Abstract
In Escherichia coli, the CpxRA two-component signal transduction system senses and responds to aggregated and misfolded proteins in the bacterial envelope. We show that CpxR-P (the phosphorylated form of the cognate response regulator) activates cpxRA expression in conjunction with RpoS, suggesting an involvement of the Cpx system in stationary-phase survival. Engagement of the CpxRA system in functions beyond protein management is indicated by several putative targets identified after a genomic screening for the CpxR-P recognition consensus sequence. Direct negative control of the newly identified targets motABcheAW (specifying motility and chemotaxis) and tsr (encoding the serine chemoreceptor) by CpxR-P was shown by electrophoretic mobility shift analysis and Northern hybridization. The results suggest that the CpxRA system plays a core role in an extensive stress response network in which the coordination of protein turnover and energy conservation may be the unifying element.
The CpxRA two-component system, together with ςE and ς32, regulates the synthesis of a number of enzymes that are involved in the folding and degradation of periplasmic proteins in Escherichia coli (4, 6, 7, 30). For example, the expression of degP (encoding a periplasmic protease) is activated by CpxRA and EςE, whereas the expression of ppiD (encoding a periplasmic peptidyl-prolyl cis-trans isomerase) is activated by CpxRA and Eς32 (4, 6, 7, 30). Moreover, CpxRA appears to play a role in sensing and responding to envelope protein distress, since it also activates the expression of ppiA (encoding another periplasmic peptidyl-prolyl cis-trans isomerase), dsbA (encoding a periplasmic disulfide oxidoreductase), and cpxP (encoding a periplasmic protein of unknown function) (4, 5, 30; for a review, see reference 33).
Certain mutant versions of the CpxA sensor kinase (CpxA*) cause a variety of seemingly unrelated phenotypes. These include a decreased ability to perform conjugation (16, 35, 36); diminished assembly of lipoprotein and OmpF in the cell envelope (19, 20); random septum positioning during cell division (29); lost ability to grow on succinate (31), l-lactose (3, 28), and l-proline (27); acquired ability to grow on l-serine (24, 25, 38); partial isoleucine and valine auxotrophy (18, 39); increased sensitivity to high temperature (16), sodium dodecyl sulfate (3), and hydrogen peroxide (8a); enhanced tolerance to high pH (5) colicins A and K (26); increased resistance to the aminoglycoside antibiotics amikacin and kanamycin (31, 40); and decreased sensitivity to CuCl2 (8a, 43). CpxA* proteins were shown to retain their kinase activity but to lack the ability to dephosphorylate CpxR-P, the phosphorylated form of the cognate response regulator (32). The resulting rise in CpxR-P levels apparently underlies the numerous aberrant CpxA* phenotypes, hinting that the Cpx regulatory system may play a physiological role that is more extensive than hitherto recognized.
Since a putative CpxR-P recognition consensus box has been reported (30), we screened E. coli promoters for the presence of this box to identify additional target operons. In this study, we present evidence that a number of genes that are not related to protein management are under the direct control of CpxR-P.
MATERIALS AND METHODS
Strains and plasmids.
The E. coli strains and plasmids used in this study are listed in Table 1. Standard molecular biological techniques were applied for their manipulation and construction (34).
TABLE 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics or genotype | Reference or source |
|---|---|---|
| Strains | ||
| BW21355 | K-12 F−rph-1 Δ(lac)X74 | 12 |
| AE2293 | K-12 F−metB+ glpK1 cpxA9 | 31 |
| JP466 | MC4100 Δfrd-101 F′ pOXgen argE::Tn10 | 30 |
| ECL3500 | BW21355 argE::Tn10 | This study |
| ECL1212 | MC4100 Δfrd-101 F′ pOXgen Δ(cpxRA)2 | 29 |
| ECL3501 | BW21355 Δ(cpxRA)2 | This study |
| ECL3502 | ECL3501 Φ(cpxR+A+-lacZ) | This study |
| ECL3503 | ECL3501 Φ(cpxRΔA+-lacZ) | This study |
| ECL3504 | ECL3501 Φ(cpxR+A*-lacZ) | This study |
| ECL3505 | ECL3501 Φ(cpxRΔA*-lacZ) | This study |
| GS015 | K-12 rpoS::Tn10 | 1 |
| ECL3506 | ECL3502 rpoS::Tn10 | This study |
| ECL3507 | ECL3503 rpoS::Tn10 | This study |
| Plasmids | ||
| pRS415 | lac-based promoter fusion vector | 37 |
| pRS415/R+A+ | pRS415 containing the cpxR+A+ operon | This study |
| pRS415/RΔA+ | pRS415 containing the cpxRΔA+ operon | This study |
| pRS415/R+A* | pRS415 containing the cpxR+A* operon | This study |
| pRS415/RΔA* | pRS415 containing the cpxRΔA* operon | This study |
| pAlter-1 | Cloning vector | Promega |
| pAlter/R+A+ | pAlter-1 containing the cpxR+A+ operon | This study |
| pAlter/RΔA+ | pAlter-1 containing the cpxRΔA+ operon | This study |
| pAlter/R+A* | pAlter-1 containing the cpxR+A* operon | This study |
| pAlter/RΔA* | pAlter-1 containing the cpxRΔA* operon | This study |
Growth media.
Cells were grown on Luria-Bertani (LB) or glucose (0.2%) minimal medium (pH 7.0) comprising 34 mM NaH2PO4, 66 mM K2HPO4, 20 mM (NH4)2SO4, 1 μM FeSO4, 30 mM MgSO4, 1 mM ZnCl2, 10 μM CaCl2, 0.3 mM isoleucine, 0.3 mM valine, and 2 mM thiamine.
Construction of the ΔcpxRA strain.
Strain BW21355, an isogenic derivative of MG1655 (12), was used for the mutant construction. First, an argE::Tn10 mutation (closely linked to cpxRA) was P1 transduced from strain JP466 (30) into strain BW21355. The transductants were selected at 37°C for tetracycline resistance (20 μg/ml) and scored for arginine (0.6 mM) auxotrophy on glucose minimal medium, resulting in the isolation of strain ECL3500. Δ(cpxRA)2 was then P1 transduced from strain ECL1212 (29) into strain ECL3500. The transductants were selected for growth on glucose minimal medium free of arginine and scored for sensitivity to tetracycline and CuCl2 (4 mM) (cpx deletion phenotype) (8a), resulting in the isolation of strain ECL3501. The presence of Δ(cpxRA)2 in ECL3501 was confirmed by PCR analysis and DNA sequencing (performed at the MicroCore Sequencing Facility of the Department of Microbiology and Molecular Genetics, Harvard Medical School).
Construction of the Φ(cpx-lacZ) reporter strains.
Strains derived from ECL3501 and bearing the various Φ(cpx-lacZ) operon fusions were constructed by PCR amplification of cpxRA+ and cpxRA* (including 309 bp upstream of the cpxR start codon) from strains BW21355 and AE2293, respectively (cpxA* allele: Leu38→Phe [TTT]) (30). For this procedure, Taq Plus Precision polymerase mix (Stratagene) was used with primers IFDRλ1 (5′-TCCCCCGGGTCGAACATATGGCTCTGCGTACTG-3′) and IFDRλ2 (5′-TACGGATCCGAAGTTTAACTCCGCTTATACAGC-3′); the PCR products (2,414 bp) were then restricted with SmaI and BamHI (New England Biolabs), the recognition sites of which are present in the primers (underlined; SmaI site in IFDRλ1 and BamHI site in IFDRλ2), and ligated into SmaI/BamHI-restricted cloning vector pAlter-1 (Promega). This procedure yielded plasmids pAlter/R+A+ and pAlter/R+A*. The recombinant plasmids were restriction mapped, and the desired clones were sequenced to confirm the cpxA* allele and the absence of PCR-introduced mutations.
To construct an in-frame deletion in cpxR, a sequence fragment (418 bp) was removed between the unique XhoI site in cpxR (bp 79) and bp 497, at which position a XhoI restriction sequence was introduced by PCR, allowing intragenic closure. First, a 1,212-bp fragment of cpxRA was PCR amplified from the BW21355 chromosome with primers CpxRXhoI (5′-GCCGCCGCTCGAGTTTACCCTGCTCTATTTG-3′) and Cpx14 (5′-GCCCATTTGCTCGGC-3′). Since primer CpxRXhoI contains an introduced XhoI restriction site at its 5′ end (underlined) and since a natural RsrII site is present in the cpxRA sequence near the 3′ end of the PCR product, this product was treated with XhoI and RsrII (New England Biolabs). The DNA fragment was then ligated to the backbone of XhoI/RsrII-restricted pAlter/R+A+ and pAlter/R+A* (a 1,630-bp fragment was released from these vectors), yielding plasmids pAlter/RΔA+ and pAlter/RΔA*, which contained a 418-bp in-frame deletion in cpxR. The identity of the plasmid constructs was confirmed by restriction analysis and DNA sequencing.
All cpx operons were released from the pAlter-based plasmids with SmaI and BamHI and ligated into SmaI/BamHI-restricted operon fusion plasmid pRS415 (36). The resulting plasmids, pRS415/R+A+, pRS415/RΔA+, pRS415/R+A*, and pRS415/RΔA*, were transformed into strain ECL3501 (30°C, LB agar with 50 μg of ampicillin/ml) and transferred to λRZ5 (13) or λRS45 (37). Selection and transduction of recombinant phages to the attB site of strain ECL3501 were carried out as previously described (37). Single-copy lysogens ECL3502, ECL3503, ECL3504, and ECL3505 (Table 1) were confirmed by PCR analysis of the attB site (11) with primers P1-attλ (5′-TCAGAACGACGTTGATCGGGCGGGGTTG-3′), P2-attP (5′-AGTTTGTCTGCAAGACTCTATGAGAAGCAG-3′), P3-attP (5′-ATGTTGCATGGTGCACTGTTTATACCAACG-3′), and P4-attλ (5′-GCGCAATGCCATCTGGTATCACTTAAAGG-3′).
To address the role of stationary-phase transcription factor ςS in cpxRA expression, an rpoS::Tn10 mutation was P1 transduced from strain GS015 (1) into strains ECL3502 and ECL3503. Transductants were selected on LB agar containing tetracycline, leading to the respective isolation of strains ECL3506 and ECL3507.
Northern analysis of motABcheAW and tsr expression.
Examination of motABcheAW and tsr expression was carried out by Northern analysis of total RNA from exponential-phase cultures (optical density at 600 nm [OD600], 0.8 to 1.0) of strains ECL3502, ECL3503, ECL3504, and ECL3505 (LB medium, 37°C). Total RNA was isolated with the RNeasy Total RNA System (Qiagen) and separated by gel electrophoresis in Tris-acetate-EDTA–agarose (1%) containing guanidine thiocyanate (20 mM). Target mRNA was hybridized with randomly labeled ([α-32P]ATP; NEN Life Science Products) DNA probes by use of the Klenow enzyme (New England Biolabs). For transcriptional analysis of motABcheAW, an 864-bp DNA fragment that covers the coding sequences of motB and cheA was PCR generated with primers MOTAB1 (5′-GAAGAGATTGAGACGCACGAAAGC-3′) and MOTAB2 (5′-TCGATTTTGAGATGGGGACGTAACG-3′). For analysis of tsr expression, an 895-bp tsr gene fragment was PCR generated with primers TSR1 (5′-CAGCGGCAGAGATCAAACGTAATTACG-3′) and TSR2 (5′-GCTTTTAATTTCACGAGCCGCCTGGG-3′). The expression of envZ (not CpxR-P regulated) was used as the internal control as previously described (30).
Analysis of CpxR-P binding to the promoter regions of motABcheAW and tsr.
The binding of CpxR or CpxR-P to the promoter regions of motABcheAW and tsr was examined by electrophoretic mobility shift analysis by a previously described method (32). A CpxR (or CpxR-P)/DNA ratio of 20 (200 pmol/10 pmol) was used in the presence of a 500-fold molar excess of competitor DNA (sheared herring sperm DNA; Promega) and a 100-fold molar excess of competitor protein (bovine serum albumin; New England Biolabs). CpxR was overproduced and purified as described before (30). CpxR was phosphorylated by incubation with acetyl phosphate (30). A 246-bp motABcheAW promoter fragment containing the putative CpxR-P recognition consensus box in the center was PCR amplified from the chromosome of strain BW21355 with primers MOT/EMS1 (5′-GGACATTGGTGCGGTTTGTTGAAAGTGG-3′) and MOT/EMS2 (5′-GCTGGAATGTTGCGCCTCACCGTATCAG-3′). A 212-bp tsr promoter fragment containing the CpxR-P recognition consensus box in the center was PCR amplified with primers TSR/EMS1 (5′-ATGTATTGATTAATAGTTGGCCGAAGCCG-3′) and TSR/EMS2 (5′-GATATGAATCACATATTTATCGTCACTTAAACG-3′). All primers were then 5′ labeled (30 min, 37°C) with [γ-32P]ATP by use of T4 polynucleotide kinase (New England Biolabs). Radiolabeled promoter DNA was PCR generated by use of the labeled primers with the previously amplified DNA fragments as templates. The labeled promoter DNA was purified (Qiagen gel purification kit) after agarose gel electrophoresis (2% SeaKem Ultrapure Agarose; FMC Corp.) and used in binding assays.
β-Galactosidase assay.
Specific β-galactosidase activities in the reporter strains grown in glucose minimal medium (20 ml, 37°C, 280 rpm) were assayed as previously described (22).
RESULTS AND DISCUSSION
CpxR-P autogenously activates cpxRA expression.
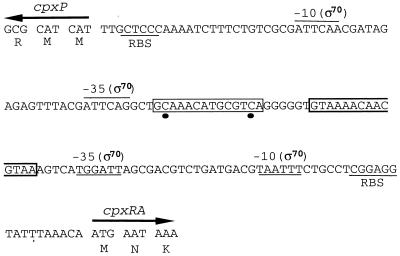
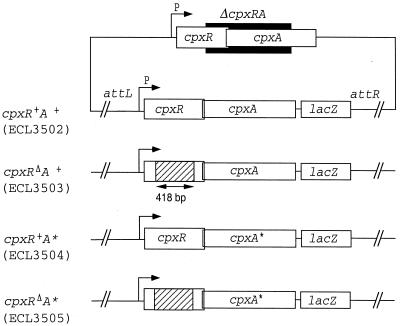
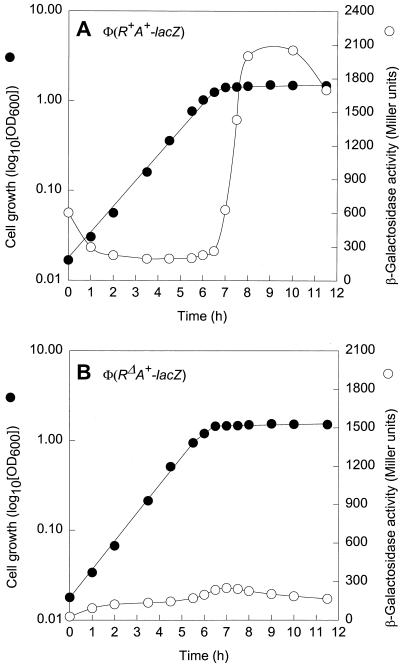
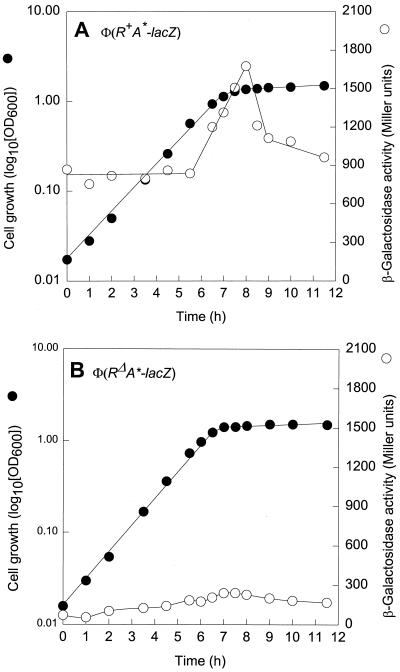
The intergenic region of the divergently transcribed cpxP and cpxRA operons contains a perfect CpxR-P binding consensus sequence, 5′-GTAAA(N)5GTAA-3′, which is located between bp 57 and 70 upstream from the CpxR translational start point (30) (Fig. 1). A second such sequence, with a 2-bp mismatch, is located between bp 77 and 90 (this study). The transcription of cpxP was shown to be activated by CpxR-P (5), but the possibility that cpxRA is also under the control of this regulator cannot be excluded. To examine this plausibility, we made two different cpxRA-lacZ constructs that express CpxR (intact or with an in-frame deletion), CpxA, and LacZ. These operon fusions were inserted at the attB site of strain ECL3501, yielding strains ECL3502 [Φ(cpxR+A+-lacZ)] and ECL3503 [Φ(cpxRΔA+-lacZ)]. In the latter fusion, cpxR sustained a 418-bp in-frame deletion (Fig. 2 and Table 1). In strain ECL3502 grown in glucose minimal medium, the Φ(cpxR+A+-lacZ) expression level (determined as specific β-galactosidase activity) rose dramatically at the onset of the stationary growth phase (typically at an OD600 of 1.0) (Fig. 3A). The in-frame deletion in cpxR (ECL3503) greatly diminished (ninefold) this increase (Fig. 3B). Thus, CpxR-P activates the expression of its own operon. Essentially the same results were obtained with cells grown in LB medium (data not shown).
FIG. 1.
Representation of the cpxP-cpxRA intergenic region. Putative recognition sequences for ς70 (single lines above or below the sequence) and ribosome binding (RBS) are shown. The perfect CpxR-P recognition consensus sequence is boxed in a bold line, whereas the consensus sequence with a 2-bp mismatch is boxed in a thin line. The consensus mismatches are indicated by black circles.
FIG. 2.
Representation of the Φ(cpx-lacZ) operon fusion constructs integrated at the λ attachment site (attB) of strain ECL3501. The black box depicts the deleted region within cpxRA. The hatched boxes depict the in-frame deletion (418 bp) within cpxR. The Φ(cpx-lacZ) constructs contain 309 bp upstream from the cpxR translational start point and include the cpxRA promoter region (P) (Fig. 1).
FIG. 3.
Effect of CpxR-P on Φ(cpxRA+-lacZ) expression. Growth curves and β-galactosidase activity profiles of strain ECL3502 [Φ(cpxR+A+-lacZ)] (A) and strain ECL3503 [Φ(cpxRΔA+-lacZ)] (B) are shown. The cells were grown in glucose minimal medium (pH 7.0) at 37°C.
RpoS activates cpxRA expression.
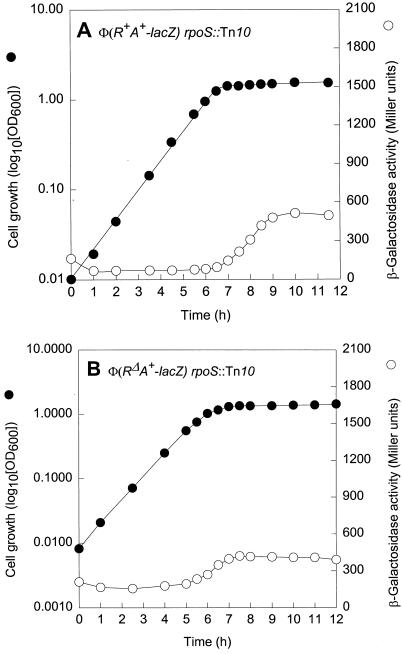
The strong increase in Φ(cpxR+A+-lacZ) expression at the onset of stationary growth (Fig. 3A) suggested an involvement of RpoS in cpxRA expression. The participation of RpoS, direct or indirect, was confirmed by a fourfold-lower stationary-phase expression level of Φ(cpxR+A+-lacZ) in strain ECL3506, bearing rpoS::Tn10 (Fig. 4A and 3A). However, in the absence of RpoS, the expression of Φ(cpxR+A+-lacZ) still rose moderately at the start of the stationary growth phase (Fig. 4A). Moreover, even in the absence of both RpoS and CpxR, a slight rise in Φ(cpxRΔA+-lacZ) expression was observed toward the end of exponential growth, suggesting the involvement of (an)other regulatory element(s) (Fig. 4B).
FIG. 4.
Effect of RpoS on Φ(cpxRA+-lacZ) expression. Growth curves and β-galactosidase activity profiles of strain ECL3506 [Φ(cpxR+A+-lacZ) rpoS::Tn10] (A) and strain ECL3507 [Φ(cpxRΔA+-lacZ) rpoS::Tn10] (B) are shown. The cells were grown in glucose minimal medium (pH 7.0) at 37°C.
Expression of cpxRA* is disproportionately enhanced during growth.
In view of the autogenous activation of cpxRA and the report that CpxA* sensor kinases are locked in the net phosphorylating mode (32), the basal level of expression of cpxRA can be expected to be elevated, possibly even during exponential growth. To test this hypothesis, we inserted two different cpxRA*-lacZ constructs that express CpxR (intact or with an in-frame deletion), CpxA* (with the cpxA* allele specifying a Leu38→Phe substitution), and LacZ. These operon fusions were inserted at the attB site in a ΔcpxRA strain (ECL3501). The resulting strains, ECL3504 [Φ(cpxR+A*-lacZ)] and ECL3505 [Φ(cpxRΔA*-lacZ), in which the R allele bears a 418-bp in-frame deletion] were then compared for their β-galactosidase activity levels during growth (Fig. 2 and Table 1). The exponential-phase expression of Φ(cpxR+A*-lacZ) (Fig. 5A) was found to be about fourfold higher than that of Φ(cpxR+A+-lacZ) (Fig. 3A). The level of Φ(cpxR+A*-lacZ) expression rose further before the end of exponential growth (typically at an OD600 of 0.6). It is unclear why the level of stationary-phase expression of Φ(cpxR+A*-lacZ) was lower than that observed in the wild-type strain. Nonetheless, the results as a whole support the notion that CpxA* causes excessive levels of CpxR-P, which may be responsible for most, if not all, of the cpxRA* phenotypes.
FIG. 5.
Effect of CpxR-P on Φ(cpxR+A*-lacZ) expression. Growth curves and β-galactosidase activity profiles of strain ECL3504 [Φ(cpxR+A*-lacZ)] (A) and strain ECL3505 [Φ(cpxRΔA*-lacZ)] (B) are shown. The cells were grown in glucose minimal medium (pH 7.0) at 37°C.
Genomic screening for CpxR-P-controlled operons.
To identify more putative CpxR-P-controlled target operons, we performed genomic scanning (Genetics Computer Group version 9.1 software) with the CpxR-P binding consensus sequence. Successful identification of additional target operons could provide more specific clues about the individual CpxA* phenotypes. About 50 consensus hits that lie within 450 bp upstream of start codons were found. The activities of some of the encoded proteins can be related to particular CpxA* phenotypes. First, the regulation of rpoH (encoding heat shock transcription factor ς32; CpxR-P box between bp 257 and 271 upstream of the rpoH start codon) can be linked to the temperature-sensitive growth condition of cpxA* mutants (16). This association is consistent with CpxR-P control of operons involved in protein rescue and/or clearance during heat stress (4, 6, 7, 30). Second, the gene product of psd (phosphatidylserine decarboxylase; CpxR-P box between bp 120 and 134 upstream of the psd start codon) catalyzes the synthesis of phosphatidylethanolamine. Lack of this phospholipid was shown to make a wild-type Cpx system hyperactive (21). A response at the level of psd expression might be a homeostatic strategy to restore a healthy membrane protein/phospholipid ratio. Third, direct regulation of the gene with accession no. U58330 (encoding a probable copper-transporting ATPase; CpxR-P box between bp 91 and 105 upstream of the U58330 start codon) may underlie the elevated resistance to CuCl2 with increasing CpxR-P levels (e.g., the CuCl2 resistance level is ECL3503 = ECL3505 < ECL3502 < ECL3504; data not shown). The functional diversity of the consensus sequence-identified operons further hints at a broad role for the Cpx system.
CpxR-P represses motility and chemotaxis genes.
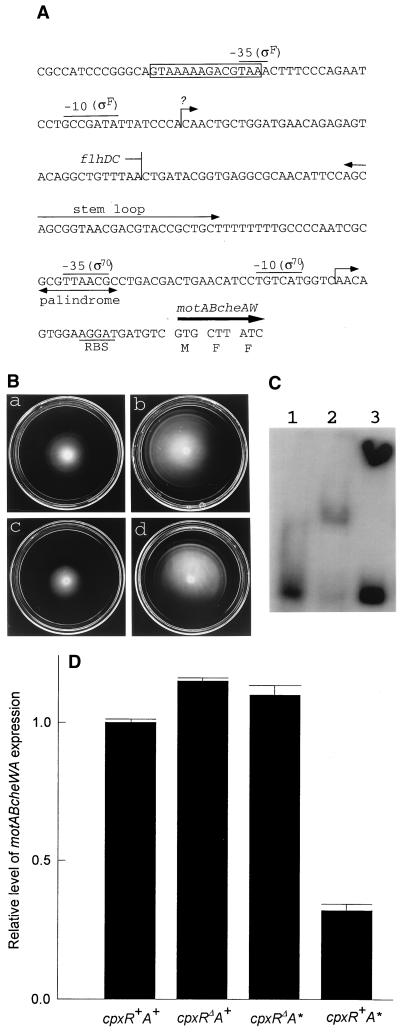
To test the usefulness of the consensus sequence screening, we analyzed Cpx control of two unanticipated target operons identified by the screening. The first was motABcheAW (specifying motility and chemotaxis). The location of the CpxR-P recognition box (Fig. 6A), overlapping the −35 site of a perfect consensus sequence for EςF (2, 14), made the regulatory involvement of CpxR-P quite plausible. We therefore compared the swarming abilities of the four Φ(cpx-lacZ) strains. Figure 6B shows that motility was negatively affected by CpxR-P. Electrophoretic mobility shift analysis showed retardation of the motABcheAW promoter DNA by CpxR in the presence of competitor DNA and protein. The CpxR regulator was more effective in the phosphorylated form (Fig. 6C), as was shown for the known target degP (32). Northern analysis supported the negative control of motABcheAW by CpxR-P (Fig. 6D). Whereas the deletion of CpxR dramatically increased the swarming rate, the effect of this deletion on the motABcheAW mRNA level was relatively small. Apparently, additional controlling elements are involved in the control of cell motility under the experimental conditions used.
FIG. 6.
Negative regulation of motABcheAW by the Cpx system. (A) Promoter region of motABcheAW (8, 14, 15). The CpxR-P recognition site is boxed. The EςF promoter and transcriptional start site are assigned on the basis of the EςF recognition consensus sequence (2, 15). RBS, ribosome binding site. (B) Swarm patterns. (a) ECL3502 [Φ(cpxR+A+-lacZ)]. (b) ECL3503 [Φ(cpxRΔA+-lacZ)]. (c) ECL3504 [Φ(cpxR+A*-lacZ)]. (d) ECL3505 [Φ(cpxRΔA*-lacZ)]. (C) Electrophoretic mobility shift analysis of the motABcheAW promoter DNA region with CpxR or CpxR-P. Lane 1, motABcheAW promoter DNA; lane 2, promoter DNA plus CpxR; lane 3, promoter DNA plus CpxR-P. (D) Profiles of expression of motABcheAW in the four Φ(cpx-lacZ) strains, as determined by Northern analysis. Error bars indicate standard deviations.
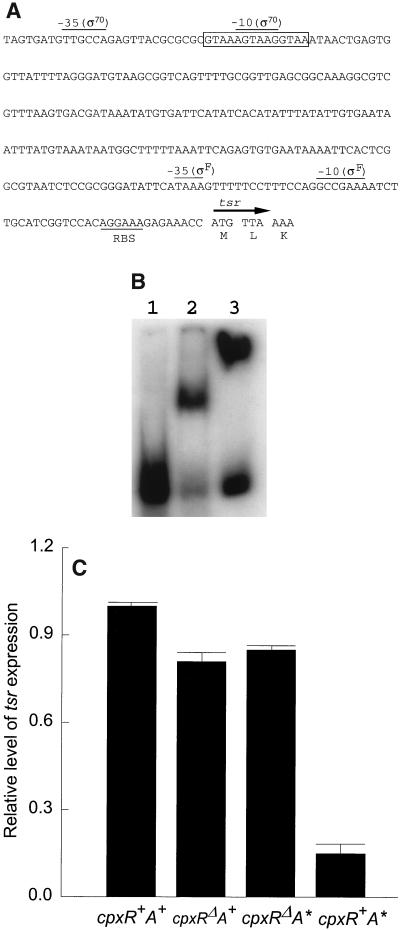
Next, the involvement of the Cpx system in the regulation of tsr (encoding the serine chemoreceptor) was examined. The tsr operon is transcribed by both EςF and Eς70 (Fig. 7A) (14). The Eς70 promoter site has not yet been defined, but a best-fitting Eς70 promoter sequence overlaps the CpxR-P recognition consensus sequence at the −10 site (Fig. 7A). Electrophoretic mobility shift analysis showed retardation of the tsr promoter DNA by CpxR in the presence of competitor DNA and protein. The CpxR regulator was again more effective in the phosphorylated form (Fig. 7B). Northern analysis showed the negative control of tsr by CpxR-P (Fig. 7C). Why the level of expression of tsr in the cpxR deletion strain did not exceed that in the wild-type strain (average of three experiments) remains unclear. It is possible that the cpxR deletion affected the synthesis or properties of other proteins involved in tsr expression. The motABcheAW and tsr operons are the first operons shown to be under the direct negative control of the Cpx regulatory pathway.
FIG. 7.
Negative regulation of tsr by the Cpx system. (A) Promoter region of tsr (14). The CpxR-P recognition site is boxed. The Eς70 promoter site is putative, but involvement of Eς70 in tsr expression has been reported (14). RBS, ribosome binding site. (B) Electrophoretic mobility shift analysis of the tsr promoter DNA region with CpxR or CpxR-P. Lane 1, tsr promoter DNA; lane 2, promoter DNA plus CpxR; lane 3, promoter DNA plus CpxR-P. (C) Profiles of expression of tsr in the four Φ(cpx-lacZ) strains, as determined by Northern analysis. Error bars indicate standard deviations.
CpxRA has a function beyond the management of periplasmic protein distress.
It is known that CpxR-P and ςE jointly activate the expression of degP (6) and that CpxR-P and ς32 jointly activate the expression of ppiD (7). Since under conditions of heat shock the expression of rpoH (encoding ς32) is activated by ςE (9, 41), an intricate regulatory circuit seems to have evolved. Our finding that CpxR-P and ςS act synergistically on cpxRA transcription expands the function of the Cpx signal transduction system into stationary-phase adaptation and suggests that starvation or energy depletion amplifies the Cpx signaling capacity. It is noteworthy that stationary-phase cells are more resistant to oxidative damage and heat shock (10). If rpoH is found to be Cpx controlled, the importance of the CpxRA system in the expression of the stress response network will become even more extensive. The fact that deleting cpxR and rpoE independently results in copper sensitivity (10a) further fuses the CpxRA and ςE pathways. In this context, it would be interesting to see whether the expression of the gene with accession no. U58330 (encoding a probable copper transporter) is also under the joint control of CpxR-P and ςE.
Although with the present knowledge it is difficult to integrate the control of motility and chemotaxis into a response network dealing with protein distress (ςE and ς32), the suppression of movement may be an energy-saving strategy during starvation. From this point of view, it is of interest to note that a recent study of Rhizobium meliloti showed that motility and chemotactic behavior are down-regulated during starvation. In that study, both flagellar maintenance and motor activity were found to be affected, but upon addition of a carbon source or chemoattractant, swarming and chemotaxis were partially restored (42).
In sum, evidence from this study suggests that the Cpx signal transduction system (Fig. 8), in conjunction with ςE and ς32, responds to a broad spectrum of adverse environmental conditions. These include heat shock, high pH (activates the Cpx pathway [5]), oxidative stress, and nutritional deprivation.
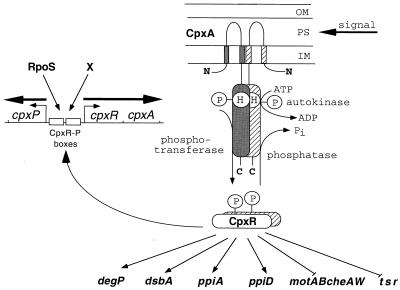
FIG. 8.
Representation of CpxRA signal transduction. CpxA and CpxR are shown as dimers. OM, outer membrane; PS, periplasmic space; IM, inner membrane; X, unknown activator. For explanations, see the text.
ACKNOWLEDGMENTS
We thank Joe Pogliano for strain JP466, Philip Silverman for strain AE2293, Gisella Storz for strain GS015, and Barry Wanner for strain BW21355. We are grateful to Jorge Membrillo-Hernandez, Philip Silverman, and Rosella Visintin for helpful discussions.
P.D.W. is a postdoctoral D. Collen Fellow of the Belgian American Educational Foundation. This work was supported by Public Health Service grants GM40993 and GM39693 from the National Institute of General Medical Sciences.
ADDENDUM
During the preparation of this manuscript, the independent discovery of the autogenous regulation of the cpxRA operon was presented by T. J. Silhavy at the 99th General Meeting of the American Society for Microbiology.
REFERENCES
- 1.Altuvia S, Almiron M, Huisman G, Kolter R, Storz G. The dps promoter is activated by OxyR during growth and by IHF and ςS in stationary phase. Mol Microbiol. 1994;13:265–272. doi: 10.1111/j.1365-2958.1994.tb00421.x. [DOI] [PubMed] [Google Scholar]
- 2.Arnosti D N, Chamberlin M J. Secondary ς factor controls transcription of flagellar and chemotaxis genes in Escherichia coli. Proc Natl Acad Sci USA. 1989;86:830–834. doi: 10.1073/pnas.86.3.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cosma C L, Danese P N, Carlson J H, Silhavy T J, Snyder W B. Mutational activation by the Cpx signal transduction pathway of Escherichia coli suppresses the toxicity conferred by certain envelope-associated stresses. Mol Microbiol. 1995;18:491–505. doi: 10.1111/j.1365-2958.1995.mmi_18030491.x. [DOI] [PubMed] [Google Scholar]
- 4.Danese P N, Silhavy T J. The ςE and the Cpx signal transduction systems control the synthesis of periplasmic protein-folding enzymes in Escherichia coli. Genes Dev. 1997;11:1183–1193. doi: 10.1101/gad.11.9.1183. [DOI] [PubMed] [Google Scholar]
- 5.Danese P N, Silhavy T J. Cpx-P, a stress-combative member of the Cpx regulon. J Bacteriol. 1998;180:831–839. doi: 10.1128/jb.180.4.831-839.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Danese P N, Snyder W B, Cosma C, Davis L J, Silhavy T J. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, Deg P. Genes Dev. 1995;9:387–398. doi: 10.1101/gad.9.4.387. [DOI] [PubMed] [Google Scholar]
- 7.Dartigalongue C, Raina S. A new heat-shock gene, ppiD, encodes a peptidyl-prolyl isomerase required for folding of outer membrane proteins in Escherichia coli. EMBO J. 1998;14:3968–3980. doi: 10.1093/emboj/17.14.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dean G E, Macnab R M, Stader J, Matsimura P, Burks C. Gene sequence and predicted amino acid sequence of the motA protein, a membrane-associated protein required for flagellar rotation in Escherichia coli. J Bacteriol. 1984;159:991–999. doi: 10.1128/jb.159.3.991-999.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a.De Wulf, P. Unpublished data.
- 9.Erickson J W, Gross C A. Identification of the ςE subunit of Escherichia coli RNA polymerase: a second alternate ς factor involved in high temperature gene expression. Genes Dev. 1989;3:1462–1471. doi: 10.1101/gad.3.9.1462. [DOI] [PubMed] [Google Scholar]
- 10.Goodrich-Blair H, Uria-Nickelson M, Kolter R. Regulation of gene expression in stationary phase. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. Austin, Tex: R. G. Landes Company and Chapman & Hall; 1996. pp. 571–583. [Google Scholar]
- 10a.Gross, C. A. Personal communication.
- 11.Haldimann A, Fisher S T, Daniels L L, Walsh C T, Wanner B L. Transcriptional regulation of the Enterococcus faecium BM4147 vancomycin resistance gene cluster by the VanS-VanR two-component regulatory system in Escherichia coli K-12. J Bacteriol. 1997;179:5903–5913. doi: 10.1128/jb.179.18.5903-5913.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jang, W., and B. L. Wanner. Unpublished data.
- 13.Jones H M, Gunsalus R P. Regulation of Escherichia coli fumarate reductase (frdABCD) operon expression by respiratory electron acceptors and the fnr gene product. J Bacteriol. 1987;169:3340–3349. doi: 10.1128/jb.169.7.3340-3349.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kundu T K, Kusano S, Ishihama A. Promoter selectivity of Escherichia coli RNA polymerase ςF holoenzyme involved in transcription of flagellar and chemotaxis genes. J Bacteriol. 1997;179:4264–4269. doi: 10.1128/jb.179.13.4264-4269.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macnab R B. Flagella and motility. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 123–145. [Google Scholar]
- 16.McEwen J, Silverman P M. Chromosomal mutations of Escherichia coli that alter expression of conjugative plasmid functions. Proc Natl Acad Sci USA. 1980;77:513–517. doi: 10.1073/pnas.77.1.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McEwen J, Silverman P M. Genetic analysis of Escherichia coli K-12 chromosomal mutants defective in expression of F-plasmid functions: identification of cpxA and cpxB. J Bacteriol. 1980;144:60–67. doi: 10.1128/jb.144.1.60-67.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McEwen J, Silverman P M. Mutations in genes cpxA and cpxB of Escherichia coli K-12 cause a defect in isoleucine and valine synthesis. J Bacteriol. 1980;144:68–73. doi: 10.1128/jb.144.1.68-73.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McEwen J, Silverman P M. Mutations in genes cpxA and cpxB alter the protein composition of Escherichia coli inner and outer membranes. J Bacteriol. 1982;151:1553–1559. doi: 10.1128/jb.151.3.1553-1559.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McEwen J, Sambucetti L, Silverman P M. Synthesis of outer membrane proteins in cpxA cpxB mutants of Escherichia coli K-12. J Bacteriol. 1983;154:375–382. doi: 10.1128/jb.154.1.375-382.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mileykovskaya E, Dowhan W. The Cpx two-component signal transduction pathway is activated in Escherichia coli mutant strains lacking phosphatidylethanolamine. J Bacteriol. 1997;179:1029–1034. doi: 10.1128/jb.179.4.1029-1034.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 23.Missiakis D, Raina S, Georgopoulos C. Heat shock regulation. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. Austin, Tex: R. G. Landes Company and Chapman & Hall; 1996. pp. 481–501. [Google Scholar]
- 24.Morris J M, Newman E B. Map location of the ssd mutation in Escherichia coli K-12. J Bacteriol. 1980;154:1504–1505. doi: 10.1128/jb.143.3.1504-1505.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newman E B, Malik N, Walker C. l-Serine degradation in Escherichia coli K-12: directly isolated ssd mutants and their intergenic revertants. J Bacteriol. 1982;150:710–715. doi: 10.1128/jb.150.2.710-715.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plate C. Mutant of Escherichia coli defective in response to colicin K and in active transport. Proc Natl Acad Sci USA. 1976;125:467–474. doi: 10.1128/jb.125.2.467-474.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plate C A, Suit J L. The eup genetic locus of Escherichia coli and its role in H+/solute symport. J Biol Chem. 1981;256:12974–12980. [PubMed] [Google Scholar]
- 28.Plate C A, Seely S A, Laffler T G. Evidence for a protonmotive force related regulatory system in Escherichia coli and its effects on lactose transport. Biochemistry. 1986;25:6127–6132. doi: 10.1021/bi00368a044. [DOI] [PubMed] [Google Scholar]
- 29.Pogliano J, Dong J M, De Wulf P, Furlong D, Boyd D, Losick R, Pogliano K, Lin E C C. Aberrant cell division and random FtsZ ring positioning in Escherichia coli cpxA* mutants. J Bacteriol. 1998;180:3486–3490. doi: 10.1128/jb.180.13.3486-3490.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pogliano J, Lynch A S, Belin D, Lin E C C, Beckwith J. Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev. 1997;11:1169–1182. doi: 10.1101/gad.11.9.1169. [DOI] [PubMed] [Google Scholar]
- 31.Rainwater S, Silverman P M. The Cpx proteins of Escherichia coli K-12: evidence that cpxA, ecfB, ssd, and eup mutations all identify the same gene. J Bacteriol. 1990;172:2456–2461. doi: 10.1128/jb.172.5.2456-2461.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raivio T L, Silhavy T J. Transduction of envelope stress in Escherichia coli by the Cpx two-component system. J Bacteriol. 1997;179:7724–7733. doi: 10.1128/jb.179.24.7724-7733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raivio T L, Silhavy T J. The ςE and Cpx regulatory pathways: overlapping but distinct envelope stress responses. Curr Opin Microbiol. 1999;2:159–165. doi: 10.1016/S1369-5274(99)80028-9. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Sambucetti L, Eoyang L, Silverman P M. Cellular control of conjugation in Escherichia coli K-12. Effect of chromosomal cpx mutations on F-plasmid gene expression. J Mol Biol. 1982;161:13–31. doi: 10.1016/0022-2836(82)90275-3. [DOI] [PubMed] [Google Scholar]
- 36.Silverman P M, Tran L, Harris R, Gaudin H M. Accumulation of the F plasmid TraJ protein in cpx mutants of Escherichia coli. J Bacteriol. 1993;175:921–925. doi: 10.1128/jb.175.4.921-925.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 38.Su H, Lang F, Newman E B. l-Serine degradation in Escherichia coli K-12: cloning and sequencing of the sdaA gene. J Bacteriol. 1989;171:5095–5102. doi: 10.1128/jb.171.9.5095-5102.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sutton A, Newman T, McEwen J, Silverman P M, Freundlich M. Mutations in genes cpxA and cpxB of Escherichia coli K-12 cause a defect in acetohydroxyacid synthase I function in vivo. J Bacteriol. 1982;151:976–982. doi: 10.1128/jb.151.2.976-982.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thorbjarnardottir S, Magnusdottir R, Eggertsson G, Kagan S, Andersson D. Mutations determining generalized resistance to aminoglycoside antibiotics in Escherichia coli. Mol Gen Genet. 1978;161:89–98. doi: 10.1007/BF00266619. [DOI] [PubMed] [Google Scholar]
- 41.Wang Q, Kaguni J M. A novel sigma factor is involved in expression of the rpoH gene of Escherichia coli. J Bacteriol. 1989;171:4248–4253. doi: 10.1128/jb.171.8.4248-4253.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei X, Bauer W D. Starvation-induced changes in motility, chemotaxis, and flagellation of Rhizobium meliloti. Appl Environ Microbiol. 1998;64:1708–1714. doi: 10.1128/aem.64.5.1708-1714.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wu, H. C. Personal communication.