Prostate cancers that benefit most from PARP inhibition have BRCA2 alterations, with BRCA2 homozygous loss associating with the longest benefit; biallelic loss of PALB2 and ATM can also sensitize to PARP inhibition.
Abstract
PARP inhibitors are approved for treating advanced prostate cancers (APC) with various defective DNA repair genes; however, further studies to clinically qualify predictive biomarkers are warranted. Herein we analyzed TOPARP-B phase II clinical trial samples, evaluating whole-exome and low-pass whole-genome sequencing and IHC and IF assays evaluating ATM and RAD51 foci (testing homologous recombination repair function). BRCA1/2 germline and somatic pathogenic mutations associated with similar benefit from olaparib; greater benefit was observed with homozygous BRCA2 deletion. Biallelic, but not monoallelic, PALB2 deleterious alterations were associated with clinical benefit. In the ATM cohort, loss of ATM protein by IHC was associated with a better outcome. RAD51 foci loss identified tumors with biallelic BRCA and PALB2 alterations while most ATM- and CDK12-altered APCs had higher RAD51 foci levels. Overall, APCs with homozygous BRCA2 deletion are exceptional responders; PALB2 biallelic loss and loss of ATM IHC expression associated with clinical benefit.
Significance:
Not all APCs with DNA repair defects derive similar benefit from PARP inhibition. Most benefit was seen among patients with BRCA2 homozygous deletions, biallelic loss of PALB2, and loss of ATM protein. Loss of RAD51 foci, evaluating homologous recombination repair function, was found primarily in tumors with biallelic BRCA1/2 and PALB2 alterations.
This article is highlighted in the In This Issue feature, p. 2659
Introduction
Metastatic castration-resistant prostate cancer (mCRPC) is enriched for genomic alterations in DNA damage repair (DDR) pathways, including homologous recombination repair (HRR) genes (1–5). DDR gene mutations (DDRm) can render mCRPC vulnerable to PARP inhibitors (PARPi; refs. 6–8). The PARPi olaparib recently received regulatory approval for the treatment of mCRPC with several DDR gene mutations, based on the results of the PROfound randomized phase III clinical trial (9, 10).
We and others have previously reported on phase II trials of different PARPi in men with mCRPC with DDRm; across these studies, BRCA2 alterations associate with higher radiologic and PSA response rates and longer progression-free survival (11–14). Similarly, subgroup analyses of the PROfound trial indicate that patients with BRCA alterations achieved the most benefit compared with patients with alterations in other genes such as ATM or CDK12 (9, 10) While collectively these trials support implementing mCRPC molecular stratification in clinical practice, further clinical qualification is needed for a more precise understanding of PARPi sensitivity in mCRPC (15, 16).
TOPARP is an investigator-initiated adaptive phase II clinical trial evaluating the antitumor activity of single-agent olaparib in mCRPC (ClinicalTrials.gov NCT01682772). The results of the second stage of this trial, TOPARP-B, confirmed the antitumor activity of olaparib in mCRPC with various DDRm (11). In TOPARP-B, patients were prospectively screened for DDRm utilizing an investigational targeted next-generation sequencing (NGS) panel. Herein, we pursued deeper molecular characterization of acquired samples, including whole-exome sequencing (WES) and low-pass whole-genome sequencing (lpWGS), and IHC and immunofluorescence (IF) assays, aiming to identify molecular features that can refine the predictive biomarker suite for patient stratification and to identify patients achieving major benefit from PARPi treatment.
Results
Patient Population and Sample Disposition for Extended Molecular Analysis
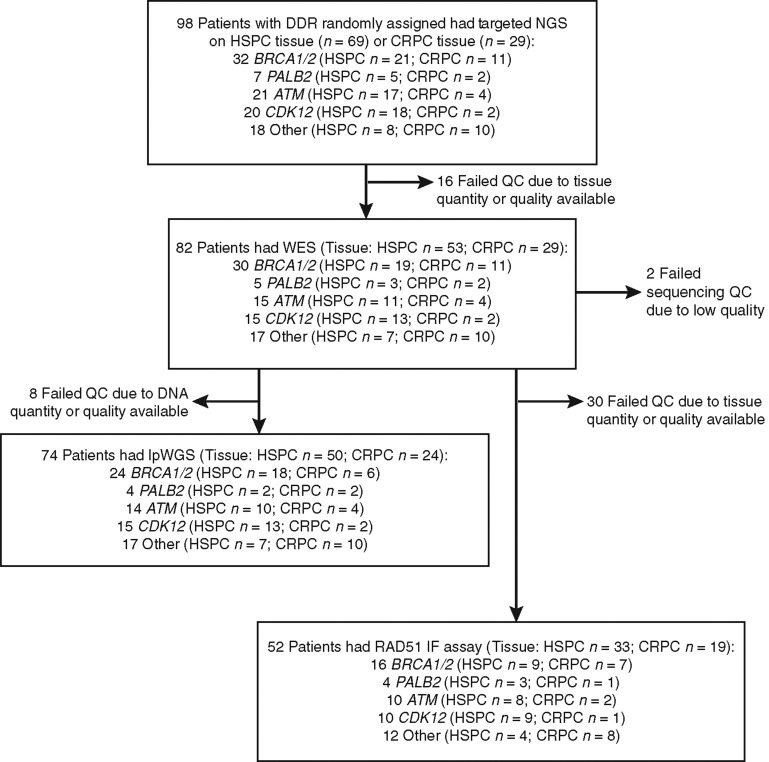
Overall, 98 men received olaparib on TOPARP-B; patient characteristics and clinical outcomes have been previously reported (Supplementary Table S1). In 69 men, a hormone treatment–naïve prostate cancer (HNPC) diagnostic biopsy was used for NGS testing, while in 29 men, a fresh mCRPC biopsy was utilized (Fig. 1). With an additional 14 months of follow-up since the primary report, 97 of 98 patients had experienced an event for radiographic progression-free survival (rPFS) analysis, with 93 of 97 patients having died. Median follow-up for those still alive was 29 months. Median rPFS and overall survival (OS) were 5.5 [95% confidence interval (95% CI), 4.6–7.5 months] and 12.8 (95% CI, 9.9–16.6) months, respectively. The BRCA1/2-altered subgroup (n = 32) had the longest median rPFS (8.4 months; 95% CI, 5.5–14.0) and OS (17.7 months; 95% CI, 9.0–22.2; Supplementary Table S2; Supplementary Fig. S1). WES was performed for 82 cases on available material from tumor biopsies (53 HNPC samples and 29 mCRPC samples) used for targeted NGS (trial prescreening phase; 2 failed sequencing quality control due to low-quality data); for the remaining cases, there was no spare tissue/DNA left, or the DNA did not pass quality controls for WES. For 74 cases, remaining DNA was also sufficient for lpWGS performed for orthogonal copy-number profiling. Predefined “qualifying” alterations detected by tumor NGS classified mCRPC into one of five subgroups: BRCA1/2, ATM, CDK12, PALB2, and “Other.” The qualifying event was an alteration detectable in germline DNA in 30 (30.6%), not detectable in germline DNA in 52 (53.1%), and a tumor homozygous deletion in 16 patients (16.3%). On the basis of the integration of targeted, WES, and lpWGS data, we identified biallelic events in the genes of interest in 64 of 98 (65.3%) cases (Supplementary Table S3). Overall, these analyses provide the deepest interrogation, to date, of mCRPC genomics in a prospective trial of PARP inhibition.
Figure 1.
Consort diagram showing the sample disposition in this study. HSPC, hormone-sensitive prostate cancer; QC, quality control.
Patterns of Response in BRCA-Altered Prostate Cancer
Of the 32 patients in the BRCA1/2 subgroup (BRCA1 n = 2, BRCA2 n = 30) identified by targeted NGS, 13 had a germline mutation and 19 a tumor-only pathogenic alteration (BRCA1 mutation n = 1, BRCA2 mutation n = 7, BRCA2 homozygous deletion n = 11). The composite response rates [primary endpoint of the TOPARP trial, including confirmed radiologic responses, PSA responses, or circulating tumor cell (CTC) count conversions] to olaparib in these BRCA1/2 subgroups were similar for those with germline mutations (10/13, 77%) and those with alterations detected only in tumor DNA (16/19, 84%; Table 1 depicts the composite response rate and the individual components).
Table 1.
Responses to olaparib (composite trial endpoint and per each of the components) based on the origin and type of the qualifying genomic alterations
| All patients, by type of response | Composite overall response by gene subgroup | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Composite overall response | RECIST 1.1 objective response | PSA fall ≥50% | CTC conversion | RECIST 1.1 or PSA response | BRCA1/2 | PALB2 | ATM | CDK12 | ||||||||||
| Resp/n | RR | Resp/n | RR | Resp/n | RR | Resp/n | RR | Resp/n | RR | Resp/n | RR | Resp/n | RR | Resp/n | RR | Resp/n | RR | |
| All randomized patients | 45/98 | 45.9 | 14/75 | 18.7 | 31/95 | 32.6 | 29/57 | 50.9 | 33/98 | 33.7 | 26/32 | 81.3 | 4/7 | 57.1 | 8/21 | 38.1 | 5/21 | 23.8 |
| By origin/type alteration | ||||||||||||||||||
| Germline mutation | 19/30 | 63.3 | 6/22 | 27.3 | 12/28 | 42.9 | 11/19 | 57.9 | 13/30 | 43.3 | 10/13 | 76.9 | 4/6 | 66.7 | 4/5 | 80 | 1/1 | 100 |
| Somatic homozygous deletion | 10/16 | 62.5 | 4/11 | 36.4 | 10/16 | 62.5 | 7/11 | 63.6 | 10/16 | 62.5 | 9/11 | 81.8 | 0/0 | 0 | 1/1 | 100 | 0 | 0 |
| Somatic mutation | 16/52 | 30.8 | 4/42 | 9.5 | 9/51 | 17.6 | 11/27 | 40.7 | 10/52 | 19.2 | 7/8 | 87.5 | 0/1 | 0 | 3/15 | 20 | 4/20 | 20 |
| Based on the evidence of biallelic loss | ||||||||||||||||||
| Biallelic hit detected | 33/64 | 51.6 | 10/47 | 21.3 | 24/64 | 37.5 | 22/39 | 56.4 | 25/64 | 39.1 | 19/24 | 79.2 | 4/4 | 100 | 5/12 | 41.7 | 4/18 | 22.2 |
| Monoallelic hit detected | 12/34 | 35.3 | 4/28 | 14.3 | 7/31 | 22.6 | 7/18 | 38.9 | 8/34 | 23.5 | 7/8 | 87.5 | 0/3 | 0 | 3/9 | 33.3 | 1/3 | 33.3 |
Abbreviations: n, patients available; Resp, number of responses; RR, response rate (%).
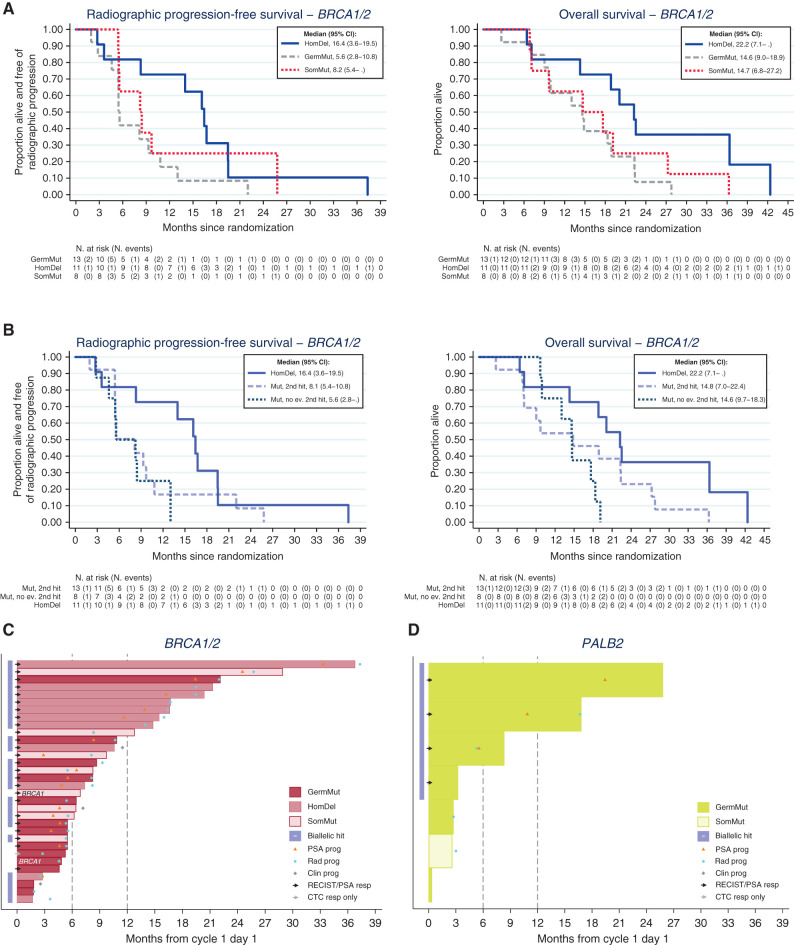
Overall, in BRCA1/2 patients, biallelic events were detected in 8 of 13 (62%) patients with germline mutations and in 16 of 19 (84%) patients with tumor-only alterations (5/8 with tumor-only mutations and all 11 with homozygous deletions that are by definition biallelic). Tumors with BRCA2 homozygous deletions (n = 11) had the best outcomes in the BRCA cohort with a median rPFS of 16.4 months versus 5.6 months for patients with deleterious BRCA1/2 germline mutations (n = 13) and versus 8.2 months for patients with BRCA1/2 somatic mutations only (n = 8; Table 1; Fig. 2A and B). Overall, 7 of 11 patients with a BRCA2 homozygous deletion were on trial for more than 1 year, with the longest responder experiencing disease progression after more than 3 years of olaparib (Fig. 2C). Median overall survival was 22.2 months from starting olaparib for the BRCA2 homozygous deletion cohort (n = 11), compared with 14.7 months for patients with deleterious BRCA1/2 germline mutations (n = 13) and 14.6 months for patients with BRCA1/2 somatic mutations only (n = 8).
Figure 2.
Outcomes with olaparib in the BRCA1/2 and PALB2 cohorts. A, Kaplan–Meier curves showing rPFS and OS in BRCA1/2 cohort depicting homozygous deletions, germline and somatic mutations: Tumors with homozygous BRCA deletions have the best outcomes. B, Kaplan–Meier curves rPFS and OS in BRCA1/2 cohort depicting outcomes in homozygous deletions and mutated genes with or without a detectable second hit. C, Swimmer plots depicting time on treatment per origin/type alterations in BRCA1/2 patients. D, Swimmer plots depicting time on treatment per origin/type alterations in PALB2 patients.
Considering all patients with biallelic loss (n = 24, mutation with a detectable second event and homozygous deletions), the median rPFS and median OS for patients with BRCA1/2 biallelic loss was 9.7 and 18.9 months, respectively, compared with a median rPFS and median OS of 5.6 and 14.6 months for those without detectable biallelic loss (n = 8, Table 1; Fig. 2B).
These data suggest that most tumors with BRCA mutations are likely to have biallelic loss, even if the most commonly used NGS assays may miss detecting some events leading to complete loss-of-function, such as complex rearrangements. Yet, our data largely refer to BRCA2 alterations, as only 2 patients had BRCA1 alterations. Patients with mCRPC with BRCA2 homozygous deletion had superior rPFS and OS outcomes from PARP inhibition, suggesting that olaparib resistance may be harder to evolve in these tumors.
Molecular Profiles in Patients Responding to PARPi beyond the BRCA Subgroup
PALB2
Among 7 patients with PALB2 mutations, 6 had germline mutations, of which 4 of 6 had a detectable second hit inducing biallelic loss. Interestingly, all 4 of 4 patients responding to olaparib in TOPARP-B, according to the composite response definition in the trial, had a germline mutation with evidence of biallelic loss, whereas in all 3 of 3 nonresponders, there was no evidence of a second detectable event based on all the NGS data analyses conducted (Table 1; Fig. 2D).
ATM
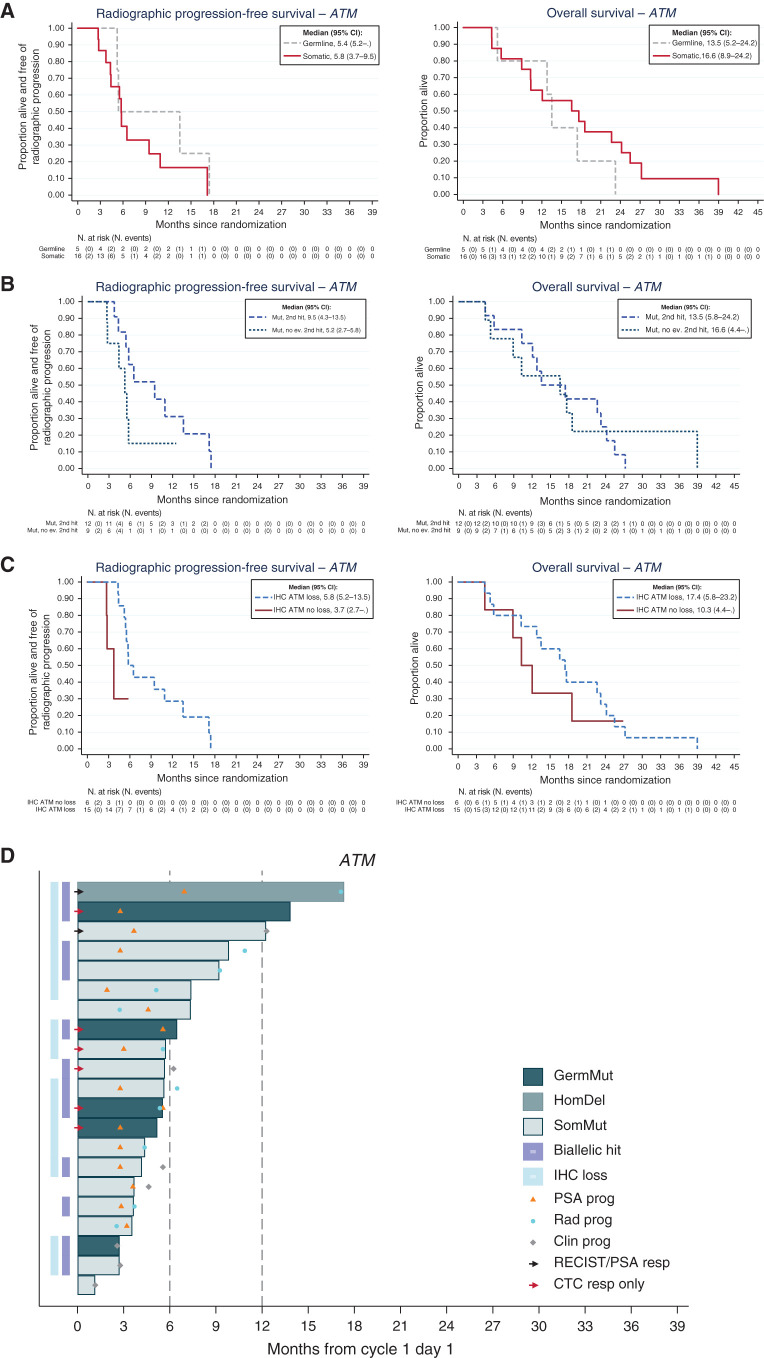
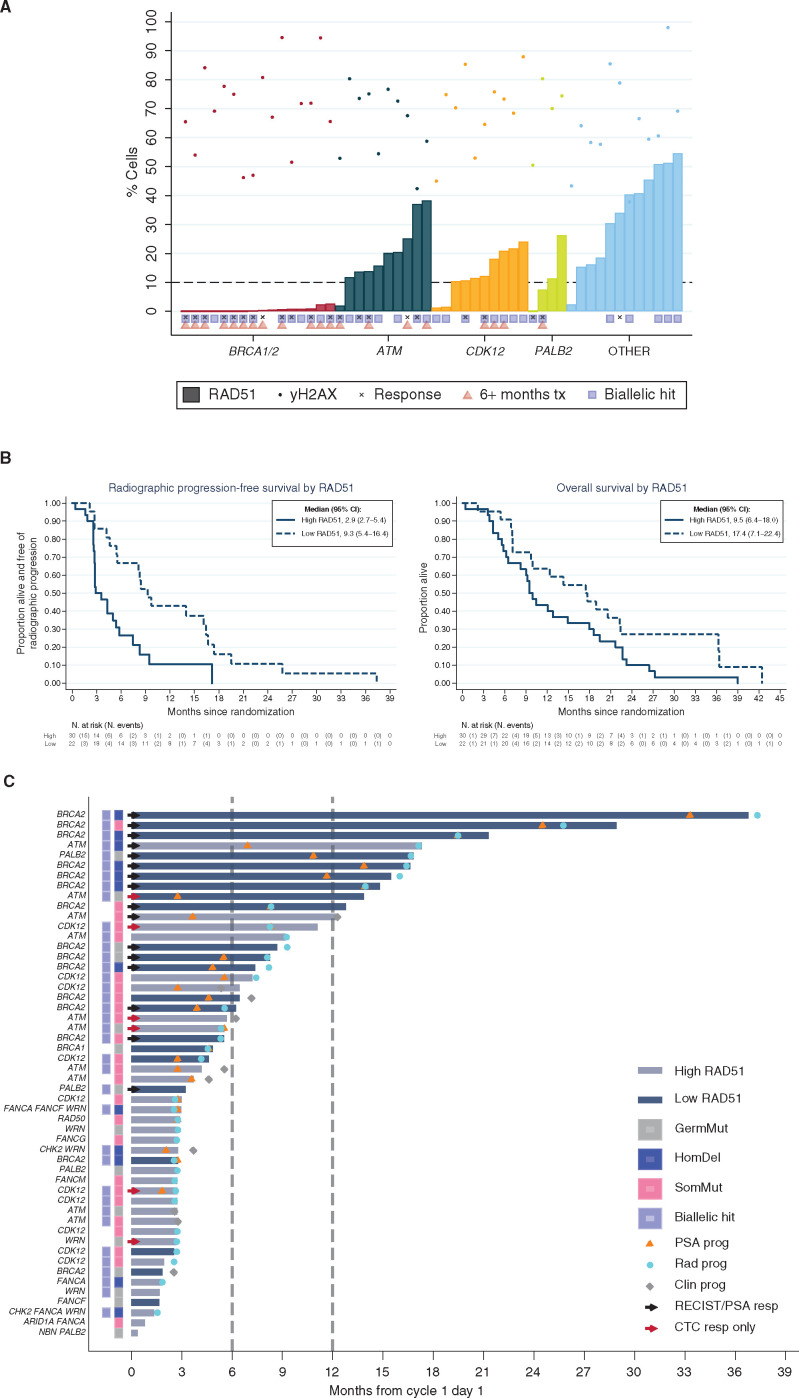
Overall, 21 men treated on TOPARP-B had ATM-altered tumors; most of these ATM aberrations were only detected in tumors (15 somatic mutations, 1 homozygous deletion), whereas 5 patients had ATM germline mutations (Fig. 3A; Supplementary Tables S1 and S3). Of 21 cases, 12 (57.1%) ATM-altered tumors had a detectable second event; these ATM-altered cases predicted to have tumor biallelic loss cases had longer rPFS (median 9.5 vs. 5.2 months; Fig. 3B), but this did not translate into improved OS (median 13.5 vs. 16.6 months; Fig. 3B). ATM protein expression by IHC was completely lost in 15 of these 21 (71%) tumors; however, 5 of these 15 IHC-negative tumors with ATM mutations had no detectable second genomic event that would cause biallelic loss (Supplementary Fig. S2; Supplementary Table S4). Interestingly, all 5 patients with germline mutations had tumors with ATM loss on IHC (compared with 10 of 16 with tumor-only mutations), with 4 of these 5 meeting the definition of response based on the composite trial endpoint compared with 4 of 16 with tumor-only mutations (Table 1). Moreover, in this ATM cohort, ATM loss of expression by IHC associated with longer rPFS (median 5.8 months vs. 3.7 months) and OS (median 17.4 months vs. 10.3 months; Fig. 3C; Supplementary Table S4). Overall, these data indicate that ATM mutations are not always associated with biallelic loss, and that ATM loss of IHC expression associates with better outcomes on olaparib, although it remains possible that other background genomic alterations in these tumors are required to sensitize to PARP inhibition (Fig. 3D).
Figure 3.
Outcomes with olaparib in ATM-altered prostate cancer indicating that complete ATM loss associates with better outcome on PARP inhibition. A, Kaplan–Meier curves depicting rPFS and OS outcomes in the prostate cancer cohort treated with olaparib and germline and somatic mutations. B, Kaplan–Meier curves depicting rPFS and OS outcomes with olaparib treatment in prostate cancers with and without second detectable genomic hits on the ATM gene. C, Kaplan–Meier curves depicting rPFS and OS outcomes in patients with prostate cancer treated with olaparib in the ATM cohort with and without ATM loss by IHC. D, Swimmer plots depicting time on treatment per origin/type alterations in the ATM gene.
CDK12
All detected CDK12 alterations were restricted to tumor-only mutations; interestingly, in 18 of 20 tumors, a second event in the same gene was detected, with most of these alterations being biallelic missense or truncating mutations. Supplementary Figure S3 summarizes outcomes on the TOPARP-B trial for patients in this CDK12 subgroup; 5 subjects were on treatment for 6 months or more, although in the majority of these olaparib was continued despite PSA progression. Overall, these data indicate that despite many CDK12-altered tumors appearing to have biallelic events, olaparib has limited antitumor activity by established response criteria in this cohort.
Other
Supplementary Figure S3 also depicts the patients whose tumors were categorized in the “Other” cohort that incorporated multiple, less common, remaining gene alterations. Three patients in this cohort were on olaparib >6 months and had deleterious CHK2, ATRX, and FANCA aberrations, with none of these having PSA progression during the 6-month period and two remaining on olaparib (CHK2, ATRX) for more than a year. Interestingly, exome-sequencing data identified concurrent deleterious PPP2R2A frameshift and POLA2 mutations in the CHK2-mutated tumor, while the ATRX-mutated tumor also had a ZMYM3 mutation that may also have affected PARPi sensitivity. No significant antitumor activity was observed among patients with germline mutations in these “Other” genomic aberrations. Overall, further data are required on the impact of CHK2, ATRX, FANCA, and other rare genomic alterations on PARP inhibition sensitivity.
Genomics of mCRPC with DDR Alterations on TOPARP-B
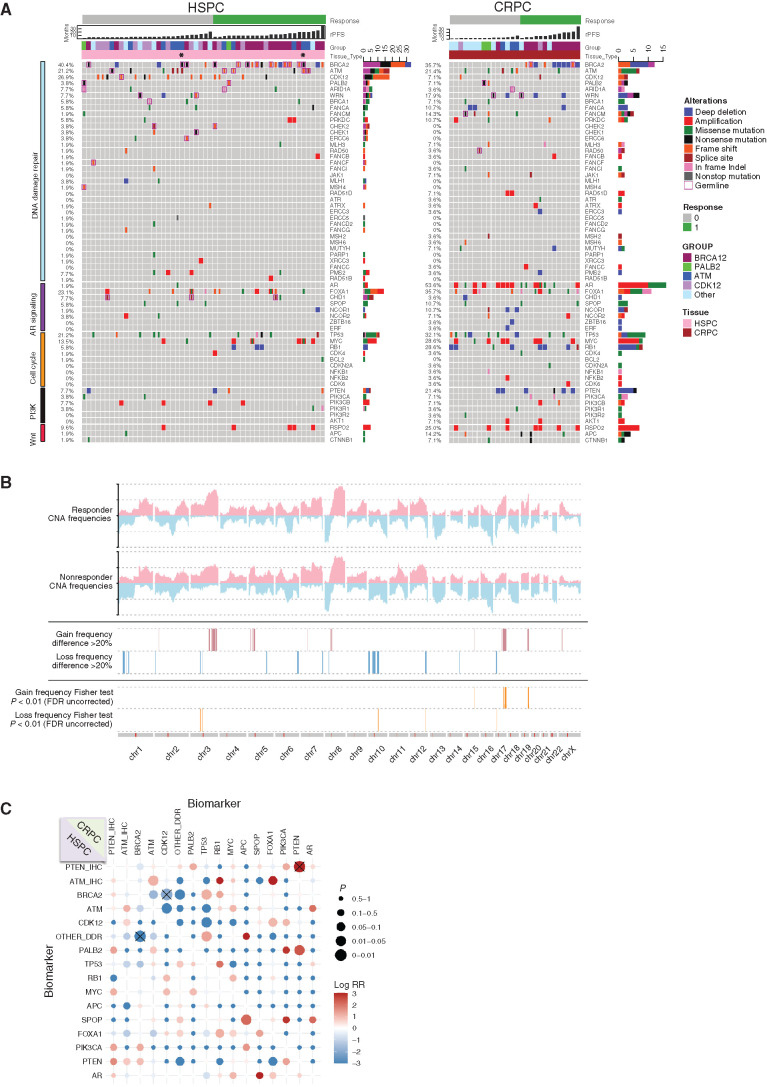
Figure 4A depicts TOPARP-B prostate cancer genomic profiles by WES ranked by binary response assessment using the predefined composite endpoint, as well as by rPFS. The genomics of diagnostic, archival samples are presented separate to those of mCRPC biopsies. Overall, genomic alterations in AR (54% vs. 2%), TP53 (mainly mutations; 32% vs. 21%), MYC (mainly amplification; 28.6% vs. 13.5%), RB1 (mainly deletions; 28.6% vs. 5.8%), PTEN (21% vs. 8%), WNT pathway aberrations including APC (14% vs. 2%) and CTNNB1 (7% vs. 2%) were more common in CRPC biopsies than diagnostic, pretreatment samples but appeared similar to that previously reported for molecularly unselected lethal prostate cancer (17, 18). Exploratory analyses comparing genomic copy-number data in responders and nonresponders identified a significant enrichment for specific genomic loci in responders (Fig. 4B), including chromosome 3q amplification in BRCA1/2-altered tumors (P < 0.01; Supplementary Fig. S4A). In the non-BRCA cohorts, those tumors responding to olaparib had significant enrichment for chromosome 15 and 19 loci gains and focal chromosome 10 locus loss (Supplementary Fig. S4B), which all also include multiple genes implicated in DNA repair (Supplementary Table S5) that warrant further study. Furthermore, because it has been suggested that some DNA repair defects co-occur with other alterations impacting PARPi sensitivity, we interrogated cooccurrence or mutual exclusivity for common prostate cancer genomic alterations in this TOPARP-B cohort; in tumors with BRCA2 and CDK12 alterations, we observed trends toward mutual exclusivity for other DDR alterations, and no significant association between ATM and TP53 alterations (Fig. 4C). Overall, these exploratory data suggest that the identified genomic loci associating with PARPi sensitivity warrant further study in independent-validation PARPi clinical trials and if validated, genes in these loci will merit further functional study.
Figure 4.
Genomic landscape of the TOPARP-B cohort. A, Oncoprint of the prostate cancer biopsies of the patients treated with olaparib on the TOPARP-B trial, separating those cases where a treatment-naïve versus castration-resistant biopsy was used in the trial for NGS. B, Copy number variation frequency plots of the APCs in the TOPARP-B patients and significant differences in the genomic copy-number profile between responders and nonresponders. C, Co-occurrence and mutually exclusive alterations plot for prostate cancer–associated genes. HSPC, hormone-sensitive prostate cancer.
Loss of RAD51 Foci as a Functional Biomarker of HRR in mCRPC
Finally, we studied γH2AX/geminin (GMN) and RAD51/GMN foci by IF in the 52 cases for whom tumor tissue from the same biopsy used for NGS was available (Supplementary Fig. S5A). In all 52 cases, γH2AX foci were detected in >40% of GMN-positive cells; inter-reader variability was low (Supplementary Fig. S5B). Overall, 22 of 52 (42%) cases were scored as RAD51 “low,” using a predefined cutoff of 10% of cells having ≥5 nuclear RAD51 foci (19–21). All 16 tested (16/16; 100%) prostate cancers with deleterious BRCA1/2 alterations had low RAD51 scores; this also included all the tumors arising with and without germline mutations and regardless of having detected a biallelic loss (Fig. 5A). Of the 4 tumors with PALB2 mutations evaluated for RAD51 foci, the 2 with low RAD51 scores were responders in the trial; both had biallelic loss; neither of the two patients with high RAD51 scores responded to olaparib with neither of these having biallelic loss (Fig. 5A). Moreover, low RAD51 foci scores associated with response to olaparib: 15 of 22 (68.2%) patients with prostate cancers with low RAD51 foci scores were responders by the trial composite response primary endpoint, compared with 7 of 30 (23.3%) patients with tumors with high RAD51 scores. Patients with low RAD51 foci scores also had longer rPFS (median 9.3 vs. 2.9 months) and OS (median 17.4 vs. 9.5 months) from initiation of olaparib therapy when compared with those with high RAD51 foci scores (Fig. 5B). These data support, for the first time, the validity of the RAD51 assay in mCRPC.
Figure 5.
Loss of RAD51 as a functional marker of HRR deficiency in prostate cancer and PARPi sensitivity. A, Percentage of GMN-positive cells positive for RAD51 and γH2AX foci per patient, sorted on the basis of the predefined subgroups per genes of interest; 10% of GMN-positive cells positive for RAD51 foci was used as the threshold to classify samples as RAD51 low versus high. B, Kaplan–Meier curves depicting rPFS and OS depending on the RAD51 assay. C, Swimmer plots depicting time on treatment in patients with high (gray) versus low (blue) RAD51 scores. Clin prog, clinical progression; PSA prog, PSA progression; Rad prog, radiologic progression.
Interestingly, ATM- and CDK12-altered prostate cancers had lower RAD51 foci scores than the prostate cancers in the “Other” DNA repair genes cohort, although these scores were higher than those in the BRCA cohort (Fig. 5A). In ATM-mutated tumors, RAD51 foci scores had a median score of 18% (interquartile range: 14–25) compared with tumors in the “Other” DDR repair gene category that had a median score of 34% (interquartile range of 16–46). Full details of the per-patient outcomes of all the TOPARP-B patients for whom sufficient tumor tissue was available for conducting this HRR function IF assay are depicted in Fig. 5C. Overall, these data indicate that RAD51 scoring identifies all BRCA1/2-mutated tumors and tumors with biallelic PALB2 loss, although not all tumors with a low RAD51 score respond to olaparib. These findings also suggest that some ATM- and CDK12-altered tumors have relatively low RAD51 scores but these are higher than those in BRCA1/2 gene prostate cancers.
Discussion
The TOPARP trial was the first to demonstrate the antitumor activity of PARP inhibition in a subset of prostate cancers with DNA repair defects (11, 14); olaparib has now been granted regulatory approval for treating mCRPC with specific, selected, DNA repair gene alterations pre- and post-chemotherapy after one next-generation hormonal agent based on the PROfound trial data (9, 10). Rucaparib has also been approved by the FDA to treat mCRPC with BRCA1/2 pathogenic alterations postchemotherapy in the United States (13). The approvals of olaparib by the FDA, and the European Medicines Agency (EMA), are quite different, however, with olaparib being approved for mCRPC with genomic alterations in 14 different DNA repair genes by the FDA but being approved only for BRCA-related cancers by the EMA. This discordance in approvals underlines why further study of biomarkers that predict clinical benefit to PARP inhibition in this disease is warranted.
In this deeper study of tissues prospectively acquired for biomarker studies in the TOPARP-B trial utilizing exome, low-pass whole-genome, and IHC studies, we now discover multiple clinically important findings that can affect patient care. First, we show that most APCs with a detectable BRCA2 alteration have biallelic loss. BRCA1/2-altered mCRPC cases all had low RAD51 scores in keeping with loss of HRR function, even for those cases where we could not detect a second inactivating event with our integrative NGS approach. Our data therefore indicate that detection of a monoallelic pathogenic mutation in BRCA2 should suffice to select patients for PARPi treatment, even in the absence of having detected a second hit by NGS, as indeed targeted panels implemented in clinical practice may not detect some events that could lead to loss-of-function, such as complex rearrangements of copy-neutral loss of heterozygosity, particularly in challenging samples with low tumor content. Moreover, we observed that BRCA2 mutations associated with similarly high response rates regardless of the germline versus somatic origin of the mutation. Of note, 30 patients in the trial had BRCA2 alterations, compared with two with BRCA1 mutations; consequently, our data for the BRCA1/2 cohort is largely related to BRCA2 alterations and extrapolation to BRCA1 alterations, which are infrequent in prostate cancer, should be avoided (22, 23).
We and others have previously reported significant variability in terms of duration of benefit in patients with mCRPC with BRCA1/2 alterations receiving olaparib or other PARPi. We hypothesized that the exact type of gene alteration may associate with different magnitudes of benefit. We now have compelling evidence that indicates that mCRPC with BRCA2 homozygous deletions have substantially longer response durations compared with tumors with frameshift or stop-gain mutations (regardless of these being germline or somatic in origin). Impressively, tumors with homozygous deletions had a median rPFS of 16.4 months, which is a considerable period of disease control for this late-stage mCRPC setting. We and others have previously reported how prostate tumors (like other cancers) with truncating mutations in BRCA1/2, PALB2, or RAD51 accumulate secondary reversion mutations while on PARPi that restore the DNA repair gene reading frame back to normal, converting a truncating mutation into an in-frame indel (24, 25); these reversion mutations associate with clinical secondary resistance to PARP inhibition (26, 27). Tumors with BRCA2 homozygous deletion, which account for approximately 5% of all metastatic prostate cancers (2), are unable to generate such secondary resistance mutations and may have exceptional responses based on these tumors finding it much more difficult to evolve resistance by reverting the homologous recombination function.
The partner and localizer of BRCA2 (PALB2) gene is much less commonly altered in APC, so very little published data are as yet available on this subset of mCRPC. The TOPARP-B trial recruited 7 patients with prostate cancers with PALB2 alterations. Surprisingly, of 7 patients with PALB2 pathogenic alterations, all but one had germline PALB2 mutations (6/7; 86%), but only four of these seven tumors had detectable biallelic loss. Overall, only the biallelic PALB2-mutated prostate cancers in this subset appeared to benefit, with all 4 men with biallelic loss mCRPC having either a PSA or RECIST response, and 3 of these being on drug >6 months. These data overall indicate that for PALB2-altered mCRPC identifying biallelic loss may be clinically important because almost half the tumors in this subset only had monoallelic loss with this group not appearing to benefit.
In this study, we tested the capacity of RAD51 foci IF to identify loss of HRR function in APC in formalin-fixed and paraffin-embedded (FFPE) tumor slides. This assay successfully identified all 16 of 16 cases with BRCA deleterious alterations as well as the PALB2-mutated tumors with biallelic loss, distinguishing the latter from PALB2-mutated tumors with monoallelic loss. This assay may have clinical utility and complement genomic testing in routine clinical practice especially when insufficient FFPE biopsy material is available for NGS, a common occurrence in both the PROfound and TRITON2 trials. Moreover, this RAD51 assay can help identify less-common genomic variants affecting HRR function that sensitize to PARP inhibition as observed for the TOPARP-B PALB2 cohort. Further clinical qualification is needed to optimally define the predictive value of this RAD51 assay for PARPi treatment.
Interestingly, the results from this RAD51 assay in the TOPARP-B cases indicate that tumors with ATM and CDK12 alterations have higher RAD51 scores than BRCA1/2-altered tumors, with some cases in these subgroups having scores at or just above the cutoff of 10% of cells having RAD51 foci. However, the responses seen in ATM- and CDK12-aberrant APC with treatment with olaparib administered for >6 months were observed in tumors with high RAD51 scores, indicating that the PARPi antitumor activity in this subgroup may not be related to complete loss of HRR function but perhaps due to the genomic instability in these tumors associated with ATM loss (28). Interestingly, unlike BRCA2 mutation–associated cancers, a significant number of the ATM-aberrant prostate cancers in the TOPARP-B trial did not have detectable biallelic loss (29). Only a small fraction of the ATM cohort had deleterious germline mutations with these tumors with germline mutations all having loss of ATM protein by IHC. The longest responding patient in this ATM cohort had homozygous deletion, and interestingly biallelic ATM mutations and ATM loss by IHC associated with better outcome. While we acknowledge the small numbers of patients (n = 21) in the TOPARP-B ATM cohort, these data indicate that patients with germline mutations and loss of ATM expression by IHC are more likely to respond and have longer duration of benefit. This is supported by the TOPARP-A data where 2 of the 3 patients with ATM-aberrant prostate cancer on olaparib for more than a year had germline deleterious mutations (14). Overall, while the response rate in this subgroup is low and exploratory analyses of the PROfound trial failed to demonstrate a significant benefit in the ATM-mutated disease subset, we have observed durable tumor responses in some prostate cancers with ATM aberrations in keeping with the preclinical genomic screen data (30). This, together with data from the PROfound trial, indicate that there is now an urgent need to integrate data from preclinical and clinical studies to better identify which ATM-aberrant prostate cancers benefit from PARP inhibition.
Most phase II/III trials of PARPi in prostate cancer conducted to date defined their patient population based on the identification of deleterious aberrations in predefined lists of genes. A major question that remains for this field is whether other relevant genomic alterations cooperate with the primary DNA repair defects, with which we have categorized these APCs, to generate sensitivity to PARPi such as olaparib. Exploratory studies described herein identify multiple putative genomic loci that statistically associate with response to olaparib with a false discovery rate of 0.01%. These loci contain multiple genes that are reported to affect DNA repair and could potentially have such a cooperative role. The chromosome 3q locus includes LRRC31 which when overexpressed is reported to inhibit DNA repair and sensitize to cell death following radiation-induced double-strand DNA breaks (31). The chromosome 10 locus includes the genes SIRT1, DNA2, and TET1, which have been implicated in DNA repair regulation, and the ubiquitin ligase HERC4, which has been previously identified as affecting PARPi sensitivity in a reported genome-wide screen (32). The chromosome 15 locus includes PARP6; ARIH1 which is implicated in DNA damage–induced translation arrest (33); C15orf60 (REC114 Meiotic Recombination Protein), which is reportedly involved in regulating DSB formation (34); and CD276 (B7-H3). The chromosome 19 locus also includes several genes implicated in the DNA damage response including AURKC, KMT5C, and POLD1. Overall, however, in light of the significant risk of false positivity in these genome-wide associations, we recommend that validation studies from other PARPi clinical trials are necessary to confirm these findings and the pursuit of wet laboratory studies of the impact of genes at these loci both as altered single or multiple genes.
We acknowledge that these analyses have limitations. First, the number of patients on this trial of the individual subsets was small; therefore, the analyses performed are largely exploratory and hypothesis-generating with any findings needing validation in larger cohorts. Second, patients were recruited to the trial based on a targeted tumor-only NGS assay, and in some cases there was insufficient material to pursue the extended deeper genomic analyses and IHC and IF assays. Despite this, however, these analyses remain the largest such deep analyses of APC samples on a prospective clinical trial of a PARPi. Finally, TOPARP-B was a phase II trial without a control treatment arm, so some of these findings may be confounded should any of these biomarkers have prognostic, rather than predictive, value; hence, validation in randomized trials is necessary.
In conclusion, our data have identified a group of exceptional responders to PARP inhibition characterized by BRCA2 homozygous deletions, and shown that BRCA-altered tumors unlike PALB2- and ATM-aberrant tumors usually have biallelic loss. These data may help refine stratification strategies in clinical practice for identifying patients with prostate cancer benefiting from this recently approved class of molecularly stratified treatment. Our study also suggests that functional assays assessing HRR, such as RAD51 foci IF, may have clinical utility for patient stratification in prostate cancer, although these now require prospective validation in larger cohorts.
Methods
Study Design, Patients, and Outcomes
The results of the TOPARP-B trial have been previously published (11). Briefly, we conducted an open-label, randomized phase II trial where patients with tumors known to have deleterious DDRm that may sensitize to PARP inhibition were randomized to receive olaparib at either 300 mg twice-daily or 400 mg twice-daily tablets. After providing written informed consent, archival primary tumor or fresh metastatic biopsies were tested using an investigational amplicon-based targeted NGS panel. Patients had to have been treated with at least one line of taxane-based chemotherapy; they received olaparib until radiographic progression, unacceptable toxicities, or withdrawal of consent. Patients treated with 300 mg twice daily were offered dose escalation to 400 mg twice daily on confirmation of radiographic progression, if clinically indicated, but no significant difference in antitumor activity was seen between the two dose levels.
The primary endpoint was confirmed tumor response, defined as a composite of: objective response by RECIST 1.1 (with PCWG2 caveats) and/or PSA decline of ≥50% from baseline and/or conversion of CTC count from ≥5 cells/7.5 mL blood at baseline to <5 cells/7.5 mL. Secondary endpoints included rPFS, defined as time from randomization to first evidence of radiographic progression (by RECIST 1.1 or bone scan as per PCWG2 criteria) or death and overall survival, defined as time from randomization to death by any cause.
The study was approved by the London, Surrey Borders, Research Ethics Committee (REC reference 11/LO/2019), and cosponsored by The Royal Marsden Hospital and The Institute of Cancer Research (ICR; London, United Kingdom).
Sequencing and Bioinformatics
DNA was extracted from the same FFPE tumor samples tested for study inclusion using the FFPE Tissue DNA Kit (QIAGEN) and quantified with the Quant-iT high-sensitivity PicoGreen Double Stranded DNA Assay Kit (Invitrogen). DNA quality control was performed by quantitative PCR using the Illumina FFPE QC Kit (WG-321–1001) according to the manufacturer's protocol as described previously (11, 14).
Libraries for WES were performed using Kapa Hyper Plus Library Prep Kits and the Agilent SureSelectXT V6 Target Enrichment Kit. Paired-end sequencing was performed using the NovaSeq 6000 S2 flow cell (2 × 100 cycles; Illumina) at the Centre for Molecular Pathology Translational Genomics Lab (RMH). FASTQ files were generated from the sequencer's output using Illumina bcl2fastq2 software (v.2.17.1.14, Illumina) with the default settings. All sequencing reads were aligned to the human genome reference sequence (GRCh37-hg19) using the BWA-MEM algorithm (v. 0.7.12). Picard tools (v.2.1.0) were used to remove PCR duplicates and to calculate sequencing metrics for quality control check. The Genome Analysis Toolkit (GATK; v. 3.5–0) was applied to realign local indels, recalibrate base scores, and identify genetic variants. Somatic point mutations and small indels were called using paired tumor–normal design using MuTect2 with stand_call_conf 30 and stand_emit_conf 30. Somatic variant was further filtered by quality PASS, does not appear in normal sample, coverage depth >10, and allele frequency >5%. By comparing tumor DNA to its matched germline DNA control, copy-number estimation was obtained through modified ASCAT2 package using (i) BAF data matrix derived from GATK variants calling and (ii) LogR data matrix of sequencing coverage at GATK variant location from Picard CalculateHsMetrics.
lpWGS was performed with libraries constructed using the NEBNext Ultra FS II DNA Kit (New England Biolabs) according to the manufacturer's protocol. Samples were pooled and run on the NextSeq (Illumina) at × 0.5 mean coverage, using the 300 cycles High Output v2.5 Kit (Illumina). BCL files were converted to FASTQ files using the bcl2fastq2 software (v.2.17.1.14, Illumina). Sequence alignments were performed using the BWA-MEM algorithm (v. 0.7.12) to the human genome reference sequence (GRCh37-hg19). Copy-number analysis (CNA) was performed using IchorCNA (35). In short, hg19 genomes (filtered centromeres) were divided into 500-kb nonoverlapping bins, and the abundance of the mapped reads was counted by HMMcopy Suite in each bin and predicted segments of CNAs. GC content and mappability bias were corrected by Loess regression and based on a panel of germline DNA sequencing from healthy donors. The maximum CNA detection was set to 20 copies.
Raw sequencing data have been deposited at the European Nucleotide Archive (ENA) with accession number: PRJEB45010.
For the purpose of this analysis, we considered “biallelic” events those cases with either: (i) two pathogenic mutations; (ii) a pathogenic mutation and a shallow deletion; (iii) a pathogenic mutation and loss-of-heterozygosity; or (iv) cases with homozygous deletions in the genes of interest, after analyzing data from the targeted, whole-exome, and low pass whole-genome sequencing and reviewing the cases manually when discordance was detected. Those cases where there was no evidence for any of those conditions were declared “not confirmed biallelic” loss (Supplementary Data File S1).
ATM IHC
ATM protein expression in the ATM group was determined by IHC staining on 3- to 4-μm–thick FFPE sections using the rabbit monoclonal anti-ATM antibody Y170 at 1:400 (catalog no. ab32420; Abcam PLC) as described previously (28). IHC slides were assessed by a pathologist, blinded to the patients' clinical characteristics, sequencing findings, and outcome data. Nuclear staining was semiquantitatively assessed using an H-score formula: three times percentage of strongly staining cells and times times percentage of moderately staining cells and percentage of weakly staining cells, giving an H-score range of 0–300. ATM negative status was considered if there was a complete absence of ATM staining or weak intensity staining in 10% or less of cancer cells (H-score ≤10).
RAD51 IF
IF for RAD51, geminin (GMN), and phospho-histone H2AX (γH2AX) was performed as described previously (19, 20), using sections of FFPE tumor biopsies. RAD51 was used as a biomarker of HRR function; GMN and γH2AX were used as quality checks to confirm that RAD51 foci were quantified in cells in the S–G2 cell-cycle phase when HRR takes place and in the presence of DNA double-strand breaks, respectively. For target antigen retrieval, sections were microwaved in DAKO Antigen Retrieval Buffer pH 9.0. Sections were permeabilized with DAKO Wash Buffer for 5 minutes, followed by five-minute incubation in blocking buffer (DAKO Wash Buffer with 1% BSA). Primary antibodies were diluted in DAKO Antibody Diluent and incubated at room temperature for 1 hour. Sections were washed and blocked again. Secondary antibodies were diluted in blocking buffer and incubated for 30 minutes at room temperature. Finally, sections were dehydrated with increasing concentrations of ethanol and mounted with DAPI ProLong Gold Antifade Reagent.
The following primary antibodies were used: rabbit anti-RAD51 (Abcam ab133534, 1:1,000), mouse anti-GMN (NovoCastra NCL-L, 1:60), rabbit anti-GMN (ProteinTech 10802–1-AP, 1:400), and mouse anti-γH2AX (Millipore #05636, 1:200). Goat anti-rabbit Alexa Fluor 568 (Invitrogen; 1:500), goat anti-mouse Alexa Fluor 488 (Invitrogen; 1:500), donkey anti-mouse Alexa Fluor 568 (Invitrogen; 1:500), and goat anti-rabbit Alexa Fluor 488 (Invitrogen; 1:500) were used as secondary antibodies. Scoring was performed by two readers independently and blinded to clinical outcome and genomics data. Scores were assessed on life images using a 60× immersion oil objective with a Nikon Eclipse Ti-E microscope. RAD51 was quantified in tumor areas by scoring the percentage of GMN-positive cells with five or more RAD51 nuclear foci. The mean of the scores obtained by the two observers was used. RAD51 scores were classified as “high” or “low” by applying a predefined cutoff of 10% (20, 21). All samples included in the analysis fulfilled the quality control criteria of having at least 40 GMN-positive cells and more than 25% of γH2AX/GMN–positive tumor cells. IF images were acquired with a 60× objective using a Nikon DS-Qi2 digital camera and generated using NIS-Elements-AR (version 4.40) software.
Statistical Analysis
All randomized patients (n = 98) were considered for this analysis, regardless of the dose group (300 mg and 400 mg) or evaluability status. Time-to-event endpoints were summarized across different gene subgroups by Kaplan–Meier curves, and median times estimated with 95% CIs. Local radiologic response assessment was used for all radiologic endpoints. For rPFS, patients alive and without radiologic progression were censored at the last scheduled disease assessment on study, at time of treatment discontinuation (in case of clinical progression not leading to death), or at time of starting a new treatment for mCRPC. Patients alive at the end of follow-up were censored for the analysis of OS. Within each of the gene subgroups, the proportion of homozygous deletions versus somatic mutations versus germline mutations were described, as well as the proportion of mutations with a demonstrated biallelic event versus mutations without confirmation of biallelic loss. Response, time on treatment, rPFS, and OS were estimated by type and origin of mutation, with time to event endpoints compared by log-rank tests. In the ATM gene subgroup, outcome of patients with ATM loss versus no loss as per IHC were also compared. The levels of RAD51 foci assessed by IF were graphically described by gene subgroup. The association of RAD51 score categories with outcomes was described graphically as above and analyzed by χ2 and log-rank tests, respectively.
Statistical analyses were conducted with the use of Stata software (version 15), on a snapshot of the data taken on September 21, 2020.
Authors' Disclosures
A. Llop-Guevara reports grants from Asociación Española Contra el Cáncer (AECC) and grants from “la Caixa” Foundation and European Institute of Innovation and Technology/Horizon 2020 during the conduct of the study; in addition, A. Llop-Guevara has a patent for PCT/EP2018/086759 pending. W. Yuan reports other support from Jilin Huarui Gene Technology Ltd outside the submitted work. V. Serra reports grants and personal fees from Instituto de Salud Carlos III and grants and personal fees from AECC during the conduct of the study; grants and personal fees from AstraZeneca, grants from Tesaro, and personal fees from Abbvie outside the submitted work; in addition, V. Serra has a patent for PCT/EP2018/086759 (WO2019122411A1) issued. C.J. Lord reports grants and personal fees from AstraZeneca outside the submitted work; in addition, C.J. Lord is an inventor on patents describing the use of DNA repair inhibitors and stands to gain from their use as part of the Institute of Cancer Research “Rewards to Inventor” scheme. C.J. Lord has received research funding from AstraZeneca, Merck KGaA, Artios, Pfizer, and has received consultancy and/or advisory fees from Artios, AstraZeneca, MerckKGaA, Tango, and GLG. C.J. Lord is also a shareholder of OviBio and Tango (issued, licensed, and with royalties paid from Institute of Cancer Research). E. Hall reports grants from Cancer Research UK, Accuray Inc., Varian Medical Systems Inc., Merck Sharp & Dohme, Janssen-Cilag, Aventis Pharma Limited (Sanofi), and grants and non-financial support from Bayer, grants and non-financial support from AstraZeneca during the conduct of the study; grants and non-financial support from Roche Products Ltd outside the submitted work. J. Mateo reports grants and non-financial support from AstraZeneca during the conduct of the study; grants and personal fees from AstraZeneca, Pfizer Oncology; personal fees from MSD, Guardant Health; personal fees and non-financial support from Roche, and personal fees and non-financial support from Janssen outside the submitted work (the authors affiliated to VHIO disclose that the institution is a joint applicant for the patent entitled Methods Based on the Detection of RAD51 Foci in Tumor Cells, which includes any patent application or patent claiming priority from European Application number EP17382884.9). J.S. de Bono reports grants and personal fees from Bayer, Pfizer, Merck Serono, AstraZeneca, and grants and personal fees from AstraZeneca during the conduct of the study; grants from MSD, Harpoon Pharmaceuticals, Taiho, CellCentric; and personal fees from Genentech/Roche, Daiichi Sankyo, SeaGen, and personal fees from Eisai outside the submitted work; in addition, J.S. de Bono has a patent for PARP inhibition in DNA repair defective cancers issued and with royalties paid (from J.S. de Bono's institution). No disclosures were reported by the other authors.
Acknowledgments
TOPARP is an investigator-initiated trial; we are grateful for the support and funding from AstraZeneca, and for the study grants from Cancer Research UK (CRUK/11/029; C12540/A12829; C12540/A13230; C12540/A20447). ICR-CTSU also receives program grant funding from Cancer Research UK (grant number: C1491/A15955, C1491/A25351). We acknowledge research funding for this work from Cancer Research UK, Prostate Cancer UK, the Movember Foundation through the London Movember Centre of Excellence (CEO13_2-002), the Prostate Cancer Foundation, and the UK Department of Health through an Experimental Cancer Medicine Centre (ECMC) grant. J.S. de Bono is a National Institute for Health Research (NIHR) senior investigator. The views expressed in this article are those of the author(s) and not necessarily those of the NHS, the NIHR, or the Department of Health; research at the Royal Marsden Hospital is supported by a Biomedical Research Centre grant. P. Rescigno was supported by a PCF Young Investigator Award 19YOUN19. The authors affiliated to VHIO acknowledge funding from “La Caixa” Foundation and European Institute of Innovation and Technology/Horizon 2020 (LCF/TR/CC19/52470003), Fundacion FERO and Fundacion Cellex. J. Mateo, A. Llop-Guevara and V. Serra received grants from Fundacion Cientifica AECC (LABAE16020PORTT, INVES20095LLOP, LABAE20019MATE) and an ERAPERMED2019-215. J. Mateo gratefully acknowledges funding from the European Union's Horizon 2020 research and innovation program (Marie Skłodowska-Curie grant 837900), Instituto de Salud Carlos III (Grant PI18/01384), CRIS Cancer Foundation (PR_TCL_2020_10) and the US Department of Defense CDMRP (Impact Award PC170510P1). S. Arce-Gallego and V. Serra were supported by Instituto de Salud Carlos III (FI19/00280, CPII19/00033).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors' Contributions
S. Carreira: Conceptualization, formal analysis, supervision, investigation, visualization, methodology, writing–original draft, writing–review and editing. N. Porta: Conceptualization, data curation, formal analysis, visualization, methodology, writing–original draft, writing–review and editing. S. Arce-Gallego: Formal analysis, investigation, visualization, methodology, writing–review and editing. G. Seed: Software, formal analysis, validation, investigation, methodology, writing–review and editing. A. Llop-Guevara: Formal analysis, investigation, writing–review and editing. D. Bianchini: Resources, project administration, writing–review and editing. P. Rescigno: Resources, project administration, writing–review and editing. A. Paschalis: Resources, writing–review and editing. C. Bertan: Investigation, methodology, writing–review and editing. C. Baker: Investigation, methodology, writing–review and editing. J. Goodall: Investigation, methodology, writing–review and editing. S. Miranda: Investigation, methodology, project administration, writing–review and editing. R. Riisnaes: Investigation, methodology, writing–review and editing. I. Figueiredo: Investigation, methodology, writing–review and editing. A. Ferreira: Investigation, methodology, writing–review and editing. R. Pereira: Investigation, methodology, writing–review and editing. M. Crespo: Investigation, methodology, writing–review and editing. B. Gurel: Investigation, writing–review and editing. D. Nava Rodrigues: Investigation, writing–review and editing. S.J. Pettitt: Writing–review and editing. W. Yuan: Software, formal analysis, writing–review and editing. V. Serra: Supervision, writing–review and editing. J. Rekowski: Formal analysis, visualization, writing–review and editing. C.J. Lord: Writing–review and editing. E. Hall: Conceptualization, formal analysis, supervision, writing–review and editing. J. Mateo: Conceptualization, formal analysis, supervision, writing–original draft, writing–review and editing. J.S. de Bono: Conceptualization, resources, formal analysis, supervision, funding acquisition, writing–original draft, project administration, writing–review and editing.
References
- 1. Sartor O, de Bono JS. Metastatic prostate cancer. N Engl J Med 2018;378:645–57. [DOI] [PubMed] [Google Scholar]
- 2. Robinson D, Van Allen EM, Wu Y-M, Schultz N, Lonigro RJ, Mosquera J-Met al. Integrative clinical genomics of advanced prostate cancer. Cell 2015;161:1215–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Armenia J, Wankowicz SAM, Liu D, Gao J, Kundra R, Reznik Eet al. The long tail of oncogenic drivers in prostate cancer. Nat Genet 2018;50:645–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Abida W, Armenia J, Gopalan A, Brennan R, Walsh M, Barron Det al. Prospective genomic profiling of prostate cancer across disease states reveals germline and somatic alterations that may affect clinical decision making. JCO Precis Oncol 2017;2017:PO.17.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chung JH, Dewal N, Sokol E, Mathew P, Whitehead R, Millis SZet al. Prospective comprehensive genomic profiling of primary and metastatic prostate tumors. JCO Precis Oncol 2019;3:PO.18.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez Eet al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005;434:913–7. [DOI] [PubMed] [Google Scholar]
- 7. Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TBet al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005;434:917–21. [DOI] [PubMed] [Google Scholar]
- 8. Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JHet al. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res 2012;72:5588–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hussain M, Mateo J, Fizazi K, Saad F, Shore N, Sandhu Set al. Survival with olaparib in metastatic castration-resistant prostate cancer. N Engl J Med 2020;383:2345–57. [DOI] [PubMed] [Google Scholar]
- 10. De Bono J, Mateo J, Fizazi K, Saad F, Shore N, Sandhu Set al. Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med 2020;382:2091–102. [DOI] [PubMed] [Google Scholar]
- 11. Mateo J, Porta N, Bianchini D, McGovern U, Elliott T, Jones Ret al. Olaparib in patients with metastatic castration-resistant prostate cancer with DNA repair gene aberrations (TOPARP-B): a multicentre, open-label, randomised, phase 2 trial. Lancet Oncol 2020;21:162–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abida W, Campbell D, Patnaik A, Shapiro JD, Sautois B, Vogelzang NJet al. NonBRCA DNA damage repair gene alterations and response to the PARP inhibitor rucaparib in metastatic castration-resistant prostate cancer: analysis from the phase II TRITON2 study. Clin Cancer Res 2020;26:2487–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Abida W, Patnaik A, Campbell D, Shapiro J, Bryce AH, McDermott Ret al. Rucaparib in men with metastatic castration-resistant prostate cancer harboring a BRCA1 or BRCA2 gene alteration. J Clin Oncol 2020;38:376372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mateo J, Carreira S, Sandhu S, Miranda S, Mossop H, Perez-Lopez Ret al. DNARepair defects and olaparib in metastatic prostate cancer. N Engl J Med 2015;373:1697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bryce AH, Sartor O, De Bono J. DNA repair and prostate cancer: a field ripe for harvest. a field ripe for harvest. Eur Urol 2020;78:486–8. [DOI] [PubMed] [Google Scholar]
- 16. Quigley DA, Dang HX, Zhao SG, Lloyd P, Aggarwal R, Alumkal JJet al. Genomic hallmarks and structural variation in metastatic prostate cancer. Cell 2018;175:889. [DOI] [PubMed] [Google Scholar]
- 17. Stopsack KH, Nandakumar S, Wibmer AG, Haywood S, Weg ES, Barnett ESet al. Oncogenic genomic alterations, clinical phenotypes, and outcomes in metastatic castration-sensitive prostate cancer. Clin Cancer Res 2020;26:3230–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mateo J, Seed G, Bertan C, Rescigno P, Dolling D, Figueiredo Iet al. Genomics of lethal prostate cancer at diagnosis and castration resistance. J Clin Invest 2020;130:1743–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Castroviejo-Bermejo M, Cruz C, Llop-Guevara A, Gutiérrez-Enríquez S, Ducy M, Ibrahim YHet al. A RAD51 assay feasible in routine tumor samples calls PARP inhibitor response beyond BRCA mutation. EMBO Mol Med 2018;10:e9172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cruz C, Castroviejo-Bermejo M, Gutiérrez-Enríquez S, Llop-Guevara A, Ibrahim YH, Gris-Oliver Aet al. RAD51 foci as a functional biomarker of homologous recombination repair and PARP inhibitor resistance in germline BRCA-mutated breast cancer. Ann Oncol 2018;29:1203–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Graeser M, McCarthy A, Lord CJ, Savage K, Hills M, Salter Jet al. A marker of homologous recombination predicts pathologic complete response to neoadjuvant chemotherapy in primary breast cancer. Clin Cancer Res 2010;16:6159–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Markowski MC, Antonarakis ES. BRCA1 versus BRCA2 and PARP inhibitor sensitivity in prostate cancer: more different than alike? J Clin Oncol 2020;38:3735–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sokol ES, Pavlick D, Khiabanian H, Frampton GM, Ross JS, Gregg JPet al. PanCancer analysis of BRCA1 and BRCA2 genomic alterations and their association with genomic instability as measured by genome-wide loss of heterozygosity. JCO Precis Oncol 2020;4:442–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lin KK, Harrell MI, Oza AM, Oaknin A, Ray-Coquard I, Tinker AVet al. BRCA reversion mutations in circulating tumor DNA predict primary and acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Discov 2019;9:210–9. [DOI] [PubMed] [Google Scholar]
- 25. Goodall J, Mateo J, Yuan W, Mossop H, Porta N, Miranda Set al. Circulating cell-free DNA to guide prostate cancer treatment with PARP inhibition. Cancer Discov 2017;7:1006–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DAet al. Resistance to therapy caused by intragenic deletion in BRCA2. Nature 2008;451:1111–5. [DOI] [PubMed] [Google Scholar]
- 27. Pettitt SJ, Frankum JR, Punta M, Lise S, Alexander J, Chen Yet al. Clinical BRCA1/2 reversion analysis identifies hotspot mutations and predicted neoantigens associated with therapy resistance. Cancer Discov 2020;10:1475–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Neeb A, Herranz N, Arce-Gallego S, Miranda S, Buroni L, Yuan Wet al. Advanced prostate cancer with ATM loss: PARP and ATR inhibitors. Eur Urol 2021;79:200–11. [DOI] [PubMed] [Google Scholar]
- 29. Marshall CH, Sokolova AO, McNatty AL, Cheng HH, Eisenberger MA, Bryce AHet al. Differential response to olaparib treatment among men with metastatic castration-resistant prostate cancer harboring BRCA1 or BRCA2 versus ATM mutations. Eur Urol 2019;76:452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McCabe N, Turner NC, Lord CJ, Kluzek K, Bialkowska A, Swift Set al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res 2006;66:8109–15. [DOI] [PubMed] [Google Scholar]
- 31. Chen Y, Jiang T, Zhang H, Gou X, Han C, Wang Jet al. LRRC31 inhibits DNA repair and sensitizes breast cancer brain metastasis to radiation therapy. Nat Cell Biol 2020;22:1276–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lodovichi S, Mercatanti A, Cervelli T, Galli A. Computational analysis of data from a genome-wide screening identifies new PARP1 functional interactors as potential therapeutic targets. Oncotarget 2019;10:2722–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. von Stechow L, Typas D, Carreras Puigvert J, Oort L, Siddappa R, Pines Aet al. The E3 ubiquitin ligase ARIH1 protects against genotoxic stress by initiating a 4EHP-mediated mRNA translation arrest. Mol Cell Biol 2015;35:1254–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Carballo JA, Panizza S, Serrentino ME, Johnson AL, Geymonat M, Borde Vet al. Budding yeast ATM/ATR control meiotic double-strand break (DSB) levels by down-regulating Rec114, an essential component of the DSB-machinery. PLoS Genet 2013;9:e1003545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Adalsteinsson VA, Ha G, Freeman SS, Choudhury AD, Stover DG, Parsons HAet al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun 2017;8:1324. [DOI] [PMC free article] [PubMed] [Google Scholar]