Abstract
Haemophilus influenzae exists as a commensal of the upper respiratory tract of humans but also causes infections of contiguous structures. We describe the identification, localization, purification, and characterization of a novel, surface-localized phosphomonoesterase from a nontypeable H. influenzae strain, R2866. Sequences obtained from two CNBr-derived fragments of this protein matched lipoprotein e (P4) within the H. influenzae sequence database. Escherichia coli DH5α transformed with plasmids containing the H. influenzae hel gene, which encodes lipoprotein e (P4), produced high levels of a membrane-associated phosphomonoesterase. The isolated ∼28-kDa enzyme was tartrate resistant and displayed narrow substrate specificity with the highest activity for arylphosphates, excluding 5-bromo-4-chloro-3-indolylphosphate. Optimum enzymatic activity was observed at pH 5.0 and only in the presence of divalent copper. The enzyme was inhibited by vanadate, molybdate, and EDTA but was resistant to inorganic phosphate. The association of phosphomonoesterase activity with a protein that has also been recognized as a heme transporter suggests a unique role for this unusual phosphohydrolase.
The bacterium Haemophilus influenzae is a gram-negative facultatively anaerobic coccobacillus and is a common commensal of the human respiratory tract. This organism is the etiologic agent of a variety of local and invasive infections in both children and adults (32). Isolates of H. influenzae are divided into encapsulated strains a through f, based on capsular polysaccharide antigenicity, and nonencapsulated strains designated nontypeable. These nonencapsulated strains account for the majority of mucosal diseases, including otitis media, sinusitis, and bronchitis. Additionally, as an opportunist in immunocompromised individuals, nontypeable H. influenzae can breach the epithelial barrier and cause septicemia, endocarditis, or pyogenic arthritis (29). While the nontypeable strains can sometimes be invasive, the encapsulated H. influenzae type b strains are responsible for the vast majority of invasive disease (33).
Identification of surface-localized macromolecular structures responsible for this pathogen’s ability to cause disease has been the aim of intensive investigation. The outer membrane of the organism possesses a number of components thought to be essential for colonization, invasion, and survival within the human host. These include the capsule, the pili, immunoglobulin protease, and certain major outer membrane proteins. Up to 36 proteins are contained within the H. influenzae envelope; 6 of these are considered abundant and are designated P1 to P6 in order of decreasing molecular weight (15). Because interest in these proteins has been for the purpose of vaccine development, much is known about their immunogenic and antigenic properties but little is known about their biological functions. Two exceptions to this are proteins P2 and P4. The P2 protein, the most prominent outer membrane protein, self-associates as a homotrimer and acts as a porin allowing the passage of molecules of 1,400 Da or less through the outer membrane (34). Outer membrane protein P4, also designated lipoprotein e (P4), is a highly conserved cationic protein found in all strains of H. influenzae. When expressed by an Escherichia coli hemA strain defective in de novo synthesis of porphyrin, H. influenzae lipoprotein e (P4), encoded by the hel gene, mediates the transport of heme through the E. coli membrane, permitting growth (20). The presence of hemin-binding motifs within lipoprotein e (P4) and the construction of H. influenzae hel mutants incapable of aerobic growth suggest that this protein plays a critical role in heme acquisition (20). While the ability to transport exogenous heme into the cell has been attributed to lipoprotein e (P4), the biochemistry of heme binding and transport has yet to be elucidated.
Acid phosphatases (EC 3.1.3.2) are ubiquitous and catalyze the hydrolysis of phosphomonoesters at an acidic pH. These enzymes participate in an assortment of essential biological functions, including the regulation of metabolism, energy conversion, and signal transduction. Studies of surface-localized acid phosphatases of both protozoan and bacterial pathogens suggest that members of a class of tartrate-resistant nonspecific acid phosphatases may play critical roles in infection and the survival of microbes within a human host. Consistent with this hypothesis, purified acid phosphatases from Leishmania donovani (23), Legionella micdadei (25), and Coxiella burnetii (1) have been shown to suppress the respiratory burst of N-formyl-Met-Leu-Phe-stimulated neutrophils. In addition, purified preparations of acid phosphatase from Francisella tularensis abrogated the respiratory burst of both internally (phorbol 12-myristate 13-acetate)- and externally (N-formyl-Met-Leu-Phe)-stimulated porcine neutrophils in a dose-dependent manner (22). We describe here the localization, purification, and characterization of a novel phosphomonoesterase of H. influenzae and its identification as the lipoprotein e (P4) encoded by the hel gene of this human commensal (21).
MATERIALS AND METHODS
Bacterial strains and materials.
H. influenzae R2866 is a previously described invasive nontypeable clinical isolate (17) which served as the source for the enzyme. This strain and other nontypeable H. influenzae strains screened for phosphomonoesterase activity, encapsulated H. influenzae a through f (ATCC 9006, 9795, 9007, 9008, 8142, and 9833), H. influenzae Rd KW20 (38), Haemophilus aegyptius strains, and Haemophilus parainfluenzae R1966 (Bossarelli) are kept as part of the Haemophilus culture collection of A. L. Smith at the University of Missouri Medical School. All strains utilized in this study were stored at −80°C in 50% defibrinated horse blood (PML Microbiologics) and 50% autoclaved skim milk. The E. coli strain used for expression of the cloned e (P4) gene, hel, was Max Efficiency DH5α competent cells (Gibco BRL). All bacteriological media were from Difco and were purchased through Fisher Scientific. All other chemicals, unless otherwise stated, were obtained from Sigma Chemical and were of the highest purity available. Ion-exchange and gel filtration chromatography resins and standards were purchased from Amersham Pharmacia Biotech. Extracti-Gel D was purchased from Pierce Chemical. Protein electrophoresis reagents were obtained from Bio-Rad. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) Perfect Protein molecular weight standards were purchased from Novagen.
Culture conditions.
All Haemophilus strains were grown in brain heart infusion (BHI) broth or on agar supplemented with hemin chloride (X factor), l-histidine, and β-NAD (V factor), each at 10 μg/ml (sBHI); all strains were also cultured on enriched chocolate agar containing 1% hemoglobin and supplement B (hCHA). Bacteria grown on solid media were incubated statically at 37°C, whereas 3-ml broth cultures were incubated at 37°C with aeration in a New Brunswick Scientific controlled-environment incubator shaker at ∼200 rpm. H. influenzae organisms used for enzyme purification were diluted from sBHI (optical density at 600 nm, ∼0.4) into sterile phosphate-buffered saline containing 0.1% gelatin and plated on enriched chocolate agar at a density that resulted in near-confluent colonies after 24 h of growth. The bacteria were harvested by scraping the culture from the agar. Colonies harvested from 40 plates (100 by 15 mm) were suspended in 25 ml of buffer A (50 mM sodium acetate [pH 5.5] containing 150 mM NaCl and 0.1 mM CuSO4).
Enzyme assays.
H. influenzae phosphomonoesterase activity was measured by a discontinuous colorimetric assay performed in microtiter wells. The 0.2-ml standard assay mixture contained 0.2 M sodium acetate (pH 5.5), 0.1 mM CuSO4, 1.0 mM p-nitrophenylphosphate (pNPP), and various amounts of enzyme. The mixtures were incubated at 37°C for 15 min with constant agitation. The reaction was stopped by addition of 100 μl of 0.5 M glycine (pH 10.0). The concentration of p-nitrophenol produced was measured with a Dynatech MR5000 microtiter plate reader at 410 nm with an extinction coefficient of 18.3 ± 0.2 mM−1 cm−1. Phosphomonoesterase was diluted in buffer A to a concentration that resulted in a linear response of the detector with increasing amounts of enzyme. One unit of enzyme activity is defined as the amount of activity required to convert 1 nmol of substrate to product per h at 37°C. Assays to determine the pH optimum of the enzyme were performed in either 0.2 M sodium acetate (pKa, 4.76), 0.2 M 2-(N-morpholino)ethanesulfonic acid (MES) (pKa, 6.21), or HEPES (pKa, 7.66) as a buffer, with the substrate at a final concentration of 2.0 mM. The substrate specificity, kinetic parameters, and protoporphyrin IX-mediated inhibition of the enzyme were determined by measuring the amount of inorganic phosphate released from phosphomonoesters (including pNPP) by the method of Lanzetta et al. (14). All substrates tested were commercially available except for p60src, a phosphorylated peptide previously described by Tian et al. (31). Substrate specificity assays were performed in 1.5-ml microcentrifuge tubes with 525 U of enzyme per reaction mixture and substrates at a 2.0 mM final concentration in a 50-μl total assay volume. Samples were incubated at 37°C in a water bath without agitation for 30 min prior to addition of the detection reagent (14). The concentration of inorganic phosphate was determined spectrophotometrically at 660 nm on a Hitachi U-2000 spectrophotometer.
Kinetic assays were performed similarly except that various amounts (2, 6, 10, 14, and 20 μl) of diluted enzyme resulting in linear spectrophotometric responses from 0.05 to 1.0 absorbance unit at 660 nm were used for each concentration of substrate tested. Samples for kinetic studies were placed on ice until time zero, at which time they were placed at 37°C. The data were analyzed with a nonlinear least-squares regression computer program (4) graciously supplied by Stephen P. J. Brookes, Carleton University, Ottawa, Canada.
Protein determination.
Protein concentrations or relative protein amounts were determined with bicinchoninic acid (BCA Protein Assay Reagent; Pierce) as described previously (28). Bovine serum albumin was used as the standard.
Phosphomonoesterase localization.
For all localization procedures, H. influenzae R2866 was cultured on enriched chocolate agar and resuspended in buffer A; all subsequent steps, unless otherwise stated, were performed at 4°C. The bacterial suspension was homogenized by repeated pipetting. Bacterial cells were broken by two cycles of a French press (Aminco Bowman) adjusted to ∼10,000 lb/in2 in a 40K rapid-fill cell with a flow rate of ∼20 drops/min. Unbroken cells and pelletable debris were removed by centrifugation at 5,000 × g for 10 min. Crude membranes were pelleted by centrifugation at 192,000 × g for 1.5 h. The membrane pellet (pellet I) was resuspended in 2 to 3 ml of buffer A and homogenized in a standard-clearance Potter-Elvehjem homogenizer with a motor-driven Wheaton overhead stirrer set at 50% speed. Any remaining particulate matter was removed from the resuspended membranes by centrifugation at 5,000 × g prior to application to a 35-ml sucrose gradient. The sucrose step gradient consisted of seven 5-ml layers of sucrose from 25 to 55% (wt/wt) in 5% increments solubilized in 50 mM sodium acetate (pH 6.0). After ∼40 h of centrifugation in a SW28 rotor at 69,000 × g, the gradient was fractionated from bottom to top into approximately 30 1.2-ml fractions with a Pharmacia peristaltic pump P1 set at maximum flow rate. Phosphomonoesterase activity and relative protein concentrations were determined as previously described. The density of each fraction was determined by the refractive index of 50 μl of each fraction measured with an American Optical ABBE refractometer.
Cytochemical localization of the phosphomonoesterase.
Cytochemical localization of the phosphomonoesterase activity on whole cells was performed by a modification of the procedure of Gomori (10). H. influenzae grown on chocolate agar was applied to carbon-coated copper grids in 50 mM sodium acetate, pH 6.0. The grids were incubated for 3 min at room temperature with filtered 5 mM pNPP in 50 mM sodium acetate buffer, pH 6.0, containing 3.3 mM lead nitrate with or without 5 mM EDTA. Following removal of the reaction solution, samples were rinsed briefly with 50 mM sodium acetate, air dried, and examined on a JEOL ×1,200 transmission electron microscope.
Phosphomonoesterase purification.
All procedures were conducted at 4°C unless otherwise noted. The crude membrane fraction (pellet I) from H. influenzae R2866 was prepared as described above. The membranes were resuspended in 17 ml of buffer A and homogenized as previously described. An equal volume of extraction buffer (buffer A containing 5 mM EDTA and 4% [vol/vol] Triton X-100) was added to the resuspended membranes. The extraction mixture was stirred for 12 h at 4°C and then centrifuged at 192,000 × g for 60 min to pellet insoluble material. The supernatant (supernatant II) containing the phosphomonoesterase was first dialyzed against 2 liters of buffer A and then applied to an SP-Sepharose cation-exchange chromatography resin (8.6 by 2.5 cm) preequilibrated with 50 mM sodium acetate (pH 6.0)–200 mM NaCl–0.1 mM CuSO4. After sample application, the column was washed with 2 volumes of equilibration buffer. Phosphomonoesterase activity was eluted from the resin in a 0.2 to 1.5 M linear NaCl gradient (400 ml) in 50 mM sodium acetate (pH 6.0), 0.1 mM CuSO4 applied to the column at ∼1 ml/min. A single peak of activity was eluted between 0.67 and 0.92 M NaCl and concentrated by ultrafiltration. A significant portion of the eluted activity bound to the ultrafiltration membrane but was recovered by overnight incubation of the stirred cell membrane in the presence of 4% Triton X-100. The recovered sample was then applied to and eluted (0.7 ml/min) from a Superose 12 gel filtration chromatography resin (HR 12/30) equilibrated in buffer A with a Pharmacia fast protein liquid chromatography (FPLC) system. Peak fractions containing phosphomonoesterase activity were pooled and concentrated with a Centricon 10 ultrafiltration unit (Millipore).
Estimation of the phosphomonoesterase molecular weight.
SDS-PAGE was performed as described by Laemmli (13) on a Hoeffer Mighty Small 250 electrophoresis unit. The polyacrylamide gels were 3% T stacking and 10% T resolving. The molecular weight of the denatured phosphomonoesterase was estimated with Novagen Perfect Protein standards.
N-terminal and cyanogen bromide peptide sequence of e (P4).
Approximately 2 μg of purified protein was subjected to gel electrophoresis on a 10% T tris-tricine polyacrylamide gel and electroblotted to a polyvinylidene difluoride membrane in 10 mM 3-[cyclohexylamino]-1-propanesulfonic acid containing 10% methanol for 3 h at 100 mA. The blotted protein was detected with 0.1% Coomassie blue R-250 in 1% acetic acid–40% methanol. The blot was destained with 50% methanol. A blotted protein was excised and further destained with 0.3% triethylamine in methanol to reduce background signals in sequencing. The sequencing of the blotted protein was performed on a Perkin-Elmer/Applied Biosystems, Inc., 492cLC protein sequencer with the cLC (capillary liquid chromatography) gas phase program. The final data were manually interpreted by the University of Missouri Protein Core Facility.
Twenty micrograms of purified phosphomonoesterase in 200 μl of buffer A containing Triton X-100 was utilized for the generation of cyanogen bromide fragments. The sample was freed of detergent by batch chromatography with Extracti-Gel D resin preequilibrated according to the manufacturer’s instructions. The recovered supernatant was dried in a Savant Speed Vac at medium heat and resuspended in 100 μl of 70% formic acid containing 5 mg of CNBr/ml. The sample was incubated for 12 h in the dark at 25°C, dried as previously described, diluted to 1 ml in H2O, dried again, and resuspended in 1× tris-tricine protein electrophoresis loading buffer. Cyanogen bromide-derived peptides were subjected to electrophoresis on a 15% T tris-tricine gel, blotted to a polyvinylidene difluoride membrane, excised, and sequenced at the University of Missouri Protein Core as previously described.
Cloning of the phosphomonoesterase (hel) gene.
DNA encoding the H. influenzae lipoprotein e (P4) was obtained from two independent sources. An E. coli clone, GHIGU90, containing a 2.4-kb hel-containing insert in pUC19 was obtained from the American Type Culture Collection (ATCC) and is one of a number of clones generated from the H. influenzae Rd KW20 genome sequencing project (8). Plasmid pJRP4 was obtained from John Mekalanos and contains a single copy of a PCR-derived hel gene from H. influenzae Rd KW20 cloned into the BamHI site of pACYC184 (20). E. coli strains containing these plasmids were propagated in Luria broth containing ampicillin (50 μg/ml) in preparation for plasmid isolation. Plasmid preparation was performed according to the Qiagen kit protocol. Cloning and restriction enzyme analysis was performed according to the method of Sambrook et al. (26). An EcoRI hel-containing fragment of pGHIGU90 was subcloned into the multiple cloning site of pBluescript KS, resulting in plasmid phel1. Similarly, a BamHI hel-containing fragment of pJRP4 was subcloned into the multiple cloning site of pBluescript KS, resulting in plasmid phel3. Both plasmids were transformed into E. coli DH5α according to the manufacturer’s protocol.
The specific phosphomonoesterase activities of E. coli clones were determined as follows. E. coli strains harboring plasmids encoding the hel gene or vector controls were grown in 25 ml of Luria broth containing ampicillin (50 μg/ml) overnight with constant aeration. Stationary-phase bacteria were pelleted at 10,000 × g and resuspended in 2 ml of buffer A. The phosphatase activity and protein concentration of the resuspended cells or isolated membranes were determined as described above. Following treatment of resuspended bacteria (1 to 3 ml) with two cycles of a French press (11,000 lb/in2) with an Aminco minicell, the E. coli membranes were isolated by ultracentrifugation at 260,000 × g with a Beckman VTi 80.
RESULTS
Detection of phosphomonoesterase activity in H. influenzae.
H. influenzae R2866 was found to harbor an abundance of cell-associated phosphomonoesterase activity when tested in the presence of 2 mM pNPP at pH 5.0 to 6.0. The average specific activity of the enzyme in bacteria cultured on hCHA was in excess of 118,000 U/mg of protein (n = 18; range, 32,000 to 202,000), a level of activity which exceeds that observed in most other acid phosphatase-producing bacterial pathogens by at least fivefold (22). As shown in Table 1, all H. influenzae strains examined (typeable and nontypeable) and two other Haemophilus species had similar levels of the enzyme activity when cultured on either sBHI or hCHA or in sBHI broth. Of nine nontypeable H. influenzae strains tested, the average phosphomonoesterase specific activity was 112,000 U/mg of protein (range, 69,000 to 217,000). The specific phosphatase activity of the typeable strains ranged from 50,000 U/mg of protein for H. influenzae type c cultured on sBHI to 280,000 U/mg of protein for H. influenzae type e cultured on hCHA. The average specific phosphatase activity for all six typeable strains tested was 104,000 U/mg of protein when the bacteria were cultured on sBHI agar and 139,000 U/mg of protein when the organisms were cultured on hCHA. This difference in specific phosphomonoesterase activity with respect to growth medium was not significant when the data were analyzed by a Student two-tailed t test (P = 0.142). Two other Haemophilus species, H. aegyptius (ATCC 33389) and H. parainfluenzae (Bossarelli), produced the enzyme at approximately 100,000 and 50,000 U/mg of protein, respectively.
TABLE 1.
Comparison of H. influenzae phosphatase activities
| Strain | Medium | Sp acta (105 U/mg) |
|---|---|---|
| H. influenzae R2866 | sBHI | 1.26 |
| H. influenzae R2866 | hCHA | 1.37 |
| H. influenzae type a | sBHI | 0.86 |
| H. influenzae type a | hCHA | 0.81 |
| H. influenzae type b | sBHI | 0.68 |
| H. influenzae type b | hCHA | 0.72 |
| H. influenzae type c | sBHI | 0.50 |
| H. influenzae type c | hCHA | 0.87 |
| H. influenzae type d | sBHI | 0.86 |
| H. influenzae type d | hCHA | 1.99 |
| H. influenzae type e | sBHI | 2.02 |
| H. influenzae type e | hCHA | 2.80 |
| H. influenzae type f | sBHI | 1.35 |
| H. influenzae type f | hCHA | 1.14 |
| H. aegyptius R3145 | hCHA | 0.90 |
| H. aegyptius R3146 | hCHA | 1.07 |
| H. parainfluenzae R1966 | hCHA | 0.59 |
Specific activity is shown as nanomoles of pNPP hydrolyzed per hour per milligram of protein.
No significant changes in the level of phosphatase activity of H. influenzae R2866 were observed by increasing or decreasing the levels of NAD, hemin, supplement B, and/or hemoglobin in either BHI agar or chocolate agar. In addition, no significant changes in specific phosphomonoesterase activity were observed during a timed growth curve assay conducted in sBHI broth (data not shown). The observed phosphomonoesterase activity in a crude, partially purified, or homogeneous state was linear with increasing amounts of sample or time of incubation and was heat labile.
Localization of H. influenzae phosphatase.
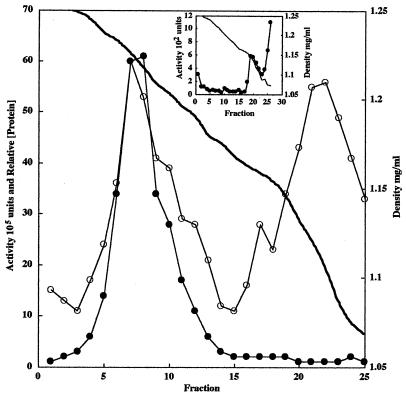
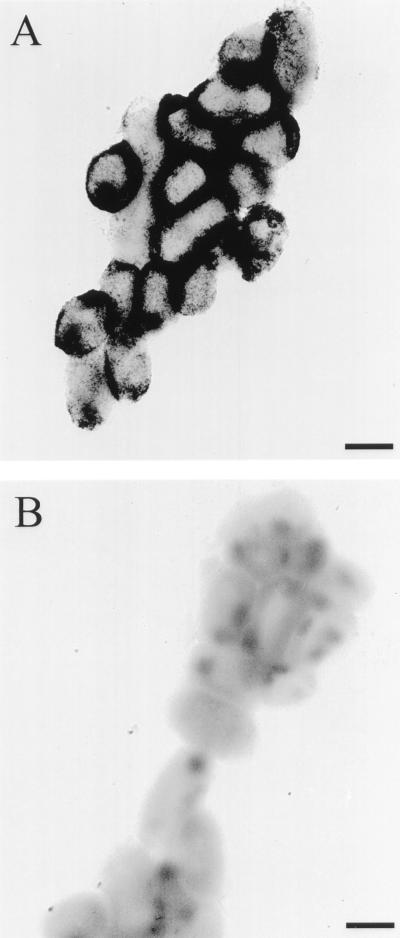
Apparent lack of enzyme crypticity, as demonstrated by a one- to twofold increase in total enzyme activity upon French press disruption of the bacteria (Table 2), suggested that the phosphomonoesterase was localized in close proximity to the cell surface. The enzyme fractionated with pelletable material after ultracentrifugation following a 2 M NaCl extraction of crude H. influenzae R2866 membranes. It was liberated to the supernatant fraction following ultracentrifugation after treatment of the membranes with 4% Triton X-100 (Table 2). These results suggested that the protein was not peripherally associated with membranes but was in intimate association with one or both membranes of this gram-negative bacterium. To discriminate between outer and inner membrane association, H. influenzae R2866 membranes were subjected to modified sucrose density ultracentrifugation as previously described (27). A typical graph of total phosphatase activity, relative protein concentration, and sucrose density as measured by refractive index for each fraction is shown in Fig. 1. The densities of all gradients ranged from 1.25 to 1.06 mg/ml of sucrose and were linear in the regions of interest. The fractions containing the highest phosphatase activity were coincident with a peak of protein associated with membranes of higher buoyant density (ρ = 1.223 ± 0.004 mg/ml of sucrose). The recovered phosphatase activity from this peak represented the majority of the total activity (>70%) loaded on the gradient. Fractions containing a protein peak indicative of inner membranes (ρ = 1.135 ± 0.036 mg/ml of sucrose) were devoid of detectable phosphatase activity. In contrast, purified phosphomonoesterase (20,000 U) run under similar conditions was found near the top of the sucrose gradient (Fig. 1, inset). Results from this control experiment suggest that the enzyme’s initial high buoyant density was not an inherent property of the protein itself but more likely a reflection of its association with the membrane. To further demonstrate the enzyme’s localization to the outer membrane, hCHA-cultured H. influenzae R2866 was subjected to a modification of the cytochemical phosphatase localization procedure of Gomori (10). By this procedure, phosphate ions liberated by enzymatic hydrolysis of substrates (phosphomonoesters) are trapped at the site of formation by lead cations present in the buffer and form highly insoluble precipitates of lead phosphate (10). The electron microscopy photomicrograph, taken at ×10,000 magnification (Fig. 2A), shows the deposition of a precipitate at the surface and surrounding intact H. influenzae R2866 cells. The observed precipitation required the concomitant presence of H. influenzae R2866, substrate, and Pb(NO3)2. The absence of any of these components completely abrogated the precipitation. In addition, the presence of divalent cations was essential, as the inclusion of 5 mM EDTA in the assay buffer prevented precipitation on H. influenzae R2866 (Fig. 2B). EDTA chelation of the lead divalent cations was unlikely, since EDTA has low affinity for this cation and formation of lead phosphate precipitate was observed in experiments with an EDTA-resistant acid phosphatase incubated under similar conditions (data not shown). The loss of enzymatic activity by removal of divalent cations was consistent with results from the characterization of the purified phosphomonoesterase. Active phosphatases were required for lead phosphate deposition, as Pseudomonas aeruginosa, which does not have detectable acid phosphatase activity when cultured on hCHA and analyzed by the standard assay, was free of precipitated material, as observed with H. influenzae R2866 (data not shown). The results of these localization studies suggest that the H. influenzae R2866 phosphomonoesterase is not only associated with the outer membrane of the organism but also accessible to substrates, necessary cofactors, and EDTA.
TABLE 2.
Summary of purification of H. influenzae phosphomonoesterase
| Purification step | Total activity (106 U) | Total protein (mg) | Sp act (104 U/mg) | Fold purification | % Yield |
|---|---|---|---|---|---|
| Pre-FPa | 12.8 | 388 | 3.3 | 1.0 | 100 |
| Post-FP | 13.1 | 250 | 5.2 | 1.6 | 102 |
| Pellet I | 12.4 | 143 | 8.6 | 2.6 | 97 |
| Supernatant II | 7.7 | 24 | 31.8 | 9.7 | 60 |
| Post-SP-Sepharose | 2.8 | 0.84 | 330 | 101 | 22 |
| Post-Superose-12 | 0.69 | 0.15 | 460 | 140 | 5 |
FP, French pressure cell treatment.
FIG. 1.
Membrane association of H. influenzae R2866 phosphomonoesterase. Crude H. influenzae R2866 membranes were isolated, subjected to sucrose gradient ultracentrifugation, and fractionated as described in Materials and Methods. The phosphomonoesterase activity (●), relative protein concentration (○), and sucrose density (line) were determined for each fraction. The inset shows localization of the purified phosphomonoesterase run under the same conditions as the enzyme associated with crude membranes.
FIG. 2.
Detection of H. influenzae R2866 phosphomonoesterase activity by transmission electron microscopy of intact bacteria (original magnification, ×10,000). H. influenzae R2866 was cultured on hCHA and exposed to buffer containing the phosphatase substrate pNPP and lead nitrate as an indicator of active phosphomonoesterases in the absence (A) or presence (B) of EDTA. Bar, 500 nm.
Phosphomonoesterase purification.
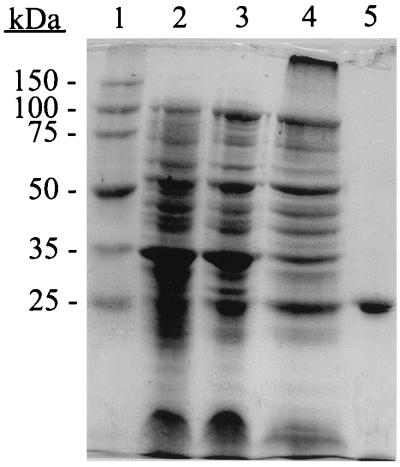
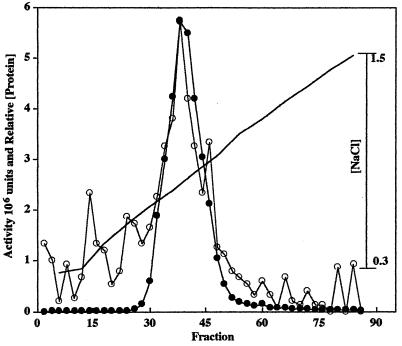
The H. influenzae R2866 phosphatase was intimately associated with the membrane fraction and was enriched two- to threefold upon the separation of membranes, pellet I, from soluble proteins following cell disruption with two cycles of a French press (Table 2). In initial attempts to solubilize the enzyme from the membrane fraction we found that inclusion of EDTA in the extraction buffer resulted in >90% loss of enzymatic activity. Extractions performed in buffer A containing 4% Triton X-100 with EDTA resulted in the liberation of 60 to 80% of total enzyme activity and a three- to fourfold increase in specific activity only after removal of EDTA by dialysis (Table 2) (see Fig. 4, lane 4). The dialyzed enzyme preparation remained in the supernatant fraction following centrifugation at 192,000 × g and was completely retained on a cation-exchange SP-Sepharose resin preequilibrated to pH 6.0 in buffer A. A single peak of phosphatase activity coincident with the major protein peak eluted between 0.67 and 0.92 M NaCl (Fig. 3). The recovered enzyme from the cation-exchange resin was enriched 100-fold over the starting material and contained approximately 22% of the total starting activity. In contrast, the phosphomonoesterase eluted in the void volume during anion-exchange chromatography on Q-Sepharose preequilibrated in 50 mM sodium acetate (pH 6, 7, or 8) (data not shown). Final purification of the enzyme was achieved by gel filtration FPLC. When individual fractions (9 to 12) from the gel filtration chromatography step were subjected to SDS-PAGE and stained with Coomassie R250 or silver stain, a single protein band with a molecular weight of approximately 28,000 was observed on the gel. These fractions were pooled, concentrated, and analyzed for phosphatase activity and protein concentration. The pooled phosphomonoesterase was enriched an additional 1.4-fold over the cation-exchange chromatography step, contained approximately 5% of starting activity, and resulted in the presence of a 28,000-MW protein band when subjected to SDS-PAGE (Fig. 4, lane 5). No difference in the molecular weight of the enzyme was observed whether electrophoresis was performed under reducing or nonreducing conditions.
FIG. 4.
SDS-PAGE separation of proteins in selected samples from the purification procedure. Lane 1, Novagen Perfect Protein standards; lane 2, 25 μg of H. influenzae R2866 cells; lane 3, 25 μg of H. influenzae R2866 cells after French press cell treatment; lane 4, 25 μg of Triton X-100 extraction supernatant postultracentrifugation; lane 5, 5-μg sample in which peak phosphomonoesterase activity was detected after Superose 12 gel filtration chromatography.
FIG. 3.
SP-Sepharose cation-exchange chromatography of Triton X-100-extracted ultracentrifugation supernatant containing enzyme activity with a 0.2 to 1.5 M NaCl linear gradient (line) as described in Materials and Methods. ●, phosphomonoesterase activity; ○, relative protein concentration.
Enzymatic properties of H. influenzae R2866 phosphomonoesterase.
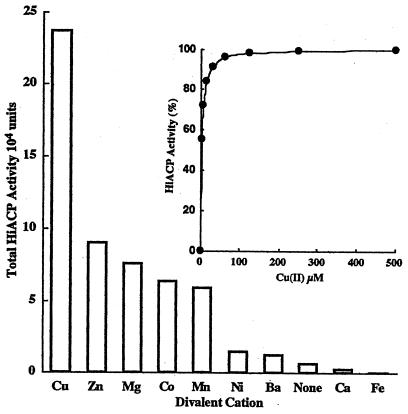
The essential conditions for optimal phosphomonoesterase activity of H. influenzae R2866 e (P4) include the presence of copper, an acidic pH, and arylphosphomonoester substrates. The presence of divalent cations for optimal phosphatase activity was inferred from the purification protocol and localization studies, as exposure of the enzyme to EDTA resulted in abrogation of phosphomonoesterase activity. To identify the essential cation(s) and to determine if loss of activity was reversible, 25,000 U of purified enzyme was dialyzed against 200 ml of 200 mM sodium acetate buffer (pH 6.0) or acetate buffer containing 2 mM EDTA at 4°C. The latter sample was then dialyzed against the acetate buffer to remove EDTA. Samples recovered from both conditions retained less than 2% of the starting enzymatic activity. When enzyme activity was assessed in assay buffer containing different cations, zinc, magnesium, and cobalt (final concentration, 1 mM), each restored less than 40% of the initial activity. The addition of copper, however, restored nearly all enzymatic activity for pNPP hydrolysis in the standard assay buffer (Fig. 5). Combinations of divalent cations, including magnesium and zinc, that are required for the activity of E. coli alkaline phosphatase (6) had no effect on the dialyzed phosphomonoesterase of H. influenzae R2866. The inclusion of cobalt had an inhibitory effect on copper-activated phosphomonoesterase activity (data not shown). The observed copper-mediated restoration of phosphomonoesterase was dependent on the presence of both substrate and native enzyme, as no increase in pNPP hydrolysis was observed in the presence of copper- and heat-inactivated enzyme (data not shown). In addition, an incremental increase in phosphatase activity was observed with increasing CuSO4 concentrations to 100 μM only in the presence of native enzyme. The observed enhancement of enzymatic activity was saturable at approximately 100 μM and remained at the maximum velocity to 500 μM CuSO4 (Fig. 5, inset).
FIG. 5.
Divalent cation requirement of H. influenzae R2866 phosphomonoesterase (Hi ACP) and copper-mediated enhancement of enzyme activity (inset). H. influenzae R2866 phosphomonoesterase (25,000 U) was dialyzed against buffer with and without EDTA at 4°C as described in Materials and Methods. Enzyme activity was assessed in the presence of the divalent cations indicated (final concentration, 1 mM). The inset shows phosphomonoesterase activity (●) of the dialyzed enzyme treated as described above in the presence of increasing amounts of CuSO4.
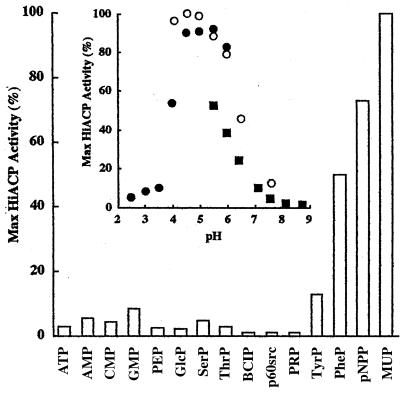
The purified H. influenzae R2866 phosphomonoesterase had narrow in vitro substrate specificity (Fig. 6). Of the 15 phosphomonoesters tested, only 3 were hydrolyzed at higher than 10% of the rate of the best identified substrate, 4-methylumbelliferyl phosphate (4MUP); none of the good substrates were of physiological significance. One physiological substrate, tyrosine phosphate, was hydrolyzed at 10% of the rate of 4MUP. Phosphomonoesterase-mediated hydrolysis of p60src, a phosphotyrosine-containing peptide, was negligible. Copper was required for hydrolysis of all substrates tested. The H. influenzae R2866 phosphomonoesterase behaved as an acid phosphatase, having maximum activity at an acidic pH in all buffers tested (Fig. 6, inset). Optimal activity with 2 mM pNPP as a substrate was achieved at pH 5.0, with approximately 50% of the activity retained at 1.5 pH units to either side of the optimum.
FIG. 6.
Substrate specificity and pH optimum (inset) of H. influenzae R2866 phosphomonoesterase (Hi ACP). The enzyme (525 U) was incubated with each indicated substrate and assessed for phosphatase activity by measuring the amount of inorganic phosphate produced, as described in Materials and Methods. PEP, phosphoenolpyruvate; GlcP, glucose 6-phosphate; SerP, O-phospho-dl-serine; ThrP, O-phospho-dl-threonine; BCIP, 5-bromo-4-chloro-3-indolylphosphate; PRP, pyridoxal 5-phosphate; TyrP, O-phospho-dl-tyrosine; PheP, phenylphosphate. The results are presented as percent activity relative to the amount of inorganic phosphate released from the phosphomonoesterase-catalyzed hydrolysis of 4MUP. Inset, pH optimum of pNPP hydrolysis in the presence of 200 mM buffer (MES [●], sodium acetate [○], or HEPES [■]).
The kinetic parameters of the H. influenzae R2866 phosphomonoesterase were determined for pNPP and 4MUP. Enzyme-mediated hydrolysis of both substrates was linear with time for at least 30 min at 37°C and was also linearly proportional to total enzyme concentration. The Kms determined for 4MUP and pNPP were 0.95 and 0.85 mM, respectively. The Vmaxs for these substrates were 234 and 172 nmol of Pi produced/h/μg for 4MUP and pNPP, respectively.
Consistent with our initial discovery of a tartrate-resistant acid phosphatase of H. influenzae R2866, the purified form of the enzyme was resistant to any inhibitory effects of sodium tartrate to a final concentration of 10 mM (Table 3). Of the common phosphomonoesterase inhibitors tested, including fluoride, phosphate, and the early transition metal oxyanions molybdate and vanadate, inclusion of only the last two anions resulted in any demonstrable inhibition of phosphatase activity in the enzyme assay at pH 5.0. The I50 (calculated concentration of inhibitor expected to inhibit enzymatic activity by 50%) for molybdate was 60 μM, while the I50 for vanadate was 297 μM when pNPP (final concentration, 2 mM) was used as a substrate. Further analysis indicated that vanadate, which is hypothesized to resemble the trigonal bipyramidal transition state formed during phosphomonoester hydrolysis (6), acted as a competitive inhibitor (final concentration, 20 μM) of the H. influenzae R2866 hydrolase. Molybdate (final concentration, 25 μM) was a noncompetitive inhibitor of the phosphatase, having no effect on the observed Km while significantly decreasing the observed Vmax. Interestingly, inorganic phosphate, when tested to a final concentration of 10 mM, had no effect on phosphomonoesterase activity. Initial studies with hydroxymercuriphenylsulfonate, which modifies cysteine residues, resulted in no observable loss of activity of the enzyme. Azide and cyanide are potent inhibitors of the copper-containing enzyme amine oxidase and bind directly to the copper cation, preventing the binding of oxygen to the oxidase (12). These compounds were used to assess the potential role of copper in the catalytic mechanism of the H. influenzae R2866 phosphomonoesterase and were found to have no inhibitory effect under the standard assay conditions. Once the phosphomonoesterase of H. influenzae was identified as lipoprotein e (P4) (see below), a protein essential to the transport of heme in Haemophilus (20), experiments to assess the effects of protoporphyrin IX on phosphatase activity were conducted. Protoporphyrin IX was identified as a competitive inhibitor of the phosphatase activity when present at a final concentration of 30 μM.
TABLE 3.
Effects of various compounds on activity of H. influenzae phosphomonoesterasea
| Inhibitor | Max concentration tested (mM) | Inhibition |
|---|---|---|
| Azide | 1 | NIb |
| Cyanide | 1 | NI |
| DEPCe | 10 | NI |
| Fluoride | 1 | NI |
| HMPSc | 1 | NI |
| Phosphate | 10 | NI |
| Tartrate | 10 | NI |
| EDTA | 1 | Id |
| Molybdate | 1 | Noncompetitive (25 μM) |
| Protoporphyrin IX | 0.08 | Competitive (30 μM) |
| Vanadate | 1 | Competitive (20 μM) |
Each inhibitor, solubilized in distilled H2O, was tested for effect on pH before use as an inhibitor.
NI, no inhibition detected.
HMPS, hydroxymercuriphenylsulfonate.
I, chelation of divalent copper.
DEPC, diethyloprocarbonate.
Identification of H. influenzae phosphomonoesterase as lipoprotein e (P4).
N-terminal amino acid analysis of the purified 28-kDa phosphomonoesterase was unsuccessful, consistent with the presence of a blocked N-terminal amino acid residue. Amino acid sequence determination from cyanogen-bromide-derived peptide fragments yielded the sequences RLGFNGVEESAFYLK for peptide 1 and LPNANYGGWE for peptide 2. Database searches showed a 100% identity in a 15-amino-acid overlap (R161 to K175) for peptide 1 and in a 10-amino-acid overlap (L237 to E246) for peptide 2 between the determined sequence and the deduced amino acid sequence of H. influenzae lipoprotein e (P4) (GenBank accession no. P26093 [11]; The Institute for Genomic Research HI0693 [8]).
To confirm that the H. influenzae lipoprotein e (P4), encoded by the hel gene, was the copper-dependent phosphomonoesterase, plasmids containing the hel gene of strain Rd KW20 were obtained from two independent sources. E. coli strains containing plasmids pGHIGU90, a high-copy-number pUC19-based plasmid, and pJRP4, a low-copy-number pACYC184-based plasmid, were obtained from the ATCC and John Mekalanos, respectively. E. coli DH5α containing pGHIGU90 had substantially higher copper-dependent specific phosphomonoesterase activity than the E. coli(pUC19) control (Table 4). To ensure that the observed activity was encoded by the hel gene, an EcoRI fragment containing the hel gene as the only open reading frame was then subcloned into pBluescript KS and transformed into E. coli DH5α, resulting in E. coli DH5α(phel1), and assessed for phosphomonoesterase activity. Levels of specific phosphomonoesterase activity similar to those observed in E. coli DH5α containing plasmid pGHIGU90 were observed in this new construct (Table 4). The activity observed in E. coli DH5α(pGHIGU90) and E. coli DH5α(phel1) was similar to that produced in H. influenzae R2866, as the activity was intimately associated with the crude membrane fraction. In addition, E. coli DH5α(phel3) containing a single copy of the hel gene obtained from plasmid pJRP4 and inserted at the BamHI site of pBluescript also produced significant levels of the copper-dependent, membrane-associated enzyme activity, consistent with our conclusion that the H. influenzae hel gene encodes the phosphomonoesterase.
TABLE 4.
Specific phosphomonoesterase activity of E. coli transformed with hel gene-bearing plasmids
| Plasmid | Fraction | Sp act (104 U/mg) |
|---|---|---|
| pUC19 | Whole cells | 0.1 |
| Membranes | 0.5 | |
| pBluescript KS | Whole cells | 0.2 |
| Membranes | 0.1 | |
| pGHIGU90 | Whole cells | 0.9 |
| Membranes | 3.0 | |
| phel1 | Whole cells | 1.8 |
| Membranes | 2.5 | |
| phel3 | Whole cells | 0.6 |
| Membranes | 1.5 |
DISCUSSION
We have demonstrated that the bacterium H. influenzae is highly enriched with a surface-localized copper-dependent phosphomonoesterase. The 28-kDa membrane-associated protein had narrow substrate specificity and a pH optimum of 5.0. It was inhibited by EDTA, molybdate, and vanadate but was resistant to tartrate, fluoride, and inorganic phosphate. Sequencing of cyanogen bromide-derived peptides from the purified protein, coupled with detection of a similar phosphomonoesterase activity in E. coli transformed with plasmids containing the hel gene, provided sufficient evidence to attribute the observed phosphomonoesterase activity to H. influenzae e (P4). These results confirm Malke’s speculation that H. influenzae e (P4) is a phosphomonoesterase, based on amino acid sequence comparison to LppC from Streptococcus equisimilis (9, 16). These results are also consistent with the suggestion by Thaller et al. and Rossolini et al. that H. influenzae e (P4) possesses phosphomonoesterase activity at an Mr of approximately 28,000 when assayed by specific staining procedures to detect phosphomonoesterase activity after SDS-PAGE and renaturation of the resolved proteins (24, 30). The work presented here represents the first demonstration of phosphomonoesterase activity of H. influenzae e (P4) in purified, native preparations of the protein. The results from this preliminary characterization of e (P4) phosphatase are also suggestive of an unusual catalytic mechanism. The concomitant presence of phosphomonoesterase activity and heme transport activity (20) within H. influenzae lipoprotein e (P4) may indicate a requisite dephosphorylation event involved in heme transport.
While a plethora of bacterial acid phosphatases have been identified and characterized, common biochemical properties and homologous amino acid sequence similarities suggest that most of these enzymes fall into one of three recognized classes (24). The class A phosphatases are secreted enzymes composed of low-molecular-weight polypeptides; they possess broad substrate specificity and are EDTA resistant. The class B enzymes are polymeric, possess broad substrate specificity, and are EDTA sensitive. The class C acid phosphatases were recently identified as a group of secreted bacterial lipoproteins endowed with phosphatase activity and are distantly related to the class B phosphatases. The classification of the class C enzymes was based on nucleotide sequence and characterization by zymogram assays, but biochemical characterization of purified preparations of these enzymes has yet to be reported. Deduced amino acid sequence of e (P4) (11) and observed acid phosphatase activity in a zymogram assay (30) are consistent with its classification as a class C phosphatase.
Characterization of the phosphatase under investigation in this study identified it as e (P4) and showed that it possessed properties of secreted bacterial lipoproteins (3), including intimate association with the bacterial outer membrane, an N terminus resistant to Edman degradation, and aggregation of protein during gel filtration chromatography. Unlike enzymes of class A or B, the e (P4) phosphatase had narrow substrate specificity, with the highest activity for arylphosphates, similar to the substrate specificities of low-molecular-weight nonspecific acid phosphatases purified from rat brain (19), human placenta (37), and bovine heart (40). Like its distantly related class B homologs, e (P4) was EDTA sensitive, had a pH optimum of 5.0 for hydrolysis, and did not catalyze the dephosphorylation of 5-bromo-4-chloro-3-indolylphosphate. While aggregation of purified enzyme prevented determination of its native molecular weight, preliminary evidence from a recombinant nonlipidated form of the enzyme suggests that it exists as a dimer when subjected to gel filtration chromatography (data not shown).
The phosphomonoesterase activity of e (P4) was unusual in its absolute requirement for copper and its lack of sensitivity to inorganic phosphate. While many phosphohydrolases are metalloenzymes, including E. coli alkaline phosphatase (6) and the purple acid phosphatases (36), none characterized to date are copper dependent. The acid phosphatases of Mycoplasma fermentans and P. aeruginosa are enhanced in the presence of copper; however, neither enzyme has an absolute requirement for the cation (7, 18). Removal of divalent cations from e (P4) by EDTA or by dialysis against acetate buffer resulted in the loss of enzymatic activity. Phosphatase activity was fully restored upon addition of copper to the native enzyme. These results suggest that the cation is not tightly bound to the protein but that its presence is required for catalytic activity. Whether catalytic or structural, the role of copper in e (P4) phosphomonoesterase-mediated catalysis has yet to be determined. Another unusual feature of the e (P4) phosphomonoesterase was its resistance to inhibition by inorganic phosphate. Typically, acid phosphatases catalyze the hydrolysis of phosphomonoesters via a two-step mechanism in which a noncovalent inorganic phosphoenzyme complex is a short-lived intermediate (35). The existence of such an intermediate has been demonstrated in some well-characterized phosphomonoesterases (36). Lipoprotein e (P4)’s lack of sensitivity to phosphate-mediated inhibition suggests that such an intermediate does not exist or that it is inaccessible to the phosphate anion. Copper dependency and lack of sensitivity to inorganic phosphate not only suggest that the e (P4) phosphomonoesterase is a unique enzyme catalytically but also may be essential properties of a membrane-bound phosphomonoesterase involved in H. influenzae heme transport.
Unlike the acid phosphatases of some bacterial pathogens (2, 5, 10), a direct role for the H. influenzae phosphomonoesterase activity in the pathogen’s survival within a host has yet to be determined. It is of considerable interest that the purified phosphatase activity is covalently associated with a protein involved in hemin transport into this heme-requiring bacterium (20). Results from previous studies have demonstrated that expression of H. influenzae lipoprotein e (P4) by an E. coli hemA mutant, defective in de novo synthesis of porphyrin, allowed aerobic growth of the mutant strain when supplied with supplemental heme. These results coupled with the construction of an H. influenzae hel mutant which was incapable of aerobic growth suggest that H. influenzae lipoprotein e (P4) is essential to the acquisition of exogenous heme. While acid phosphatase LppC of S. equisimilis has significant homology to H. influenzae e (P4), an E. coli hemA strain expressing this phosphatase was unable to grow aerobically in the presence of exogenously supplied heme, suggesting that phosphomonoesterase activity alone is not sufficient for heme acquisition. Possible roles of heme transport-associated phosphomonoesterase activity include substrate modification and mediation of the phosphorylation state of other components of a putative heme transport complex. Data in support of substrate modification were provided by the observation that protoporphyrin IX was a competitive inhibitor of enzyme activity, suggesting that not only do the heme binding motif, KVAFDH, and phosphatase activity reside within the same covalent structure, they also may be in close proximity to one another.
Alternatively, the e (P4)-associated phosphomonoesterase may function as a phosphorylation state regulator in a heme uptake system, such as the hemopexin transport complex recently identified in H. influenzae. Wong et al. described isolation of a hemopexin binding receptor complex from H. influenzae composed of 57-, 38-, and 29-kDa proteins, of which the last had a blocked N terminus (39). We, like others (20), believe this 29-kDa component to be lipoprotein e (P4). While the lack of broad substrate specificity limits the number of hypothesized biological roles for the enzyme, it does provide the inherent property of narrow specificity, suggesting that perhaps an outer membrane-localized and specific substrate does exist and that its phosphorylation state is essential to heme transport.
Production of a soluble overexpressed phosphomonoesterase is in progress so that we may begin to investigate this protein’s interesting enzymology and its possible interactions with surface-localized Haemophilus components and to delineate its structure. In addition, experiments with site-directed mutations in the hel gene are in progress and may help elucidate the role of the phosphomonoesterase in the transport of heme, in pathogenesis, and ultimately in the growth and survival of H. influenzae.
ACKNOWLEDGMENTS
We thank the University of Missouri Electron Microscopy Facility, School of Veterinary Medicine, and the University’s Protein Core Facility for use of their electron microscopy suite and sequencing of the peptides, respectively. We also thank Olen Brown and Richard Finkelstein for use of their FPLC system, including chromatography resins and French press, respectively. We are also grateful to Mark Kuhlenschmidt (University of Illinois College of Veterinary Medicine, Urbana, Ill.) for his many helpful suggestions in this endeavor and to Frederick Greenaway (Clark University, Department of Chemistry, Worcester, Mass.) for his helpful suggestions regarding the potential role of copper in enzyme activity. We also thank Gian Rossolini (Dipartimento di Biologia Molecolare, Sezione di Micobiologia Università di Siena, Siena, Italy) for help in acquiring recently published references and Leah Cohn for her many helpful suggestions in the preparation of the manuscript.
This work was supported by National Institute of Health grants T32 AI07276 (T.J.R.) and F32 AI 10053 (D.L.C.) and by the University of Missouri Research Board (A.L.S.).
REFERENCES
- 1.Baca O G, Roman M J, Glew R H, Christner R F, Buhler J E, Aragon A S. Acid phosphatase activity in Coxiella burnetii: a possible virulence factor. Infect Immun. 1993;61:4232–4239. doi: 10.1128/iai.61.10.4232-4239.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bliska J B, Clemens J C, Dixon J E, Falkow S. The Yersinia tyrosine phosphatase: specificity of a bacterial virulence determinant for phosphoproteins in the J774a.1 macrophage. J Exp Med. 1992;176:1625–1630. doi: 10.1084/jem.176.6.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun V, Wu H C. Lipoproteins, structure, function, biosynthesis and model for protein export. New Comp Biochem. 1994;27:319–341. [Google Scholar]
- 4.Brooks S P J. A simple computer program with statistical tests for the analysis of enzyme kinetics. BioTechniques. 1992;13:906–911. [PubMed] [Google Scholar]
- 5.Chhatwal G S, Walker M J, Yan H, Timmis K N, Guzman C A. Temperature dependent expression of an acid phosphatase by Bordetella bronchiseptica: role in intracellular survival. Microb Pathog. 1997;22:257–264. doi: 10.1006/mpat.1996.0118. [DOI] [PubMed] [Google Scholar]
- 6.Coleman J E, Besman M J. Phosphomonoesterases. In: Neuberger A, Brocklenhurst K, editors. Hydrolytic enzymes. New York, N.Y: Elsevier Science; 1987. pp. 377–406. [Google Scholar]
- 7.Domenich C E, Teresita A L, Salvano M A, Garrido M N. Pseudomonas aeruginosa acid phosphatase: activation by divalent cations and inhibition by aluminum ion. FEBS Lett. 1992;299:96–98. doi: 10.1016/0014-5793(92)80108-s. [DOI] [PubMed] [Google Scholar]
- 8.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, McKenney K, Sutton G, FitzHugh W, Fields C, Gocayne J D, Scott J, Shirley R, Liu L I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 9.Gase K, Liu G, Bruckmann A, Steiner K, Ozegowski J, Malke H. The lppC gene of Streptococcus equisimilis encodes a lipoprotein that is homologous to the e (P4) outer membrane protein from Haemophilus influenzae. Med Microbiol Immunol. 1997;186:63–73. doi: 10.1007/s004300050047. [DOI] [PubMed] [Google Scholar]
- 10.Gomori G. Microscopic histochemistry: principles and practice. Chicago, Ill: University of Chicago Press; 1952. [Google Scholar]
- 11.Green B A, Farley J E, Quinn-Dey T, Deich R A, Zlotnick G W. The e (P4) outer membrane protein of Haemophilus influenzae: biologic activity of anti-e serum and cloning and sequencing of the structural gene. Infect Immun. 1991;59:3191–3198. doi: 10.1128/iai.59.9.3191-3198.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He Z, Zou Y, Greenaway F T. Mechanism-based inactivation of porcine kidney diamine oxidase by 1,4-diamino-2-butene. Arch Biochem Biophys. 1995;319:185–195. doi: 10.1016/0167-4838(95)00158-q. [DOI] [PubMed] [Google Scholar]
- 13.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–695. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 14.Lanzetta P A, Alvarez L J, Reinach P S, Candia O A. An improved assay for nanomole amounts of inorganic phosphate. Anal Biochem. 1979;100:95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- 15.Loeb M R, Smith D H. Outer membrane protein composition in disease isolates of Haemophilus influenzae: pathogenic and epidemiological implications. Infect Immun. 1980;30:709–717. doi: 10.1128/iai.30.3.709-717.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malke H. Cytoplasmic membrane lipoprotein LppC of Streptococcus equisimilis functions as an acid phosphatase. Appl Environ Microbiol. 1998;64:2439–2442. doi: 10.1128/aem.64.7.2439-2442.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nizet V, Colina K F, Almquist J R, Rubens C E, Smith A L. Virulent nonencapsulated Haemophilus influenzae. J Infect Dis. 1996;173:180–186. doi: 10.1093/infdis/173.1.180. [DOI] [PubMed] [Google Scholar]
- 18.Noda M, Shibata K, Sawa Y, Shimokoube H, Watanabe T. Purification and characterization of an acid phosphatase from Mycoplasma fermentans. Microbiol Immunol. 1994;38:103–107. doi: 10.1111/j.1348-0421.1994.tb01750.x. [DOI] [PubMed] [Google Scholar]
- 19.Okada M, Owada K, Nakagawa H. [Phosphotyrosine] protein phosphatase in rat brain: a major phosphotyrosine protein phosphatase is a 23 kD protein distinct from acid phosphatase. Biochem J. 1986;289:155–162. doi: 10.1042/bj2390155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reidl J, Mekalanos J J. Lipoprotein e(P4) is essential for hemin uptake by Haemophilus influenzae. J Exp Med. 1996;183:621–629. doi: 10.1084/jem.183.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reilly T J, Chance D L, Smith A L. Abstracts of the 98th General Meeting of the American Society for Microbiology. 1998. Identification and partial purification and characterization of an outer membrane phosphomonoesterase from a nontypeable Haemophilus influenzae; p. 300. Washington, D.C. [Google Scholar]
- 22.Reilly T J, Baron G S, Nano F E, Kuhlenschmidt M S. Characterization and sequencing of a respiratory burst-inhibiting acid phosphatase from Francisella tularensis. J Biol Chem. 1996;271:10973–10983. doi: 10.1074/jbc.271.18.10973. [DOI] [PubMed] [Google Scholar]
- 23.Remaley A T, Kuhnst D B, Basford R E, Glew R H, Kaplan S S. Leishmanial phosphatase blocks neutrophil O2− production. J Biol Chem. 1984;259:11173–11175. [PubMed] [Google Scholar]
- 24.Rossolini G M, Schippa S, Riccio M L, Berlutti F, Macaskie L E, Thaller M C. Bacterial nonspecific acid phosphohydrolases: physiology, evolution, and use as tools in microbial biotechnology. Cell Mol Life Sci. 1998;54:833–850. doi: 10.1007/s000180050212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saha A K, Dowling J N, LaMarco K L, Das S, Remaley A T, Olomu N, Pope M T, Glew R H. Properties of an acid phosphatase from Legionella micdadei which blocks superoxide anion production by human neutrophils. Arch Biochem Biophys. 1985;243:150–160. doi: 10.1016/0003-9861(85)90783-0. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 27.Schnaitman C A. Protein composition of the cell wall and cytoplasmic membrane of Escherichia coli. J Bacteriol. 1970;104:890–901. doi: 10.1128/jb.104.2.890-901.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith P K, Krohn R I, Hermanson G T, Mallia A K, Gartner F H, Provenzano M D, Fujimoto E K, Goeke N M, Olson B J, Klenk D C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1995;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 29.St. Geme J W. Progress towards a vaccine for nontypeable Haemophilus influenzae. Ann Med. 1996;28:31–37. doi: 10.3109/07853899608999071. [DOI] [PubMed] [Google Scholar]
- 30.Thaller M C, Schippa S, Rossolini G M. Conserved sequence motifs among bacterial eukaryotic and archaeal phosphatases that define a new phosphohydrolase superfamily. Protein Sci. 1998;7:1647–1652. doi: 10.1002/pro.5560070722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian Z, Gu C, Roeske R W, Zhou M, Van Etten R L. Synthesis of phosphotyrosine-containing peptides by solid-phase method. Int J Peptide Protein Res. 1993;42:155–158. doi: 10.1111/j.1399-3011.1993.tb00491.x. [DOI] [PubMed] [Google Scholar]
- 32.Turk D C. Pathogenicity of Haemophilus influenzae. J Med Microbiol. 1984;18:1–16. doi: 10.1099/00222615-18-1-1. [DOI] [PubMed] [Google Scholar]
- 33.Turk D C. Clinical importance of Haemophilus influenzae—1981. In: Sell S H, Wright P F, editors. Haemophilus influenzae: epidemiology, immunology and prevention of disease. New York, N.Y: Elsevier Biomedical; 1982. pp. 3–9. [Google Scholar]
- 34.Vachon V D, Lyew J, Coulton J W. Transmembrane permeability channels across the outer membrane of Haemophilus influenzae type b. J Bacteriol. 1985;162:918–925. doi: 10.1128/jb.162.3.918-924.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vincent J B, Crowder M W, Averill B A. Evidence for a phosphoryl-enzyme intermediate in phosphate ester hydrolysis by purple acid phosphatase from bovine spleen. J Biol Chem. 1991;266:17737–17740. [PubMed] [Google Scholar]
- 36.Vincent J B, Crowder M W, Averill B A. Hydrolysis of phosphate monoesters: a biological problem with multiple chemical solutions. Trends Biochem Sci. 1992;17:105–110. doi: 10.1016/0968-0004(92)90246-6. [DOI] [PubMed] [Google Scholar]
- 37.Waheed A, Laidler P M, Wo Y Y P, Van Etten R L. Purification and physiochemical characterization of a human placental acid phosphatase possessing phosphotyrosyl protein phosphatase activity. Biochemistry. 1988;27:4265–4273. doi: 10.1021/bi00412a010. [DOI] [PubMed] [Google Scholar]
- 38.Wilcox K W, Smith H O. Isolation and characterization of mutants of Haemophilus influenzae deficient in an adenosine 5′-triphosphate-dependent deoxyribonuclease activity. J Bacteriol. 1975;122:443–453. doi: 10.1128/jb.122.2.443-453.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong J C, Patel Y R, Kendall D, Whitby P W, Smith A, Holland J, Williams P. Affinity, conservation, and surface exposure of hemopexin binding proteins in Haemophilus influenzae. Infect Immun. 1995;63:2327–2333. doi: 10.1128/iai.63.6.2327-2333.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Z Y, Van Etten R L. Purification and characterization of a low molecular weight acid phosphatase-A phosphotyrosyl protein phosphatase from bovine heart. Arch Biochem Biophys. 1990;282:39–49. doi: 10.1016/0003-9861(90)90084-c. [DOI] [PubMed] [Google Scholar]