Abstract
The ccr gene, encoding crotonyl coenzyme A (CoA) reductase (CCR), was cloned from Streptomyces cinnamonensis C730.1 and shown to encode a protein with 90% amino acid sequence identity to the CCRs of Streptomyces collinus and Streptomyces coelicolor. A ccr-disrupted mutant, S. cinnamonensis L1, was constructed by inserting the hyg resistance gene into a unique BglII site within the ccr coding region. By use of the ermE* promoter, the S. collinus ccr gene was expressed from plasmids in S. cinnamonensis C730.1/pHL18 and L1/pHL18. CCR activity in mutant L1 was shown to decrease by more than 90% in both yeast extract-malt extract (YEME) medium and a complex fermentation medium, compared to that in wild-type C730.1. Compared to C730.1, mutants C730.1/pHL18 and L1/pHL18 exhibited a huge increase in CCR activity (14- and 13-fold, respectively) in YEME medium and a moderate increase (3.7- and 2.7-fold, respectively) in the complex fermentation medium. In the complex fermentation medium, S. cinnamonensis L1 produced monensins A and B in a ratio of 12:88, dramatically lower than the 50:50 ratio observed for both C730.1 and C730.1/pHL18. Plasmid (pHL18)-based expression of the S. collinus ccr gene in mutant L1 increased the monensin A/monensin B ratio to 42:58. Labeling experiments with [1,2-13C2]acetate demonstrated the same levels of intact incorporation of this material into the butyrate-derived portion of monensin A in both C730.1 and mutant C730.1/pLH18 but a markedly decreased level of such incorporation in mutant L1. The addition of crotonic acid at 15 mM led to significant increases in the monensin A/monensin B ratio in C730.1 and C730.1/pHL18 but had no effect in S. cinnamonensis L1. These results demonstrate that CCR plays a significant role in providing butyryl-CoA for monensin A biosynthesis and is present in wild-type S. cinnamonensis C730.1 at a level sufficient that the availability of the appropriate substrate (crotonyl-CoA) is limiting.
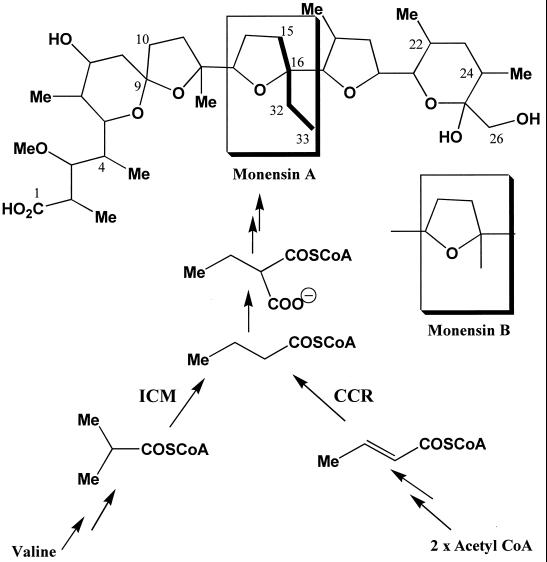
Polyketide synthases (PKSs) produce natural products such as erythromycin, pikromycin, and rifamycin by catalyzing successive decarboxylative condensations with malonyl coenzyme A (CoA) and methylmalonyl-CoA and an appropriate starter unit (2, 22, 28). Malonyl-CoA is likely derived from the carboxylation of acetyl-CoA, while a variety of different pathways give rise to methylmalonyl-CoA (18, 24). A number of streptomycete PKSs, such as those involved in monensin, FK520, tylosin, and niddamycin production, also use ethylmalonyl-CoA at a specific stage in polyketide chain assembly (6, 8, 14, 18). Ethylmalonyl-CoA is likely derived from the carboxylation of butyryl-CoA. For monensin and FK520, either methylmalonyl-CoA or ethylmalonyl-CoA can be used at the same stage in elongation, presumably reflecting a relaxed substrate specificity for the corresponding acyltransferase domain of the PKS (3, 6, 9). Thus, fermentations of Streptomyces cinnamonensis produce a mixture of monensins A and B (Fig. 1) in a ratio presumably dependent upon the relative concentrations of ethylmalonyl-CoA and methylmalonyl-CoA. Genetic manipulation of the pathways that play an important role in butyryl-CoA production should significantly alter the monensin A/monensin B ratio.
FIG. 1.
Role of CCR in providing a butyryl-CoA precursor for monensin A biosynthesis.
Stable isotope incorporation experiments have indicated the presence of at least two pathways for butyryl-CoA production in streptomycetes. One pathway involves isomerization of the valine catabolite isobutyryl-CoA to form butyryl-CoA and is catalyzed by coenzyme B12-dependent isobutyryl-CoA mutase (ICM) (18, 29). The second pathway involves the condensation of two acetate units and is thought to culminate in the reduction of crotonyl-CoA to butyryl-CoA, catalyzed by crotonyl-CoA reductase (CCR) (25). This enzyme was first purified from Streptomyces collinus, and the corresponding gene, ccr, was shown to be located within a set of primary metabolic genes involved in acetate assimilation in S. collinus (25). A similar set of genes was recently identified from sequencing of the Streptomyces coelicolor chromosome (Fig. 2). Subsequently, ccr homologs were observed within the biosynthetic gene clusters of tylosin, niddamycin, and coronafacic acid, all natural products made with an ethylmalonyl-CoA precursor (8, 17, 22). Despite these observations, the role of these homologs or ccr itself in providing butyryl-CoA for polyketide biosynthesis by the corresponding producing organisms has yet to be established.
FIG. 2.
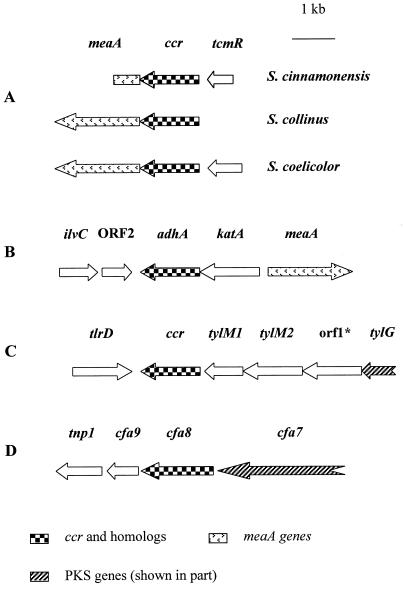
Comparison of genetic organization surrounding ccr and its homologs in different bacteria. (A) S. cinnamonensis, S. collinus, and S. coelicolor. (B) M. extorquens. (C) Streptomyces fradiae. (D) Pseudomonas syringae. ORFs and their orientations are indicated by arrows. ccr encodes CCR, meaA encodes a coenzyme B12-dependent mutase, tcmR encodes a product with homology to the tetracenomycin C transcriptional repressor, adhA encodes alcohol dehydrogenase, katA encodes catalase, ilvC encodes acetohydroxy acid isomeroreductase, tlrD is a Tyr (tylosin resistance)-encoding gene, tylM1 encodes methyltransferase, tylM2 encodes glycosyltransferase, tylG encodes tylosin PKS, cfa7 encodes coronafacic acid PKS, cfa8 encodes oxidoreductase, cfa9 encodes thioesterase, and tnp1 encodes transposase.
In the current study, S. cinnamonensis ccr has been cloned, sequenced, and shown to be located within a conserved set of primary metabolic genes. This gene, rather than any putative ccr homologs located within the monensin PKS gene cluster, is primarily responsible for CCR activity in S. cinnamonensis and has a significant role in producing butyryl-CoA for monensin A biosynthesis. Heterologous expression of S. collinus ccr in S. cinnamonensis produces increased levels of CCR but does not change the monensin A/monensin B ratio. This latter observation contrasts the recent observation that heterologous expression of the S. collinus ccr gene is necessary for the production of 6-desmethyl-6-ethylerythromycin in a Saccharopolyspora erythraea strain expressing a hybrid PKS which contains a methylmalonyl-CoA–ethylmalonyl-CoA acyltransferase switch (22).
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. S. cinnamonensis C703.1 was kindly provided by Eli Lilly & Company. Escherichia coli XL1-Blue and ET12567 were grown at 37°C in Luria-Bertani medium supplemented with either ampicillin (100 μg/ml) or apramycin (50 μg/ml) when necessary (20). Cultures of S. cinnamonensis and Streptomyces lividans were grown in YEME medium (13) at 30°C for isolation of genomic and plasmid DNAs, preparation of protoplasts, CCR enzyme assay, and fatty acid analysis. R2YE medium (13) was used for preparation of Streptomyces spore suspensions and for regeneration of protoplasts after transformations.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| S. cinnamonensis | ||
| C730.1 | Wild type | Eli Lilly & Company |
| L1 | ccr-disrupted mutant | This work |
| C730.1/pHL18 | C730.1 carrying pHL18 | This work |
| L1/pHL18 | L1 carrying pHL18 | This work |
| S. lividans 1326 | SLP2 SLP3 | 13 |
| E. coli | ||
| XL1-Blue | F′::Tn10 proA+B+ lac1q Δ(lacZ)M15 | Stratagene |
| ET12567 | F−dam-13::Tn9 dcm-6 | 16 |
| Plasmids | ||
| pUC119 | High-copy-number E. coli vector, Apr | New England Biolabs |
| pKC1139 | Streptomyces-E. coli bifunctional vector, Amrrepts | 5 |
| pIJ963 | pUC18 derivative with 1.7-kb hyg gene fragment | 15 |
| pZYB3 | pET3C with 1.4-kb S. collinus ccr fragment | 25 |
| pSE34 | pWHM3 with ermE* promoter | Pfizer Inc. |
| pHL1 | pUC119 with 5.7-kb PstI insert containing S. cinnamonensis ccr | This work |
| pHL7 | pUC119 with 3.0-kb PstI-NcoI fragment from pHL1 | This work |
| pHL9 | pHL7 with hyg inserted in the ccr coding region | This work |
| pHL18 | pSE34 with 1.9-kb XbaI-HindIII S. collinus ccr fragment from pZYB3 | This work |
| pHL19 | pKC1139 with 4.7-kb ccr::hyg fragment from pHL9 | This work |
Apr, ampicillin resistance; Amr, apramycin resistance; repts, temperature-sensitive replicon.
DNA isolation, amplification, and manipulation.
Streptomyces genomic DNA was prepared following standard protocols (13). Alkaline lysis was used to isolate plasmid DNA from Streptomyces strains (13). E. coli plasmid DNA was prepared with a Sigma P-MINI kit. Oligonucleotides for PCR were obtained from Gibco BRL (Gaithersburg, Md.). PCR amplifications were carried out with a GeneAmp 2400 PCR system from Perkin-Elmer (Branchburg, N.J.). General DNA manipulations were performed following standard protocols (20).
Hybridizations.
For Southern hybridization, genomic DNA from S. cinnamonensis C730.1 was completely digested with a range of restriction endonucleases. The fragments, after separation by agarose gel electrophoresis, were transferred to nylon membranes (20). For colony hybridization, E. coli colonies were replica transferred to nylon membranes. The cells were then lysed, and the DNA was denatured, neutralized, and immobilized (20). The radiolabeled probe was prepared with [α-32P]dCTP (3,000 Ci/mmol; Amersham), a random-primer labeling kit from Strategene (La Jolla, Calif.), and a 1.4-kb S. collinus ccr gene fragment from pZYB3 (25). Conditions for prehybridization and hybridization were slightly modified from standard protocols (20), with final washes for Southern hybridization and colony hybridization being carried out at 30°C in 1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and at 65°C in 0.5× SSC, respectively.
Nucleotide sequence analysis.
Fragments of S. cinnamonensis genomic DNA containing ccr were subcloned into pUC119, and the corresponding plasmids were recovered from E. coli XL1-Blue. DNA sequencing was carried out at the Biopolymer Laboratory at the University of Maryland, Baltimore. Sequence data were analyzed with MacVector software (version 6.01; Oxford Molecular Ltd.). The nucleotide and deduced amino acid sequences were compared with those in public sequence databases by use of the BLAST family of programs (1).
Transformations.
Preparation and transformation of competent E. coli cells were performed by standard methods (20). Streptomyces protoplasts were transformed in the presence of 25% polyethylene glycol (13). To transform S. cinnamonensis, plasmid DNA prepared from S. lividans 1326 or E. coli ET12567 was used.
Insertional inactivation of S. cinnamonensis ccr.
A shuttle vector (pKC1139) containing the temperature-sensitive Streptomyces origin of replication from pSG5 was used to construct ccr gene disruption plasmids (5). A 1.7-kb BglII hyg (hygromycin resistance) gene fragment was excised from a pIJ963 (15) derivative in which the BamHI site had been destroyed. This hyg gene fragment was inserted in the coding region of S. cinnamonensis ccr. The primers used for PCR identification of the double-crossover mutant were CCR1 (5′-AAGCAGGGCGACAACGTTCGGATCC-3′) and CCR2 (5′-GATGTCGATCGGGATCCACTTGGGG-3′).
Enzyme assay for CCR.
The CCR activity of the S. cinnamonensis cultures grown in either YEME medium or the production medium (described below) was analyzed as previously described (25).
Fatty acid analysis.
S. cinnamonensis cultures were grown in YEME medium at 30°C and 300 rpm for 24 h, and the fatty acids were extracted and analyzed as described previously (27).
Production and quantitation of monensins A and B.
A two-stage fermentation process was used for monensin production. In the first stage, S. cinnamonensis spore suspensions were inoculated into a seed medium consisting of glucose (2.5%), soybean meal (1.5%), CaCO3 (0.3%), FeSO4 · 7H2O (0.03%), and MnCl2 · 4H2O (0.003%); incubations were carried out at 30°C and 300 rpm for 30 h. In the second stage, a 5% inoculum of the seed cultures was transferred to a production medium, which was the same as the seed medium but which contained 5% glucose. Fermentations were carried out at 30°C and 300 rpm for 6 days. For feeding studies with either crotonic acid or butyric acid, the compounds were added as stock solutions of 1 M and pH 7.0 in equal portions at 48, 60, and 72 h during fermentation to final concentrations of 15 mM. Monensins A and B were isolated from the whole broth by the standard method (18), which involves homogenization with methanol and extraction with chloroform. The crude extracts containing the antibiotics were finally dissolved in methanol and analyzed by high-pressure liquid chromatography (HPLC) with a C18 column and a refractive index detector (3). The mobile phase used in HPLC analysis was composed of methanol and water (90:10). Monensin A/monensin B ratios were determined as the ratios of the corresponding peak areas. Total monensin titers were quantified by comparison of the sum of monensin A and B peak areas against the standard curve obtained with the commercial monensin A product from Sigma.
Isotope labeling experiments with [1,2-13C2]acetate.
Conditions for the production and preliminary extraction of monensins A and B used in these studies were identical to those described above. A 1:3 mixture of [1,2-13C2]acetate and unlabeled acetate was prepared at neutral pH and added batchwise at 48, 60, and 72 h during the production phase of S. cinnamonensis fermentations to a final concentration of 30 mM. Monensin A was purified from organic extracts of fermentations of S. cinnamonensis C730.1 (400 ml), C730.1/pHL18 (400 ml), and S. cinnamonensis L1 (600 ml) following standard procedures (18). The 13C {1H} NMR spectra of monensin A purified from each fermentation were recorded on a 300-MHz spectrometer.
Nucleotide sequence accession number.
The sequence of S. cinnamonensis ccr and the meaA fragment reported here has been deposited in the GenBank database under accession no. AF178673.
RESULTS
Cloning and sequence analysis of S. cinnamonensis ccr.
Southern analysis with a 1.4-kb NdeI-BamH fragment of pZYB3 (containing the entire S. collinus ccr gene) as a probe revealed a single 5.7-kb hybridizing fragment from a PstI digest of S. cinnamonensis C730.1 genomic DNA. A 3.0-kb region of this fragment was sequenced and shown to contain one complete (encoding 453 amino acids) and two incomplete (encoding 167 and 228 amino acids) open reading frames (ORFs), all transcribed in the same direction (Fig. 2). Sequence analysis showed that the complete ORF designated ccr encodes a CCR with the highest predicted amino acid sequence identity (90%) and similarity (93%) to the CCR of S. collinus (25) and the putative CCR of S. coelicolor (GenBank accession no. AL035161). S. cinnamonensis ccr has an meaA gene (encoding a putative coenzyme B12-dependent mutase) (12) located downstream and a tcmR-homologous gene (encoding a putative transcriptional regulator) (10) located upstream (Fig. 2). Sequencing data available to date are consistent with this organization of genes in S. collinus and S. coelicolor (12). The requirement of ccr and meaA for efficient growth of S. collinus on acetate has also been established (12).
Targeted disruption of S. cinnamonensis ccr.
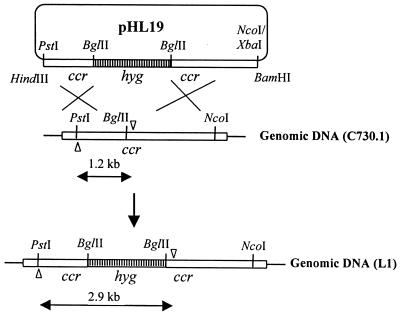
An insertional inactivation strategy was used to disrupt S. cinnamonensis ccr. A 2.7-kb NcoI-XbaI DNA segment was removed from pHL1 (a pUC119 derivative carrying the cloned 5.7-kb PstI S. cinnamonensis ccr gene fragment), and the resulting linearized plasmid was blunt ended and religated to yield pHL7. To construct pHL9, the 1.7-kb BglII hyg gene fragment described in Materials and Methods was inserted into a unique BglII site within the ccr coding region of pHL7. The orientation of this fragment in pHL9 is such that both hyg and ccr are transcribed in the same direction. The 4.7-kb BamHI-HindIII ccr::hyg fragment was excised from pHL9 and subcloned into pKC1139 to generate pHL19 (Fig. 3). S. cinnamonensis protoplasts were transformed with pHL19 isolated from E. coli ET12567, and colonies resistant to both hygromycin and apramycin (Hmr Amr) were obtained. Mutants in which a single crossover between pHL19 and the S. cinnamonensis chromosome had occurred were selected by cultivating one of the Hmr Amr transformants at 39°C in the presence of the two antibiotics. The genotype of one such single-crossover mutant was confirmed by PCR. Following three rounds of propagation of this single-crossover mutant in the absence of any antibiotics at 30°C, colonies resistant to only hygromycin (Hmr Ams) were obtained. One such colony, designated L1, was confirmed as the desired ccr-disrupted mutant by both PCR analysis (Fig. 3) and a CCR enzyme assay (see below).
FIG. 3.
Strategy used for the creation of the ccr-disrupted mutant S. cinnamonensis L1 (see text for details). Arrowheads indicate the positions of the PCR primers used for confirmation of the double-crossover mutants. With primers CCR1 (▿) and CCR2 (▵), a 2.9-kb fragment from S. cinnamonensis L1 and a 1.2-kb fragment from S. cinnamonensis C730.1 were amplified, respectively.
Expression of the S. collinus ccr gene in S. cinnamonensis C730.1 and L1.
The S. collinus ccr gene was excised as a 1.9-kb XbaI-HindIII fragment from pZYB3 (25) and subcloned into pSE34 (Table 1) to produce pHL18. In pHL18, ccr is expressed from the strong constitutive ermE* promoter (4). pHL18 was introduced into S. lividans 1326. Plasmid DNA isolated from S. lividans transformants was used to transform S. cinnamonensis C730.1 and L1, generating the ccr-overexpressing mutant C730.1/pHL18 and the complementation mutant L1/pHL18. The overproduction of CCR in these mutants was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis (data not shown) and by a CCR enzyme assay (see below).
CCR activity in S. cinnamonensis cell extracts.
The effects of disruption of the chromosomal ccr gene and plasmid-based expression of ccr on CCR activity in S. cinnamonensis were evaluated with cell extracts. This analysis revealed that cloned ccr was responsible for the majority of the CCR activity under two different growth conditions and that significant increases in CCR activity in either S. cinnamonensis strain (C730.1 and L1) could be accomplished by plasmid-based expression of S. collinus ccr. In YEME medium, CCR activity in S. cinnamonensis C730.1 (wild type) was fivefold higher than that previously reported for cell extracts of S. collinus (Table 2) (25). Only 5% of this activity could be detected in cell extracts of S. cinnamonensis L1, while 14- and 13-fold increases in CCR activity were observed for S. cinnamonensis C730.1/pHL18 and L1/pHL18, respectively. In the complex fermentation medium used for monensin production, the levels of CCR activity in S. cinnamonensis C730.1 were 2.5- to 4.4-fold higher than even the levels observed for the same strain grown in YEME medium. At day 5 of fermentation, the levels of CCR activity were 60% higher than the levels of CCR activity obtained by expression of S. collinus ccr from the ermE* promoter in S. erythraea (22). CCR activity in S. cinnamonensis L1 grown in the same complex fermentation medium was again 90 to 95% lower than that observed with strain C730.1. The highest levels of CCR activity were observed for C730.1/pHL18 and L1/pHL18 grown in complex fermentation medium. The activities were 3.7- and 2.7-fold higher than those observed for C730.1 grown under the same conditions.
TABLE 2.
Role of CCR in providing butyryl-CoA for monensin A biosynthesisa
| S. cinnamonensis strain | CCR activity (mU/mg)b in:
|
Monensin titer (mg/liter) | Monensin A/monensin B ratio (%)d | [1,2-13C2]acetate labeling of the following monensin A unite (%):
|
|||
|---|---|---|---|---|---|---|---|
| YEME medium (48 h) | CFMc on day:
|
||||||
| 2 | 5 | C2 | C4 | ||||
| C730.1 | 5.7 | 14.4 | 25.1 | 373 | 50/50 | 0.8 ± 0.2 | 0.4 ± 0.1 |
| C730.1/pHL18 | 80.7 | 53.1 | 90.9 | 315 | 48/52 | 0.8 ± 0.2 | 0.4 ± 0.1 |
| L1 | 0.3 | 1.3 | 1.2 | 445 | 12/88 | 0.9 ± 0.2 | ∼0.1 |
| L1/pHL18 | 71.1 | 39.4 | 68.8 | 327 | 42/58 | ND | ND |
All experiments except for [1,2-13C2]acetate labeling were carried out in either duplicate or triplicate. ND, not determined.
One unit of CCR activity is defined as the amount of enzyme required to catalyze the oxidation of 1 μmol of NADPH per min in the presence of crotonyl-CoA.
CFM, complex fermentation medium used for monensin production (see Materials and Methods).
Triplicate HPLC analyses revealed a maximum 1% variation in the monensin A/monensin B ratio in each experiment.
C2 and C4 units are the carbons of monensin A derived from malonyl-CoA and ethylmalonyl-CoA, respectively.
Effect of CCR on monensin A/monensin B ratios.
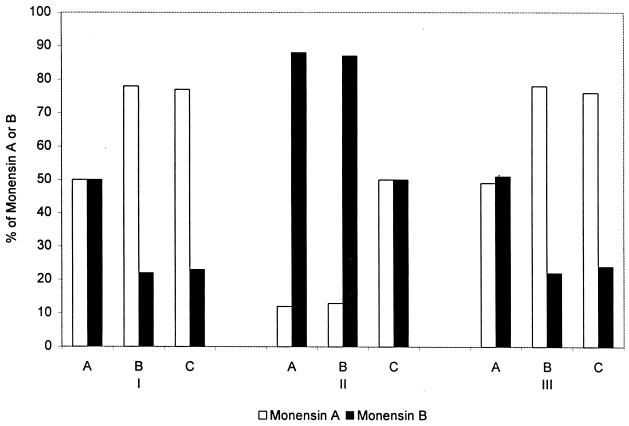
The role of CCR in producing butyryl-CoA for monensin A biosynthesis was investigated by analyzing the relative amounts of monensins A and B made by the different S. cinnamonensis strains. It was predicted that any significant changes in the amount of butyryl-CoA formed in S. cinnamonensis as a result of changes in the levels of CCR activity would be reflected in this ratio. In the complex fermentation medium, S. cinnamonensis C730.1 produced almost equal amounts of monensins A and B (Table 2 and Fig. 4). No significant change in this ratio was seen when the CCR expression plasmid was introduced into this strain. In contrast, the ccr-disrupted mutant (S. cinnamonensis L1) produced predominantly monensin B, with monensin A representing less than 15% of the total monensins. This monensin A/monensin B ratio returned to almost that observed in the wild type when the CCR expression plasmid was introduced into this mutant (Table 2). In these experiments, disruption and/or overexpression of ccr caused no more than 20% changes in the amounts of total monensins.
FIG. 4.
Relative amounts of monensins A and B produced by S. cinnamonensis C730.1 (I), L1 (II), and C730.1/pHL18 (III) grown in a complex fermentation medium (A) and medium supplemented with crotonic acid (B) or butyric acid (C).
Incorporation of [1,2-13C2]acetate into monensin A.
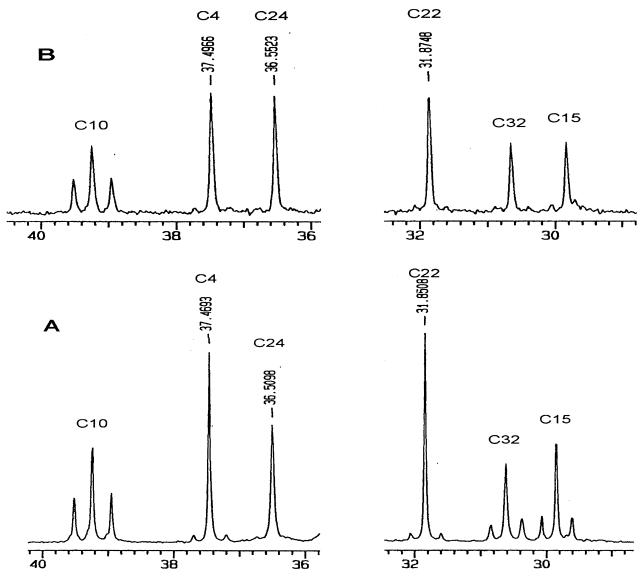
The decreased monensin A/monensin B ratio observed in fermentations of S. cinnamonensis L1 clearly suggests a significant role of CCR in producing a butyryl-CoA precursor for monensin A biosynthesis. A decrease in the labeling of the butyrate-derived positions of monensin A by labeled acetyl-CoA would also be expected because crotonyl-CoA, the substrate for CCR, is thought to be derived from acetyl-CoA via a reversal of the fatty acid β-oxidation pathway (25). This prediction was confirmed by carrying out monensin A [1,2-13C2]acetate incorporation experiments with S. cinnamonensis C730.1, L1, and C730.1/pHL18. In S. cinnamonensis C730.1, approximately 0.8% ± 0.2% of the acetate-derived positions of monensin A were labeled intact by the dually labeled acetate (Table 2 and Fig. 5). Labeling of 0.4% ± 0.1% of the butyrate-derived position of monensin A (C15, C16, C32, C33) was observed in this experiment. Thus, under the fermentation conditions used, approximately 50% of the butyrate-derived position of monensin A is derived from the acetate pool in S. cinnamonensis. Very similar labeling of the acetate- and butyrate-derived positions was seen with S. cinnamonensis C730.1/pHL18. This result is entirely consistent with the observation that the increased levels of CCR activity in this strain do not increase the monensin A/monensin B ratio (Table 2). In S. cinnamonensis L1, similar levels of labeling of the acetate-derived positions of monensin A were observed. However, in this experiment, only low-level (approximately 0.1%) intact labeling of the butyrate-derived position by dually labeled acetate was observed (Table 2 and Fig. 5). Thus, in this mutant, very little of butyrate-derived portion of monensin A is derived from the acetate pool and must be generated from some other pathway, such as isomerization of the valine catabolite isobutyryl-CoA.
FIG. 5.
Partial 13C NMR analyses of monensin A from S. cinnamonensis C730.1 (A) and L1 (B) fermentations carried out in the presence of [1,2-13C2]acetate. In both fermentations, the 13C natural-abundance signal at 39.2 ppm for C-10 of monensin A (derived from C-2 of an acetate precursor) is flanked by a doublet, indicating intact incorporation of labeled acetate into C-9 and C-10 (Fig. 1). Similar intact incorporation was observed for the other acetate-derived positions of monensin A. Intact incorporation of acetate into the butyrate-derived positions of monensin A was clearly observed for S. cinnamonensis C730.1 and less so for S. cinnamonensis L1, as evidenced by doublets around the natural-abundance C-15 and C-32 signals. Similar observations were made with the signals for C-16 and C-33. C-4, C-24 and C-22 of monensin A are derived from propionate precursors.
These results confirm that cloned S. cinnamonensis ccr plays a major role in the formation of butyryl-CoA from acetyl-CoA (Fig. 1). Deletion of this gene would be predicted to yield a strain producing substantially less monensin A. This prediction closely fits the observation of a monensin A level decrease in such a strain (Table 2) and indicates that the loss of butyryl-CoA formation via CCR is not significantly compensated for by increased flux from alternative pathways.
Effect of crotonic acid and butyric acid on monensin A/monensin B ratios.
The increased levels of CCR activity in S. cinnamonensis C730.1/pHL18 did not significantly alter the monensin A/monensin B ratio or increase the intact incorporation of dually labeled acetate into the butyrate-derived position of monensin A, suggesting that some additional factor controlled the concentrations of butyryl-CoA. The possibility that under the growth conditions used the crotonyl-CoA substrate for CCR was limiting was investigated by carrying out fermentations in the presence of crotonic acid and butyric acid. The addition of either crotonic acid or butyric acid to S. cinnamonensis C730.1 led to a significant increase in the monensin A/monensin B ratio (Fig. 4). A similar result was observed with S. cinnamonensis C730.1/pHL18. S. cinnamonensis L1, in contrast, showed an increase in the monensin A/monensin B ratio when grown in the presence of butyric acid but not crotonic acid (Fig. 4). These results demonstrate that the concentration of crotonyl-CoA is a limiting factor in monensin A biosynthesis in the wild-type strain and further verify a physiological role of the cloned S. cinnamonensis ccr gene in converting this crotonyl-CoA to butyryl-CoA.
Effect of CCR on fatty acid profiles.
It has previously been shown that exogenously supplied labeled butyric acid can be used intact, presumably as a coenzyme A thioester, as a starter unit for straight-chain fatty acid biosynthesis (26, 27). In addition, butyryl-CoA has recently been shown to be an efficient substrate for the 3-ketoacylsynthase III which is thought to initiate both straight-chain and branched-chain fatty acid biosynthesis in streptomycetes (11). Alterations in the concentration of the butyryl-CoA pool through changes in the levels of CCR activity in S. cinnamonensis were reflected in changes in the monensin A/monensin B ratio. The amount of even-carbon-number straight-chain fatty acids relative to that of other fatty acids might also be predicted to reflect changes in the concentration of the butyryl-CoA pool. The ratios of palmitate to the other fatty acids, however, were the same in fatty acid analyses of S. cinnamonensis C730.1, C730.1/pHL18, and L1.
DISCUSSION
A number of important macrolide and polyether polyketides, such as tylosin and monensin A, contain ethyl side chains derived from a butyryl-CoA precursor (8, 9). In some cases, analogs with either a methyl or an allyl side chain have also been isolated (6, 9). As these analogs typically exhibit differences in biological activity, it is relevant to understand factors controlling the ratio of the different analogs produced within a fermentation. For S. cinnamonensis, the ratio of the ethyl side chain product monensin A to the methyl side chain product monensin B appears to be dictated by the levels of the carboxylated butyryl-CoA precursor, ethylmalonyl-CoA, relative to those of methylmalonyl-CoA. In this study, it has been demonstrated that factors which affect the concentrations of butyryl-CoA within a fermentation lead to predictable changes in the monensin A/monensin B ratio.
The enzyme CCR, putatively involved in the final step of a pathway leading to butyryl-CoA from acetyl-CoA, was first identified in S. collinus (25). Initially, it was suggested that CCR was responsible for providing butyryl-CoA for fatty acid biosynthesis. While disruption of the corresponding ccr gene resulted in a decreased ability to grow on acetate as a sole carbon source, a fatty acid profile of the mutant demonstrated no increase in the straight-chain fatty acid palmitate relative to the branched-chain fatty acids (12). A similar lack of change in fatty acid profiles was obtained in the current study with S. cinnamonensis L1. Disruption of the ccr gene in S. cinnamonensis, however, did lead to a significant reduction in the monensin A/monensin B ratio, indicating a significantly decreased butyryl-CoA concentration. Butyryl-CoA has recently been shown to be used efficiently in vitro by β-ketoacyl acyl carrier protein synthase III, the enzyme which initiates fatty acid biosynthesis (11). Furthermore, the exogenous addition of labeled butyrate (4.3 mM) to streptomycetes fermentations leads to efficient intact incorporation into the straight-chain fatty acid palmitate (27). The fact that a loss of CCR activity in S. cinnamonensis L1 decreases the monensin A/monensin B ratio but not the ratio of palmitate to the branched-chain fatty acids may be a result of the differential timing of fatty acid and polyketide biosynthesis during growth. Alternatively, these observations may indicate that in the absence of exogenously supplied butyrate, acetyl-CoA is the major precursor used to initiate palmitate biosynthesis.
The current study clearly demonstrates that CCR plays a major role in providing butyryl-CoA for monensin A biosynthesis and that the S. cinnamonensis ccr gene, primarily responsible for this enzyme activity, is located within the same sets of homologous genes observed in S. coelicolor and S. collinus (12). In all of these organisms, ccr is located upstream of meaA, another gene required for efficient growth on acetate. In all three organisms, the cloned ccr gene is not clustered with any polyketide biosynthetic genes. Thus, it appears that ccr and meaA are part of an operon involved in primary metabolism and as such may be present in many if not all streptomycetes (a related set of genes also appears to be involved in primary metabolism in Methylobacterium extorquens [7, 21]). Apparent homologs of S. cinnamonensis ccr have been identified in the tylosin (ccr; 79% identity and 85% similarity), coronafacic acid (cfa8; 34% identity and 50% similarity), and niddamycin biosynthetic gene clusters (8, 17, 22) (Fig. 2). All of these polyketides, like monensin A, have an ethyl side chain derived from a butyryl-CoA precursor. It is currently unknown what role these ccr homologs play in providing butyryl-CoA and whether the producing organisms also contain a second copy of ccr clustered with meaA. A ccr homolog making a relatively minor contribution to the production of butyryl-CoA might similarly be located within the monensin biosynthetic gene cluster. Consistent with this possibility are the observations that in S. cinnamonensis L1, residual CCR activity can be detected and dually labeled acetate is still incorporated into the butyrate-derived position of monensin A (albeit at an efficiency markedly lower than that in S. cinnamonensis C730.1).
The ability of S. cinnamonensis L1 to produce some levels of monensin A despite a significant decrease in CCR activity demonstrates that under the fermentation conditions used, another pathway or pathways can contribute to the production of butyryl-CoA. Furthermore, it is evident that in S. cinnamonensis L1, the loss of butyryl-CoA via CCR is not significantly compensated for by an increase in flux through alternate pathways. The most likely alternate pathway for butyryl-CoA formation is isomerization of the valine catabolite isobutyryl-CoA, catalyzed by ICM. Consistent with this hypothesis is the observation that the growth of S. cinnamonensis L1 in a chemically defined medium with valine as a major component results in a mixture of monensins A and B, while almost exclusively monensin B is produced if alternative amino acids, such as isoleucine, are used (14a). The icm gene has recently been identified from S. cinnamonensis (29), and it should be possible to unequivocally demonstrate the role of ICM in monensin A biosynthesis by deletion of the gene in S. cinnamonensis L1.
Based on enzyme assays of cell extracts, S. cinnamonensis C730.1 grown in the monensin production medium exhibits surprisingly high levels of CCR. Accordingly, the amount of monensin A made relative to monensin B in this strain is limited less by CCR activity than by availability of the crotonyl-CoA substrate. Thus, no significant increase in the ratio of monensin A to monensin B is observed with an increase in the levels of CCR in S. cinnamonensis C730.1/pHL18. On the other hand, the addition of crotonic acid to this strain and the wild-type strain but not S. cinnamonensis L1 results in significant increases in the monensin A/monensin B ratio. A similar effect has recently been observed for FK520-producing Streptomyces hygroscopicus var ascomyceticus, where the addition of 5.6 mM crotonic acid reduces the level of a major analog impurity (FK523) from 6.7 to 2.5% (23). Thus, this organism, like S. cinnamonensis, appears to have CCR present at sufficient levels that the availability of the crotonyl-CoA substrate contributes to the FK520/FK523 ratio. Indeed, detectable levels of CCR activity have been observed in this organism, and PCR has been used to identify a 384-bp fragment which encodes a putative CCR with 88% amino acid sequence identity to the S. collinus CCR (19, 23). In contrast, S. erythraea cell extracts contain no clearly detectable CCR activity, and no evidence of a ccr gene in this organism can be found by Southern hybridization with the S. collinus ccr gene as a probe (22). In S. erythraea EAT4, the production of 6-ethylErA (the ethyl analog of erythromycin) is dependent upon either the addition of butyric acid or the expression of the S. collinus ccr gene (22). Thus, in S. erythraea the availability of the butyryl-CoA precursor for polyketide biosynthesis is limited by the levels of the enzyme CCR and not the substrate crotonyl-CoA (despite the fact that this organism is known to contain an ICM for converting the valine catabolite isobutyryl-CoA to butyryl-CoA [27]).
In conclusion, the monensin A/monensin B ratio produced by S. cinnamonensis is dependent upon both the levels of CCR activity and the substrate for this enzyme, crotonyl-CoA. Manipulation of the levels of both of these factors through either genetic approaches or changes in the components of the fermentation medium can be used to alter this ratio in a predictable fashion.
ACKNOWLEDGMENTS
We are grateful to Stewart Campbell at Insmed Pharmaceuticals for the use of a refractive index detector for HPLC analysis of monensin production. Eli Lilly & Company kindly provided S. cinnamonensis C730.1.
Financial support was provided by the National Institutes of Health (grant GM50542).
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Meyers W, Lipman D J. Basic local alignment search tools. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.August P R, Lang T, Yoon Y J, Ning S, Muller R, Yu T W, Taylor M, Hoffman D, Kim C G, Zhang X, Hutchinson C R, Floss H G. Biosynthesis of the ansamycin antibiotic rifamycin: deductions from the molecular analysis of the rif biosynthetic cluster of Amycolatopsis mediterranei. Chem Biol. 1998;5:69–79. doi: 10.1016/s1074-5521(98)90141-7. [DOI] [PubMed] [Google Scholar]
- 3.Beran M, Zima J. Determination of monensins A and B in the fermentation broth of Streptomyces cinnamonensis by high performance liquid chromotography. Chromatographia. 1993;35:206–208. [Google Scholar]
- 4.Bibb M J, Janssen G R, Ward J M. Cloning and analysis of the promoter region of the erythromycin gene (ermE) of Streptomyces erythraeus. Gene. 1985;38:215–226. doi: 10.1016/0378-1119(85)90220-3. [DOI] [PubMed] [Google Scholar]
- 5.Bierman M, Logan R, O’Brien K, Seno E T, Rao R N, Schoner B E. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 6.Byrne K M, Shafiee A, Nielsen J B, Arison B, Monaghan R L, Kaplan L. The biosynthesis and enzymology of an immunosuppressant, immunomycin, produced by Streptomyces hygroscopicus var. ascomyceticus. Dev Ind Microbiol. 1993;32:29–44. [Google Scholar]
- 7.Chistoserdova L V, Lidstrom M E. Molecular characterization of a chromosomal region involved in the oxidation of acetyl-CoA to glyoxylate in the isocitrate-lyase-negative methylotroph Methylobacterium extorquens AM1. Microbiology. 1996;142:1459–1468. doi: 10.1099/13500872-142-6-1459. [DOI] [PubMed] [Google Scholar]
- 8.Gandecha A R, Large S L, Cundliffe E. Analysis of four tylosin biosynthetic genes from the tylM region of the Streptomyces fradiae genome. Gene. 1997;184:197–203. doi: 10.1016/s0378-1119(96)00595-1. [DOI] [PubMed] [Google Scholar]
- 9.Gorman M, Chamberlin J W, Hamill R L. Monensin, a new biologically active compound. V. Compounds related to monensin. Antimicrob Agents Chemother. 1967;7:363–367. [PubMed] [Google Scholar]
- 10.Guilfoile P G, Hutchinson C R. The Streptomyces glaucescens TcmR protein represses transcription of the divergently oriented tcmR and tcmA genes by binding to an intergenic operator region. J Bacteriol. 1992;174:3659–3666. doi: 10.1128/jb.174.11.3659-3666.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han L, Lobo S, Reynolds K A. Characterization of β-ketoacyl acyl carrier protein synthase III from Streptomyces glaucescens: its role in the initiation of fatty acid biosynthesis. J Bacteriol. 1998;180:4481–4486. doi: 10.1128/jb.180.17.4481-4486.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han L, Reynolds K A. A novel alternate anaplerotic pathway to the glyoxylate cycle in streptomycetes. J Bacteriol. 1997;179:5157–5164. doi: 10.1128/jb.179.16.5157-5164.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Smith J M, Ward J M, Schrempf H S. Genetic manipulation of streptomycetes: a laboratory manual. Norwich, United Kingdom: John Innes Institute; 1985. [Google Scholar]
- 14.Kakavas S J, Katz L, Stassi D. Identification and characterization of the niddamycin polyketide synthase genes from Streptomyces caelestis. J Bacteriol. 1997;179:7515–7522. doi: 10.1128/jb.179.23.7515-7522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.Liu, H., and K. A. Reynolds. Unpublished results.
- 15.Lydiate D J, Ashby A M, Henderson D J, Kieser H M, Hopwood D A. Physical and genetic characterization of chromosomal copies of the Streptomyces coelicolor minicircle. J Gen Microbiol. 1989;135:941–955. [Google Scholar]
- 16.MacNeil D J, Gewain K M, Ruby C L, Dezeny G, Gibbons P H, MacNeil T. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel intergration vector. Gene. 1992;111:61–68. doi: 10.1016/0378-1119(92)90603-m. [DOI] [PubMed] [Google Scholar]
- 17.Rangaswamy V, Mitchell R, Ullrich M, Bender C. Analysis of genes involved in biosynthesis of coronafacic acid, the polyketide component of the phytotoxin coronatine. J Bacteriol. 1998;180:3330–3338. doi: 10.1128/jb.180.13.3330-3338.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reynolds K, O’Hagan D, Gani D, Robinson J A. Butyrate metabolism in streptomycetes. Characterization of a vicinal interchange rearrangement linking isobutyrate and butyrate in Streptomyces cinnamonensis. J Chem Soc Perkin Trans 1. 1988;1988:3195–3207. [Google Scholar]
- 19.Reynolds K A, Demain A L. Rapamycin, FK506, and ascomycin-related compounds. In: Strohl W R, editor. Biotechnology of antibiotics. 2nd ed. New York, N.Y: Marcel Dekker, Inc.; 1997. pp. 497–521. [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 21.Smith L M, Meijer W G, Dijkhuizen L, Goodwin P. A protein having similarity with methylmalonyl CoA mutase is required for assimilation of methanol and ethanol by Methylobacterium extorquens AM1. Microbiology. 1996;142:657–684. doi: 10.1099/13500872-142-3-675. [DOI] [PubMed] [Google Scholar]
- 22.Stassi D L, Kakavas S J, Reynolds K A, Gunawardana G, Swanson S, Zeidner D, Jackson M, Liu H, Buko A, Katz L. Ethyl-substituted erythromycin derivatives produced by directed metabolic engineering. Proc Natl Acad Sci USA. 1998;95:7305–7309. doi: 10.1073/pnas.95.13.7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun W-S, Salmon P, Wilson J, Connors N. Crotonic acid directed biosynthesis of the immunosuppressants produced by Streptomyces hygroscopicus var. ascomyceticus. J Ferment Bioeng. 1998;86:261–265. [Google Scholar]
- 24.Tang L, Zhang Y X, Hutchinson C R. Amino acid catabolism and antibiotic synthesis: valine is a source of precursors for macrolide biosynthesis in Streptomyces ambofaciens and Streptomyces fradiae. J Bacteriol. 1994;176:6107–6119. doi: 10.1128/jb.176.19.6107-6119.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wallace K K, Bao Z, Dai H, Digate R, Schuler G, Speedie M K, Reynolds K A. Purification of crotonyl CoA reductase from Streptomyces collinus and cloning, sequencing and expression of the corresponding gene in Escherichia coli. Eur J Biochem. 1995;233:954–962. doi: 10.1111/j.1432-1033.1995.954_3.x. [DOI] [PubMed] [Google Scholar]
- 26.Wallace K K, Lobo S, Han L, McArthur H A I, Reynolds K A. In vivo and in vitro effect of thiolactomycin on fatty acid biosynthesis in Streptomyces collinus. J Bacteriol. 1997;179:3884–3891. doi: 10.1128/jb.179.12.3884-3891.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wallace K K, Zhao B, McArthur H A I, Reynolds K A. In vivo analysis of straight-chain and branched-chain fatty acid biosynthesis in three actinomycetes. FEMS Microbiol Lett. 1995;131:227–234. doi: 10.1111/j.1574-6968.1995.tb07781.x. [DOI] [PubMed] [Google Scholar]
- 28.Xue Y, Zhao L, Liu H-W, Sherman D H. A gene cluster for macrolide antibiotic biosynthesis in Streptomyces venezuelae: architecture of metabolic diversity. Proc Natl Acad Sci USA. 1998;95:12111–12116. doi: 10.1073/pnas.95.21.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zerbe-Burkhardt K, Ratnatilleke A, Philippon N, Birch A, Leiser A, Vrijbloed J W, Hess D, Hunziker P, Robinson J A. Cloning, sequencing, expression, and insertional inactivation of the gene for the large subunit of the coenzyme B12-dependent isobutyryl-CoA mutase from Streptomyces cinnamonensis. J Biol Chem. 1998;273:6508–6517. doi: 10.1074/jbc.273.11.6508. [DOI] [PubMed] [Google Scholar]