Stromal metabolite C-6-S induced macrophage M2 polarization and tumor immune exclusion via coactivating JAK/STAT3 and Hedgehog pathways. Combining anti–PD-1 and targeting of the “immunometabolism checkpoint” C-6-S showed promise as an effective immunotherapy strategy for microsatellite-stable colorectal cancer.
Abstract
Metastatic microsatellite-stable (MSS) colorectal cancer rarely responds to immune checkpoint inhibitors (ICI). Metabolism heterogeneity in the tumor microenvironment (TME) presents obstacles to antitumor immune response. Combining transcriptome (The Cancer Genome Atlas MSS colorectal cancer, n = 383) and digital pathology (n = 96) analysis, we demonstrated a stroma metabolism–immune excluded subtype with poor prognosis in MSS colorectal cancer, which could be attributed to interaction between chondroitin-6-sulfate (C-6-S) metabolites and M2 macrophages, forming the “exclusion barrier” in the invasive margin. Furthermore, C-6-S derived from cancer-associated fibroblasts promoted co–nuclear translocation of pSTAT3 and GLI1, activating the JAK/STAT3 and Hedgehog pathways. In vivo experiments with C-6-S–targeted strategies decreased M2 macrophages and reprogrammed the immunosuppressive TME, leading to enhanced response to anti–PD-1 in MSS colorectal cancer. Therefore, C-6-S–induced immune exclusion represents an “immunometabolic checkpoint” that can be exploited for the application of combination strategies in MSS colorectal cancer ICI treatment.
Introduction
Immunotherapy with immune checkpoint inhibitors (ICI) targeting PD-1/PD-L1 has revolutionized the treatment for cancer (1). A pioneering clinical trial in 2015 investigating PD-1 inhibitor efficacy highlighted high microsatellite instability (MSI-H) as a pan-solid tumor biomarker (1). However, only approximately 5% of metastatic colorectal cancer are MSI-H, while the remaining 95% low microsatellite instability (MSI-L)/patients with microsatellite-stable (MSS) metastatic colorectal cancer barely respond to ICIs monotherapy (2, 3).
Recent advances identified the reasons for MSS colorectal cancer immunotherapy resistance: inadequate antigen release and presentation, poor immunogenicity, and an immune-cold or immune-excluded tumor microenvironment (TME; ref. 2). The abundant immunosuppressive cells, including tumor-associated macrophages (TAM), myeloid-derived suppressor cells (MDSC), regulatory T cells, and microenvironment factors like VEGFA and TGFβ (2), could exclude CD8+ T-cell infiltration and impair antitumor response. ICIs combined with VEGFR2 blockade has shown enhanced efficacy in MSS colorectal cancer preclinical models (4). Thus, elucidating crucial immune evasive factors in the TME may facilitate solving the obstacles of ICI treatment in MSS colorectal cancer.
Immunometabolism represents the interaction between metabolism and the immune TME (5). Metabolic flux in the TME, including deprivation of metabolic substrates, accumulation of metabolic waste and metabolism activity in various cell types, has crucial effects on the antitumor response (5). Immune regulatory metabolic genes, enzymes, and metabolites are recognized as “immunometabolic checkpoints” (6). Recently, targeting an immunometabolic checkpoint through glutamine enzyme inhibition was found to suppress glutamine circuits of tumor cells while upregulating oxidative metabolism in effector T cells (7). Metabolic antagonists may sensitize the TME to immunotherapy by triggering divergent metabolic programs that amplify the function of immune cells and reshape the immune TME with low toxicity and high efficiency (6). At present, immunometabolism strategies, such as glucose metabolism inhibitor Metformin, glutamine pathway inhibitor CB-839, adenosine inhibitor oleclumab, and IDO1/IDO2 inhibitors, combined with anti–PD-1/PD-L1 are under investigation in preclinical or clinical research (5, 8). Metabolism interventions have great potential in tackling the difficulties of poor MSS colorectal cancer immunotherapy response.
We identified a metabolic-immune subtype with the worst prognosis in MSS colorectal cancer, characterized by abundant stroma and high-level immune evasion. Furthermore, we discovered chondroitin sulfate (CS), specifically chondroitin-6-sulfate (C-6-S), as the key factor of poor prognosis. CS, a type of glycosaminoglycans (GAG), is a linear acidic polysaccharide comprised of repeating disaccharides (9). CS has shown anti-inflammatory activity in osteoarthritis and proinvasive effect in tumors (9, 10). However, the role of CS in the TME and immunotherapy is unclear. Here, we targeted C-6-S as an immunometabolic checkpoint through a metabolism-immune microenvironment exploration workflow, which may contribute to improving MSS colorectal cancer ICIs strategies.
Materials and Methods
Human and mouse cell lines
We obtained murine colon carcinoma cell line CT26 from the National Collection of Authenticated Cell Cultures (NCACC). Cells were cultured in RPMI1640 medium (Solarbio, 31800) supplemented with 10% FBS (Solarbio, S9030). Murine MC38 colon cancer cell line was from BNCC and cultured in DMEM (Solarbio, 12100) with 10% FBS (Solarbio, S9030). Human monocytic cell line THP-1 was from NCACC and cultured with RPMI1640 medium (Solarbio, 31800) supplemented with 10% FBS (Solarbio, S9030). When indicated, THP-1 monocytes were incubated with phorbol 12-myristate 13-acetate (PMA; 100 ng/mL; Selleck, S7791) for 24 hours to differentiate into macrophages. Murine macrophage cell line RAW264.7 was from NCACC and cultured in DMEM (Solarbio, 12100) with 10% FBS (Solarbio, S9030). All cell lines were cultured at 37°C in a humidified atmosphere with 5% CO2. Authentication for all cell lines was acquired from the manufacturers and reauthenticated with short tandem repeat test within the past year. Cells were cultured for approximately 3 months for experiments and were regularly tested for Mycoplasma with PCR method. No more than 10 passages for THP-1 and RAW264.7 cells and no more than 20 passages for CT26 and MC38 cells were used.
Study cohorts
We performed transcriptomic analysis in The Cancer Genome Atlas (TCGA) colorectal cancer cohort. The cohort was filtered with tissue type (primary tumor). MSI-L/MSS samples are similar to proficient mismatch repair (pMMR) status, while MSI-H are similar to deficient mismatch repair status (11). After filtering samples with MSI status (MSI-L/MSS), 383 patients were selected and the cohort is referred to TCGA MSS colorectal cancer cohort (Supplementary Table S1). We assembled a clinical cohort with 112 colorectal cancer patient's paraffin-embedded surgical tumor specimens from Nanfang Hospital (Guangzhou, P.R. China; n = 35), Guangzhou First People's Hospital (Guangzhou, P.R. China; n = 74), and Zhujiang Hospital (Guangzhou, P.R. China; n = 3). Patients were diagnosed with colorectal cancer and received surgery in 2014–2018. Signed informed consents were obtained from all the patients for the use of clinical information and tissue samples. Paraffin-embedded tissue sections from primary tumors were collected and stored at 4°C until analysis. Tissue sections were observed under microscopy and tumor area was calculated as tumor cells–infiltrated area/total area × 100%. Tissue sections with more than 20% tumor area were qualified to perform further analysis. MMR status was defined on the basis of MSH2, MSH6, PMS2, and MLH1 protein expression via immunohistochemistry (IHC). pMMR status was considered as MSS, according to previous report (11). A total of 96 patients with pMMR were included for further investigation, among which 30 patients were from Nanfang Hospital (Guangzhou, P.R. China), 63 from Guangzhou First People's Hospital (Guangzhou, P.R. China), and 3 from Zhujiang Hospital (Guangzhou, P.R. China). The study was conducted in accordance with the Declaration of Helsinki. The use of human tissue samples and clinical data was approved by the ethics committee of Nanfang Hospital (Guangzhou, P.R. China).
DNA MMR status classification
DNA MMR system is constituted by four MMR genes and their proteins (MLH1, MSH2, MSH6, and PMS2). For collected colorectal cancer samples, IHC of the four MMR proteins was performed as described in the IHC assays. One pathologist blind to data analysis interpreted the results. Normal colonic crypt epithelium adjacent to tumor, lymphoid cells, and stroma cells were used as internal positive control. Any positive staining of tumor cells is considered positive. If the expression of all four MMR indicators is normal, the sample is classified as proficient MMR/MSS; if the expression of one or more indicators is missing, then the sample is classified as deficient MMR/MSI-H.
Calculation of metabolism and immune characteristics score
The metabolism pathways were obtained from Kyoto Encyclopedia of Genes and Genomes (KEGG) database (https://www.genome.jp/kegg/; ref. 12) and curated on the basis of a previous report (13). Metabolism-related pathways with gene set size of 3–200 were selected, resulting in 113 metabolism pathways with 1,784 metabolic genes. GAGs metabolism could be divided into CS, heparan sulfate, and keratan sulfate metabolism according to the KEGG database. Metabolism pathway gene sets, including subsets from GAGs metabolism, are provided in Supplementary Table S2. Considering cell type–specific expression signature and dominant cell types in colorectal cancer, we applied curated immune and microenvironment cell gene sets of 25 cell types reported by previous study (14), which were filtered from CIBERSORT and MCP-counter. We integrated gene sets from Msigdb (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp) and previous research to generate TME gene sets [including hypoxia, reactive oxygen species, angiogenesis, lymphangiogenesis (15) and stroma (16) pathways] and immune phenotype regulatory factor gene sets including MHC machinery BioCarta, IFNG signaling (BioCarta), CD8 T effector, immune checkpoints (17), cytokines related to immune response (Reactome), TGFβ (17), angiogenesis, stroma, and DNA replication (Gene Ontology) pathways. In addition, immune response and evasion-related mechanic pathways (18), including nonredundant 24 HALLMARK pathways, 21 IPA pathways (http://www.ingenuity.com), adenosine, immungentic cell death, and NOS1 pathways were obtained. Tumor molecular function portrait (19), including oncogenic pathways (20), pro- and antitumor microenvironmental factors and stromal network, based on single-sample gene set enrichment analysis (ssGSEA) scores was performed. Transcripts per kilobase million (TPM) values were used, unless specified, to perform ssGSEA via GSVA R package (21). Enrichment scores of each pathway for each patient were calculated for further investigation.
Clustering of metabolism subtypes
We used K-means clustering algorithms and the ConsensusClusterPlus R package (22) to identify metabolism subtypes. To achieve stable clustering, we performed 1,000 iterations with 80% resampling (k = 2–6). Cluster stability was evaluated with cumulative distribution function and delta area plot. k = 3 was chosen on the basis of clustering stability and clinical significance. Samples were reordered according to K-means clustering results. Metabolism pathway scores were scaled before heatmap graphing using ComplexHeatmap R package (23).
Metabolism subtype–specific pathway and category activation ratio
To select subtype-specific pathways, we first applied Boruta algorithms based on a random forest model. With metabolism subtype as classification indicator, featured pathway selection was carried out using Boruta R package (24) (maxRuns = 200). Comparisons of metabolism pathway scores [stroma metabolism (SM) subtype vs. Others, nucleotide metabolism (NM) subtype vs. Others, energy metabolism (EM) subtype vs. Others] were conducted using the Limma R package (25). log2 (fold change) > 0 and FDR < 0.05 was considered significant upregulation. The top 10 significantly upregulated metabolism pathways that passed the Boruta importance test in each subtype were defined as subtype-specific metabolism pathways (3 for SM subtype, 9 for NM subtype, and 10 for EM subtype). On the basis of information in the KEGG database, pathways were classified into 11 metabolism categories (Supplementary Table S2). Categories with three or more pathways (n = 9) were used in metabolism category activation ratio analysis. Metabolism category activation ratios were calculated as the proportion of subtype-specific upregulated pathways in each metabolism category for each subtype.
Immune response and evasion score
T cell–inflamed gene expression profile (GEP; ref. 26) reflects IFNγ signaling related antitumor immune response. High GEP was reported to predict immunotherapy efficacy in melanoma and non–small cell lung cancer (26). In this study, we calculated the GEP score as the average expression of 18 genes in the signature using TPM values with log2 transformation. T-cell dysfunction and exclusion (TIDE) score (27) describes immune evasion; high TIDE scores predict immunotherapy resistance. Standardized data were generated using the gene expression median as the normalization control and uploaded to obtain TIDE score for tumor samples (http://tide.dfci.harvard.edu/log/). Cytotoxicity score was defined by the average expression values of GZMA and PRF1 (28).
Digital pathology analysis
Image analysis processing included the following steps: (i) Two regions of interest (ROI) in each histologic slide were manually outlined, namely the tumor core (TC) and the invasive margin (IM). The TC is the main area of the tumor tissue, and the IM is a 500 μm wide band-shaped area at the interface between tumor and non-tumor tissue, according to previous study (29). (ii) Open-source software QuPath (v0.2.2, https://github.com/qupath/qupath; ref. 30) was used to automatically detect all cell structures in the ROI, and the staining intensity threshold was set to recognize positive-staining cell. In general, dark brown is considered as strong positive, brown-yellow as moderate positive, light yellow as weak positive, and blue nuclei as negative. The detection results were manually checked. (iii) Output the density of positive cells (positive cell number per mm2) or H-Score [1× percentage of weak positive cells (%) + 2× percentage of moderate positive cells (%) + 3× percentage of strong positive cells (%)] for further analysis. (iv) Use QuPath's built-in random forest cell recognition module when necessary, and train the random forest model to identify cells in the ROI, using annotated tumor cells and stromal cells regions as input. Each slide was manually checked and a suitable classification model was applied. The steps above were supervised by two pathologists. As for immune phenotype, high CD8+ TC and high CD8+ IM is described as “hot”; low CD8+ TC and low CD8+ IM is “cold,” and high CD8+ IM but low CD8+ TC is “excluded” phenotype. A total of 70th percentile of CD8+ cell density was used as the cutoff to determine high/low grouping. The digitized tissue slides at 40× magnification were acquired using Aperio GT450 digital pathology scanner and analysis system (Leica). Because of automated focusing errors, insufficient staining and limited clinical specimen tissues, the tissue sections that failed to produce qualified digital slides were marked as NA values.
IHC assays
Tissue samples were collected and formalin fixed and paraffin embedded. A total of 4-μm tissue sections were used for immunostaining as reported previously (31). Briefly, after antigen retrieval, samples were blocked with BSA (Solarbio, 9048-46-8) for 1 hour, and then incubated with primary antibodies overnight at 4°C. Tissue sections were then washed in PBS and incubated with horseradish peroxidase–labeled goat anti-rabbit IgG antibody (Bioss, bs-0295G) for 1 hour at room temperature. 3,3′-Diaminobenzidine staining and hematoxylin counterstaining were applied and mounted. Primary antibodies were used as follows: C-6-S (Millipore, MAB2035, RRID: AB_11214309, 1:300), CD8 (Proteintech, 66868-1-Ig, RRID: AB_2882205, 1:3,000), CD163 (Proteintech, 16646-1-AP, RRID: AB_2756528, 1:800), CD206 (Proteintech, 60143-1-Ig, RRID: AB_2144924, 1:8,000), αSMA (Proteintech, 14395-1-AP, RRID: AB_2756528, 1:8,000), GLI1 (Proteintech, 66905-1-Ig, RRID: AB_2882232, 1:1,000), pSTAT3 (Cell Signaling Technology, 9145, RRID: AB_2491009, 1:800), MLH1 (Proteintech, 11697-1-AP, RRID: AB_2145604, 1:200), MSH2 (Proteintech, 60161-1-Ig, RRID: AB_10666855, 1:200), MSH6 (Proteintech, 66172-1-Ig, RRID: AB_2881567, 1:400), PMS2 (Proteintech, 66075-1-Ig, RRID: AB_11182595, 1:400).
Primary cultured fibroblasts
Fresh human colorectal cancer specimens and the adjacent non-tumor tissues were collected to isolate cancer-associated factors (CAF) and NFs, respectively. Murine CAFs were isolated from CT26 subcutaneous tumors. Tumor or non-tumor tissues were diced into approximately 1 mm3 with a razor blade and were digested with a solution of 5 mg/mL DNase I (Meilunbio, MB3069) + 20 mg/mL collagenase IV (Biofroxx, 2091MG100) + 20 mg/mL hyaluronidase (Solarbio, H8030) for 3–4 hours at 37°C on a rotating platform. Then cells were resuspended, filtered through a cell strainer (75 μm), and rinsed with PBS. Cells were collected, seeded into 24-well plates, and were cultured in DMEM (Solarbio, 12100) supplemented with 10% FBS (Solarbio, S9030) and 1% antibiotic-antimycotic (Solarbio, P8420). Primary fibroblasts were purified from other cell populations by differential adhesion and serial passage. Cell identity was confirmed by αSMA immunofluorescence (IF) staining. All primary fibroblasts in experiments were below 10 passages.
Primary mouse bone marrow–derived macrophages, mouse peritoneal macrophages, and human colorectal cancer TAMs
Six- to 10-week-old C57BL/6 mice were euthanatized to obtain femurs. The femurs were dissected with scissors and the muscles attached to the bone were removed. Bone marrow was flushed out using DMEM (Solarbio, 12100) and placed into sterile tube. The bone marrow was then homogenized with plastic pipette and the primary progenitor cell suspension was generated. Then the cell suspensions were incubated in DMEM (Solarbio, 12100) with addition of 10% FBS (Solarbio, S9030), 100 U/mL penicillin-streptomycin (Solarbio, P1400) and 10% L929 cell-conditioned medium, for 7 days. L929 cell-conditioned medium was generated through cultivation of L929 cells in RPMI1640 medium (Solarbio, 31800) with 10% FBS (Solarbio, S9030) for 10 days, which contains macrophage colony-stimulating factor. Differentiation of macrophages was determined using anti-F4/80 (MultiSciences, 70-AM048010-20, Clone:BM8.1) with flow cytometry as described previously (32). Primary peritoneal macrophages (PM) were obtained by flushing the mouse peritoneal cavity with PBS. Cells were washed with PBS twice and cultured in DMEM (Solarbio, 12100) for 1 hour at 37°C and 5% CO2. PBS was then used to remove nonadherent cells. The purity of macrophages was tested with anti-F4/80 (MultiSciences, 70-AM048010-20, Clone:BM8.1) by flow cytometry (purity >80%). Human TAMs were isolated from fresh colorectal cancer tissues as described previously (33). After digestion, cell suspensions were obtained and placed in a 15 mL tube with 5 mL 45% Percoll (Solarbio, P8370) in the middle and 5 mL 60% at the bottom, which were then centrifuged at 800 × g for 30 minutes. Cells from the interphase were then isolated with CD14 isolation kit (Miltenyi Biotec, 130-097-052) based on the manufacturer's instructions.
Three-dimensional cocultivation assays
Collagen type I (3 mg/mL; Solarbio, C8062), 1 mol/L NaOH solution (Acmec, S41251), 10× PBS (Servicebio, G0002) solution, and dH2O were mixed in a volume ratio of 40:1:6:13 to prepare a pH neutral collagen working solution. Aliquots of 300 μL were used to prepare three-dimensional (3D) collagen. After the primary CAF cells and macrophages were counted, 3.0 × 105 cells of each were mixed in equal proportions, dissolved in 3D collagen, and placed in 37°C and 5% CO2 atmosphere. After gelation, the 3D collagen system was covered with culture medium to construct a 3D coculture model. IF was used to detect the expression and localization of related indicators.
Coculture system for CAFs and macrophages
In the co-culture model, fibroblasts and macrophages were cultured in a chamber (JET Biofil, TCS016012). A total of 2 × 105 adherent fibroblasts were added to the upper layer, and 2 × 105 macrophages were cultured in the lower layer. Co-cultivation lasted for continuous 48 hours. The following reagents were added as indicated: Surfen (Millipore, S6951, 20 μmol/L), chondroitinase ABC (Ch-ABC; Yuanye Bio-Technology, S31309, 0.2 U), Stattic (Selleck, S7024, 2.5 μmol/L), and Vismodegib (Targetmol, T2590, 0.05 μmol/L). The lower layer of macrophages was collected for RNA extraction, and M1 and M2 phenotype polarization indicators were detected via qRT-PCR and flow cytometry.
IF assays
The IF was performed as described previously (31). The samples were incubated with the primary antibody at 4°C overnight. Then the secondary antibody Alexa fluor 647-labeled goat anti-mouse IgG(H+L) (Beyotime, A0473, RRID: AB_2891322, 1:500), Alexa Fluor 488-labeled goat anti-rabbit IgG(H+L) (Beyotime, A0423, RRID: AB_2891323, 1:500), Cy3-labeled goat anti-rat IgG(H+L) (Beyotime, A0507, 1:500) or Alexa Fluor 594 AffiniPure donkey anti-goat IgG (H+L) (Yeasen, 34312ES60, 1:200) was used for incubation for 1 hour. Next, the samples were incubated with the methanol dilution of DAPI (Beyotime, C1002, 1:1,000) at room temperature for 5 minutes. The images were taken with a fluorescence and laser confocal microscope (Nikon ECLIPSE Ti2) and were analyzed using ImageJ software (34). Radial fluorescence intensity analysis was performed using Plot Profile, a plug-in of ImageJ software, to observe fluorescence colocalization. The following antibody concentrations were used: C-6-S (Millipore, MAB2035, RRID: AB_11214309, 1:200), EPCAM (Proteintech, 21050-1-AP, RRID: AB_10693684, 1:100), αSMA (Novus Biologicals, NB300-978SS, 1:200), CD163 (Proteintech, 16646-1-AP, RRID: AB_2144924, 1:200), CD8 (Novus Biologicals, NB200-578, RRID: AB_10003082, 1:200), CD206 (Cell Signaling Technology, 91992, RRID: AB_2800175, 1:200), GLI1 (Proteintech, 66905-1-Ig, RRID: AB_2882232, 1:200), pSTAT3 (Cell Signaling Technology, 9145, RRID: AB_2491009, 1:200).
qRT-PCR
According to the manufacturer's instructions, total RNA from tissue samples was extracted with TRIzol reagent (Invitrogen). The reverse transcription kit HiScript II Q RT SuperMix for qPCR (R222-01, Vazyme, Nanjing) was used to synthesize cDNA from total RNA. Real-time qPCR was performed using the LightCycler 480 system Version 1.5 (Roche). The indicator gene expression was scaled using GAPDH expression as control. The 2–ΔΔCt method was used to calculate the expression fold change. Each qRT-PCR experiment was independently repeated in triplicate. The primer sequences used for qRT-PCR in this study are shown in the Supplementary Table S3.
Western blotting and coimmunoprecipitation
Western blotting (WB) was performed as described previously (31). Immunoblots were detected with fluorophore-conjugated goat anti-rabbit or anti-mouse secondary antibodies by an Odyssey imaging system (LI-COR). Antibodies used were as follows: GLI1 (Proteintech, 66905-1-Ig, RRID: AB_2882232, 1:1,000), pSTAT3 (Cell Signaling Technology, 9145, RRID: AB_2491009, 1:1,000). For the immunoprecipitation (IP) experiment, FLAG-GLI plasmid (GeneCopoeia, EX-F0407-Lv242) was transiently transfected into macrophages using Lipofectamine 2000 (Invitrogen) as described previously (35). Following 24 hours, the medium was replaced and treated with C-6-S (1 mg/mL for RAW264.7 and 2.5 mg/mL for THP-1) for another 24 hours. FLAG tag antibody (Cell Signaling Technology, 14793, AB_2572291) was used to pull down FLAG-GLI1 protein and its binding protein complex. pSTAT3 antibody (Cell Signaling Technology, 9145, RRID: AB_2491009) was used to pull down the complex of its binding protein to determine the interaction between the two transcription factors. Briefly, after rinsing with cold PBS, cells were lysed in IP lysis buffer (Meilunbio, MB9900). Then, Protein A+G Sepharose Beads (7Sea biotech) and the primary IP antibody were added and placed on a vibration platform overnight at 4°C to precipitate the immune complexes. Samples were rinsed with IP lysis buffer five times, boiled and eluted in SDS-PAGE buffer (LEAGENE, PE0025) to carry out further WB analysis. Equal amounts of protein were electrophoresed on SDS-PAGE and then transferred to nitrocellulose membrane. The immunoblots were blocked with 5% skim milk powder and detected by using enhanced chemiluminescence reagent. Antibodies were used as follows: GLI1 (Proteintech, 66905-1-Ig, RRID: AB_2882232) pulldown (1:50), WB (1:1,000); pSTAT3 (Cell Signaling Technology, 9145, RRID: AB_2491009) pulldown (1:50), WB (1:1,000); FLAG (Cell Signaling Technology, 14793, RRID: AB_2572291) pulldown (1:50), WB (1: 1,000).
In vitro and in vivo flow cytometry analysis
In the in vitro experiments, cells were incubated with the antibody conjugated with fluorescence in 100 μL of FACS staining buffer [1×PBS (Servicebio, G0002) containing 1% BSA (Solarbio, 9048-46-8)] and were protected from light and incubated for 30 minutes. In the in vivo flow cytometry analysis, the subcutaneous tumor was removed and then mechanically separated with scissors in sterile PBS. The tumor tissues were passed through a 75 μm cell strainer to obtain a single-cell suspension. After resuspending in PBS containing 0.5% BSA, mouse tumor tissue monocyte separation medium kit (Solarbio, P3970) was used to extract monocyte macrophages according to the manufacturer's instructions. Cells were incubated with appropriate antibodies used for cell labeling for 30 minutes. Use the following antibodies: PE rat monoclonal CD206 antibody (eBioscience, 12-2061-82, MR6F3, RRID: AB_2637421, 0.125 μg/test), Super Bright 436 rat monoclonal PD-L1 antibody (eBioscience, 62-5982-80, MIH5, RRID: AB_2637417, 0.25 μg/test), PE-Cy7 mouse monoclonal F4/80 antibody (MultiSciences, 70-AM048010-20, Clone:BM8.1, 0.25 μg/test), APC mouse monoclonal CD11c antibody (MultiSciences, 70-AM011C05-20, 0.125 μg/test), FITC anti-human CD14 antibody (BioLegend, 301803, M5E2, RRID: AB_314185, 5 μL per million cells), PerCP/Cyanine5.5 anti-human CD86 antibody (BioLegend, 305419, IT2.2, RRID: AB_1575070, 5 μL per million cells), PE anti-human CD163 antibody (BioLegend, 333605, GHI/61, RRID: AB_1134005, 5 μL per million cells). A FACS Aria II (BD Biosciences) was used to detect fluorescence, and FlowJo X (v10.6.2) software was used to analyze the data.
In vivo mouse studies
All animal experiments were performed in accordance with the protocol approved by the Ethics Committee of Nanfang Hospital of Southern Medical University (Guangzhou, P.R. China). Female BALB/c mice (6–7 weeks of age) and C57BL/6 mice (7–8 weeks of age) were obtained from the Experimental Animal Center, Nanfang Hospital (Guangzhou, P.R. China), Southern Medical University (Guangzhou, P.R. China). The mice were maintained at 22°C–24°C temperature, 60 ± 10% humidity, with the 12-hour light/dark cycle, under pathogen-free conditions. Standard rodent laboratory diet and water ad libitum were provided. Murine CT26 colorectal cancer cell line was obtained from the NCACC (Shanghai, P.R. China), which was reported to be MSS (36). A total of 1 × 106 CT26 cells were injected into the right thigh of the mice (marked as day 0), and the tumor size was measured every other day with calipers. Using the following formula to calculate the tumor volume (mm3): tumor volume = (π)/6 × L × W, where L is the long axis size and W is the vertical size. Seven days after tumor implantation, the mice were randomly divided into experimental groups. The following 2-week treatments were given: Surfen (Selleck, S6951), 1 mg/kg, intraperitoneal injection, days 7–13 for 7 consecutive days; anti–PD-1 (BioXcell, BE0146), 12.5 mg/kg, i.p., twice a week, that is, on days 10, 13, 17, 20; mFOLFOX6, oxaliplatin (Sanofi) 6 mg/kg followed 2 hours later by 5-fluorouracil (Xudong) 5 mg/kg and leucovorin (Yaoyou) 90 mg/kg, all injected intraperitoneally, once a week, that is on day 8 and day 15; Regorafenib (Bayer, BAY73-4506), dissolved in 0.5% methylcellulose (aladin, 9004-65-3), administered orally, daily for 2 weeks. In addition, azoxymethane (Sigma-Aldrich, A5486)/dextran sulfate sodium (MP Biomedicals, 9011-18-1) [azoxymethane/dextran sulfate sodium (AOM/DSS)] induced colorectal cancer mice model were established as reported previously (37). Briefly, C57BL/6 mice were treated with a single intraperitoneal injection of 10 mg/kg AOM, followed by 6 days 2% DSS drinking water and then 16 days normal water for three cycles. After that, mice were randomized into experimental groups and given a 2-week treatment. Surfen or anti–PD-1 was applied when indicated (Surfen: 1 mg/kg, i.p., everyday; anti–PD-1: 12.5 mg/kg, i.p., twice a week). When the treatment completed, mice were then sacrificed for analysis. The investigators were blind to the treatment groups during the experiment and outcome assessment. The mice were monitored daily and euthanized by cervical dislocation when showing any sign of discomfort.
Statistical analysis
The χ2 test was used to test the relationship between clinical information and metabolic subtypes. The Kolmogorov–Smirnov test was used to confirm whether the data follow a normal distribution. Independent-sample t tests, paired-sample t tests, one-way ANOVA, Wilcoxon tests, and Kruskal–Wallis tests were used to compare continuous variables where appropriate. Correlation coefficients were calculated using Pearson and Spearman rank test. The GSVA R package (21) was used to generate ssGSEA scores for indicated gene set in each patient. Wald statistical test and generalized linear model was used for differential gene analysis via the DESeq2 R package (38). Differential ssGSEA scores were analyzed using the Limma package (25). The Kaplan–Meier method was used for survival analysis to generate survival curves, and the log-rank test was applied to determine the statistical significance. A univariate and multivariate Cox proportional hazard regression model was used to calculate the HR and 95% confidence intervals. Multivariate logistic regression was performed with treatment response as a binary outcome. When indicated, the survminer package was used to determine the optimal threshold based on the maximum rank statistic, which was then used to divide patients into high and low expression groups. R packages ggplot2, ComplexHeatmap (23), clusterProfiler (39), and survival were used to analyze data and generate plots. Gene set enrichment analysis (GSEA) java software was used for gene enrichment analysis. The Benjamini-Hochberg test was used to adjust the P value to reduce the false positive rate. All tests are bilateral, and P < 0.05 is considered significant. Statistical tests were performed using R software (version 3.6.2, http://www.R-project.org) or GraphPad Prism 8.0.
Data and code availability
The datasets used in the current study, including TCGA cancer cohorts, GSE39582 (40), GSE17536 (41), GSE33113 (42), GSE81861 (43), and GSE35602 (44), are available in TCGA database (tcga-data.nci.nih.gov/tcga) or the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo). Datasets referring to cohorts receiving ICI treatment were acquired through public database or appropriate request to authors of previous research. The urothelial cancer cohort (17), receiving anti–PD-L1 drugs (atezolizumab), can be downloaded from http://research-pub.gene.com/IMvigor210CoreBiologies. The data and code that support the findings of this study are included in the article and Supplementary Data.
Results
Comprehensive transcriptomic analyses identify an SM subtype with poor prognosis in MSS colorectal cancer
To systematically interpret the interaction of metabolism and the immune TME, we profiled the transcriptome of 14 types of solid tumors in TCGA database. There was a general correlation between metabolic pathways activation and immune infiltration (Supplementary Fig. S1A; Supplementary Tables S1, S2, and S4). Within colorectal cancer, the MSI-L/MSS subgroup had a higher proportion of metabolic-immune correlation (MSI-L/MSS vs. MSI-H: 54% vs. 18.7%; Supplementary Fig. S1A), indicating key roles of metabolic factors in regulating the immune TME.
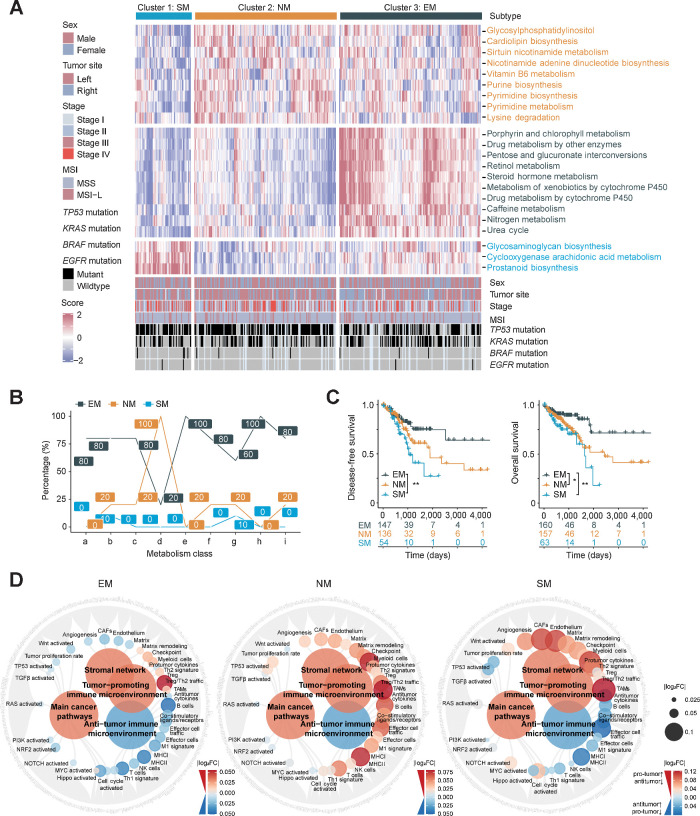
To unravel metabolic heterogeneity, we performed unsupervised clustering of TCGA MSS colorectal cancer cohort and identified three clusters with distinct metabolism characteristics (Fig. 1A; Supplementary Fig. S1B–S1D). On the basis of importance score from the Boruta algorithm (24) and subtype-specific expression, 22 feature pathways were profiled. Cluster 1 displayed high GAGs biosynthesis, cyclooxygenase arachidonic acid metabolism and prostaglandin biosynthesis; Cluster 2 highly expressed pyrimidine synthesis and metabolism; Cluster 3 was characterized by nitrogen metabolism and urea cycle pathways (Fig. 1A; Supplementary Table S5). Metabolic activation ratio analysis revealed a prominent activation (100%) of NM in Cluster 2 and EM and xenobiotic biodegradation in Cluster 3 (Fig. 1B). Thus, we named Cluster 2 and Cluster 3 as NM subtype and EM subtype, respectively. In contrast, Cluster 1 exhibited minimum activation (≤10%) in any of metabolic categories (Fig. 1B). We found Cluster 1 overlapped with the subtypes with abundant stroma infiltration and poor prognosis in published CCMS, CRCA, CCS, Stroma Contribution Subtype, and CMS (45), suggesting that Cluster 1 was associated with stroma remolding (Supplementary Fig. S1E; Supplementary Table S6). Moreover, the feature metabolic pathways of Cluster 1 were more highly enriched in colorectal cancer stroma than epithelial tissue [GSE35602 (44)], based on which we named it SM subtype (Supplementary Fig. S1F). Metabolic subtypes showed significant prognosis difference, among which SM subtype was the worst and EM subtype was better [disease-free survival (DFS): EM vs. SM, P = 0.0056; overall survival (OS): EM vs. NM, P = 0.016; OS: EM vs. SM, P = 0.0022; Fig. 1C]. The reproducibility of our clustering and prognosis value were validated by external cohorts [GSE39582 (40), GSE17536 (41); Supplementary Fig. S1G–S1I].
Figure 1.
Comprehensive transcriptomic metabolism pathway-based clustering in MSS colorectal cancer. A, Metabolic pathway scores and unsupervised k-means clustering were performed in TCGA colorectal cancer cohort (n = 472). Patients with MSS colorectal cancer (n = 383) were filtered and three metabolic subtypes are shown with the specifically upregulated metabolic pathways. B, The proportions (%) of significantly upregulated pathways (log2 FC > 0, FDR < 0.05) in each metabolism categories among metabolic subtypes (a, Carbohydrate metabolism; b, Lipid metabolism; c, Amino acid metabolism; d, NM; e, EM; f, Metabolism of other amino acids; g, Glycan biosynthesis and metabolism; h, Xenobiotics biodegradation and metabolism; i, Metabolism of cofactors and vitamins). C, Kaplan–Meier plot showing DFS (left) and OS (right) among metabolic subtypes. P value was evaluated by log-rank test. D, Molecular function portrait (including oncogenic pathways, pro- and anti-TME factors and stromal network) based on ssGSEA score difference among subtypes (EM vs. Others, NM vs. Others, SM vs. Others). Antitumor effect is marked with blue and protumor factors with red, and the absolute value of log2 FC was shown with color and size changes. **, P < 0.01; *, P < 0.05.
In terms of clinicopathologic characteristics, there were no significant differences in sex, tumor site, stage, or KRAS, BRAF and EGFR mutations among the three subtypes (Supplementary Fig. S2A). Furthermore, we applied molecular function portrait (ref. 19; including oncogenic pathways, protumor and antitumor microenvironmental factors and stromal network) to characterize the three subtypes. Most oncogenic pathways were downregulated in the EM subtype, while cell cycle, TP53, and proliferation scores were upregulated in the NM subtype (Fig. 1D). Multiple oncogenic pathways, including Hippo, NOTCH, and Wnt pathways, were amplified in the SM subtype, indicating a high degree of malignancy (Fig. 1D). The SM subtype also featured simultaneous activation of both protumor and antitumor microenvironment factors, along with stromal network dysregulation (Fig. 1D). GSEA showed that TNFα inflammatory and angiogenesis pathways were enriched in the SM subtype (Supplementary Fig. S2B and S2C). Taken together, we identified distinct metabolism subtypes in MSS colorectal cancer, among which the SM subtype, with distinct TME and stromal remolding, had the worst prognosis and requires further investigation.
CS metabolism and immune microenvironment cross-talk in the MSS colorectal cancer SM subtype
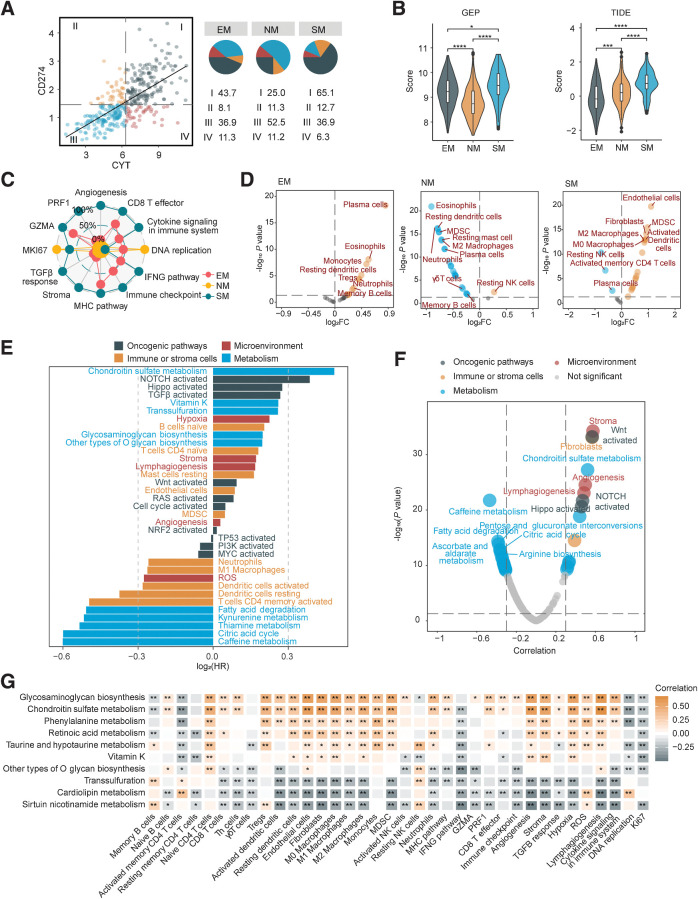
Potential immunotherapy response can be marked by a high cytolytic activity (CYT) score (represented by PRF1 and GZMA) and PD-L1. We observed 65.1% of SM subtype patients highly expressed CYT and PD-L1, implying the SM subtype could potentially benefit from immunotherapy despite the poor prognosis (Fig. 2A). We next applied GEP and TIDE scores (26, 27) and found that the SM subtype exhibited high GEP and TIDE score simultaneously (Fig. 2B). Both antitumor immune response and immune evasion factors may be activated in the SM subtype.
Figure 2.
Chondroitin sulfate metabolism and immune microenvironment crosstalk in MSS colorectal cancer SM subtype. A, Classification of patients with TCGA MSS colorectal cancer based on median CYT score and CD274 (PD-L1) expression (log2 (TPM+1)). The proportions (%) of the four groups in each metabolic subtype were shown on the right. I, CYT-high, CD274-high; II, CYT-low, CD274-high; III, CYT-low, CD274-low; IV, CYT-high, CD274-low. B, Violin plots presenting GEP (left) and TIDE (right) scores among metabolic subtypes. The Wilcoxon test was performed to assess significance. C, Radar chart presenting the ssGSEA score of immune regulatory factors. Scores were rescaled into 0%–100% with simple linear conversion. D, Differential analysis of infiltration scores of immune and TME cells (n = 25) between metabolic subtypes (SM vs. Others, NM vs. Others, EM vs. Others). E, Univariate Cox regression analysis for OS of metabolic pathways, including 25 TME cell types, 10 oncogenic pathways and 5 TME factors. F, Volcano plot, the X axis represents the Pearson correlation coefficient between factors and TIDE score. G, Top 10 metabolic pathways with highest HR in univariate Cox model for OS and their correlation with immune regulatory factors. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, P > 0.05.
In general, successful antitumor immune response requires seven steps in the cancer-immunity cycle (27, 46), in which immune evasion could occur at any step. We assessed seven regulatory factors: (i) antigen presentation molecules and costimulators; (ii) IFNγ signaling; (iii) cytolytic activity; (iv) immune checkpoint expression; (v) cytokine and chemokines; (vi) tumor proliferation marker Ki-67, cell cycle and DNA replication; (vii) TME TGFβ signaling, stroma remodeling and angiogenesis. Most MHC molecules and costimulatory molecules were upregulated in EM and SM subtypes (Supplementary Fig. S3A and S3B). IFNγ signaling, CYT and most immune checkpoints were elevated in the SM subtype (Supplementary Fig. S3C–S3E). As for cytokine and chemokines, the EM subtype highly expressed CCL28 (Supplementary Fig. S3F), which is associated with transportation of lymphocytes (47). We noticed CSF1/CSF1R axis and CCL2 were enriched in the SM subtype, indicating the relevance to macrophage recruitment and M2 polarization (Supplementary Fig. S3G and S3H; ref. 48). Immune exclusion-related TME factors also enhanced in the SM subtype included TGFβ signaling, stroma remodeling, and angiogenesis score (Fig. 2C; Supplementary Fig. S3I). In addition, we evaluated the variety and abundance of tumor-infiltrated immune cells in the TME. The EM subtype displayed increased infiltration of plasma cells, most immune cell infiltration was decreased in the NM subtype, and the SM subtype displayed observed an increase of stromal cells (CAFs, endothelial cells) and immunosuppressive myeloid cells (M2 macrophages, MDSCs), which is closely related to immune exclusion (Fig. 2D; Supplementary Fig. S3J; Supplementary Table S7). Therefore, we extended its definition to be the SM–immune excluded (SM-IE) subtype.
To determine key factors causing immune exclusion in the SM-IE subtype, we comprehensively evaluated oncogenic pathways, metabolic pathways, and TME cells and factors through univariate Cox model (top five factors with highest or lowest HR value selected; Fig. 2E). Correlation coefficient values with TIDE score were regarded as another parameter (Fig. 2F). We found that none of the TME factors or cells were significant in both tests, while the oncogenic NOTCH pathway was a risk factor for prognosis and positively correlated with TIDE score. In terms of metabolism, only CS metabolism gained significance, with a higher effect size compared with the NOTCH pathway (Fig. 2E and F; Supplementary Table S8). Multivariate Cox regression analysis confirmed that CS metabolism was a risk factor for OS, independent of age, stage, and oncogenic pathways, which was also supported by colorectal cancer external cohorts [GSE33113 (42); Supplementary Table S9].
We then evaluated the correlations between the top 10 metabolic pathways with highest HR and immune TME factors. Results demonstrated a close relationship between CS metabolism and M2 macrophages, CAFs, and immune exclusion-related TME factors (angiogenesis and stroma; Fig. 2G; Supplementary Table S10). Collectively, these results suggested immune exclusion in MSS colorectal cancer SM-IE subtype, and CS metabolism may be intimately intertwined with an immune suppressive TME.
An immune exclusion barrier is defined by C-6-S and M2 macrophages in the MSS colorectal cancer invasive margin
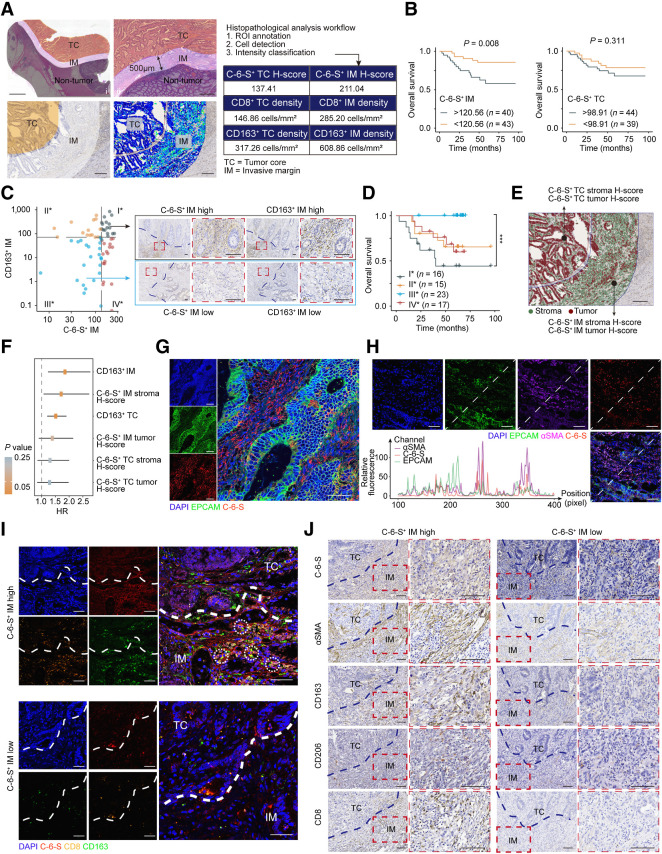
It is reported that CS subtype C-6-S, not chondroitin-4-sulfate (C-4-S), markedly increases in colorectal cancer (49). To determine spatial patterns of C-6-S and immune cells in the TME, we carried out digital pathology analysis on 96 patients with MSS colorectal cancer in a multicenter clinical cohort (see clinicopathologic characteristics in Supplementary Table S11). QuPath (30) was used for cell detection and cell intensity classifications (Supplementary Tables S12 and S13). The distribution of C-6-S, CD8+ T cells (CD8), and M2 macrophages (CD163) were analyzed in TC and IM (Fig. 3A). Interestingly, C-6-S+ IM (P = 0.008), but not C-6-S+ TC (P = 0.311), predicted poor prognosis of MSS colorectal cancer (Fig. 3B). IM is the boundary compartment between tumor and normal tissue, reshaped by chemokines, cytokines, and TME cells like CAFs and macrophages (29). These results suggested the interplay between C-6-S and immune cells located in the IM.
Figure 3.
An “exclusion barrier” was constructed by C-6-S and M2 macrophages in MSS CRC invasive margin. A, Schematic of digital pathology analysis. Left: i, Manual delineation of TC and IM. ii, Enlarged images of TC and IM region. iii-iv, Region delineation (iii) and digital identification of cell structure and stain intensity (iv). Right: Example of digital pathology analysis workflow and output. Scale bar: 2 mm in (i), 500 μm in (ii) and 200 μm for (iii) and (iv). B, Kaplan–Meier plots for the 5-year survival of MSS colorectal cancer in C-6-S+IM or C-6-S+TC low/high subgroups. The survminer R package determined the optimal cutoffs. P value was determined by log-rank test. C and D, Left, patients were grouped by C-6-S+ IM and CD163+ IM optimal cutoffs for OS. Right, representative IHC images of group I* and III* (C). Scale bar: 100μm. Overall survival (D) of four subgroups is shown. P value was determined by Log-rank test. E and F, Recognition of tumor and stroma cells using QuPath software and the H-score of C-6-S+ stroma and C-6-S+ tumor in IM and TC (E). Scale bar: 200 μm. Univariate Cox regression analysis for 5-year OS (F). The HR and 95% confidence intervals are shown. IF detected EPCAM and C-6-S (G) or EPCAM, αSMA and C-6-S (H) in human colorectal cancer specimens. Nuclei are shown in blue (DAPI). EPCAM (tumor cell marker) is in green, αSMA (CAF marker) is in purple and C-6-S is in red. The fluorescent intensity at each position along the indicated diagonal were quantified (H, bottom left). Scale bar: 50 μm. I, IF assay with C-6-S (red), CD8 (orange), and CD163 (green) in IM and TC regions. White circles highlight the adjacent position of CD8+ T cells (CD8) and M2 macrophages (CD163). Scale bar: 50 μm. J, Representative IHC images displaying the differential expression of C-6-S, αSMA, CD163, CD206 and CD8 in C-6-S+ IM stroma, stratified by H-score high/low groups. Scale bar: 100 μm. ***, P < 0.001.
As for immune cells, CD8 and CD163 presented TC-high/IM-high, TC-low/IM-low and TC-low/IM-high patterns (Supplementary Fig. S4A and S4B). Correlation analysis between C-6-S and immune cells revealed coexpression of CS metabolism and CD163 in MSS colorectal cancer transcriptome (Supplementary Fig. S4C). Moreover, digital pathology analysis of C-6-S+ IM and CD163+ IM stratified prognostic risk, as the C-6-S+ IM high/CD163+ IM high subgroup had the worst prognosis (I* vs. III*: P = 0.0003; Fig. 3C and D). The IM region in MSS colorectal cancer may therefore be collaboratively shaped by metabolites C-6-S and M2 macrophages.
C-6-S can be expressed on tumor cell membranes or secreted by CAFs, participating in signal transduction and extracellular matrix (ECM) construction (49). We used a Random Forest model to recognize tumor cells and stroma cells, calculating a C-6-S+ stroma H-score and C-6-S+ tumor H-score in IM and TC (Fig. 3E). Univariate Cox analysis revealed that C-6-S+ stroma H-score in IM was a risk factor predicting poor prognosis in MSS colorectal cancer (Fig. 3F). Subsequently, we analyzed single-cell RNA sequencing of colorectal cancer [GSE81861 (43)], finding a significant upregulation of CS metabolism pathway in CAFs compared with other cells (Supplementary Fig. S4D). IF showed that C-6-S was mainly located in the tumor stroma, distinct from EPCAM, the colorectal cancer epithelial marker (Fig. 3G). Both human colorectal cancer specimens and mouse subcutaneous tumor exhibited a synchronized trend between αSMA and C-6-S measured by IF intensity (Fig. 3H; Supplementary Fig. S4E). Human and mouse-derived CAFs had higher expression of 6S-related sulfotransferases CHST3 and CHST7 but a reduction or no difference in 4S-related genes CHST11 and CHST13 (Supplementary Fig. S4F). These results suggested that CAFs are an important source of C-6-S in colorectal cancer.
Furthermore, we explored the relationship between C-6-S and immune cells. IF showed that in C-6-S–high tumor margin, CD163+ M2 macrophages accumulated, while CD8+ T cells were closely associated with M2 macrophages and surrounded by C-6-S, suggesting that CAF-derived C-6-S was correlated with M2 macrophage and T-cell distribution (Fig. 3I). On the basis of CD8 IHC, tumors can be classified as “cold” (TC-low/IM-low), “hot” (TC-high/IM-high), and “immune excluded” (TC-low/IM-high) subtype (50). A high C-6-S+ stroma H-score in IM, rather than TC, correlated with higher CD8+ IM (P < 0.01), CD163+ IM (P < 0.05), and CD206+ IM (P < 0.05) and a higher proportion of CD8 immune-excluded tumors (TC-low/IM-high, stroma H-score high vs. low: 48.15 vs. 30.67% immune-excluded; Fig. 3J; Supplementary Fig. S4G). However, C-6-S+ IM tumor cells failed to similarly correlate with immune infiltration (Supplementary Fig. S4H). Taken together, C-6-S+ CAFs in IM mediated the CD8+ T cell–excluded phenotype, and the “exclusion barrier” was characterized by C-6-S and M2 macrophages.
To summarize a general process, we established a “metabolism-immune microenvironment” exploration workflow: (i) profiling metabolism-immune infiltration correlations; (ii) transcriptome clustering of metabolic subtypes and differential analysis of the immune TME; (iii) digital pathology analysis of metabolites and immune cell spatial patterns; (iv) identifying interactions between metabolites and immune phenotypes.
CAF-derived C-6-S induces macrophage M2 polarization and causes immune exclusion
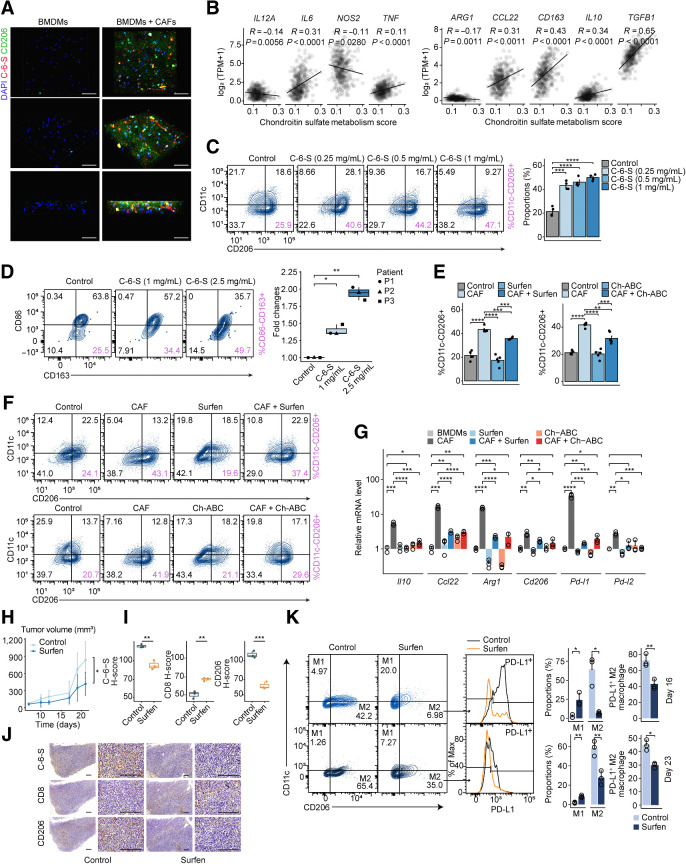
CAFs and TAMs are the main stromal components regulating the immune TME. Evidence indicates their cell–cell communication is critical to tumor progression and drug resistance (51). 3D coculture assays showed that incubation with CAFs could significantly increase C-6-S in the ECM, along with CD206 or CD163 on macrophages, in both mouse bone marrow–derived macrophages (BMDM) and PMA-induced human THP-1 macrophages, indicating that C-6-S may constitute the bridge between CAFs and TAMs (Fig. 4A; Supplementary Fig. S5A). We therefore investigated the underlying mechanism of their interaction. In TCGA MSS colorectal cancer cohort, we observed an inverse correlation between CS metabolism pathway activity and expression of M1 macrophage markers (IL12A, NOS2), but a positive correlation with most of the M2 markers (CCL22, CD163, IL10, and TGFB1;Fig. 4B).
Figure 4.
CAF-derived C-6-S induces M2 macrophage polarization and causes immune exclusion. A, IF staining showing C-6-S in ECM and CD206 in BMDMs colcultured with CAFs in a 3D system. Nuclei are shown in blue (DAPI), CD206 (M2-type marker) is in green, and C-6-S is in red. Scale bar: 50 μm. B, The correlation between CS metabolism ssGSEA scores and M1-type markers (left), M2-type markers (right) in TCGA MSS colorectal cancer cohort. Mouse PMs (C) and human colorectal cancer TAMs (D) were treated with various concentrations of C-6-S. Flow cytometry detected the CD11c−CD206+ or CD86−CD163+ M2 macrophages. Bar graphs or boxplot show the summarized data. Student t test was used to calculate P values. Mouse PMs with or without CAF cocultivation and CS antagonists (Surfen and Ch-ABC) were detected with flow cytometry (E and F) and BMDMs were detected with qPCR (G). H–K, CT26 tumor–bearing mice were randomized to control or Surfen group (n = 5 for each group). Tumor growth curves from day 0–21, and P value by Student t test on day 21 (H). Representative IHC images of C-6-S, CD8, and CD206 in each group on day 16 (J), along with statistical plots (I). Scale bar: 200 μm. On day 16 (top) and day 23 (bottom), macrophages isolated from each group were analyzed for CD11c, CD206, and PD-L1 expression, and the M1/M2 type and PD-L1+ proportions were summarized (K). Statistics were calculated with 3 (D and G), 4 (C), or 5 (E) samples. Student t test was performed in C–E, G, I, and K. Data are presented as mean ± SD. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, P > 0.05.
Flow cytometry analysis revealed that with C-6-S treatment, the proportion of M2 (CD11c−CD206+) macrophages was increased, both in mouse PMs and RAW264.7 cells (Fig. 4C; Supplementary Fig. S5B). Human colorectal cancer–isolated TAMs showed different baseline proportions of M2 (CD86−CD163+; Supplementary Fig. S5C), but a similar trend was observed, indicated by fold changes (Fig. 4D). Also, PD-L1+M2 macrophages (CD11c−CD206+PD-L1+) increased with C-6-S treatment (Supplementary Fig. S5B). Moreover, we found C-6-S supplementation in BMDMs, RAW264.7 and PMA-induced THP-1 macrophages could reduce M1 macrophage factors, such as IL12A and IL6, but upregulated M2 markers IL10, CCL22, ARG1, CD163, and CD206, and immune checkpoints PD-L1 and PD-L2 (Supplementary Fig. S5D–S5F; Supplementary Table S3). Consistent with previous studies, C-4-S supplementation promoted inflammatory macrophages (Supplementary Fig. S5G and S5H; ref. 52).
To determine the M2-induced effect of CS, we conducted a rescue experiment with GAGs antagonists after C-6-S supplementation. Ch-ABC is an enzyme that digests extracellular CS glycans (53). Another GAGs antagonist, bis-2-methyl-4-amino-quinolyl-6-carbamide (Surfen), can bind sulfated GAGs in a charge density-dependent manner (9, 54). Toxicology studies in mice proved it well tolerated in administration (54). We found both Ch-ABC and Surfen could reverse the C-6-S–induced effects on M2 polarization (Supplementary Fig. S6A–S6E), and partially inhibit the CAF-mediated M2 polarization (Fig. 4E–G; Supplementary Fig. S6F–S6I), suggesting CS plays a role in CAF-induced M2 polarization.
By secreting C-6-S into the ECM, CAFs program macrophages toward the M2 type and reshape the IM microenvironment with upregulation of immunosuppressive factors and immune checkpoints, resulting in an immune-excluded phenotype. Next, to determine whether targeting C-6-S could reverse the immune-excluded TME in vivo, we treated MSS colorectal cancer CT26 tumor–bearing mice with Surfen. Seven days after tumor implantation (day 0), low-dose Surfen (1 mg/kg) was applied by intraperitoneal injection for 7 consecutive days (days 7–13). The infiltration of CD8+T and M1/M2-type macrophages were detected on days 16 and 23. Surfen treatment inhibited tumor growth (Fig. 4H). IHC showed an increased CD8+ T-cell infiltration and decreased accumulation of CD206+ M2 macrophages in the Surfen-treated group on day 16 (Fig. 4I and J). We also noted an increase of M1 macrophages (CD11c+CD206−, gated on F4/80+ cells), while M2 macrophages (CD11c−CD206+, gated on F4/80+ cells) and PD-L1+ M2 macrophages (gated on F4/80+CD11c−CD206+ cells) were diminished (Fig. 4K). Notably, Surfen treatment achieved a long-lasting effect, as we could still observe a lower proportion of M2 macrophages and PD-L1+M2 macrophages 10 days after the end of treatment (day 23; Fig. 4K). The long-lasting effect in the TME provides a better “soil” for immune cells to migrate and infiltrate, which may facilitate immunotherapy to take effect. Altogether, CAF-derived C-6-S induced M2 polarization and targeting C-6-S with Surfen could subvert the immune-excluded TME in MSS colorectal cancer.
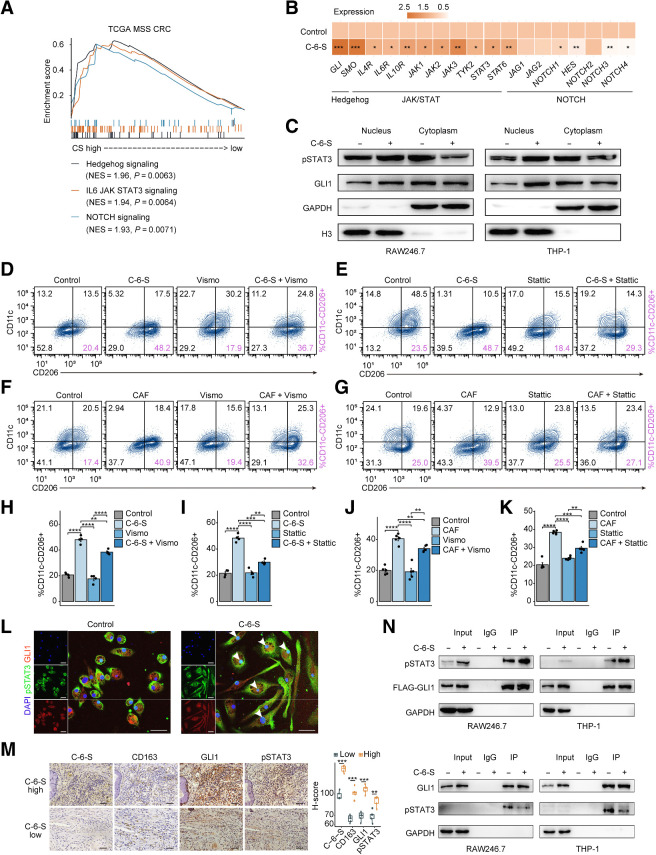
CAF-derived C-6-S educates M2 macrophages via coactivating JAK/STAT3 and Hedgehog pathways
Many oncogenic signaling pathways also participate in immune evasion outside their tumor-promoting function (18). To unveil the potential pathways regulating the C-6-S–mediated immune TME, we analyzed oncogenic and immune evasion gene sets that involve tumor proliferation, stemness, metabolism, and microenvironment. Considering the candidate pathways expressed in the SM subtype and their relevance to CS metabolism and M2 macrophages, we speculated that JAK/STAT3, Hedgehog, and NOTCH pathways were most likely to mediate the TME remodeling (Fig. 5A; Supplementary Fig. S7A and S7B). Subsequently, in vitro C-6-S treatment caused higher upregulation of genes in the JAK/STAT and Hedgehog pathways than those in the NOTCH pathway (Fig. 5B). WB analysis confirmed that C-6-S induced the nuclear translocation of the key transcription factors of the JAK/STAT and Hedgehog pathways, pSTAT3 and GLI1 (Fig. 5C). Using multiple macrophage cell models, we detected that, in PCR and flow cytometry, C-6-S or CAF-induced high M2 markers and PD-L1 expression were partly rescued when treated with STAT3 inhibitor Stattic or Hedgehog pathway inhibitor Vismodegib (Fig. 5D–K; Supplementary Fig. S8A–S8H) and completely inhibited when using both inhibitors (Supplementary Fig. S8E). This suggested that M2 macrophages induced by CAFs and C-6-S relied on the activation of the JAK/STAT3 and Hedgehog pathways. Recently, coexpression of pSTAT3 and GLI1 was found to be associated with poor prognosis in triple-negative and HER2-positive breast cancer (55). However, their collaboration in regulating immune cell functions was largely unknown. Through our IF colocalization assay, we found C-6-S treatment could not only promote the nuclear translocation of pSTAT3 and GLI1 in macrophages but also increase their intranuclear colocalization (Fig. 5L; Supplementary Fig. S8I). The coexpression of GLI1 and pSTAT3, along with CD163, was confirmed in MSS colorectal cancer clinical samples (n = 10) categorized by high/low C-6-S expression (Fig. 5M). A coimmunoprecipitation (Co-IP) assay demonstrated that C-6-S enhanced the protein–protein binding between pSTAT3 and GLI1 (Fig. 5N). In general, we found C-6-S promoted the coactivation of pSTAT3 and GLI1 and activated the JAK/STAT3 and Hedgehog pathways, thereby inducing M2 polarization and PD-L1 expression of macrophages.
Figure 5.
CAF-derived C-6-S educates M2 macrophages via coactivating JAK/STAT3 and Hedgehog pathways. A, Patients with TCGA MSS colorectal cancer were grouped by the median ssGSEA score of CS metabolism pathway. The enrichment of Hedgehog, JAK/STAT3, and NOTCH pathways by GESA are shown. B, The mRNA expression of marker genes in Hedgehog, JAK/STAT3, and NOTCH pathway of PMA-induced THP-1 following 24 hours of C-6-S treatment. C, The expression of transcription factors of the JAK/STAT pathway (pSTAT3) and Hedgehog pathway (GLI1) detected by WB. RAW264.7 and THP-1 cells were cocultured with C-6-S for 24 hours, and the nucleus and cytoplasm were separated. D–K, Flow cytometry detected the expression of CD11c and CD206 in mouse PMs with or without C-6-S, CAFs, Vismodegib, and Stattic. The proportions of M2-type macrophage were calculated (D–G). Bar graphs of the summary data are shown (H–K). L, Expression and intracellular localization of GLI1 and pSTAT3 in mouse BMDMs with or without C-6-S treatment. Nuclei are shown in blue (DAPI), pSTAT3 is in green, and GIL1 is in red. Scale bar: 50 μm. M, IHC images demonstrating staining intensity of CD163, GIL1, and pSTAT3 in C-6-S+ IM high and low patients (n = 10). Statistical data are shown in the right. Scale bar: 50 μm. N, Co-IP assay detecting the protein–protein binding of p-STAT3 and GLI1. Top, FLAG-GLI1 antibody coprecipitating p-STAT3. Bottom, p-STAT3 antibody coprecipitating GLI1. Input, protein expression in cell lysates detected by WB. IgG, negative control. IP, expression of compound coprecipitated by FLAG-GLI1 or p-STAT3 antibody. Statistics were calculated with 4 (H and I) or 5 (J and K) samples. Student t test was performed in B, H–K, and M. Data are presented as mean ± SD. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, P > 0.05.
Blockade of C-6-S enhances the anti–PD-1 response of MSS colorectal cancer in vivo
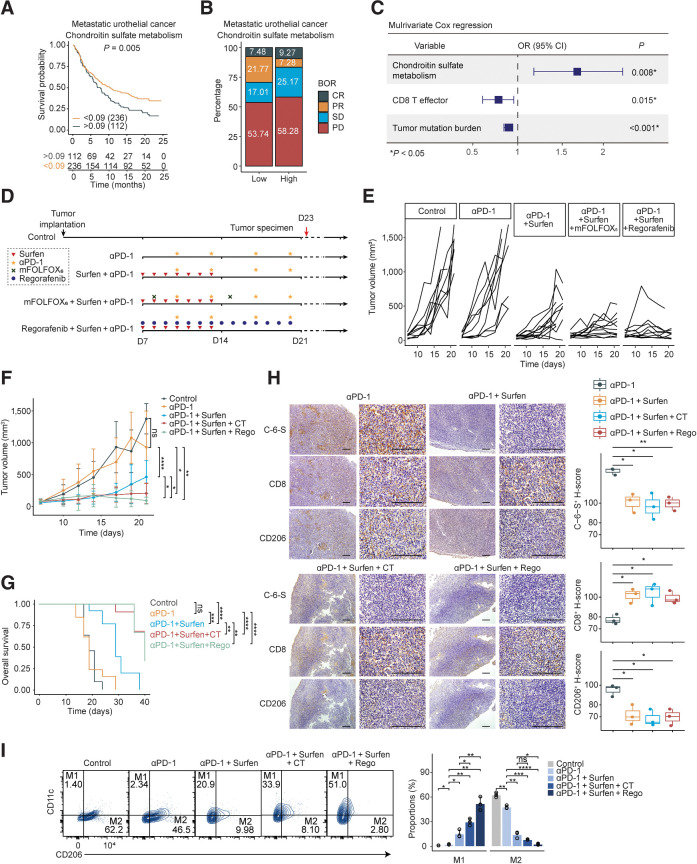
We have found that Surfen could suppress M2 polarization and improve the immune TME. We then explored the role of CS metabolism as a predictive marker. We found CS metabolism correlated with a worse prognosis in urothelial cancer (P = 0.005; Fig. 6A) cohorts treated with ICIs (17). Moreover, a higher stable disease/progressive disease (SD/PD) or PD rate was found in enhanced CS metabolism subgroups (Fig. 6B), and multivariate logistic regression validated that CS metabolism has additional predictive value independent of established biomarkers (Fig. 6C). These data reinforced the hypothesis that targeting CS may reverse immune evasion and sensitize MSS colorectal cancer to immunotherapy. Accordingly, we intended to use CS antagonist Surfen to enhance immunotherapy efficacy.
Figure 6.
Blockade of C-6-S enhances the anti–PD-1 response of MSS colorectal cancer in vivo. A–C, The predictive value of CS metabolism pathway ssGSEA score for immunotherapy efficacy and patients' survival in urothelial cancer cohort from Mariathasan and colleagues (17). A, Kaplan–Meier survival curves for patients with high/low CS metabolism score. The optimal cutoff of continuous variables was defined by survminer R package. B, Bar plots presenting the ICIs response stratified by the median CS metabolism pathway score. C, Multivariate Cox logistic regression analysis with PR/CR and SD/PD as binary outcome. D, The in vivo experiment overview. Mice subcutaneous tumor model was established using CT26 colon cancer cell line. On day 7 after tumor implantation, the mice were randomized into five groups (control, anti–PD-1, Surfen+anti–PD-1, mFOLFOX6+Surfen+anti–PD-1, Regorafenib+Surfen+anti–PD-1, n = 11–13 for each group). E and F, Tumor growth curves on days 0–21 (E) and intergroup difference analysis on day 19 or 21 (F). G, The overall survival of mice in each group up to day 40. The endpoint was established at tumor volume ≥ 1,000 mm3. H, Subcutaneous tumor IHC of C-6-S, CD8, and CD206 in experimental groups on day 23. Statistical analyses are shown on the right. Scale bar: 200 μm. I, On day 23, macrophages isolated from each group were detected for M1 and M2-type markers by flow cytometry. CT, chemotherapy regimen, mFOLFOX6; Rego, Regorafenib. Statistics were calculated with three samples (H and I). Student t test was performed in F, H, and I. Log-rank test was performed in G. Data are presented as mean ± SD. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, P > 0.05.
Combination of immunotherapy with chemotherapy or anti-angiogenesis therapy has achieved preliminary progress (2). We conducted in vivo experiments with Surfen in combination with anti–PD-1, mFOLFOX6+anti–PD-1, or Regorafenib+anti–PD-1 (Fig. 6D). Surfen plus anti–PD-1 administration significantly suppressed tumor growth (day 19, P < 0.0001) and prolonged survival (P < 0.001; Fig. 6E–G), while anti–PD-1 monotherapy failed to improve survival of mice, consistent with previous research (56). Surfen plus anti–PD-1 promoted CD8+ T cells infiltration (Fig. 6H) and decreased M2 macrophages (Fig. 6H and I). The triple combination regimen (mFOLFOX6/Regorafenib+Surfen+anti–PD-1) effectively reversed the increase of M2 macrophages (Fig. 6I) and stimulated CD8+ T cells in the TME (Fig. 6H), further inhibited tumor growth (day 21, P < 0.05 for both regimens) and prolonged the survival (P < 0.0001 for both regimens; Fig. 6E–G).
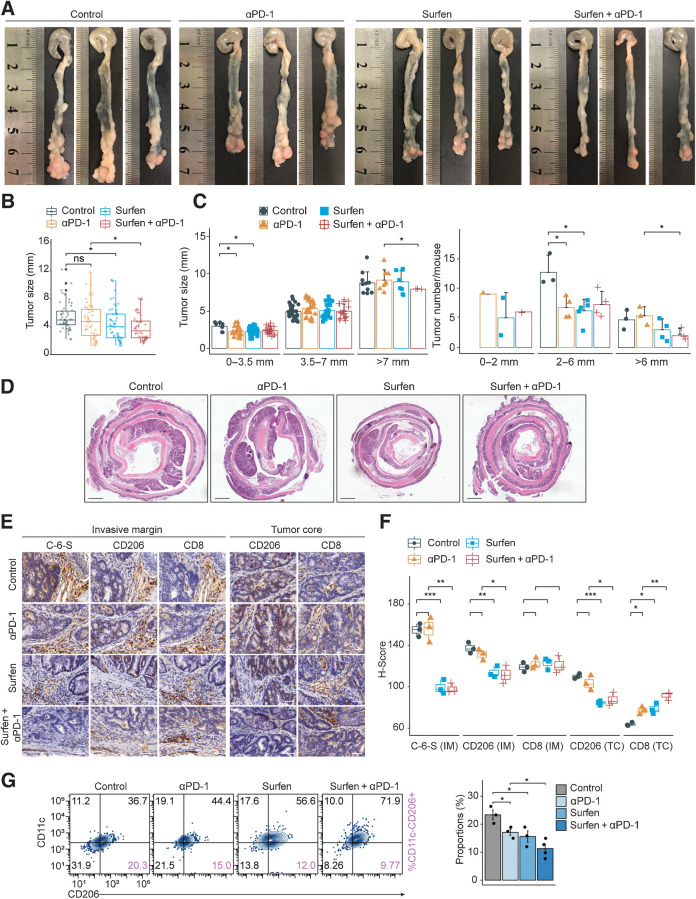
Furthermore, an AOM/DSS-induced colorectal cancer mouse model (37), which could reflect the pathologic process of colorectal cancer, was utilized to validate treatment efficacy. We found that anti–PD-1 could reduce the size and the number of small tumors but failed to inhibit large ones (Fig. 7A–C). Surfen plus anti–PD-1 treatment significantly reduced the size and the number of the large tumors (>6–7 mm), compared with anti–PD-1 (Fig. 7A–C). Hematoxylin and eosin (H&E) staining demonstrated a decrease of adenocarcinoma lesions in the combination group (Fig. 7D). IHC indicated that anti–PD-1 could slightly increase CD8+ T-cell infiltration, and Surfen could significantly lower CD206+ M2 macrophages both in tumor margin and core (Fig. 7E and F). Importantly, Surfen+anti–PD-1 significantly decrease M2 proportions (F4/80+CD11c−CD206+; Fig. 7G) and increase CD8+ T cells in the TC (Fig. 7E and F). To sum up, Surfen markedly enhanced anti–PD-1 response in MSS colorectal cancer. Triple combination regimen with chemotherapy or Regorafenib may achieve better therapeutic outcome, emerging as a promising combinatorial strategy.
Figure 7.
Targeting C-6-S combined with anti–PD-1 strategy in AOM/DSS-induced colorectal cancer model. Establishment of AOM/DSS-induced colorectal cancer model followed by 2-week drug therapeutic regimen (Control, anti–PD-1, Surfen, Surfen+anti–PD-1; n = 3–5 for each group). A, Representative images of colon from each group. B and C, Tumor size and number were compared between groups. D, H&E staining was applied to observe the pathologic state. Scale bar: 1 mm. IHC of C-6-S, CD206, and CD8 was performed (E), and H-score were compared in IM and TC region among groups (F). Scale bar: 100 μm. G, Macrophages isolated from each group were analyzed for CD11c−CD206+ M2 macrophages (n = 3–4 for each group). Statistics were calculated with three samples (F). Student t test was performed in B, C, F, and G. Data are presented as mean ± SD. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, P > 0.05.
At the pan-cancer level, GAGs metabolism also showed a positive correlation with immune evasion (Supplementary Fig. S9A). An SM-like subtype, characterized by GAGs metabolism, was identified in pancreatic adenocarcinoma (PAAD, Cluster A; Supplementary Fig. S9B and S9C) and stomach adenocarcinoma (STAD, Cluster B; Supplementary Fig. S9D and S9E) cohorts, and both displayed a poor prognosis (Supplementary Fig. S9F and S9G). This supports the pan-cancer immunomodulatory effect of GAGs metabolism, especially in stroma-rich tumors like STAD and PAAD. Our immunometabolism strategy in MSS colorectal cancer may serve to enhance immunotherapy strategies in stroma-involved tumors.
Discussion
The emergence of immunotherapy provides new perspectives for the application of metabolic agents. Apart from killing tumor cells, metabolic agents can also modulate immune cell function, and thus reduce the toxicity and boost the efficacy of combination therapy (5). It is worth noting that there is a stronger relationship between metabolism and the immune TME in patients with MSS colorectal cancer, rather than in MSI-H cancers. Here, we found the immunometabolic checkpoint C-6-S shaped an immune evasive TME in MSS colorectal cancer. Targeting C-6-S can reverse M2 polarization of TAMs and reduce PD-L1 expression, consequently remodeling a better immune TME and enhance ICI efficacy. The exploration of immunometabolic checkpoints is a promising direction for MSS colorectal cancer therapeutic strategies.
The discovery of C-6-S as an immunometabolic checkpoint was based on comprehensive transcriptome and digital pathology analysis. The spatial pattern of oxygen, nutrients, and metabolites among TC and IM profoundly influence the fate of tumor cells and immune TME formation (29). Through spatial pattern assessment, we observed C-6-S+ IM can predict poor OS in MSS colorectal cancer. Moreover, patients could be further stratified by C-6-S+ IM and CD163+ IM markers, indicating a potential immunometabolic combination predictor. The microenvironment rich in C-6-S promoted the M2 TAM phenotype and thus induced T-cell exclusion. The evidence highlighted the function of C-6-S in promoting M2 polarization of TAMs and collaboratively formed an “exclusion barrier” in IM.
CAFs help shape an immunosuppressive TME by means of physical barriers and secretory factors like TGFβ (51). Elevated infiltration of CAFs and M2 TAMs were both found in the SM subtype, indicating their joint effect in shaping the immune TME. Previous studies suggested that collaboration between CAFs and TAMs can accelerate tumor progression and drug resistance (51). In this study, we discovered: (i) C-6-S+ CAFs in IM largely accounted for the prognostic role of C-6-S+ IM; (ii) C-6-S+ CAFs in IM were linked to M2 TAM recruitment and CD8+ T-cell exclusion; and (iii) CS antagonists blocked the CAF-induced M2 polarization. These results suggested that C-6-S acts as a messenger between CAFs and TAMs. Targeting C-6-S may break down the communication and improve the immune TME.
It can be inferred from previous studies that C-4-S and C-6-S affected the NFκB pathway in macrophages (52). Mixtures of C-4-S and C-6-S in gradient concentrations influence the expression of polarization markers in BMDMs (52). In general, the effects of CS sulfated metabolites on macrophages are still under debate, and how CS works to remodel the immune TME remains largely unknown. We here uncovered the crucial role of C-6-S in regulating M2 polarization of TAMs, with the JAK/STAT3 and Hedgehog pathways as the key routes. Hedgehog and JAK/STAT3 inhibitors have great potential in treating basal cell carcinoma and triple-negative breast cancer (55). The JAK/STAT and Hedgehog pathways regulate not only tumor cell biological processes, but also immune cells functions, which make them potential targets for immunotherapy (18). However, JAK/STAT3 inhibitor monotherapy failed in clinical trials, possibly attributed to the interplay of the key transcription factors of the two pathways, pSTAT3 and GLI1 (55, 57). PSTAT3 and GLI1 were reported to share the same promoter binding sites or form complex downstream regulatory transcriptional activity (57). We showed that the C-6-S–mediated M2 phenotype greatly depended on the synchronized nuclear translocation and protein–protein binding of pSTAT3 and GLI1. Therefore, the combined intervention in the JAK/STAT3 and Hedgehog pathways may reverse a TME dominated by M2 TAMs and sensitize the TME to ICIs.
Many clinical trials are investigating the combination of anti-metabolism and immunotherapy strategies (5). We identified CS metabolism as a novel predictive indicator for ICI treatment. Moreover, small doses of Surfen therapy were capable of breaking the “malignant homeostasis” and constructing a favorable TME for immunotherapy in MSS colorectal cancer. Anti-angiogenesis brings benefits for patients with multi-line colorectal cancer in clinical application, but represented by REGONIVO regimens, anti-angiogenic drugs combined with ICIs still have a long way to go (2, 58). We noticed the SM subtype was also enriched in angiogenesis pathways. The interweaving of SM and angiogenesis may cause ineffective immunotherapy in MSS colorectal cancer (58). In our study, Surfen combined with Regorafenib and anti–PD-1 can further reduce tumor volume, confirming our hypothesis. FOLFOX is a widely recognized cornerstone of advanced colorectal cancer treatment (59), and as collaborative treatments with ICIs, chemotherapy drugs such as oxaliplatin have been confirmed to induce immunogenic cell death (59). However, the MODUL trial generated disappointing results, probably attributed to the suppressive TME (60). We found that Surfen alongside chemotherapy and ICIs can prolong survival in vivo, implicating the dual effects of increased immunogenicity and decreased immunosuppression. An anti-metabolism strategy targeting C-6-S serves to enhance the immune TME, broadening the path of immunotherapy in problematic MSS colorectal cancer.
As multi-omics technology evolves, immunometabolism has stepped into a new era (61), with two central concepts taking shape to drive future research. First, the integration of multi-omics is getting increasing attention, as transcriptome-based metabolic pathways assessment, which characterizes the global variation of metabolic enzymes, becomes the key process to investigate tumor metabolism heterogeneity. Second, spatial patterns of immunometabolism require special attention. By integrating metabolic pathway activity and spatial patterns data, we proposed and implemented the preliminary “metabolism-immune microenvironment” exploration workflow. Identification of the C-6-S immunometabolic checkpoint in MSS colorectal cancer proved it practical and brings about further perspectives for immunometabolism studies.
Limitations of the study
Although C-6-S was found mainly derived from CAFs and distributed in colorectal cancer stroma, it was also expressed on the tumor cell surface. The function of C-6-S in different forms requires more exploration. As for the validation of the stromal metabolite C-6-S polarizing macrophages toward the M2 phenotype, results in THP-1 cells should be further validated with a primary macrophage model and in vivo experiments, which could better represent the situation in the TME. In addition, it remained to be investigated how C-6-S induced coactivation of pSTAT3 and GLI1. Finally, the therapeutic efficacy of anti-CS metabolism combined with ICIs would need further investigation in preclinical and clinical research.
Authors' Disclosures
No disclosures were reported.
Acknowledgments
This work was supported by National Natural Science Foundation of China (grant number 82073325, to M. Shi; grant number 82103015, to Q. Huang), Ministry of Health of China|Wu Jieping Medical Foundation (grant number 320675020201018), and Chinese Society of Clinical Oncology Foundation (grant number Y-BMS2019-074, to M. Shi). The funders had no role in study design, data collection, analysis or interpretation and the final decision to publish the work. We are very thankful to Yanqing Ding from Department of Pathology, Nanfang Hospital (Guangzhou, P.R. China) for professional help in histopathology analysis. We are also grateful to Leica Biosystems for providing Aperio GT450 in automated, high-capacity digital pathology slide scanning.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Immunology Research Online (http://cancerimmunolres.aacrjournals.org/).
Authors' Contributions
Q. Wu: Conceptualization, data curation, software, formal analysis, validation, investigation, visualization, writing–original draft, project administration, writing–review and editing. Q. Huang: Conceptualization, formal analysis, funding acquisition, validation, investigation, methodology, writing–original draft, project administration, writing–review and editing. Y. Jiang: Formal analysis, investigation, visualization, writing–original draft. F. Sun: Investigation, visualization, methodology, writing–original draft. B. Liang: Data curation, formal analysis, validation, investigation, visualization, methodology. J. Wang: Data curation, formal analysis, validation, investigation, methodology. X. Hu: Validation, investigation. M. Sun: Validation, investigation. Z. Ma: Validation, investigation. Y. Shi: Validation, investigation. Y. Liang: Resources, methodology. Y. Tan: Resources, methodology. D. Zeng: Software, investigation, methodology. F. Yao: Validation, investigation. X. Xu: Validation, investigation. Z. Yao: Validation, investigation. S. Li: Resources, validation. X. Rong: Resources, validation, methodology. N. Huang: Resources, validation, methodology. L. Sun: Resources, validation, methodology. W. Liao: Conceptualization, resources, supervision, methodology. M. Shi: Conceptualization, resources, supervision, funding acquisition, validation, investigation, methodology, writing–review and editing.
References
- 1. Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 2015;372:2509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ghiringhelli F, Fumet JD. Is there a place for immunotherapy for metastatic microsatellite stable colorectal cancer? Front Immunol 2019;10:1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Corcoran RB, Grothey A. Efficacy of immunotherapy in microsatellite-stable or mismatch repair proficient colorectal cancer-fact or fiction? JAMA Oncol 2020;6:823–4. [DOI] [PubMed] [Google Scholar]
- 4. Kim CG, Jang M, Kim Y, Leem G, Kim KH, Lee H, et al. VEGF-A drives TOX-dependent T cell exhaustion in anti-PD-1-resistant microsatellite stable colorectal cancers. Sci Immunol 2019;4:eaay0555. [DOI] [PubMed] [Google Scholar]
- 5. Li X, Wenes M, Romero P, Huang SC, Fendt SM, Ho PC. Navigating metabolic pathways to enhance antitumour immunity and immunotherapy. Nat Rev Clin Oncol 2019;16:425–41. [DOI] [PubMed] [Google Scholar]
- 6. Mazumdar C, Driggers EM, Turka LA. The untapped opportunity and challenge of immunometabolism: a new paradigm for drug discovery. Cell Metab 2020;31:26–34. [DOI] [PubMed] [Google Scholar]
- 7. Leone RD, Zhao L, Englert JM, Sun IM, Oh MH, Sun IH, et al. Glutamine blockade induces divergent metabolic programs to overcome tumor immune evasion. Science 2019;366:1013–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Long GV, Dummer R, Hamid O, Gajewski TF, Caglevic C, Dalle S, et al. Epacadostat plus pembrolizumab versus placebo plus pembrolizumab in patients with unresectable or metastatic melanoma (ECHO-301/KEYNOTE-252): a phase 3, randomised, double-blind study. Lancet Oncol 2019;20:1083–97. [DOI] [PubMed] [Google Scholar]
- 9. Logun MT, Wynens KE, Simchick G, Zhao W, Mao L, Zhao Q, et al. Surfen-mediated blockade of extratumoral chondroitin sulfate glycosaminoglycans inhibits glioblastoma invasion. FASEB J 2019;33:11973–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Henrotin Y, Marty M, Mobasheri A. What is the current status of chondroitin sulfate and glucosamine for the treatment of knee osteoarthritis? Maturitas 2014;78:184–7. [DOI] [PubMed] [Google Scholar]
- 11. Taieb J, Shi Q, Pederson L, Alberts S, Wolmark N, Van Cutsem E, et al. Prognosis of microsatellite instability and/or mismatch repair deficiency stage III colon cancer patients after disease recurrence following adjuvant treatment: results of an ACCENT pooled analysis of seven studies. Ann Oncol 2019;30:1466–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2017;45:D353–D61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rosario SR, Long MD, Affronti HC, Rowsam AM, Eng KH, Smiraglia DJ. Pan-cancer analysis of transcriptional metabolic dysregulation using The Cancer Genome Atlas. Nat Commun 2018;9:5330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xiao Y, Ma D, Zhao S, Suo C, Shi J, Xue MZ, et al. Multi-omics profiling reveals distinct microenvironment characterization and suggests immune escape mechanisms of triple-negative breast cancer. Clin Cancer Res 2019;25:5002–14. [DOI] [PubMed] [Google Scholar]
- 15. Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011;27:1739–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bareche Y, Buisseret L, Gruosso T, Girard E, Venet D, Dupont F, et al. Unraveling triple-negative breast cancer tumor microenvironment heterogeneity: towards an optimized treatment approach. J Natl Cancer Inst 2020;112:708–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y, et al. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018;554:544–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Roelands J, Hendrickx W, Zoppoli G, Mall R, Saad M, Halliwill K, et al. Oncogenic states dictate the prognostic and predictive connotations of intratumoral immune response. J Immunother Cancer 2020;8:e000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bagaev A, Kotlov N, Nomie K, Svekolkin V, Gafurov A, Isaeva O, et al. Conserved pan-cancer microenvironment subtypes predict response to immunotherapy. Cancer Cell 2021;39:845–65. [DOI] [PubMed] [Google Scholar]
- 20. Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, et al. Oncogenic signaling pathways in The Cancer Genome Atlas. Cell 2018;173:321–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 2010;26:1572–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016;32:2847–9. [DOI] [PubMed] [Google Scholar]
- 24. Degenhardt F, Seifert S, Szymczak S. Evaluation of variable selection methods for random forests and omics data sets. Brief Bioinform 2019;20:492–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017;127:2930–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med 2018;24:1550–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Narayanan S, Kawaguchi T, Yan L, Peng X, Qi Q, Takabe K. Cytolytic activity score to assess anticancer immunity in colorectal cancer. Ann Surg Oncol 2018;25:2323–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kather JN, Suarez-Carmona M, Charoentong P, Weis CA, Hirsch D, Bankhead P. Topography of cancer-associated immune cells in human solid tumors. Elife 2018;7:e36967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bankhead P, Loughrey MB, Fernandez JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: open source software for digital pathology image analysis. Sci Rep 2017;7:16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang Q, Li S, Hu X, Sun M, Wu Q, Dai H, et al. Shear stress activates ATOH8 via autocrine VEGF promoting glycolysis dependent-survival of colorectal cancer cells in the circulation. J Exp Clin Cancer Res 2020;39:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jun JI, Lau LF. CCN1 is an opsonin for bacterial clearance and a direct activator of Toll-like receptor signaling. Nat Commun 2020;11:1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Su S, Liu Q, Chen J, Chen J, Chen F, He C, et al. A positive feedback loop between mesenchymal-like cancer cells and macrophages is essential to breast cancer metastasis. Cancer Cell 2014;25:605–20. [DOI] [PubMed] [Google Scholar]
- 34. Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, et al. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 2017;18:529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. He W, Liang B, Wang C, Li S, Zhao Y, Huang Q, et al. MSC-regulated lncRNA MACC1-AS1 promotes stemness and chemoresistance through fatty acid oxidation in gastric cancer. Oncogene 2019;38:4637–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Castle JC, Loewer M, Boegel S, de Graaf J, Bender C, Tadmor AD, et al. Immunomic, genomic and transcriptomic characterization of CT26 colorectal carcinoma. BMC Genomics 2014;15:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Parang B, Barrett CW, Williams CS. AOM/DSS model of colitis-associated cancer. Methods Mol Biol 2016;1422:297–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marisa L, de Reyniès A, Duval A, Selves J, Gaub MP, Vescovo L, et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med 2013;10:e1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen MS, Lo YH, Chen X, Williams CS, Donnelly JM, Criss ZK 2nd, et al. Growth factor-independent 1 is a tumor suppressor gene in colorectal cancer. Mol Cancer Res 2019;17:697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. de Sousa EMF, Colak S, Buikhuisen J, Koster J, Cameron K, de Jong JH, et al. Methylation of cancer-stem-cell-associated Wnt target genes predicts poor prognosis in colorectal cancer patients. Cell Stem Cell 2011;9:476–85. [DOI] [PubMed] [Google Scholar]
- 43. Li H, Courtois ET, Sengupta D, Tan Y, Chen KH, Goh JJL, et al. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat Genet 2017;49:708–18. [DOI] [PubMed] [Google Scholar]
- 44. Nishida N, Nagahara M, Sato T, Mimori K, Sudo T, Tanaka F, et al. Microarray analysis of colorectal cancer stromal tissue reveals upregulation of two oncogenic miRNA clusters. Clin Cancer Res 2012;18:3054–70. [DOI] [PubMed] [Google Scholar]
- 45. Isella C, Terrasi A, Bellomo SE, Petti C, Galatola G, Muratore A, et al. Stromal contribution to the colorectal cancer transcriptome. Nat Genet 2015;47:312–9. [DOI] [PubMed] [Google Scholar]
- 46. Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity 2013;39:1–10. [DOI] [PubMed] [Google Scholar]
- 47. Shang L, Fukata M, Thirunarayanan N, Martin AP, Arnaboldi P, Maussang D, et al. Toll-like receptor signaling in small intestinal epithelium promotes B-cell recruitment and IgA production in lamina propria. Gastroenterology 2008;135:529–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Szekely B, Bossuyt V, Li X, Wali VB, Patwardhan GA, Frederick C, et al. Immunological differences between primary and metastatic breast cancer. Ann Oncol 2018;29:2232–9. [DOI] [PubMed] [Google Scholar]
- 49. Pudelko A, Wisowski G, Olczyk K, Kozma EM. The dual role of the glycosaminoglycan chondroitin-6-sulfate in the development, progression and metastasis of cancer. FEBS J 2019;286:1815–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Galon J, Bruni D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat Rev Drug Discov 2019;18:197–218. [DOI] [PubMed] [Google Scholar]
- 51. Komohara Y, Takeya M. CAFs and TAMs: maestros of the tumour microenvironment. J Pathol 2017;241:313–5. [DOI] [PubMed] [Google Scholar]
- 52. Hatano S, Watanabe H. Regulation of macrophage and dendritic cell function by chondroitin sulfate in innate to antigen-specific adaptive immunity. Front Immunol 2020;11:232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cooney CA, Jousheghany F, Yao-Borengasser A, Phanavanh B, Gomes T, Kieber-Emmons AM, et al. Chondroitin sulfates play a major role in breast cancer metastasis: a role for CSPG4 and CHST11 gene expression in forming surface P-selectin ligands in aggressive breast cancer cells. Breast Cancer Res 2011;13:R58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schuksz M, Fuster MM, Brown JR, Crawford BE, Ditto DP, Lawrence R, et al. Surfen, a small molecule antagonist of heparan sulfate. Proc Natl Acad Sci U S A 2008;105:13075–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Doheny D, Sirkisoon S, Carpenter RL, Aguayo NR, Regua AT, Anguelov M, et al. Combined inhibition of JAK2-STAT3 and SMO-GLI1/tGLI1 pathways suppresses breast cancer stem cells, tumor growth, and metastasis. Oncogene 2020;39:6589–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Liu L, Mayes PA, Eastman S, Shi H, Yadavilli S, Zhang T, et al. The BRAF and MEK inhibitors dabrafenib and trametinib: effects on immune function and in combination with immunomodulatory antibodies targeting PD-1, PD-L1, and CTLA-4. Clin Cancer Res 2015;21:1639–51. [DOI] [PubMed] [Google Scholar]
- 57. Sirkisoon SR, Carpenter RL, Rimkus T, Anderson A, Harrison A, Lange AM, et al. Interaction between STAT3 and GLI1/tGLI1 oncogenic transcription factors promotes the aggressiveness of triple-negative breast cancers and HER2-enriched breast cancer. Oncogene 2018;37:2502–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Arai H, Battaglin F, Wang J, Lo JH, Soni S, Zhang W, et al. Molecular insight of regorafenib treatment for colorectal cancer. Cancer Treat Rev 2019;81:101912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bains SJ, Abrahamsson H, Flatmark K, Dueland S, Hole KH, Seierstad T, et al. Immunogenic cell death by neoadjuvant oxaliplatin and radiation protects against metastatic failure in high-risk rectal cancer. Cancer Immunol Immunother 2020;69:355–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schmoll HJ, Arnold D, de Gramont A, Ducreux M, Grothey A, O'Dwyer PJ, et al. MODUL-a multicenter randomized clinical trial of biomarker-driven maintenance therapy following first-line standard induction treatment of metastatic colorectal cancer: an adaptable signal-seeking approach. J Cancer Res Clin Oncol 2018;144:1197–204. [DOI] [PubMed] [Google Scholar]
- 61. Artyomov MN, Van den Bossche J. Immunometabolism in the single-cell era. Cell Metab 2020;32:710–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used in the current study, including TCGA cancer cohorts, GSE39582 (40), GSE17536 (41), GSE33113 (42), GSE81861 (43), and GSE35602 (44), are available in TCGA database (tcga-data.nci.nih.gov/tcga) or the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo). Datasets referring to cohorts receiving ICI treatment were acquired through public database or appropriate request to authors of previous research. The urothelial cancer cohort (17), receiving anti–PD-L1 drugs (atezolizumab), can be downloaded from http://research-pub.gene.com/IMvigor210CoreBiologies. The data and code that support the findings of this study are included in the article and Supplementary Data.