Activation of Txnip-p53-p21 axis boosts quiescence and survival of Dnmt3a-mutant hematopoietic progenitors during IFNγ challenge, elucidating functional and molecular basis of clonal hematopoiesis.
Abstract
Clonal hematopoiesis (CH) refers to the age-related expansion of specific clones in the blood system, and manifests from somatic mutations acquired in hematopoietic stem cells (HSCs). Most CH variants occur in the gene DNMT3A, but while DNMT3A-mutant CH becomes almost ubiquitous in aging humans, a unifying molecular mechanism to illuminate how DNMT3A-mutant HSCs outcompete their counterparts is lacking. Here, we used interferon gamma (IFNγ) as a model to study the mechanisms by which Dnmt3a mutations increase HSC fitness under hematopoietic stress. We found Dnmt3a-mutant HSCs resist IFNγ-mediated depletion, and IFNγ-signaling is required for clonal expansion of Dnmt3a-mutant HSCs in vivo. Mechanistically, DNA hypomethylation–associated overexpression of Txnip in Dnmt3a-mutant HSCs leads to p53 stabilization and upregulation of p21. This preserves the functional potential of Dnmt3a-mutant HSCs through increased quiescence and resistance to IFNγ-induced apoptosis. These data identify a previously undescribed mechanism to explain increased fitness of DNMT3A-mutant clones under hematopoietic stress.
Significance:
DNMT3A mutations are common variants in clonal hematopoiesis, and recurrent events in blood cancers. Yet the mechanisms by which these mutations provide hematopoietic stem cells a competitive advantage as a precursor to malignant transformation remain unclear. Here, we use inflammatory stress to uncover molecular mechanisms leading to this fitness advantage.
See related commentary by De Dominici and DeGregori, p. 178.
This article is highlighted in the In This Issue feature, p. 171
INTRODUCTION
Hematopoietic stem cells (HSCs) randomly acquire somatic mutations with age (1). The vast majority of these mutations are inconsequential, but some variants provide a fitness advantage to HSC clones, allowing them to become disproportionally represented in the blood (2). This state of abnormal HSC expansion without overt hematologic disease is known as clonal hematopoiesis (CH), and is associated with increased risk of cardiovascular events and blood cancers (3–5). Approximately 50% of all genetic variants in CH occur in the gene DNMT3A (6), which encodes a de novo DNA methyltransferase enzyme responsible for establishing new DNA methylation patterns during development and stem cell fate decisions (7, 8). DNMT3A is also recurrently mutated in blood diseases such as acute myeloid leukemia (AML; ref. 9) and myelodysplastic syndromes (MDS; ref. 10) where it functions as an initiating mutation (11). Emerging evidence indicates somatic DNMT3A variants can be acquired early in life (12, 13), before becoming almost ubiquitous in the aging population (14).
Mouse models show Dnmt3a loss-of-function skews HSC fate decisions toward self-renewal, facilitating clonal expansion in the bone marrow (BM; ref. 15, 16). However, analysis of DNA methylation and gene expression patterns in mouse (15, 17) and human (17, 18) systems have yet to identify a unifying molecular mechanism to explain the increased fitness of these mutant HSCs. The competitive advantage of Dnmt3a-mutant HSCs is exacerbated by hematopoietic stress such as serial transplantation (19) and microbial infection (20). In addition to genetics, environmental factors likely contribute to clonal selection in CH. In particular, chronic inflammation has been proposed as an environmental pressure that selects for certain clones in CH (21, 22). We recently showed ulcerative colitis patients have a higher incidence of DNMT3A-mutant CH associated with increased serum IFNγ (23). Here, we used IFNγ as a model of inflammatory stress to study the molecular mechanisms that empower Dnmt3a-mutant HSCs with a fitness advantage. We found Dnmt3a-mutant HSCs specifically resist IFNγ-mediated depletion, and IFNγ signaling is required for clonal expansion of Dnmt3a-mutant HSCs in vivo. Complementary genomic techniques identified a Txnip–p53–p21 pathway that preserves the functional potential of Dnmt3a-mutant HSCs under conditions of inflammatory stress, which was validated by functional genetic rescue experiments in vivo. These findings highlight a novel mechanistic basis to explain the increased fitness of Dnmt3a-mutant HSCs, and supports rationale for developing interventions to mitigate expansion of premalignant clones as a method of blood cancer prevention.
RESULTS
DNMT3A-Mutant HSCs Are Insensitive to the Deleterious Effects of IFNγ In Vivo
Mouse genetic models were generated to represent the spectrum of DNMT3A mutations found in human CH; Dnmt3a heterozygous (Vav-Cre;Dnmt3afl/+ = Dnmt3aHET) and homozygous (Vav-Cre;Dnmt3afl/fl = Dnmt3aKO) hematopoietic loss-of-function, and a knockin model (24) analogous to the point mutation most prevalent in AML (Vav-Cre;Dnmt3aR878H/+ = Dnmt3aR878). Most DNMT3A variants in human CH are missense heterozygous loss-of-function (6). However, we and others have shown that Dnmt3aHET HSCs produce only modest phenotypes in mice (25, 26), likely because these variants provide very minor fitness advantages to HSCs (2) that require the extended lifespan of humans to manifest measureable phenotypes. In mice, complete loss of Dnmt3a function is required to generate robust phenotypes. Inclusion of the Dnmt3aR878 model (although this variant is less prevalent in CH than AML) allows for the most complete analysis of Dnmt3a mutations in HSC function.
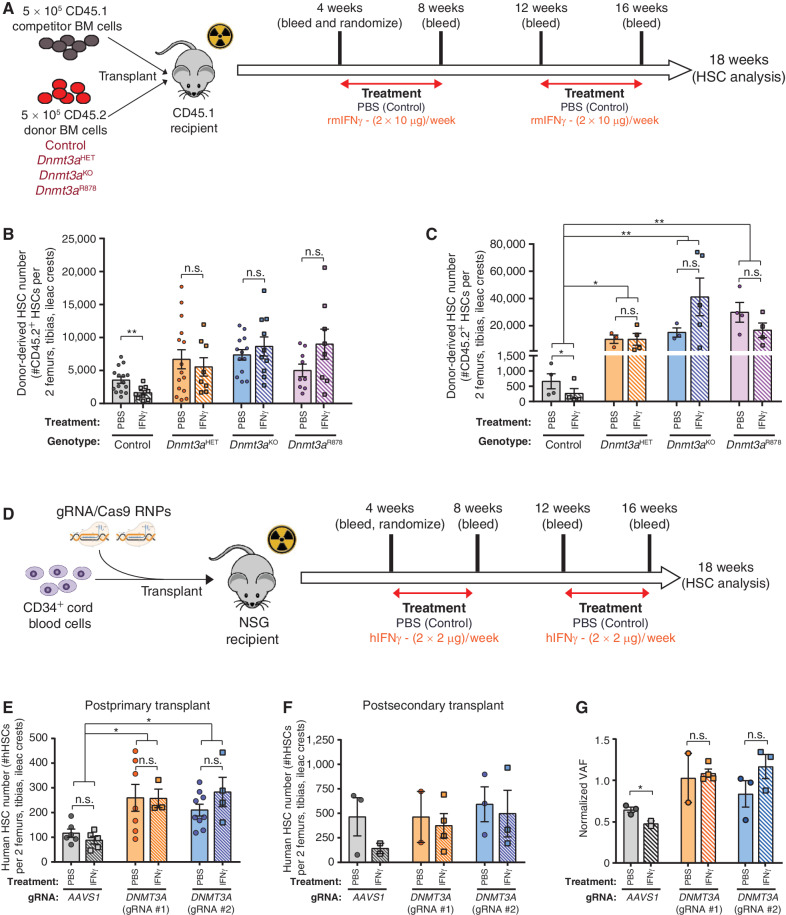
To examine how Dnmt3a mutations impact the HSC response to IFNγ, hematopoietic chimeras were created by transplanting recipient mice (CD45.1) with 5.0 × 105 BM cells from control (Vav-Cre;Dnmt3a+/+), Dnmt3aHET, Dnmt3aKO, or Dnmt3aR878 mice in competition with 5.0 × 105 BM cells from congenic wild-type (CD45.1) mice. Recipients were treated with PBS (control) or recombinant mouse IFNγ (Fig. 1A). IFNγ treatment had minimal impact on peripheral blood engraftment (Supplementary Fig. S1A and S1B). Control HSCs (Supplementary Fig. S1C) were reduced following IFNγ treatment in relation to PBS-treated mice (Fig. 1B). In contrast, Dnmt3aHET, Dnmt3aKO, and Dnmt3aR878 HSCs (Supplementary Fig. S1D) were resistant to depletion in response to IFNγ (Fig. 1B; Supplementary Fig. S1E). To determine the functional consequences of prior IFNγ exposure on long-term HSC function, 200 donor-derived HSCs (CD45.2+Lineage−c-Kit+EPCR+CD48−CD150+) were purified from primary transplants and transferred to secondary recipients along with 2.5 × 105 fresh BM competitor cells. The trend toward impaired peripheral blood engraftment of IFNγ-treated control HSCs was not observed from IFNγ-treated Dnmt3a-mutant HSCs (Supplementary Fig. S1F). While IFNγ was detrimental to the self-renewal of control HSCs, clonal expansion of Dnmt3aHET, Dnmt3aKO, and Dnmt3aR878 HSCs was unaffected (Fig. 1C).
Figure 1.
DNMT3A-mutant HSCs are insensitive to the deleterious effects of IFNγ in vivo. A, Schematic of competitive transplant where recipients were challenged with IFNγ. B, Number of donor-derived HSCs (CD45.2+Lineage−c-Kit+EPCR+CD48−CD150+) in BM of recipients 18 weeks postprimary transplant (n = 8–15). C, Quantification of donor-derived HSCs in BM of recipients 18 weeks postsecondary transplant (n = 3–5). D, Schematic of transplantation of CRISPR-edited human CD34+ cord blood cells into NSG mice. E, Number of human HSCs (mCD45−hCD45+Lineage−CD38−CD34+CD45RA−CD90+CD49f+) in BM of NSG recipients 18 weeks posttransplant (n = 5–9). F, Number of human HSCs in BM of NSG recipients 18 weeks postsecondary transplant (n = 2–4). G, Relative clone size of CRISPR-edited human cells postsecondary transplant (hCD45+ BM). VAF postsecondary transplant was normalized to that of pretransplant CD34+ cells. One-way ANOVA; *, P < 0.05; **, P < 0.01. Data represent mean ± SEM.
To determine whether human HSCs showed a similar phenotype, cord blood CD34+ cells were nucleofected with Cas9-RNPs (27) complexed with guide RNA (gRNA) targeting DNMT3A or the inert AAVS1 locus (negative control). Edited CD34+ cells were transplanted into NSG mice, then randomized to receive either recombinant human (hIFNγ) or vehicle (PBS; Fig. 1D). As with mice, treatment with hIFNγ did not influence peripheral blood engraftment of human cells (Supplementary Fig. S2A and S2B). Analysis of the BM (Supplementary Fig. S2C) showed the increased abundance of DNMT3A-targeted HSCs was not influenced by IFNγ treatment (Fig. 1E). As the self-renewal phenotype of murine Dnmt3a-mutant HSCs is exacerbated by serial transplant, we attempted to perform secondary transfer of cord blood cells that has not previously been reported. We found transfer of 1 × 106 hCD34+ cells from primary recipients via intratibial injection was required for robust engraftment in secondary recipients. Again, while peripheral blood engraftment was not different between genotypes (Supplementary Fig. S2D), the trend toward reduced self-renewal of IFNγ-treated AAVS1-targered HSCs was not observed in IFNγ-treated DNMT3A-targeted HSCs (Fig. 1F). Analysis of CRISPR genome edit variant allele frequency (VAF) revealed that DNMT3A mutations maintained their clone size, whereas AAVS1 mutations were depleted over serial transplantation (Supplementary Fig. S2E) and by IFNγ treatment (Fig. 1G). Cumulatively, these data suggest that Dnmt3a-mutant HSCs are specifically resistant to IFNγ-mediated depletion.
DNMT3A-Mutant HSCs Are Resistant to IFNγ-Induced Apoptosis
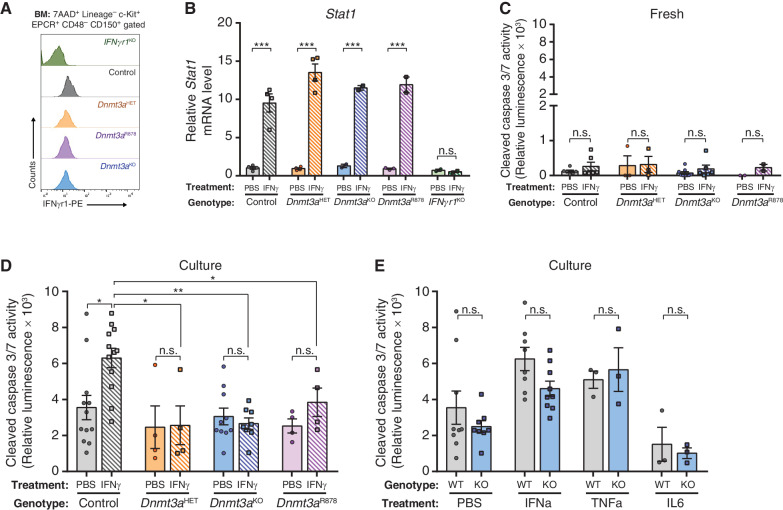
One of the major mechanisms of inflammation-mediated attrition of HSCs is sensitization to stress-induced apoptosis (28). To examine this, mice were injected with two doses of recombinant cytokines 24 hours apart. HSCs were purified 2 hours after the second dose, then apoptosis was examined immediately after purification (fresh) or after a 16-hour culture period. All HSC genotypes express comparable levels of cell surface IFNgr1 (Fig. 2A) and showed equivalent activation of the IFNγ target gene Stat1 (Fig. 2B). Thus, any differences should not result from the inability of the mutant HSCs to sense or respond to IFNγ. There were no differences in apoptosis (cleaved Caspase 3/7 activity) between freshly isolated HSCs of any genotype (Fig. 2C). While IFNγ-treated control HSCs showed significantly elevated apoptosis after culture, this response was absent in Dnmt3a-mutant HSCs (Fig. 2D). Moreover, the apoptotic resistance phenotype of Dnmt3a-mutant HSCs to IFNγ was not observed after exposure to related proinflammatory cytokines (Fig. 2E). These results demonstrate that Dnmt3a mutations render HSCs specifically insensitive to IFNγ-induced apoptosis.
Figure 2.
DNMT3A-mutant HSCs are resistant to IFNg-induced apoptosis. A, Cell surface protein expression (MFI) of IFNgr1 on HSCs (Lineage−c-Kit+EPCR+CD48−CD150+) measured by flow cytometry. B, Relative mRNA level of Stat1 in HSCs 2-hours after in vivo exposure to IFNγ (n = 2–4) measured by qPCR. C, Cleaved caspase 3/7 activity of HSCs freshly purified from mice treated with PBS or IFNγ (n = 2–9). D, Cleaved caspase 3/7 activity in HSCs purified from mice treated with PBS or IFNγ after a 16-hour culture period (n = 4–12). E, Cleaved caspase 3/7 activity in HSCs (WT = Control; KO = Dnmt3aKO) from mice treated with indicated cytokines or PBS after a 16-hour culture period (n = 3–9). One-way ANOVA; *, P < 0.05; **, P < 0.01; ***, P < 0.001. Data represent mean ± SEM.
IFNγ Signaling Is Required for Clonal Expansion of Dnmt3a-Mutant HSCs
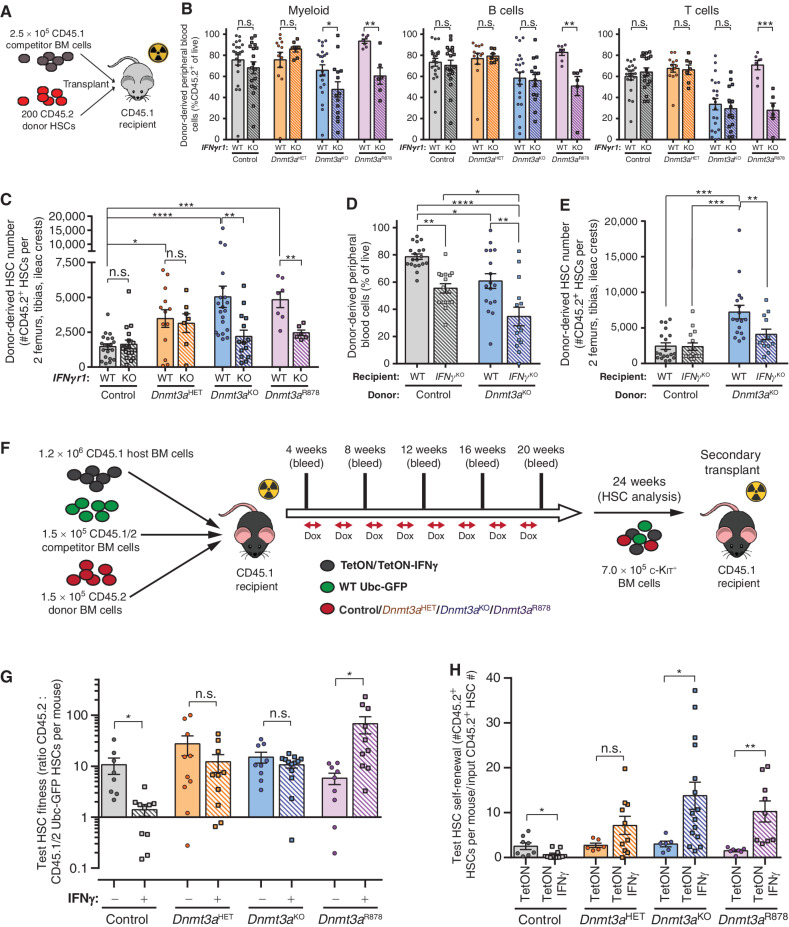
To study the cell-intrinsic role of IFNγ-signaling in HSC function, Dnmt3a-mutant mice were crossed to mice lacking the IFNγ receptor (IFNgr1−/− = IFNgr1KO). In unperturbed adult (∼12-week old) mice, genetic loss of IFNgr1 normalized the increased abundance of Dnmt3a-mutant HSCs in the BM (Supplementary Fig. S3A and S3B). HSC function was assessed by competitive transplantation of 200 phenotypically defined (Lineage−c-Kit+EPCR+CD48−CD150+) HSCs against 2.5 × 105 congenic BM cells (Fig. 3A). While loss of IFNgr1 had no effect on blood contribution from control HSCs, Dnmt3aKO and Dnmt3aR878H HSCs lacking IFNgr1 showed significantly reduced myeloid cell output (Fig. 3B). Loss of IFNgr1 also hindered lymphoid production from Dnmt3aR878H HSCs (Fig. 3B). Analysis of recipient mice BM 18 weeks posttransplant showed loss of IFNgr1 ameliorated the clonal expansion of Dnmt3aKO and Dnmt3aR878H HSCs while having no effect on the self-renewal of control HSCs (Fig. 3C). These data show IFNγ signaling is required for the enhanced self-renewal of Dnmt3a-mutant HSCs. IFNγ signaling does not appear a universal CH selection mechanism as genetic deletion of IFNgr1 from Vav-Cre;Tet2fl/fl (= Tet2KO) HSCs had no effect on their competitive advantage in analogous transplantation experiments (Supplementary Fig. S3C and S3D).
Figure 3.
IFNγ signaling is required for clonal expansion of Dnmt3a-mutant HSCs. A, Schematic of competitive HSC transplantation. B, Donor-derived chimerism in peripheral blood lineages 16 weeks posttransplant (n = 6–21). C, Number of donor-derived HSCs (CD45.2+Lineage−c-Kit+EPCR+CD48−CD150+) in BM of recipients 18 weeks posttransplant (n = 6–21). D, Donor-derived chimerism in peripheral blood 16 weeks posttransplant in wild-type (WT) or IFNγKO recipients (n = 14–19). E, Number of donor-derived HSCs in BM of recipients 18 weeks posttransplant (n = 14–19). F, Schematic showing CH competition model. G, Test HSC fitness ratio 24 weeks posttransplant in CH competition model (n = 8–13). H, Number of test HSCs (CD45.2+Lineage−c-Kit+EPCR+CD48−CD150+Ubc-GFP−) generated in BM of secondary recipients 18 weeks posttransplant per input HSC from primary transplant shown in G (n = 7–15). One-way ANOVA (D and E) or two-tailed t test (B, C, G, and H; data are compared for treatment/condition relative to parental genotype); *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Data represent mean ± SEM.
If Dnmt3a-mutant HSCs require IFNγ signaling for clonal expansion, then reducing systemic IFNγ levels could represent a targetable approach for impeding DNMT3A-mutant CH. To examine this, 200 control or Dnmt3aKO HSCs were transplanted into wild-type (WT) or IFNγ-deficient (IFNg−/−) mice with 2.5 × 105 BM cells matched to recipient genotype. Loss of systemic IFNγ reduced blood chimerism of both control and Dnmt3aKO cells compared with transplantation into IFNγ-replete environments (Fig. 3D). BM analysis posttransplant showed comparable numbers of control HSCs in WT and IFNγ−/− recipients. Conversely, Dnmt3aKO HSCs were significantly reduced in IFNγ−/− recipients (Fig. 3E), suggesting that both intrinsic IFNγ signaling and extrinsic IFNγ stimulation are required for clonal expansion of Dnmt3a-mutant HSCs.
Dnmt3a Mutations Confer HSCs a Fitness Advantage under Repeated IFNγ Exposure
Although the competitive advantage of Dnmt3a-mutant HSCs during inflammation has been implied (20, 23), no direct experiments have been performed to quantify fitness between mutant and WT HSCs in response to IFNγ. We created a novel model to examine HSC competition under chronic IFNγ exposure. Mice bearing a doxycycline-inducible IFNγ allele were crossed to Rosa26rtTA-M2 mice on an IFNgr1−/− background (tetO_IFNg; Rosa26rtTA-M2; IFNgr1KO = “TetON-IFNg”), so the resultant BM can overexpress IFNγ in response to doxycycline, but remain insensitive to the effects of this IFNγ overexpression and serve as a consistent internal control. Titration of doxycycline concentration in chow (Supplementary Fig. S4A) identified a dose that approximated serum levels of IFNγ in mice infected with the Mycobacterium avium pathogen (29). To model a 10% VAF for CH variants, chimeras were created by transplanting a 1:1:8 ratio of test BM (1.5 × 105 CD45.2 - control, Dnmt3aHET, Dnmt3aKO, Dnmt3aR878), wild-type competitor BM (1.5 × 105 CD45.1/2 Ubc-GFP), and TetON-IFNg or control (Rosa26rtTA-M2; IFNgr1KO = “TetON”) BM (1.2 × 106 CD45.1). Four weeks posttransplant, recipients were placed on weekly cycles on 1,250 ppm doxycycline chow (Fig. 3F), mimicking episodic inflammation. Chronic IFNγ exposure did not significantly alter blood chimerism (Supplementary Fig. S4B) for each donor cell genotype relative to engraftment in TetON chimeras (Supplementary Fig. S4C). HSC fitness was calculated as the ratio of test CD45.2: CD45.1/2 Ubc-GFP HSCs per mouse (Supplementary Fig. S4D). A ratio of 1 indicates equal competition. Control test HSCs possessed an advantage without IFNγ due to the described fitness deficit of Ubc-GFP HSCs (30). However, this advantage was diminished under chronic IFNγ (Fig. 3G). Conversely, fitness of Dnmt3a-mutant HSCs was generally unaffected by chronic IFNγ, and was in fact enhanced for Dnmt3aR878 HSCs (Fig. 3G). Serial transplantation was performed by transferring 7.0 × 105 c-Kit-enriched BM cells from primary recipients into secondary mice to preserve clonal composition (Fig. 3F). In secondary recipients, there was a marked reduction in blood output of control HSCs previously exposed to IFNγ (Supplementary Fig. S4E). In contrast, IFNγ-exposed Dnmt3a-mutant cells showed significantly increased blood engraftment in secondary transplant (Supplementary Fig. S4E), showing a fitness advantage over WT cells previously exposed to chronic IFNγ. Self-renewal of Dnmt3a-mutant HSCs with prior IFNγ treatment was enhanced postsecondary transplant (Fig. 3H; Supplementary Fig. S4F).
Dnmt3a-Mutant HSCs Retain Quiescence after IFNγ Challenge
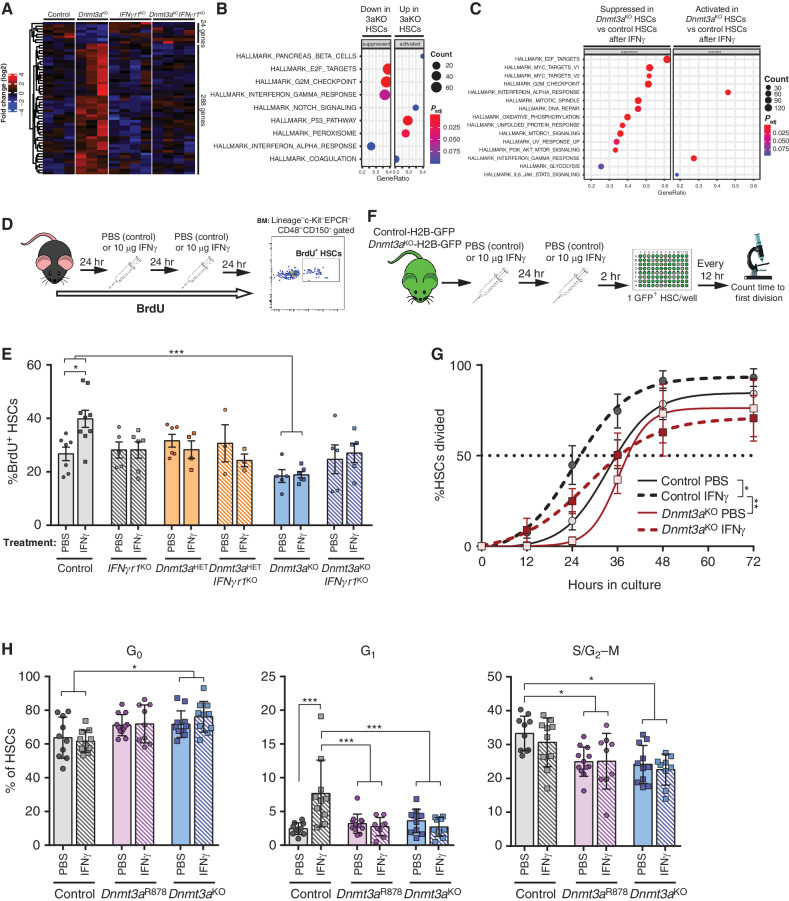
To investigate the molecular mechanisms underlying the fitness advantage of Dnmt3a-mutant HSCs under IFNγ, RNA-seq was performed on HSCs from mice at baseline and after a 2-hour IFNγ treatment to examine gene expression. In unmanipulated mice, there were few significantly differentially expressed genes (DEG) between control and Dnmt3aKO HSCs (Fig. 4A), consistent with prior studies (15). In support of the functional data, genetic deletion of IFNgr1 in Dnmt3aKO HSCs normalized gene expression, including the previously reported (15) upregulation of HSC “stemness” genes (Supplementary Fig. S5A). Control and Dnmt3aKO HSCs displayed a similar transcriptional response to IFNγ (Supplementary Fig. S5B). In HSCs at baseline, gene-set enrichment analysis (GSEA) revealed suppression of cell-cycle networks (E2F targets, G2–M checkpoint) in Dnmt3aKO HSCs, whereas p53 pathway targets were activated (Fig. 4B). Comparison of IFNγ-stimulated HSCs showed a more robust IFN response in Dnmt3aKO HSCs, but cell cycle–related genesets remained suppressed (Fig. 4C), suggesting Dnmt3aKO HSCs may have a different proliferative response to IFNγ exposure.
Figure 4.
Dnmt3a-mutant HSCs retain quiescence after IFNγ challenge. A, Expression pattern of differentially expressed genes (DEGs; P < 0.05, fold-change >2) between control and Dnmt3aKO HSCs across IFNgr1KO and Dnmt3aKOIFNgr1KO HSCs. B, Gene-set enrichment analysis (GSEA) comparison of control and Dnmt3aKO HSCs at baseline showing suppression of cell cycle–related genesets in Dnmt3aKO HSCs and activation of p53 targets. C, GSEA comparison of control and Dnmt3aKO HSC after acute IFNγ challenge. D, Schematic of BrdU incorporation assay to assess HSC proliferative responses. E, Percentage of BrdU+ HSCs (Lineage−c-Kit+EPCR+CD48−CD150+) from indicated genotypes after 72-hour timecourse and treatment with either PBS or IFNγ (n = 3–9). F, Experimental workflow for single HSC division assays using H2B-GFP-labeled HSCs. G, Time to first division of HSCs (GFP+Lineage−c-Kit+EPCR+CD48−CD150+) in response to PBS or IFNγ treatment (mean ± SEM plus line of best fit, n = 5–6). H, Flow cytometric Ki67/7AAD cell-cycle analysis of HSCs (Lineage−c-Kit+EPCR+CD48−CD150+) from PBS- or IFNγ-treated mice following 26-hour culture to stimulate proliferation (n = 9–12). One-way ANOVA (E and H) or two-way ANOVA (G); *, P < 0.05; ***, P < 0.001. Data represent mean ± SEM.
Loss of quiescence is a major factor leading to erosion of HSC self-renewal (31, 32). As IFN stimulation is known to induce HSC proliferation (29, 33), we hypothesized that Dnmt3a–mutant HSCs may resist the detrimental effects of IFNγ exposure by maintaining a quiescent state. To examine HSC proliferation in vivo, mice were labeled with BrdU and treated with PBS (control) or IFNγ (Fig. 4D). Control HSCs were stimulated to proliferate in response to IFNγ (Fig. 4E). Proliferation of IFNgr1KO HSCs was not different than control HSCs in mice treated with PBS, and remained unchanged following IFNγ treatment. However, Dnmt3aKO HSCs were more quiescent at baseline, and their proliferation was unaltered in response to IFNγ (Fig. 4E). This suggests an uncoupling of the molecular response of Dnmt3aKO HSCs to IFNγ exposure with the normal functional effect of increased proliferation. Deletion of IFNgr1 in Dnmt3aKO HSCs restored their proliferation rate at baseline to that of control HSCs (Fig. 4E). Dnmt3aKOIFNgr1KO HSCs remained insensitive to IFNγ, suggesting the effect is cell intrinsic. Dnmt3aHET HSCs were not more quiescent than the control HSCs at baseline, but like Dnmt3aKO HSCs were resistant to IFNγ-induced proliferation (Fig. 4E).
To track HSC division on a clonal level, a histone labeling strategy was used. Control (Vav-Cre;Dnmt3a+/+; Rosa26rtTA-M2; Col1a1tetO_H2B-GFP) and Dnmt3aKO (Vav-Cre;Dnmt3afl/fl; Rosa26rtTA-M2; Col1a1tetO_H2B-GFP) mice were administered doxycycline chow for three weeks to label cells with H2B-GFP, then withdrawn from doxycycline for one week prior to experimentation to “wash out” the H2B-GFP label. H2B-GFP is quickly lost from proliferative hematopoietic cells, while being retained by the quiescent HSCs. The purpose of using H2B-GFP labeling in this experiment was to aid microscopic visualization of single HSCs. H2B-GFP labeled mice were treated with PBS (control) or IFNγ, then individual H2B-GFP+ HSCs were purified and monitored in vitro (Fig. 4F). Time to first division was not different between PBS-treated control and Dnmt3aKO HSCs. While IFNγ treatment accelerated time to first division for control HSCs, Dnmt3aKO HSCs were unaffected (Fig. 4G), again indicating Dnmt3a-mutant HSCs resist proliferation following IFNγ exposure. To more specifically determine whether Dnmt3aKO HSCs were restricted at defined stages in cell cycle, Ki67/DAPI staining was performed on BM from PBS- or IFNγ-treated mice. Analysis was performed on both freshly isolated BM (Supplementary Fig. S5C), and also after HSCs were purified and cultured for 26 hours to stimulate proliferation. While there were no significant cell-cycle differences in freshly analyzed HSCs (Supplementary Fig. S5D), Dnmt3aR878 and Dnmt3aKO HSCs showed a delayed exit from G0 after a 26-hour culture period with concomitant deceases of HSCs in G1 and S/G2–M (Fig. 4H). This suggests quiescence-enforcing mechanisms may delay the proliferative activation of Dnmt3a-mutant HSCs following IFNγ exposure.
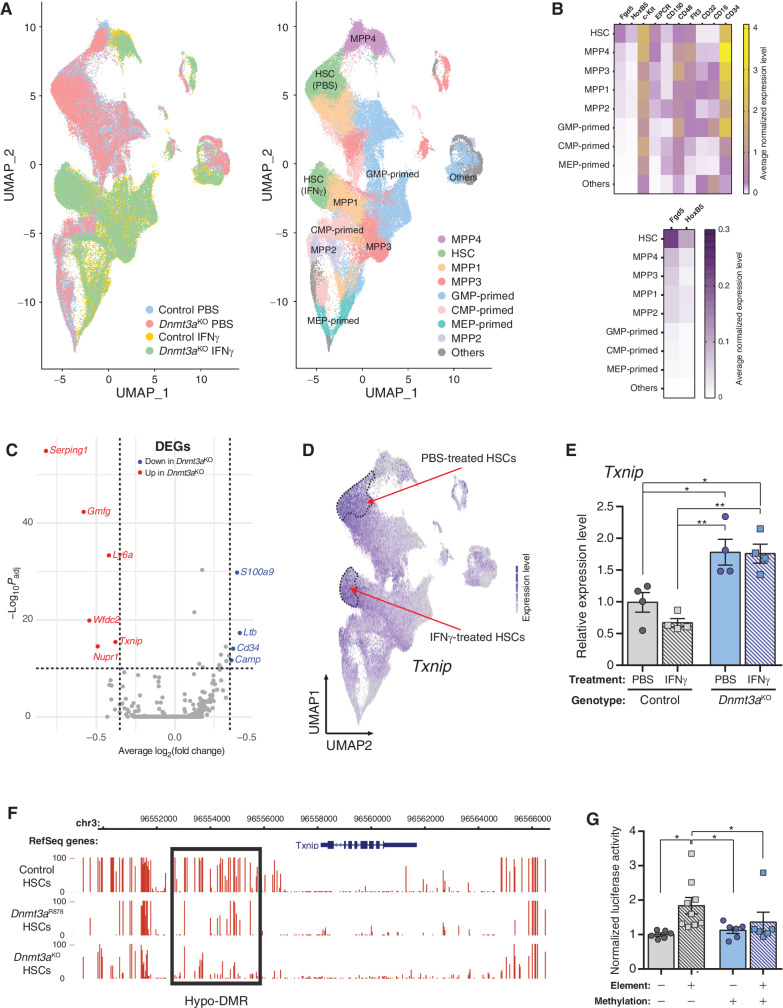
Txnip Is a DNA Methylation–Sensitive Gene Involved in HSC Stress Response
The single-cell division assay also revealed another interesting observation: while virtually all IFNγ-treated control HSCs had divided by 72 hours in culture, approximately 30% of Dnmt3aKO HSCs had not divided by this timepoint, suggesting some clonal heterogeneity even within this highly purified cell population. To investigate this, single-cell RNA-seq was performed on PBS- and IFNγ-treated control and Dnmt3aKO Lineage−c-Kit+EPCR+ cells. UMAP clustering showed treatment was a stronger influence on transcriptome than genotype (Fig. 5A, left). Assignment of cell identities (Fig. 5B) resolved two clusters of HSCs, with both genotypes grouped together but separated by treatment (Fig. 5A, right). Similar to the bulk HSC RNA-seq data, analysis of single-cell transcriptomic data identified only approximately 50 DEGs in the HSC clusters between genotypes and treatments. GSEA comparison of PBS-treated HSCs replicated many of the findings of bulk RNA-seq such as downregulation of “E2F targets” and “MYC targets” in Dnmt3aKO HSCs, whereas analysis of IFNγ-treated cells confirmed the increased IFNγ signaling in Dnmt3aKO HSCs (Supplementary Fig. S5E). The functional differences of Dnmt3aKO HSCs were not due to the inability to upregulate immediate early response genes and myeloid differentiation factors in response to IFNγ (Supplementary Fig. S5F), but the single-cell RNA-seq was not able to resolve discrete Dnmt3aKO HSC subpopulations which may be primed for differential IFNγ responses as suggested from the single HSC division assay (Fig. 4G).
Figure 5.
Txnip is a DNA methylation–sensitive gene involved in HSC stress response. A, UMAP clustering of single-cell RNA-seq data of control and Dnmt3aKO Lineage− c-Kit+EPCR+ hematopoietic cells after treatment with PBS or IFNγ. Left, clustering of cells by genotype and treatment. Right, assignment of cell identities to cell clusters. Two clusters of HSCs are identified, which represents HSCs from both genotypes separated by treatment. B, Marker gene expression used to assign identities to cell clusters. C, Volcano plot of differentially expressed genes (DEG) between IFNγ-treated control and Dnmt3aKO HSCs. D,Txnip expression in UMAP-clustered scRNA-seq data. Dashed lines outline HSC clusters. E,Txnip expression in HSCs purified from mice treated with PBS or IFNγ (n = 4). F, DNA methylation profile of Txnip locus in HSCs determined by WGBS. Height of red bar indicates methylation level of individual CpG. Black box denotes hypomethylated DMR in Dnmt3a-mutant HSCs. G, Luciferase activity of Txnip regulatory element in methylation-sensitive reporter assay. Firefly luciferase activity was measured 48 hours posttransfection and normalized to Renilla luciferase levels (n = 6–9). One-way ANOVA; *, P < 0.05; **, P < 0.01. Data represent mean ± SEM.
Thioredoxin interacting protein (Txnip) was one of the few genes upregulated specifically in Dnmt3aKO HSCs (Fig. 5C), and its expression pattern was HSC-predominant (Fig. 5D). This gene was of interest due to its described role in HSC function (34), cell-cycle regulation (35), and stress response (36). Targeted qPCR confirmed the upregulation of Txnip in Dnmt3aKO HSCs at baseline, and while there was a trend for reduced Txnip in IFNγ-treated control HSCs, there was no diminution of the levels in IFNγ-treated Dnmt3aKO HSCs (Fig. 5E). To determine whether the gene expression changes of Dnmt3a-mutant HSCs under IFNγ stress were associated with DNA methylation differences, whole genome bisulfite sequencing (WGBS) was performed on cells from TetON-IFNγ chimeric mice postprimary transplant. WGBS was performed on donor-derived (CD45.2+) BM cells as we have previously shown that the DNA methylation changes in Dnmt3a-mutant HSCs manifest more prominently in their differentiated progeny (15). Dnmt3aKO cells showed global hypomethylation, whereas the overall genomic DNA methylation profile of Dnmt3aHET cells was virtually indistinguishable from control cells (Supplementary Fig. S6A). Exposure to chronic IFNγ did not dramatically alter the methylome of any genotype (Supplementary Fig. S6B). Identification of differentially methylated regions (DMR) showed virtually all differences in DNA methylation were driven by genotype, rather than treatment (Supplementary Fig. S6C–S6E). This suggests the differences of Dnmt3a-mutant HSCs in response to IFNγ were not associated with DNA methylome remodeling. However, it has been shown that focal changes in DNA methylation in DNMT3A-mutant samples may have important biological consequences (18, 37). Dnmt3a-mutant cells showed a hypomethylated DMR in the Txnip regulatory region (Supplementary Fig. S6F), although the DNA methylation level was not altered by IFNγ treatment, suggesting this loss of methylation was a canonical consequence to Dnmt3a loss-of-function. The same region was also hypomethylated specifically in Dnmt3aKO and Dnmt3aR878 HSCs (Fig. 5F). As this genomic region overlapped with ENCODE active enhancer histone marks, the DMR was cloned into a methylation-sensitive reporter assay. In the absence of DNA methylation, the DMR functioned as an active regulatory element (Fig. 5G). Artificial methylation of the reporter plasmid with the bacterial CpG methyltransferase M.SssI ablated this regulatory element activity (Fig. 5G). These observations suggest Txnip may be a DNA methylation–sensitive gene that primes the stress response of Dnmt3a-mutant HSCs.
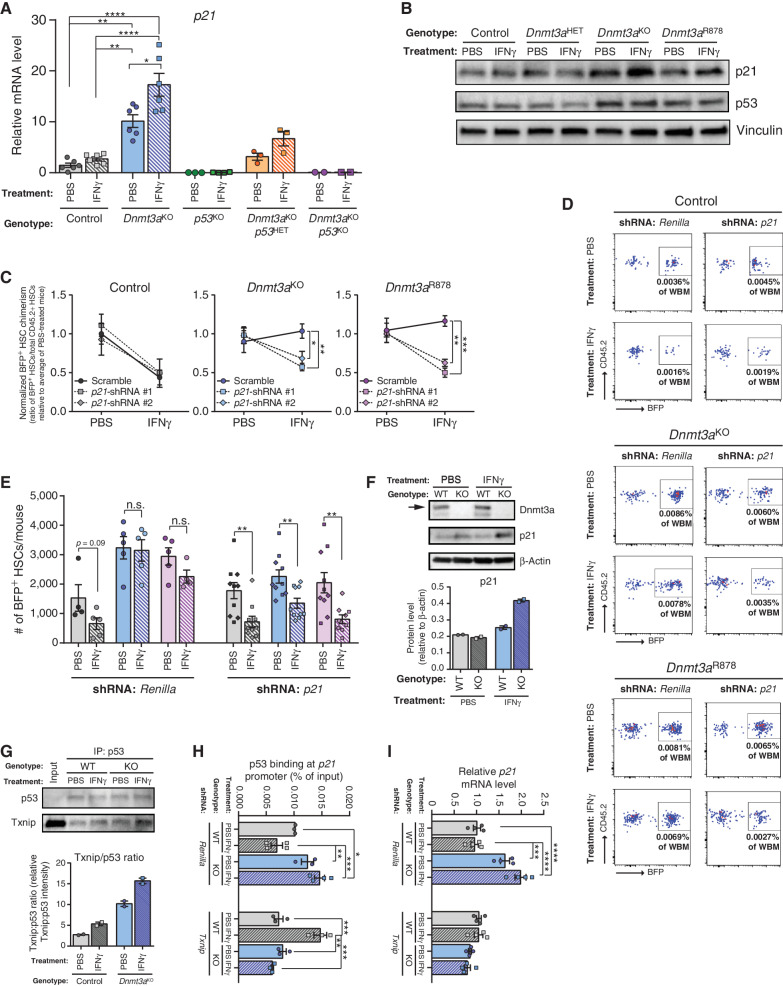
Txnip Upregulates p21 in Dnmt3a-Mutant HSCs in Response to IFNγ
Txnip is a thiol-oxidoreductase that controls cellular redox signaling to protect cells from oxidative stress, and regulates HSC quiescence under stress conditions (34) by stabilizing p53 protein via direct interaction (38). H2DCFDA staining was performed in HSCs to correlate Txnip upregulation in Dnmt3aKO with reactive oxygen species (ROS; Supplementary Fig. S7A). While IFNγ-treatment increased ROS in control HSCs, this response was absent in Dnmt3aHET and Dnmt3aKO HSCs (Supplementary Fig. S7B). Given the activated p53 gene expression signature in naïve Dnmt3a-mutant HSCs (Fig. 4B), we hypothesized Txnip may function in the response of Dnmt3a-mutant HSCs to IFNγ by regulating p53 and its targets. p53 has diverse functions in HSC responses to inflammation (28), which can be broadly classified as survival, apoptosis, and cell-cycle depending on the context. To examine p53 target genes that might explain the functional differences of Dnmt3a-mutant HSCs to IFNγ, gene expression analysis was performed on HSCs from mice treated with IFNγ. There were no IFNγ-induced differences in transcript levels of p53 itself, or major p53 targets for prosurvival (Bcl2, Mcl1) or proapoptosis (Bax, Puma) across HSC genotypes (Supplementary Fig. S7C). However, the p53 target gene Cdkn1a (p21) was significantly upregulated in Dnmt3aKO HSCs at baseline and further increased after IFNγ treatment (Fig. 6A). A similar expression profile was observed in DNMT3A-edited cord blood CD34+ cells after transplant (Supplementary Fig. S7D). Hematopoietic progenitor–enriched BM cells (Lineage−c-Kit+) from Dnmt3a-mutant mice also showed upregulation of p21 associated with p53 stabilization following IFNγ (Fig. 6B; Supplementary Fig. S7E). Dnmt3aKO mice were crossed to p53−/− mice and the progeny treated with PBS or IFNγ. Gene expression analysis showed the upregulation of p21 in Dnmt3aKO HSCs was p53-dependent (Fig. 6A).
Figure 6.
Txnip upregulates p21 in Dnmt3a-mutant HSCs in response to IFNγ. A,p21 mRNA levels in HSCs (Lineage−c-Kit+EPCR+CD48−CD150+) from mice treated with PBS or IFNγ (two doses, cells purified 2 hours after the second dose) measured by qPCR (n = 2–6). B, Protein levels measured by Western blot in hematopoietic progenitor cells (Lineage−c-Kit+) from mice treated with PBS or IFNγ (two doses, cells purified 2 hours after the second dose). Representative of two independent experiments. C, Normalized level of BFP+ cell chimerism in donor-derived HSC (CD45.2+Lineage−c-Kit+EPCR+CD48−CD150+) pool 18 weeks posttransplant. Ratio for each mouse is normalized to average BFP+ HSC chimerism in PBS treatment for individual shRNAs for each genotype to account for differences in transduction efficiency (n = 4–5 per group). D, Representative flow cytometry plots from mice transplanted with shRNA-transduced hematopoietic progenitor cells. Plots show BFP+ HSC chimerism for each genotype and treatment from Lineage−c-Kit+EPCR+CD48−CD150+-gated BM. E, Number of BFP+ donor-derived HSCs (BFP+CD45.2+Lineage−c-Kit+EPCR+CD48−CD150+) in BM of recipients 18 weeks posttransplant. Data for two independent shRNAs targeting p21 are compiled, denoted by square (p21-shRNA #1) or diamond (p21-shRNA #2) shapes. F, Protein levels measured by western blot in 32D cells (WT = control; KO = Dnmt3aKO) following 24-hour IFNg (10 ng/mL) or PBS treatment. Arrow indicates full-length Dnmt3a. Representative of two independent experiments, quantification shown underneath. G, Western blot analysis of 32D cell lysate (treated with PBS or IFNg for 24h; WT = control, KO = Dnmt3aKO) following immunoprecipitation with p53 antibody. Representative of two independent experiments, quantification shown underneath. H, Chromatin-immunoprecipitation qPCR analysis of p53 enrichment at p21 promoter in 32D cells (WT = control, KO = Dnmt3aKO) transduced with shRNA targeting Renilla (control) or Txnip. Chromatin was isolated following a 24-hour exposure of IFNγ (10 ng/mL) or PBS (n = 3–4). I, qPCR for p21 expression levels in the same cells used in H. One-way ANOVA (A, C, H, I) or two-tailed t test (E, data are compared for IFNγ relative to PBS within each genotype/shRNA); *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Data represent mean ± SEM.
p21 is a potent cell-cycle inhibitor (39), thus we hypothesized that p21 upregulation may be the downstream effector that preserves the quiescence of Dnmt3a-mutant HSCs under IFNγ challenge. To test this, c-Kit-enriched BM from control, Dnmt3aKO and Dnmt3aR878 (the Dnmt3a-mutant strains with robust HSC phenotypes) mice was transduced with lentiviruses expressing one of two shRNAs targeting p21 (Supplementary Fig. S7F) or a nontargeting Renilla control shRNA. 1.5 × 105 transduced c-Kit+ cells were transplanted in competition with congenic 2.5 × 105 BM cells, and then the recipients were treated with PBS (control) or IFNγ (Fig. 1A). In terms of HSC clonal expansion, the reduction of Renilla shRNA-transduced control HSCs following IFNγ treatment was completely paralleled in the same HSCs transduced with p21-targeting shRNAs (Fig. 6C and D). That is, p21 knockdown had no influence on the response of normal HSCs to IFNγ. In contrast, the increased chimerism of Dnmt3aKO and Dnmt3aR878 HSCs after IFNγ (Fig. 6C) was completely reversed following p21 knockdown (Fig. 6C–E). These data suggest that p21 has genotype-specific functions in the stress response of HSCs to inflammation.
Biochemical studies were performed to examine the role of Txnip and p53 in the regulation of p21 in Dnmt3a-mutant cells. For experiments requiring greater input than can be obtained from primary HSCs, we used the mouse myeloid progenitor 32D cell line as a surrogate model. After CRISPR/Cas9-mediated Dnmt3a deletion, we confirmed these cells replicated the IFNγ-dependent increase in p21 seen in Dnmt3a-mutant BM cells (Fig. 6F). As Txnip has been reported to stabilize p53 protein in hematopoietic stress conditions, protein interaction between Txnip and p53 was examined by immunoprecipitation. This showed an IFNγ-dependent increase in binding of Txnip to p53 specifically in Dnmt3a-null 32D cells (Fig. 6G). As p53 regulates p21 expression by transcriptional activation, ChIP-qPCR was performed for p53 occupancy at the well-annotated binding site in the p21 promoter. WT and Dnmt3a-null 32D cells were transduced with lentiviral shRNAs targeting Txnip (Supplementary Fig. S7F) or Renilla control, then treated with either PBS (control) or IFNγ. In Renilla-transduced cells, there was increased binding of p53 to the p21 promoter in Dnmt3a-null 32D cells at baseline, which increased further after IFNγ treatment (Fig. 6H). In contrast, there was reduced p53 binding at the p21 promoter in WT 32D cells treated with IFNγ. Knockdown of Txnip normalized binding of p53 to the p21 promoter in Dnmt3a-null 32D cells to approximate WT 32D cells, and was unchanged in these cells after IFNγ (Fig. 6H). This suggests that in response to IFNγ, Txnip binds p53 in Dnmt3a-mutant cells, leading to p53 stabilization and increased binding to the p21 promoter. The subsequent increased p21 expression (Fig. 6I) hinders proliferation of Dnmt3a-mutant cells to preserve functional potential. We did note that p53 binding was significantly increased at the p21 promoter in WT 32D cells following Txnip knockdown which was the opposite response to Renilla-transduced cells (Fig. 6H). However, this did not translate to increased p21 transcription (Fig. 6I) and implies a fundamental difference in the molecular response to IFNγ between control and Dnmt3a-mutant cells.
Txnip Preserves Functional Integrity of Dnmt3a-Mutant HSCs via p21
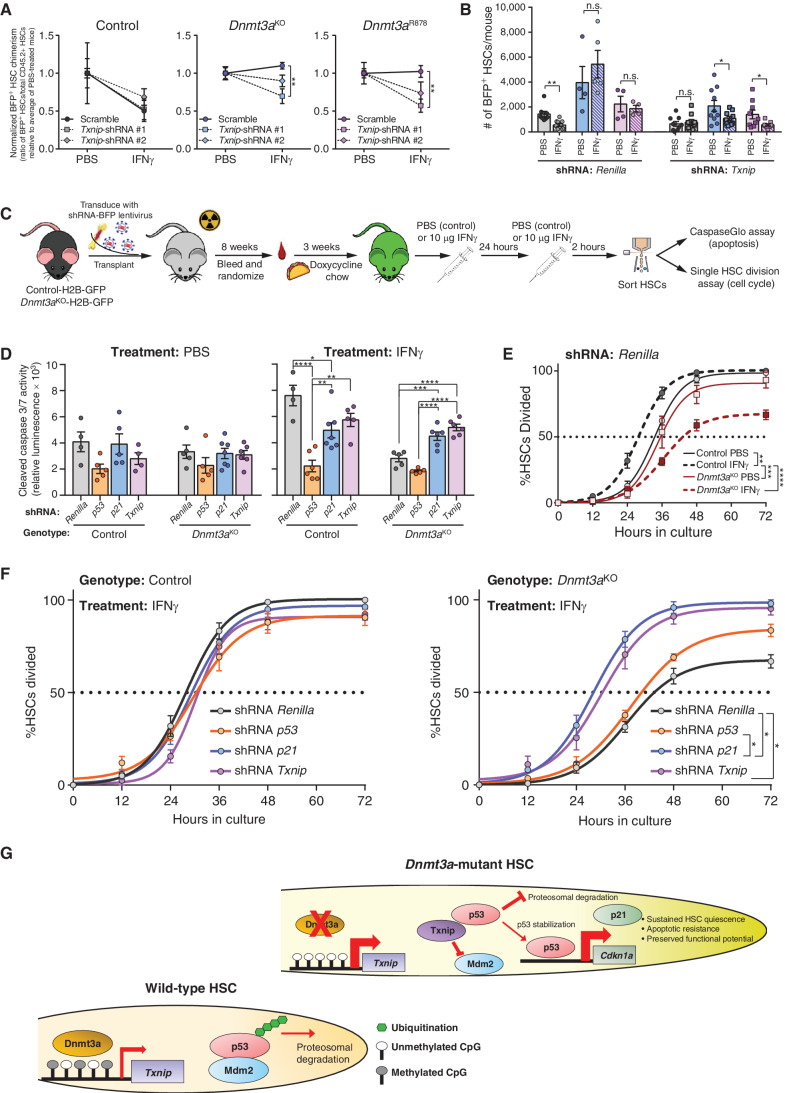
To validate Txnip as the upstream regulator of p21 in the response of Dnmt3a-mutant HSCs to IFNγ, shRNA transplants were performed to reduce expression of Txnip as previously described for p21. Similar to inhibition of p21, Txnip knockdown did not influence the impaired self-renewal of control HSCs exposed to IFNγ (Fig. 7A). However, Txnip shRNA–transduced Dnmt3aKO or Dnmt3aR878 HSCs phenocopied p21 knockdown whereby the competitive advantage and clonal expansion of these HSCs under IFNγ challenge was mitigated by Txnip inhibition (Fig. 7B). p21 expression was reduced in Txnip shRNA-transduced Dnmt3aKO or Dnmt3aR878 HSCs (Supplementary Fig. S7G), implicating Txnip as an important regulator of p21 in these mutant HSCs under IFNγ challenge.
Figure 7.
Txnip preserves functional integrity of Dnmt3a-mutant HSCs via p21. A, Normalized level of BFP+ cell chimerism in donor-derived HSC (CD45.2+Lineage−c-Kit+EPCR+CD48−CD150+) pool 18 weeks posttransplant. Ratio for each mouse is normalized to average BFP+ HSC chimerism in PBS treatment for individual shRNAs for each genotype to account for differences in transduction efficiency (n = 4–5 per group). B, Number of BFP+ donor-derived HSCs (BFP+CD45.2+Lineage−c-Kit+EPCR+CD48−CD150+) in BM of recipients 18 weeks posttransplant. Data for two independent shRNAs targeting Txnip are compiled, denoted by square (Txnip-shRNA #1) or diamond (Txnip-shRNA #2) shapes. C, Schematic of functional genetic rescue by lentiviral shRNA transduction in H2B-GFP–labeled HSCs. D, Cleaved caspase 3/7 activity of shRNA-transduced HSCs (GFP+BFP+Lineage−c-Kit+EPCR+CD48−CD150+) purified from mice treated with PBS or IFNγ (two doses, cells purified 2 hours after the second dose). Apoptosis was quantified following a 16-hour culture period (n = 4–7). Data for two independent shRNAs targeting p53, p21 and Txnip are compiled. E, Time to first division of H2B-GFP–labeled HSCs (GFP+BFP+Lineage−c-Kit+EPCR+CD48−CD150+) of indicated genotypes transduced with Renilla-targeting shRNA (control) from mice treated with PBS or IFNγ (n = 4). F, Time to first division of H2B-GFP–labeled control or Dnmt3aKO HSCs (GFP+BFP+Lineage−c-Kit+EPCR+CD48−CD150+) transduced with shRNA targeting indicated genes from mice treated with IFNγ (n = 4). G, Schematic illustrating Txnip-p53-p21 axis in the preservation of Dnmt3a-mutant HSCs. One-way ANOVA (A), two-tailed t test (B, data are compared for IFNγ relative to PBS within each genotype/shRNA), two-way ANOVA (D and E) or one-way ANOVA with Tukey multiple test correction (F); *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Data represent mean ± SEM.
To further the mechanistic connection between Txnip-p53-p21 in response to IFNγ, a genetic experiment was performed by transducing H2B-GFP labeled control and Dnmt3aKO HSCs with lentiviral shRNAs (Supplementary Fig. S7F), followed by BM transplantation. After stable engraftment, recipients were acutely treated with either PBS or IFNγ (Fig. 7C). HSC functional responses to IFNγ in terms of apoptosis and cell cycle were examined by cleaved caspase 3/7 activity and single HSC division assay, respectively. There were no significant differences in apoptosis between any shRNA-transduced HSCs from PBS-treated mice after culture (Fig. 7D), similar to previous observations (Fig. 2D). HSCs transduced with the control Renilla shRNA recapitulated the native setting with Dnmt3aKO HSCs resistant to IFNγ-induced apoptosis (Fig. 7D). Knockdown of p53 reduced apoptosis in both IFNγ-treated control and Dnmt3aKO HSCs, likely because the genetic inhibition nonspecifically altered a myriad of p53-related cellular functions, including induction of apoptosis (40). More specific effects were observed from genetic inhibition of p21 and Txnip. While p21 and Txnip inhibition was protective for control HSCs following IFNγ exposure, knockdown of these genes in Dnmt3aKO HSCs increased IFNγ-induced apoptosis to approximate the levels observed in control HSCs (Fig. 7D). These data again highlight genotype-specific HSC responses and implicate a protective function for Txnip and p21 in preventing stress-induced apoptosis in Dnmt3a-mutant HSCs in response to IFNγ.
In terms of cell cycle, IFNγ treatment of control HSCs transduced with Renilla shRNA reduced time to first division, unlike Dnmt3aKO HSCs (Fig. 7E), analogous to the native setting (Fig. 4G). In PBS-treated mice, Txnip inhibition had minimal effect on time to first division in both HSC genotypes compared with respective Renilla controls (Supplementary Fig. S7H). In IFNγ-treated mice, while Txnip inhibition in control HSCs had no effect compared with Renilla-shRNA, Txnip inhibition in Dnmt3aKO HSCs restored their IFNγ-induced proliferative response (Fig. 7F). Analogous results were observed from p21-shRNA–expressing IFNγ-treated Dnmt3aKO HSCs (Fig. 7F). Cumulatively, these data support a model whereby loss of DNA methylation at Txnip promoter in Dnmt3a-mutant HSCs, leads to increased expression and primes an inflammatory stress response. When Dnmt3a-mutant HSCs are challenged with IFNγ, Txnip stabilizes p53 through direct protein interaction, leading to upregulation of p21 which helps prevent IFNγ-induced proliferation and apoptosis (Fig. 7G), providing the fitness advantage required to establish CH.
DISCUSSION
Because the identification of DNMT3A mutations in blood malignancies, a unifying mechanism of how these variants provide HSCs with a competitive advantage has proved elusive. Here, we used IFNγ as a selective pressure to identify molecular pathways that increase fitness of Dnmt3a-mutant HSCs. IFNγ signaling is required for clonal expansion of Dnmt3a-mutant HSCs in vivo, and chronic IFNγ provides an environment that selects these HSCs. Functionally, Dnmt3a-mutant HSCs resist IFNγ-induced apoptosis and preserve quiescence, allowing them to gain clonal dominance.
This study identified a Txnip–p53–p21 mechanism that protects Dnmt3a-mutant HSCs from the deleterious effects of IFNγ. While chronic IFNγ exposure did not dramatically alter the DNA methylome, Txnip upregulation in Dnmt3a-mutant HSCs was associated with a hypomethylated regulatory element. This reinforces previous findings that DNMT3A loss-of-function does not lead to global DNA methylation changes (15, 16, 19), yet focal methylation alterations appear physiologically relevant (17, 18, 37, 41). While there were few adaptive molecular changes in Dnmt3a-mutant HSCs in response to IFNγ, hypomethylation and upregulation of Txnip may prime Dnmt3a-mutant HSCs to respond differently to inflammation. Our data show under IFNγ challenge, upregulation of Txnip leads to p53 stabilization via direct interaction, with consequent upregulation of p21 preserving functional integrity of Dnmt3a-mutant HSCs. Furthermore, dissecting each component of this Txnip–p53–p21 pathway revealed the relative importance and functional contribution of each factor. Although p53 and p21 have a plethora of functions in HSCs, we demonstrate a genotype-specific effect in preserving the functional integrity of Dnmt3a-mutant HSCs in response to IFNγ. But while the importance of the Txnip–p53–p21 mechanism was functionally validated in vivo, this pathway cannot be the only molecular mechanism that increases the fitness of Dnmt3a-mtuant HSCs. p53 has a multitude of context-dependent activities in HSCs which may not be inextricably linked to the presented mechanism. Indeed, constitutive p53 activity impairs hematopoietic homeostasis and causes HSC depletion (42). In this study, while Txnip and p21 shRNAs rescued the apoptosis and cell-cycle phenotypes of Dnmt3a-mutant HSCs in response to IFNγ, p53 shRNAs largely did not. Moreover, only a fraction of Txnip appears to bind p53, suggesting Txnip has p53-independent actions. These data highlight the complexity of the mechanistic regulation of the HSC response to inflammation and how CH mutations can corrupt normal pathways.
Recent studies have identified a role for DNMT3A in the response of HSCs to chronic inflammation. The fitness advantage of Dnmt3a-mutant HSCs was quantified here using a novel transgenic IFNγ overexpression mouse model that enabled direct comparison between HSC genotypes. The IFNγ exposure regimen used in this study revealed an enforced quiescence phenotype in Dnmt3a-mutant HSCs by both in vivo BrdU labeling, cell-cycle analysis and in vitro single-cell division assay. We propose that enhanced quiescence-enforcing mechanisms, such as upregulation of p21, in Dnmt3a-mutant HSCs protect them from extrinsic forces that normally stimulate HSC proliferation and lead to functional decline. Cell-cycle analysis showed Dnmt3a-mutant HSCs show a delayed exit from G0 in response to IFNγ. While p21 is more typically thought to function as a regulator of the G1–S phase of cell cycle, p21 also causes cell-cycle arrest in G0 in different settings (43, 44), again reinforcing the idea that these mechanisms are highly context-dependent. Moreover, there are likely many other molecular barriers that impede cell-cycle entry of Dnmt3a-mutant HSCs, one possibility being reduced metabolic activity suggested by GSEA of IFNγ-treated HSCs (Fig. 4C). Importantly, this study demonstrates the first in vivo evidence that human DNMT3A-mutant HSCs are sustained in response to episodic IFNγ exposure. The relatively small survival benefit observed in this model supports the premise that the fitness advantage conveyed by DNMT3A mutations in HSCs requires cumulative selection pressure over decades to manifest human CH. One caveat to these studies that must be acknowledged is the use of irradiation to condition the hosts, a process that induces inflammation, but appropriate controls were included in each experiment.
Despite tremendous advances in understanding the genetic basis of MDS and AML, cure rates have not substantially improved in the last 40 years (45). To our knowledge, this is one of the first studies to link changes in an environmental selection pressure with a fitness advantage for HSCs bearing a specific mutation. Targeting the mechanisms that allow DNMT3A-mutant HSCs to gain clonal dominance may highlight interventions that can selectively inhibit their growth and diminish future risk of malignant transformation.
METHODS
Mice and Transplantation
The Institutional Animal Care and Use Committee at Washington University School of Medicine approved all animal procedures. Mice were housed in specific pathogen-free conditions at Washington University School of Medicine on a 12:12-hour light:dark cycle in temperature- and humidity-controlled rooms. Donor mice were typically 12 weeks old for experimentation. Both male and female mice were used. All mice were from C57Bl/6 background. Dnmt3afl/fl, Dnmt3afl/R878, and Tet2fl/fl mice were crossed to Vav-Cre strain. Recipient mice (C57Bl/6 CD45.1, The Jackson Laboratory strain #002014) mice were approximately 8 weeks old and transplanted by retro-orbital injection after a split dose (4 hours apart) of 10.5 Gy irradiation (Cesium-137). For general HSC-competitive transplants, 200 phenotypically defined HSCs (donors, CD45.2) were transplanted along with 2.5 × 105 whole BM (WBM) cells from wild-type (CD45.1) mice. For the competitive transplants challenged with stressors, 5 × 105 WBM donor cells (CD45.2) were cotransplanted with 5 × 105 WBM competitors (CD45.1). Recipients were randomized 4 weeks posttransplant and treated with PBS (control) or two consecutive doses of 10ug recombinant mouse IFNγ (mIFNγ; retro-orbital injection). Recipients were sacrificed 18-weeks posttransplant for HSC analysis. For CH competition model, 1.2 × 106 WBM (CD45.1) from Rosa26M2rtTA; IFNgr1KO (= TetON; control) or tetO_IFNg; Rosa26rtTA-M2; IFNgr1KO (= TetON-IFNg) were cotransplanted with 1.5 × 105 WBM cells from CD45.2 (donors) and 1.5 × 105 CD45.1/2 Ubc-GFP cells (competitors) into CD45.1 recipients. This models human CH with a 10% VAF of the test genotype. Recipients were fed 1,250 ppm doxycycline chow (TestDiet #1818439–203) on alternate weeks during 5 to 20 weeks posttransplant. This level of doxycycline chow was titrated to induce a physiologically relevant serum IFNγ level (∼200 pg/mL). HSC quantification was performed 24 weeks posttransplant, and 7 × 105 c-Kit–enriched chimeric BM was transplanted into secondary recipients via retro-orbital injection. HSC quantification of secondary transplants was analyzed 18 weeks posttransplant. For all mouse transplants, experiments were repeated across at least two independent cohorts. Final graphs represent compiled data across multiple cohorts to control for experimental variability. The following mouse strains were used - Vav-Cre (46), Dnmt3afl/fl (47), Dnmt3afl/R878 (24), Tet2fl/fl (48), p53−/− (49), Ubc-GFP (The Jackson Laboratory strain #004353), tetO_IFNg (The Jackson Laboratory strain #009344), Rosa26rtTA-M2 (The Jackson Laboratory strain #006965); IFNgr1−/−( = IFNgr1KO; The Jackson Laboratory strain #003288), IFNg−/−( = IFNgKO; The Jackson Laboratory strain # 002287), and Col1a1tetO-H2B-GFP ( = H2B-GFP; The Jackson Laboratory strain #016836). For studies requiring single HSC division assays, H2B-GFP labeling of HSCs was induced with a 3-week course of 10,000 ppm doxycycline chow, followed by a 1-week chase period prior to isolation of HSCs. The purpose of H2B-GFP in this experiment was simply a label to more easily visualize HSCs during microscopic evaluation.
Human Cord Blood Transplantation
Deidentified cord blood specimens were obtained from the St. Louis Cord Blood Bank. Because samples were anonymized, no member of the study team had access to information that allowed the specimens to be linked to identifiable individuals, and the provider of the specimens was not involved in the design or conduct of the research, these experiments were deemed “nonhuman studies” by the Washington University Human Research Protection Office (HRPO). Mononuclear cells were isolated by Ficoll gradient according to standard procedures. CD34+ cells were isolated using magnetic enrichment (Miltenyi Biotec #130–100–453) and cultured overnight in SFEMII media (StemCell Technologies #09605) supplemented with 50 U/mL penicillin–streptomycin (Fisher Scientific #MT30002CI), 50 ng/mL human stem cell factor (SCF; Miltenyi Biotec #130–096–695), 50 ng/mL human thrombopoietin (TPO; Miltenyi Biotec #130–094–013), and 50 ng/mL human FLT3 L (Miltenyi Biotec #130–096–479). Enrichment efficiency was confirmed by flow cytometry.
For each sublethally irradiated (200 rads) NSG (The Jackson Laboratory strain #005557) recipient, 7 × 104 live cells were transplanted via intratibial injection. Engraftment was determined 4 weeks posttransplantation, and recipients were randomized for IFNγ treatment. Two consecutive doses (24 hours apart) of recombinant human IFNγ (hIFNγ, 2 μg) were retro-orbitally injected per week during two treatment blocks (week 5–8 and week 13–16). Volume-equivalent PBS was injected as a control. Recipients were sacrificed 18 weeks posttransplant for HSC analysis. Approximately 3 × 104 hCD45+ cells were sorted from each recipient's BM to quantify posttransplant VAF. For secondary transplantation, hCD34+ cells were enriched using anti-human CD34 antibody from pooled samples per group. 1 × 106 hCD34+ cells were transplanted into secondary recipients via intratibial injection. Recipients were sacrificed 18 weeks posttransplantation for HSC analysis and VAF quantification.
CRISPR and VAF Determination
Single guide RNAs (gRNA) targeting DNMT3A (exon 12 and exon 14) or AAVS1 were designed using the UCSC Genome Brower (https://genome.ucsc.edu) with the following sequences – AAVS1: GGGGCCACTAGGGACAGGAT; DNMT3A exon 12 (1012): TCCCCAGCATCGGACCCCAC; DNMT3A exon 14 (1014): CAGGCGTGGTAGCCACAGTG. Gene-knockout was performed with the CRISPR-RNP system (IDT) via nucleoporation using Neon Transfection Kit following the manufacturer's instructions. Briefly, 1.5 μg of CRISPR-Cas9 (NLS, 2, IDT) and 1 uμg of gRNA were nucleofected into 2 × 105 CD34+ cells. Cells were incubated at 37°C with 5% CO2 overnight postnucleofection and recovered for an additional 24 hours in fresh media. 5 × 104 edited cells were collected to determine CRISPR/Cas9 targeting efficiency using PCR amplicon–based deep sequencing. CRISPresso 2.0 was used to identify mutations and quantify VAF. The following primers were used to generate amplicons from genomic DNA for VAF quantification - AAVS1-forward: CACTCTTTCCCTACACGACGCTCTTCCGATCTACAGGAGGTGGGGGTTAGAC with AAVS1-reverse: GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTCCCCTATGTCCACTTCAGGA; DNMT3A-1012-forward: CACTCTTTCCCTACACGACGCTCTTCCGATCTCACCTCGTACTCTGGCTCGT with DNMT3A-1012-reverse: GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTCAGGAATGAATGCTGTGGAA; and DNMT3A-1014-forward: CACTCTTTCCCTACACGACGCTCTTCCGATCTCACCTCGTACTCTGGCTCGT with DNMT3A-1014-reverse: GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTCAGGAATGAATGCTGTGGAA.
Cell Purification and Flow Cytometry
BM cells were isolated from iliac crests, femurs, and tibias of mice. Cells were stained in Hanks Balanced Salt Solution (HBSS, Corning #21021CV) containing 100 Units/mL penicillin/streptomycin (Fisher Scientific #MT30002CI), 10 μmol/L HEPES (Life Technologies #15630080) and 2% Serum Plus II (Sigma #14009C) at a density of 1.0 × 108/mL. Staining was performed for >20 minutes at 4°C with desired antibodies. For cell sorting, BM was enriched with anti-mouse CD117-conjugated microbeads (Miltenyi Biotec, #130–0910224) using AutoMacs Pro Separator (Miltenyi Biotec), then stained with appropriate antibody cocktails. For typical mouse HSC sorting and analysis, staining panel included: CD45.1-FITC (clone A20; BioLegend #110706), CD45.2-BV421 (clone 104; BioLegend #109831), B220-APCcy7 (clone RA3–6B2; BioLegend #103224), Gr-1-APCcy7 (clone RB6–8C5; BioLegend #108424), Mac-1-APCcy7 (clone M1/70; BioLegend #101226), CD3e-APCcy7 (clone 145–2C11; BioLegend #100330), Ter119-APCcy7 (clone TER-119; BioLegend #116223), CD48-PECy7 (clone HM48–1; BioLegend #103424), CD150-PE (clone TC15–12F12.2; BioLegend #115904), c-Kit-BV605 (clone 2B8; BioLegend #105847), EPCR-APC (clone RCR-16; BioLegend #141506). For BrdU analysis, mice were injected intraperitoneally with a single dose of 3.33 mg BrdU (Sigma #B5002) in 500 μL PBS, then supplemented with 0.8 mg/mL BrdU in drinking water over a 72-hour timecourse. At 24 and 48 hours, mice were given either PBS (control) or 10 μg recombinant mouse IFNγ (rmIFNg) by retro-orbital injection. At 72 hours, mice were sacrificed for BrdU analysis. BM from each mouse was enriched with anti-CD117 microbeads as above, then stained with the following antibodies: B220-APCCy7 (clone RA3–6B2; BioLegend #103224), Gr-1-APCcy7 (clone RB6–8C5; BioLegend #108424), Mac-1-APCcy7 (clone M1/70; BioLegend #101226), CD3e-APCcy7 (clone 145–2C11; BioLegend #100330), Ter119-APCcy7 (clone TER-119; BioLegend #116223), CD48-PECy7 (clone HM48–1; BioLegend #103424), CD150-BV421 (clone TC15–12F12.2; BioLegend #115925), c-Kit-FITC (clone 2B8; BD #561680), EPCR-PE (clone 1560; BD #566337). After antibody staining, samples were processed with the BrdU-APC flow kit (BD #552598) as per manufacturer instructions. For measurement of ROS levels in HSCs, 10 million BM cells or 500 cultured HSCs stained with a HSC flow panel were incubated with 5 μmol/L CM-H2DCFDA (Thermo Fisher) at 37°C for 30 minutes, and mean fluorescence intensity (MFI) was quantified. For determination of cell-cycle stages in HSCs, 10 million BM cells or 500 cultured HSCs stained with a HSC flow panel were fixed, permeabilized, and stained with anti-Ki67-BV605 (clone 16A8; BioLegend #652413; 1:50 dilution) at 4°C for 1 hour. DNA content was determined by 7AAD (BioLegend #420404, at 1:10 dilution).
For mouse peripheral blood analysis, red blood cells were lysed then samples were stained with the following antibodies: CD45.1-FITC (clone A20; BioLegend #110706), CD45.2-BV421 (clone 104; BioLegend #109831), B220-PECy7 (clone RA3–6B2; BioLegend #103222), B220-APC (clone RA3–6B2; BioLegend #103212), Gr-1-PECy7 (clone RB6–8C5; BioLegend #108416), Mac-1-PECy7 (clone M1/70; BioLegend #101216), CD3e-APC (clone 145–2C11; BioLegend #100312).
For peripheral blood analysis from NSG transplants, blood was collected as above then samples were stained with the following antibodies: anti-mouse CD45-BV605 (clone 30-F11; BioLegend #103139), anti-human CD45-APC (clone 2D1; BioLegend #368512), anti-human CD3-FITC (clone OKT3; BioLegend #317306), anti-human CD19-PECy7 (clone HIB19; BioLegend #302215), anti-human CD33-BV421 (clone WM53; BD #565949). For BM analysis of cord blood–transplanted NSG mice, BM was collected as above then samples were stained with the following antibodies - anti-mouse CD45-BV605 (clone 30-F11; BioLegend #103139), anti-human CD45-APC (clone 2D1; BioLegend #368512), anti-human CD34-PE (clone 561; BioLegend #343606), anti-human CD90-PECy7 (clone 5E10; BioLegend #328124), anti-human CD45RA-BV421 (clone HI100; BioLegend #304130), anti-human CD38-FITC (clone HB7; Invitrogen #11–0388–42), anti-human CD49f-APCcy7 (clone GoH3; BioLegend #313628), and anti-human Lineage cocktail-BV510 (BioLegend #348807).
All antibodies were used at 1:100 dilution unless stated otherwise. Dead cells were excluded with 7AAD (BioLegend #420404, used at 1:100 dilution). Cell sorting was performed using MoFlo (Beckman Coulter) and FACS Aria II (BD) flow cytometers. Flow cytometric cell analysis was performed using Attune NxT (Thermo Fisher Scientific) and FACS Aria II (BD) flow cytometers. Acquired flow cytometry data were analyzed with FlowJo software (Tree Star).
Lentiviral Production and Transduction
293T cells were cotransfected with the packaging plasmids pMD.G, psPAX2 and with an shRNA-containing plasmid (BFP-(miR-E)-PGK-Puro) using PEI-based transfection protocol (Polysciences #23966–1). shRNAs were designed according to published protocols and the following 22-mer target sequences were used for final experimentation – p21 shRNA#1: TTTAAATAACTTTAAGTTTGGA; p21 shRNA#2: TTAAATAACTTTAAGTTTGGAG; p53 shRNA#1: TTACACATGTACTTGTAGTGGA; p53 shRNA#2: TAAAATAGGAAATTGATATATA; Txnip shRNA#1: TAATCATACAAAAAGATACACA; Txnip shRNA#2: TTAATCATACAAAAAGATACAC. The negative control shRNA targeting Renilla luciferase was subcloned from Addgene plasmid #111170. Transfections for lentiviral production were performed in 150 × 25 mm tissue culture dishes (Falcon #353025) when 293T cells reached >85% confluency. Forty-eight hours posttransfection, 293T cell supernatants were collected and concentrated by centrifugation at 76,000 × g for 1.5 hours at 4°C. For lentiviral transduction, c-Kit+ BM cells were purified using magnetic enrichment and adjusted to 5 × 105 cells/100 mL in Stempro-34 medium (Gibco #10639011) supplemented with 100 Units/mL penicillin/streptomycin (Fisher Scientific #MT30002CI), 2 mmol/L l-glutamine (Gibco #25030–081), 100 ng/mL murine stem cell factor (SCF; BioLegend #579704), 100 ng/mL murine thrombopoietin (TPO; PeproTech, #315–14), 50 ng/mL murine Flt3 L (PeproTech, #250–31L), 5 ng/mL murine IL3 (Miltenyi Biotec #130–099–510), 4 μg/mL polybrene (Sigma; #TR-1003-G), and spin-infected with lentivirus preparations at 250 × g for 2 hours in a 96-well plate. Twenty-four hours posttransduction, 1.6 × 105 transduced cells were transplanted into lethally irradiated mice (following a split dose of 10.5 Gy of irradiation) by retro-orbital injection.
Single-Cell Division Assay
Control-H2B-GFP (Vav-Cre; Dnmt3a+/+; Rosa26M2rtTA; Col1a1tetO-H2B-GFP) and Dnmt3aKO–H2B-GFP (Vav-Cre; Dnmt3afl/fl; Rosa26M2rtTA; Col1a1tetO-H2B-GFP) mice were fed 10,000 ppm doxycycline chow (TestDiet #1815461–203) for 21-days. A 7-day recovery period was given prior to two-consecutive doses of rmIFNγ (10 μg) or PBS (24 hours apart). Mice were sacrificed 2 hours post second dose. Single immunophenotypically-defined HSC (Lineage− c-Kit+ EPCR+ CD150+ CD48− GFPhi) were sorted into individual wells of 96-well plates containing complete Stempro-34 media described above. For single-cell division assay post-shRNA transductions, c-Kit+ cells were enriched with AutoMacs Pro Separator from three Control-H2B-GFP and three Dnmt3aKO–H2B-GFP mice and transduced with shRNA-containing viral particles. Twenty-four hours posttransduction, 1.6 × 105 transduced cells (pooled by genotype) were transplanted into recipients. When stable engraftment was achieved at 8 weeks posttransplant, recipients were fed 10,000 ppm doxycycline chow to label HSCs with GFP. After a 7-day recovery period, two doses of rmIFNγ (10 γg) or PBS were injected into recipients and 2 hours posttreatment, single immunophenotypically defined HSCs were sorted into 96-well plates. Single HSCs were cultured in Stempro-34 medium and cell division behaviors were monitored. H2B-GFP allowed accurate identification of HSCs under a fluorescent microscope. Cell division was monitored at 12-hour intervals (60 hours postsorting was not evaluated). The latency to first division was determined as the time when 50% of trackable cells achieved their first division. Trackable cells were defined as cells that died postdivision and cells that survived throughout the 72-hour period.
CaspaseGlo Assay
Five-hundred HSCs were purified from mice treated with two consecutive doses of PBS or rmIFNγ (10 μg) and sorted into white-walled 96-well plates (Corning #3610) containing complete Stempro-34 media. For measuring apoptosis in freshly purified HSCs, cleaved caspase 3/7 activity (CaspaseGlo 3/7 assay; Promega # G8090) was measured immediately postsorting. For measuring apoptosis in stressed HSCs, the CaspaseGlo assay was employed after a 16-hour culture period. CaspaseGlo assay was carried out according to manufacturer instructions. Cleaved caspases 3/7 activity was measured by luminescence intensity using BioTek Synergy H1 Hybrid Plate Reader.
Serum IFNg Determination
Sera were isolated from peripheral blood using microtainer tubes with serum separator additive (BD #365967). Serum IFNγ levels were quantified using the Mouse IFNg ELISA Kit II (BD #558258).
Quantitative Real-time PCR
Total RNA was extracted using the Nucleospin RNA XS kit (Macherey-Nagel #740902–250) and reverse-transcribed with the SuperScript VILO kit (Invitrogen #11754–050). Quantification was performed with TaqMan Master Mix (Applied Biosystems #4304437), the desired gene-specific probe (FAM-MGB; Applied Biosystems) and an 18S probe (VIC-MGB; Applied Biosystems). Transcript expression levels were normalized to endogenous 18S, and fold-changes were determined using the ΔΔCt method.
RNA Sequencing
HSCs were purified from biological replicates of mice (pooled WBM from two male and two female 12-week-old mice) treated twice with either PBS or recombinant mouse IFNγ (rmIFNγ) 24 hours apart. HSCs were purified 2-hours after the second dose. A NucleoSpin RNA XS kit (Macherey-Nagel #740902.250) was used to isolate RNA. Library preparation, sequencing, and alignment were performed by the Genome Technology Access Center (Washington University). The SMARTer Ultra Low RNA kit (ClonTech) was used to prepare the libraries from 3 to 5 ng of total RNA. Sequencing was performed with an Illumina NextSeq 3000. Reads were aligned with STAR version 2.5.4b with Gencode release M20 (GRCm38.p6) genome assembly. Unambiguous read counts were calculated by HTSEQ-count version 0.6.0 with mode “intersection-strict.” Expression data were imported into Noiseq v2.28.0 for differential gene expression analysis with TMM normalization and batch correction. Generalized linear models were then created to test for gene/transcript level differential expression. Differentially expressed genes (DEG) were then filtered for unadjusted P values <0.05. Gene-set enrichment analysis (GSEA) was performed with fGSEA v1.10.0.
Single-Cell RNA Sequencing
Two male and two female mice per genotype (control, Dnmt3aKO) were treated with two doses of PBS (control) or rmIFNγ (10 μg) 24 hours apart, then sacrificed 2 hours after the second injection. BM was isolated from individual mice and 17,000 Lineage– c-Kit+ EPCR+ BM cells per mouse were purified by flow cytometry. Cells were resuspended at 1,200 cells per μL in PBS + 0.04% BSA, with cell concentration verified by flow cytometric counting. cDNA was prepared after the GEM generation and barcoding, followed by the GEM-RT reaction and bead cleanup steps. Purified cDNA was amplified for 11 to 13 cycles before being cleaned up using SPRIselect beads. Samples were then run on a Bioanalyzer to determine the cDNA concentration. GEX libraries were prepared as recommended by the 10x Genomics Chromium Single Cell 3′ Reagent Kits v3 user guide with appropriate modifications to the PCR cycles based on the calculated cDNA concentration. For sample preparation on the 10x Genomics platform, the Chromium Single Cell 3′ GEM, Library and Gel Bead Kit v3 (PN-1000075), Chromium Chip B Single Cell Kit, 48 rxns (10x Genomics PN-10000153), and Chromium Dual Index Kit TT Set A (PN-1000215) were used. The concentration of each library was accurately determined through qPCR utilizing the KAPA library Quantification Kit according to the manufacturer's protocol (KAPA Biosystems/Roche) to produce cluster counts appropriate for the Illumina NovaSeq6000 instrument. Normalized libraries were sequenced on a NovaSeq6000 S4 Flow Cell using the XP workflow and a 28 × 10 × 10 × 150 sequencing recipe according to manufacturer protocol. A median sequencing depth of 50,000 reads per cell was targeted for each library. Single-cell RNA-seq data were demultiplexed and aligned to the Genome Reference Consortium mouse genome (mm10). The aligned data were then annotated and UMI-collapsed using Cellranger (v3.1.0, 10x Genomics). Cells with more than 10% mitochondrial gene expression and with top 5% unique feature counts were excluded. This resulted in the following number of unique cells that passed quality control and were examined in downstream analysis: Control-PBS: 26,199; Control-IFNγ: 19,829; Dnmt3aKO-PBS: 11,804; Dnmt3aKO-IFNγ: 20,396. Principal component analysis was performed to reduce data using Seurat 3.0. Clusters were identified applying the dimensional reduction techniques (tSNE and UMAP) using the same package. Expression levels of conventional cell surface markers were examined to annotate clusters. Differentially expressed genes were identified within the cluster of interest.
Whole Genome-Wide Bisulfite Sequencing
At the conclusion of the CH competition model transplant with chronic IFNγ (primary transplant), 3 × 105 donor-derived (CD45.2+) BM cells per genotype per treatment were sorted from pooled recipient BM cells. Genomic DNA (gDNA) was extracted using PureLink Genomic DNA Mini Kit (Invitrogen). 250 ng of gDNA for each sample, spiked in with lambda control, was fragmented using Covaris. The fragmented DNA was then subjected to an overnight cytosine-to-thymine conversion using EZ DNA Methylation-Gold Kit (Zymo Research). The CT converted samples were used for library construction with Accel-NGS Methyl-Seq DNA Library Prep Kit, indexed with Methyl-Seq Dual Indexing kit. The sequencing was performed on NovaSeqS4 300XP. Raw reads were quality and adapter trimmed using cutadapt (v2.4) with paramteters -q 15,10 –minimum-length 36. Trimmed reads were aligned using bismark (v0.19.0) with bowtie2 and options -X 1000 –score_min L,0,-0.6, -N 0. After removal of PCR duplicates from aligned reads, methylation levels were calculated using bismark_methylation_extractor. CpG methylation files were loaded in R (v 3.6.3) and transformed into a BS object using bsseq. Differentially methylated loci (DML) were determined using the DMLtest function of DSS after with option smoothing = TRUE. Differentially methylated regions (DMR) were calculated from the result of DMLtest using the callDMR function of DSS with option p.threshold = 1e-5. Resultant DMRs were further filtered to remove any DMRs with fewer than 4 CGs or a methylation difference between conditions of less than 20%. Bed formatted coordinates of DMRs were extracted from filtered lists of DMRs. Methylation levels were transformed into bigwig format and plotted at DMRs using deeptools. Average methylation levels in all DMR comparisons were determined using the bedtools map function with options -o mean and compiled into a table. The table was plotted as a heatmap using the pheatmap package (v1.0.12) in R with clustering across the DMRs using the default for the hclust function.
In Vitro Methylation-Sensitive Reporter Assay
The hypomethylated DMR upstream region of Txnip (-3283 to -1793bp) was amplified from wild-type mouse gDNA using primers Upstream_Txnip_F (AAATTTAAAAGGTACCGGGTAGTCTCAAATCCCACAGACTCA) and Upstream_Txnip_R (TTTAAATTTTGAGCTCCCAGAAAGTTAACTAACTAGGCGTCA) and ligated to the pGL4.23 [luc2] vector (Promega) using KpnI and SacI restriction enzyme sites. The bacterial CpG methyltransferase M.SssI (New England Biolabs) was used to methylate the element-containing construct and vector only according to manufacturer's instruction. In vitro methylation efficiency was determined by restriction enzyme digestion using BstUI that only recognize unmethylated CpG but is not able to cut methylated CpG sites. pGL4.23-based plasmids and pGL4.75 [hRluc] (Promega) (9:1) were transiently transfected into 3T3 cells using PEI-based transfection protocol (Polysciences #23966–1). pGL4.74 (hRluc/TK) contains Renilla luciferase and was used to control for transfection efficiency. Transfections were performed in triplicates. Luciferase assays were carried out 48 hours posttransfection using the Dual-Glo luciferase assay system (Promega) according to manufacturer's instructions.
In Vitro IFNγ Exposure
32D cells were cultured in RPMI1640 medium supplemented with 2 mmol/L L-glutamine, 10 mmol/L HEPES and 100 U/mL Penicillin–streptomycin and incubated at 37°C with 5% CO2 until reaching 100% confluency. Thirty-six hours postconfluency, 2.6 × 106 cells were seeded into a new flask at the density of 2.6 × 105 cells/ml with 10 ng/mL recombinant mouse IFNγ or volume-equivalent PBS. Cells were collected 24 hours postexposure for protein or chromatin harvest.
Chromatin Immunoprecipitation and Real-time PCR
Chromatin was extracted, sheared and isolated from 5 × 106 32D cells using SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling Technology) following manufacturer instruction. Chromatin shearing efficiency was examined by agarose gel electrophoresis. Ten micrograms of prepared chromatin was used to precipitate p53-bound gDNA using p53 antibody (Cell Signaling Technology #32532). An IgG control antibody was also applied to each sample as isotype control. The precipitated gDNA was then eluted and subjected to qPCR using gene-specific primers for p21 promoter (forward: CTTTCTGGCCTTCAGGAACA; reverse: GCTGGTAGTTGGGTATCATCAG). For positive controls, all samples were immunoprecipitated with H3 antibody (Cell Signaling Technology #4620) and amplified with Mouse RPL30 Intron 2 Primers (Cell Signaling Technology #7015) to ensure the quality of chromatin was comparable. The enrichment of p53 was determined as 2(Ct_input –Ct_sample).
Immunoprecipitation and Western Blotting
Thirty to 50 μg of protein samples were separated in precasted 4%–15% gradient SDS gels (Bio-Rad, #456–1084) and transferred to polyvinylidene difluoride membranes (Millipore #IPVH00010). Membranes were subsequently probed with antibodies to detect p21 (Santa Cruz Biotechnology, # sc-6264), p53 (Cell Signaling Technology #32532), Txnip (Cell Signaling Technology #32532), β-Actin (Santa Cruz Biotechnology, #SC-47778) and Vinculin (Santa Cruz Biotechnology #sc-73614). Detection was performed using horseradish peroxidase–conjugated secondary mouse or rabbit antibody and chemiluminescence HRP substrate (Millipore #WBKLS0100). For protein coimmunoprecipitation, total protein was extracted from 10 × 106 32D cells using Cell Lysis Buffer (Cell Signaling Technology #9803) following manufacturer's instructions. Four-hundred micrograms of freshly prepared total protein was then incubated with p53 antibody (1:50 dilution; Cell Signaling Technology #32532) at 4°C overnight. Antibody-bound lysates were captured by protein G beads. The lysates were then eluted and prepared for Western blot analysis. Forty micrograms of pooled total protein was also loaded on Western blot as 10% input control. p53-bound proteins were examined to detect p53 and Txnip.
Statistical Analysis
Student t test and one-way ANOVA were used for statistical comparison where appropriate. Significance is indicated using the following convention: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. All graphs represent mean ± SEM. unless otherwise indicated.
Data Reporting
No statistical methods were used to predetermine sample size. Group sizes were based on historical data outcomes from our laboratories. The experiments were randomized where stated. Investigators were not blinded to allocation during experiments and outcome assessment.
Data Availability
Raw data of bulk RNA-seq, single-cell RNA-seq and whole-genome bisulfite-sequencing are available in the Gene Expression Omnibus (GEO) repository under accession number GSE168807 (http://www.ncbi.nlm.nih.gov/geo/). Previously generated WGBS data (19) used to generate Fig. 5F are available under accession number GSE98191. There are no restrictions on data availability.
Supplementary Material
Acknowledgments
We thank all members of the Challen laboratory for ongoing contributions and critical discussion. We thank the Alvin J. Siteman Cancer Center at Washington University for use of the Siteman Flow Cytometry Core, supported in part by NCI Grant CA91842 and NIH WLC6313040077. We thank the Genome Technology Access Center and McDonnell Genome Institute at Washington University for genomic analysis, partially supported by NCI Grant CA91842 and by ICTS/CTSA NIH Grant UL1TR000448. This work was supported by the NIH (DK102428 and DK124883 to G.A. Challen; HL136333 and, HL134880 to K.Y. King), the Edward P. Evans Foundation, Gabrielle's Angel Foundation, Gabrielle's Angel Foundation, and The Longer Life Foundation (to G.A. Challen). C.R. Zhang was supported by an American Society of Hematology post-doctoral scholar award and Edward P. Evans Center for MDS Post-Doctoral Fellowship. E.L. Ostrander was supported by NIH 5T32CA113275 and NIH F31DK114951. C. Mallaney was supported by NIH T32HL007088, and NIH DK111058. H. Celik was supported by an Edward P. Evans Foundation Young Investigator Award. G.A. Challen is a scholar of the Leukemia and Lymphoma Society.
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
Authors’ Disclosures
K.Y. King reports grants from NIH during the conduct of the study. G.A. Challen reports grants from NIDDK, NHLBI, NCI, Edward P. Evans Foundation, Gabrielle's Angel Foundation, American Society of Hematology, and grants from Leukemia and Lymphoma Society during the conduct of the study. No disclosures were reported by the other authors.
Authors’ Contributions
C.R. Zhang: Conceptualization, resources, data curation, software, formal analysis, validation, investigation, visualization, methodology. E.L. Ostrander: Data curation, validation, investigation, methodology. O. Kukhar: Investigation. C. Mallaney: Investigation. J. Sun: Investigation. E. Haussler: Investigation. H. Celik: Investigation. W.K. Koh: Investigation. K.Y. King: Conceptualization, resources. P. Gontarz: Software, formal analysis, methodology. G.A. Challen: Conceptualization, formal analysis, supervision, funding acquisition, writing-original draft, project administration, writing–review and editing.
References
- 1. Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell 2012;150:264–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Watson CJ, Papula AL, Poon GYP, Wong WH, Young AL, Druley TE, et al. The evolutionary dynamics and fitness landscape of clonal hematopoiesis. Science 2020;367:1449–54. [DOI] [PubMed] [Google Scholar]
- 3. Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 2014;371:2488–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Genovese G, Kahler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 2014;371:2477–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med 2014;20:1472–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Challen GA, Goodell MA. Clonal hematopoiesis: mechanisms driving dominance of stem cell clones. Blood 2020;136:1590–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999;99:247–57. [DOI] [PubMed] [Google Scholar]
- 8. Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol 2003;23:5594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med 2010;363:2424–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Walter MJ, Ding L, Shen D, Shao J, Grillot M, McLellan M, et al. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia 2011;25:1153–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature 2014;506:328–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Van Egeren D, Escabi J, Nguyen M, Liu S, Reilly CR, Patel S, et al. Reconstructing the lineage histories and differentiation trajectories of individual cancer cells in myeloproliferative neoplasms. Cell Stem Cell 2021;28:514–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Williams N, Lee J, Mitchell E, Moore L, Baxter EJ, Hewinson J, et al. Life histories of myeloproliferative neoplasms inferred from phylogenies. Nature 2022;602:162–8. [DOI] [PubMed] [Google Scholar]
- 14. Young AL, Challen GA, Birmann BM, Druley TE. Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat Commun 2016;7:12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet 2012;44:23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Challen GA, Sun D, Mayle A, Jeong M, Luo M, Rodriguez B, et al. Dnmt3a and Dnmt3b have overlapping and distinct functions in hematopoietic stem cells. Cell Stem Cell 2014;15:350–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Izzo F, Lee SC, Poran A, Chaligne R, Gaiti F, Gross B, et al. DNA methylation disruption reshapes the hematopoietic differentiation landscape. Nat Genet 2020;52:378–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Spencer DH, Russler-Germain DA, Ketkar S, Helton NM, Lamprecht TL, Fulton RS, et al. CpG island hypermethylation mediated by DNMT3A is a consequence of AML progression. Cell 2017;168:801–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jeong M, Park HJ, Celik H, Ostrander EL, Reyes JM, Guzman A, et al. Loss of Dnmt3a immortalizes hematopoietic stem cells in vivo. Cell Rep 2018;23:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hormaechea-Agulla D, Matatall KA, Le DT, Kain B, Long X, Kus P, et al. Chronic infection drives Dnmt3a-loss-of-function clonal hematopoiesis via IFNgamma signaling. Cell Stem Cell 2021;28:1428–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cai Z, Kotzin JJ, Ramdas B, Chen S, Nelanuthala S, Palam LR, et al. Inhibition of inflammatory signaling in Tet2 mutant preleukemic cells mitigates stress-induced abnormalities and clonal hematopoiesis. Cell Stem Cell 2018;23:833–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Abegunde SO, Buckstein R, Wells RA, Rauh MJ. An inflammatory environment containing TNFalpha favors Tet2-mutant clonal hematopoiesis. Exp Hematol 2018;59:60–5. [DOI] [PubMed] [Google Scholar]
- 23. Zhang CRC, Nix D, Gregory M, Ciorba MA, Ostrander EL, Newberry RD, et al. Inflammatory cytokines promote clonal hematopoiesis with specific mutations in ulcerative colitis patients. Exp Hematol 2019;80:36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Loberg MA, Bell RK, Goodwin LO, Eudy E, Miles LA, SanMiguel JM, et al. Sequentially inducible mouse models reveal that Npm1 mutation causes malignant transformation of Dnmt3a-mutant clonal hematopoiesis. Leukemia 2019;33:1635–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cole CB, Russler-Germain DA, Ketkar S, Verdoni AM, Smith AM, Bangert CV, et al. Haploinsufficiency for DNA methyltransferase 3A predisposes hematopoietic cells to myeloid malignancies. J Clin Invest 2017;127:3657–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Celik H, Mallaney C, Kothari A, Ostrander EL, Eultgen E, Martens A, et al. Enforced differentiation of Dnmt3a-null bone marrow leads to failure with c-Kit mutations driving leukemic transformation. Blood 2015;125:619–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gundry MC, Brunetti L, Lin A, Mayle AE, Kitano A, Wagner D, et al. Highly efficient genome editing of murine and human hematopoietic progenitor cells by CRISPR/Cas9. Cell Rep 2016;17:1453–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pietras EM, Lakshminarasimhan R, Techner JM, Fong S, Flach J, Binnewies M, et al. Re-entry into quiescence protects hematopoietic stem cells from the killing effect of chronic exposure to type I interferons. J Exp Med 2014;211:245–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Baldridge MT, King KY, Boles NC, Weksberg DC, Goodell MA. Quiescent haematopoietic stem cells are activated by IFN-gamma in response to chronic infection. Nature 2010;465:793–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Faltusova K, Szikszai K, Molik M, Linhartova J, Paral P, Sefc L, et al. Stem cell defect in ubiquitin-green fluorescent protein mice facilitates engraftment of lymphoid-primed hematopoietic stem cells. Stem Cells 2018;36:1237–48. [DOI] [PubMed] [Google Scholar]
- 31. Wilson A, Laurenti E, Oser G, van der Wath RC, Blanco-Bose W, Jaworski M, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 2008;135:1118–29. [DOI] [PubMed] [Google Scholar]
- 32. Foudi A, Hochedlinger K, Van Buren D, Schindler JW, Jaenisch R, Carey V, et al. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat Biotechnol 2009;27:84–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Essers MA, Offner S, Blanco-Bose WE, Waibler Z, Kalinke U, Duchosal MA, et al. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature 2009;458:904–8. [DOI] [PubMed] [Google Scholar]
- 34. Jeong M, Piao ZH, Kim MS, Lee SH, Yun S, Sun HN, et al. Thioredoxin-interacting protein regulates hematopoietic stem cell quiescence and mobilization under stress conditions. J Immunol 2009;183:2495–505. [DOI] [PubMed] [Google Scholar]
- 35. Kamitori K, Yamaguchi F, Dong Y, Hossain A, Katagi A, Noguchi C, et al. Both Ser361 phosphorylation and the C-arrestin domain of thioredoxin interacting protein are important for cell cycle blockade at the G1/S checkpoint. FEBS Open Bio 2018;8:1804–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jung H, Kim DO, Byun JE, Kim WS, Kim MJ, Song HY, et al. Thioredoxin-interacting protein regulates haematopoietic stem cell ageing and rejuvenation by inhibiting p38 kinase activity. Nat Commun 2016;7:13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ketkar S, Verdoni AM, Smith AM, Bangert CV, Leight ER, Chen DY, et al. Remethylation of Dnmt3a (-/-) hematopoietic cells is associated with partial correction of gene dysregulation and reduced myeloid skewing. Proc Natl Acad Sci U S A 2020;117:3123–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jung H, Kim MJ, Kim DO, Kim WS, Yoon SJ, Park YJ, et al. TXNIP maintains the hematopoietic cell pool by switching the function of p53 under oxidative stress. Cell Metab 2013;18:75–85. [DOI] [PubMed] [Google Scholar]
- 39. Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer 2009;9:400–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moll UM, Wolff S, Speidel D, Deppert W. Transcription-independent pro-apoptotic functions of p53. Curr Opin Cell Biol 2005;17:631–6. [DOI] [PubMed] [Google Scholar]
- 41. Jeong M, Sun D, Luo M, Huang Y, Challen GA, Rodriguez B, et al. Large conserved domains of low DNA methylation maintained by Dnmt3a. Nat Genet 2014;46:17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pant V, Quintas-Cardama A, Lozano G. The p53 pathway in hematopoiesis: lessons from mouse models, implications for humans. Blood 2012;120:5118–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu Z, Liu H, Yuan X, Wang Y, Li L, Wang G, et al. Downregulation of Pim-2 induces cell cycle arrest in the G0/G1 phase via the p53-non-dependent p21 signaling pathway. Oncol Lett 2018;15:4079–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gulappa T, Reddy RS, Suman S, Nyakeriga AM, Damodaran C. Molecular interplay between cdk4 and p21 dictates G0/G1 cell cycle arrest in prostate cancer cells. Cancer Lett 2013;337:177–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Greenberg P, Cox C, LeBeau MM, Fenaux P, Morel P, Sanz G, et al. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood 1997;89:2079–88. [PubMed] [Google Scholar]
- 46. Georgiades P, Ogilvy S, Duval H, Licence DR, Charnock-Jones DS, Smith SK, et al. VavCre transgenic mice: a tool for mutagenesis in hematopoietic and endothelial lineages. Genesis 2002;34:251–6. [DOI] [PubMed] [Google Scholar]
- 47. Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 2004;429:900–3. [DOI] [PubMed] [Google Scholar]
- 48. Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 2011;20:11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA Jr, Butel JS, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 1992;356:215–21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data of bulk RNA-seq, single-cell RNA-seq and whole-genome bisulfite-sequencing are available in the Gene Expression Omnibus (GEO) repository under accession number GSE168807 (http://www.ncbi.nlm.nih.gov/geo/). Previously generated WGBS data (19) used to generate Fig. 5F are available under accession number GSE98191. There are no restrictions on data availability.