LH-induced decrease in cGMP that causes resumption of meiosis in rat oocytes. Development 141: 3594–3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shuhaibar LC, Robinson JW, Vigone G, Shuhaibar NP, Egbert JR, Baena V, Uliasz TF, Kaback D, Yee SP, Feil R, Fisher MC, Dealy CN, Potter LR, and Jaffe LA. 2017. Dephosphorylation of the NPR2 guanylyl cyclase contributes to inhibition of bone growth by fibroblast growth factor. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson JW, Egbert JR, Davydova J, Schmidt H, Jaffe LA, and Potter LR. 2017. Dephosphorylation is the mechanism of fibroblast growth factor inhibition of guanylyl cyclase-B. Cell Signal 40: 222–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otto NM, McDowell WG, Dickey DM, and Potter LR. 2017. A Glutamate-Substituted Mutant Mimics the Phosphorylated and Active Form of Guanylyl Cyclase-A. Mol Pharmacol 92: 67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Potter LR 2011. Regulation and therapeutic targeting of peptide-activated receptor guanylyl cyclases. Pharmacology & Therapeutics 130: 71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pattanaik P, Fromondi L, Ng KP, He J, and van den Akker F. 2009. Expression, purification, and characterization of the intra-cellular domain of the ANP receptor. Biochimie 91: 888–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson JW, and Potter LR. 2012. Guanylyl cyclases a and B are asymmetric dimers that are allosterically activated by ATP binding to the catalytic domain. Science signaling 5: ra65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burczynska B, Duda T, and Sharma RK. 2007. ATP signaling site in the ARM domain of atrial natriuretic factor receptor guanylate cyclase. Mol Cell Biochem 301: 93–107. [DOI] [PubMed] [Google Scholar]
- 26.Joubert S, Jossart C, McNicoll N, and De Lean A. 2005. Atrial natriuretic peptide-dependent photolabeling of a regulatory ATP-binding site on the natriuretic peptide receptor-A. FEBS J 272: 5572–5583. [DOI] [PubMed] [Google Scholar]
- 27.Kornev AP, Haste NM, Taylor SS, and Eyck LF. 2006. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc Natl Acad Sci U S A 103: 17783–17788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim J, Ahuja LG, Chao FA, Xia Y, McClendon CL, Kornev AP, Taylor SS, and Veglia G. 2017. A dynamic hydrophobic core orchestrates allostery in protein kinases. Sci Adv 3: e1600663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hammarén HM, Ungureanu D, Grisouard J, Skoda RC, Hubbard SR, and Silvennoinen O. 2015. ATP binding to the pseudokinase domain of JAK2 is critical for pathogenic activation. Proc Natl Acad Sci U S A 112: 4642–4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeqiraj E, Filippi BM, Goldie S, Navratilova I, Boudeau J, Deak M, Alessi DR, and van Aalten DM. 2009. ATP and MO25alpha regulate the conformational state of the STRADalpha pseudokinase and activation of the LKB1 tumour suppressor. PLoS Biol 7: e1000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murphy JM, Zhang Q, Young SN, Reese ML, Bailey FP, Eyers PA, Ungureanu D, Hammaren H, Silvennoinen O, Varghese LN, Chen K, Tripaydonis A, Jura N, Fukuda K, Qin J, Nimchuk Z, Mudgett MB, Elowe S, Gee CL, Liu L, Daly RJ, Manning G, Babon JJ, and Lucet IS. 2014. A robust methodology to subclassify pseudokinases based on their nucleotide-binding properties. Biochem J 457: 323–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mishra V, Goel R, and Visweswariah SS. 2018. The regulatory role of the kinase-homology domain in receptor guanylyl cyclases: nothing ‘pseudo’ about it! Biochem Soc Trans. [DOI] [PubMed] [Google Scholar]
- 33.Potter LR 2005. Domain analysis of human transmembrane guanylyl cyclase receptors: implications for regulation. Frontiers in bioscience : a journal and virtual library 10: 1205–1220. [DOI] [PubMed] [Google Scholar]
- 34.Shi F, Telesco SE, Liu Y, Radhakrishnan R, and Lemmon MA. 2010. ErbB3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation. Proc Natl Acad Sci U S A 107: 7692–7697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ungureanu D, Wu J, Pekkala T, Niranjan Y, Young C, Jensen ON, Xu CF, Neubert TA, Skoda RC, Hubbard SR, and Silvennoinen O. 2011. The pseudokinase domain of JAK2 is a dual-specificity protein kinase that negatively regulates cytokine signaling. Nat Struct Mol Biol 18: 971–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Antos LK, and Potter LR. 2007. Adenine nucleotides decrease the apparent Km of endogenous natriuretic peptide receptors for GTP. American journal of physiology. Endocrinology and metabolism 293: E1756–1763. [DOI] [PubMed] [Google Scholar]
- 37.Knape MJ, Ballez M, Burghardt NC, Zimmermann B, Bertinetti D, Kornev AP, and Herberg FW. 2017. Divalent metal ions control activity and inhibition of protein kinases. Metallomics 9: 1576–1584. [DOI] [PubMed] [Google Scholar]
- 38.Dickey DM, Edmund AB, Otto NM, Chaffee TS, Robinson JW, and Potter LR. 2016. Catalytically Active Guanylyl Cyclase B Requires Endoplasmic Reticulum-mediated Glycosylation, and Mutations That Inhibit This Process Cause Dwarfism. J Biol Chem 291: 11385–11393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bryan PM, Smirnov D, Smolenski A, Feil S, Feil R, Hofmann F, Lohmann S, and Potter LR. 2006. A sensitive method for determining the phosphorylation status of natriuretic peptide receptors: cGK-Ialpha does not regulate NPR-A. Biochemistry 45: 1295–1303. [DOI] [PubMed] [Google Scholar]
- 40.Potthast R, Abbey-Hosch SE, Antos LK, Marchant JS, Kuhn M, and Potter LR. 2004. Calcium-dependent dephosphorylation mediates the hyperosmotic and lysophosphatidic acid-dependent inhibition of natriuretic peptide receptor-B/guanylyl cyclase-B. The Journal of biological chemistry 279: 48513–48519. [DOI] [PubMed] [Google Scholar]
- 41.Joubert S, McNicoll N, and De Lean A. 2007. Biochemical and pharmacological characterization of P-site inhibitors on homodimeric guanylyl cyclase domain from natriuretic peptide receptor-A. Biochem Pharmacol 73: 954–963. [DOI] [PubMed] [Google Scholar]
- 42.Chinkers M, and Wilson EM. 1992. Ligand-independent oligomerization of natriuretic peptide receptors. Identification of heteromeric receptors and a dominant negative mutant. The Journal of biological chemistry 267: 18589–18597. [PubMed] [Google Scholar]
- 43.Jaleel M, Saha S, Shenoy AR, and Visweswariah SS. 2006. The kinase homology domain of receptor guanylyl cyclase C: ATP binding and identification of an adenine nucleotide sensitive site. Biochemistry 45: 1888–1898. [DOI] [PubMed] [Google Scholar]
- 44.Robinson MJ, Harkins PC, Zhang J, Baer R, Haycock JW, Cobb MH, and Goldsmith EJ. 1996. Mutation of position 52 in ERK2 creates a nonproductive binding mode for adenosine 5’-triphosphate. Biochemistry 35: 5641–5646. [DOI] [PubMed] [Google Scholar]
- 45.Iyer GH, Garrod S, Woods VL, and Taylor SS. 2005. Catalytic independent functions of a protein kinase as revealed by a kinase-dead mutant: study of the Lys72His mutant of cAMP-dependent kinase. J Mol Biol 351: 1110–1122. [DOI] [PubMed] [Google Scholar]
- 46.Kornev AP, Taylor SS, and Ten Eyck LF. 2008. A helix scaffold for the assembly of active protein kinases. Proc Natl Acad Sci U S A 105: 14377–14382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor SS, and Kornev AP. 2011. Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem Sci 36: 65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu J, Yu H, Kornev AP, Zhao J, Filbert EL, Taylor SS, and Shaw AS. 2011. Mutation that blocks ATP binding creates a pseudokinase stabilizing the scaffolding function of kinase suppressor of Ras, CRAF and BRAF. Proc Natl Acad Sci U S A 108: 6067–6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu J, Stites EC, Yu H, Germino EA, Meharena HS, Stork PJ, Kornev AP, Taylor SS, and Shaw AS. 2013. Allosteric activation of functionally asymmetric RAF kinase dimers. Cell 154: 1036–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor SS, Shaw AS, Kannan N, and Kornev AP. 2015. Integration of signaling in the kinome: Architecture and regulation of the αC Helix. Biochim Biophys Acta 1854: 1567–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Manning G, Whyte DB, Martinez R, Hunter T, and Sudarsanam S. 2002. The protein kinase complement of the human genome. Science 298: 1912–1934. [DOI] [PubMed] [Google Scholar]
- 52.Baude EJ, Arora VK, Yu S, Garbers DL, and Wedel BJ. 1997. The cloning of a Caenorhabditis elegans guanylyl cyclase and the construction of a ligand-sensitive mammalian/nematode chimeric receptor. J Biol Chem 272: 16035–16039. [DOI] [PubMed] [Google Scholar]
- 53.Lowe DG, Chang MS, Hellmiss R, Chen E, Singh S, Garbers DL, and Goeddel DV. 1989. Human atrial natriuretic peptide receptor defines a new paradigm for second messenger signal transduction. Embo J 8: 1377–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Biswas KH, Shenoy AR, Dutta A, and Visweswariah SS. 2009. The evolution of guanylyl cyclases as multidomain proteins: conserved features of kinase-cyclase domain fusions. J Mol Evol 68: 587–602. [DOI] [PubMed] [Google Scholar]
- 55.Singh S, Lowe DG, Thorpe DS, Rodriguez H, Kuang WJ, Dangott LJ, Chinkers M, Goeddel DV, and Garbers DL. 1988. Membrane guanylate cyclase is a cell-surface receptor with homology to protein kinases. Nature 334: 708–712. [DOI] [PubMed] [Google Scholar]
- 56.Chinkers M, and Garbers DL. 1989. The protein kinase domain of the ANP receptor is required for signaling. Science 245: 1392–1394. [DOI] [PubMed] [Google Scholar]
- 57.Eyers PA, and Murphy JM. 2016. The evolving world of pseudoenzymes: proteins, prejudice and zombies. BMC Biol 14: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Antos LK, Abbey-Hosch SE, Flora DR, and Potter LR. 2005. ATP-independent activation of natriuretic peptide receptors. The Journal of biological chemistry 280: 26928–26932. [DOI] [PubMed] [Google Scholar]
- 59.Foster DC, and Garbers DL. 1998. Dual role for adenine nucleotides in the regulation of the atrial natriuretic peptide receptor, guanylyl cyclase-A. J Biol Chem 273: 16311–16318. [DOI] [PubMed] [Google Scholar]
- 60.Bhandari R, Suguna K, and Visweswariah SS. 1999. Guanylyl cyclase C receptor: regulation of catalytic activity by ATP. Biosci Rep 19: 179–188. [DOI] [PubMed] [Google Scholar]
- 61.Yamazaki A, Yu H, Yamazaki M, Honkawa H, Matsuura I, Usukura J, and Yamazaki RK. 2003. A critical role for ATP in the stimulation of retinal guanylyl cyclase by guanylyl cyclase-activating proteins. J Biol Chem 278: 33150–33160. [DOI] [PubMed] [Google Scholar]
- 62.Gazzano H, Wu HI, and Waldman SA. 1991. Activation of particulate guanylate cyclase by Escherichia coli heat-stable enterotoxin is regulated by adenine nucleotides. Infect Immun 59: 1552–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koller KJ, de Sauvage FJ, Lowe DG, and Goeddel DV. 1992. Conservation of the kinaselike regulatory domain is essential for activation of the natriuretic peptide receptor guanylyl cyclases. Mol Cell Biol 12: 2581–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bartels CF, Bukulmez H, Padayatti P, Rhee DK, van Ravenswaaij-Arts C, Pauli RM, Mundlos S, Chitayat D, Shih LY, Al-Gazali LI, Kant S, Cole T, Morton J, Cormier-Daire V, Faivre L, Lees M, Kirk J, Mortier GR, Leroy J, Zabel B, Kim CA, Crow Y, Braverman NE, van den Akker F, and Warman M. L. a.. 2004. Mutations in the transmembrane natriuretic peptide receptor NPR-B impair skeletal growth and cause acromesomelic dysplasia, type Maroteaux. Am J Hum Genet 75: 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dickey DM, Otto NM, and Potter LR. 2017. Skeletal overgrowth-causing mutations mimic an allosterically activated conformation of guanylyl cyclase-B that is inhibited by 2,4,6,-trinitrophenyl ATP. J Biol Chem 292: 10220–10229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Robinson JW, Dickey DM, Miura K, Michigami T, Ozono K, and Potter LR. 2013. A human skeletal overgrowth mutation increases maximal velocity and blocks desensitization of guanylyl cyclase-B. Bone 56: 375–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Champion HC, Bivalacqua TJ, Hyman AL, Ignarro LJ, Hellstrom WJ, and Kadowitz PJ. 1999. Gene transfer of endothelial nitric oxide synthase to the penis augments erectile responses in the aged rat. Proceedings of the National Academy of Sciences of the United States of America 96: 11648–11652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Domino SE, Tubb DJ, and Garbers DL. 1991. Assay of guanylyl cyclase catalytic activity. Methods Enzymol 195: 345–355. [DOI] [PubMed] [Google Scholar]
- 69.Schreiner GF, and Protter AA. 2002. B-type natriuretic peptide for the treatment of congestive heart failure. Curr Opin Pharmacol 2: 142–147. [DOI] [PubMed] [Google Scholar]
- 70.Schäfer HJ, Coskun U, Eger O, Godovac-Zimmermann J, Wieczorek H, Kagawa Y, and Grüber G. 2001. 8-N(3)-3’-biotinyl-ATP, a novel monofunctional reagent: differences in the F(1)- and V(1)-ATPases by means of the ATP analogue. Biochem Biophys Res Commun 286: 1218–1227. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. 8-azido-2’/3’-biotinyl-ATP binding.
Fig. S2. ATP-agarose interaction assay.
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.