Abstract
The HER3/ERBB3 receptor is an oncogenic receptor tyrosine kinase that forms heterodimers with EGFR family members and is overexpressed in numerous cancers. HER3 overexpression associates with reduced survival and acquired resistance to targeted therapies, making it a potential therapeutic target in multiple cancer types. Here, we report on immunogenic, promiscuous MHC class II–binding HER3 peptides, which can generate HER3-specific CD4+ Th1 antitumor immune responses. Using an overlapping peptide screening methodology, we identified nine MHC class II–binding HER3 epitopes that elicited specific Th1 immune response in both healthy donors and breast cancer patients. Most of these peptides were not identified by current binding algorithms. Homology assessment of amino acid sequence BLAST showed >90% sequence similarity between human and murine HER3/ERBB3 peptide sequences. HER3 peptide–pulsed dendritic cell vaccination resulted in anti-HER3 CD4+ Th1 responses that prevented tumor development, significantly delayed tumor growth in prevention models, and caused regression in multiple therapeutic models of HER3-expressing murine tumors, including mammary carcinoma and melanoma. Tumors were robustly infiltrated with CD4+ T cells, suggesting their key role in tumor rejection. Our data demonstrate that class II HER3 promiscuous peptides are effective at inducing HER3-specific CD4+ Th1 responses and suggest their applicability in immunotherapies for human HER3-overexpressing tumors.
Introduction
Oncodrivers are proteins overexpressed in tumor cells that promote proliferation and growth, but counteract cellular senescence, contributing to tumor malignancy. Such oncodriver addiction in tumor cells makes these proteins a promising target for developing new immunotherapies relevant to a large number of patients. Human ERBB3 receptor tyrosine kinase 3 (HER3/ERBB3) is a member of the ERBB family of growth receptors that has several ligands, including heregulin and neuregulin. Although lacking intrinsic kinase activity, HER3 is a critical heterodimerization partner for other members of the family (EGFR and HER2) contributing to the growth, proliferation, and survival of tumor cells (1). HER3 is an established oncodriver in multiple cancers because HER3 overexpression has been detected in breast, melanoma, colorectal, prostate, lung, and ovarian cancers (2). The HER2–HER3 heterodimer is responsible for the most potent ligand-induced tyrosine phosphorylation and downstream signaling via the PI3K–AKT pathway, utilizing the six p85-binding motifs present in HER3 intracellular domain (3, 4). In HER2-overexpressing (HER2pos) breast cancer, HER3 is identified as one of the most prominent inducers of therapy escape, leading to trastuzumab resistance and hyperactivation of PI3K–AKT-mediated signaling (3). Transcriptional/translational upregulation of HER3 is linked to resistance to MEK/RAF inhibitors in melanoma (5), castration-resistant prostate cancer (6), platinum-resistant ovarian cancer (7), and EGFR TKI-resistant non–small cell lung cancer (NSCLC; refs. 8, 9). In triple-negative breast cancer (TNBC), HER3 overexpression has been independently identified as a prognostic marker of poor survival (10, 11), and combined antagonism of EGFR and HER3 enhances responses to PI3K–AKT inhibitors in TNBC preclinical and clinical samples (12). Multiple monoclonal antibodies targeting HER3 are currently being tested at various stages of preclinical and clinical studies; however, therapeutic potential of a HER3-specific immunotherapy has not been comprehensively tested. Developing HER3-specific cellular immunotherapy can be a novel and efficient treatment strategy for multiple cancer types overexpressing HER3 in order to improve patient prognosis and survival.
Although current immunotherapies focus on cytotoxic CD8+ T-cell activity, “helper signals” from CD4+ T cells have been deemed essential for the proliferation, recruitment, and effector function of CD8+ T cells in tumors (13, 14). Help from CD4+ T cells is necessary for memory T-cell survival during recall expansion (15), and IL-2 secreted from CD4+ T cells in the tumor microenvironment has been shown to increase CD8+ T-cell proliferation and granzyme B production (16). Absence of CD4+ T-cell help affects survival and clonal expansion of CD8+ T cells, and defective recall responses by CD8+ T cells have been reported in CD4−/− mice (17). In a head and neck cancer model, HER3-specific helper T-cell responses induce cytolytic CD4+ T-cell activity against tumor cells in an HLA-DR–restricted manner (18). These studies highlight the potential for developing CD4+ T cell–based immunotherapy for cancer, and as previously observed by our lab and others, oncodrivers represent an excellent choice for developing such therapeutic strategies (19).
A previous study from our lab demonstrates gradual loss of HER3-specific CD4+ Th1 immune responses in the peripheral circulation of TNBC patients compared with healthy donors and other breast cancer subtypes, and this loss of immune response negatively associates with patient outcome (20). In the current study, we investigated whether promiscuous MHC class II HER3 peptides could be identified to develop an immunotherapy strategy for HER3-overexpressing cancers. Herein, we present a peptide screening methodology for identifying promiscuous class II epitopes that are capable of inducing tumor-specific CD4+ T-cell responses. We identified nine immunogenic class II epitopes and investigated the therapeutic efficacy of HER3 peptide–pulsed type I DC (HER3-DC1) treatment in multiple preclinical models of breast cancer and melanoma and in both preventive and therapeutic settings.
Materials and Methods
HER3 expression in cancer and correlation with survival
The Cancer Genome Atlas (TCGA) RNA sequencing (RNA-seq) expression data (FPKM) for HER3 (ERBB3) in 12 types of cancers were downloaded from the Genomic Data Commons Data Portal (GDC; https://portal.gdc.cancer.gov, RRID:SCR_014514). Project IDs included in analysis were as follows: TCGA-BLCA, bladder urothelial carcinoma (n = 408); TCGA-BRCA, breast invasive carcinoma (n = 1092); TCGA-COAD, colon adenocarcinoma (n = 456); TCGA-HNSC, head and neck squamous cell carcinoma (n = 501); TCGA-LUAD, lung adenocarcinoma (n = 515); TCGA-LUSC, lung squamous cell carcinoma (n = 501); TCGA-OV, ovarian serous cystadenocarcinoma (n = 376); TCGA-PAAD, pancreatic adenocarcinoma (n = 177); TCGA-PRAD, prostate adenocarcinoma (n = 496); TCGA-SKCM, skin cutaneous melanoma (n = 468); and TCGA-STAD, stomach adenocarcinoma (n = 380). Only samples with available sequence reads were chosen for further analysis. HER3 mRNA expression in primary and normal tissues was assessed and graphed as a boxplot using a customized R script (Supplementary File). For tumor samples, only data from primary tissues were used. An unpaired t test was used for differential analysis of the data set.
Survival plots were generated by the cBioPortal (https://www.cbioportal.org, RRID:SCR_014555) survival tool. Samples were sorted in the descending order, according to the median value of the HER3 gene expression, and the list was divided into two groups in the middle of the list: High expression was defined as the top 50% of the list and low expression as the bottom 50% of the list.
Peptide library generation
The amino acid sequence for native human HER3 protein (1,342 amino acids, Uniprot Accession number P21860) was divided into two peptide libraries: the extracellular domain (ECD, aa 20–643) and intracellular domain (ICD, aa 665–1,342). Libraries consisted of peptide fragments 15 amino acids in length, with a 10-amino acid overlap between adjacent sequences, generating 123 ECD and 134 ICD peptides (GenScript). BRAF-negative control peptide was purchased from GenScript. Peptides were reconstituted according to solubility instructions (GenScript) to 1 mg/mL concentrations. ICD peptides p37, p54, and p55 were unable to be synthesized. Identified immunogenic human HER3 peptides were used for testing in preclinical mouse models as well, due to significant sequence homology.
Human samples
Autologous monocyte fractions were obtained from leukapheresis products from the peripheral blood of normal donors and breast cancer patients. Normal donor samples (n = 11) were purchased through the Cell Therapies Core at H. Lee Moffitt Cancer Center and Research Institute, which performed the elutriation to generate monocyte- and lymphocyte-rich fractions. Breast cancer patient elutriation fractions were obtained under the Moffitt Cancer Center (MCC) 19000 protocol and collected during 2017–2021. Inclusion criteria included samples from newly diagnosed, operable breast cancer patients with: (i) incident invasive primary ductal breast cancer prior to surgery with Her2/neu overexpression via IHC; (ii) age over 18; and (iii) consent to participate. Patients with bilateral breast cancer were not excluded. Exclusion criteria included: (i) patients with no plan for definitive surgery; (ii) no detectable residual breast disease after diagnostic biopsy; and (iii) patients unable to receive neoadjuvant herceptin-based chemotherapy. The control group was composed of unaffected, seemingly healthy individuals with inclusion criteria: (i) no prior history of systemic cancer treatment (i.e., chemotherapy or hormone therapy); (ii) age over 18; (iii) consent to participate; and (iv) BIRADS 1–3 breast imaging within the past 12 months. Exclusion criteria included: (i) active diagnosis of autoimmune disease (i.e., rheumatoid arthritis, lupus, Sjogren syndrome) and (ii) systemic steroid use within the past 12 months.
Elutriation fractions were washed in PBS (cat. #SH30028LS, Fisher Scientific) and lysed with ACK RBC lysis buffer containing NH4Cl (0.15M, cat. #A649-500; Fisher Scientific), KHCO3 (10 mmol/L, cat. #P184-500, Fisher Scientific) and Na2EDTA (0.5 mmol/L; cat. #E7889–100 mL, Sigma-Aldrich), with pH 7.2, when necessary to remove remaining RBCs. Processed fractions were stored in liquid nitrogen until further use.
Breast cancer patient (n = 10) and healthy control (n = 6) samples were used for in vitro validation of identified HER3 class II peptides. All clinical samples were obtained by informed written consent from the subjects, following an Institutional Review Board–approved protocol. All research involving human subjects was performed in accordance with the Declaration of Helsinki and was approved by the IRB at Advarra.
Human DC generation
Monocytes were differentiated into dendritic cells (DC) through the addition of recombinant human (rh)GM-CSF (50 ng/mL; cat. #215-GM-050, R&D Systems) and rhIL4 (10 ng/mL; cat. #204-IL-050, R&D Systems) and cultured in macrophage serum-free media (cat. #12065074; Thermo Fisher Scientific) for 24 hours at 37°C and 5% CO2. DCs were then pulsed with HER3 ECD or ICD peptides (10 μg/mL), a negative control (BRAF class II p8), or left unpulsed. After 24 hours, DCs were matured and polarized to a type I phenotype (DC1) with the addition of rhIFN-γ (1,000 IU/mL; cat. #285IF-100, R&D Systems), followed by LPS (0.2 μg/mL; cat. #L4391-1MG, Sigma-Aldrich) 6 hours prior to harvest. Mature DC1 cells were harvested after 72 hours of culture. Immature DCs (iDC) remained in rhGM-CSF and rhIL4 for 48 hours prior to harvesting. DCs were preserved in 10% DMSO (cat. #D5879-500 mL, VWR) and 90% human AB serum (cat. #S11550, Atlanta Biologicals) and stored in liquid nitrogen until use.
Culture medium
Complete media consisted of RPMI-1640 growth media (cat. #MT-10-040-CM, Corning) supplemented with 10% heat-inactivated fetal bovine serum (FBS; cat. #S11550 Atlanta Biologicals), 0.1 mmol/L nonessential amino acids (cat. #25-025-CI, Corning), 1 mmol/L sodium pyruvate (cat. #MT-25-000-CI, Corning), 100 mg/mL streptomycin, 100 U/mL penicillin (cat. #MT-30-002-CI, Fisher Scientific), 50 mg/mL gentamycin (cat. #15750-060, Gibco), 0.5 mg/mL amphotericin B (cat. #400-104, GeminiBio), and 0.05 mmol/L 2-mercaptoethanol (cat. #21985-023, Invitrogen).
Peptide screening
Mature HER3-DC1 were cocultured in complete media with autologous lymphocytes at a 1:10 ratio in a 12-well tissue culture plate and incubated at 37°C and 5% CO2. The lymphocyte-rich apheresis fractions were processed as described above. After 24 hours, rhIL2 (5 IU/mL) was added to induce the proliferation of CD4+ T cells. Following 8 to 10 days of coculture, T cells were harvested and restimulated in a 96-well plate with iDCs (105 T cells with 104 iDCs for 10:1 ratio) pulsed with the corresponding HER3 class II peptide, a negative class II control (BRAF class II p8, 10 μg/mL), or unpulsed iDCs.
Culture supernatants were collected after 24 hours, and IFN-γ production was measured using a Human Quantikine IFN-γ ELISA kit (cat. #SIF50, R&D Systems) after 1:20 or 1:30 sample dilution. The screening process proceeded using a 10-peptide pool, 5-peptide pool, and individual peptide (peptide concentration 10 μg/mL throughout the screening sequence) sequential scheme (Supplementary Fig. S1A) for both ECD and ICD libraries, separately. Libraries were first screened in pools of 10 peptides and run on three independent donor samples. The 10-peptide pools that showed ≥1.5-fold increase in IFN-γ production in two of the three donors were further pursued in 5-peptide pools. Common positive 5-peptide pools were then tested as individual peptides and screened in additional donor samples to identify peptides that could reproducibly induce an increased CD4+ Th1 immune response. Due to the variability in donor sample size and the progressive nature of the peptide screening methodology, the sample number tested for each identified peptide was inconsistent; however, individual peptide candidates were screened across a minimum of five donor samples, with a threshold response rate of ≥1.5-fold to be considered immunogenic.
Whole HER3 ECD/ICD protein restimulation
Immunogenicity of the identified HER3 class II epitopes was confirmed through sensitization of CD4+ T cells with HER3-DC1 as described above, followed by restimulation with iDCs pulsed with the corresponding HER3 class II peptide, a negative peptide control (BRAF class II p8), native HER3 ECD (cat. #NBP2-52128-0.05 mg, Novus Biologicals) or ICD (cat. #10201-H20B1, SinoBiological) whole protein sequence, or a negative whole protein control (Hemocyanin-Keyhole Limpet Native protein; cat. #SRP6195, Sigma). The same number of T cells, HER3-DC1, and iDCs were used for this experiment and 10 μg/mL of whole protein and/or peptide was used throughout this experiment. Culture supernatants were collected after 24 hours, and IFN-γ production was measured using the Human Quantikine IFN-γ ELISA kit after 1:20 or 1:30 sample dilution.
ELISPOT assays
To evaluate anti-HER3 CD4+ Th1 immune responses in breast cancer patients, IFN-γ production was compared between healthy controls and patient samples using the human IFN-γ ELISPOT kit (cat. #HIFNgp-1M/10, Cellular Technologies Limited). ELISPOT plates precoated with human IFN-γ capture antibody were incubated with nine HER3 class II peptides (4 μg/well), media only (untreated/negative control), or anti-human CD3 (Orthoclone OKT3, cat. #73337989, Johnson and Johnson, treated/positive control, 15 ng/mL). Cryopreserved PBMCs were plated (2 × 105 cells/well) in CTL-Test medium supplemented with 1% L-glutamine (provided with the kit) and incubated at 37°C, 5% CO2 for 48 hours. Per manufacturer's protocol, plates were washed, and detection antibody (anti-human IFN-γ Biotin; 100 mg/mL) was added to each well. After incubation at room temperature for 2 hours, 1:1,000-diluted streptavidin-AP was added and incubated for 30 minutes. Plates were washed again before substrate solution was added, and plates were incubated for 15 minutes to allow for color development. Plates were washed with tap water, dried overnight at room temperature, and spot-forming cells (SFC) were counted using an automated reader (Immunospot Cellular Technology Limited). Because inter-replicate variability in ELISPOT was low, an empiric method of determining antigen-specific responses was used as previously shown (21). In brief, positive response to an individual class II HER3 peptide was defined as (i) threshold minimum of 20 SFC/2 × 105 cells in experimental wells after subtracting unstimulated background; and (ii) approximately 2-fold increase of antigen-specific SFCs over background. Three metrics of CD4+ Th1 response were defined for each group: (i) overall anti-HER3 responsivity (proportion of patients responding to ≥1 peptide), (ii) response repertoire (mean number of reactive peptides), and (iii) cumulative response across nine identified class II HER3 peptides (total SFC/1 × 106 cells).
HLA typing of donor samples
Lymphocyte fractions of healthy donors (n = 11) used in peptide screening HER3 ECD and ICD libraries were sent for HLA-DRB1, HLA-DQB1, and HLA-DPB1 allele typing (American Red Cross). Alleles expressed by each donor were cross-referenced with the CD4+ Th1 response induced by each identified HER3 class II peptide. Each HER3 peptide tested against a donor expressing a particular HLA allele was indicated with a (+) if ≥1 instance of this allele demonstrated a peptide-specific Th1 immune response (IFN-γ production increased ≥1.5-fold compared to negative control) to the corresponding peptide. Because HER3 ECD and ICD libraries were screened using different donor samples, this resulted in two groupings.
NetMHCIIpan 4.0 prediction algorithm
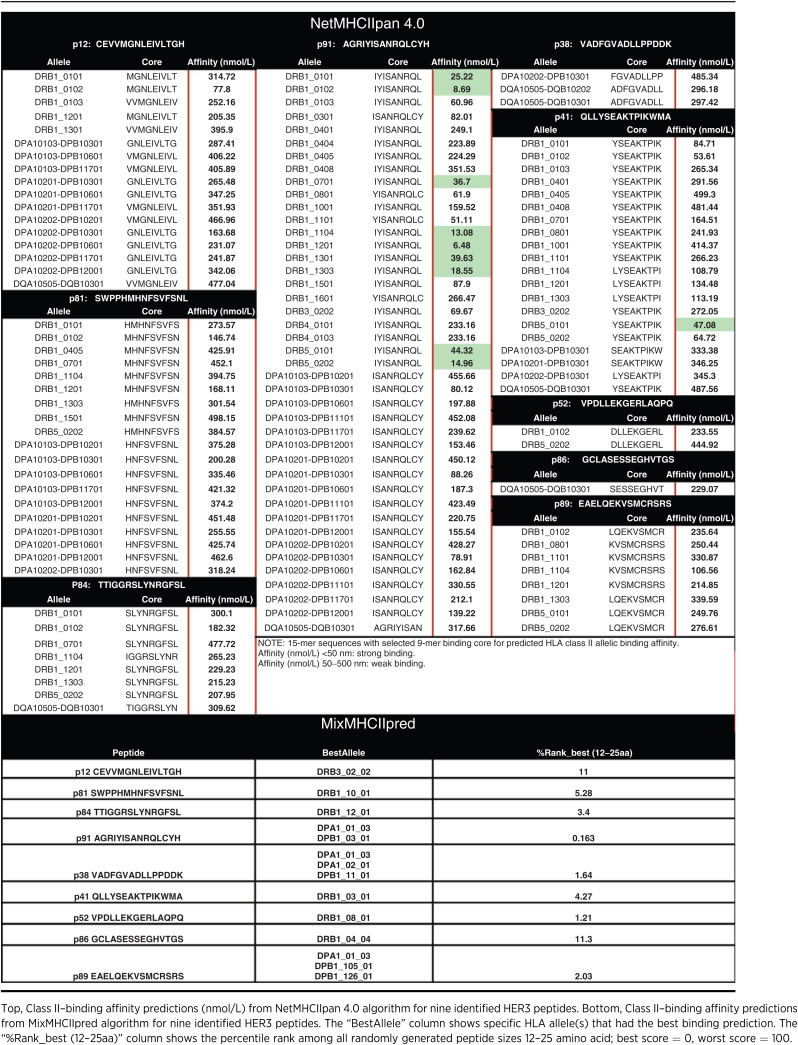
Epitope prediction was done using NetMHCIIpan 4.0 (22) as implemented by www.iedb.org (23). A series of 15-mer peptides were serially generated (one amino acid difference) from the HER3 protein. Each peptide was then predicted for its binding affinity (nmol/L) with each HLA allele type by identifying a 9-mer binding core. Peptide candidates with affinity <500 nmol/L were labeled WB (weak binding) and <50 nmol/L as SB (strong binding).
MixMHCIIpred prediction algorithm
Epitope prediction was done using the MixMHCIIpred algorithm (24, 25). A series of random 12–25 amino acid (aa) peptides were generated from the HER3 protein sequence by the algorithm. A percentile rank was provided for each of the nine HER3 peptides out of all 12–25aa random peptides, for each of the 38 HLA allele types. Best score for percentile rank was 0 and the worst score was 100. The best alleles with the highest binding affinity for each peptide were also identified.
Cell lines and reagents
The human TNBC cell lines MDA-MB-231 (ATCC HTB-26, RRID:CVCL_0062), MDA-MB-468 (ATCC HTB-132, RRID:CVCL_0419), HCC1143 (ATCC CRL-2321, RRID:CVCL_1245), Hs578T (ATCC HTB-126, RRID:CVCL_0332), BT-549 (ATCC HTB-122, RRID:CVCL_1092), SK-BR-3 (ATCC HTB-30, RRID: CVCL_0033) and mouse breast cancer cell lines 4T1 (ATCC CRL-2539, RRID:CVCL_0125) and EO771 (ATCC CRL-3461, RRID:CVCL_GR23) were obtained from the American Type Culture Collection. The TUBO murine breast cancer cell line (a kind gift from Dr. Wei Zen Wei, Wayne State University) was cloned from a spontaneous mammary tumor in BALB/c mice transgenic for the rat HER2/neu gene (BALB-HER2/neuT; ref. 26). The M05 cell line was a kind gift from Dr. Shari Pilon-Thomas (Moffitt Cancer Center), and cells were grown in media containing 0.8 mg/mL G418 (neomycin; cat. #30234CR, Corning; refs. 27, 28). M05 is derived from the B16 murine melanoma cell line, transfected to express ovalbumin, and transgene expression was maintained using G418 as the selection antibiotic in the culture media. Cells were grown at 37°C in a humidified 5% CO2 incubator in complete media. All cell lines used in the study tested negative for mycoplasma using a mycoplasma kit (PlasmoTest, cat. #rep-pt1, InvivoGen). Cell lines at passages 2–5 were used for all experiments. For all in vitro experiments, recombinant human and mouse IFN-γ were purchased from R&D Biosystems (human: cat. #285IF-100; mouse: cat. #485-MI-100).
BLASTP analysis
Amino acid sequence homology between identified human HER3 peptides and murine Erbb3 sequence (UniProtKB; Q61526) was evaluated by using the BLASTP program (http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&LINK_ LOC=blasthome RRID:SCR_001010). The percentage of amino acid sequence homology has been reported.
Western blots
For protein expression, cells were seeded (4 × 105 cells/well for human TNBC cells, 2 × 105 cells/well for mouse TNBC cells) in six-well plates and cell lysates were prepared in RIPA buffer (cat. #20-188, EMD Millipore) containing protease inhibitor cocktail (10 μL/mL; cat. #P8340-1ML, Sigma-Aldrich) and phosphatase inhibitor (100 μL/mL; cat. #A32957, Pierce) by incubating for 20 minutes at 4°C. Cell lysates were centrifuged at 14,000 × g for 20 minutes at 4°C, and total protein in the supernatant was collected and stored at −86°C until further use. Protein concentration was measured using Bradford protein assay (cat. #5000006, Bio-Rad). For Western blotting, 20 μg protein was resolved on 4% to 12% SDS-PAGE (GenScript) and transferred onto PVDF membranes (cat. #IPVH00010, Millipore) using eBlot L1 wet transfer system (GenScript). Membranes were blocked with 5% BSA/TBS-T for 1 hour at room temperature, and then incubated with following primary antibodies overnight at 4°C: HER3 (cat. #12708S), phospho-Stat1 (Tyr701; 58D6; cat. #9167S, Cell Signaling Technology), cleaved caspase-3 (Asp175; cat. #9661S, Cell Signaling Technology), phospho-p44/42 MAPK (Erk1/2; Thr202/Tyr204; cat. N#9101S, Cell Signaling Technology; RRID:AB_2315112), phospho-Akt (Ser473; cat. #9271S, Cell Signaling Technology; RRID:AB_329825; all at 1:1,000 dilution), and β-actin (C4; cat. #sc-47778, Santa Cruz Biotechnology; RRID:AB_2714189; 1:3,000 dilution). Membranes were probed with anti-rabbit IgG HRP-linked secondary antibody (cat. #7074S, Cell Signaling Technology, RRID:AB_2099233) or goat anti-mouse IgG (H + L)–HRP conjugated secondary antibody (cat. #172-1011, Bio-Rad, RRID:AB_11125936; 1:3,000 dilution) for 1 hour at room temperature. Protein expression was detected with ECL Western blotting substrate (cat. #32106, Pierce) using an AmershamTM Imager 600 image acquisition system. Analysis and quantification of Western blot images were performed using ImageJ software (http://rsb.info.nih.gov/ij/, RRID:SCR_003070).
Immunofluorescence
Cells were seeded (4 × 105 cells/well for human cells (MDA-MB-468, SK-BR-3), 2 × 105 cells/well for mouse cells (4T1, TUBO, and M05) in six-well plates, each well containing three 12-mm round glass coverslips (cat. #12-545-80, Fisher Scientific). After cells reached 70% to 80% confluence, cells were washed twice in PBS, fixed with 4% paraformaldehyde for 15 minutes at room temperature, and washed three times with PBS. Cells were permeabilized with 0.02% Triton X-100 (cat. #T8787—50 mL, Sigma; in PBS) for 10 minutes at room temperature, washed three times with PBS, and blocked with 5% BSA/PBS blocking buffer for 1 hour at room temperature. Cells were incubated with primary anti-HER3 (cat. #12708S, Cell Signaling Technology) in 3% BSA/PBS (1:500 dilution) overnight at 4°C. The next day, cells were washed in PBS three times and incubated in Alexa Fluor 594-conjugated goat-anti-rabbit secondary antibody (1:5,000 dilution; cat. #8889S, Cell Signaling Technology) and FITC-conjugated goat anti-mouse secondary antibody (1:5,000 dilution; cat. #115-095-003 Jackson ImmunoResearch Inc, RRID: AB_2338589) for 1 hour at room temperature. Cells were washed three times in PBS, and the coverslips were mounted onto sterile glass slides using VECTASHIELD Antifade Mounting Medium with DAPI (cat. #H-1200, Vector Laboratories). Slides were sealed with clear nail varnish and allowed to cure overnight at 4°C in the dark. Immunofluorescence images were obtained using Zeiss Apotome.2 fluorescence microscope (Carl Zeiss Inc.).
Mouse DC generation
Bone marrow was harvested from 6- to 8-week-old BALB/c (RRID:IMSR_ORNL:BALB/cRl) and C57BL/B6 (RRID: IMSR_ JAX:000664) mice (purchased from Charles River Laboratories), as described previously (29). Briefly, femurs and tibias were harvested from mice, bone marrow cells were flushed to obtain a single-cell suspension in PBS, and red blood cells were lysed using ACK lysis buffer. Cells (2 × 106 cells/mL) were then cultured in complete media containing recombinant human Flt3L (25 ng/mL; cat. #10778-670, VWR/PeproTech) and recombinant mouse IL6 (30 ng/mL; cat. #406ml025, R&D Systems), and incubated for 6 days at 37°C and 5% CO2. On day 6, cells were harvested, washed with RPMI-1640, and cultured with recombinant mouse GM-CSF (50 ng/mL; cat. #415-ML-050, R&D Systems) and recombinant mouse IL4 (10 ng/mL; cat. #404ml050, R&D Systems) overnight for DC differentiation. The cells were matured with DC1-polarizing cytokines: CPG/ODN1826, a TLR9 agonist (10 ng/mL; cat. #NC9685794, InVivoGen), and LPS, a TLR4 agonist (20 ng/mL). DCs were pulsed with the nine immunogenic class II human HER3 peptides (10 μg/mL) 6 to 8 hours later and harvested the following morning prior to injection into mice.
Mouse models
Female BALB/c and C57BL/6 mice (6–8 weeks old) were housed at the Animal Research Facility of the H. Lee Moffitt Cancer Center and Research Institute. The study protocol was designed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the NIH. The protocol was reviewed and approved by the Institutional Animal Care and Use Committee at the University of South Florida. Mice were observed daily and were euthanized by CO2 inhalation at the end of the study or if a solitary subcutaneous tumor exceeded endpoint (250 mm2 for 4T1 and 400 mm2 for TUBO and M05), following the American Veterinary Medical Association Guidelines. All efforts were made to minimize suffering.
Preventive models
BALB/c and C57BL/6 mice (n = 8–10/group) were vaccinated with either unpulsed mature DC1 cells or HER3-DC1 cells subcutaneously (1 × 106 cells/mouse) twice a week for a total of six doses. Two weeks after the last DC injection, naïve and vaccinated BALB/c mice received either 4T1 (50,000 cells/mouse) or TUBO (250,000 cells/mouse) tumor cells, and C57BL/6 mice received M05 (300,000 cells/mouse) tumor cells, administered subcutaneously on the opposite flank to the vaccination site. Tumor size was measured using caliper and recorded every 2 to 3 days. Mouse tumor area was determined by the formula length × width (mm2). Each experiment was performed three times.
Therapeutic models
For therapeutic models, either 4T1 (50,000 cells/mouse) or TUBO (250,000 cells/mouse) were injected subcutaneously in BALB/c mice, and M05 cells (300,000 cells/mouse) were injected in C57BL/6 mice (n = 8–10 mice/group). After 7 to 10 days (7 days for 4T1 and TUBO, 10 days for M05) when tumors were palpable, mice received intratumoral injection of either unpulsed mature DC1 cells or HER3-DC1 (2 × 106 cells/mouse for 4T1 and 1 × 106 cells/mouse for TUBO and M05, in 50 μL PBS) once (TUBO and M05) or twice (4T1) a week for a total of six doses. Control mice received PBS intratumoral injection. Tumor growth was measured with caliper and recorded every 2 to 3 days, and tumor area was determined by the formula length × width (mm2). Each experiment was performed three times.
CD4+ T-cell depletion in therapeutic models
BALB/c mice were injected intraperitoneally with monoclonal CD4 depletion antibody [InVivoMAb, anti-mouse CD4 (GK1.5); cat. #BE0003-1, Bio X Cell; RRID:AB_1107636; 300 μg/mouse] starting 3 days before subcutaneous TUBO tumor cell injection (250,000 cells/mouse) and was continued twice a week until endpoint. When tumors were palpable, mice were randomized into two groups. One group of mice continued receiving CD4 depletion antibody only, and the other group received once-weekly intratumoral HER3-DC1 (1 × 106 cells/mouse, in 50 μL PBS) along with CD4 depletion antibody. Another group of TUBO tumor–bearing mice received intratumoral HER3-DC1 only, and the control group of mice received PBS intratumorally. Tumor growth was measured with caliper and recorded every 2 to 3 days, and tumor area was determined by the formula length × width (mm2).
Flow cytometry
For functional analyses, mice (n = 3/group) were sacrificed 2 weeks after the last HER3-DC1 injection, and tumors, spleen, and lymph nodes (tumor-draining and nondraining) were collected under sterile conditions for in vitro assays. Single-cell suspensions were prepared from the tumor samples by enzymatic digestion with HBSS (Fisher Scientific; cat. #MT-21-022-CM), containing 1 mg/mL collagenase (cat. #C9891 and C-5138), 0.1 mg/mL DNase I (cat. #DN25), and 2.5 U/mL hyaluronidase (cat. #H-6254-1G; all purchased from Millipore Sigma), by constant stirring for 2 hours at room temperature. Tumor digests were strained through a 100-μm cell strainer, and ACK lysis buffer was used to remove red blood cells from tumor digests. Resulting cell suspensions were further strained through a 70-μm and a 30-μm cell strainer to generate single-cell suspension. Splenocytes and lymph nodes were processed in PBS. Samples were macerated using the back of a syringe plunger in a 10-cm culture dish, followed by red blood cell lysis using ACK lysis buffer. After lysis, cell suspensions were strained through a 70-μm and a 30-μm cell strainer sequentially. Cell suspensions were centrifuged at 1,500 rpm for 5 minutes, and cells were resuspended in complete media and counted before plating.
To analyze distribution and phenotype of immune cell populations, 1 × 106 cells were incubated with Live/Dead Zombie near IR (cat. #423106, BioLegend) for 30 minutes at room temperature. After washing once with PBS, cells were stained with the following mouse antibodies for surface expression analysis: CD3-APC (cat. #553066, Clone 145-2C11, BD Biosciences), CD45-PE/Cy7 (cat. #103114, Clone 30-F11, BioLegend), or CD45-BUV395 (cat. #564279, Clone 30-F11, BD Biosciences), CD4-BUV805 (cat. #564922, Clone GK1.5, BioLegend), CD8-Pacific Blue (cat. #558106, Clone 53–6.7, BD Biosciences), CD44-FITC (cat. #553133, Clone IM7, BD Biosciences), CD62L-BUV395 (cat. #740218, Clone MEL-14, BD Biosciences). Cells were incubated in the antibody solution made in staining buffer, which was made with PBS (without calcium or magnesium, 100 mL/L; cat. #14200166, GIBCO), sodium azide (1 g/L; cat. #S2002, Sigma) and BSA (1% final W/V; cat. #BP1605-100, Fisher Scientific) in water, for 20 minutes on ice and protected from light, following the manufacturer's instructions. Flow cytometry acquisition was performed using an LSRII (BD Biosciences) cytometer, and FACS data analysis was performed with FlowJo software (FlowJo, RRID:SCR_008520).
Functional restimulation/coculture assays
To investigate antigen specificity following HER3-DC1 administration in mice, spleens and lymph nodes were harvested 2 weeks after the last vaccination, processed as stated above, and splenocytes from control and vaccinated/treated mice were plated in 48-well tissue culture plates at 2 × 106 cells/mL in 1% FBS-supplemented RPMI and rested for 24 hours at 37°C and 5% CO2. After 24 hours, cells were either pulsed (2 μg/mL) with individual HER3 class II immunogenic peptides, a negative class II control peptide, or left unpulsed. For lymph nodes, lymphocytes were plated in a 96-well plate with DCs, previously matured as described above, and pulsed with HER3 class II peptides, a negative class II control (OT-II), or left unpulsed (lymphocyte:DC ratio 10:1). Following 72 hours of incubation, culture supernatant was collected, and IFN-γ secretion was measured after 1:20 dilution of the supernatant, using mouse IFN-γ Quantikine ELISA Kit (cat. #PMIF00, R&D Systems). The same protocol was followed for all mouse models included in the study.
Intracellular staining for IFN-γ secretion
Tumors were collected (n = 3–5/group) from control and HER3-DC1–treated TUBO tumor–bearing mice. Tumor-infiltrating lymphocytes (TIL) were isolated following the protocol described above and cocultured with mature DC1 cells pulsed with individual HER3 peptides (10:1 TIL:DC; i.e., 106 TIL:105 DC in 1 mL total volume). Intracellular staining was performed using the BD Cytofix/Cytoperm Plus Fixation/Permeabilization Kit with BD GolgiPlug Protein transport inhibitor containing Brefeldin A (cat. #555028, BD Biosciences). Briefly, 6 hours after TIL:DC coculture, GolgiPlug was added to inhibit intracellular protein transport (1 μL/106 cells) for 12 hours. Cells were harvested the next day, and surface staining with CD45-BUV395 (cat. #564279, Clone 30-F11, BD Biosciences), CD4-BUV805 (cat. #612900, Clone GK1.5, BD Biosciences), and CD8-Pacific Blue (cat. #558106, Clone 53–6.7, BD Biosciences) was performed as described above. Cells were fixed and permeabilized following the manufacturer's protocol and were stained for intracellular IFN-γ-PE (cat. #554412, Clone XMG1.2, BD Biosciences). Acquisition was performed using an LSRII (BD Biosciences) cytometer, and FACS data analysis was performed with FlowJo software (FlowJo).
Statistical analyses
To compare immune response generated by the peptides, ELISA data were analyzed by multiple t test, without correction for multiple comparisons, using GraphPad Prism software (RRID:SCR_002798). Each row was analyzed individually, without assuming consistent standard deviation. Data are represented as mean ± SEM. For analyzing immune response across HLA alleles, statistical significance was determined using Mann–Whitney test (ns, P > 0.05), and data are represented as mean ± SEM. To compare tumor growth between groups, data were analyzed using multiple t test without correction for multiple comparisons. Each row was analyzed individually, without assuming a consistent standard deviation, and data are represented as mean ± SEM. A log-rank (Mantel–Cox) test was used to determine differences between the survival curves. Unpaired two-tailed t test was performed to analyze Western blot data. For all analyses, significance threshold was considered as *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Results
HER3 is overexpressed in multiple cancers
HER3 is an established oncodriver that contributes to the growth, proliferation, and survival of cancer cells (30, 31). Analysis of HER3 RNA-seq expression data in 12 cancers from the GDC demonstrated significantly increased HER3 expression in bladder (P = 0.0045), breast (P = 1.15E–12), lung adenocarcinoma (P = 1.77E–19), prostate (P = 6.63E–11), and stomach cancers (P = 1.31E–5; Fig. 1A). Although not statistically significant, a similar trend of high HER3 expression was also noted in pancreatic cancer (P = 0.1843). Conversely, HER3 mRNA expression in tumor tissues was significantly lower than healthy tissues in HNC (P = 7.27E–7) and lung squamous cell carcinoma (P = 1.82E–7), indicating cancer type specificity. Due to lack of data on normal tissue HER3 mRNA expression in TCGA database, comparison of ovarian serous cystadenocarcinoma and melanoma could not be performed; and no change in HER3 expression was noted in colon adenocarcinoma (P = 0.1259) or esophageal cancer (P = 0.758). We used the cBioPortal survival tool to further investigate if elevated/suppressed HER3 expression correlated with overall patient survival. Whereas in breast cancer high versus low HER3 expression had no statistically significant correlation with overall survival (P = 0.182), high HER3 expression showed a significantly negative correlation with overall survival in melanoma (P = 0.0246; Fig. 1B–C). Although HER3 mRNA expression was not significantly different between normal pancreas and pancreatic adenocarcinoma, a significant negative correlation was observed between high HER3 expression and overall survival (P = 6.248E–3) in pancreatic cancer (Supplementary Fig. S1B). A positive correlation between high HER3 expression and overall survival was indicated in head and neck squamous cell carcinoma (P = 3.913E–3; Supplementary Fig. S1C). We did not observe a statistically significant correlation between HER3 expression and overall survival in any of the other cancers investigated (Supplementary Fig. S1D–S1K). These results suggested an impact of increased expression of HER3 in specific malignancies.
Figure 1.
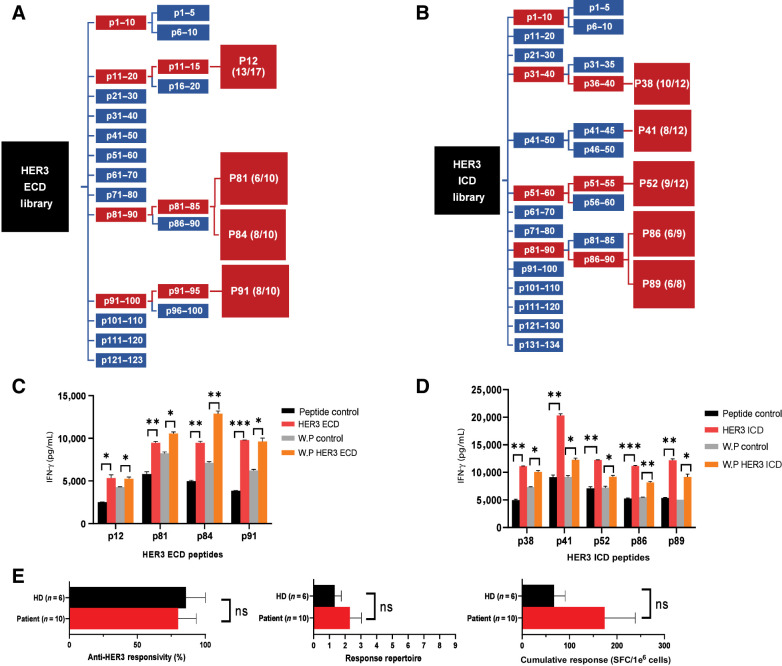
HER3 expression in cancer, and HER3 peptide screening of ECD and ICD class II peptide libraries. A, Expression of HER3 mRNA in normal (N) and tumor (T) tissues obtained from RNA-seq data from the GDC across cancer types (see Materials and Methods). B, Correlation between percentage of HER3 expression and overall patient survival (in months) in breast cancer. Samples were sorted in the descending order of HER3 expression and put into two groups: high HER3 (red) and low HER3 (blue). C, Correlation of overall patient survival with high HER3 (red) versus low HER3 (blue) expression in melanoma. P value indicated in individual graphs. D and E, IFN-γ production at each screening step for sample 1 [normal donor (ND) 8; D] and sample 2 (ND 9; E) when stimulated with ECD peptides. F and G, IFN-γ production at each screening step for sample 3 (ND 3; F) and sample 4 (ND 5; G) upon stimulation with ICD peptides. D–G, IFN-γ response to negative peptide control (black) compared with HER3 peptides (red) with an immunogenic response threshold of ≥1.5-fold increase. Data represented as mean ± SEM with statistical significance determined using a multiple t test without correction for multiple comparisons. Each row was analyzed individually, without assuming consistent SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Identification of HER3 class II peptide epitopes
To identify immunogenic class II epitopes from the HER3 protein, human DCs were pulsed with overlapping 15-mer peptides from a HER3 library, rapidly matured into a type I phenotype (DC1), and cocultured with autologous naïve CD4+ T cells (Supplementary Fig. S2). Following restimulation of peptide-primed CD4+ T cells with the matching class II peptide or negative control, supernatants were collected to measure peptide-specific Th1 responses by IFN-γ secretion. Peptide libraries were sequentially screened in pools of 10-peptides, 5-peptides, and individual peptides based on significant fold increases in IFN-γ compared with control (see Materials and Methods and Supplementary Fig. S1A). Both ECD and ICD HER3 peptide libraries were screened separately on three healthy donor samples with two representative screenings reported for ECD (samples 1–2; Fig. 1D and E) and ICD libraries (sample 3–4; Fig. 1F and G) to identify immunogenic HER3 peptides and confirm reproducibility of peptide-specific Th1 immune responses. Screening of HER3 ECD 10-peptide pools revealed p11–20, p81–90, and p91–100 as inducing a significant increase in Th1 immune responses compared with the negative control (Fig. 1D and E). Further breakdown into 5-peptide pools showed p11–15, p81–85, and p91–95 had a comparable significant increase in IFN-γ. Lastly, the corresponding 15 individual peptides were screened, identifying peptides p12 (P = 0.0281), p81 (P = 0.0041), p84 (P = 0.0023), and p91 (P = 0.0161) in sample 1 and likewise, p12 (P = 0.0052), p81 (P = 0.0021), p84 (P = 0.0009), and p91 (P = 0.0012) in sample 2. Similarly, p31–40, p51–60, and p81–90 demonstrated significant increases in Th1 immune responses when screening HER3 ICD 10-peptide pools (Fig. 1F and G). Of the three donor samples used in HER3 ICD 10-peptide pool screening, the first donor sample demonstrated an overall lack in response to pool p41–50 for both the control- and peptide-stimulated T cells (Supplementary Fig. S3A). Prior to continued screening of HER3 ICD 10-peptide pools, all peptides within the pool p41–50 were screened individually, with p41 inducing a significant increase (P = 0.0158) in IFN-γ (Supplementary Fig. S3B) and was included as an epitope candidate. To validate this discovery, individual peptides from two additional unresponsive pools were screened in the ECD (Supplementary Fig. S3C–S3D) and ICD (Supplementary Fig. S3E and S3F) libraries; no other positive peptides were identified. 5-Peptide pools were then screened, which revealed p36–40, p51–55, and p86–90 as common positive pools. Thirteen peptides were then screened individually, in addition to the five individual peptides within pool p56–60, which showed a significant increase in IFN-γ in sample 3. Overall, five individual HER3 ICD peptides demonstrated a common significant increase in IFN-γ compared with the negative control: p38 (P = 0.0055), p52 (P = 0.0119), p86 (P = 0.0020), and p89 (P = 0.0028) in sample 3 and, similarly, p38 (P = 0.0043), p41 (P = 0.0124), p52 (P = 0.0018), p86 (P = 0.0016), and p89 (P = 0.0012) in sample 4. Taken together, four HER3 ECD and five HER3 ICD peptides were identified as potential class II epitopes through the sequential peptide screening of HER3 ECD and ICD peptide libraries: HER3aa56–70, HER3aa401–415, HER3aa416–430, and HER3aa451–465, HER3aa850– 864, HER3aa865–879, HER3aa920–934, HER3aa1090–1104, and HER3aa1105–1119 (Supplementary Table S1). Screening schema for the ECD and ICD libraries at each step are shown in Fig. 2A–B. The nine HER3 class II peptides were recognized as candidate HER3 epitopes and screened across 6–13 additional samples (Supplementary Table S1) to confirm the reproducibility of the peptide-specific CD4+ Th1 immune responses (Supplementary Fig. S4A–S4F).
Figure 2.
HER3 class II peptides demonstrate anti-HER3 immune responses in vitro. A and B, ECD and ICD peptide libraries were screened sequentially in 10-peptide pools, 5-peptide pools, and individual peptides with an immunogenic response threshold of ≥1.5-fold increase in IFN-γ production between peptide and control restimulated CD4+ T cells (see Fig. 1), ultimately identifying four ECD and five ICD HER3 class II peptides. Each schematic is representative of the combined responses across samples used in the peptide screening (n = 3), indicating reproducible significant immunogenic response compared with the class II control in ≥2 samples and number of donor responses in parentheses (red). C and D, Peptide-primed CD4+ T cells were restimulated with matching class II peptide (HER3 ECD or HER3 ICD), class II–negative control (peptide control), whole HER3 domain protein (WP HER3 ECD or WP HER3 ICD), or whole protein control (WP control). Data represented as mean ± SEM with statistical significance determined using multiple t test without correction for multiple comparisons. Each row was analyzed individually, without assuming a consistent SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001. E, PBMCs from healthy donors (HD, black bar, n = 6) or breast cancer patients (Patient, red bar, n = 10) were individually stimulated with the nine HER3 class II peptides and analyzed via IFN-y ELISPOT. Left, percentage of subjects responding to ≥1 HER3 peptide (anti-HER3 responsivity). Middle, mean number of peptides inducing anti-HER3–specific immunity (response repertoire). Right, total IFN-γ spots (mean total SFC/1e6 cells) from stimulation with HER3 peptides (cumulative response). Data represented as mean ± SEM with statistical significance determined using Mann–Whitney test. ns, not significant.
HER3 epitopes induce HER3-specific Th1 immune responses in vitro
To confirm the identified HER3 peptides were competent epitopes in vitro, HER3-peptide-primed CD4+ T cells were restimulated with the corresponding whole HER3 ECD or ICD protein. HER3 ECD peptides demonstrated a significant increase in IFN-γ production when peptide-primed CD4+ T cells were restimulated with the matching class II peptide [(p12, P = 0.0181), (p81 = 0.0075), (p84, P = 0.0025), (p91, P = 0.00004)], and responses were comparable to that of restimulation with the full HER3 ECD protein domain [(p12, P = 0.0417), (p81, P = 0.0121), (p84, P = 0.0036), p91 (P = 0.0155); Fig. 2C]. HER3 ICD peptide–primed CD4+ Th1 cells restimulated with the matching HER3 class II peptides similarly showed a significant response compared with the peptide negative control [(p38 (P = 0.0012), (p41, P = 0.0021), (p52, P = 0.0035), (p86, P = 0.0008), (p89, P = 0.0021)], which was analogous to that of peptide-primed CD4+ Th1 cells restimulated with the full HER3 ICD protein domain [(p38, P = 0.0131), (p41, P = 0.0191), (p52, P = 0.0430), (p86, P = 0.0072), (p89, P = 0.0159); Fig. 2D].
To determine if the identified HER3 peptides induced comparable anti-HER3 Th1 immune responses in healthy donor and breast cancer patient samples in vitro, PBMCs from healthy donor controls (n = 6) and breast cancer patients (n = 10) were sensitized to the nine HER3 peptides, and IFN-γ responses were evaluated by ELISPOT (Fig. 2E). There was no significant difference between healthy donor controls and breast cancer patients in anti-HER3 responsiveness (P = 0.0820), response repertoire (P = 0.4717), or cumulative responses (P = 0.3132). Together, these results indicate CD4+ T cells specific to the nine HER3 class II peptide candidates could reproducibly elicit the desired HER3-specific Th1 immune response in different donor and breast cancer patient samples and were competent epitopes in vitro.
HER3 class II peptides represent promiscuous epitopes
To evaluate if the identified HER3 class II peptides were restricted to specific HLA alleles, donor samples used in both ECD and ICD peptide library screenings were analyzed for MHC class II HLA allele expression (Supplementary Table S2). A total of 35 different alleles were expressed across HLA-DR (13 alleles), HLA-DP (11 alleles), and HLA-DQ (11 alleles), with minimal overlap in expression across donor samples (≤3 samples expressed the same HLA allele). Allele expression was cross-referenced with the corresponding HER3 ECD and HER3 ICD (Supplementary Table S3) peptide–specific Th1 immune responses. Together, these data confirmed high variability in HLA expression across samples and demonstrates the promiscuous binding of identified HER3 peptides across multiple MHC II HLA alleles, indicating the application of identified HER3 epitopes for a HER3-DC1 vaccine with capabilities for widespread application.
Class II predictive algorithm did not identify HER3 peptides as candidate epitopes
MHC II prediction algorithm, NetMHCIIpan 4.0, was used to predict class II peptides from the full HER3 protein sequence, and predicted results were compared with those obtained from the experimental peptide screening. NetMHCIIpan predicted approximately 458 class II peptide sequences (15-mer) with >700 different 9-mer binding cores exhibiting strong binding affinity (<50 nmol/L; Table 1). HER3 ECD p91 was the only peptide with predicted strong binding affinity with multiple HLA alleles. The remaining HER3 peptides were predicted to establish only weak binding interactions (50–500 nmol/L) with approximately 1–19 different HLA alleles. However, the MixMHCIIpred algorithm predicted a strong binding affinity for all nine HER3 peptides, with p91 showing the strongest binding affinity to most of the 38 tested HLA alleles (Table 1). Most of the peptides showed strong binding affinity toward a handful of HLA alleles, although each showed significant immunologic responses when tested experimentally. These results indicate the inability of the NetMHCIIpan prediction algorithm, as well as inconsistencies between algorithms, to identify the majority of HER3 class II peptides as candidate epitopes from the full HER3 protein and supports the indispensability of an empirical approach to epitope identification for tumor antigens.
Table 1.
Prediction of binding affinity from algorithms for identified HER3 peptides.
Antigen-specific responses and delayed tumor growth by preventive vaccination
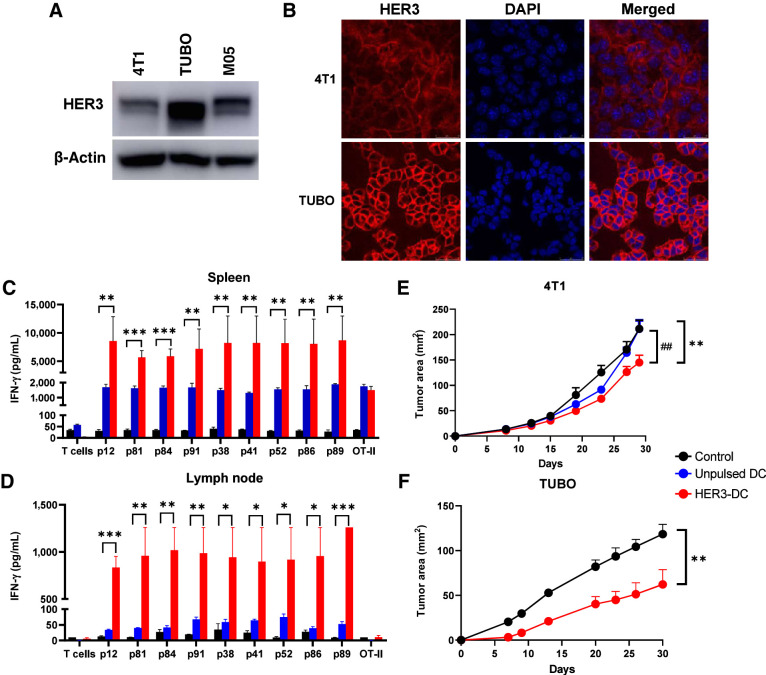
To investigate the antitumor immune response generated by the nine identified human HER3 peptides, we investigated both preventive and therapeutic efficacy of HER3 peptide–pulsed DC1 (HER3-DC1) in preclinical murine models. Because we observed high HER3 expression in breast cancer and melanoma compared with normal tissues in our TCGA analysis, we chose murine mammary carcinoma models 4T1 and TUBO, which represent human TNBC and HER2pos breast cancer, respectively, and the M05 murine melanoma model. Western blots showed HER3 protein expression in all three cell lines, with TUBO cells showing the highest and 4T1 showing the lowest HER3 expression (Fig. 3A). Immunofluorescence staining showed HER3 expression in 4T1 and TUBO cells, primarily confined to the cellular surface of both cell lines (Fig. 3B). In human TNBC and HER2pos breast cancer cells, we observed HER3 expression, although the expression level varied considerably across cell lines (Supplementary Fig. S5A and S5B). To test the human peptides in murine models, we tested the amino acid sequence similarity between the identified human HER3 peptides and full-length murine Erbb3 amino acid sequence using the BLASTP program. We observed 100% sequence homology between murine Erbb3 and human HER3 peptides ECD p12, ECD p81, ECD p84, ICD p38, and ICD p86; 93% sequence homology with ICD p41, ICD p52, and ICD p89; and 87% sequence homology with ECD p91 (Supplementary Table S4).
Figure 3.
HER3-DC1 vaccination elicits peptide-specific immune response and delays tumor growth. A, Immunoblotting of murine tumor cell lines 4T1, TUBO, and M05 to detect HER3. β-Actin: loading control. B, Immunofluorescence for HER3 (red) and nucleus (DAPI, blue) in 4T1 and TUBO murine mammary tumor cells (image magnification: 1,200×). C and D, Individual HER3 peptide–specific immune responses in spleens (C) and lymph node–derived immune cells (D) from control (black), unpulsed mature DC1 (blue), and HER3-DC1 (red) vaccinated BALB/c mice (n = 3). Spleens were processed, and splenocytes were restimulated with the HER3 peptides for 72 hours to detect IFN-γ by ELISA. Lymph node–derived lymphocytes were cocultured with DC1 pulsed with individual HER3 peptides for 72 hours to detect IFN-γ by ELISA. E, Tumor growth after 4T1 tumor challenge in control (black), unpulsed mature DC1 (blue), and HER3-DC1 (red) vaccinated mice (n = 7–10 mice/group). Mice were challenged 2 weeks after the last vaccination and were monitored until endpoint. *, control versus HER3-DC1; #, unpulsed DC1 versus HER3-DC1. F, TUBO tumor growth in control (black) and HER3-DC1 (red) vaccinated mice (n = 7–10 mice/group). Mice were challenged 2 weeks after the last vaccination. Data represented as mean ± SEM with statistical significance determined using multiple t test without correction for multiple comparisons. Each row analyzed individually, without assuming a consistent SD. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ##, P ≤ 0.01.
Next, we evaluated the preventive efficacy of HER3-DC1 in BALB/c mice by vaccination with either unpulsed DC1 or HER3-DC1, followed by 4T1 or TUBO tumor challenge. To investigate immune response and antigen specificity of the response to HER3 peptides in murine models, splenocytes from vaccinated and naïve control mice were restimulated with the HER3 peptides for 72 hours ex vivo, followed by an IFN-γ ELISA using culture supernatants. We observed significantly higher IFN-γ secretion from HER3-DC1–vaccinated mice compared with control mice (Fig. 3C; Supplementary Fig. S5C). No significant difference in splenocyte IFN-γ secretion from unpulsed DC1 vaccinated mice compared with controls was observed, suggesting immune responses were specifically stimulated by HER3 peptides and was not a result of nonspecific immune stimulation by mature, nonprimed DC1 vaccination.
Similarly, IFN-γ in supernatants from cocultures of the lymph node–derived lymphocytes with DC1 pulsed with cognate HER3 peptides showed significantly increased IFN-γ secretion from HER3-DC1–vaccinated BALB/c mice compared with naïve controls (Fig. 3D; Supplementary Fig. S5D). These data suggest that the development of systemic immune responses in HER3-DC1–vaccinated mice is HER3 peptide–specific and not induced by unpulsed mature DC1.
Two weeks after the last vaccination, HER3-DC1–vaccinated or unvaccinated control mice were challenged with either 4T1 or TUBO tumor to compare preventive efficacy of HER3-DC1 vaccine. HER3-DC1–vaccinated mice challenged with 4T1 showed significantly delayed tumor growth (P = 0.007) and extended survival, whereas there was no significant difference in tumor growth and survival between control and unpulsed DC1-vaccinated mice (P = 0.9782; Fig. 3E; Supplementary Fig. S5E). Tumor growth in HER3-DC1–vaccinated mice was also significantly delayed compared with the unpulsed DC1 mice (P = 0.0086), suggesting the role of anti-HER3 Th1 response in preventing tumor growth over unpulsed DC1 vaccine.
In the TUBO model, we observed significantly delayed tumor growth in HER3-DC1–vaccinated mice compared with the unvaccinated controls (P = 0.0191; Fig. 3F). Despite clear preventive benefit and systemic immune response after HER3-DC1 vaccination, the NetMHCIIPan4.0 algorithm did not predict binding affinity of any of the nine HER3 peptides (Supplementary Table S5), further highlighting the limitation of false-negative predictions by algorithms. These data suggest HER3-DC1 vaccination stimulates HER3 antigen–specific immune responses and offers preventive benefit to delay tumor growth in both TNBC and HER2pos breast cancer models.
Intratumoral HER3 DCs delay tumor growth and enhance immune infiltration
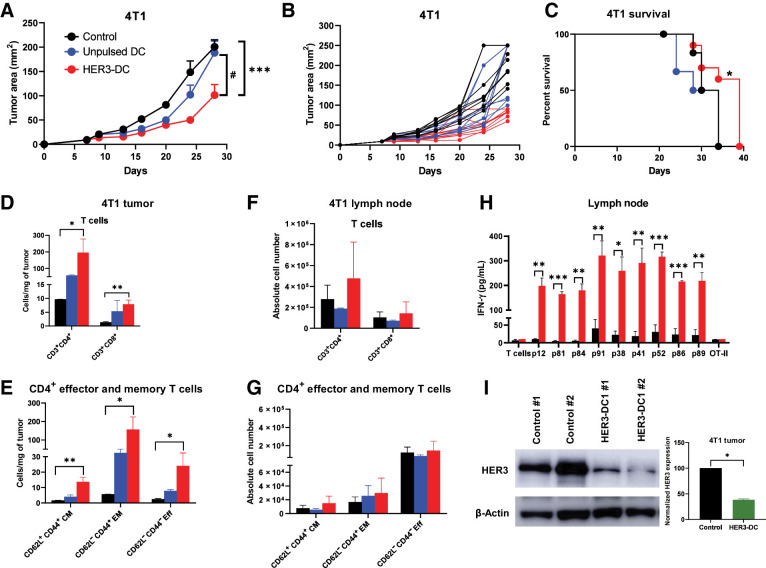
Next, we evaluated the therapeutic efficacy of HER3-DC1 in the aggressive murine mammary carcinoma model 4T1 (32), which mimics human TNBC. Intratumoral HER3-DC1 administration significantly delayed tumor growth compared with controls (P = 0.0019) and unpulsed DC1-treated mice (P = 0.0183), indicating therapeutic benefit of HER3-DC1 (Fig. 4A and B), and no significant difference was noted in tumor growth between control and unpulsed DC1–treated groups. We also observed a significant increase in survival rate in the HER3-DC1–treated group compared with control mice (P = 0.0327; Fig. 4C). Compared with the intratumoral route of delivery, subcutaneous HER3-DC administration did not show therapeutic benefit in the 4T1 model (Supplementary Fig. S5F and S5G).
Figure 4.
Intratumoral HER3-DC1 administration elicits peptide-specific immune responses and delays tumor growth. A, Tumor growth in the 4T1 murine mammary carcinoma model. BALB/c mice bearing subcutaneous 4T1 tumors received either intratumoral PBS (black), unpulsed mature DC1 (blue), or HER3 peptide–pulsed DC1 (red; n = 10 mice/group), starting on day 7 when tumors were palpable. Tumor growth was monitored until endpoint and was compared between control and HER3-DC1, as well as between unpulsed DC1 and HER3-DC1. *, control versus HER3-DC1; #, unpulsed DC1 versus HER3-DC1. B, Individual tumor growth for each mouse from control (black)-, unpulsed DC1 (blue)–, and HER3-DC1 (red)–treated groups. C, Percent survival in the 4T1 mouse model. Control, black; unpulsed DC1, blue; HER3-DC1, red. D, Intratumoral CD3+CD4+ and CD3+CD8+ T-cell infiltration per milligram of tumor in control (black)-, unpulsed DC1 (blue)–, and HER3-DC1 (red)–treated mice. Absolute number of immune cells was compared between control and HER3-DC1 groups. E, Frequency of CD62L+CD44+ central memory (CM), CD62L−CD44+ effector memory (EM), and CD62L−CD44− effector (Eff) T-cell populations within intratumoral CD4+ cells between control- (black) and HER3-DC1–treated (red) tumors. The unpulsed DC1 (blue) group was not included in any statistical analyses. F, Absolute number of CD3+CD4+ and CD3+CD8+ T cells in lymph nodes of control (black)-, intratumoral unpulsed DC1 (blue)–, and HER3-DC1 (red)–treated mice. Cell numbers were compared in control versus HER3-DC1 groups. Data shown are the representative from three independent experiments. G, Absolute numbers of CD4+ CM, EM, and Eff T-cell populations in lymph nodes of control (black), unpulsed DC1 (blue), and HER3-DC1 (red) mice. Data shown are the representative from three independent experiments. H, Lymphocytes from the lymph nodes of control and treated mice were cocultured with DC1 pulsed with individual HER3 or OT-II (negative control) peptides. Culture supernatants were collected after 72 hours, and IFN-γ was measured by ELISA (control: black bar; HER3-DC1: red bar). I, Total protein isolated from in vivo tumor samples was analyzed by Western blotting to compare HER3 protein expression after intratumoral HER3-DC1 (green) administration with respect to the control (black). β-Actin: loading control. Data represented as mean ± SEM with statistical significance determined using multiple t test without correction for multiple comparisons. Each row was analyzed individually, without assuming a consistent SD. A log-rank (Mantel–Cox) test was used to determine differences between the survival curves. Unpaired two-tailed t test was performed to analyze Western blot data. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; #, P ≤ 0.01.
We then investigated changes in the intratumoral immune landscape after intratumoral delivery of HER3-DC1, as well as in the tumor-draining lymph nodes, by flow cytometry. The flow gating strategy to identify CD4+ and CD8+ TILs and their subtypes is shown in Supplementary Fig. S6. Flow analysis revealed significantly higher CD4+ (P = 0.03) and CD8+ (P = 0.0042) T-cell infiltration per milligram of tumor in HER3-DC1–treated mice compared with control tumors (Fig. 4D; Supplementary Fig. S7A). We observed a significant increase in the CD62L−CD44+ effector memory (EM, P = 0.0304), CD62L+CD44+ central memory (CM, P = 0.0057), and CD62L−CD44− effector (P = 0.0412) T-cell populations in HER3-DC1–treated tumors compared with controls (Fig. 4E; Supplementary Fig. S7B). We then compared immune phenotypes of cells isolated from the draining lymph nodes of the HER3-DC1 versus control mice and observed no statistically significant increases in total CD4+ (P = 0.6492) and CD8+ (P = 0.7699) populations in the HER3-DC1 group compared with controls (Fig. 4F), and no statistically significant increases in CD4+ CM, EM, and effector T-cell abundance in the HER3-DC1 group compared with the controls was seen (Fig. 4G).
To evaluate systemic immune responses after intratumoral HER3-DC1 administration, lymph nodes harvested from control and treated mice were cocultured with DC1 cells pulsed with individual HER3 peptides or OT-II (as negative control) for 72 hours. ELISA with culture supernatants revealed significantly higher IFN-γ secretion in the HER3-DC1 group, compared with the control, for all nine peptides (Fig. 4H). Lastly, we investigated HER3 protein expression from in vivo tumor samples by immunoblotting to determine if intratumoral DC administration altered receptor protein expression in tumor cells. Total HER3 protein expression was significantly reduced in HER3-DC1–treated tumors compared with control tumors (P = 0.0369; Fig. 4I), suggesting molecular cross-talk between HER3-DC1–induced immune responses and oncodriver signaling can affect receptor protein expression in 4T1 tumor cells.
HER3-DC1 delays tumor growth and enhances immune infiltration in the TUBO model
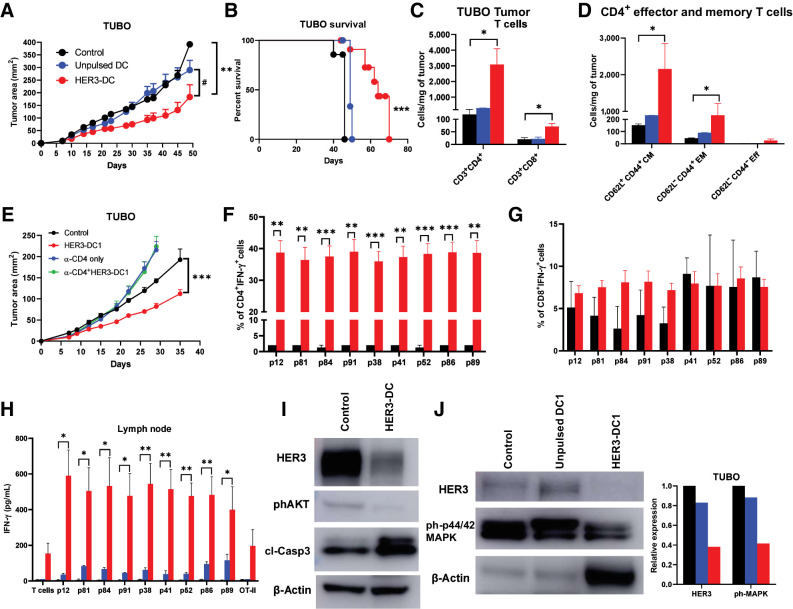
HER3 is the most potent dimerization partner of HER2 that facilitates downstream signaling, contributing to the growth and proliferation of tumor cells. Hyperactivation of HER3 has also been identified as one of the primary mechanisms of therapeutic resistance to HER2-targted therapies in HER2pos breast cancer (33, 34). Therefore, we investigated the therapeutic efficacy of HER3-DC1 in a HER2-resistant TUBO murine mammary tumor model (35). We observed significantly delayed tumor growth in mice that received intratumoral HER3-DC1 compared to control- (P = 0.0028) and unpulsed DC1-treated mice (P = 0.0185). Tumor regression occurred in ∼30% of HER3-DC1–treated mice, and growth was significantly delayed in the remaining mice, compared with the controls (Fig. 5A; Supplementary Fig. S8A). This resulted in a significant improvement in survival of HER3-DC1–treated mice compared with the control (median survival 64 days vs. 46 days, P = 0.0002) and unpulsed DC1-treated groups (median survival 64 days vs. 49 days, P = 0.0015; Fig. 5B).
Figure 5.
Intratumoral HER3-DC1 delays tumor growth and enhances immune infiltration in a HER2pos TUBO therapeutic model in a CD4-dependent manner. A, Tumor growth in the TUBO murine mammary carcinoma model. BALB/c mice were injected with TUBO tumor cells, and on day 7, mice received either PBS control (black), unpulsed mature DC1 (blue), or HER3-DC1 (red) intratumorally once weekly for six doses (n = 10 mice/group). Tumor growth was monitored until endpoint and compared in control versus HER3-DC1 (*) and unpulsed DC1 versus HER3-DC1 (#) mice. B, Percent survival in TUBO mouse model. Control: black; unpulsed DC1: blue; HER3-DC1: red. C, CD3+CD4+ and CD3+CD8+ T cells per milligram of tumors from mice (A) after intratumoral DC injection was compared between control (black) and HER3-DC1 (red) groups. No statistical analyses were performed for the unpulsed DC1 (blue) mice (n = 3/group). D, Abundance of CD4+ central memory (CD62L+CD44+ CM), effector memory (CD62L−CD44+ EM), and effector (CD62L−CD44− Eff) T cells in control (black) versus HER3-DC1 mice (red) per milligram of tumor tissue. Data shown are the representative from three independent experiments. E, Tumor growth of TUBO tumors after CD4 depletion. BALB/c mice were injected with anti-CD4 antibodies 3 days before subcutaneous TUBO tumor injection. When tumors were palpable, mice received either PBS control (black), intratumoral HER3-DC1 once weekly (red) for six doses, CD4 depletion antibody alone (blue; continued twice weekly until endpoint), or HER3-DC1 (green) with CD4 depletion. Tumor growth was monitored until endpoint. F and G, Percentage of CD4+IFN-γ+ (F) and CD8+IFN-γ+ (G) TILs in the tumors from control (black) versus HER3-DC1 (red) mice from E. H, Coculture of the lymph node immune cells with HER3 peptide–pulsed DC1 for 72 hours to detect IFN-γ via ELISA. Control: black bar; unpulsed DC1: blue bar; HER3-DC1: red bar. I, Western blot for HER3, phosphorylated AKT (phAKT), and cleaved caspase-3 (clCasp-3) with total protein isolated from control- and HER3-DC1–treated TUBO tumors. β-Actin: loading control. J, Western blot for HER3 and phosphorylated p44/42 MAPK (ph-p44/42 MAPK) from control-, unpulsed DC1–, and HER3-DC1–treated TUBO tumors. β-Actin: loading control. Data represented as mean ± SEM with statistical significance determined using multiple t test without correction for multiple comparisons. Each row was analyzed individually, without assuming a consistent SD. A log-rank (Mantel–Cox) test was used to determine differences between the survival curves. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; #, P ≤ 0.01.
To evaluate immune infiltration into the tumor microenvironment after HER3-DC1 intratumoral delivery, we collected tumors from control- and HER3-DC1–treated mice and analyzed immune cell abundance by flow cytometry. Compared with control tumors, we observed significantly higher infiltration of CD4+ (P = 0.0449) and CD8+ (P = 0.0201) T cells in tumors from HER3-DC1–treated mice (Fig. 5C; Supplementary Fig. S8B). Within the CD4+ population, we noted significantly amplified abundance of CD62L+CD44+ CM (P = 0.0446) and CD62L−CD44+ EM (P = 0.0458), but not CD62L−CD44− effector T cells (Fig. 5D). We also observed significantly increased CD8+ T cells, specifically CD62L+CD44+ CM (P = 0.0131), and CD62L−CD44+ EM T cells (P = 0.033) in tumors from HER3-DC1–treated mice compared with the controls (Supplementary Fig. S8C). When CD4+ T cells were selectively depleted, tumor growth was comparably faster than the control group, and the antitumor effect of HER3-DC1 was completely abolished (Fig. 5E), highlighting the necessity of CD4+ T cells for functional activity of the class II peptide–pulsed HER3-DC1. When TILs from these mice were cocultured with DC1 pulsed with individual HER3 peptides, a significantly higher percentage of CD4+IFN-γ+ cells in HER3-DC1–treated TILs were noted in response to each of the HER3 peptides compared with the untreated controls. These data indicate HER3-specific IFN-γ secretion by CD4+ T cells following HER3-DC1 administration (Fig. 5F; Supplementary Fig. S8D). On the contrary, no statistically significant difference was noted in the percentage of CD8+IFN-γ+ cells between groups (Fig. 5G). In HER3-DC1–treated lymph nodes, we observed minimal, but not statistically significant, increase in the number of CD4+ and CD8+ T cells (Supplementary Fig. S8E). Together, these data suggest intratumoral HER3-DC1 can stimulate CD4+ immune responses, which, in turn, can activate CD8+ T cells.
Coculture of the lymph node immune cells with HER3 peptide–pulsed DC1 for 72 hours confirmed antigen specificity, with significantly augmented IFN-γ secretion in the HER3-DC1–treated group compared with controls (Fig. 5H). Similarly, elevated IFN-γ secretion was noted when HER3-DC1 splenocytes were restimulated with HER3 peptides, compared with the controls (Supplementary Fig. S8F).
Next, we examined whether HER3-DC1 intratumoral administration alters the molecular identity of TUBO tumor cells. Western blots on tumor cell lysates revealed downregulation of HER3 protein in HER3-DC1–treated tumors compared with controls. We also observed diminished phosphorylated AKT in HER3-DC1 tumors, along with an upregulation of apoptosis marker, cleaved caspase-3, compared to the control tumors (Fig. 5I). Downregulation of phosphorylated p44/42 MAPK in HER3-DC1 tumors was also noted, whereas no difference in HER3 and phospho-p44/42MAPK expression was observed in tumors from unpulsed DC1-treated mice compared to the controls (Fig. 5J). These data suggest HER3-DC1–induced Th1 immune responses can modulate molecular signaling to interfere with oncodriver expression and activation and induce tumor cell apoptosis.
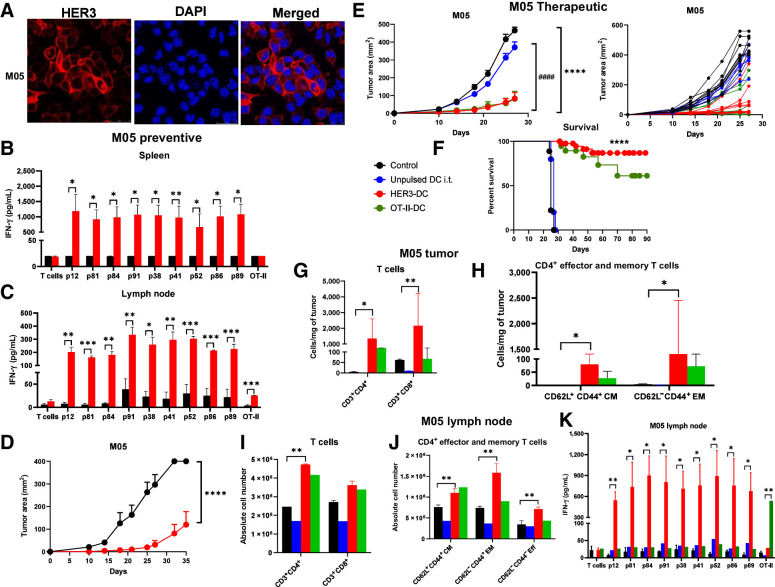
HER3-DC1 vaccination prevents tumor development in the M05 model
Because TCGA and cBioPortal analyses indicated high HER3 expression in skin cutaneous melanoma compared with normal tissue and a negative correlation between high HER3 expression and overall patient survival, we tested the preventive and therapeutic efficacy of HER3-DC1 in an M05 murine melanoma model, derived from the original B16 melanoma cell line transfected with the ovalbumin gene (36). Immunoblotting of M05 cell lysates showed moderate HER3 protein expression (Fig. 3B), whereas substantial surface expression of HER3 was detected by immunofluorescence (Fig. 6A). Therefore, we tested the preventive efficacy of HER3-DC1 in C57BL/6 mice. Compared with controls, splenocytes from HER3-DC1–vaccinated mice showed significantly enhanced peptide-specific immune responses when restimulated with HER3 peptides (Fig. 6B). Similarly, when immune cells isolated from the lymph nodes of vaccinated mice were cocultured with HER3 peptide–pulsed DC1 ex vivo, we observed significantly higher IFN-γ secretion compared with naïve controls (Fig. 6C). When naïve and vaccinated mice were challenged with M05 tumor 2 weeks after the last vaccination, tumor growth was significantly delayed, and survival was improved in vaccinated mice compared with naïve controls (P = 0.0004; Fig. 6D; Supplementary Fig. S8G). These data suggest HER3-DC1 vaccination induces peptide-specific, systemic immune responses in C57BL/6 mice.
Figure 6.
Vaccination prevents tumor development in a preventive HER3+ melanoma model and diminishes tumor growth and improves survival in a therapeutic HER3+ melanoma model. A, Immunofluorescence for HER3 (red) surface expression in M05 cells. DAPI (blue): nucleus (image magnification: 1,200×). B, HER3 peptide–specific immune responses (n = 3–5/group). Splenocytes from control (black) and vaccinated (red) mice were restimulated with individual HER3 peptides for 72 hours to detect IFN-γ by ELISA. C, Lymph node lymphocytes from control (black) and HER3-DC1 (red) vaccinated mice were cocultured with individual HER3 peptide–pulsed DCs to detect antigen-specific immune response in HER3-DC1 vaccinated mice by ELISA. D, Preventive model: Two weeks after the last vaccine, C57BL/6 mice (n = 10 mice/group) were challenged with M05 tumor cells. Tumor growth was monitored until endpoint in control (black) versus HER3-DC1 vaccinated (red) mice. E, Therapeutic setting: C57BL/6 mice were injected subcutaneously with the M05 murine melanoma cells in the left flank, and upon palpable tumor formation on day 10, mice were randomized into four groups (n = 10 mice/group). Tumor growth was monitored in mice receiving PBS (black), unpulsed DC1 (blue), HER3-DC1 (red), and OT-II peptide–pulsed DC1 (green) intratumorally once weekly for 6 weeks. Tumor growth was compared between control and HER3-DC1 (*) and unpulsed DC1 vs. HER3-DC1 (#) groups. Individual tumor growth curve for each mouse shown on the right. F, Percent survival in M05 mouse model for control (black)-, unpulsed DC1 (blue)–, HER3-DC1 (red)–, and OT-II-DC1 (green)–treated mice. G, Intratumoral infiltration of CD3+CD4+ and CD3+CD8+ T cells was compared between control (black) and HER3-DC1 (red) mice from E. H, CD4+ central memory (CD62L+CD44+ CM) and effector memory (CD62L−CD44+ EM) T-cell infiltration per milligram of tumors in control (black) versus HER3-DC1 (red) mice from E. I, Absolute number of CD3+CD4+ and CD3+CD8+ T cells in the lymph nodes of control- versus HER3-DC1–treated mice from E. J, Abundance of CD4+ CM, EM, and effector (Eff) T cells in the lymph nodes of control versus treated mice. For G–J, Unpulsed DC1 (blue) and OT-II-DC1 (green) groups were not included in the statistical analyses. K, Lymph node lymphocytes from control and treated mice (E) were cocultured with individual HER3 peptide–pulsed DC1 for 72 hours to detect IFN-γ by ELISA. Responses were compared between control and HER3-DC1 groups for HER3 peptides, and control versus OT-II-DC1 groups to OT-II peptide–pulsed DCs. Data shown are the representative from three independent experiments and are represented as mean ± SEM with statistical significance determined using multiple t test without correction for multiple comparisons. Each row was analyzed individually, without assuming a consistent SD. A log-rank (Mantel–Cox) test was used to determine differences between the survival curves. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001; ####, P ≤ 0.0001.
Intratumoral HER3-DC1 diminishes tumor growth in the M05 melanoma model
Next, we investigated the therapeutic efficacy of intratumoral HER3-DC1 treatment in a subcutaneous M05 murine melanoma model. An OT-II–pulsed DC1 group was included as a positive control, since the M05 cell line expresses ovalbumin. Compared with controls and unpulsed DC1-treated mice, HER3-DC1 treatment significantly delayed tumor growth (Fig. 6E; control vs. HER3-DC1, P ≤ 0.000001; unpulsed DC1 vs. HER3-DC1, P < 0.0001). HER3-DC1 treatment was comparable with OT-II-DC1 treatment in terms of tumor growth and tumor regression. We observed tumor regression in 20% of HER3-DC1–treated mice, whereas tumor growth was significantly slower in 40% of the remaining mice, with a steady growth in the rest. This resulted in significantly improved survival in HER3-DC1–treated mice compared with controls (P ≤ 0.0001, median survival control = 25 days, HER3-DC1 = undefined; Fig. 6F). We next assessed how intratumoral HER3-DC1 treatment modulated the tumor immune landscape. Like the murine mammary carcinoma models, we observed significantly increased CD4+ T-cell infiltration in HER3-DC1 mice compared with the controls (P = 0.0339). A significantly enhanced CD8+ T-cell infiltration was also noted in the HER3-DC1–treated group, which could contribute to the antitumor efficacy of the intratumoral HER3-DC1 observed (P = 0.0079; Fig. 6G). For CD4+ T-cell subsets, EM (P = 0.0208) and CM (P = 0.0199) populations were significantly enhanced in the HER3-DC1 group, with no differences in effector T-cell populations (Fig. 6H).
We investigated the systemic immune responses mediated by intratumoral HER3-DC1 treatment. Lymph node immune cells from control and treated mice were assessed for lymphoid markers. As previously seen in the breast cancer models, we observed significantly enhanced CD4+ T cells in HER3-DC1–treated mice (P = 0.0085), which was comparable to the positive control OT-II-DC1 group; however, no significant difference was noted in the frequency of CD8+ T cells (Fig. 6I). Within the CD4+ population, we noted significantly increased EM (P = 0.0058), CM (P = 0.0052), and effector (P = 0.0016) T cells (Fig. 6J). We evaluated IFN-γ secretion by immune cells residing in the lymph node of treated and control mice. As previously seen in the breast cancer models, we noted significantly enhanced IFN-γ secretion in the HER3-DC1 group compared with the control, when lymph node immune cells were cocultured with HER3 peptide–pulsed DCs (Fig. 6K). In the OT-II-DC1–treated group, a significant positive response was noted only when cocultured with the OT-II peptide–pulsed DC1 (P = 0.00567), but not the HER3 peptides, indicating the antigen specificity of the response. We also detected significantly higher IFN-γ secretion from splenocytes isolated from the HER3-DC1–treated M05 mice compared with controls (Supplementary Fig. S8H). Together, these data suggest HER3-DC1 induces potent antitumor immune response in a HER3+ murine melanoma model, resulting in tumor regression/delayed growth and improved survival by enhancing intratumoral and systemic CD4+ T cells and secretion of IFN-γ.
Discussion
HER3/ERBB3, a member of the ERBB family of growth receptors, has gained momentum as a therapeutic target, owing to its multifaceted role in tumor development, growth, and therapy resistance (1). In TCGA, HER3 expression was found to predict worse survival in melanoma patients. Interestingly, in response to BRAF/MEK inhibitor treatment, melanoma cells are shown to adapt and escape therapy by upregulating HER3 expression via the FOXP3 transcription factor (5). DC1 pulsed with promiscuous MHC class II HER3 peptides demonstrated a dramatic therapeutic effect in a murine melanoma model that expressed increased HER3, suggesting the possibility of clinically developing a HER3 immunotherapy in melanoma patients expressing HER3, especially those treated with BRAF/MEK inhibitors to possibly prevent escape through HER3.
We did not find a significant impact of high HER3 expression on breast cancer survival, which could be attributed to the small sample size. However, high HER3 expression has been previously identified as a prognostic marker of poor overall and disease-free survival in TNBC (10) and has been shown to contribute to therapy resistance (37). Upregulated HER3 expression and activation have also been documented as one of the primary compensatory mechanisms allowing therapy escape in HER2pos breast cancer via restimulation of the PI3K/AKT pathway (38). We utilized a HER2pos mammary tumor model resistant to HER2-targeted therapies, which was sensitive to HER3-mediated CD4+ Th1 immune response. This raises the possibility that HER3 peptides can be used to overcome resistant HER2 populations.
CD4+ T cells are critical players in regulating antitumor immune responses, and Th1 cells secreting IFN-γ contribute to elimination of tumors through multiple mechanisms, including antibody class switching, CD8+ T-cell help, modulation of innate effectors, such as macrophages and NK cells, and maturation of DCs (39). This study showed increased production of IFN-γ by CD4+ Th1 cells in both human CD4+ T cells ex vivo and in murine preclinical models of mammary carcinoma and melanoma. CD4+ T cells accumulated in the tumors of mice treated with HER3 peptide–pulsed DC1, suggesting a contribution of the CD4+ response in tumor regression. We validated the significance of CD4+ T cells in stimulating antitumor immune responses by class II peptide–pulsed DC1 treatment. HER3-DC1 intratumoral administration significantly enhanced CD4+ T-cell infiltration, and restimulation with HER3 peptide–pulsed DC resulted in increased intracellular IFN-γ secretion by those CD4+ TILs. This observation highlights the CD4-dependent mechanism of action of the HER3-DC1.
In the 4T1 and TUBO models, HER3-DC1 intratumoral injection and subsequent Th1 immune responses result in delayed tumor growth and downregulation of HER3 expression in tumor cells, which indicates possible correlation at the mechanistic level between HER3 expression and the antitumor effect. We observed significantly decreased total HER3 protein in the HER3-DC1 tumors obtained from both 4T1 and TUBO models, suggesting that intratumoral delivery of HER3-DC1 may induce cellular cross-talk downstream and inhibit HER3 protein expression, which contributes to tumor growth and proliferation. For TUBO tumors, we observed HER3-DC1 tumors had downregulation of phospho-AKT and phospho-p44/42MAPK, the primary signaling effectors downstream of HER3, suggesting HER3-DC1 may interfere with activation and function of HER3 in tumors. Also, prominent upregulation of the apoptosis marker cleaved caspase-3 indicated intratumoral cell death due to HER3-DC1. The Th1 cytokine IFN-γ can downregulate expression of HER family of receptor proteins in vitro (35) and, hence, HER3-DC1 treatment may reduce HER3 expression by CD4+ Th1 stimulation.
The peptide library screening method described here identified promiscuous class II epitopes that were not clearly predicted by current binding algorithms. Only ECD p91 was predicted to have high binding affinity, whereas none of the remaining eight peptides were indicated to be high-affinity binders. Previous reports have shown high rates of false negatives for immunogenic peptide epitopes that are intermediate- or low-affinity MHC binders (40–43). The MixMHCIIpred algorithm predicted strong binding affinity of all nine peptides identified empirically. However, it should be considered that binding affinity alone is not the only predictor of immunologic response generated by immunogenic peptides, and hence, the large number of random peptides generated by the algorithm that showed a strong binding affinity will still need to be tested individually and experimentally for immune responses in vitro, to avoid false-positive epitopes. Absence of defined cutoff percentile rank also leads to subjective interpretation of strong versus weak binding. Our identified peptides showed promiscuous binding across multiple MHC II HLA alleles, suggesting broad application across patients, whereas MixMHCIIpred algorithm showed variation of binding affinity of the peptides across HLA alleles. Lastly, the stark difference of predictive accuracy between the two algorithms used in this study further highlights the efficiency of our empirical approach to accurately identify immunologically responsive peptides. MHC I prediction algorithms have shown success in identifying immunogenic class I epitopes that can elicit potent CD8+ T-cell responses in vivo; however, the identification of MHC class II epitopes that activate CD4+ T cells has been less successful (24, 41–46). This discrepancy can be attributed to the highly polymorphic nature of MHC II molecules, which results in different binding patterns across the diverse HLA alleles (43–45, 47–49). Unlike MHC I, MHC II molecules consist of an open binding groove, allowing for longer peptides of 9–25 amino acid length to bind at different locations across the groove's open surface (43, 45, 46, 48). Our peptide screening attempted to circumvent several limitations in MHC II prediction algorithms to identify immunogenic class II epitopes for use in therapeutic development. First, a library of 15-mer peptides was generated for the protein of interest, with a 10-amino acid overlap between adjacent sequences. This design aimed to account for differences in the binding pattern across the MHC II open binding groove. Second, screening the full peptide library sequentially in pools of 10 peptides, 5 peptides, and individual peptides rapidly highlighted regions within the protein that may harbor immunogenic peptide sequences. Screening the full library essentially surveys all regions of the protein for epitope candidates, minimizing the probability of false negatives. Third, peptide candidates were screened across numerous donor samples to ensure the peptide-specific immune responses were reproducible and demonstrated promiscuous binding for widespread therapeutic application. Lastly, a key benefit to this methodology is that it takes an entirely experimental approach to epitope identification by recapitulating the in vivo processes of antigen processing and presentation, T-cell priming, and immune activation to isolate peptides capable of acting as competent epitopes in vitro and in vivo. Because this overlapping peptide library can be created and rapidly tested, it offers a novel avenue to MHC II epitope discovery against other known oncodrivers. We have, thus, begun to interrogate such oncodrivers for CD4+ Th1 peptide epitopes. Our study may also open the avenue for potential use of the DC vaccine platform to generate oncodriver-specific CD4+ T cells for novel adoptive T-cell therapies. In summary, using an overlapping 15-mer peptide library derived from the HER3 oncodriver, we identified promiscuous MHC II epitopes capable of driving an anti-HER3 CD4+ Th1 response that can have therapeutic impact in breast cancer and melanoma, and can be developed for cancer therapy in these diseases.
Authors' Disclosures
K.N. Kodumudi reports a patent for PCT/US2020/050689 pending to Moffitt Cancer Center. K.S.M. Smalley reports personal fees from Elsevier outside the submitted work. P.A. Forsyth reports personal fees from AbbVie Inc., Bayer, Bristol Myers Squibb, Boehringer Ingelheim, Inovio, NCI–NIH–NCRI, Novocure, Novellus, Physical Sciences Oncology Network, Tocagen, Ziopharm, and grants from CDMRP, Department of Defense, Moffitt Cancer Center of Excellence, NIH/NCI, Pfizer, and State of FL Bankhead Coley outside the submitted work. H. Han reports personal fees from Lilly and grants from Department of Defense outside the submitted work. H. Soliman reports personal fees from PUMA, Novartis, Eisai, Seattle Genetics, and AstraZeneca outside the submitted work. B.J. Czerniecki reports grants from CDMRP DOD during the conduct of the study; other support from ImmunoRestoration outside the submitted work; and a patent for identification of immunogenic MHC class II peptides for immune-based therapy (patent# 10,829,538) issued to the University of Pennsylvania. No disclosures were reported by the other authors.
Supplementary Material
Acknowledgments
This work was supported by Department of Defense Award W81XWH-19-1-0675, awarded to B.J. Czerniecki, P.A. Forsyth, P.C. Rodriguez, and P. Kalinski. This work was also supported by Department of Defense Award W81XWH-16-1-0385, awarded to B.J. Czerniecki and G.K. Koski, and Pennies in Action, awarded to B.J. Czerniecki and G.K. Koski. This work was supported in part by the Flow Cytometry Core, Analytic Microscopy Core Facility, Cell Therapies Core, and Vivarium Services Core at the H. Lee Moffitt Cancer Center and Research Institute, an NCI-designated Comprehensive Cancer Center (P30-CA076292).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Immunology Research Online (http://cancerimmunolres.aacrjournals.org/).
Authors' Contributions
A. Basu: Conceptualization, formal analysis, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. G.K. Albert: Conceptualization, formal analysis, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. S. Awshah: Validation, investigation, methodology, writing–review and editing. J. Datta: Conceptualization, formal analysis, validation, investigation, methodology, writing–review and editing. K.N. Kodumudi: Conceptualization, formal analysis, supervision, validation, investigation, visualization, methodology, project administration, writing–review and editing. C. Gallen: Investigation, methodology, writing–review and editing. A. Beyer: Investigation. K.S.M. Smalley: Writing–review and editing. P.C. Rodriguez: Writing–review and editing. D.R. Duckett: Writing–review and editing. P.A. Forsyth: Writing–review and editing. A. Soyano: Writing–review and editing. G.K. Koski: Writing–review and editing. R.L.B. Costa: Writing–review and editing. H. Han: Writing–review and editing. H. Soliman: Writing–review and editing. M.C. Lee: Writing–review and editing. P. Kalinski: Writing–review and editing. B.J. Czerniecki: Conceptualization, resources, supervision, funding acquisition, validation, writing–original draft, project administration, writing–review and editing.
References
- 1. Haikala HM, Janne PA. Thirty years of HER3: from basic biology to therapeutic interventions. Clin Cancer Res 2021;27:3528–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ocana A, Vera-Badillo F, Seruga B, Templeton A, Pandiella A, Amir E. HER3 overexpression and survival in solid tumors: a meta-analysis. J Natl Cancer Inst 2013;105:266–73. [DOI] [PubMed] [Google Scholar]
- 3. Gala K, Chandarlapaty S. Molecular pathways: HER3 targeted therapy. Clin Cancer Res 2014;20:1410–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pinkas-Kramarski R, Soussan L, Waterman H, Levkowitz G, Alroy I, Klapper L, et al. Diversification of Neu differentiation factor and epidermal growth factor signaling by combinatorial receptor interactions. EMBO J 1996;15:2452–67. [PMC free article] [PubMed] [Google Scholar]
- 5. Abel EV, Basile KJ, Kugel CH 3rd, Witkiewicz AK, Le K, Amaravadi RK, et al. Melanoma adapts to RAF/MEK inhibitors through FOXD3-mediated upregulation of ERBB3. J Clin Invest 2013;123:2155–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang Z, Karthaus WR, Lee YS, Gao VR, Wu C, Russo JW, et al. Tumor microenvironment-derived NRG1 promotes antiandrogen resistance in prostate cancer. Cancer Cell 2020;38:279–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bezler M, Hengstler JG, Ullrich A. Inhibition of doxorubicin-induced HER3–PI3K–AKT signalling enhances apoptosis of ovarian cancer cells. Mol Oncol 2012;6:516–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Recondo G, Bahcall M, Spurr LF, Che J, Ricciuti B, Leonardi GC, et al. Molecular mechanisms of acquired resistance to MET tyrosine kinase inhibitors in patients with MET exon 14-mutant NSCLC. Clin Cancer Res 2020;26:2615–25. [DOI] [PubMed] [Google Scholar]
- 9. Lee Y, Ma J, Lyu H, Huang J, Kim A, Liu B. Role of erbB3 receptors in cancer therapeutic resistance. Acta Biochim Biophys Sin 2014;46:190–8. [DOI] [PubMed] [Google Scholar]
- 10. Bae SY, Choi YL, Kim S, Kim M, Kim J, Jung SP, et al. HER3 status by immunohistochemistry is correlated with poor prognosis in hormone receptor-negative breast cancer patients. Breast Cancer Res Treat 2013;139:741–50. [DOI] [PubMed] [Google Scholar]
- 11. Ogden A, Bhattarai S, Sahoo B, Mongan NP, Alsaleem M, Green AR, et al. Combined HER3-EGFR score in triple-negative breast cancer provides prognostic and predictive significance superior to individual biomarkers. Sci Rep 2020;10:3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tao JJ, Castel P, Radosevic-Robin N, Elkabets M, Auricchio N, Aceto N, et al. Antagonism of EGFR and HER3 enhances the response to inhibitors of the PI3K–Akt pathway in triple-negative breast cancer. Sci Signal 2014;7:ra29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Laidlaw BJ, Craft JE, Kaech SM. The multifaceted role of CD4(+) T cells in CD8(+) T cell memory. Nat Rev Immunol 2016;16:102–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Basu A, Ramamoorthi G, Albert G, Gallen C, Beyer A, Snyder C, et al. Differentiation and regulation of TH cells: a balancing act for cancer immunotherapy. Front Immunol 2021;12:669474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Novy P, Quigley M, Huang X, Yang Y. CD4 T cells are required for CD8 T cell survival during both primary and memory recall responses. J Immunol 2007;179:8243–51. [DOI] [PubMed] [Google Scholar]
- 16. Bos R, Sherman LA. CD4+ T-cell help in the tumor milieu is required for recruitment and cytolytic function of CD8+ T lymphocytes. Cancer Res 2010;70:8368–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shedlock DJ, Shen H. Requirement for CD4 T cell help in generating functional CD8 T cell memory. Science 2003;300:337–9. [DOI] [PubMed] [Google Scholar]
- 18. Kumai T, Ohkuri T, Nagato T, Matsuda Y, Oikawa K, Aoki N, et al. Targeting HER-3 to elicit antitumor helper T cells against head and neck squamous cell carcinoma. Sci Rep 2015;5:16280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lowenfeld L, Mick R, Datta J, Xu S, Fitzpatrick E, Fisher CS, et al. Dendritic cell vaccination enhances immune responses and induces regression of HER2(pos) DCIS independent of route: results of randomized selection design trial. Clin Cancer Res 2017;23:2961–71. [DOI] [PubMed] [Google Scholar]
- 20. Fracol M, Datta J, Lowenfeld L, Xu S, Zhang PJ, Fisher CS, et al. Loss of anti-HER-3 CD4+ T-helper type 1 immunity occurs in breast tumorigenesis and is negatively associated with outcomes. Ann Surg Oncol 2017;24:407–17. [DOI] [PubMed] [Google Scholar]
- 21. Datta J, Rosemblit C, Berk E, Showalter L, Namjoshi P, Mick R, et al. Progressive loss of anti-HER2 CD4(+) T-helper type 1 response in breast tumorigenesis and the potential for immune restoration. Oncoimmunology 2015;4:e1022301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reynisson B, Barra C, Kaabinejadian S, Hildebrand WH, Peters B, Nielsen M. Improved prediction of MHC II antigen presentation through integration and motif deconvolution of mass spectrometry MHC eluted ligand data. J Proteome Res 2020;19:2304–15. [DOI] [PubMed] [Google Scholar]
- 23. Vita R, Overton JA, Greenbaum JA, Ponomarenko J, Clark JD, Cantrell JR, et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res 2015;43:D405–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Racle J, Michaux J, Rockinger GA, Arnaud M, Bobisse S, Chong C, et al. Robust prediction of HLA class II epitopes by deep motif deconvolution of immunopeptidomes. Nat Biotechnol 2019;37:1283–6. [DOI] [PubMed] [Google Scholar]
- 25. Moore TV, Nishimura MI. Improved MHC II epitope prediction—a step towards personalized medicine. Nat Rev Clin Oncol 2020;17:71–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rovero S, Amici A, Di Carlo E, Bei R, Nanni P, Quaglino E, et al. DNA vaccination against rat her-2/Neu p185 more effectively inhibits carcinogenesis than transplantable carcinomas in transgenic BALB/c mice. J Immunol 2000;165:5133–42. [DOI] [PubMed] [Google Scholar]
- 27. Falo LD, Kovacsovicsbankowski M, Thompson K, Rock KL. Targeting antigen into the phagocytic pathway in-vivo induces protective tumor-immunity. Nat Med 1995;1:649–53. [DOI] [PubMed] [Google Scholar]
- 28. Kodumudi KN, Weber A, Sarnaik AA, Pilon-Thomas S. Blockade of myeloid-derived suppressor cells after induction of lymphopenia improves adoptive T cell therapy in a murine model of melanoma. J Immunol 2012;189:5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cintolo JA, Datta J, Xu S, Gupta M, Somasundaram R, Czerniecki BJ. Type I-polarized BRAF-pulsed dendritic cells induce antigen-specific CD8+ T cells that impact BRAF-mutant murine melanoma. Melanoma Res 2016;26:1–11. [DOI] [PubMed] [Google Scholar]
- 30. Mishra R, Patel H, Alanazi S, Yuan L, Garrett JT. HER3 signaling and targeted therapy in cancer. Oncol Rev 2018;12:355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Majumder A, Sandhu M, Banerji D, Steri V, Olshen A, Moasser MM. The role of HER2 and HER3 in HER2-amplified cancers beyond breast cancers. Sci Rep 2021;11:9091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pulaski BA, Ostrand-Rosenberg S. Mouse 4T1 breast tumor model. Curr Protoc Immunol 2001;Chapter 20:Unit 20.2. [DOI] [PubMed] [Google Scholar]
- 33. Yang L, Li Y, Shen E, Cao F, Li L, Li X, et al. NRG1-dependent activation of HER3 induces primary resistance to trastuzumab in HER2-overexpressing breast cancer cells. Int J Oncol 2017;51:1553–62. [DOI] [PubMed] [Google Scholar]
- 34. Sergina NV, Rausch M, Wang D, Blair J, Hann B, Shokat KM, et al. Escape from HER-family tyrosine kinase inhibitor therapy by the kinase-inactive HER3. Nature 2007;445:437–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jia Y, Kodumudi KN, Ramamoorthi G, Basu A, Snyder C, Wiener D, et al. Th1 cytokine interferon gamma improves response in HER2 breast cancer by modulating the ubiquitin proteasomal pathway. Mol Ther 2021;29:1541–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu H, Weber A, Morse J, Kodumudi K, Scott E, Mullinax J, et al. T cell mediated immunity after combination therapy with intralesional PV-10 and blockade of the PD-1/PD-L1 pathway in a murine melanoma model. PLoS One 2018;13:e0196033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sinevici N, Ataeinia B, Zehnder V, Lin K, Grove L, Heidari P, et al. HER3 differentiates basal from claudin type triple negative breast cancer and contributes to drug and microenvironmental induced resistance. Front Oncol 2020;10:554704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chakrabarty A, Sanchez V, Kuba MG, Rinehart C, Arteaga CL. Feedback upregulation of HER3 (ErbB3) expression and activity attenuates antitumor effect of PI3K inhibitors. Proc Natl Acad Sci U S A 2012;109:2718–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Castro F, Cardoso AP, Goncalves RM, Serre K, Oliveira MJ. Interferon-gamma at the crossroads of tumor immune surveillance or evasion. Front Immunol 2018;9:847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mazor R, Tai CH, Lee B, Pastan I. Poor correlation between T-cell activation assays and HLA-DR binding prediction algorithms in an immunogenic fragment of Pseudomonas exotoxin A. J Immunol Methods 2015;425:10–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dhanda SK, Karosiene E, Edwards L, Grifoni A, Paul S, Andreatta M, et al. Predicting HLA CD4 immunogenicity in human populations. Front Immunol 2018;9:1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Soria-Guerra RE, Nieto-Gomez R, Govea-Alonso DO, Rosales-Mendoza S. An overview of bioinformatics tools for epitope prediction: implications on vaccine development. J Biomed Inform 2015;53:405–14. [DOI] [PubMed] [Google Scholar]
- 43. Wang P, Sidney J, Dow C, Mothe B, Sette A, Peters B. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput Biol 2008;4:e1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jurtz V, Paul S, Andreatta M, Marcatili P, Peters B, Nielsen M. NetMHCpan-4.0: improved peptide-MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J Immunol 2017;199:3360–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chaves FA, Lee AH, Nayak JL, Richards KA, Sant AJ. The utility and limitations of current Web-available algorithms to predict peptides recognized by CD4 T cells in response to pathogen infection. J Immunol 2012;188:4235–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Andreatta M, Trolle T, Yan Z, Greenbaum JA, Peters B, Nielsen M. An automated benchmarking platform for MHC class II binding prediction methods. Bioinformatics 2018;34:1522–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kropshofer H, Singer T. Overview of cell-based tools for preclinical assessment of immunogenicity of biotherapeutics. J Immunotoxicol 2006;3:131–6. [DOI] [PubMed] [Google Scholar]
- 48. Nielsen M, Lund O, Buus S, Lundegaard C. MHC class II epitope predictive algorithms. Immunology 2010;130:319–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang L, Udaka K, Mamitsuka H, Zhu S. Toward more accurate pan-specific MHC-peptide binding prediction: a review of current methods and tools. Brief Bioinform 2012;13:350–64. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



![Figure 1. HER3 expression in cancer, and HER3 peptide screening of ECD and ICD class II peptide libraries. A, Expression of HER3 mRNA in normal (N) and tumor (T) tissues obtained from RNA-seq data from the GDC across cancer types (see Materials and Methods). B, Correlation between percentage of HER3 expression and overall patient survival (in months) in breast cancer. Samples were sorted in the descending order of HER3 expression and put into two groups: high HER3 (red) and low HER3 (blue). C, Correlation of overall patient survival with high HER3 (red) versus low HER3 (blue) expression in melanoma. P value indicated in individual graphs. D and E, IFN-γ production at each screening step for sample 1 [normal donor (ND) 8; D] and sample 2 (ND 9; E) when stimulated with ECD peptides. F and G, IFN-γ production at each screening step for sample 3 (ND 3; F) and sample 4 (ND 5; G) upon stimulation with ICD peptides. D–G, IFN-γ response to negative peptide control (black) compared with HER3 peptides (red) with an immunogenic response threshold of ≥1.5-fold increase. Data represented as mean ± SEM with statistical significance determined using a multiple t test without correction for multiple comparisons. Each row was analyzed individually, without assuming consistent SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/314e/9414303/59579a6903d7/108fig1.jpg)