Abstract
New approaches beyond PD-1/PD-L1 inhibition are required to target the immunologically diverse tumor microenvironment (TME) in high-grade serous ovarian cancer (HGSOC). In this study, we explored the immunosuppressive effect of B7-H3 (CD276) via the CCL2–CCR2–M2 macrophage axis and its potential as a therapeutic target. Transcriptome analysis revealed that B7-H3 is highly expressed in PD-L1–low, nonimmunoreactive HGSOC tumors, and its expression negatively correlated with an IFNγ signature, which reflects the tumor immune reactivity. In syngeneic mouse models, B7-H3 (Cd276) knockout (KO) in tumor cells, but not in stromal cells, suppressed tumor progression, with a reduced number of M2 macrophages and an increased number of IFNγ+CD8+ T cells. CCL2 expression was downregulated in the B7-H3 KO tumor cell lines. Inhibition of the CCL2–CCR2 axis partly negated the effects of B7-H3 suppression on M2 macrophage migration and differentiation, and tumor progression. In patients with HGSOC, B7-H3 expression positively correlated with CCL2 expression and M2 macrophage abundance, and patients with B7-H3–high tumors had fewer tumoral IFNγ+CD8+ T cells and poorer prognosis than patients with B7-H3–low tumors. Thus, B7-H3 expression in tumor cells contributes to CCL2–CCR2–M2 macrophage axis–mediated immunosuppression and tumor progression. These findings provide new insights into the immunologic TME and could aid the development of new therapeutic approaches against the unfavorable HGSOC phenotype.
Introduction
Ovarian cancer is the eighth leading cause of cancer death in women worldwide, with an estimated figure of more than 200,000 deaths per year (1). In addition to conventional surgery and chemotherapy, the introduction of anti-VEGF and PARP inhibitors as therapeutic modalities has remarkably prolonged the duration of progression-free survival (PFS), even though progression remains inevitable in the majority of cases (2, 3). The efficacy of PD-1/PD-L1 inhibition therapy is limited in ovarian cancer, as evidenced by clinical trials, and immunotherapy has not yet become the standard treatment option (4–6).
Of all the histologic types of ovarian cancer, high-grade serous ovarian cancer (HGSOC) remains the most common and the most aggressive (7, 8). Although the tumor microenvironment (TME) of HGSOC varies widely, it is well characterized by four subtypes based on gene expression profiles: differentiated, immunoreactive, mesenchymal, and proliferative subtypes (9, 10). The immunoreactive subtype is characterized as tumors with high expression of IFNγ and PD-L1, a representative antitumor cytokine and an immunomodulatory molecule upregulated in response to IFNγ, respectively, and patients have a better prognosis (10–12). The mesenchymal type is characterized as epithelial–mesenchymal transition–high, immunosuppressive tumors with poor prognosis (10, 13, 14). The differentiated and proliferative subtypes consist of immune "desert" tumors (14). PD-1/PD-L1 inhibition therapy is relatively effective in treating PD-L1–high immunoreactive tumors. However, nonimmunoreactive phenotypes, which comprise a majority of HGSOC tumors, respond poorly to the treatment (4). To elucidate what contributes to the differences in the TME, and new immunologic approaches to treat nonimmunoreactive phenotypes other than PD-1/PD-L1 inhibition, are required.
B7-H3 is a transmembrane protein from the B7 family; it was first reported in 2001 (15). Developments in therapies targeting B7-H3, including antibody–drug conjugates, radioimmunotherapy, and chimeric antigen receptor T cells, which focus on its high selective expression in tumor tissues, highlight the potential of B7-H3 as a therapeutic target (16–18). B7-H3 seems to play complex roles in modulating the TME. Not only does it exert its immunomodulatory effects by directly acting on target cells as an immune checkpoint (15, 19, 20), it is also involved in the intracellular signaling of cancer cells, such as the STAT3 pathway (21–23). Its contribution to immunosuppressive TME, other than as an immune checkpoint molecule, remains to be fully elucidated.
Here, we demonstrate that B7-H3 expression is upregulated in the nonimmunoreactive tumors of HGSOC and that B7-H3 contributes to ovarian cancer progression via CCL2–CCR2–M2 macrophage axis–mediated immunosuppression, in both mouse syngeneic ovarian cancer models and HGSOC cases. Our results not only augment the current understanding of the immunologic heterogeneity of HGSOC but also provide a foundation for stratification of the immunosuppressive HGSOC phenotype with a B7-H3–high, M2 macrophage–rich TME and facilitate the development of new treatment strategies other than PD-1/PD-L1 inhibition that can specifically benefit patient groups with extremely poor prognosis.
Materials and Methods
Study approval
This study was approved by the Kyoto University Graduate School and Faculty of Medicine Ethics Committee (reference number G531) and conforms to the Declaration of Helsinki. Informed consent was obtained from all participants via an opt-in approach (wherein participants signed a printed informed consent document) or an opt-out approach (wherein participants were informed about the study through the website). All animal studies were approved by the Kyoto University Animal Research Committee.
Human samples
Formalin-fixed, paraffin-embedded (FFPE) primary site tumor specimens (n = 62) and fresh primary site tumor samples (n = 28) were collected from patients with HGSOC, who underwent primary surgery at the Kyoto University Hospital (Kyoto, Japan) between 1998 and 2015. Patients without primary site resection surgery or those who received chemotherapy prior to surgery were excluded. Blood samples from patients with HGSOC (n = 23) and healthy female donors (n = 10) were also collected between 2012 and 2020. FFPE samples were stored at room temperature, and fresh tumor samples and blood samples were stored at −80°C until use.
Bioinformatics analysis
Gene expression data of 30 HGSOC specimens from the Kyoto University Hospital (Kyoto, Japan) that were partially deposited in the Gene Expression Omnibus (RRID: SCR_005012, accession numbers: GSE39204 and GSE55512) were classified into the four transcriptome subtypes (24), according to the Classification of Ovarian Cancer (CLOVAR) subtype signatures (10). Differentially expressed genes (DEG) between the immunoreactive (n = 8) and nonimmunoreactive (n = 22) subtypes were extracted using the Samroc method as described previously (25). The gene expression profiles from The Cancer Genome Atlas-Ovarian Cancer (TCGA-OV) RNA-sequencing dataset (n = 263) from TCGA Data Portal (RRID: SCR_003193, illuminahiseq_rnaseqv2_Level_3_RSEM_genes_normalized_data files obtained on October 19, 2015) was used for comparison. Single-sample gene set enrichment analysis (26), obtained from GenePattern software (RRID:SCR_003201, version 3.5.0), was performed to evaluate the IFNγ signature activity scores using the previously reported gene set (27) using R software (RRID: SCR_001905, version 3.6.1).
Cell culture and transfection
The OV2944-HM-1 (HM-1) mouse ovarian cancer cell line was purchased from RIKEN BioResource Center (RRID: SCR_003250, catalog no. RCB1483; RRID: CVCL_E954; Tsukuba, Japan) in January 2003. HM-1 cells were cultured and maintained in minimal essential medium (MEM)-alpha (catalog no. 12571063, Thermo Fisher Scientific) supplemented with 10% (volume for volume) heat-inactivated FBS (catalog no. S1810, Biowest) and penicillin–streptomycin (100 IU/mL penicillin and 100 μg/mL streptomycin; catalog no. 26253-84, Nacalai Tesque) in an atmosphere of 5% CO2 at 37°C. The ID8 mouse ovarian cancer cell line was kindly provided by Katherine Roby of The University of Kansas Medical Center (Kansas City, KS; RRID: CVCL_IU14) in September 2009. Human ovarian cancer cell lines OVCAR3 (RRID: CVCL_0465) and OVCA420 (RRID: CVCL_3935) were kindly provided by Susan K. Murphy of Duke University (Durham, NC) in June 2007. Both ID8 and human ovarian cancer cells were cultured and maintained in RPMI1640 medium (Invitrogen) supplemented with 10% FBS and penicillin–streptomycin in an atmosphere of 5% CO2 at 37°C. All cell line–based experiments were performed before passage 20. All cell lines were regularly tested for Mycoplasma contamination (date of last check: April 12, 2021) using a Mycoplasma PCR Detection Kit (catalog no. G238; Applied Biological Materials). Human cell lines (OVCAR3 and OVCA420) were reauthenticated on April 14, 2021, by short tandem repeat analysis.
For CRISPR/Cas9-mediated Cd276 knockout (B7-H3 KO), target sequences were determined as described previously (16). Three single-guide RNAs (sgRNA) and a control sgRNA were synthesized by FASMAC. A mixture of the RNAs and Guide-it recombinant Cas9 (catalog no. 632641, Takara Bio Inc.) was transfected into the HM-1 and ID8 cells using the TransIT-X2 Dynamic Delivery System (catalog no. MIR6003, Mirus Bio). The cells were incubated with rabbit anti-CD276 (1:1,000 dilution, clone: EPNCIR122, catalog no. ab134161; RRID: AB_2687929, Abcam) for 20 minutes at 4°C and with Goat Anti-Rabbit IgG H&L (Alexa Fluor 647; 1:2,000 dilution, catalog no. ab150079; RRID:AB_2722623, Abcam) for 10 minutes at 4°C. Alexa Fluor 647–negative cell populations were sorted by flow cytometry using BD FACSAria II Cell Sorter (RRID: SCR_018934, BD Biosciences). The purity of Alexa Fluor 647–negative cells was >99%. Single cells were isolated by limiting dilution and cultured again on 96-well plates (catalog no. 655180, Greiner Bio-One) for 8 days under the same conditions described in the section above, followed by expansion. Finally, B7-H3 expression deficiency was confirmed using FACS analysis and quantitative PCR (qPCR).
B7-H3–knockdown HGSOC cells, OVCAR3-sh-B7-H3 and OVCA420-sh-B7-H3, and their control cells were generated by lentiviral-mediated transfection with short hairpin RNAs (shRNA) targeting B7-H3 or a nonsilencing control (sh1, catalog no. TRCN0000128599; sh2, catalog no. TRCN0000436667; sh control, catalog no. SHC002; Sigma-Aldrich). The sgRNA and shRNA sequences are listed in Supplementary Table S1.
Mice
Five- to 6-week-old female C57BL6 (strain code: 027) and B6C3F1 (C57BL6×C3/He F1, strain code: 031) mice were purchased from CLEA Japan and used in immunocompetent mouse experiments. Five- to 6-week-old female BALB/c-nu (Foxn1nu, strain code: 194) mice were purchased from CLEA Japan and used in immunodeficient mouse experiments. The C57BL6 background Cd276−/− (B7-H3 KO) mice were kindly provided by Brad St. Croix of the NCI (Bethesda, MD). Animals were maintained under specific pathogen–free conditions.
Tumor models
A total of 1 × 106 HM-1-B7-H3 KO cells or control cells were inoculated intradermally into the shaved abdominal lesion of B6C3F1 and nude mice. A total of 5 × 105 HM-1-B7-H3 KO or control cells were injected into the abdominal cavity of B6C3F1 or nude mice (intraperitoneal model). Similarly, 5 × 106 ID8 B7-H3 KO or control cells were injected into the abdominal cavity of C57BL6 or nude mice (intraperitoneal model). Intradermal tumor size was measured using calipers and calculated as follows: volume = length × width × height × π/6. In the intraperitoneal models, tumor growth was evaluated on the basis of changes in body weight, which reflect the amount of ascites and omental tumor weight. Survival analysis was also performed in these models. Intradermal tumor size and body weight were measured once or twice a week. Intradermal tumors of HM-1-B7-H3 KO or control cells at days 10, 12, and 25 were used for RNA sequencing, flow cytometry, IHC, and ELISA, respectively. Intraperitoneal tumors of ID8-B7-H3 KO or control cells at days 63 and 67 were used for ELISA and flow cytometry, respectively. For B7-H3 KO mouse studies, only Cd276+/+ and Cd276−/− littermates derived from Cd276 heterozygous intercrosses were used for comparison. The CCR2 antagonist (RS504393: 2 mg/kg body weight; catalog no. 17330, Cayman Chemical) treatment was initiated a day after tumor cell inoculation and was administered intraperitoneally daily. Mice were euthanized before becoming moribund.
Cell treatments
OVCAR3 and OVCA420 human ovarian cancer cells (1 × 105) were cultured in 6-well plates (catalog no. 657160, Greiner Bio-One) as described in the “Cell culture and transfection” section above and treated with 20 ng/mL recombinant IFNγ (catalog no. 570202, BioLegend; ref. 28) or 0.1% BSA (catalog no. 7284, Sigma-Aldrich) as a control the following day. Cells were analyzed for the expression of B7-H3 (CD276), PD-L1 (CD274), and PD-L2 (PDCD1LG2) using qPCR and flow cytometry (as described below) after treatment for 24 and 48 hours, respectively.
For STAT3 inhibition, 1 × 105 cells were treated with the STAT3 inhibitor C188-9 (catalog no. S8605, Selleck Chemicals) for the mouse cell lines HM-1 and ID8 (29) or with Stattic (catalog no. ab120952, Abcam) for the human cell lines OVCAR3 and OVCA420 (30) at 1.25 and 5 μmol/L for 24 hours, respectively. Ccl2 and CCL2 expression levels were analyzed using qPCR as described below.
Tumor cell proliferation assay
HM-1-B7-H3 KO cells, ID8-B7-H3 KO cells, or their respective control cells were seeded in 96-well plates at 1 × 103 cells/well (Greiner Bio-One) and incubated for 3 days in an atmosphere of 5% CO2 at 37°C. The number of viable cells in each well was examined every 24 hours by absorbance at 450 nm using Cell Count Reagent SF (catalog no. 07553-44, Nacalai Tesque) and iMark microplate reader (catalog no. 168-1130J, Bio-Rad). Absorbance values at 72 hours were compared using Microsoft Excel (RRID: SCR_016137).
Macrophage and monocyte selection
For tumor-derived macrophage selection, single cells from the tumors were prepared as described in the “Flow cytometry” section. F4/80-positive (F4/80+) macrophages were positively selected using anti-Mouse F4/80 PE antibody (clone: BM8, catalog no. 123109; RRID: AB_893498, BioLegend), EasySep Mouse PE positive selection kit II (catalog no. 17666, Stemcell Technologies), and EasySep Magnet (catalog no. 18000, Stemcell Technologies). The purity of F4/80+ cells was >90% as confirmed by flow cytometry. For mouse monocyte selection, single cells from the bone marrow of the femur and tibia of B6C3F1 or C57BL6 non–tumor-bearing mice were collected by mechanical dissociation. For human monocyte selection, peripheral blood mononuclear cells (PBMC) of healthy donors were separated using Leucosep (catalog no. 227290, Greiner) and Lymphosep (catalog no. 1692254, MP Biomedicals). Ly6C-positive mouse monocytes and CD14-positive human monocytes were positively selected by EasySep Magnet as described above using anti-Mouse Ly6C PE antibody (clone: HK1.4, catalog no. 128007; RRID: AB_1186133, BioLegend) and anti-Human CD14 PE antibody (clone: M5E2, catalog no. 301806; RRID: AB_314188, BioLegend). The purity of Ly6C-positive and CD14-positive cells was >90%, respectively, as confirmed by flow cytometry.
RNA extraction and reverse transcription qPCR
Human (OVCAR3 and OVCA420) and mouse (HM-1 and ID8) cells or macrophages isolated from the tumors of HM-1-B7-H3 KO or control cells (as described in the preceding section) were lysed in RLT buffer (catalog no. 1015750, Qiagen) containing 1% 2-mercaptoethanol (catalog no. 1610710, Bio-Rad). Intradermal tumors of HM-1-B7-H3 KO cells or control cells were treated in a bead homogenizer with RLT buffer and centrifuged at 16,000 ×g for 1 minute at 4°C. Total RNA was extracted from the lysates using RN easy Mini Kit (catalog no. 74104, Qiagen).
A ReverTra Ace qPCR RT Kit (catalog no. FSQ-101, TOYOBO) was used for cDNA synthesis. Gene amplification was performed using PowerUp SYBR Green Master Mix (catalog no. A25742, Thermo Fisher Scientific) on a StepOne Plus real-time PCR system (Applied Biosystems). Fifty nanograms of RNA was used as the template for PCR to detect the relative expression levels of CD276, CD274 (PD-L1), PTCD1LG2 (PD-L2), and CCL2 for human cells, and Cd276, Ccl2, Arg1, Il10, and Ifng for mouse cells; the primer sequences are listed in Supplementary Table S1. Relative expression levels were calculated using the 2−ΔΔCt method. GAPDH/Gapdh was used for normalization.
Immunostaining and evaluation
FFPE sections of the 62 HGSOC samples and 3 to 6 mouse tumors of HM-1-B7-H3 KO cells or control cells were immunostained as described previously (31). Epitope retrieval was performed at 120°C for 10 minutes using citrate buffer at pH 6.0 (B7-H3) or Tris-EDTA buffer at pH 9.0 (αSMA, CD206, CD8, and IFNγ). The kits, antibodies, and dilutions used are listed in Supplementary Table S2. Tumor cell B7-H3 expression was evaluated as described previously (32) and scored as follows: 0, negative or very weak; 1, weak; 2, moderate, and 3, strong (16). Scores of 0/1 and 2/3 were defined as B7-H3 low and B7-H3 high, respectively. Tumor-infiltrating CD206+ cells (200×) were counted from five fields, and the average number of cells was calculated. For the evaluation of CD8+ and IFNγ+CD8+ T cells, the number of cells in the representative CD8+ cell–rich field was counted at 200× magnification. The sensitivity and specificity of the IFNγ antibody was confirmed using appendix (catalog no. CS802695, OriGene) and tonsil (kindly provided by Kyoto University Department of Otolaryngology, Head and Neck Surgery, Kyoto, Japan) tissue as positive controls (Supplementary Fig. S1A and S1B). Slides were analyzed using a BZ 9000 fluorescence microscope (RRID: SCR_015486, Keyence).
Flow cytometry
IFNγ- or BSA-treated OVCAR3 and OVCA420 cells, HM-1-B7-H3 KO cells and ID8-B7-H3 KO cells or their respective control cells, and OVCAR3-sh-B7-H3 and OVCA420-sh-B7-H3 or their respective control cells were collected from the culture plates using trypsin (catalog no. T4549, Sigma-Aldrich) and incubated for 20 minutes at 4°C with the corresponding antibodies listed in Supplementary Table S2. Tumors of HM-1-B7-H3 KO cells, ID8-B7-H3 KO cells, or their control cells were minced into thin pieces and dissociated in 100 μg/mL of DNase I (catalog no. 11284932001, Roche) and 1 mg/mL of collagenase IV (catalog no. CLS4, Worthington) in RPMI1640 medium. The tissues were incubated for 1 hour at 37°C with agitation. Single cells were prepared through a 70 μm nylon mesh strainer (catalog no. 542070, Greiner Bio-One). FcR blocking for leukocytes was performed with FcR Blocking Reagent (for mouse samples: catalog no. 130-092-575, RRID: AB_2892833; for human samples: catalog no. 130-059-901, RRID:AB_2892112, Miltenyi Biotec) for 10 minutes at 4°C. Referring to ref. 33, tumor-infiltrating immune cells were stained to detect CD4+ T cells (CD3+CD4+CD8−), CD8+ T cells (CD3+CD4−CD8+), M2 macrophages (CD45+F4/80+CD206+), M1 macrophages (CD45+F4/80+CD80+ or CD45+F4/80+CD86+), and dendritic cells (DC, CD45+CD11c+MHC class 2+) with the antibodies listed in Supplementary Table S2 for 30 minutes at 4°C. Gating strategies for each cell subset are shown in Supplementary Fig. S2. For IFNγ staining, single cells from the tumor were incubated for 4 hour with eBioscience Cell Stimulation Cocktail (catalog no. 00-4970-93, Invitrogen) and for 2 hours with Brefeldin A (catalog no. B7651, Sigma-Aldrich). BD Cytofix/Cytoperm Fixation/Permeabilization Kit (catalog no. 554714, BD Biosciences) was used for intracellular staining. Nonviable cells were stained with 7-amino-actinomycin D solution (catalog no. 51-68981E, BD Biosciences) or DAPI solution (catalog no. D1306, Life Technologies) and gated out. MACS Quant Analyzer 10 (Miltenyi Biotec) and FlowJo (RRID: SCR_008520, FlowJo LLC) were used for analysis.
T-cell proliferation assay
For T-cell selection, single cells from the spleens of B6C3F1 mice bearing tumors of HM-1-B7-H3 KO cells or control cells harvested at day 12 were collected by mechanical dissociation. Red blood cells were lysed with Pharm Lyse buffer (catalog no. 555899, BD Biosciences). T cells were negatively selected using mouse Pan-T-cell isolation kit II (catalog no. 130-095-130, Miltenyi Biotec), an LS column (catalog no. 130-042-401, Miltenyi Biotec), and QuadroMACS Separator (catalog no. 130-090-976, Miltenyi Biotec). The purity of CD3+ cells was >90% as confirmed by flow cytometry.
T cells were labeled using 10 mmol/L carboxyfluorescein succinimidyl ester (CFSE; catalog no. 600121, Cayman Chemical). T cells (1 × 105) were cocultured with F4/80+ macrophages (obtained as described in the “Macrophage and monocyte selection” section above) at 2:1, 4:1, and 8:1 ratios with RPMI1640 medium supplemented with 10% FBS, 2 mmol/L of l-glutamine (catalog no. 25030-081, Gibco), and 50 μmol/L of 2-mercaptoethanol for 72 hours in an atmosphere of 5% CO2 at 37°C. During incubation, T cells were activated with Dynabeads Mouse T-activator CD3/28 (1:100 dilution, catalog no. 11453D, Thermo Fisher Scientific). Activated T cells without macrophages were used as a positive control. After 72 hours, T-cell proliferation was examined using flow cytometry according to the decrease of the CFSE fluorescence intensity. MACS Quant Analyzer 10 (Miltenyi Biotec) and FlowJo (FlowJo) were used for analysis. The culture supernatants were collected and used for ELISA.
ELISAs
Mouse IFNγ levels in the cell culture supernatant of T cells cocultured with macrophages were measured using Mouse IFNγ Quantikine ELISA Kit (catalog no. MIF00, R&D Systems). HM-1-B7-H3 KO cells, ID8-B7-H3 KO cells or their control cells, and OVCAR3-sh-B7-H3 and OVCA420-sh-B7-H3 or their control cells were seeded at a density of 1 × 105 with 500 μL of the corresponding medium (see “Cell culture and transfection” section) in 24-well plates (catalog no. 662160, Greiner Bio-One). Cell culture supernatants were collected after 24 hours with a 0.45 μm Millex filter (catalog no. SLHVR33RS, Merck Millipore) and used for ELISA. These supernatants were also used for migration and monocyte differentiation assays (described in detail below) as the tumor cell–conditioned medium (TCM). Human and mouse tumor tissues were treated in a bead homogenizer, and sonicated in 1× RIPA buffer (catalog no. 89900, Thermo Fisher Scientific) containing a protease inhibitor cocktail (catalog no. 03969-21, Nacalai Tesque) and a phosphatase inhibitor cocktail (catalog no. 07575-51, Nacalai Tesque), followed by centrifugation at 16,000 ×g for 5 minutes at 4°C. Human tumor lysates were used for B7-H3 and CCL2 detection at 75 and 200 μg as protein amount, respectively, and 20 μg as protein amount of mouse tumor lysates were used for CCL2 detection. Mouse CCL2 protein levels in culture supernatants and tumor lysates were measured using the Mouse CCL2/JE/MCP1 DuoSet kit (catalog no. DY479-05, R&D Systems). Human CCL2 protein levels in culture supernatants, tumor lysates, and serum samples were measured using Human CCL2/MCP-1 Quantikine ELISA Kit (catalog no. DCP00, R&D Systems). Human B7-H3 protein levels in tumor lysates were measured using Human B7-H3 Quantikine ELISA Kit (catalog no. DB7H30, R&D Systems). The absorbance readings at 570 nm were subtracted from the readings taken at 450 nm using an iMark microplate reader (Bio-Rad) to establish standard curves for calculation of the concentration. Microsoft Excel was used for analysis.
Immunoblotting
Nuclear proteins were collected from HM-1-B7-H3 KO cells, ID8-B7-H3 KO cells or their control cells, and OVCAR3-sh-B7-H3 and OVCA420-sh-B7-H3 or their control cells using NE-PER nuclear and cytoplasmic extraction reagents (catalog no. 78833, Thermo Fisher Scientific). Protein (20 μg) of HM-1-B7-H3 KO cells or their control cells, and 10 μg protein of the other cells were loaded onto 8% acrylamide gels. Proteins were subsequently separated using SDS-PAGE and transferred onto polyvinylidene difluoride membranes (catalog no. 1620177, Bio-Rad). After blocking for 1 hour with Blocking One-P (catalog no. 05999-84, Nacalai Tesque), the membranes were then immunoblotted with the antibodies listed in Supplementary Table S2 at the indicated dilutions. The membranes were incubated with the primary antibody overnight at 4°C and with the secondary antibody for 1 hour at room temperature. Bands were visualized using ChemiDoc XRS+ Systems (Bio-Rad). Signals were quantified using Image Lab 2.0 (RRID: SCR_014210, Bio-Rad).
RNA sequencing
Total RNA from HM-1-B7-H3 KO cells, ID8-B7-H3 KO cells or their control cells, or intradermal tumors of HM-1-B7-H3 KO cells or their control cells at day 10 of tumor growth were extracted as described in the “RNA extraction and reverse transcription qPCR” section and used for RNA sequencing. Sequencing was performed on an Illumina NovaSeq 6000 platform (RRID: SCR_016387, Illumina) to generate 100 bp paired-end reads. Raw reads were trimmed using Trim Galore (RRID: SCR_011847), and the resulting reads were aligned to the mouse reference genome GRCm38/mm10 using STAR (RRID: SCR_004463). Differential expression analysis was performed using DESeq2 (RRID: SCR_015687) between control (n = 3) and B7-H3 KO (n = 3) groups of the ID8 and HM-1 cell lines, and HM-1 intradermal tumors (processed as indicated in above sections). After excluding the genes showing low expression and low fold change between groups (“baseMean” > 1000; |log2FoldChange| > 2; Padj < 0.01), the remaining genes were considered as DEGs and listed in Supplementary Tables S3–S5. RNA-sequencing data have been deposited in the Gene Expression Omnibus database, under accession number GSE174137.
In vitro M2 macrophage generation
Mouse and human monocytes (obtained as described in the “Macrophage and monocyte selection” section) were cultured in 6-well plates (catalog no. 657160, Greiner Bio-One) at 3 × 106 cells/well and 6 × 105 cells/well, respectively, with RPMI1640 medium supplemented with 40 ng/mL of MCSF (mouse: catalog no. 576402, human: catalog no. 574802, BioLegend) and 40 ng/mL of IL6 (mouse: catalog no. 216-16, PeproTech, human: catalog no. 570802, BioLegend) for 6 to 7 days in an atmosphere of 5% CO2 at 37°C and used for migration assays. After incubation, >70% of the cultured mouse cells were F4/80+ and CD206+, and >85% of the cultured human cells were CD206+ as confirmed by flow cytometry.
Migration assays
Monocytes or the generated M2 macrophages (mouse: 1 × 105 cells, human: 5 × 104 cells, isolated as described in the “Macrophage and monocyte selection” and “In vitro M2 macrophage generation” sections, and pretreated with 2 μmol/L RS504393 as described in “Tumor models”) or DMSO was plated in the upper compartment of 8 μm cell culture insert (catalog no. 353097, Corning), and 500 μL of TCM of HM-1-B7-H3 KO cells, ID8-B7-H3 KO cells or their control cells, and OVCA420-sh-B7-H3 or their control cells (generated as described in the “ELISAs” section) was added to the lower compartment. Plates were incubated for 1 hour in an atmosphere of 5% CO2 at 37°C, and migrated cells were counted using CountBright Absolute Counting beads (catalog no. 36950, Thermo Fisher Scientific).
Monocyte differentiation assay
Mouse bone marrow monocytes (isolated and cultured as described in the “Macrophage and monocyte selection” section) were plated in the upper compartment of 0.4 μm cell culture inserts (catalog no. 353095, Corning) at a density of at 1 × 105, and 500 μL of TCM of HM-1-B7-H3 KO cells, ID8-B7-H3 KO cells or their control cells (generated as described in the “ELISAs” section) supplemented with 20 μg/mL anti-mouse CCL2 (catalog no. AB-479-NA, RRID:AB_354366; R&D Systems) or goat IgG control (catalog no. AB-108-C, RRID:AB_354267; R&D Systems) was added to the lower compartment. Plates were incubated for 72 hours in an atmosphere of 5% CO2 at 37°C, and F4/80+CD206+ cells were then evaluated using flow cytometry, as described above.
Statistical analysis
At least three in vitro experiments and at least two in vivo experiments were independently performed. Data from one representative experiment are presented. For mouse experiments, cages of mice were randomly allocated to the experimental groups. Sample sizes were chosen to assure reproducibility of the experiments in accordance with the replacement, reduction, and refinement principles of the animal ethics regulation. Grouped data are shown as the average ± SEM in all figures except non-normally distributed data for clinical samples, which shows the median with 95% confidence intervals. The investigator was not blinded to the group allocation. A priori power analysis was not performed for human samples. Survival curves were constructed using the Kaplan–Meier method. Prognostic factors for PFS and overall survival (OS) of patients with HGSOC were assessed by univariate and multivariate analysis using the Cox proportional hazards regression model. Statistical analysis was performed using R software and GraphPad Prism 7 (RRID: SCR_002798, GraphPad Software), and the appropriate tests are indicated in the figure legends. Significance was set as *, P < 0.05; **, P <0.01; and ***, P <0.001.
Results
B7-H3 expression is upregulated in the nonimmunoreactive TME of HGSOC
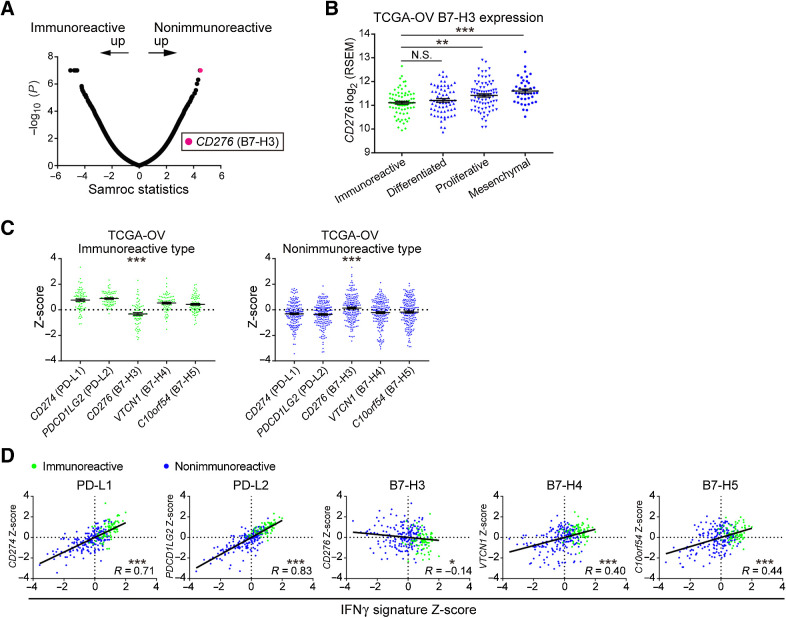
We first identified the genes acting antagonistic to the immunoreactive TME in HGSOC based on our RNA expression array data. CD276 (B7-H3) was the most upregulated gene in the nonimmunoreactive subtype (Fig. 1A). Analysis of TCGA-OV revealed that B7-H3 expression was higher in the proliferative and mesenchymal subtypes than in the immunoreactive subtypes (P = 0.004 and P < 0.001; Fig. 1B). Among the T cell–suppressive immune checkpoint molecules of the B7 family, only B7-H3 expression was lower in the immunoreactive subtype and higher in the nonimmunoreactive subtype (both P < 0.001; Fig. 1C).
Figure 1.
CD276 (B7-H3) expression is upregulated in the nonimmunoreactive TME of HGSOC. A, Microarray analysis of HGSOC clinical samples from Kyoto University (n = 30). The immunoreactive (n = 8) and nonimmunoreactive (n = 22) subtypes were compared using the Samroc method. The magenta dot represents CD276 (B7-H3). B,CD276 expression among the molecular subtypes of 263 HGSOC samples from TCGA-OV. Data are presented as the mean ± SEM; **, P < 0.01; ***, P < 0.001; N.S., not significant by one-way ANOVA with Tukey multiple comparison test. C, Gene expression comparison of T cell–suppressive B7 family molecules in the immunoreactive and nonimmunoreactive subtypes of TCGA-OV. Data are presented as the mean ± SEM after Z-score normalization. ***, P < 0.001 by one-way ANOVA. D, Pearson correlation analysis of the IFNγ signature activity scores and expression of T cell–suppressive B7 family molecules in TCGA-OV. Values after Z-score normalization are plotted. *, P < 0.05; ***, P < 0.001. (Green and blue dots represent the immunoreactive and nonimmunoreactive subtypes, respectively, in B–D.
A clear positive correlation was observed between the IFNγ signature, which is a major characteristic of the immunoreactive tumors and can be used as an index for predicting the therapeutic efficacy of immune checkpoint inhibitor (ICI) treatment (27), and the other B7 family molecules. By contrast, the IFNγ signature negatively correlated with B7-H3 expression (P = 0.020; Fig. 1D). Based on the response to IFNγ, we performed qPCR or flow cytometry and observed significantly increased expression of PD-L1 and PD-L2 and unchanged or slightly decreased expression of B7-H3 in HGSOC cell lines (Supplementary Fig. S3A and S3B). Moreover, PD-L1 expression negatively correlated with B7-H3 expression (Supplementary Fig. S3C). These results indicate that B7-H3 expression is associated with the nonimmunoreactive TME and that B7-H3 is involved in immunosuppression, albeit via a different mechanism than that of PD-L1.
B7-H3 mediates murine ovarian cancer growth via immune system regulation
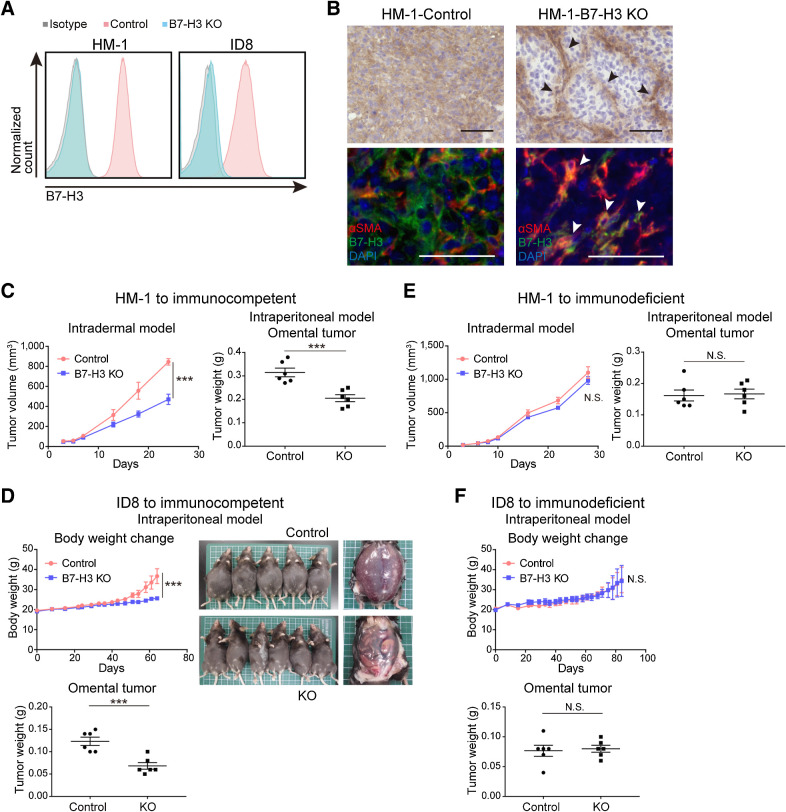
To investigate the function of B7-H3 in tumor cells, we generated three B7-H3 KO HM-1 and ID8 mouse ovarian cancer cell lines using CRISPR/Cas9. Both cell lines express B7-H3 and are resistant to PD-1/PD-L1 inhibition therapy (34, 35). We confirmed B7-H3 KO by flow cytometry and immunostaining (Fig. 2A and B; Supplementary Fig. S4A). B7-H3 KO did not affect cell proliferation in vitro (Supplementary Fig. S4B). In immunocompetent mice, B7-H3 KO delayed the growth of HM-1 intradermal and intraperitoneal tumors, as well as ID8 intraperitoneal tumors (Fig. 2C and D; Supplementary Fig. S4C). B7-H3 suppression significantly prolonged the survival of mice with ID8 tumors, a trend that was also observed with HM-1 tumors (Supplementary Fig. S5A). In contrast, in immunodeficient mice, tumor growth and survival were not affected by B7-H3 expression in either HM-1 or ID8 cells (Fig. 2E and F; Supplementary Fig. S5B). These data suggest that B7-H3 is involved in tumor development via immune-related mechanisms. Next, to verify the role of B7-H3 in stromal cells, we injected ID8 cells into Cd276 KO (B7-H3 KO) mice and observed that B7-H3 expression did not significantly affect tumor development (Supplementary Fig. S6). Overall, these results indicate that B7-H3 expression in tumor cells is important for tumor progression.
Figure 2.
B7-H3 contributes to tumor progression via an immune-related mechanism in murine ovarian cancer models. A, Flow cytometry of B7-H3 KO HM-1 and ID8 and their respective controls. Blue, pink, and gray histograms represent B7-H3 KO cells, B7-H3 control cells, and the isotype control, respectively. B, Representative B7-H3 IHC (top) and immunofluorescence (bottom) images of controls and HM-1 B7-H3 KO tumors for the indicated markers. αSMA-positive fibroblasts represented by black and white arrowheads. Scale bar, 50 μm. Positive cells were stained brown in IHC, and red (αSMA), green (B7-H3), and blue (DAPI) in immunofluorescence images. C, Intradermal tumor volume (left, n = 6) and omental tumor weight at day 10 (right, n = 6) of immunocompetent mice injected with HM-1 B7-H3 KO or control cells. D, Changes in body weight (top, n = 5–6) and omental tumor weight at day 52 (bottom, n = 6) of immunocompetent mice intraperitoneally injected with ID8 B7-H3 KO or control cells. E, Intradermal tumor volume (left, n = 5) and omental tumor weight at day 10 (right, n = 6) of immunodeficient mice injected with HM-1 B7-H3 KO or control cells. F, Changes in body weight (top, n = 6) and omental tumor weight at day 52 (bottom, n = 6) of immunodeficient mice intraperitoneally injected with ID8 B7-H3 KO or control cells. C–F, Data are presented as the mean ± SEM; ***, P < 0.001; N.S., not significant, unpaired t test.
B7-H3 suppression reduces M2 macrophages and increases T cell–produced IFNγ in the TME
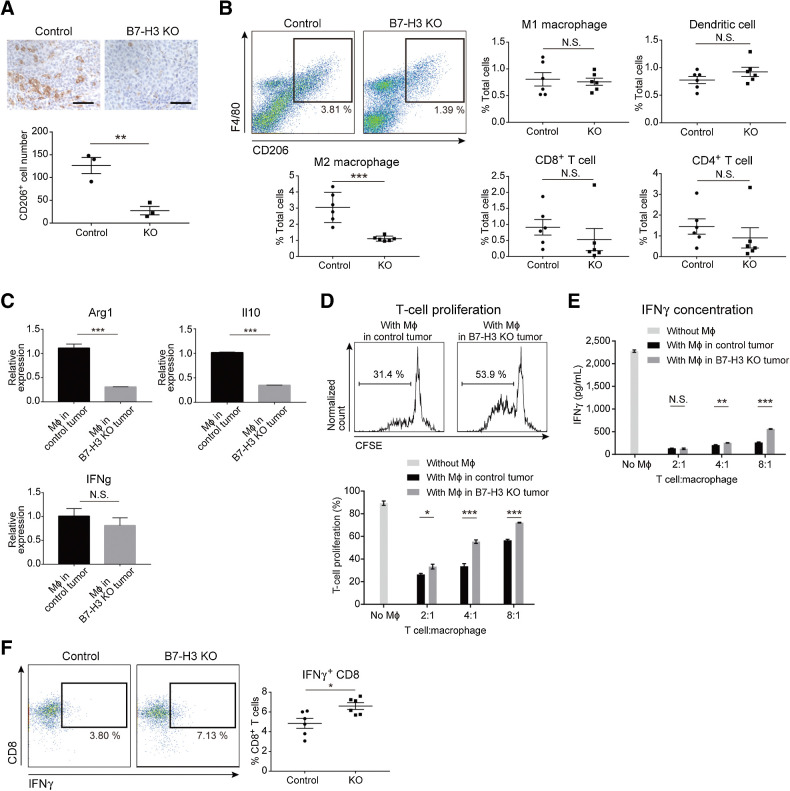
First, we confirmed a direct T cell–suppressive effect of tumor cell B7-H3 in vitro (Supplementary Fig. S7A–S7C), which has been reported in several studies (19, 20). Next, we evaluated tumor-infiltrating immune cells. Although there was no difference in the number of CD8+ T cells, CD206+ M2 macrophages decreased in HM-1 B7-H3 KO tumors (Fig. 3A; Supplementary Fig. S8A). Flow cytometry showed that F4/80+CD206+ M2 macrophages decreased in HM-1 B7-H3 KO tumors, whereas M1 macrophages, CD8+ T cells, CD4+ T cells, and DCs were unchanged (Fig. 3B). ID8 tumors similarly showed a decrease in M2 macrophages but no change in M1 macrophages (Supplementary Fig. S8B). These results suggest that B7-H3 KO tumors are not only associated with a decreased number of M2 macrophages within tumor tissues, but also alters macrophage polarization (i.e., a decrease in the immunosuppressive fraction).
Figure 3.
B7-H3 suppression in tumor cells decreases the number of intratumoral M2 macrophages and increases IFNγ production of CD8+ T cells. A, IHC of CD206 in the B7-H3 KO intradermal HM-1 tumors and controls (n = 3). CD206+ cells were stained brown. Scale bar, 50 μm. B, Flow cytometry of immune cells from HM-1 tumors. Density plots (top left) show the M2 macrophages at day 12. Each dot represents live CD45+ cells, and the boxed area represents F4/80+CD206+ M2 macrophages. The percentage of positive cells relative to the total cells is plotted (n = 6). C, qPCR for the indicated genes in F4/80+ macrophages (Mϕ) isolated from the HM-1 intradermal tumors (n = 5). D, T-cell proliferation in presence of HM-1 tumor–derived macrophages. Top histograms show the percentage of proliferating T cells cocultured (4:1) with macrophages from control or B7-H3 KO tumors. Bottom bar graph shows the percentage of proliferating T cells cocultured with various ratios of macrophages (n = 5). E, IFNγ levels in the supernatants of cocultures in D (n = 5). F, Flow cytometry of IFNγ+CD8+ T cells from HM-1 tumors. Each dot represents live CD45+CD3+ cells, and the boxed area represents IFNγ+CD8+ T cells. The percentage of IFNγ+CD8+ T cells relative to total CD8+ T cells is plotted (n = 6). A–F, Data are presented as the mean ± SEM; *, P < 0.05; **, P < 0.01; ***, P < 0.001; N.S., not significant, unpaired t test.
In macrophages from the B7-H3 KO tumors, we observed decreased expression of the M2 markers Arg1 and Il10 but unchanged expression of the M1 marker Ifng (Fig. 3C). T-cell proliferation assays with CD3/CD28-activated T cells cocultured with tumor-derived macrophages showed that macrophages from the B7-H3 KO tumors had reduced ability to inhibit T-cell proliferation compared with those from control tumors (Fig. 3D), and IFNγ production in coculture supernatants was increased (Fig. 3E). We also observed an increased number of IFNγ+CD8+ T cells in B7-H3 KO tumors (Fig. 3F). These results suggest that B7-H3 suppression in tumor cells reduces immunosuppression in the TME by not only attenuating direct T-cell inhibition, but also reducing the number of M2 macrophages, which may contribute to the increased IFNγ production by CD8+ T cells and enhanced antitumor immunity.
B7-H3 suppression downregulates CCL2 production in tumor cells, partly via the STAT3 pathway
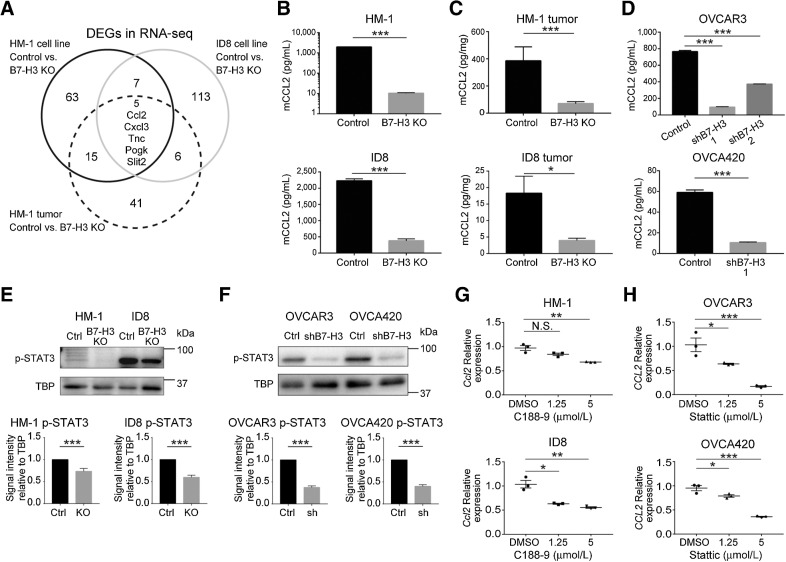
Next, we elucidated the mechanism by which B7-H3 suppression in tumor cells reduced M2 macrophage infiltration. Using RNA sequencing, we found 90, 131, and 67 DEGs in the HM-1 and ID8 cell lines and HM-1 intradermal tumors relative to controls, respectively (Supplementary Tables S3–S5). We identified Ccl2 as a common downregulated gene in the KO groups (Fig. 4A). We also confirmed that changes in expression of several chemokines related to T-cell trafficking were not consistent between the cell lines and the B7-H3 KO tumors (Supplementary Fig. S9). A reduction in CCL2 protein in the B7-H3 KO groups was confirmed in both culture supernatants and tumor lysates from HM-1 and ID8 cells (Fig. 4B and C). Then, we investigated whether B7-H3 suppression also altered CCL2 in human HGSOC cell lines (Supplementary Fig. S10) and confirmed a decrease in CCL2 in the culture supernatants of the shB7-H3 cells (Fig. 4D).
Figure 4.
B7-H3 suppression downregulates CCL2 production, in part, via the STAT3 pathway in ovarian cancer cells. A, Differential gene expression based on RNA sequencing (RNA-seq) between controls (n = 3) and B7-H3 KO (n = 3) groups in HM-1 and ID8 cells and HM-1 intradermal tumors. CCL2 protein in the culture supernatants (B) and tumors derived from HM-1 B7-H3 KO (top), ID8 B7-H3 KO (bottom), and their respective controls (n = 5; C). D, CCL2 protein in the culture supernatants of the OVCAR3 shB7-H3 (top), OVCA420 shB7-H3 (bottom), and their respective controls (n = 5). Representative Western blot analysis of nuclear phosphorylated STAT3 (p-STAT3) in HM-1 and ID8 (E) and OVCAR3 and OVCA420 (F) cells. Bottom bar graphs show the signal intensity of p-STAT3 relative to the TBP control in HM-1 and ID8 cells (both n = 10) and OVCAR3 and OVCA420 cells (both n = 7). qPCR for expression of Ccl2 in HM-1 and ID8 cells (G) and CCL2 in OVCAR3 and OVCA420 cells (H) treated with 1.25 and 5 μmol/L of the STAT3 inhibitors C188-9 (G) and Stattic (H), respectively. DMSO was used as the control (n = 3). Data are presented as the mean ± SEM; *, P < 0.05; **, P < 0.01; ***, P < 0.001; N.S., not significant, unpaired t test in B, C, D (OVCA420), E, and F and one-way ANOVA with Tukey multiple comparisons test in D (OVCAR3), G, and H.
To investigate the mechanism by which B7-H3 suppression induced CCL2 downregulation, we examined STAT3 (21–23) and NFκB (36) pathways, which have been reported as downstream pathways of B7-H3. Western blots showed that phosphorylated STAT3 was decreased in the B7-H3–suppressed cells (Fig. 4E and F). In contrast, phosphorylated p65, a major factor in the classical NFκB pathway, was decreased in ID8 and OVCAR3 cell lines; thus, consistent results were not observed among cell lines (Supplementary Fig. S11A and S11B). Next, we examined whether the decrease in phosphorylated STAT3 contributed to CCL2 downregulation. Treatment with STAT3 inhibitors C188-9 for mouse cells (29) and Stattic for human cells (30) led to a decrease in Ccl2 and CCL2 expression, respectively (Fig. 4G and H). These results indicate that B7-H3 suppression downregulates the production of CCL2 partly via the STAT3 pathway.
The CCL2–CCR2–M2 macrophage axis is involved in B7-H3–mediated tumor growth
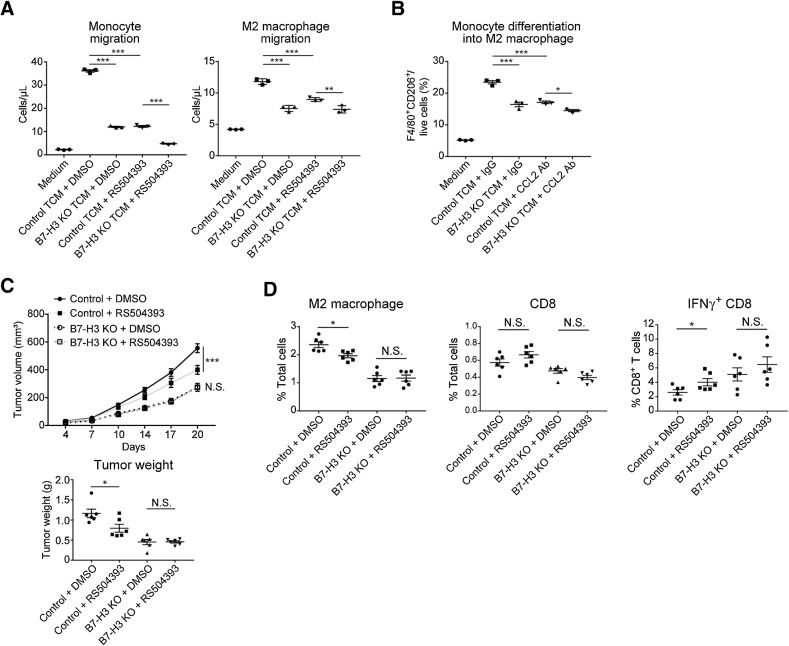
We confirmed whether the CCL2–CCR2 axis contributed to the effects of B7-H3 suppression on M2 macrophage migration and differentiation both in vitro and in vivo. Compared with controls, the migration of monocytes and M2 macrophages was reduced in the B7-H3 KO TCM, and pretreatment of target cells with a CCR2 inhibitor (RS504393) partially eliminated this effect (Fig. 5A; Supplementary Fig. S12A and S12B). M2 macrophage polarization was significantly reduced in B7-H3 KO TCM, and this effect was partially abrogated by CCL2 antibodies (Fig. 5B; Supplementary Fig. S12C). These results indicate that B7-H3 suppression in tumor cells reduces monocyte migration and macrophage differentiation into the M2 phenotype in vitro, and that the CCL2–CCR2 axis is responsible for these effects.
Figure 5.
The CCL2–CCR2–M2 macrophage axis contributes to B7-H3–mediated tumor progression. A, Chemotaxis of mouse monocytes (left) and generated M2 macrophages (right) in response to HM-1 control or B7-H3 KO TCM. Monocytes or M2 macrophages were pretreated with 2 μmol/L RS504393 (CCR2 antagonist) or DMSO before plating. MEM-Alpha was used as the negative control (n = 3). B, Differentiation of mouse monocytes into M2 macrophages in response to HM-1 control or B7-H3 KO TCM supplemented with 20 μg/mL anti-CCL2 or control IgG. MEM-Alpha was used as the negative control (n = 3). Ab, antibody. C, Tumor growth in mice intradermally injected with HM-1 B7-H3 KO cells or control cells and treated with RS504393 (2 mg/kg body weight) or DMSO daily following tumor inoculation. D, Flow cytometric analysis of the treated tumors in C (n = 6). Data are presented as the mean ± SEM (*, P < 0.05; **, P < 0.01; ***, P < 0.001; N.S., not significant, one-way ANOVA with Tukey multiple comparisons test in A and B, and unpaired t test in C and D).
In the HM-1 model, CCR2 inhibition suppressed tumor growth in controls but did not affect B7-H3 KO tumors (Fig. 5C). Flow cytometry showed that CCR2 inhibition reduced the number of M2 macrophages and increased the number of IFNγ+CD8+ T cells in control tumors but had no effect on B7-H3 KO tumors (Fig. 5D). The number of total CD8+ T cells was not changed by the treatment in either control or B7-H3 KO tumors. These results indicate that CCL2 downregulation induced by B7-H3 suppression contributes to the reduction of M2 macrophages and the increase in IFNγ+CD8+ T cells in vivo, and that the CCL2–CCR2 axis and M2 macrophages partly contribute to B7-H3–mediated tumor progression.
Poor prognosis of B7-H3–high HGSOC with an M2 macrophage–rich, IFNγ+CD8+ T cell–sparse TME
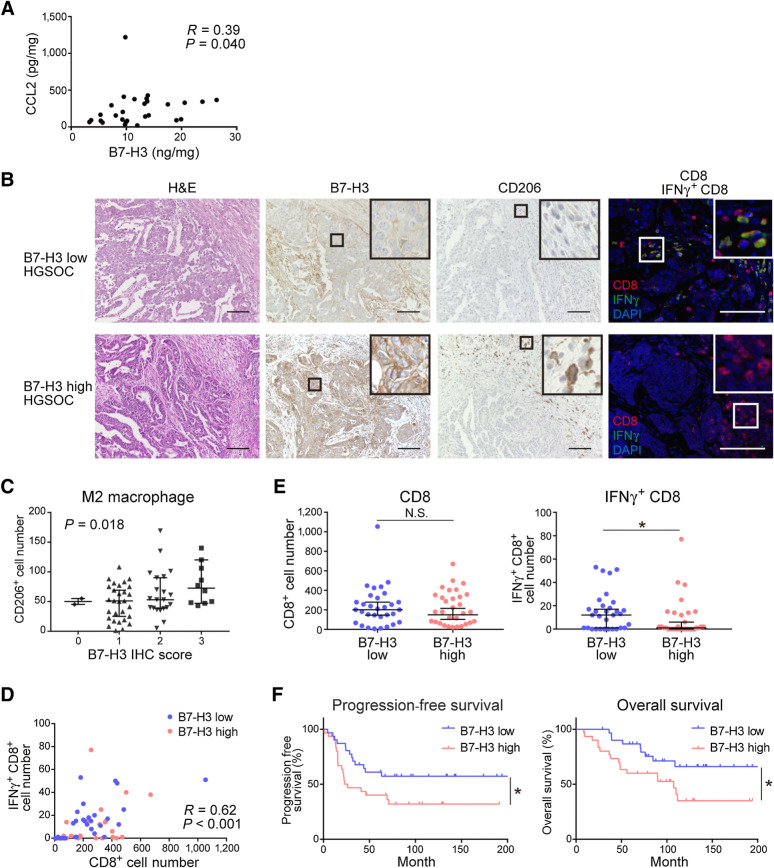
Finally, we investigated the relationships among B7-H3 and CCL2, M2 macrophages, CD8+ T cells, and IFNγ+CD8+ T cells in HGSOC clinical samples. B7-H3 and CCL2 protein in HGSOC primary tumors positively correlated (P = 0.040; Fig. 6A). Serum CCL2 was elevated in patients with HGSOC compared with those in healthy donors (P < 0.001; Supplementary Fig. S13A); however, there was no significant correlation between serum CCL2 and tumor B7-H3 (P = 0.089; Supplementary Fig. S13B).
Figure 6.
B7-H3 expression associates with an M2 macrophage–rich, IFNγ+CD8 T cell–poor TME and a poor prognosis in patients with HGSOC. A, Correlation between CCL2 expression and B7-H3 protein in primary HGSOC tumors (n = 28) analyzed by Spearman correlation. B, Representative images of HGSOC samples showing B7-H3 expression and CD206+, CD8+, and IFNγ+CD8+ cells. Cells were stained with hematoxylin and eosin (H&E) or immunostained with the corresponding antibodies. Scale bar, 100 μm. Positive cells were stained brown in IHC, and red (CD8), green (IFNγ), and blue (DAPI) in immunofluorescence slides. The boxed area represents the zoomed image shown in the top right corner. C, Correlation between the infiltration of CD206+ cells and B7-H3 expression in the corresponding primary tumors of HGSOC (n = 62) analyzed using Jonckheere–Terpstra test. Data are presented as the median with 95% confidence intervals. D, Correlation between tumor-infiltrating CD8+ and IFNγ+CD8+ T cells (n = 62) analyzed using Spearman correlation. Blue and pink dots represent B7-H3–low and B7-H3–high expression cases, respectively. E, Comparisons of the number of tumor-infiltrating CD8+ T cells (left) and IFNγ+CD8+ T cells (right) in B7-H3–low and B7-H3–high groups by immunostaining (n = 62). Data are presented as the median with 95% confidence intervals; *, P < 0.05; N.S., not significant, Mann–Whitney U test. F, PFS (left) and OS (right) of patients with HGSOC (n = 62). Patients were classified into a B7-H3–low group (n = 31) and a B7-H3–high group (n = 31). *, P < 0.05; N.S., not significant, log-rank test.
FFPE specimens from the primary tumor site of HGSOC cases were evaluated for tumor cell B7-H3 and CD206+ cells by IHC and CD8+ and IFNγ+CD8+ cells by immunofluorescence (Fig. 6B). A positive correlation between B7-H3 expression and the number of CD206+ M2 macrophages was observed (P = 0.018; Fig. 6C). The number of CD8+ T cells and IFNγ+CD8+ cells positively correlated (P < 0.001; Fig. 6D). There was no difference in the number of CD8+ T cells in the B7-H3–high and B7-H3–low groups; however, the number of IFNγ+CD8+ cells was significantly lower in B7-H3–high versus B7-H3–low tumors (P = 0.047; Fig. 6E). Patient characteristics are shown in Supplementary Table S6. Higher B7-H3 expression was a potential poor prognostic factor for both PFS and OS in the univariate analysis, although it was not identified as an independent prognostic factor in the multivariate analysis (Fig. 6F; Supplementary Table S7). These results show the immunosuppressive properties of B7-H3 in relation to M2 macrophages and IFNγ+CD8+ T cells in patients with HGSOC.
Discussion
In this study, we investigated B7-H3 upregulation in the nonimmunoreactive subtype of HGSOC and hypothesized that B7-H3 exerted immunosuppressive effects via a mechanism distinct from that of other B7 family members, such as PD-L1, overexpressed in immunoreactive tumors. We showed that B7-H3, which has been mainly reported to directly modulate T-cell function (15, 19, 20), similar to other immune checkpoint molecules, is involved in indirect T-cell suppression via the CCL2–CCR2–M2 macrophage axis.
Several studies have investigated the relationship between B7-H3 and M2 macrophages. B7-H3 promotes the in vitro differentiation of macrophages into the M2 phenotype in cocultures of HepG2 hepatoma cells and THP-1 macrophages (37) and cultures of PBMC-derived monocytes in colon cancer cell culture supernatants (38); however, the underlying mechanism has not yet been clarified. In an immunocompetent transgenic mouse model of head and neck cancer, anti–B7-H3 treatment reduces the number of immature myeloid cells, including macrophages, in the tumor and activates antitumor immunity (32). However, whether the decreased number of macrophages was an effect of anti–B7-H3 on tumor cells or on macrophages expressing B7-H3 was not clarified. To the best of our knowledge, this is the first study to demonstrate that CCL2 downregulation following B7-H3 suppression in tumor cells contributes to the reduction in M2 macrophages and the subsequent improvement in antitumor responses both in vitro and in vivo. Using clinical tumor samples, we also clarified the relationship of B7-H3 with CCL2 and M2 macrophages.
The immunologic and nonimmunologic roles of B7-H3 have been reported previously (39). In this study, the difference in tumor growth between the control and B7-H3 KO tumors observed in immunocompetent mice was not observed in immunodeficient mice, suggesting that immunologic effects have a greater influence on tumor growth than nonimmunologic effects. B7-H3 has been reported to exert direct immunomodulatory effects mainly on T cells (15, 20) and natural killer (NK) cells (40). However, the results of our tumor inoculation experiments in immunodeficient nude mice with normal NK-cell function suggest that the effect of B7-H3 on NK cells is not significant.
We confirmed both the direct and indirect immunosuppressive effects of B7-H3 on T cells. The increased number of IFNγ+CD8+ T cells in B7-H3 KO tumors and B7-H3-low HGSOC cases, both of which were not accompanied by changes in the number of CD8+ T cells, suggests that B7-H3 is not involved in the recruitment of T cells; instead, it attenuates T-cell activity at the tumor site either directly or indirectly through M2 macrophages. Several studies have reported T cell–suppressive functions for M2 macrophages (41, 42). Although it remains unclear which between the direct and indirect effects contribute more to T-cell suppression, the fact that CCR2 blocking in vivo decreased the number of M2 macrophages and increased that of IFNγ+CD8+ T cells and inhibited tumor growth indicates that the M2 macrophage–mediated indirect effects significantly affect tumor growth.
In tumor tissues, B7-H3 is expressed not only in tumor cells but also in other stromal cells (16, 20). Although B7-H3 expression in antigen-presenting cells has been reported to regulate T-cell function (15, 19), its role in other normal cells in tumor tissues remains to be elucidated. Our results showed that B7-H3 expression in stromal cells does not significantly affect tumor growth. This result is similar to that reported by a study on MC38 mouse colon cancer cells, in which no difference in tumor growth was observed in the presence or absence of stromal B7-H3 (16). In contrast, another study on E.G7-OVA mouse lymphoma cells showed delayed tumor growth in the absence of stromal B7-H3 (43). These contrasting results suggest that the effect of stromal B7-H3 expression on tumor growth may vary depending on tumor type.
To date, the B7-H3 receptor remains unknown, and it is unclear how B7-H3 acts among cells or within B7-H3–expressing cells. The spontaneous dimerization of B7-H3 in vitro suggests that B7-H3 among neighboring cells may transmit signals to each other (44). IL20RA has been identified as a binding partner of B7-H3 (45). The involvement of B7-H3 in the activation of intracellular signals, such as STAT3 in tumor cells, has also been revealed (21–23). We found decreased nuclear phosphorylated STAT3 following B7-H3 suppression and CCL2 downregulation by STAT3 inhibitors in tumor cells. These findings indicate that B7-H3 partly acts via STAT3–CCL2 intracellular signaling in the cancer cells expressing B7-H3 or among neighboring cancer cells. Further elucidation of the molecular mechanism underlying B7-H3–mediated signaling, including the identification of its receptors, is thus required.
In TCGA-OV, we found that B7-H3 expression negatively correlated with an IFNγ signature and PD-L1 expression. Nonimmunoreactive, B7-H3–high, and PD-L1–low HGSOC tumors are not expected to benefit from ICI treatment, and therefore, require a different treatment strategy than that used for immunoreactive tumors. M2 macrophages promote tumor progression in ovarian cancer (46, 47), and the abundance of M2 macrophages is an important distinctive characteristic of cancer subtypes with poor prognosis (48, 49). We found a positive correlation between B7-H3 expression and the number of M2 macrophages in HGSOC cases. CCR2 inhibition had no therapeutic effect on B7-H3–suppressed tumors with sparse M2 macrophages, whereas in B7-H3–expressing tumors with abundant M2 macrophages, it exerted a mild therapeutic effect, suggesting that M2 macrophages are potential therapeutic targets in patients with B7-H3–high, M2 macrophage–rich tumors. Ongoing clinical trials targeting macrophages are expected to be effective in such tumor subtypes (50). Our findings also confirm those of a previous study which stated that anti–B7-H3 treatment reduces immature myeloid cells, including macrophages (32), suggesting that B7-H3 suppression could be effective in reducing the number of M2 macrophages. In addition to its previously reported clinical applications, including as a direct inhibitor of T cells and a pan-tumor antigen with high selectivity, B7-H3 may be a promising therapeutic target in cancers with an M2 macrophage–mediated immunosuppressive TME.
There are several limitations to this study. We did not identify the upstream regulators of B7-H3 expression, and hence the fundamental causes that shape the differential immunologic TME. B7-H3 upregulation in tumor cells was not observed by the addition of any cytokines, including IFNγ in our in vitro experiments. The relationship between B7-H3 expression and genetic factors, such as tumor mutation burden (TMB), remains unknown. Verhaak and colleagues report that the differentiated type has lower TMB and aneuploidy than other subtypes, whereas the proliferative type has fewer germline BRCA 1/2 mutations than other subtypes (10). Desbois and colleagues found no differences in TMB or BRCA mutations, homologous recombination deficiency status, or microsatellite instability status among the molecular subtypes of HGSOC (14). Therefore, it remains unclear when and how B7-H3 expression and the immunoreactive TME are determined. The identification of the definitive drivers is warranted in future studies.
In conclusion, we revealed the involvement of B7-H3 in the CCL2–CCR2–M2 macrophage axis and in the immunosuppressive TME in HGSOC. Our findings further the understanding of the immunologic TME and demonstrate the promising potential of B7-H3 as a therapeutic target for the B7-H3–high, M2 macrophage–rich, unfavorable phenotype of HGSOC.
Authors' Disclosures
T. Miyamoto reports other support from Japan Society for the Promotion of Science outside the submitted work. R. Murakami reports grants from the Grants-in-Aid for Strategic Medical Science Research (grant numbers 18K16769 and 20K18210) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan during the conduct of the study. J. Hamanishi reports grants from Ono Pharmaceutical, Sumitomo Dainippon Pharma, and MSD outside the submitted work. K. Yamaguchi reports grants from Bayer outside the submitted work. N. Matsumura reports personal fees from Takara Bio Inc. and AstraZeneca, and grants from AstraZeneca outside the submitted work. M. Mandai reports grants and personal fees from Chugai Pharmaceutical; personal fees from Takeda Pharmaceutical and AstraZeneca; and grants from MSD and Tsumura & Co. outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
T. Miyamoto: Conceptualization, resources, data curation, software, formal analysis, investigation, visualization, writing–original draft, project administration. R. Murakami: Conceptualization, resources, data curation, software, formal analysis, funding acquisition, visualization, writing–original draft, project administration. J. Hamanishi: Conceptualization, validation, project administration, writing–review and editing. K. Tanigaki: Methodology, writing–review and editing. Y. Hosoe: Investigation, writing–review and editing. N. Mise: Methodology, writing–review and editing. S. Takamatsu: Resources, software, formal analysis, visualization, writing–review and editing. Y. Mise: Methodology, writing–review and editing. M. Ukita: Methodology, writing–review and editing. M. Taki: Methodology, writing–review and editing. K. Yamanoi: Methodology, writing–review and editing. N. Horikawa: Methodology, writing–review and editing. K. Abiko: Conceptualization, validation, project administration, writing–review and editing. K. Yamaguchi: Conceptualization, validation, project administration, writing–review and editing. T. Baba: Conceptualization, supervision, project administration, writing–review and editing. N. Matsumura: Conceptualization, supervision, project administration, writing–review and editing. M. Mandai: Supervision, writing–review and editing.
Supplementary Material
Acknowledgments
This work was supported by Grants-in-Aid for Strategic Medical Science Research (grant numbers 18K16769 and 20K18210) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. The authors thank Editage (www.editage.com) for English language editing.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Immunology Research Online (http://cancerimmunolres.aacrjournals.org/).
References
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. [DOI] [PubMed] [Google Scholar]
- 2. Ray-Coquard I, Pautier P, Pignata S, Perol D, Gonzalez-Martin A, Berger R, et al. Olaparib plus bevacizumab as first-line maintenance in ovarian cancer. N Engl J Med 2019;381:2416–28. [DOI] [PubMed] [Google Scholar]
- 3. Gonzalez-Martin A, Pothuri B, Vergote I, DePont Christensen R, Graybill W, Mirza MR, et al. Niraparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med 2019;381:2391–402. [DOI] [PubMed] [Google Scholar]
- 4. Matulonis UA, Shapira-Frommer R, Santin AD, Lisyanskaya AS, Pignata S, Vergote I, et al. Antitumor activity and safety of pembrolizumab in patients with advanced recurrent ovarian cancer: results from the phase II KEYNOTE-100 study. Ann Oncol 2019;30:1080–7. [DOI] [PubMed] [Google Scholar]
- 5. Moore KN, Bookman M, Sehouli J, Miller A, Anderson C, Scambia G, et al. LBA31 primary results from IMagyn050/GOG 3015/ENGOT-OV39, a double-blind placebo (pbo)-controlled randomised phase III trial of bevacizumab (bev)-containing therapy ± atezolizumab (atezo) for newly diagnosed stage III/IV ovarian cancer (OC). Ann Oncol 2020;31:S1161–S2. [Google Scholar]
- 6. Disis ML, Taylor MH, Kelly K, Beck JT, Gordon M, Moore KM, et al. Efficacy and safety of avelumab for patients with recurrent or refractory ovarian cancer: phase 1b results from the JAVELIN solid tumor trial. JAMA Oncol 2019;5:393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Izar B, Tirosh I, Stover EH, Wakiro I, Cuoco MS, Alter I, et al. A single-cell landscape of high-grade serous ovarian cancer. Nat Med 2020;26:1271–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Armstrong DK, Alvarez RD, Bakkum-Gamez JN, Barroilhet L, Behbakht K, Berchuck A, et al. Ovarian cancer, version 2.2020, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw 2021;19:191–226. [DOI] [PubMed] [Google Scholar]
- 9. Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011;474:609–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Verhaak RG, Tamayo P, Yang JY, Hubbard D, Zhang H, Creighton CJ, et al. Prognostically relevant gene signatures of high-grade serous ovarian carcinoma. J Clin Invest 2013;123:517–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chin CD, Fares CM, Campos M, Chen HW, Shintaku IP, Konecny GE, et al. Association of PD-L1 expression by immunohistochemistry and gene microarray with molecular subtypes of ovarian tumors. Mod Pathol 2020;33:2001–10. [DOI] [PubMed] [Google Scholar]
- 12. Mandai M, Hamanishi J, Abiko K, Matsumura N, Baba T, Konishi I. Dual faces of IFNgamma in cancer progression: a role of PD-L1 induction in the determination of pro- and antitumor immunity. Clin Cancer Res 2016;22:2329–34. [DOI] [PubMed] [Google Scholar]
- 13. Taki M, Abiko K, Baba T, Hamanishi J, Yamaguchi K, Murakami R, et al. Snail promotes ovarian cancer progression by recruiting myeloid-derived suppressor cells via CXCR2 ligand upregulation. Nat Commun 2018;9:1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Desbois M, Udyavar AR, Ryner L, Kozlowski C, Guan Y, Durrbaum M, et al. Integrated digital pathology and transcriptome analysis identifies molecular mediators of T-cell exclusion in ovarian cancer. Nat Commun 2020;11:5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chapoval AI, Ni J, Lau JS, Wilcox RA, Flies DB, Liu D, et al. B7-H3: a costimulatory molecule for T cell activation and IFN-gamma production. Nat Immunol 2001;2:269–74. [DOI] [PubMed] [Google Scholar]
- 16. Seaman S, Zhu Z, Saha S, Zhang XM, Yang MY, Hilton MB, et al. Eradication of tumors through simultaneous ablation of CD276/B7-H3-positive tumor cells and tumor vasculature. Cancer Cell 2017;31:501–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Modak S, Zanzonico P, Grkovski M, Slotkin EK, Carrasquillo JA, Lyashchenko SK, et al. B7H3-directed intraperitoneal radioimmunotherapy with radioiodinated omburtamab for desmoplastic small round cell tumor and other peritoneal tumors: results of a phase I study. J Clin Oncol 2020;38:4283–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Du H, Hirabayashi K, Ahn S, Kren NP, Montgomery SA, Wang X, et al. Antitumor responses in the absence of toxicity in solid tumors by targeting B7-H3 via chimeric antigen receptor T cells. Cancer Cell 2019;35:221–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Prasad DV, Nguyen T, Li Z, Yang Y, Duong J, Wang Y, et al. Murine B7-H3 is a negative regulator of T cells. J Immunol 2004;173:2500–6. [DOI] [PubMed] [Google Scholar]
- 20. Hofmeyer KA, Ray A, Zang X. The contrasting role of B7-H3. Proc Natl Acad Sci U S A 2008;105:10277–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu H, Tekle C, Chen YW, Kristian A, Zhao Y, Zhou M, et al. B7-H3 silencing increases paclitaxel sensitivity by abrogating Jak2/Stat3 phosphorylation. Mol Cancer Ther 2011;10:960–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tekle C, Nygren MK, Chen YW, Dybsjord I, Nesland JM, Maelandsmo GM, et al. B7-H3 contributes to the metastatic capacity of melanoma cells by modulation of known metastasis-associated genes. Int J Cancer 2012;130:2282–90. [DOI] [PubMed] [Google Scholar]
- 23. Lin L, Cao L, Liu Y, Wang K, Zhang X, Qin X, et al. B7-H3 promotes multiple myeloma cell survival and proliferation by ROS-dependent activation of Src/STAT3 and c-Cbl-mediated degradation of SOCS3. Leukemia 2019;33:1475–86. [DOI] [PubMed] [Google Scholar]
- 24. Murakami R, Matsumura N, Mandai M, Yoshihara K, Tanabe H, Nakai H, et al. Establishment of a novel histopathological classification of high-grade serous ovarian carcinoma correlated with prognostically distinct gene expression subtypes. Am J Pathol 2016;186:1103–13. [DOI] [PubMed] [Google Scholar]
- 25. Broberg P. Statistical methods for ranking differentially expressed genes. Genome Biol 2003;4:R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009;462:108–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017;127:2930–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Abiko K, Mandai M, Hamanishi J, Yoshioka Y, Matsumura N, Baba T, et al. PD-L1 on tumor cells is induced in ascites and promotes peritoneal dissemination of ovarian cancer through CTL dysfunction. Clin Cancer Res 2013;19:1363–74. [DOI] [PubMed] [Google Scholar]
- 29. Hox V, O'Connell MP, Lyons JJ, Sackstein P, Dimaggio T, Jones N, et al. Diminution of signal transducer and activator of transcription 3 signaling inhibits vascular permeability and anaphylaxis. J Allergy Clin Immunol 2016;138:187–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schust J, Sperl B, Hollis A, Mayer TU, Berg T. Stattic: a small-molecule inhibitor of STAT3 activation and dimerization. Chem Biol 2006;13:1235–42. [DOI] [PubMed] [Google Scholar]
- 31. Hamanishi J, Mandai M, Iwasaki M, Okazaki T, Tanaka Y, Yamaguchi K, et al. Programmed cell death 1 ligand 1 and tumorinfiltrating CD8 T lymphocytes are prognostic factors of human ovarian cancer. Proc Natl Acad Sci U S A 2007;104:3360–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mao L, Fan TF, Wu L, Yu GT, Deng WW, Chen L, et al. Selective blockade of B7-H3 enhances antitumour immune activity by reducing immature myeloid cells in head and neck squamous cell carcinoma. J Cell Mol Med 2017;21:2199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee C, Jeong H, Bae Y, Shin K, Kang S, Kim H, et al. Targeting of M2-like tumor-associated macrophages with a melittin-based pro-apoptotic peptide. J Immunother Cancer 2019;7:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mulati K, Hamanishi J, Matsumura N, Chamoto K, Mise N, Abiko K, et al. VISTA expressed in tumour cells regulates T cell function. Br J Cancer 2019;120:115–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Peng J, Hamanishi J, Matsumura N, Abiko K, Murat K, Baba T, et al. Chemotherapy induces programmed cell death-ligand 1 overexpression via the nuclear factor-kappaB to foster an immunosuppressive tumor microenvironment in ovarian cancer. Cancer Res 2015;75:5034–45. [DOI] [PubMed] [Google Scholar]
- 36. Wang R, Ma Y, Zhan S, Zhang G, Cao L, Zhang X, et al. B7-H3 promotes colorectal cancer angiogenesis through activating the NF-kappaB pathway to induce VEGFA expression. Cell Death Dis 2020;11:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kang FB, Wang L, Li D, Zhang YG, Sun DX. Hepatocellular carcinomas promote tumor-associated macrophage M2-polarization via increased B7-H3 expression. Oncol Rep 2015;33:274–82. [DOI] [PubMed] [Google Scholar]
- 38. Mao Y, Chen L, Wang F, Zhu D, Ge X, Hua D, et al. Cancer cell-expressed B7-H3 regulates the differentiation of tumor-associated macrophages in human colorectal carcinoma. Oncol Lett 2017;14:6177–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kontos F, Michelakos T, Kurokawa T, Sadagopan A, Schwab JH, Ferrone CR, et al. B7-H3: an attractive target for antibody-based immunotherapy. Clin Cancer Res 2021;27:1227–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Castriconi R, Dondero A, Augugliaro R, Cantoni C, Carnemolla B, Sementa AR, et al. Identification of 4Ig-B7-H3 as a neuroblastoma-associated molecule that exerts a protective role from an NK cell-mediated lysis. Proc Natl Acad Sci U S A 2004;101:12640–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Van de Velde LA, Subramanian C, Smith AM, Barron L, Qualls JE, Neale G, et al. T cells encountering myeloid cells programmed for amino acid-dependent immunosuppression use rictor/mTORC2 protein for proliferative checkpoint decisions. J Biol Chem 2017;292:15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Benner B, Scarberry L, Suarez-Kelly LP, Duggan MC, Campbell AR, Smith E, et al. Generation of monocyte-derived tumor-associated macrophages using tumor-conditioned media provides a novel method to study tumor-associated macrophages in vitro. J Immunother Cancer 2019;7:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee YH, Martin-Orozco N, Zheng P, Li J, Zhang P, Tan H, et al. Inhibition of the B7-H3 immune checkpoint limits tumor growth by enhancing cytotoxic lymphocyte function. Cell Res 2017;27:1034–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vigdorovich V, Ramagopal UA, Lazar-Molnar E, Sylvestre E, Lee JS, Hofmeyer KA, et al. Structure and T cell inhibition properties of B7 family member, B7-H3. Structure 2013;21:707–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Husain B, Ramani SR, Chiang E, Lehoux I, Paduchuri S, Arena TA, et al. A platform for extracellular interactome discovery identifies novel functional binding partners for the immune receptors B7-H3/CD276 and PVR/CD155. Mol Cell Proteomics 2019;18:2310–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hensler M, Kasikova L, Fiser K, Rakova J, Skapa P, Laco J, et al. M2-like macrophages dictate clinically relevant immunosuppression in metastatic ovarian cancer. J Immunother Cancer 2020;8:e000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Maccio A, Gramignano G, Cherchi MC, Tanca L, Melis L, Madeddu C. Role of M1-polarized tumor-associated macrophages in the prognosis of advanced ovarian cancer patients. Sci Rep 2020;10:6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tekpli X, Lien T, Rossevold AH, Nebdal D, Borgen E, Ohnstad HO, et al. An independent poor-prognosis subtype of breast cancer defined by a distinct tumor immune microenvironment. Nat Commun 2019;10:5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang Q, Hu B, Hu X, Kim H, Squatrito M, Scarpace L, et al. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell 2017;32:42–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. DeNardo DG, Ruffell B. Macrophages as regulators of tumour immunity and immunotherapy. Nat Rev Immunol 2019;19:369–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.