DNA and RNA sequencing of paired primary tumor/metastases from 381 patients with metastatic breast cancer shed light on enriched genomic alterations, subtype switching, and immune cell composition, and identified new candidate biomarkers of premature lethality.
Abstract
AURORA aims to study the processes of relapse in metastatic breast cancer (MBC) by performing multi-omics profiling on paired primary tumors and early-course metastases. Among 381 patients (primary tumor and metastasis pairs: 252 targeted gene sequencing, 152 RNA sequencing, 67 single nucleotide polymorphism arrays), we found a driver role for GATA1 and MEN1 somatic mutations. Metastases were enriched in ESR1, PTEN, CDH1, PIK3CA, and RB1 mutations; MDM4 and MYC amplifications; and ARID1A deletions. An increase in clonality was observed in driver genes such as ERBB2 and RB1. Intrinsic subtype switching occurred in 36% of cases. Luminal A/B to HER2-enriched switching was associated with TP53 and/or PIK3CA mutations. Metastases had lower immune score and increased immune-permissive cells. High tumor mutational burden correlated to shorter time to relapse in HR+/HER2− cancers. ESCAT tier I/II alterations were detected in 51% of patients and matched therapy was used in 7%. Integration of multi-omics analyses in clinical practice could affect treatment strategies in MBC.
Significance:
The AURORA program, through the genomic and transcriptomic analyses of matched primary and metastatic samples from 381 patients with breast cancer, coupled with prospectively collected clinical data, identified genomic alterations enriched in metastases and prognostic biomarkers. ESCAT tier I/II alterations were detected in more than half of the patients.
This article is highlighted in the In This Issue feature, p. 2659
INTRODUCTION
Despite an improvement in survival rates following advances in early detection and treatments, breast cancer remains one of the leading causes of cancer-related mortality among women (1). Metastases are the main cause of death for patients, highlighting the need for treatment strategies to avoid metastatic relapse and to improve the outcome of patients with de novo metastatic breast cancer (MBC). So far, progress in the treatment of MBC has mainly occurred through the conducting of empirical clinical trials in which patients are segregated by breast cancer subtyping, namely HR+/HER2−, HER2+, or triple-negative breast cancer (TNBC). Consequently, treatment decisions are dictated by these limited subtypes, and the lines of therapy rely on minimal biological data (e.g., presence or absence of hormone receptors, HER2 status). As an example, CDK 4/6 inhibitors are now routinely added to first- or second-line endocrine therapies for ER+/HER2− disease, dual HER2 blockade is combined with chemotherapy as first-line treatment of HER2+ breast cancer, and PD-1/PD-L1 checkpoint inhibitors are added to first-line chemotherapy in relapsed PD-L1+ TNBC (2). These treatment algorithms, while improving outcomes for some patients, have also led to marked increases in treatment costs, and contrast with growing knowledge of the molecular, genetic, and immunologic complexity of the disease, which can be captured by novel technologies such as targeted gene sequencing (TGS) of tissue and cell-free DNA (cfDNA).
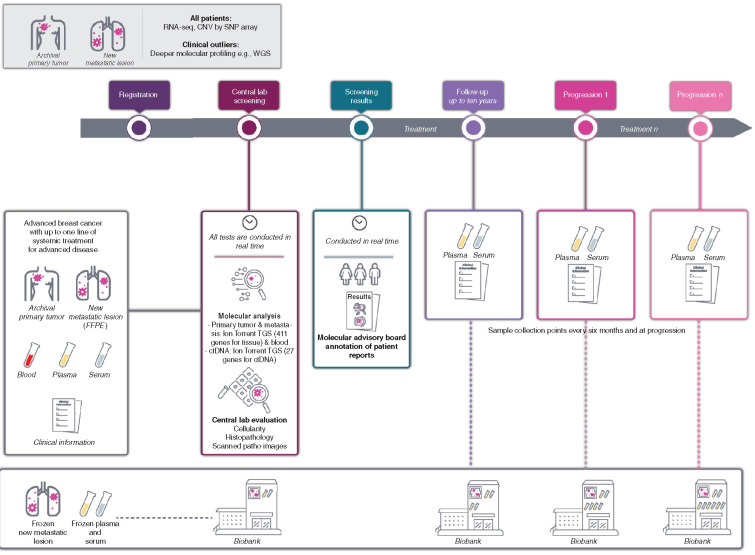
The Breast International Group (BIG) is conducting Aiming to Understand the Molecular Aberrations in Metastatic Breast Cancer (AURORA; NCT02102165), a molecular screening program that aims to improve the understanding of MBC through the extensive profiling of paired primary tumors and metastatic samples, as well as cfDNA extracted from plasma, collected from at least 1,000 patients with MBC (Fig. 1). The feasibility and logistics of this pan-European effort had been previously assessed in a pilot study that involved four centers in four European countries (3). Taking advantage of the large number of paired primary and metastatic samples obtained in the program, with the analyses described in this article we aimed to: (i) identify molecular alterations enriched in the early phases of metastatic disease; (ii) describe variations in gene expression between primary samples and their paired metastasis; (iii) correlate genomic and transcriptomic markers with outcome; (iv) evaluate the contribution of molecular profiling to the management of patients with MBC. The analyses were performed using genomic (TGS), transcriptomic, and clinical data from the first 381 patients included in AURORA. To our knowledge, this is the largest study to date collecting paired samples from patients with MBC, and the largest dataset of RNA sequencing (RNA-seq) in MBC.
Figure 1.
Study design. Illustration of the design of the AURORA molecular screening program including the baseline and longitudinal collections of samples as well as the clinical data. ctDNA, circulating tumor DNA.
RESULTS
By February 28, 2018, 381 patients had been included in 51 centers in nine European countries (Belgium, Germany, Iceland, Italy, Luxembourg, Spain, Sweden, Switzerland, United Kingdom). Two patients were excluded for violation of the eligibility criteria, leaving 379 patients for the current analysis. An additional four paired samples were excluded after a careful review of the clinical data suggested the possibility of unmatched primary tumors and metastases (e.g., due to the presence of a second primary breast or other tumor; Supplementary Fig. S1). On the basis of the IHC status of the primary tumor, 247 (65%) patients had HR+/HER2− breast cancer, 72 (19%) had TNBC, and 60 (16%) had HER2+ breast cancer. The majority of patients were treatment-naïve for MBC at inclusion (n = 274; 72%) but most had received some form of systemic therapy in the (neo)adjuvant setting. A sizable fraction of the cohort (n = 87; 23%) presented with de novo MBC and the metastasis site was sampled prior to any therapy in 54 cases (14% of the whole cohort). Liver was the most frequent site of metastatic biopsies (n = 152; 40%) followed by lymph nodes (n = 104; 27%), then skin and soft tissue (n = 52; 14%). Patients and tumor characteristics are provided in Supplementary Table S1 and sites of metastatic biopsies are illustrated in Supplementary Fig. S2.
The current analysis includes data from matched pairs from 242 patients with TGS, 152 patients with RNA-seq, and 67 patients with copy-number variations (CNV) by single nucleotide polymorphism (SNP) arrays. Details are available in the CONSORT diagram (Supplementary Figs. S1 and S3). Data from cfDNA are available for 99 patients, as this analysis only started later after a protocol amendment.
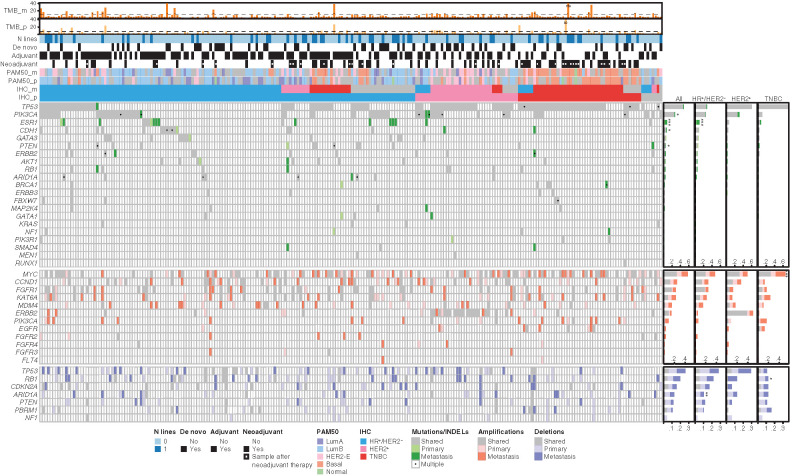
The first objective of AURORA is to study the genomic alterations driving metastatic relapse and progression of MBC. Driver genes potentially enriched in MBC when compared with primary tumors were identified in large studies by comparing genomic data from retrospectively collected samples with genomic data generated mostly from primary tumors. Compared with these cohorts, AURORA has a large number of primary/metastasis pairs collected for the majority of patients before any treatment for MBC or after only one line of therapy for some. This should allow the investigation of molecular alterations involved in metastatic relapse without the potential impact of therapy in the metastatic setting. Figure 2 illustrates, for the cohort of paired samples with TGS (n = 242), IHC subtypes, intrinsic subtypes (based on PAM50 derived from RNA-seq data), treatment history, tumor mutational burden (TMB), somatic single nucleotide variants (SNV), short indels and CNVs. To identify the most common cancer genes in the AURORA cohort, we applied the dN/dS algorithm that compares the normalized ratio of nonsynonymous to synonymous mutations, to quantify selection in cancer genomes. Using this approach, a total of 21 cancer genes were found to be under positive selection cohort-wise [in primary tumors (Supplementary Fig. S4A) and/or metastases (Supplementary Fig. S4B)]. Fourteen (67%) of these genes overlapped with those previously identified by the Martincorena dN/dS analysis of The Cancer Genome Atlas (TCGA) breast cohort (n = 702; ref. 4) and seven additional genes (ERBB3, ESR1, FBXW7, GATA1, KRAS, MEN1, NF1) were identified as significantly mutated (Supplementary Fig. S4C). There is evidence supporting these genes being drivers, and the differences we observe between the two cohorts likely reflect the unique composition of AURORA. The dN/dS analyses also provided evidence that somatic mutations in GATA1 (q = 0.02 for missense mutations in the primary tumors) and MEN1 (q = 0.01 for metastatic samples) are under positive selection. These are known cancer genes but have not been identified through the analysis of large series of primary and/or metastatic breast cancer to date. GATA1 encodes a transcription factor and has been associated with the induction of epithelial-to-mesenchymal transition in breast cancer (5). MEN1 is a tumor suppressor gene associated with multiple endocrine neoplasia 1 syndrome. MEN1 germline mutations are linked to an increase in breast cancer risk (6). The most prevalent point mutations in primary tumors and/or metastases were found in TP53 followed by PIK3CA, ESR1, CDH1, and GATA3. The most prevalent copy-number gains were in MYC, CCND1, FGFR1, KAT6A, MDM4 and ERBB2; the most prevalent losses were in TP53 and RB1.
Figure 2.
Repertoire of somatic gene alterations. Oncoplot of the relevant genomic alterations in the set of 242 patients with available Target Gene Sequencing (TGS) data for primary and metastatic samples. From top to bottom, the oncoplot includes three sections: tumor mutational burden (TMB), clinical data, and genomic alterations. TMB section shows the bar plots of TMB in primary and metastatic samples. Dashed lines refer to the TMB threshold used to define high-TMB patients based on the 90th percentile of the TMB distribution (corresponding to 8 for primary and 11 for metastatic samples). Clinical data section includes information about the number of treatment lines for metastatic disease, de novo metastatic disease, adjuvant and neoadjuvant therapy, and molecular subtype information in primary and metastatic samples by PAM50 and IHC. Genomic alterations are classified as shared (if present in both primary and metastatic samples), primary (private to primary sample), and metastatic (private to metastatic samples). Genomic alterations include driver mutations (single-nucleotide variants and insertions/deletions) in driver genes, amplifications in oncogenes, and deletions in tumor suppressor genes. On the right, the bar plots summarize, for each gene, the frequency of shared, private to primary, and private to metastatic events. The asterisks refer to genes showing significant difference in terms of alteration frequency in metastatic compared to primary samples (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Genes (SNVs) with significant positive selection on missense mutations and/or truncating substitutions on the dN/dS analysis are represented in the figure. We have included in the figure CNVs of genes known as breast cancer drivers.
We then compared the prevalence of molecular alterations from primary and metastatic samples found in our dataset with those enriched in the TCGA breast cancer cohort (7) that used whole-exome sequencing (WES) and the METABRIC cohort (8) that used TGS. We wanted to assess if the driver alterations putatively responsible for metastatic relapse were comparable in other series. We first identified the point mutations enriched in patients with a breast cancer relapse in TCGA and METABRIC by comparing the genomic landscape of patients without a relapse to those with a relapse (Supplementary Fig. S5A and S5B). TP53 mutations were more prevalent in AURORA whereas PIK3CA mutations were less prevalent (Supplementary Fig. S5C). These differences are possibly explained by the characteristics of AURORA that included only patients with a confirmed metastatic relapse and complete clinicopathologic annotation.
The analyses of paired primary–metastasis samples in AURORA allow us to directly examine changes emerging within each cancer over the course of its evolution to study the molecular alterations enriched in early MBC. The median TMB was higher in the metastatic samples when compared with the paired primary samples (P < 10−9; Fig. 2; Supplementary Fig. S6A). This finding is consistent with the fact that somatic mutation accumulation is a continuous process. The difference did not reach statistical significance for TNBC (Supplementary Fig. S6B), probably reflecting the fact that TNBC relapses more rapidly, so there is less time to acquire enough additional mutations within the scope of the small TGS panel to see a difference in most cases. We also looked at median TMB in the de novo and not de novo cases (Supplementary Fig. S6C and S6D). The median TMB in not de novo cases was higher when compared with de novo cases, and the difference was statistically significant (P = 0.046).
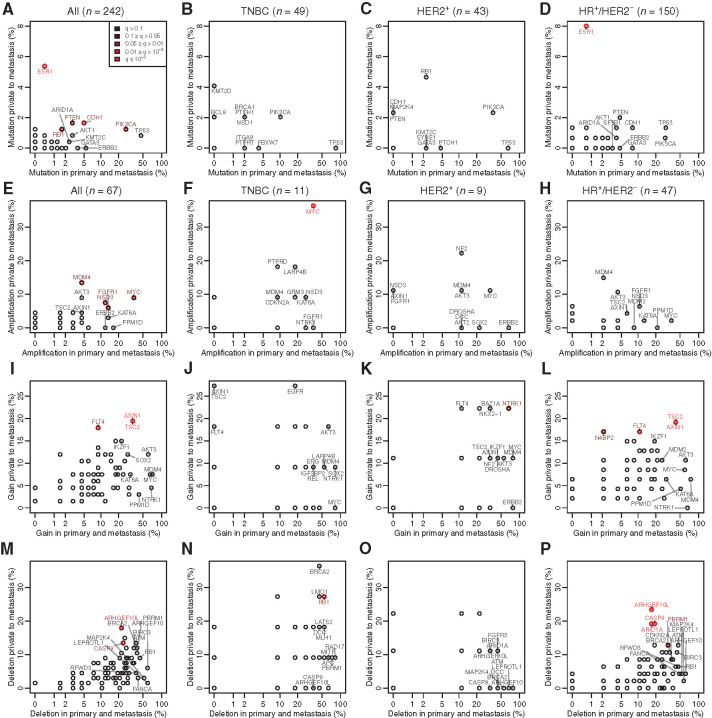
In the full population of patients with paired TGS data (n = 242), 88% of point mutations in the driver genes were shared between the primary sample and the metastasis (86% in HR+/HER2−, 93% in TNBC, and 87% in HER2+). At least one driver point mutation acquired in the metastases was identified in 10% of cases (10% in HR+/HER2−, 12% in HER2+, and 6% in TNBC, Supplementary Fig. S7A–S7D). At least one acquired CNV detected by ASCAT (n = 67) was found in 31% of cases (30% in HR+/HER2−, 43% in HER2+, and 27% in TNBC; Supplementary Fig. S7A–S7D). Point mutations in the following genes were enriched in the metastatic samples: ESR1, PTEN, CDH1, PIK3CA, and RB1 (Fig. 3A–D). In matched pairs of primary and metastatic samples (n = 67) analyzed by SNP arrays, copy number (CN) gains (including amplifications; Fig. 3E–L) of MDM4, MYC, NSD3, FGFR1, AXIN1, TSC2, FLT4, NTRK1, and N4BP2 and deletions (Fig. 3M–P) of ARHGEF10L, CASP9, RB1, ARID1A, and PBRM1 were more frequent in the metastatic samples. The same analyses were performed for the subset of patients with de novo MBC. Similar alterations were found between the primaries of de novo and not de novo patients. We did not observe in the metastasis of de novo patients the enrichment of alterations that we observed for non–de novo patients, in particular ESR1 mutations. However, the small number of patients in the de novo group does not allow drawing firm conclusions (Supplementary Figs. S8A–S8P, S9A–S9P, S10A–S10P, S11A–S11P).
Figure 3.
Comparison of the truncal aberrations with those private to the metastasis. All plots are on paired samples, each point representing the percentage of tumors with an aberration common between the primary tumor and the metastasis versus the percentage of tumors with an aberration found only in the metastasis. Mutations are shown (A, all subtypes; B, TNBC; C, HER2+; D, HR+/HER2−) as well as CN amplifications (normalize CN > 4; E, all subtypes; F, TNBC; G, HER2+; H, HR+/HER2−), gains (normalized CN > 1.5; I, all subtypes; J, TNBC; K, HER2+; L, HR+/HER2−) and deletions (normalized CN < 1.5; M, all subtypes; N, TNBC; O, HER2+; P, HR+/HER2−). The points are colored in function of their q-values, which assess whether a given aberration is more often private to the mutation than expected by the play of chance, corrected for multiple testing by panel.
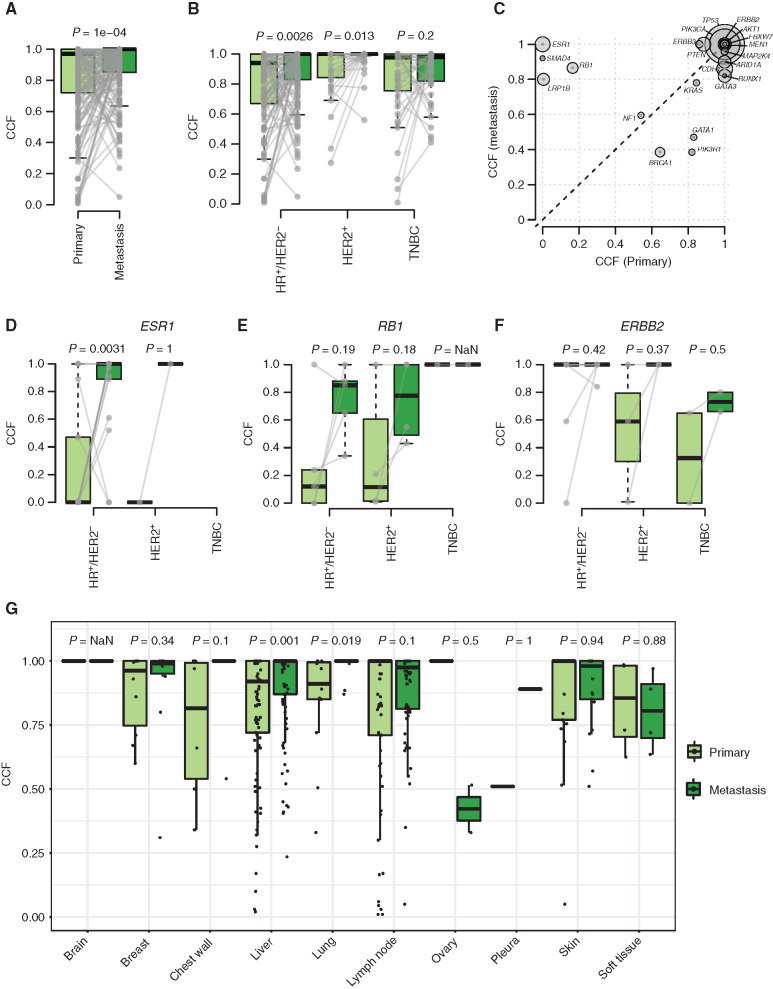
We then studied the evolution of the clonal composition from the primary tumors to their paired metastases (n = 242). We found an increase in the median cancer cell fraction (CCF) for point mutations between the primary samples and their paired metastases, suggesting an overall increase in clonality in metastatic versus primary samples (Fig. 4A). The increase was statistically significant for the HR+/HER2− and HER2+ subtypes but not TNBC (Fig. 4B). Genes with the most significant increase were ESR1, SMAD4, RB1, and LRP1B (Fig. 4C). An increase in clonality was seen in genes with potential clinical impact such as ESR1 in HR+/HER2− MBC (endocrine resistance), RB1 in HR+/HER2− and HER2+ MBC (resistance to CDK4/6 inhibitors) and ERBB2 in all subtypes (endocrine resistance, resistance to anti-HER2 therapies, sensitivity to certain anti-HER2 tyrosine kinase inhibitors; Fig. 4D–F). The increase in median CCF was consistent by metastatic biopsy site when considering the most frequent biopsy sites (Fig. 4G). Supplementary Figure S12 highlights the changes between primary and paired metastasis in the CCF of driver and other genes.
Figure 4.
CCF changes between primary and metastatic samples. Box plots showing the distribution of median CCF by patient in paired primary and metastatic samples (A) and stratified by subtype (HR+/HER2−, HER2+, TNBC; B). Gray lines refer to paired samples. C, Median CCF by driver genes in metastatic (y-axis) versus primary (x-axis) samples. The size of the circles refers to each gene alteration frequency. Box plots of the distribution of CCF for driver mutations in ESR1 (D), RB1 (E), and ERBB2 (F), stratified by subtype. G, Distribution of median CCF in paired primary and metastatic samples by biopsy site. P values are estimated by paired Wilcoxon–Mann–Whitney test.
As homologous recombination deficiency (HRD)–related genes (BRCA1/2 and others) have therapeutic importance in MBC (9), we applied SigMA to estimate the mutational signature associated with HRD and the change between primary tumors and their paired metastasis. The SigMA computational tool allows to detect the mutational signature associated with HRD (10) using TGS (see Supplementary Methods). Three different callers were used (mva, mva_strict, and ml). Only samples with enough mutations as per SigMA were selected: 251 samples in total from 181 patients. There were paired samples from 70 (38.7%) patients, 95 patients with metastatic samples only, and 16 patients with primary tumors only. Tumor samples harboring BRCA1 or BRCA2 mutations (only pathogenic and likely pathogenic variants were considered) had a higher likelihood of being flagged by SigMA, and the difference was statistically significant for both germline and somatic mutations (Supplementary Fig. S13A). There was a trend toward a higher prevalence of the HRD signature in TNBC that was statistically significant with the ml caller on metastatic samples (Supplementary Fig. S13B). The HRD signature was enriched in metastatic samples compared with primary tumors for all subtypes combined; this difference was statistically significant with the ml caller (Supplementary Fig. S13C).
We then aimed to describe variations in gene expression between primary samples and their paired metastasis. RNA-seq data were available for 152 pairs of primary tumors and metastases. First, we studied the correlation between mutations in selected driver genes (ESR1 and ERBB2) and their level of expression. ESR1 mutations in the metastatic samples were associated with a higher level of expression of ESR1 mRNA (P < 0.001) and a higher probability of a Luminal B subtyping (Supplementary Fig. S14A–S14E). Similarly, HR+/HER2− tumors (primary and metastasis) harboring ERBB2-activating mutations had a higher expression of ERBB2 mRNA (P = 0.039 for primary tumors and P = 0.0042 for metastases) albeit lower than HER2+ tumors, and were more likely to be classified as HER2-enriched (HER2-E; P = 0.014 for the primary tumors and P = 0.0042 for the metastases; Supplementary Fig. S14F–S14J).
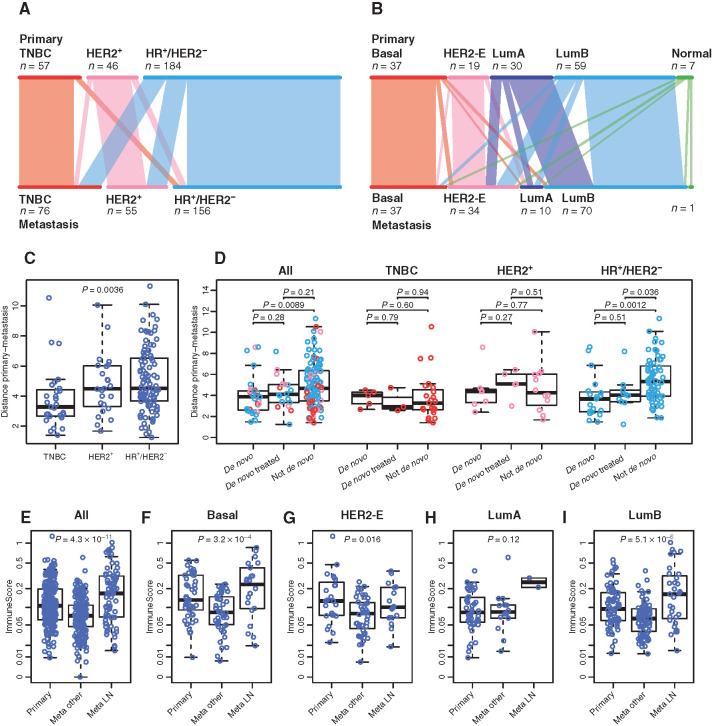
Previous reports have shown that the IHC and intrinsic subtypes do not completely overlap (11), bringing potential clinical implications. Furthermore, the intrinsic subtype can change between the primary tumor and the metastasis (12). Beyond IHC subtype switching (Fig. 5A), we wanted to study the molecular determinants of intrinsic subtype switching by assessing mutations and clonal evolution associated with this phenomenon. The prevalence of the intrinsic subtypes of the primary tumors in our cohort (n = 211) as determined using RNA-seq is as follows: 22% Luminal A, 38% Luminal B, 11% HER2-E, 25% Basal, 4% Normal. Intrinsic subtype switching in the patients with paired RNA-seq data (n = 152) was seen in 55 (36%) cases. Almost all Luminal A primaries switched in metastatic samples (90% of cases) and there was subtype switching from Normal, Luminal (A or B), and Basal to HER2-E in n = 18 cases (Fig. 5B; Supplementary Fig. S15). We focused on the 14 cases switching from Luminal A or B to HER2-E because of the prevalence in our cohort and potential clinical implications. For the cases with available TGS data (n = 13), higher prevalence of TP53 and/or PIK3CA mutations was found in these tumors compared with Luminal A or B cases not switching, in both the metastatic and primary samples (Supplementary Fig. S16). To assess whether subtype switching was related to tumor heterogeneity and change in clonality under therapy pressure, we compared the median CCF between Luminal A or B primary tumors and their paired metastases switching to the HER2-E subtype. No decrease in median CCF could be identified in our cohort (Supplementary Fig. S17), which is in line with previous reports (13).
Figure 5.
RNA-seq of paired primary tumors and metastatic samples. A and B, Subtype switching, on IHC subtypes (A) or on PAM50, estimated from RNA-seq (B). C, Distribution of the distances between primaries and metastases in term of expression of the PAM50 genes, in function of the clinical subtype. D, Similar comparison as C, but between untreated de novo metastatic patients, treated de novo metastatic patients and patients with a later relapse. E–I, Difference in immune signal between primary and metastasis across PAM50 subtypes. “Meta LN” are lymph node metastases; “Meta other” are all other metastases.
The analyses of RNA-seq data from matched pairs using uniform manifold approximation and projection (UMAP) for dimension reduction show that Basal tumors cluster by pairs in contrast to the Luminal and HER2-E subtypes, owing to closer gene expression profiles in the primaries and metastases of TNBC (Supplementary Fig. S18A–S18L). The gene expression differences were significantly larger in HR+/HER2− MBC when compared with the other subtypes. Gene expression differences were also larger between primary samples and their paired metastasis when compared with paired samples from cases of de novo MBC. Statistical significance was however only reached in HR+/HER2− MBC. This finding remained true in the HR+/HER2− subtype after comparison with de novo MBC pairs that had metastatic tissue collection after one line of therapy for metastatic disease (Fig. 5C and D). A longer time to relapse was associated with HR+/HER2− MBC and larger gene expression differences (Supplementary Fig. S19A and S19B). These findings may indicate that adjuvant endocrine therapy exerts the major influence on these gene expression differences that were seen between primary tumor and paired metastasis, suggesting an adaptive transcriptional reprogramming associated with endocrine resistance. Another explanation is a longer time before relapse, allowing the accumulation of more mutations.
Immune biomarkers such as tumor-infiltrating lymphocytes (TIL) are prognostic biomarkers in early breast cancer and are being investigated as predictive biomarkers (14). Immune checkpoint inhibitors have been approved in combination with chemotherapy in metastatic PD-L1+ TNBC and are investigated in other subtypes such as Luminal B. We investigated immune signatures in paired primary tumors and metastases by site, as well as the difference in the immune composition between paired primary tumors and metastases by site. Expression of the immune module score (15) was lower in metastatic samples but not in lymph node metastases (Fig. 5E–I). We looked at lymph node metastases separately because of the potential bias related to cellular composition. When performing the same analysis by metastatic site, expression of the immune signature was higher in skin metastases compared with other metastatic sites and lower in liver metastases (Supplementary Fig. S20). Using CIBERSORT (16) we studied the immune cell composition in primary tumors and their paired metastasis. Immune cell populations increasing in metastatic samples were: Mast cell activated, Myeloid dendritic cell resting, natural killer (NK) cell–activated, T-cell regulatory, Macrophage M1, and Macrophage M2 (Supplementary Fig. S21A). Immune cell populations decreasing in metastatic samples were T-cell CD4+ memory activated and T follicular helper (Supplementary Fig. S21A). Correlation between the expression of the immune score and the immune cell composition by site of metastasis is seen in Supplementary Fig. S21B.
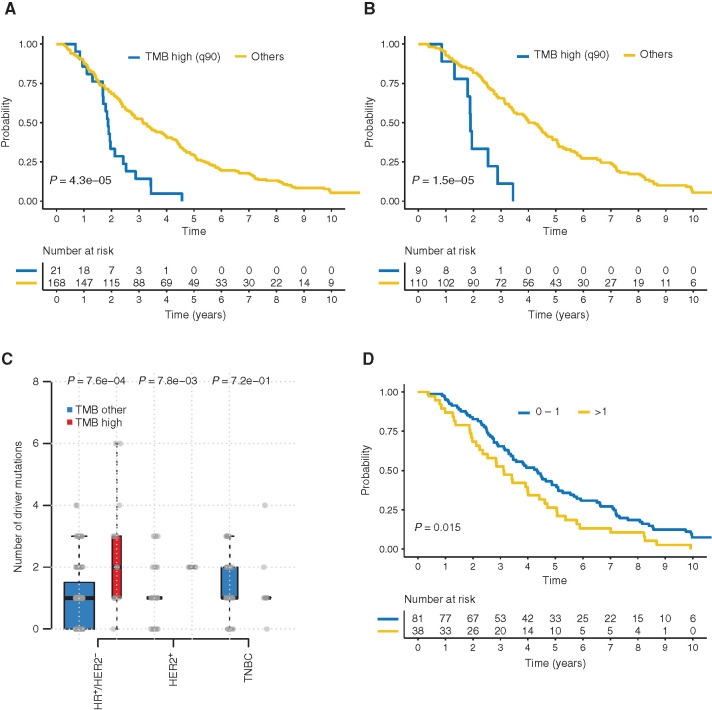
The next objective was to identify genomic markers that could predict the outcome for patients: overall survival (OS) from the diagnosis of MBC to death; and/or time to relapse (TTR) from the diagnosis of primary breast cancer to the metastatic relapse. Median duration of follow-up was 847 days (range, 2–2,082 days). Two hundred fifty-nine patients had experienced at least one event of disease progression and 209 patients had died. Of those, 126 patients had ER+/HER2− breast cancer, 57 had TNBC, and 26 had HER2+ breast cancer. In a recent study utilizing TGS, a higher number of mutations were associated with a trend toward worse survival in HR+/HER2− and HER2+ MBC (17). We tested whether high TMB as defined by the upper 90th percentile (see Supplementary Methods) was associated with patient outcomes. We had seen that median TMB was significantly higher in paired metastatic samples compared with primary tumors in HR+/HER2− and HER2+ MBC but not in TNBC (Supplementary Fig. S6B). High TMB in primary tumors was associated with a shorter TTR (Fig. 6A), and this was statistically significant for the HR+/HER2− (Fig. 6B) subtype but not for HER2+ nor TNBC (Supplementary Fig. S22A and S22B). High TMB in primary tumors was also associated with a statistically significant shorter OS and there was a trend toward shorter OS for high TMB in the metastatic samples (Supplementary Fig. S22C and S22D). This was driven by the HR+/HER2− subtype as the difference was not statistically significant in HER2+ and in TNBC (Supplementary Fig. S22E–S22J). In multivariate analysis controlling for other covariates such as primary tumor size, nodal status, and tumor grade (see Supplementary methods), high TMB in HR+/HER2− primary tumors remained an independent predictor of short TTR (P = 0.0012) and OS (P = 0.0088). The association between high TMB and TTR in HR+/HER2− breast cancer was not found when tested in the TCGA cohort (Supplementary Fig. S23A). We therefore hypothesized that the TTR finding in HR+/HER2− could be driven by the number of driver mutations in the primary tumor. High TMB was indeed associated with a significantly higher number of drivers in HR+/HER2− primary tumors in our dataset (Fig. 6C) and in TCGA (Supplementary Fig. S23B). A higher number of drivers in HR+/HER2− primary tumors was found to be associated with a shorter TTR (Fig. 6D), but TMB remained an independent predictor in the multivariate model when adjusting for the number of driver mutations (P < 0.001). We then looked at the association between mutations and outcome overall and by subtype, and two genes were associated with worse outcome: TP53 and LRP1B (Supplementary Fig. S24A–S24D). Mutations in LRP1B were correlated with worse OS (P = 0.0028, FDR = 0.037, Supplementary Fig. S25A). This was true for shared and acquired mutations (P = 0.0017, Supplementary Fig. S25B). LRP1B is a putative tumor suppressor and a member of the low-density lipoprotein (LDL) receptor family.
Figure 6.
TMB and patient outcome. A, TTR by TMB in all subtypes primary samples. B, TTR by TMB in HR+/HER2− primary samples. C, TMB and number of drivers correlation HR+/HER2−. D, TTR by number of drivers HR+/HER2− primary samples.
Finally, we investigated the clinical utility of molecular profiling of MBC because patients were enrolled prospectively and results reported on a sample-by-sample basis. We classified the molecular alterations identified by TGS using the ESMO Scale of Clinical Actionability for molecular Targets (ESCAT; ref. 18). Many but not all drivers selected for the breast cancer ESCAT tiers classification are detected and reported to the treating physicians in AURORA. We found that at least one Tier I or II alteration was identified for 51% of patients, or 36% if we exclude ERRB2 amplification that is a standard-of-care biomarker (Table 1). The choice of therapy remained at the discretion of the treating physician. Because these genomic results are generated in a research environment, the treating physician should have ensured that the results are confirmed using conventional/approved genetic tests, prior to the introduction of any clinical action. We queried the clinical data of this cohort of 379 patients to estimate the rate of therapy matched to a genomic alteration. We have identified 102 patients (27%) treated with targeted therapies. Of those, we have excluded patients treated with a targeted therapy without a proven genomic biomarker and patients treated with a targeted therapy while we did not identify the matching genomic alteration in AURORA. Therapy matched to a genomic alteration was prescribed for 25 patients (7%; Supplementary Table S2).
Table 1.
Actionability of alterations as per ESCAT
| Readiness of use in clinical practice | ESCAT for alterations in breast cancer | Prevalence in the AURORA population | Cumulative prevalence in the AURORA population | |
|---|---|---|---|---|
| Tier I | Targets ready for implementation in routine clinical decisions | ERBB2 amplification (IA), germline BRCA1/2 mutations (IA), PIK3CA mutations (IA), MSI (IC), TRK fusions (IC) | 143 (38%) | 143 (38%) |
| Tier II | Investigational targets likely to define patients who benefit from a targeted drug, but additional data needed | PTEN loss (IIA), ESR1 mutations (IIA), AKT1 mutations (IIB), ERBB2 mutations (IIB) | 65 (17%) | 193 (51%) |
| Tier III | Clinical benefit previously demonstrated in other tumor type or for similar molecular targets | Somatic BRCA1/2 mutations (IIIA), MDM2 amplification (IIIA), ERBB3 mutations (IIIB) | 35 (13%) | 206 (54%) |
| Tier IV | Preclinical evidence of actionability | ARID1A/B, ATM/ATR/PALB2, CDH1, IGF1R, INPP4B loss, MAP2K4/MAP3K1, MT4, MYC, NF1, PIK3R1, RUNX1/CBFB, SF3B1, TP53 (IVA) | 233 (61%) | 313 (83%) |
| Tier V | Evidence supporting co-targeting approaches | |||
| Tier X | Lack of evidence of actionability | FGFR1 amplification, CCND1 amplification | 99 (26%) | 328 (86%) |
NOTE: We have reproduced the different molecular alterations and their level of evidence as per ESCAT (18). The molecular alterations reported on a sample-per-samples basis in AURORA are in bold.
Data from the cfDNA analyses of the baseline plasma sample were available for 99 patients. Thirty-nine patients had no mutations detected in cfDNA. At least one mutation was detected in 60% of cases (Supplementary Fig. S26A). Out of 77 genomic alterations detected in one or both tissue samples, 31 (40%) were not found in cfDNA (Supplementary Fig. S26B). The variants found in a tissue sample and not detected in cfDNA had a statistically significant lower variant allele frequency (VAF) in the tissue samples (primary and/or metastatic sample, Mann–Whitney U test P < 0.001 for both primary and metastatic samples, Supplementary Fig. S26C). Tier I or II mutations as per ESCAT (PIK3CA, ESR1, AKT1, ERBB2) were detected in cfDNA but in none of the tissue samples (primary tumor nor metastasis) in 11 cases (11%; Supplementary Fig. S26D).
DISCUSSION
Recent studies aimed to describe the genomic landscape of MBC (19–21) and identify potentially acquired driver genes, largely by inter-dataset comparisons with series of largely primary tumors (7, 8, 22). The design of AURORA, based on the prospective collection of paired primary and metastatic samples in patients who are treatment-naïve for MBC or after just one line of therapy, is built to allow the unbiased characterization of molecular alterations causing metastatic relapse or acquired early in the course of metastatic disease. The majority of driver point mutations were shared (88%), and only a minority of patients (10%) had at least one mutation private to the metastatic sample. This is in contrast to other studies that collected paired samples and that reported a higher rate of acquired genomic alterations ranging from 45% (23) to 73% (24). Fully concordant alterations between the primary and metastatic samples were found in only 18% of cases in another study (17). An explanation could be the clinical setting, as patients in AURORA are enrolled early in the course of MBC while acquired driver alterations occurred late in the metastatic process (23). It was recently reported that mutations private to metastasis are less associated with the metastatic spread and are accumulating under the pressure of therapy (25). The heterogeneity of NGS techniques [whole-genome sequencing (WGS), WES, TGS] could also contribute to this difference.
Molecular alterations found to be enriched in AURORA metastatic samples could contribute to new drug development for MBC. Several are involved in cancer epigenetics (MYC amplification, NSD3 amplification, PBRM1 deletion, and ARID1A deletion), and this field is emerging as a contributor to endocrine resistance (21, 26). Agents targeting cancer epigenetics (bromodomain and extra-terminal domain inhibitors, histone deacetylase inhibitors, and EZH2 inhibitors) are currently in development in combination with endocrine therapy. AXIN1 (WNT pathway) and NSD3 (27) are involved in stemness, another process that promotes therapy resistance. Agents targeting stem cells are now in clinical development (gamma-secretase inhibitors, LGR5 inhibitors), and a study testing AL101 (gamma-secretase inhibitor) has just started recruitment in metastatic TNBC (NCT04461600). Enriched alterations also include biomarkers associated with innate or acquired resistance to therapies available in the clinic such as PTEN loss (28), RB1 loss-of-function alterations (29) and FGFR1 amplification (30) for CDK4/6 inhibitors, PBRM1 loss-of-function alterations (31, 32) and MDM4 amplification (33) for immune checkpoint inhibitors. The higher prevalence of the HRD signature in metastatic disease may widen the target population beyond germline BRCA1 and BRCA2 mutation carriers that could benefit from treatment with PARP inhibitors. This reinforces the case of other biomarkers such as those investigated in the TBCRC 048 trial (9).
The largest dataset to date of RNA-seq of the paired primary tumors and metastases allowed us to study intrinsic subtype switching that is gaining clinical implications. Indeed, intrinsic subtyping, especially for the Luminal and HER2-E subtypes, is emerging as a predictor for a number of targeted therapies in different settings: Everolimus (34) and CDK4/6 inhibitors in HR+/HER2− MBC (35), CDK4/6 inhibitors (36) and anti-HER2 therapies (37) in HER2+. Treatment refinement and de-escalation based on the incorporation of the intrinsic subtype are being included in prospective clinical trials and follow single sample studies, while a study has shown intrinsic subtype switching between primary tumor and paired metastasis in 31% of cases including 14.3% of luminal A or B tumors converting to HER2-E. The authors also suggested that expression of FGFR4 could be driving the HER2-E phenotype (12). This finding was not confirmed in AURORA where we found an association of subtype switching from Luminal A/B to HER2-E with TP53 and/or PIK3CA mutations. We could not demonstrate that subtype switching was related to tumor heterogeneity and change in clonality under therapy pressure. The UMAP analyses allowed us to postulate that adjuvant endocrine therapy may drive the differential gene expression between primary and metastatic samples in HR+/HER2− tumors. Because most patients with TNBC and HER2+ tumors have received therapy prior to the development of metastatic relapse, future analyses should study the mutational footprints of various therapeutics as in Pich and colleagues (38).
Comparing immune signatures between primary tumors and paired metastases as well as computing the differential immune cell composition in primary tumors and in different metastatic sites can contribute to the understanding of immune evasion and the planning of future trials testing immunotherapy. Our larger cohort adds to the finding of a smaller cohort (n = 11; ref. 39) by demonstrating that the immune signal is lower in metastases (excluding lymph nodes). This finding was not true for lymph node metastases where expression of the immune signatures could be influenced by the presence of immune cells. Interestingly, this may partly explain what was seen in the phase III clinical trial of the combination of chemotherapy and atezolizumab in the first-line setting of metastatic TNBC (40) and the phase II study of pembrolizumab monotherapy in metastatic TNBC (41) that showed an increased clinical benefit with immunotherapy among patients with lymph node–only metastatic disease. The low expression of immune signatures in liver metastases mirrors the recent finding about reduced efficacy of immune checkpoint inhibitors in patients with liver metastases (42). We found increases and decreases in some immune cell populations leading to a more immune-permissive microenvironment in the metastatic samples, in line with studies of residual primary tumors after neoadjuvant chemotherapy (43).
While a higher TMB in MBC has been associated with a worse outcome (19), AURORA is to our knowledge the first study to demonstrate that high TMB associates with a shorter TTR after treatment for HR+/HER2− early breast cancer. This finding remained significant after taking into consideration the number of genomic drivers, highlighting the possibility of incorporating this biomarker in the design of trials testing novel adjuvant therapies in this poor-prognosis population. Correlation of outcomes with individual genes has associated LRP1B with shorter OS, particularly when the mutation occurred in the metastases. Confounders are however to be acknowledged as the LRP1B variants are not in known functional domains and the very large size of the gene (91 exons and a mRNA size > 16 kb; ref. 44) makes it a probable proxy of TMB.
Molecular profiling of metastatic cancer has entered the clinic with alpelisib in HR+/HER2− MBC with PIK3CA mutations (45) and PARP inhibitors in MBC with germline BRCA1/2 mutations (46). Other targets are emerging but some reputed scientific societies still do not recommend routine molecular profiling in the management of MBC (47). This is largely due to the lack of approved biomarker-driven targeted therapies in MBC other than anti-HER2 therapies. Despite the fact that 51% (193) of patients had at least one Tier I or II alteration as per ESCAT (83% of patients if we include Tier III and IV alterations currently investigated in clinical trials), only 7% of patients in this cohort had received matched therapy at the data cutoff. This can be explained by drug availability, as the recruitment in AURORA started in 2014 while the first regulatory approval for a targeted therapy matched to a molecular alteration in MBC (excluding HER2+) only occurred in 2018, and access to clinical trials and new targeted agents is very heterogeneous in different countries. The results of TGS of the baseline cfDNA in AURORA expanded the landscape of actionable mutations (11% of cases with alterations not identified in a tissue samples). The clinical utility of therapy decided based on the results of cfDNA analyses has been demonstrated in settings such as PIK3CA mutations in HR+/HER2− MBC (48). Actionable alterations can, however, be missed in plasma (40% of alterations in AURORA), possible due to lower VAF in tissue samples for variants that were missed. Liquid biopsies are therefore complementary to tissue biopsies for the management of patients with advanced cancer, but current sensitivity of detection is not yet ready to forego tissue biopsy of metastatic disease for full molecular understanding.
There are limitations to be recognized. AURORA has so far reported genomic results of TGS while other studies have utilized more extensive profiling such as WES and WGS. The AURORA biobank of formalin-fixed, paraffin-embedded (FFPE) and frozen samples should allow more in-depth analyses. The exclusion of variants with a VAF < 10% hinders the discovery potential of this study. This cutoff was set based on the algorithms used by the sequencing service provider and is considered to be conservative as results are reported to clinicians. Calling variants as low as 1% if the variant is seen at a higher VAF in the matched sample was used to describe mutations enriched in the metastases. We would still, however, miss subclonal alterations that might have a VAF < 10% in both samples. Also, the majority of samples being core biopsies and collected from one metastatic site, the enrichment findings could be confounded with intratumoral heterogeneity. Furthermore, while AURORA requires the collection of primary tumors and a paired metastasis, other samples such as a second primary tumor, pre- or post-neoadjuvant therapy samples, or local relapses are not collected and could create a gap in the understanding of the metastatic process.
Beyond the findings discussed in this article, the AURORA initiative is providing the basis for future research in MBC, by allowing future research on residual samples and unexploited fresh-frozen metastatic samples and serial collection of plasma and serum samples currently stored in the centralized AURORA biobank. The curated clinical database as well as the central storing of pathology high-resolution scanned images, and the collection of clinical data, including the identification of outlier patients—exceptional responders or with highly resistant disease—that will be studied in-depth, could allow the generation of hypotheses for novel therapeutic strategies.
METHODS
The objectives, study design, and eligibility criteria of AURORA have been previously described (49). The study aims to include at least 1,000 patients with MBC at the diagnosis of metastatic disease or after one line of therapy. Collected samples include FFPE tissue from the primary tumor, FFPE, and fresh-frozen samples from a metastatic biopsy, whole blood, plasma, and serum. At the central lab (OncoDNA) and after pathology review and standard IHC for estrogen receptor (ER), progesterone receptor (PR), and HER2, TGS is performed on DNA extracted from the primary tumor, the FFPE metastatic sample (gene list in Supplementary Table S3), whole blood and baseline plasma sample (gene list in Supplementary Table S4). Results are reported on sample-by-sample basis to the investigators after annotation by a molecular advisory board (MAB). RNA-seq and CNV analyses using SNP arrays are performed on the available tumor samples in batches (at EUROFINS). The oncoplot included CNVs as determined by TGS and SNP arrays. Discordances were seen mainly in genes involving deletions (Supplementary Methods and Supplementary Fig. S27). Liver genes were excluded for transcriptomic analyses (supplementary methods and supplementary file—liver genes). Indeed, visualization of the biopsy site of the samples using UMAP showed that liver samples were clustered together (Supplementary Fig. S28A–S28V). Residual FFPE tumor samples, frozen tumor samples and plasma and serum samples collected serially (every 6 months and at every disease progression for up to 10 years) were shipped in batches and stored in the study biobank (IBBL, Dudelange, Luxembourg). In the first version of the protocol, a patient was considered included only if TGS was successful on both primary and metastatic tumor samples. The protocol was amended following a high rate of screen failures to allow inclusion of patients if one of the two tissue samples was successfully sequenced. The amendment also introduced the sample-by-sample concomitant TGS of cfDNA extracted from the baseline plasma sample. A cohort capped at 100 patients with bone-only metastases was introduced; patients were allowed to provide only material from the primary tumor and were considered included if TGS of the primary tumor was successful. The AURORA study recruited patients at 51 sites in 11 European countries. A dedicated IT platform served for the logistics, collection of data on the disease and samples, pathology images scanning and for reporting of the genomic results. The study is conducted in collaboration with Institut Jules Bordet (IJB) and Frontier Science (FS). IJB's Clinical Trials Support Unit holds the clinical database for AURORA, in which clinical data including information on previous and subsequent treatments as well as survival follow-up for up to 10 years are collected. The MAB is composed of experts in molecular biology, genetics, drug development, pathology, and bioinformatics; it convenes remotely every two weeks and provides variant annotations and clinical implications that are included in a consolidated molecular report. The study was approved by the ethical committees of the participating institutions and is performed in accordance with relevant guidelines and procedures. Participating patients provided written informed consent.
Data Availability
Instructions to access the manuscript processed data for reproducibility purposes are available at the webpage https://aurora.bigagainstbreastcancer.org/ and can be obtained upon signature of an appropriate data transfer agreement subject to applicable laws. Instructions to access processed or raw manuscript data to perform original research are also available on the webpage and investigators can contact aurora.researchproposals@bigagainstbc.org for enquiries. Access to data for research will be granted upon review of a project proposal and endorsement by the study Steering Committee, and after entering into an appropriate data access agreement subject to applicable laws.
Authors' Disclosures
P. Aftimos reports personal fees from Boehringer Ingelheim, Macrogenics, from Roche, Novartis, Servier, Amcure, G1 Therapeutics, Radius, Deloitte, Synthon, Amgen, Gilead, nonfinancial support from MSD, Amgen, Pfizer, Roche; and grants from Roche outside the submitted work. M. Oliveira reports grants from Genentech and grants from Breast Cancer Research Foundation during the conduct of the study; grants, personal fees, and non-financial support from Roche and Novartis; grants and personal fees from SeaGen, AstraZeneca, GlaxoSmithKline, and Puma Biotechnology; personal fees from Pfizer; Guardant Health, and MSD; grants from Immunomedics, Boehringer Ingelheim, Sanofi, Cascadian Therapeutics, Bayer, Piqur, Zenith Epigenetics; non-financial support from Eisai; and grants and non-financial support from Pierre Fabre outside the submitted work. A. Irrthum reports grants from Breast Cancer Research Foundation (BCRF), Fondation Cancer (Luxembourg), National Lottery (Belgium), NIF Foundation, Barrie and Deena Webb; and other support from Candriam, Fondation Futur 21, Sogerim, Think Pink Belgium (SMART Fund), Fund Friends of BIG, managed by the King Baudouin Foundation, and grants from Foundation against Cancer (Belgium) during the conduct of the study. D. Fumagalli reports grants from Breast Cancer Research Foundation, Fondation Cancer Luxembourg, National Lottery Belgium, NIF Foundation, Barrie and Deena Webb, Foundation against Cancer Belgium, Roche/Genentech, AstraZeneca, Novartis, Pfizer, Servier, GSK, Tesaro, Biovica, and grants from Sanofi outside the submitted work; and other support from Candriam, Fondation Futur 21, Sogerim, Think Pink Belgium (SMART Fund), and other support from Fund Friends of BIG (managed by King Baudouin Foundation) during the conduct of the study. C. Sotiriou reports personal fees from Seattle Genetics, Puma, Amgen, Merck & Co Inc., Eisai, Prime Oncology, Exact Sciences, Roche, other support from Genentech, and other support from Pfizer outside the submitted work. E. Nili Gal-Yam reports personal fees from Pfizer, Roche, Novartis, and Eli Lilly outside the submitted work. M.E. Robson reports other support from AstraZeneca, Merck, and other support from Pfizer outside the submitted work; and Medscape, Physican's Education Resources, Change Healthcare: Honoraria. A. Di Leo reports personal fees from Amgen, Astrazeneca, Athenex, Bayer, Daiichi-Sankyo, Eisai, Genentech, Genomic Health, Ipsen, Lilly, Novartis, Pfizer, Pierre Fabre, Puma Biotechnology, Roche, Seattle Genetics, and personal fees from Sellas Life Sciences Group outside the submitted work. E.M. Ciruelos reports personal fees from Roche, Pfizer, Novartis, A Zeneca, and personal fees from Lilly outside the submitted work. E. de Azambuja reports other support from Pierre Fabre, Lilly, Novartis, Roche, Seattle Genetics, Zodiacs, and other support from Libbs outside the submitted work. G. Viale reports personal fees from Roche Genentech, MSD Oncology, Daichii Sankyo, Dako Agilent, Pfizer, Ventana, and Menarini outside the submitted work. G. Curigliano reports personal fees from Pfizer, Lilly, Novartis, Astra Zeneca, Daichii Sankyo, Merck, Veracyte, and Ellipsis outside the submitted work. J.M. Bliss reports grants from Breast International Group (BIG) during the conduct of the study; grants and non-financial support from AstraZeneca, grants and non-financial support from Merck Sharpe & Dohme, grants and non-financial support from Puma Biotechnology, grants and non-financial support from Clovis Oncology, grants and non-financial support from Pfizer, grants and non-financial support from Janssen-Cilag, grants and non-financial support from Novartis, grants and non-financial support from Roche, and grants and non-financial support from Eli Lilly outside the submitted work. J.S. Reis-Filho reports personal fees from Goldman Sachs; personal fees and other support from Repare Therapeutics, Paige.AI, Volition RX, Grupo Oncoclinicas; other support from Roche Tissue Diagnostics, Genentech, Roche, Novartis, In vicro, and personal fees from Eli Lilly outside the submitted work. M. Colleoni reports other support from BIG during the conduct of the study; grants from Roche outside the submitted work. M. Balic reports non-financial support from Amgen; grants, personal fees, and non-financial support from Eli Lilly, Novartis, and Pfizer; personal fees and non-financial support from Pierre FABRE, Astra Zeneca, MSD, Roche, Astra Zeneca, and personal fees and non-financial support from Seagen during the conduct of the study and outside the submitted work. F. Cardoso reports other support from payment for the Institution for the Clinical Trial during the conduct of the study; personal fees from Amgen, Astellas/Medivation, AstraZeneca, Celgene, Daiichi-Sankyo, Eisai, GE Oncology, Genentech, GlaxoSmithKline, Macrogenics, Medscape, Merck-Sharp, Merus BV, Mylan, Mundipharma, Novartis, Pfizer, Pierre-Fabre, Prime Oncology, Roche, Samsung, Sanofi, personal fees from Seagen Bioepis, and personal fees from Teva outside the submitted work. J. Albanell reports grants from GEICAM - BIG during the conduct of the study; personal fees from Genomic Health, Exact Sciences, Palex; grants and personal fees from Roche, Daiichi-Sankyo, Seagen, Pfizer, Lilly, and personal fees from Novartis outside the submitted work; in addition, J. Albanell has a patent for 20382681.3 pending and a patent for EP14382288 licensed and with royalties paid from Biocartis; and Stock options with Inbiomotion. A. Gombos reports other support from Lilly, Astra Zeneca, Daiichi Sankyo, and other support from Pfizer outside the submitted work. H. Wildiers reports grants from Roche; other support from Immutep Pty, MSD, Astrazenca Ireland, Daiichi, AbbVie, Lilly, PSI CRO AG, KCE, Eisai, AstraZeneca, Roche, Lilly, Congres care, Pfizer, Ariez, Sirtex, Roche, Pfizer, TRM Oncology, Orion Corporation, The Planning Shop, Novartis, Biocartes, and other support from Puma Biotech outside the submitted work. A. Guerrero-Zotano reports grants from Pfizer during the conduct of the study; personal fees from AstraZeneca, Roche, Novartis, Exact Sciences, Daichi-Sankio, and personal fees from Merck outside the submitted work. R. Greil reports other support from Celgene, Roche, Merck, Takeda, AstraZeneca, Novartis, Amgen, BMS, MSD, Sandoz, Abbvie, Gilead, and other support from Daiichi Sankyo outside the submitted work. S. Loibl reports grants and other support from Abbvie and Bayer; grants, non-financial support, and other support from Amgen, AstraZeneca, Celgene, Novartis, BMS, Roche, Samsung, Seagen; personal fees from Chugai; other support from Daiichi-Sankyo, Eirgenix, GSK, Ipsen, Lilly, Merck; non-financial support and other support from Pfizer; and grants and non-financial support Immunomedics/Gilead; and grants and non-financial support from Vifor outside the submitted work; in addition, S. Loibl has a patent for EP18209672.7 issued, a patent for EP21152186.9 pending, a patent for EP15702464.7 issued, and a patent for VM Scope GmbH licensed. B. Linderholm reports personal fees from AstraZeneca, Pfizer, Merck, Eli Lilly, Pierre Fabre, and Daiichi Sankyo/UCB Japan outside the submitted work. G. Zoppoli reports other support from Thermo Fisher Scientific outside the submitted work. P.L. Bedard reports grants from AstraZeneca, BMS, GSK, Merck, Lilly, Amgen, Sanofi, Roche/Genentech, grants from Bicara Therapeutics, Zymeworks, Servier, Mersana, Immunomedics, Novartis, and grants from Pfizer outside the submitted work; and uncompensated advisory for SeaGen, Lilly, Merck, Amgen, Gilead, BMS, Sanofi, Roche/Genentech. S. Loi reports other support from research funding to institution from Novartis, Bristol Meyers Squibb, Merck, Puma Biotechnology, Eli Lilly, Nektar Therapeutics Astra Zeneca, Roche-Genentech, and Seattle Genetics, consultant (paid to her institution) to Aduro Biotech, Novartis, GlaxoSmithKline, Roche-Genentech, Astra Zeneca, Silverback Therapeutics, G1 Therapeutics, PUMA Biotechnologies, Pfizer, Seattle Genetics and Bristol Meyers Squibb; and personal fees from Scientific Advisory Board Member of Akamara Therapeutics outside the submitted work; and acted as consultant (not compensated) to Seattle Genetics, Novartis, Bristol Meyers Squibb, Merck, AstraZeneca and Roche-Genentech; in addition, S. Loi is supported by the National Breast Cancer Foundation of Australia Endowed Chair and the Breast Cancer Research Foundation, New York. D.A. Cameron reports other support from BIG during the conduct of the study; in addition, D.A. Cameron chairs the Breast International Group, the sponsor of this study. D.A. Cameron receive no payment for this role, nor any financial or other rewards for this role. N. Harbeck reports personal fees from AstraZeneca, Daiichi-Sankyo, Lilly, MSD, Novartis, Pierre Fabre, Pfizer, Roche, Sandoz/Hexal, and personal fees from Seattle Genetics outside the submitted work. M. Lasa Montoya reports grants from Breast Cancer Research Foundation (BCRF), Fondation Cancer (Luxembourg), National Lottery (Belgium), NIF Foundation, Barrie and Deena Webb; other support from Candriam, Fondation Futur 21, Sogerim, Think Pink Belgium (SMART Fund), Fund Friends of BIG, managed by the King Baudouin Foundation, and grants from Foundation against Cancer (Belgium) during the conduct of the study. M. Brandão reports other support from Breast International Group during the conduct of the study; personal fees and non-financial support from Roche/GNE outside the submitted work. C. Caballero reports grants from Breast Cancer Research Foundation, Fondation Cancer (Luxembourg), National Lottery (Belgium), NIF Foundation, Barrie and Deena Webb, Candriam, Fondation Futur 21, Think Pink Belgium (SMART fund), Fund Friends of BIG, managed by the King Baudoin Foundation, and grants from Fondation Contre le Cancer (Belgique) during the conduct of the study; and grants from Roche/Genentech outside the submitted work. L.R. Yates reports grants from Wellcome Trust during the conduct of the study. M. Benelli reports personal fees from Novartis outside the submitted work. M.J. Piccart reports personal fees from Oncolytics, grants and personal fees from AstraZeneca; personal fees from Camel IDS, Debiopharm, Immunomedics, Lilly; grants and personal fees from MSD, Novartis, Pfizer, Roche-Genentech; grants from Radius, Servier, Synthon; personal fees from Menarini, Odonate; Seattle Genetics, Immutep, Seagen, and personal fees from NBE Therapeutics outside the submitted work. No disclosures were reported by the other authors.
Supplementary Material
File with the supplementary material & methods, supplementary tables and supplementary figure.
Supplementary file with the listing of the liver genes excluded from transcriptomic analyses.
Acknowledgments
The authors thank all the patients who participated in the program and their families.
Current or former study Steering Committee members:
P. Aftimos, M. Oliveira, M. Piccart, J. Albanell, M. Balic, P. Bedard, M. Benelli, K. Benn, J. Bliss, C. Caballero, F. Cardoso, D. Cameron, E. Ciruelos, M. Colleoni, G. Curigliano, N. Davidson, E. De Azambuja, A. Di Leo, C. Duhem, D. Fumagalli, N. Harbeck, C. Herremans, F. Hilbers, A. Irrthum, O. Johannsson, S. Knox, B. Linderholm, S. Loi, S. Loibl, S. Marreaud, J.-L. Martinez, E. Nil Gal-Yam, D. Rea, J. Reis-Filho, E. Scheepers, C. Sotiriou, G. Viale, D. Venet, A. Vingiani, L. Yates, G. Zoppoli, H. Azim, R. Bartsch, P. Campbell, E. Carrasco, P. Dinh, V. Golfinopoulos, T. Goulioti, C. Herremans, M. Maetens, C. Tryfonidis, D. Zardavas, C. Fallato, F. Daly.
Local PIs participating in the program (who are neither authors not part of other committees):
Austria: R. Bartsch, C. Singer; Belgium: J.L. Canon, L. Dirix, F. Duhoux, G. Jerusalem, K. Papadimitriou, D. Taylor; Germany: C. Cirkel, S. Kümmel, S. Seiler, C. Jackisch Italy: F. Artioli, A. Ballestrero, A. Bernardo, L. Biganzoli, G. Bisagni, E. Cretella, F. Giovanardi, L. Moscetti, A. Musolino, E. Seles; Spain: B. Bermejo, L. Calvo Martinez, L. Garcia Estevez, J.A. Garcia Saenz, M. Gonzalez Cao, S. Gonzalez-Santiago, E. Martinez de Dueñas, S. Morales, V. Iranzo, M. Ruiz Borrego, S. Servitja Tormo; Sweden: M. Ekholm, H. Lindman; Switzerland: S. Aebi, C. Leo, M. Rabaglio, M. Schwitter; United Kingdom: J. Abraham, A. Armstrong, U. Barthakur, J. Braybrooke, S. Chan, D. Davies, J. Macaskill, I. MacPherson, A. Thomson, A. Tutt.
The participating groups: Austrian Breast & Colorectal Cancer Study Group (ABCSG); Centre Hospitalier de Luxembourg (CHL); European Organisation for Research and Treatment of Cancer- Breast Cancer Cooperative Group (EORTC BCG); German Breast Group (GBG); Grupo Espanol de Investigacion en Cancer de Mama (GEICAM); Gruppo Oncologico Italiano di Ricerca Clinica (GOIRC); Icelandic Breast Cancer Group (IBCG); International Breast Cancer Study Group (IBCSG); National Cancer Research Institute Breast Clinical Studies Group (NCRI-BCSG); Swedish Association of Breast Oncologists (SABO); Spanish Breast Cancer Study Group (SOLTI); Westdeutsche Studien Gruppe (WSG).
The members of the study Data Monitoring Committee: M. Castiglione, S. Litière, F. Orsi.
Current or former members of the Molecular Advisory Board: P. Aftimos, F. Hilbers, A. Irrthum, E. Nil Gal-Yam, M. Oliveira, A. Vingiani, M. Robson, T. Severson.
Breast International Group Headquarters members: P. Boussis, C. Cameron, E. Ceysens, S. Corachan, M. Debiasi, K. Delobelle, E. De Tayrac, K. Engelen, N. Garg, B. Guillet-Revol, B. Herpoel, A. Karabogdan, B. Jacques, M. Jooris, D. Moreno, S. Massey, G. Questiaux, C. Saravia, S. Schmitz, C. Schurmans, O. Spagnolo, C. Straehle, E. Treillard, V. Van der Veeken, I. Van Der Straten, A. Raimbault, A. Arahmani.
Frontier Science Scotland members: I. Bradbury.
Institut Jules Bordet's Clinical Trials Support Unit (IJB/CTSU) members: E. Agostinetto, B. Andre, M. Bajji, M. Brandao, C. de Angelis, C. De Greef, S. Guillaume, M. Franzoi, M.-P. Gauthier, M. Monkangwo, J. Ndozeng, V. Van Dooren, M.-L. Vicente, D. Martins Branco.Other collaborators: Bioinformatics Unit Hospital of Prato, Breast Cancer Translational Research Laboratory (BCTL), Clinigen Clinical Supplies Management, EUROFINS, Europa Donna, Integrated BioBank of Luxembourg (IBBL), OncoDNA.
This work was supported by Breast Cancer Research Foundation (BCRF; ELFF-16-001, ELFF-17-003, ELFF-19-001, ELFF-20-001, SPEC-18-004 and BCRF-19-186) as the main funder, Barrie and Dena Webb, Fondation Cancer (Luxembourg, FC/2014/01, FC/2019/05), Foundation NIF, Fondation contre le Cancer (Belgium, C/2020/1429), National Lottery (Belgium), Think Pink Belgium (SMART Fund), and many individual donors. AURORA has also been supported by the Fund Friends of BIG, managed by the King Baudouin Foundation.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors' Contributions
P. Aftimos: Conceptualization, data curation, formal analysis, supervision, investigation, writing–original draft, writing–review and editing. M. Oliveira: Data curation, formal analysis, supervision, writing–review and editing. A. Irrthum: Data curation, formal analysis, validation, methodology, writing–original draft, writing–review and editing. D. Fumagalli: Conceptualization, resources, writing–original draft, project administration, writing–review and editing. C. Sotiriou: Conceptualization, formal analysis, supervision, investigation, writing–review and editing. E. Nili Gal-Yam: Data curation, formal analysis, validation, writing–review and editing. M.E. Robson: Investigation, writing–review and editing. J. Ndozeng: Data curation, writing–review and editing. A. Di Leo: Supervision, investigation, writing–review and editing. E.M. Ciruelos: Investigation, writing–review and editing. E. de Azambuja: Supervision, investigation, writing–review and editing. G. Viale: Conceptualization, writing–review and editing. E.D. Scheepers: Data curation, formal analysis, writing–review and editing. G. Curigliano: Investigation, writing–review and editing. J.M. Bliss: Investigation, writing–review and editing. J.S. Reis-Filho: Investigation, writing–review and editing. M. Colleoni: Investigation, writing–review and editing. M. Balic: Investigation, writing–review and editing. F. Cardoso: Investigation, writing–review and editing. J. Albanell: Investigation, writing–review and editing. C. Duhem: Investigation, writing–review and editing. S. Marreaud: Writing–review and editing. D. Romagnoli: Data curation, formal analysis, writing–original draft, writing–review and editing. B. Rojas: Investigation, writing–review and editing. A. Gombos: Investigation, writing–review and editing. H. Wildiers: Investigation, writing–review and editing. A. Guerrero-Zotano: Investigation, writing–review and editing. P. Hall: Investigation, writing–review and editing. A. Bonetti: Investigation, writing–review and editing. K.F. Larsson: Investigation, writing–review and editing. M. Degiorgis: Investigation, writing–review and editing. S. Khodaverdi: Investigation, writing–review and editing. R. Greil: Investigation, writing–review and editing. A. Sverrisdóttir: Investigation, writing–review and editing. M. Paoli: Data curation, formal analysis, writing–original draft, writing–review and editing. E. Seyll: Data curation, writing–review and editing. S. Loibl: Investigation, writing–review and editing. B. Linderholm: Investigation, writing–review and editing. G. Zoppoli: Investigation, writing–review and editing. N.E. Davidson: Resources, writing–review and editing. O.T. Johannsson: Investigation, writing–review and editing. P.L. Bedard: Supervision, writing–review and editing. S. Loi: Supervision, writing–review and editing. S. Knox: Supervision, writing–review and editing. D.A. Cameron: Resources, supervision, investigation, writing–review and editing. N. Harbeck: Investigation, writing–review and editing. M. Lasa Montoya: Resources, supervision, project administration, writing–review and editing. M. Brandão: Data curation, validation, writing–review and editing. A. Vingiani: Investigation, writing–review and editing. C. Caballero: Funding acquisition, project administration, writing–review and editing. F.S. Hilbers: Conceptualization, data curation, formal analysis, supervision, validation, writing–original draft, writing–review and editing. L.R. Yates: Data curation, formal analysis, supervision, writing–review and editing. M. Benelli: Resources, data curation, software, formal analysis, validation, methodology, writing–original draft, writing–review and editing. D. Venet: Data curation, software, formal analysis, validation, writing–original draft, writing–review and editing. M.J. Piccart: Conceptualization, resources, supervision, funding acquisition, investigation, writing–review and editing.
References
- 1. Caswell-Jin JL, Plevritis SK, Tian L, Cadham CJ, Xu C, Stout NKet al. Change in survival in metastatic breast cancer with treatment advances: meta-analysis and systematic review. JNCI Cancer Spectrum 2018;2:pky062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cardoso F, Paluch-Shimon S, Senkus E, Curigliano G, Aapro MS, André Fet al. 5th ESO-ESMO international consensus guidelines for advanced breast cancer (ABC 5). Ann Oncol 2020;31:1623–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Maetens M, Brown D, Irrthum A, Aftimos P, Viale G, Loibl Set al. The AURORA pilot study for molecular screening of patients with advanced breast cancer-a study of the breast international group. NPJ Breast Cancer 2017;3:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Martincorena I, Raine KM, Gerstung M, Dawson KJ, Haase K, Van Loo Pet al. Universal patterns of selection in cancer and somatic tissues. Cell 2017;171:1029–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li Y, Ke Q, Shao Y, Zhu G, Li Y, Geng Net al. GATA1 induces epithelial-mesenchymal transition in breast cancer cells through PAK5 oncogenic signaling. Oncotarget 2015;6:4345–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dreijerink KMA, Goudet P, Burgess JR, Valk GD, International Breast Cancer in MEN1 Study Group. Breast-cancer predisposition in multiple endocrine neoplasia type 1. N Engl J Med 2014;371:583–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. METABRIC Group, Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OMet al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012;486:346–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tung NM, Robson ME, Ventz S, Santa-Maria CA, Nanda R, Marcom PKet al. TBCRC 048: phase II study of olaparib for metastatic breast cancer and mutations in homologous recombination-related genes. J Clin Oncol 2020;38:4274–82. [DOI] [PubMed] [Google Scholar]
- 10. Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou Xet al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016;534:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheang MCU, Martin M, Nielsen TO, Prat A, Voduc D, Rodriguez-Lescure Aet al. Defining breast cancer intrinsic subtypes by quantitative receptor expression. Oncologist 2015;20:474–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cejalvo JM, Martínez de Dueñas E, Galván P, García-Recio S, Burgués Gasión O, Paré Let al. Intrinsic subtypes and gene expression profiles in primary and metastatic breast cancer. Cancer Res 2017;77:2213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lluch A, González-Angulo AM, Casadevall D, Eterovic AK, Martínez de Dueñas E, Zheng Xet al. Dynamic clonal remodelling in breast cancer metastases is associated with subtype conversion. Eur J Cancer 2019;120:54–64. [DOI] [PubMed] [Google Scholar]
- 14. Criscitiello C, Esposito A, Trapani D, Curigliano G. Prognostic and predictive value of tumor infiltrating lymphocytes in early breast cancer. Cancer Treat Rev 2016;50:205–7. [DOI] [PubMed] [Google Scholar]
- 15. Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol 2017;18:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol 2018;1711:243–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van Geelen CT, Savas P, Teo ZL, Luen SJ, Weng C-F, Ko Y-Aet al. Clinical implications of prospective genomic profiling of metastatic breast cancer patients. Breast Cancer Res 2020;22:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Condorelli R, Mosele F, Verret B, Bachelot T, Bedard PL, Cortes Jet al. Genomic alterations in breast cancer: level of evidence for actionability according to ESMO Scale for Clinical Actionability of molecular Targets (ESCAT). Ann Oncol 2019;30:365–73. [DOI] [PubMed] [Google Scholar]
- 19. Bertucci F, Ng CKY, Patsouris A, Droin N, Piscuoglio S, Carbuccia Net al. Genomic characterization of metastatic breast cancers. Nature 2019;569:560–4. [DOI] [PubMed] [Google Scholar]
- 20. Angus L, Smid M, Wilting SM, van Riet J, Van Hoeck A, Nguyen Let al. The genomic landscape of metastatic breast cancer highlights changes in mutation and signature frequencies. Nat Genet 2019;51:1450–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Razavi P, Chang MT, Xu G, Bandlamudi C, Ross DS, Vasan Net al. The genomic landscape of endocrine-resistant advanced breast cancers. Cancer Cell 2018;34:427–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. The Oslo Breast Cancer Consortium (OSBREAC), Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman Cet al. The landscape of cancer genes and mutational processes in breast cancer. Nature 2012;486:400–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yates LR, Knappskog S, Wedge D, Farmery JHR, Gonzalez S, Martincorena Iet al. Genomic evolution of breast cancer metastasis and relapse. Cancer Cell 2017;32:169–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Siegel MB, He X, Hoadley KA, Hoyle A, Pearce JB, Garrett ALet al. Integrated RNA and DNA sequencing reveals early drivers of metastatic breast cancer. J Clin Invest 2018;128:1371–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hu Z, Li Z, Ma Z, Curtis C. Multi-cancer analysis of clonality and the timing of systemic spread in paired primary tumors and metastases. Nat Genet 2020;52:701–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xu G, Chhangawala S, Cocco E, Razavi P, Cai Y, Otto JEet al. ARID1A determines luminal identity and therapeutic response in estrogen-receptor-positive breast cancer. Nat Genet 2020;52:198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jeong G-Y, Park MK, Choi H-J, An HW, Park Y-U, Choi H-Jet al. NSD3-induced methylation of H3K36 activates NOTCH signaling to drive breast tumor initiation and metastatic progression. Cancer Res 2021;81:77–90. [DOI] [PubMed] [Google Scholar]
- 28. Costa C, Wang Y, Ly A, Hosono Y, Murchie E, Walmsley CSet al. PTEN loss mediates clinical cross-resistance to CDK4/6 and PI3Kα inhibitors in breast cancer. Cancer Discov 2020;10:72–85. [DOI] [PubMed] [Google Scholar]
- 29. Fry DW, Harvey PJ, Keller PR, Elliott WL, Meade M, Trachet Eet al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol Cancer Ther 2004;3:1427–38. [PubMed] [Google Scholar]
- 30. Formisano L, Lu Y, Servetto A, Hanker AB, Jansen VM, Bauer JAet al. Aberrant FGFR signaling mediates resistance to CDK4/6 inhibitors in ER+ breast cancer. Nat Commun 2019;10:1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu X-D, Kong W, Peterson CB, McGrail DJ, Hoang A, Zhang Xet al. PBRM1 loss defines a nonimmunogenic tumor phenotype associated with checkpoint inhibitor resistance in renal carcinoma. Nat Commun 2020;11:2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhou H, Liu J, Zhang Y, Huang Y, Shen J, Yang Yet al. PBRM1 mutation and preliminary response to immune checkpoint blockade treatment in non-small cell lung cancer. NPJ Precis Oncol 2020;4:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kato S, Goodman A, Walavalkar V, Barkauskas DA, Sharabi A, Kurzrock R. Hyperprogressors after immunotherapy: analysis of genomic alterations associated with accelerated growth rate. Clin Cancer Res 2017;23:4242–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Prat A, Brase JC, Cheng Y, Nuciforo P, Paré L, Pascual Tet al. Everolimus plus exemestane for hormone receptor-positive advanced breast cancer: a PAM50 intrinsic subtype analysis of BOLERO-2. Oncologist 2019;24:893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Prat A, Chaudhury A, Solovieff N, Paré L, Martinez D, Chic Net al. Correlative biomarker analysis of intrinsic subtypes and efficacy across the MONALEESA phase III studies. J Clin Oncol 2021;39:1458–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Finn RS, Dering J, Conklin D, Kalous O, Cohen DJ, Desai AJet al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res 2009;11:R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fumagalli D, Venet D, Ignatiadis M, Azim HA, Maetens M, Rothé Fet al. RNA sequencing to predict response to neoadjuvant anti-HER2 therapy: a secondary analysis of the NeoALTTO Randomized Clinical Trial. JAMA Oncol 2017;3:227–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pich O, Muiños F, Lolkema MP, Steeghs N, Gonzalez-Perez A, Lopez-Bigas N. The mutational footprints of cancer therapies. Nat Genet 2019;51:1732–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Szekely B, Bossuyt V, Li X, Wali VB, Patwardhan GA, Frederick Cet al. Immunological differences between primary and metastatic breast cancer. Ann Oncol 2018;29:2232–9. [DOI] [PubMed] [Google Scholar]
- 40. Schmid P, Adams S, Rugo HS, Schneeweiss A, Barrios CH, Iwata Het al. Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N Engl J Med 2018;379:2108–21. [DOI] [PubMed] [Google Scholar]
- 41. Adams S, Schmid P, Rugo HS, Winer EP, Loirat D, Awada Aet al. Pembrolizumab monotherapy for previously treated metastatic triple-negative breast cancer: cohort A of the phase II KEYNOTE-086 study. Ann Oncol 2019;30:397–404. [DOI] [PubMed] [Google Scholar]
- 42. Yu J, Green MD, Li S, Sun Y, Journey SN, Choi JEet al. Liver metastasis restrains immunotherapy efficacy via macrophage-mediated T cell elimination. Nat Med 2021;27:152–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. DeNardo DG, Brennan DJ, Rexhepaj E, Ruffell B, Shiao SL, Madden SFet al. Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov 2011;1:54–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu C-X, Musco S, Lisitsina NM, Yaklichkin SY, Lisitsyn NA. Genomic organization of a new candidate tumor suppressor gene, LRP1B. Genomics 2000;69:271–4. [DOI] [PubMed] [Google Scholar]
- 45. André F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HSet al. Alpelisib for PIK3CA -mutated, hormone receptor–positive advanced breast cancer. N Engl J Med 2019;380:1929–40. [DOI] [PubMed] [Google Scholar]
- 46. Robson M, Im S-A, Senkus E, Xu B, Domchek SM, Masuda Net al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med 2017;377:523–33. [DOI] [PubMed] [Google Scholar]
- 47. Mosele F, Remon J, Mateo J, Westphalen CB, Barlesi F, Lolkema MPet al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol 2020;31:1491–505. [DOI] [PubMed] [Google Scholar]
- 48. Juric D, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo Het al. Alpelisib + fulvestrant for advanced breast cancer: subgroup analyses from the phase III SOLAR-1 trial [abstract]. In:Proceedings of the 2018 San Antonio Breast Cancer Symposium; 2018 Dec 4–8; San Antonio, TX. Philadelphia (PA): AACR; 2019. Abstract nr GS3-08. [Google Scholar]
- 49. Zardavas D, Maetens M, Irrthum A, Goulioti T, Engelen K, Fumagalli Det al. The AURORA initiative for metastatic breast cancer. Br J Cancer 2014;111:1881–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
File with the supplementary material & methods, supplementary tables and supplementary figure.
Supplementary file with the listing of the liver genes excluded from transcriptomic analyses.
Data Availability Statement
Instructions to access the manuscript processed data for reproducibility purposes are available at the webpage https://aurora.bigagainstbreastcancer.org/ and can be obtained upon signature of an appropriate data transfer agreement subject to applicable laws. Instructions to access processed or raw manuscript data to perform original research are also available on the webpage and investigators can contact aurora.researchproposals@bigagainstbc.org for enquiries. Access to data for research will be granted upon review of a project proposal and endorsement by the study Steering Committee, and after entering into an appropriate data access agreement subject to applicable laws.