ZNF384 fusion oncoproteins are shown to drive lineage aberrant leukemia by epigenetic remodeling and deregulation of hematopoietic transcription factors. ZNF384 leukemias are sensitive to FLT3 inhibition in vivo.
Abstract
ZNF384-rearranged fusion oncoproteins (FO) define a subset of lineage ambiguous leukemias, but their mechanistic role in leukemogenesis and lineage ambiguity is poorly understood. Using viral expression in mouse and human hematopoietic stem and progenitor cells (HSPC) and a Ep300::Znf384 knockin mouse model, we show that ZNF384 FO promote hematopoietic expansion, myeloid lineage skewing, and self-renewal. In mouse HSPCs, concomitant lesions, such as NRASG12D, were required for fully penetrant leukemia, whereas in human HSPCs, expression of ZNF384 FO drove B/myeloid leukemia, with sensitivity of a ZNF384-rearranged xenograft to FLT3 inhibition in vivo. Mechanistically, ZNF384 FO occupy a subset of predominantly intragenic/enhancer regions with increased histone 3 lysine acetylation and deregulate expression of hematopoietic stem cell transcription factors. These data define a paradigm for FO-driven lineage ambiguous leukemia, in which expression in HSPCs results in deregulation of lineage-specific genes and hematopoietic skewing, progressing to full leukemia in the context of proliferative stress.
Significance:
Expression of ZNF384 FO early in hematopoiesis results in binding and deregulation of key hematopoietic regulators, skewing of hematopoiesis, and priming for leukemic transformation. These results reveal the interplay between cell of origin and expression of ZNF384 FO to mediate lineage ambiguity and leukemia development.
This article is highlighted in the In This Issue feature, p. 171
INTRODUCTION
Acute leukemia arises from acquisition of germline and/or somatic genetic alterations that perturb multiple cellular pathways culminating in an arrest of hematopoietic development and excessive cellular proliferation, resulting in an expansion of immature blast cells that overrun the bone marrow (1). Diagnosis of acute leukemias into acute lymphoblastic leukemia (ALL) or acute myeloid leukemia (AML) is primarily based on cell morphology and immunophenotype, with molecular genetic features defining subtypes of leukemia within each lineage. This subtyping is critical for risk stratification and assignment of therapy, which rests largely on nonselective cytotoxic agents agnostic to specific genomic alterations (2). Lineage ambiguous leukemias are characterized by the presence of immunophenotypic markers of multiple lineages, and include entities such as mixed phenotype acute leukemia (MPAL), in which leukemias most commonly express myeloid (e.g., MPO) and B- or T-lymphoid lineage markers, and early T-cell precursor leukemia (ETP ALL; refs. 3–6). Such lineage ambiguous leukemias pose a diagnostic and therapeutic dilemma, as diagnosis rests on immunophenotyping without recourse to underlying biologic drivers.
Several recurring genomic alterations have been identified that define entities of lineage ambiguous leukemia that transcend strict immunophenotypic criteria (7, 8). Rearrangements of ZNF384 define a group of lineage aberrant leukemias, diagnosed either as B-ALL (commonly with aberrant expression of myeloid markers such as CD13 or CD33; approximately 5% of B-ALL cases) or B/myeloid MPAL in children (with expression of myeloperoxidase, MPO; approximately 50% of B/myeloid MPAL cases; refs. 7, 9–12). Expression of MPO is commonly the only feature resulting in diagnosis of MPAL rather than B-ALL; otherwise ZNF384-rearranged (ZNF384r) leukemias exhibit homogeneous immunophenotype and concomitant genomic alterations (7).
ZNF384 rearrangements result in the expression of chimeric fusion oncoproteins (FO), in which full-length ZNF384 is the C-terminal partner, juxtaposed to a range of partners at the N terminus, most commonly EP300, TCF3 and TAF15, but also ARID1B, ATP5C1, BMP2K, CLTC, CREBBP, EWSR1, NIPBL, SMARCA2, SMARCA4, and SYNRG (Supplementary Table S1; Supplementary Fig. S1A). Common concomitant genetic alterations in ZNF384r leukemia include alterations of Ras pathway genes in 35% to 50% of cases (NRAS, KRAS, PTPN11), regulators of B lymphoid development in 85% of cases (ETV6, IKZF1, PAX5, VPREB1), and genes encoding chromatin modifiers in 81% to 83% of cases (KMT2D, SETD2, CREBBP, KDM6A; refs. 11–16). The gene expression profile of ZNF384r leukemia is homogeneous irrespective of fusion partner or leukemia lineage, with reduced expression of genes related to B-cell development (VPREB1 and IKZF1), cell cycle (TGFB2 and E2F1), oxidative phosphorylation, and DNA repair, and increased expression of hematopoietic stem and myeloid progenitor signature genes (SALL4 and RUNX2), FLT3 (without FLT3 mutation), JAK–STAT signaling, leukocyte adhesion and differentiation genes, and apoptosis pathway genes (7, 17, 18).
ZNF384 is a ubiquitously expressed transcription factor with multiple splicing isoforms that encode proteins containing five to eight Cys2His2 zinc fingers, which mediate binding to AT-rich DNA minor grooves (19). The roles of ZNF384 and the mouse ortholog, Zfp384 (hereafter referred to as Znf384), in hematopoiesis are unknown; however, these genes are expressed at low levels in mouse or human HSCs, with increased expression in subsequent developmental stages. Znf384 knockout mice appear to have normal hematopoiesis, with increased bone formation, decreased arthritis, and decreased effects of influenza infection compared with wild-type mice, attributed to deregulated expression of direct ZNF384 transcriptional targets (20–24).
Overexpression of ZNF384 FO transform NIH3T3 cells (12, 25, 26), and mouse transplantation experiments have shown that expression of EP300::ZNF384 may lead to leukemia development in mice with incomplete penetrance (12, 27). ZNF384 FO, like wild-type ZNF384, localize in the nucleus outside nucleoli and are able to bind the wild-type consensus sequence (27), with varied effect on transcriptional regulation that may be dependent on cofactor interaction (12, 25, 26). In addition, ZNF384 FO activate transcription of the most highly upregulated genes in ZNF384r patients (7, 14, 28, 29). One postulated mechanism is interaction with EP300, a transcriptional scaffold and mediator of histone 3 lysine 27 acetylation, which could lead to enhanced transcriptional activation observed when EP300 and EP300::ZNF384 were coexpressed in transactivation experiments (29).
These data suggest that expression of selected ZNF384 FO may transform mouse hematopoietic cells, potentially through altered histone acetylation. However, these studies do not provide insight into the mechanistic basis of the striking lineage ambiguity characteristic of ZNF384r leukemia. To address this central question, we developed multiple models of ZNF384r leukemia utilizing viral expression of ZNF384 FO in mouse and human hematopoietic stem and progenitor cells (HSPCs) and an engineered mouse model enabling conditional expression of Ep300::Znf384. We describe the use of these models to examine the role of ZNF384 FO, concomitant genetic alterations, and cell of origin in hematopoietic transformation, lineage skewing, and leukemia formation. To gain mechanistic insight into the basis of downstream gene deregulation, we integrated multiple approaches to examine wild-type and ZNF384 FO chromatin occupancy, histone acetylation, chromatin accessibility, and gene expression. Finally, we utilized a xenograft model of ZNF384r leukemia to examine the potential for targeted therapeutic approaches.
RESULTS
Expression of ZNF384 FO Increased Self-Renewal and Altered the Immunophenotype of Mouse Lineage-Negative Cells
To fully understand the genomic landscape and FO structure of ZNF384r leukemia, we interrogated a large genomic dataset of ZNF384r B-ALL (N = 46) or B/myeloid MPAL (N = 14) compiled from 8 prior genomic studies of newly diagnosed and relapsed ALL (10, 11, 30–33) and MPAL (7, 8; Supplementary Table S1; Supplementary Fig. S1A) and confirmed recurring concomitant lesions: deletion of ETV6 (46%), deletion of CDKN2A/B (23%), and Ras pathway mutations (48%; Supplementary Fig. S1B; Supplementary Table S2). Fusion expression was validated in patient samples by RT-PCR and immunoblotting for a subset of samples (Supplementary Fig. S2A and S2B).
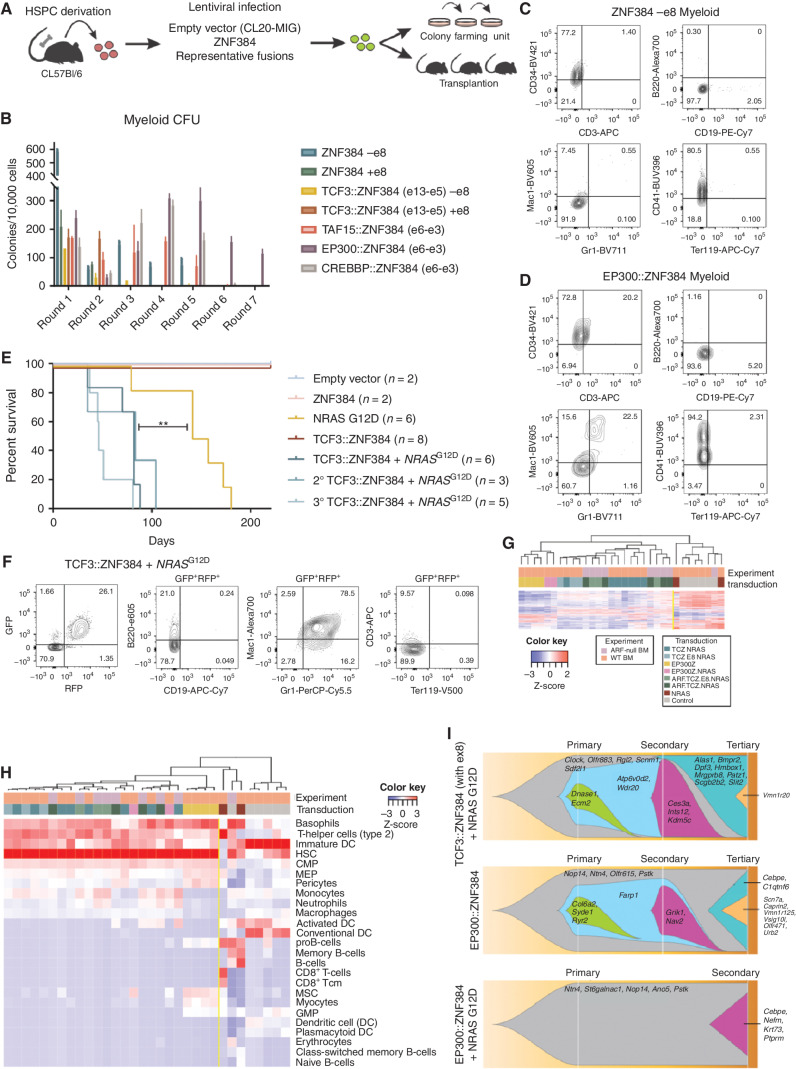
To determine the effect of ZNF384 fusions on hematopoietic self-renewal, wild-type ZNF384 and representative fusions were expressed in mouse bone marrow HSPCs by lentiviral transduction followed by in vitro colony-forming assays (Fig. 1A). When cultured in myeloid differentiation conditions, TAF15::ZNF384, EP300::ZNF384, and CREBBP::ZNF384 expressing cells were able to sustain serial replating (Fig. 1B). EP300::ZNF384–expressing cells were able to replate up to seven rounds with retention of an immature myeloid phenotype showing higher expression of CD34 compared with cells expressing wild-type ZNF384 (Fig. 1C and D). In contrast, TCF3::ZNF384–expressing cells had an initially high colony-forming ability but failed to replate after the second round (Fig. 1B). These cells exhibited high expression of ITGAM (Mac1) and Ly6G/C (Gr1), indicating they were myeloid progenitors (Supplementary Fig. S3A). In lymphoid growth conditions, only CREBBP::ZNF384 promoted serial replating (Supplementary Fig. S3B). These data support the notion that expression of ZNF384 FO transforms mouse HSPCs, with variable immunophenotype dependent upon fusion partner.
Figure 1.
Mouse HSPC transformation by expression of ZNF384 FO. A, The workflow for colony-forming assays and transplantation assays. Bone marrow was isolated from mice, enriched for HSPC, transduced with representative fusions, and sorted for colony-forming assays (CFU) or transplanted directly into mice. B, CFU of lineage-negative bone marrow cells expressing wild-type or FO, with or without exon 8, grown in myeloid differentiation conditions. Columns show means of three replicates ± SD. C, Cells harvested from CFU after two or more replatings were subjected to flow cytometry and representative immunophenotype is shown of wild-type cells (D) or EP300::ZNF384–expressing cell. E, Survival curves in primary, secondary, and tertiary recipients. Two-sided log-rank Mantel–Cox test, **, P = 0.008, F, Immunophenotyping from a representative mouse (ID 1723) showing tumors harvested from bone marrow expressing Mac1 and Gr1. G, Unsupervised hierarchical clustering of tumors generated in mice. Abbreviations: TCZ.NRAS: TCF3::ZNF384 + NRASG12D; TCZ.E8.NRAS: TCF3::ZNF384 (with exon 8) + NRASG12D; EP300Z.NRAS: EP300::ZNF384 + NRASG12D; ARF.TCZ.NRAS: ARF-null TCF3::ZNF384 + NRASG12D; ARF.TCZ.E8.NRAS: ARF-null TCF3::ZNF384 (with exon 8) + NRASG12D; NRAS: NRASG12D. H, Unsupervised hierarchical clustering xCell analysis showing hematopoietic cell-type signature enrichment. Color legend is the same from G. I, Fishplot of three mouse tumors subjected to exome sequencing showing mutational evolution.
ZNF384 FO Cooperate with NRASG12D to Drive AML In Vivo
To investigate the leukemic potential of ZNF384 FO, mouse HSPCs were harvested, transduced with representative fusions, and transplanted into lethally irradiated recipient mice (Fig. 1A; Supplementary Tables S3 and S4). Cells expressing TCF3::ZNF384, EP300::ZNF384, or TAF15::ZNF384 and NRASG12D produced an aggressive leukemia with a median latency of 82 days (Fig. 1E; Supplementary Fig. S3C), with leukocytosis and multiorgan dissemination (Supplementary Fig. S3D and S3E). Leukemia cells expressed the myeloid markers MPO, Mac1, and Gr1, but were negative for CD19, CD3, and Ly76 (TER119; Fig. 1F; Supplementary Fig. S4A–S4C). A subset of tumors was serially transplantable with reduced latency (Fig. 1E). Of the FO tested, only EP300::ZNF384 was able to promote formation of serially transplantable leukemia without concomitant genomic alterations (Supplementary Fig. S4D). Mice transplanted with cells expressing NRASG12D alone developed hematopoietic malignancies but with longer latency and greater lineage variability (mast cell, T-ALL, lymphoma) compared with the ZNF384r/ NRASG12D tumors (Fig. 1E; Supplementary Tables S3 and S4). Thus, expression of ZNF384 FO results in an engraftment disadvantage, but when coexpressed with NRASG12D, drives myeloid leukemia across multiple fusion partners.
Combining ARF Loss with ZNF384 Rearrangements and NRASG12D Reduced the Latency of AML In Vivo
Deletion of CDKN2A/B, encoding the INK4A, INK4B, and ARF tumor suppressors and cell-cycle regulators, is common in ALL and ZNF384r leukemia, and the central role of ARF in the pathogenesis of ALL has been demonstrated in B-ALL models (34, 35). Analysis of ZNF384r leukemia whole-genome sequencing data indicated that the minimal region of deletion was CDKN2A. Accordingly, we transduced and transplanted ARF-null or wild-type HSPCs with TCF3::ZNF384 and/or NRASG12D. ARF-null–derived tumors were of myeloid lineage, but with a shorter latency (median 29 days) than ARF wild-type TCF3::ZNF384/NRASG12D tumors (Supplementary Fig. S5A–S5E; Supplementary Tables S3 and S4). Tumors arising from ARF-null/NRASG12D cells were CD3-positive thymic lymphomas and B-cell lymphomas with a median latency of 42 days (Supplementary Tables S3 and S4). Whole-exome sequencing (WES) of representative tumors from each experiment revealed no secondary lesions that were predicted to contribute to tumorigenesis and no structural variations were identified, suggesting that additional acquired genomic lesions are not required for leukemogenesis and the tumors reported are driven by the lesions modeled (Supplementary Table S5). Thus, in this model, concomitant lesions perturbing tumor suppression and signaling promote ZNF384 FO–mediated leukemogenesis.
In contrast, we observed no differences in immunophenotype, proliferation, cytokine independence, and cell cycling in vitro or survival in vivo between ZNF384 FO (with or without expression NRASG12D) and ZNF384 wild-type or empty vector–transduced ARF-null pre-B cells (Supplementary Fig. S6A–S6F). Thus, although ZNF384 FO may drive a B-lineage leukemia, transformation of HSPCs but not pre-B cells upon directed FO expression supports HSPCs as the cell of origin for ZNF384r leukemia.
ZNF384r Mouse Tumors Exhibit Myeloid Progenitor Cell Signatures
Transcriptome sequencing–based gene expression profiling of these models identified two clusters of samples: ZNF384 FO–expressing tumors and non-FO tumors or untransduced control cells (Fig. 1G). Furthermore, xCell analysis (36), a gene signature–based method that can determine cell-type enrichment, showed that ZNF384 FO tumors have enriched expression of HSC, and less strongly, myeloid signatures, with repression of lymphoid and erythroid signatures (Fig. 1H). Notably, tumors expressing EP300::ZNF384 have a higher enrichment for pro-B cell and MSC signatures and lower myeloid signatures compared with other ZNF384 tumors. Nonfusion (NRASG12D or ARF-null driven) tumors had enriched expression of lymphoid (B-cell or T-cell) signatures. Gene-set enrichment analysis (GSEA) demonstrated similarity of the gene expression profile of mouse ZNF384 FO leukemia to human ZNF384r leukemia, demonstrating fidelity of this model to human disease (Supplementary Fig. S6G). Whole-exome sequencing of mouse tumors did not identify mutations shared between tumors (Fig. 1I).
Expression of Ep300::Znf384-V5-HA in Hematopoietic Cells Drives Hematopoietic Lineage Skewing and Primes For Leukemic Transformation
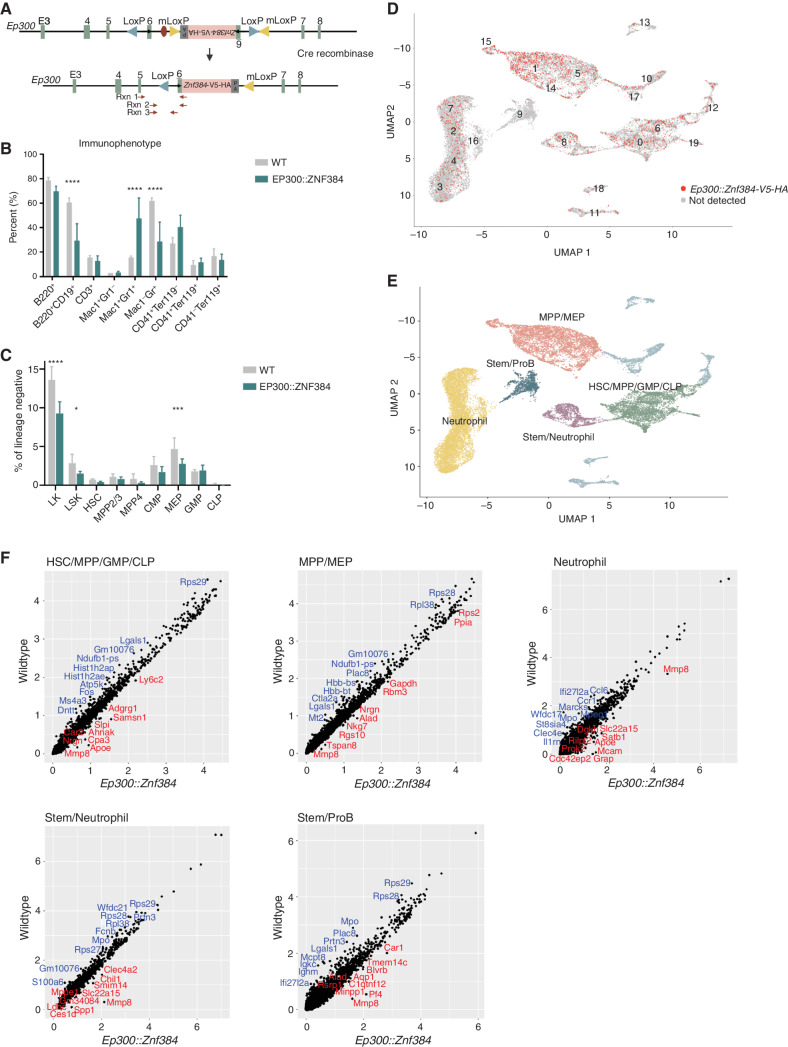
Although lentiviral overexpression of ZNF384 FO in mouse HSPCs resulted in the development of leukemia, a limitation of this method is the lack of FO expression from the endogenous promoter. To examine the effect of FO expression at endogenous levels on normal hematopoietic development and leukemogenesis, we generated a mouse model by knock-in of a floxed, inverted Ep300-exon6::Znf384-V5-HA cDNA minigene into intron 5 of Ep300 (Fig. 2A). In the presence of Cre recombinase, the minigene was flipped into the correct orientation leading to the expression of Ep300::Znf384 fusion gene and disruption of full-length Ep300. We crossed the Ep300Znf384-V5-HA mouse with the Vav-Cre strain to induce recombination in the hematopoietic system, and validated by PCR of peripheral blood (Supplementary Fig. S7A; ref. 37). Notably, the percentage of CD19+ B cells was significantly reduced in the peripheral blood of 2- to 6-month-old mice compared with WT controls (Fig. 2B). In addition, these mice had an increased frequency of Mac1/Gr1–double-positive cells and a reduction of Mac1-negative, Gr1-positive cells. Detailed immunophenotypic characterization of medullary hematopoiesis showed a similar number of granulocyte–monocyte progenitor (GMP; lin−SCA1−CKIT+CD34+CD16/32hi) cells in Ep300::Znf384 and wild-type mice, but reduced frequencies of lineage-negative, cKIT-positive (LK); lineage-negative, SCA1-positive, cKIT-positive (LSK); and megakaryocyte erythroid progenitor (MEP; lin−SCA1−CKIT+CD34−CD16/32−) cells in the FO-expressing bone marrow (Fig. 2C).
Figure 2.
Mice expressing Ep300::Znf384-V5-HA show skewing of hematopoiesis with immature myeloid cell expansion. A, A schematic representation of the reverse-orientation minigene that was knocked into intron 6 of Ep300. Upon Cre-driven recombination, the minigene is flipped and replaces endogenous exon 6. The primers used for PCR validation are shown in red arrows. B, Immunophenotype of the peripheral blood, determined by flow. Columns show means of four replicates ± SD. ****, P < 0.0001; ***, P = 0.0008. C, HSPC compartment characterization by flow analysis of bone marrow cells. Lineage-negative, Kit+ (LK); lineage-negative, Sca1+, Kit+ (LSK); hematopoietic stem cell (HSC); multipotent progenitor 2 and 3 (MPP2/3); multipotent progenitor 4 (MPP4); common myeloid progenitor (CMP); megakaryocyte erythroid progenitor (MEP); granulocyte monocyte progenitor (GMP). ***, P = 0.0008; *, P = 0.0460. D, Uniform Manifold Approximation and Projection (UMAP) dimensionality reduction showing wild-type and EP300::ZNF384 cells, with red indicating detectable Ep300::ZNF384-V5-HA expression. E, UMAP showing that similar clusters were grouped together to make five partitions. F, Gene expression was compared between wild-type and EP300::ZNF384-V5-HA samples in the HSC/MPP/GMP/CLP partition, MPP/MEP partition, neutrophil partition, stem/neutrophil partition or stem/pro B partition.
With aging, Ep300::Znf384 mice, but not control mice, developed colitis and subsequent rectal prolapse due to Helicobacter spp. infection (Supplementary Fig. S7B), suggesting an impaired immune response, which was supported by bone marrow myeloid hyperplasia and reduction of lymphoid cells. Expression of NRASG12D in Ep300::Znf384 bone marrow cells prior to transplantation altered the immunophenotype but did not drive leukemia (Supplementary Fig. S7C). To characterize the effects of FO expression on hematopoietic development, we performed single-cell transcriptome sequencing (scRNA-seq) on 6-week-old Znf384 FO and wild-type bone marrow, fractionated into equal numbers of unselected and lineage-negative cells to enrich for the stem and progenitor populations. Twenty transcriptional clusters were observed in both wild-type and FO-positive samples, with expression of Ep300::Znf384 observed in all clusters except cluster 9, which lacks expression of Ep300 (Fig. 2D; Supplementary Fig. S7D). Using data from scRNAseq (38, 39) and ImmGen (40), we grouped clusters into five partitions: hematopoietic stem cell (HSC)/multipotent progenitor (MPP)/granulocyte macrophage progenitor (GMP)/common lymphoid progenitor (CLP), MPP/megakaryocyte erythroid progenitor (MEP), neutrophil, stem/neutrophil, and stem/pro B (Fig. 2E). Comparison of partitions revealed changes in transcriptional programs associated with FO expression (Supplementary Table S6). In the HSC/MPP/GMP/CLP partition (clusters 0, 6, and 19), Ep300::Znf384 mouse cells had lower expression of CD34 cell (Fos, Romo1, and ribosomal genes) and granulocyte signature genes (Elane, Prtn3, and Ccr2) and increased expression of hematopoietic lineage-specifying transcription factors, such as Gata3 and Sox3 (Fig. 2F; Supplementary Table S7). The MPP/MEP partition (clusters 1, 5, 14, and 15) in FO mice had reduced expression of CD34+ cell and GMP signature genes (Prtn3, Mpo, and Lgals9), and the neutrophil partition (clusters 2, 3, 4, 7, and 16) had increased expression of hematopoietic lineage-specifying transcription factors (Gata2, Klf4, Spi1, Runx1) and decreased expression of dendritic, monocyte, neutrophil, and macrophage signature genes (Lst1, Mpeg1, Ccr1, S100A8, and Cd33). In the stem/neutrophil partition (cluster 8), Ep300::Znf384 mouse cells had lower expression of HSC and GMP signature genes (Mpo, Ctsg, and Prtn3) and higher expression of myeloid lineage signature genes (Nfam1, Dok2, and Lsp1). We also observed an enrichment for decreased expression of MYC target genes and increased expression of ZNF384 target genes in multiple clusters, consistent with our previously reported chromatin immunoprecipitation sequencing (ChIP-seq) data (7). Thus, FO cells in the immature partitions had reduced expression of stem and progenitor signatures and increased expression of myeloid lineage signatures, whereas the mature neutrophil partition has lower mature myeloid signatures and increased immature signatures, indicating that FO expressing cells are transitioning from stem and progenitor cells toward myeloid lineage but have reduced maturation.
Expression of ZNF384 FO in Human HSPCs Drives Expansion of Immature Myeloid Cells and Biphenotypic Disease In Vivo
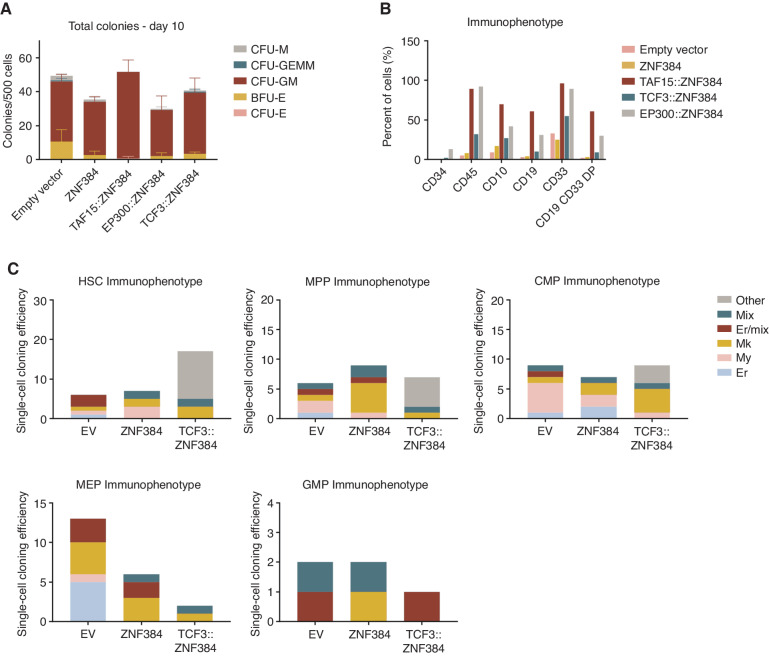
The results of modeling ZNF384 FO in mouse HSPCs show roles in transformation and hematopoietic lineage skewing but did not directly recapitulate lineage ambiguity characteristic of human leukemia. Thus, to examine the effect of ZNF384 FO on human hematopoiesis, we transduced human cord blood CD34+ cells with lentivirus expressing wild-type ZNF384, TAF15::ZNF384, TCF3::ZNF384, EP300::ZNF384, or GFP only (Supplementary Fig. S8A). Upon seeding in methylcellulose differentiation assays, there were no differences in colony number or morphology; however, there were immunophenotypic differences in the cells expressing FO compared with controls (Fig. 3A–C). Cells expressing FO had a biphenotypic expansion of CD19, CD33 double-positive cells with high expression of CD45. The immunophenotype differed slightly based on fusion partner, with TCF3::ZNF384–expressing cells having a smaller population of biphenotypic cells (Fig. 3B; Supplementary Fig. S8B).
Figure 3.
Human CD34 cells expressing ZNF384 FO exhibit immature myeloid cell expansion. A, CFU of CD34 human cord blood cells expressing empty vector, wild-type, or FO. Columns show means of two replicates ± SD. Colonies were scored based on morphology; macrophage (CFU-M), granulocyte, erythrocyte, macrophage, megakaryocyte (CFU-GEMM), granulocyte, macrophage (CFU-GM), burst forming unit-erythroid (BFU-E), or erythroid (CFU-E). B, Cells harvested from CFU were subjected to flow cytometry. FO-expressing cells have an expansion of CD45- and CD33-positive cells compared with controls. FO samples also have CD19, CD33 double-positive cells. C, Cloning efficiency and lineage outcomes of single cells from indicated genotypes. Data in B and C represent cells pooled from replicates for analysis.
To examine the effects of ZNF384 FO more directly on specific human HSPC populations, we utilized the single-cell differentiation assay developed by Notta and colleagues (Supplementary Fig. S8A; ref. 41). Human CD34+ cells were purified into the following stem and progenitor population: HSC (CD34+CD38−CD90+CD45RA−CD49f+), MPP (CD34+CD38−CD90−CD45RA−CD49f−), common myeloid progenitor (CMP; CD34+CD38+CD7/10−FLT3+CD45RA−CD71−), MEP (CD34+CD38+CD7/10−FLT3−CD45RA−CD71−), and GMP (CD34+CD38+CD7/10−FLT3+CD45RA+; Supplementary Fig. S9A), followed by transduction with empty vector, wild-type ZNF384, or TCF3::ZNF384 and single GFP+ cells were dispensed into 96-well plates by FACS to enable a myeloid–erythroid differentiation assay. After two weeks in culture, the immunophenotype of the colonies that formed was assessed to determine the lineage potential of each cell (Supplementary Fig. S9B). On the basis of expression of myeloid and erythroid markers, colonies could be scored as erythroid, myeloid, megakaryoblast, erythroid/megakaryoblast, or a mixture of erythromyeloid cells. HSCs that expressed TCF3::ZNF384 had an increased colony-forming efficiency compared with empty vector and wild-type ZNF384–transduced cells (Fig. 3C). MPP and CMP cells had similar number of colonies between each group, whereas MEP and GMP cells expressing the FO had a reduced colony-forming efficiency. In addition, the majority of colonies that arose from HSC, MPP, or CMP cells expressing TCF3::ZNF384 were not able to be classified into the designated myeloid/erythroid categories and lacked expression of the lineage-associated markers GlyA, CD41, CD11b, or CD15 but were positive for CD45 and CD33 (Supplementary Fig. S9C). This suggests that early stem and progenitor cells are more permissive than later progenitors to transformation by TCF3::ZNF384, and that transformed cells exhibit limited myeloid differentiation.
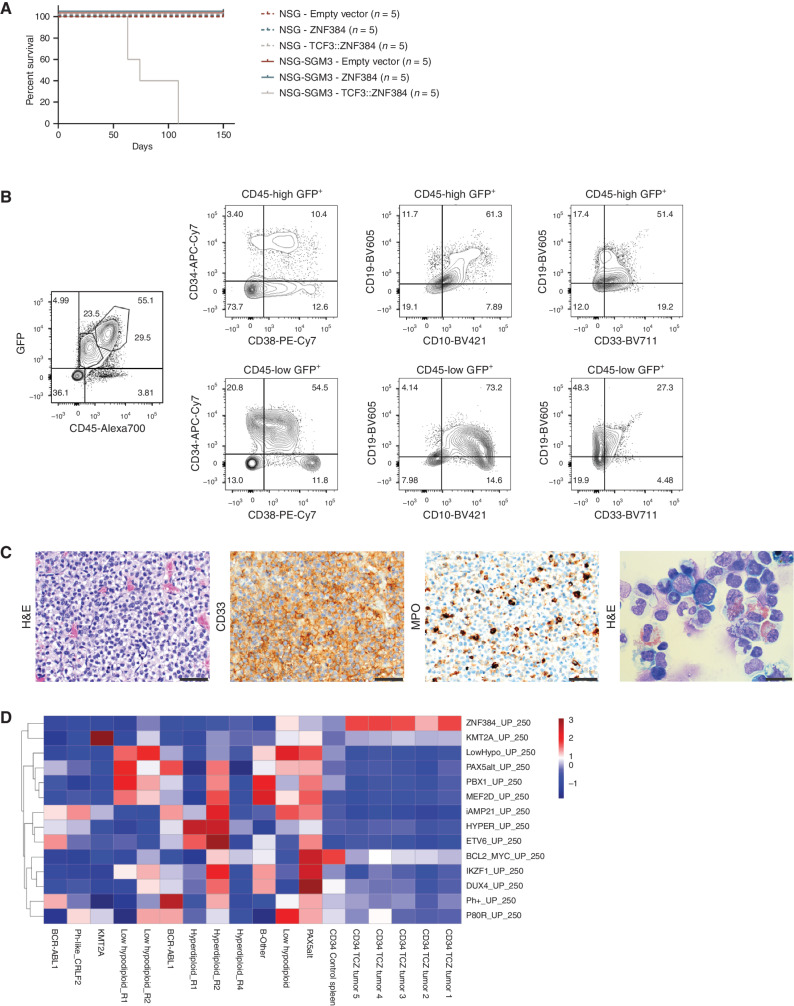
Next, we transplanted CD34+ cells transduced with lentiviral vectors expressing wild-type ZNF384, TAF15::ZNF384, TCF3::ZNF384, EP300::ZNF384, or empty vector into sublethally irradiated NOD-scid Il2Rγnull (NSG) or NSG-SGM3 [NSG expressing human stem cell factor (SCF), granulocyte–macrophage colony-stimulating factor (GM-CSF), and IL3] mice (Supplementary Table S8; ref. 42). NSG-SGM3 mice transplanted with cells expressing TCF3::ZNF384, but not wild-type ZNF384 or the other ZNF384 FO developed lethal disease with a median latency of 74 days (Fig. 4A) due to expansion of hematopoietic cells expressing CD33, CD19, and MPO, consistent with the immunophenotype of human B/myeloid MPAL (Fig. 4B and C; ref. 2). NSG mice transplanted with TCF3::ZNF384–expressing cells showed engraftment that did not develop disease.
Figure 4.
Human CD34 cells expressing TCF3::ZNF384 (TCZ) drive B/myeloid leukemia in vivo. A, Survival curves in primary recipients transplanted with indicated cells. B, Immunophenotyping from a representative mouse showing tumors express CD34, CD38, CD33, and CD19. C, Hematoxylin and eosin (H&E) staining (left), CD33 staining (middle), MPO staining (right) of sternal section and cytospin of bone marrow cells. Scale bars represent 50 µm for all except for bone marrow cytospin (scale bar, 20 µm). D, Single-sample GSEA displaying relative enrichment score (Z-score) for B-ALL subtype-specific gene lists. Experimental tumors are enriched for ZNF384r gene set. Patients 1–12 (non-ZNF384r) were included for heterogeneity, which is required for analysis. Adjacent samples labeled R1, R2, or R4 are relapsed samples from the same patient. TCZ, TCF3::ZNF384.
RNA was isolated from sorted GFP+ spleen cells of 5 leukemic mice and one empty vector–infected mouse. RNA-sequencing and GSEA comparison of engrafted tumors and representative primary human leukemias showed enrichment of the primary human ZNF384r leukemia transcriptomic signature in the human CD34 TCF3::ZNF384 leukemia expression profiles (Fig. 4D). Thus, in human HSPCs, expression of TCF3::ZNF384 is sufficient to generate B/myeloid hematopoietic disease that recapitulates human ZNF384r leukemia.
Deregulation of Hematopoietic Gene Expression Programs by ZNF384 FO
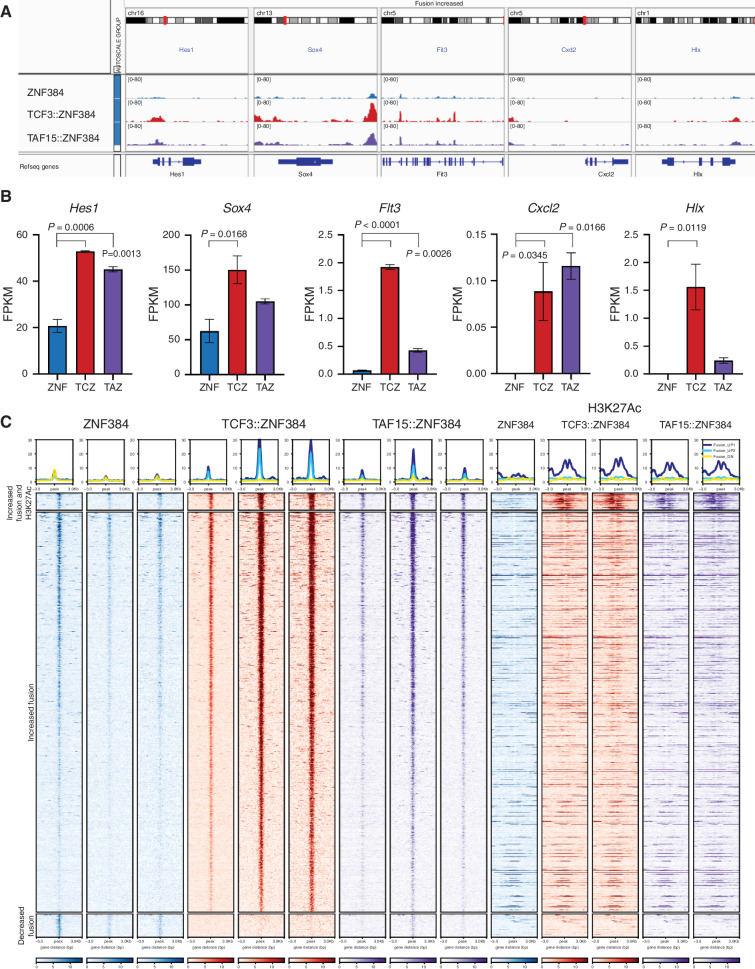
To further examine the mechanistic basis of hematopoietic cell transformation, we utilized integrated analysis of gene expression, chromatin occupancy, and protein interactions in mouse pre-B cells expressing ZNF384 FO. We previously showed that TCF3::ZNF384-HA and TAF15::ZNF384-HA had increased binding at 2,298 genomic loci and decreased binding at 495 loci compared with wild-type ZNF384 (7). We analyzed the 2,298 regions for motif enrichment that identified ZNF384 and RUNX1/2/3–binding motifs, AAAAAA and AACCAC, respectively (Supplementary Fig. S10A). GREAT analysis was performed to predict the cis-regulatory function of these regions and showed an enrichment for genes in pathways related to hematopoiesis, proliferation, and signaling (Supplementary Fig. S10B; Supplementary Tables S9 and S10; ref. 43). One key pathway is stem cell maintenance; we confirmed differential binding and observed a subset of these genes had increased gene expression (Fig. 5A and B). Deregulation of FLT3 is a hallmark of ZNF384r leukemia and has been implicated as a therapeutic target (Supplementary Fig. S10C; refs. 7, 44). Our data revealed increased binding of the FO and increased expression of Flt3 in ZNF384 FO–expressing cells, suggesting that deregulation is due to binding by ZNF384 FO (Fig. 5A and B). We also observed differential binding at and gene activation by the FO of genes that are normally highly expressed at specific hematopoietic cell types but not pre-B cells, such as Cxcl2 that is normally expressed in granulocytes (Fig. 5A and B). Hlx, which encodes a homeobox transcription factor that is highly expressed in AML, had the highest fold-change increase of expression in the FO-expressing cells compared with wild-type cells (Fig. 5A and B; ref. 45). Accordingly, many of these genes have elevated expression in ZNF384r leukemia (Supplementary Fig. S10C; ref. 10).
Figure 5.
ZNF384 FO binding to chromatin is associated with H3K27 acetylation and increased expression of hematopoietic regulators. A, Representative genomic regions with increased binding of the FO compared with wild-type. B, Representative expression in fragments per kilobase million (FPKM) of genes with differential binding. Columns show means of two replicates ± SD. ZNF, wild-type ZNF384; TCZ, TCF3::ZNF384; TAZ, TAF15::ZNF384. C, Heat map showing the ChIP-seq signal, centered on ZNF384 peaks, of wild-type ZNF384 compared with TCF3::ZNF384 and TAF15::ZNF384; and H3K27Ac signal, centered on ZNF384 peaks (top). Peaks with increased binding of FO compared with wild-type proteins separated by regions with increased H3K27Ac or regions with same H3K27Ac (middle). Peaks with decreased binding of the FO compared with wild-type proteins (bottom).
We next performed ChIP-seq of the histone marks H3K27Ac, H3K18Ac, and H3K4me1 to investigate whether expression of ZNF384 FO alters the chromatin landscape. We found no global differences in H3K4me1, H3K18Ac, or H3K27Ac. However, we identified a subset of 47 regions that had increased binding of the FO and increased H3K27Ac (Fig. 5C). In contrast to most ZNF384 FO–binding sites that are also bound by wild-type ZNF384, but less avidly, these 47 regions were only bound by ZNF384 FO as well as being decorated by H3K27Ac, were often intergenic and intronic, and have variable effect on nearby gene expression. We extended these analyses by performing H3K27Ac HiChIP to determine whether changes in H3K27Ac were accompanied by changes in chromatin topology but did not identify any differences in looping of H3K27Ac-marked regions (Supplementary Fig. S10D), suggesting that these changes were directly mediated by ZNF384 FO binding without altering genome organization.
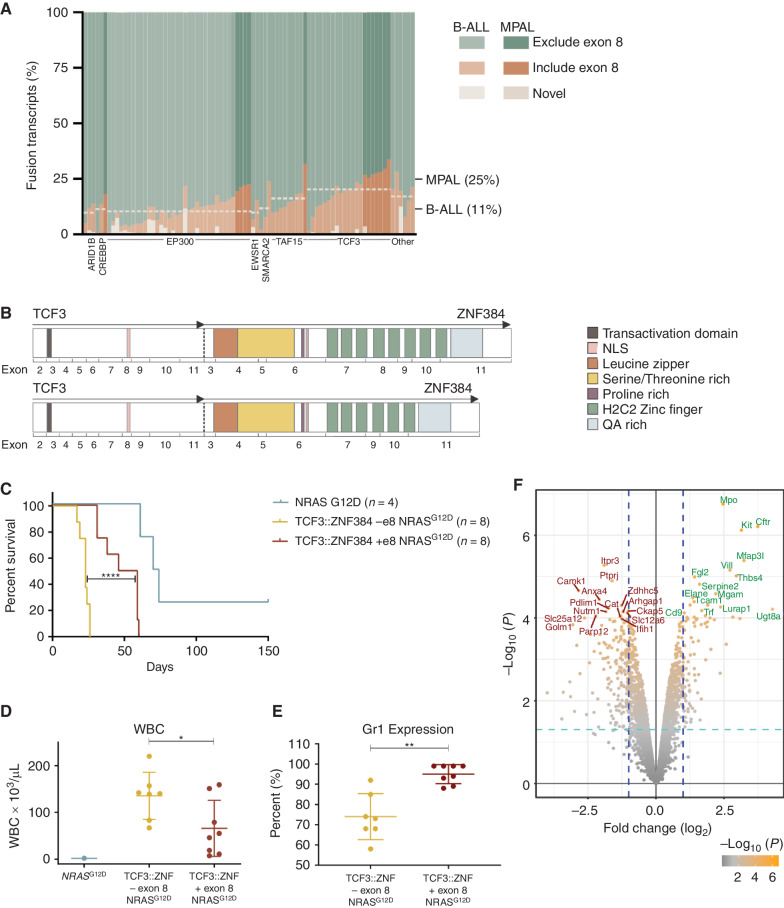
ZNF384 Exon 8 Utilization Influences Chromatin Occupancy and Gene Deregulation
Alternate splicing results in ZNF384 FO isoforms that retain or exclude ZNF384 exon 8, with isoforms lacking exon 8 comprising up to 90% of transcripts in human leukemia. The highest rates of inclusion were observed in TCF3::ZNF384 leukemia and MPAL cases (Fig. 6A), with no significant difference in isoform usage between immunophenotypic subpopulations in xenografts or the EP300::ZNF384 cell line JIH-5 (Supplementary Fig. S11A and S11B). Exclusion of exon 8 results in in-frame splicing with loss of zinc fingers 4 and 5 (Fig. 6B). To examine the role of exon 8 in leukemogenesis, we transduced and transplanted mouse HSPCs with NRASG12D and TCF3::ZNF384 with exon 8 or without exon 8. Retention of exon 8 was associated with longer leukemia latency, lower blood leukocyte count, and higher Gr1 expression, indicating a more immature phenotype, compared with tumors without exon 8 (Fig. 6C–E). RNA-seq–based gene expression profiling demonstrated downregulation of immune response genes and upregulation of cell-cycle arrest and antiproliferative pathways in TCF3::ZNF384 with exon 8 tumors (Fig. 6F). One of the top upregulated genes in tumors with exon 8 was Kit, which is a marker of stem cells and an important receptor for cytokine signaling. The most significantly upregulated gene was Mpo, the marker used to diagnose MPAL (Fig. 6F).
Figure 6.
The effect of TCF3::ZNF384 alternative splicing isoforms. A, Relative abundance of ZNF384 splice isoforms in ZNF384r B-ALL and MPAL. Each column represents a case and shows the cumulative abundance of all isoforms as a proportion within each case. The mean inclusion of exon 8 based on N-terminal partner is marked by the horizontal line across cases including that partner. TCF3::ZNF384 fusions have a higher mean exon 8 inclusion rate. Light cases were diagnosed as B-ALL and darker cases as MPAL, with the mean exon 8 inclusion for each diagnosis marked on the left. MPAL cases have a higher average exon 8 inclusion rate. B, A schematic representation of TCF3::ZNF384 mRNA splicing isoforms that differ in exon 8 inclusion. C, Survival curves in primary recipients transplanted with indicated cells. Two-sided log-rank Mantel–Cox test; ****, P < 0.0001. D, White blood cell (WBC) count at death in mice transplanted with indicated cells. The mean expression is shown by the horizontal line in the scatter dot plot and the error bars represent the SD. *, P = 0.0315. E, Percent of cells expressing Gr1 as determined by flow analysis. The mean expression is shown by the horizontal line in the scatter dot plot and the error bars represent the SD; **, P = 0.0042. F, A volcano plot displaying −log10P value by log2 fold change to compare gene expression of tumors driven by TCF3::ZNF384 with exon 8 to TCF3::ZNF384 without exon 8.
ChIP-seq for TCF3::ZNF384 showed that the isoform retaining exon 8 bound to 1,128 more sites than the isoform lacking exon 8 (Supplementary Fig. S12A). The regions with increased binding by the exon 8 retaining isoform were often flanked by regions of decreased binding of full-length isoform compared with the isoform lacking exon 8 (Supplementary Fig. S12B), with enrichment for genes involved in RNA stability or lymphocyte differentiation (Supplementary Fig. S12C).
GREAT analysis also revealed that genes exhibiting increased binding by the TCF3::ZNF384 isoform retaining exon 8 are critical for normal myelopoiesis, as evidenced by prior data showing inactivation of these genes is associated with impaired neutrophil or granulocyte differentiation in knockout mouse models (e.g., Crebbp, Ikzf3, Jak3, Zfp36l2). Thus, differential isoform usage, which is associated with variation in the complement of C2H2 zinc fingers, is associated with human leukemia phenotype, variation in the latency of experimental leukemia, and mechanistically altered chromatin occupancy and hematopoietic gene expression.
The Protein Interactome of ZNF384 FO
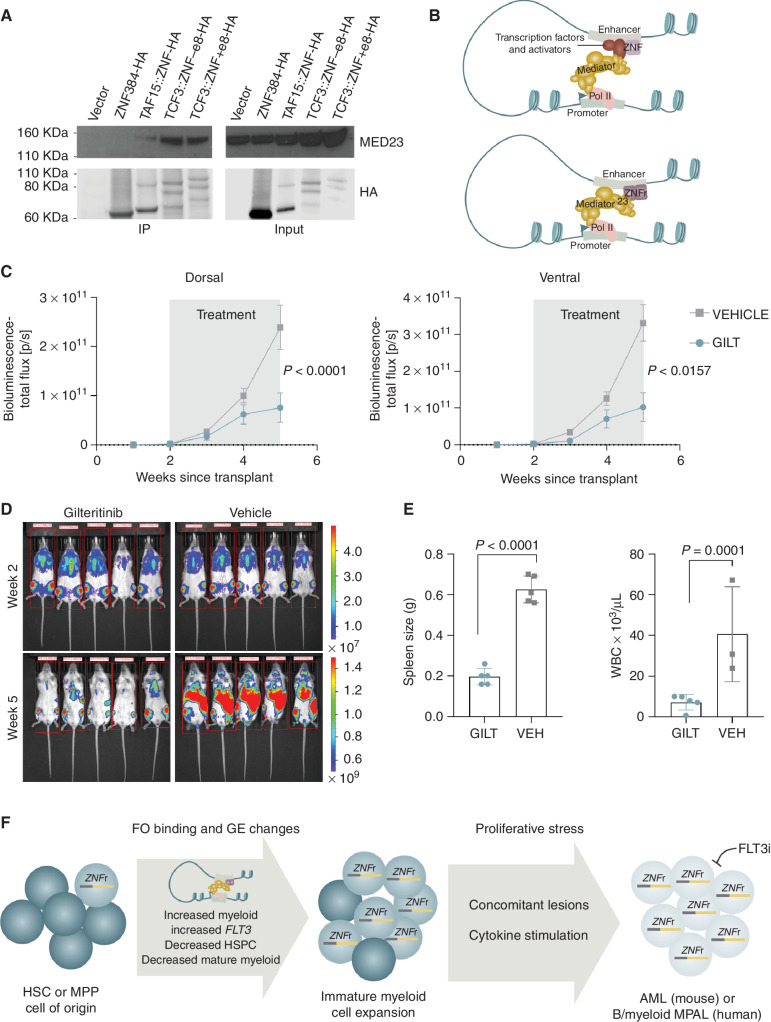
To examine the role of altered ZNF384 protein–protein interactions in leukemogenesis, we compared the protein interactome of wild-type ZNF384, TCF3::ZNF384, and TAF15::ZNF384 by anti-HA pull down of HA-tagged ZNF384 or FO expressed in HEK293T cells and rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME). RIME utilizes crosslinking and chromatin shearing prior to immunoprecipitation to identify protein interactions that occur on chromatin, either directly with the wild-type or fusion ZNF384 protein, or indirectly as part of a larger complex. The majority of proteins identified interacted in a complex with both wild-type and FO, although the abundance was often lower in wild-type immunoprecipitations (Supplementary Table S11). However, ZNF384 FO, but not wild-type ZNF384, directly or indirectly interacted with 74 proteins, many of which are involved in the preinitiation complex (PIC) and include the mediator and cohesin complexes, chromatin-modifying enzymes including ARID1A and KMT2A, RNA polymerase subunits, E3-ubiquitin ligases, and transcription factors including TRPS1 and FOXP1 (Supplementary Table S12). One of the top unique identified interactors with a ZNF384 FO protein complex that was confirmed by coimmunoprecipitation was MED23 (Fig. 7A), which connects the mediator complex to sequence-specific transcription factors and has been shown to play an important role in hematopoiesis and AML (46–48). By directly interacting with MED23, ZNF384 FO can engage with the mediator complex without the need of other transcription factors or activators, which leads to the observed increased chromatin occupancy and transcriptional activation at known ZNF384-binding sites (Fig. 7B).
Figure 7.
Proposed model of ZNF384 FO mechanism and in vivo sensitivity to gilteritinib. A, Co-immunoprecipitation assay using 293T cells and HA antibody. Pull-down lysates were blotted with anti-MED23 or anti-HA to show that ZNF384 FOs, but not ZNF384, interact with MED23. White bars between images delineate samples run on separate gels. B, Proposed model showing that ZNF384 binds to enhancer regions and through interactions with transcription factors and activators can interact with the Mediator complex to initiate transcription (top). In contrast, ZNF384 FO directly interact with MED23 of the Mediator complex, which bypasses the need to interact with additional transcription factors and activators to initiate transcription (bottom). C, MPAL patient-derived xenograft expressing TCF3::ZNF384 were randomized to receive 20 mg/kg of gilteritinib (GILT) or vehicle (VEH) once daily from week 2 to week 5. Tumor burden was monitored by bioluminescent imaging. D, Ventral bioluminescent imaging of indicated treatment groups at beginning (top) and end (bottom) of treatment. E, Spleen weight at time of death (left) or white blood cell (WBC; right) count at time of death in mice with indicated treatment. The mean expression is shown by the bar and the error bars represent the SD. F, Graphical summary of the results from this study. ZNF384 rearrangements arise in an early HSC or multi-potent progenitor (MPP). Through increased binding of ZNF384 FO at wild-type target genes (related to normal hematopoietic development), gene expression (GE) is deregulated, likely through a stabilized Mediator complex. Changes in GE programs leads to an expansion of immature myeloid cells that are primed for leukemic transformation in the presence of proliferative stress. In addition, ZNF384r leukemia shows in vivo sensitivity to FLT3 inhibition.
In Vivo Sensitivity to FLT3-Targeted Therapy
Because of the high expression of nonmutated FLT3 in ZNF384r patients (7, 44), evidence that expression of ZNF384 FO leads to increased Flt3 expression and that FO directly bind Flt3 in vitro, we examined the therapeutic potential of FLT3 inhibition by assessing the in vivo sensitivity of a luciferase-marked patient-derived xenograft (PDX) model of TCF3::ZNF384 to the FLT3 inhibitor gilteritinib (49). FLT3 was expressed in this model at RNA and protein level. Engrafted NSG-SGM3 mice were randomized to gilteritinib (20 mg/kg/day) or vehicle and commenced therapy at 2 × 109 bioluminescence flux (p/s). Vehicle-treated mice were moribund at 5 weeks posttransplant, whereas gilteritinib-treated mice had a reduced tumor burden (Fig. 7C and D), with a marked reduction in spleen weight and leukocytosis at study completion (Fig. 7E). This striking reduction of tumor burden without toxicity from a single agent suggests FLT3-targeted therapy may be an effective approach for treating ZNF384r leukemia.
DISCUSSION
Genomic analyses of large cohorts of B-ALL and MPAL cases have defined a subtype of leukemia that is driven by rearrangements of ZNF384 and characterized by striking lineage ambiguity in which tumor cells expressing both B-cell and myeloid markers, often with multiple immunophenotypically distinct but genetically identical subclones in individual cases (7, 11, 12, 18). Although identification of ZNF384 rearrangements in HSPCs isolated from primary human leukemia samples and plasticity of leukemia cell immunophenotype in xenografts suggested that these tumors arise in an early stem or progenitor cell, the mechanistic basis of ZNF384 FO in driving lineage ambiguous leukemia, and importance of cell of origin had not been directly examined (7). By utilizing multiple complementary mouse and human experimental models, and downstream genomic, epigenomic, and proteomic findings, we provide several important conceptual insights into the role of ZNF384 FO in leukemogenesis (Fig. 7F).
Here we have shown that expression of ZNF384 FO promotes expansion of immature myeloid cells in vitro and leukemia development in vivo. Interestingly, we showed differences in transformation characteristics dependent on cell-of-origin, fusion partner, and splicing isoform usage. In mouse models, HSPCs, but not pre-B cells, were permissive to transformation by the FO and in human models, early stem and progenitors were transformed by FO expression but more mature progenitors were not affected, supporting an early HSPC as the cell of origin. In addition, leukemias that arose in mouse cells were of myeloid lineage and required a second hit (e.g., expression of NRASG12D); however, human cell derived leukemias arose from FO expression alone and recapitulated human B/myeloid MPAL. Fusion partners also influenced leukemia penetrance and phenotype, with EP300::ZNF384 mouse tumors expressing a more lymphoid signature compared with the other FO-driven tumors. In CD34 transplants, only TCF3::ZNF384 expression was able to promote leukemogenesis. Finally, comparison of TCF3::ZNF384 splicing isoforms revealed that the isoform lacking exon 8 drove a more aggressive leukemia compared with the full-length isoform.
The mouse models developed here demonstrated the transforming role of ZNF384 FO; however, concomitant genomic alterations were required for leukemogenesis. Although ZNF384 FO alone did not drive leukemia upon expression in mouse HSPCs, the expression of these FO restricted the lineage of leukemia to myeloid phenotype. In addition, we observed distinct differences in transformation between HSPCs and pre-B cells, which are otherwise permissive to other types of B-ALL, supporting the notion that the cell of origin is a hematopoietic stem or progenitor. The conditional Ep300::Znf384 knock-in model enabled expression of Ep300::Znf384 in hematopoietic cells at an endogenous level and allowed investigation of the early stages of transformation in vivo that demonstrated lineage skewing. Collectively, these experiments model the early gene expression and hematopoietic differentiation changes that occur when ZNF384 FO are expressed in HSPCs, which primes cells for leukemogenesis upon acquisition of additional mutations.
Modeling in human CD34+-derived HSPCs enabled several observations that provided further mechanistic insight, notably expansion of immature myeloid cells and a restriction of cell types permissive to transformation by ZNF384 FO. In addition, this model provided insights that were unique to the human system. We demonstrated that early HSPC populations were more permissive to transformation compared with mature progenitors, and that ZNF384 FO expression promoted expansion of a biphenotypic population. In vitro, we showed that FO expression promotes a B/myeloid immunophenotype, and specifically that HSCs and MPPs, were susceptible to transformation, demonstrating the direct role of ZNF384 FO in driving lineage ambiguous leukemia. Most importantly, we directly demonstrated the capability of ZNF384 FO to drive development of a bona fide B/myeloid MPAL that recapitulated several key features of the human disease.
Mechanistic studies revealed that ZNF384 FO bind to canonical ZNF384 sites with higher avidity and a subset of these sites are patterned with H3K27 acetylation, suggestive of aberrant enhancer activation. Integrated transcriptomic and epigenomic profiling provided insights into how the FO drives lineage ambiguity. Rather than a massive shift in the genomic sites of chromatin occupancy from wild-type to FO, most sites bound by FO showed increased occupancy compared with wild-type ZNF384, consistent with the observation that a diverse set of truncated N-terminal partners typically fuse with full-length ZNF384. However, the FO did exhibit binding of a subset of new sites with evidence of enhancer formation. Moreover, genes with increased binding of FO and increased expression compared with wild-type were enriched for hematopoietic differentiation gene sets attesting to the direct role of ZNF384 FO in driving lineage aberrancy. Further mechanistic insight into deregulating transcription was provided by chromatin-based proteomic studies that implicate acquisition of FO-specific interactions with multiple components of the core enhancer and transcriptional machinery. FO interacts directly with MED23 of the Mediator complex, eliminating the need of additional cofactors to initiate interaction with Mediator, which would lead to a more stable complex that results in increased chromatin occupancy of ZNF384 FO and increased transcription of target genes.
Although the ZNF384 FO defines a discrete subset of leukemia, there is nuance with some preferential associations of fusion partner and ZNF384 isoform usage according to leukemia lineage. Patients preferentially express the ZNF384 fusion isoform lacking exon 8, with a mean of 14% of reads including exon 8. However, when stratified by fusion partner or leukemia lineage, we observed an enrichment of exon 8 inclusion in patients with TCF3::ZNF384 fusions (20% of reads) or in patients diagnosed as MPAL (25% of reads). Experimentally, we showed that the FO isoform with exon 8 drove leukemia with a more immature phenotype and increased expression of Mpo compared with the isoform lacking exon 8, which supports the increased diagnosis of MPAL in patients expressing the isoform including exon 8. Our experiments highlight this variable facility of the different FO in inducing transformation, and differences in gene expression and chromatin occupancy associated with the FO.
Overexpression of nonmutated FLT3 has been described in ZNF384r leukemias and one patient has been successfully treated with a FLT3 inhibitor, sunitinib (7, 44). Here we have shown that FO binds directly to Flt3 and that expression of FO results in overexpression of Flt3, providing the first evidence into the mechanism of elevated FLT3 expression in ZNF384r leukemia. Furthermore, inhibition of FLT3 using gilteritinib in a PDX model of TCF3::ZNF384–driven leukemia led to a remarkable reduction of disease burden, implicating FLT3 as a promising target in ZNF384r leukemia.
These results show the critical importance of cooperation between the cell of origin, specific FO, secondary hits, pressure to expand, and recipient microenvironment in driving a lineage ambiguous leukemia.
METHODS
Patients and Cell Lines
Human Samples.
Genomic and xenografting studies using human leukemia or normal samples were approved by the St Jude Children's Hospital Institutional Review Board. The studies were conducted in accordance with the Declaration of Helsinki, and all subjects and/or their guardians provided written informed consent and/or assent to the use of samples for research. Genomic data were combined from previously published studies (7, 8, 10, 11, 30–33) that included children with diagnosis and/or relapsed ALL and available leukemia samples treated on St Jude, Children's Oncology Group or adult cooperative group or center (ECOG-ACRIN, MD Anderson Cancer Center and the Alliance – Cancer and Leukemia Group B) studies (71 samples); and MPAL samples collected as part of an international cooperative effort (15 samples). Samples studied included diagnosis and relapse leukemia and remission bone marrow or peripheral blood samples. Samples were purified by density-gradient centrifugation and cryopreserved until thawing and extraction of nucleic acids and/or xenografting. Deidentified human cord blood samples were obtained from Duke University - Carolinas Cord Blood Bank (Durham, NC) with written informed consent obtained from the donors. Cord blood from multiple donors was pooled and processed 24 to 48 hours postdelivery. Mononuclear cells were enriched using Accu-Prep (Accurate Chemical) and SepMate-50 tubes (STEMCELL Technologies). CD34-positive selection was performed using CD34 Microbead kit UltraPure (Miltenyi Biotec).
Cell Lines.
Mouse ARF-null pre-B-cell cultures were established from bone marrow for each experiment as described (35, 50). Briefly, whole bone marrow was isolated from 8-week-old, female, ARF-null C57BL/6 mice and seeded on irradiated T220–29 stromal cells in RPMI 1640 supplemented with 5% FCS, penicillin, streptomycin, l-glutamine; IL7 (10 ng/mL; PeproTech); and 55 µmol/L β-mercaptoethanol (Gibco). Media was changed every 3 days and pre-B cells were removed from stromal cells on day 7, and verified as Mycoplasma spp. negative using MycoALERT (Lonza). Upon establishment of stable ARF-null pre-B cultures, cells, and viral supernatants (pCL20c MSCV-ires-GFP lentivirus expressing the wild-type or fusion genes of interest or pCL20-NRAS G12D-ires-RFP) were added to RetroNectin-coated plates (Takara) and centrifuged at 1,500 × g for 90 minutes at room temperature. Cells were maintained at 37°C for 48 hours and transduced GFP-positive or GFP/RFP double-positive cells were obtained via FACS. All ARF-null pre-B-cell cultures were grown at 37°C with 8% CO2 and maintained in RPMI-1640 supplemented with 10% FCS, penicillin, streptomycin, l-glutamine; IL7 (10 ng/mL; PeproTech); and 55 µmol/L β-mercaptoethanol (Gibco).
HEK293T cells (ATCC) and NIH3T3 cells (ATCC) were grown at 37°C with 5% CO2 and maintained in DMEM supplemented with 10% FCS, penicillin, streptomycin, and glutamine.
Cloning
Lentiviral Cloning.
RNA isolated from patient samples was used to make cDNA with the Superscript III First-Strand Synthesis System (Invitrogen). ZNF384 (with or without exon 8), EP300::ZNF384 (with the breakpoint of EP300 exon 6 fused in frame to exon 3 of ZNF384), TAF15::ZNF384 (exon 6 – exon 3), TCF3::ZNF384 (exon 13 – exon 5; with or without exon 8), and CREBBP::ZNF384 (exon 6 – exon 3) breakpoints were validated and full-length transcripts were amplified by PCR using Phusion High-Fidelity DNA Polymerase (New England BioLabs) and the primers listed in the Supplementary Table S13. Amplified products were purified using the Wizard SV Gel and PCR Clean-up system (Promega) and validated by Sanger sequencing. Full-length transcripts were cloned into the pCL20c MSCV-ires-GFP lentiviral vector. RNA encoding NRASG12D was amplified, purified, and cloned into pCL20c-MSCV-ires-mRFP. pCL20 MSCV-ires-cre plasmid (a gift from C. Sherr) was digested with EcoRI/NotI to remove Cre and GFP or mRFP were ligated into the plasmid. An in-frame HA (hemagglutinin) epitope tag was added directly after the start codon (EP300::ZNF384, TAF15::ZNF384) or directly before the stop codon (ZNF384, TCF3::ZNF384) by mutagenesis using the QuikChange II XL or NEB Q5 Site-Directed Mutagenesis kit (Agilent). All constructs were verified by restriction enzyme digestion and Sanger sequencing.
Lentivirus Production.
HEK293T cells were cultured in DMEM supplemented with 10% FBS; 1× penicillin, streptomycin, and l-glutamine (Gibco). Lentivirus was produced by transfecting HEK293T cells with pCAG RTR, pCAG HIV, and the envelope pHDM-VSV-G (a gift from the laboratory of J. Dick), to infect mouse lineage-negative or human CD34+ cells) or CAG4-Eco (St. Jude's Vector Core Laboratory, to infect ARF-null mouse pre-B cells) using Fugene HD (Promega) according to manufacturer's recommendations. Cell culture medium was changed 18 hours posttransfection. Ecotropic virus production was in DMEM, 10% FBS, and HDMG virus production was in Iscove modified Dulbecco medium (IMDM), 10% FBS. Viral supernatant was collected and filtered 42 hours and 66 hours after transfection. Lentivirus with the pHDM-VSV-G envelope was concentrated 100-fold by ultracentrifugation at 100,000 × g for 90 minutes. Virus was titered by transduction of NIH3T3 cells followed by flow cytometry to measure the proportion of GFP+ or mRFP+ cells and used fresh for transduction of hematopoietic cells.
Experimental Mouse Modeling
Animals.
Mice were housed in the American Association for Accreditation of Laboratory Animal Care–accredited facility of St Jude Children's Research Hospital. All experiments were approved and in compliance with the SJCRH Institutional Animal Care and Use Committee (IACUC)-approved protocol in accordance with NIH guidelines.
Mouse Colony-forming Unit Assays.
Bone marrow cells were harvested from 6–8 week old wild-type C57BL/6 mice (Jackson Laboratory), and lineage-negative HSPCs were isolated using the EasySep Mouse Hematopoietic Progenitor Cell Isolation Kit (StemCell Technologies) according to the manufacturer's recommendations. HSPCs were cultured for 48 hours in IMDM with 20% FBS supplemented with recombinant mouse stem cell factor (SCF; 50 ng/mL), FLT-3 ligand (40 ng/mL), IL6 (30 ng/mL), IL3 (20 ng/mL), and IL7 (10 ng/mL; PeproTech). Cells and viral supernatants (pCL20c MSCV-ires-GFP lentivirus expressing the wild-type or fusion genes of interest) were added to RetroNectin-coated plates (Takara) and centrifuged at 1,500 × g for 90 minutes at room temperature. Cells were maintained at 37°C for 48 hours and transduced GFP-positive cells were obtained via FACS. For clonogenic assays, 10,000 cells were plated in triplicate in Methocult M3231 (StemCell Technologies) with the appropriate factors (SCF, 100 ng/mL; FLT-3 ligand, 10 ng/mL; IL7, 20 ng/mL) for mouse lymphoid progenitor cells or in M3534 (StemCell Technologies) with the addition of GM-CSF (10 ng/mL) for mouse myeloid progenitor cells. Colonies were scored 7–10 days later. For replating, 1 × 104 cells were cultured in identical conditions, with colonies counted between days 7 to 10. Colony identity was confirmed via morphologic analysis of Wright—Giemsa–stained cytospin preparations and by flow cytometric analysis of a panel of lineage markers: B220-Alexa700 (eBioscience, 56–0452–82, RA3–6B2, 1:100); Gr1-BV711 (BioLegend, 108443, RB6–8C5, 1:100); TER119-APC-Cy7 (BD Biosciences, 560509, TERR-119, 1:200); CD3-APC (BD Biosciences, 553066, 145–2C11, 1:100); Mac1-BV605 (BioLegend, 101237, M1/70, 1:50); CD41-BUV396 (BD Biosciences, 564056, MWReg30, 1:100); CD34-BV421 (BD Biosciences, 562608, RAM34, 1:100); and CD19-PE-Cy7 (BD Biosciences, 552854, 1D3, 1:100).
Donor cells were obtained for transplant experiments from female 6- to 8-week-old C57BL/6 mice (Jackson Laboratory) or ARF-null C57BL/6 mice using pretreatment with 5-fluorouracil at 150 mg/kg by intraperitoneal injections and bone marrow cells were harvested 5 days later (51). Cells were treated with RBC Lysis Buffer (BioLegend) according to the manufacturer's recommendations. Cells were maintained in IMDM with 20% FBS supplemented with recombinant mouse SCF (50 ng/mL), FLT-3 ligand (50 ng/mL), IL3 (10 ng/mL), IL6 (10 ng/mL), IL7 (10 ng/mL; PeproTech). Cells were added to non-tissue culture–treated plates that were prepared with RetroNectin (Takara). Viral supernatants (pCL20c MSCV-ires-GFP lentivirus expressing the wild-type or fusion genes of interest or pCL20-NRAS G12D-ires-RFP) were added to the wells and the plates were centrifuged at 1,500 × g for 90 minutes at room temperature. Cells were collected 24 hours later and 2 × 105 cells were transplanted by tail vein injection into lethally irradiated (1,100 Rad) female 8- to 12-week-old C57BL/6 mice (Jackson Laboratory). A portion of cells were retained in culture and the proportion of GFP-positive or GFP+RFP+ cells was determined by flow cytometry 48 to 72 hours posttransduction. Animals that became moribund were euthanized and blood, bone marrow, and spleen were analyzed for evidence of leukemia, using morphology, flow cytometry, and histopathologic analysis. For secondary transplantation, sub-lethally irradiated (550 Rad) 8- to 12-week-old C57BL/6 mice were injected with 5 × 105 FACS, GFP-positive or GFP, RFP double-positive bone marrow cells from the primary leukemic mice. The same monitoring, euthanasia, and tissue analysis was used for secondary transplant recipients.
For pre-B-cell transplantation, 5 × 105 cells were transplanted by tail vein injection into sub-lethally irradiated (550 Rad) female 8- to 12-week-old C57BL/6 mice (Jackson Laboratory) and monitored for disease development. Animals that became moribund were euthanized and blood, bone marrow, and spleen were analyzed for evidence of leukemia, using morphology, flow cytometry, and histopathologic analysis.
Ep300::Znf384 Conditional Mouse Model.
The Ep300::Znf384 conditional mouse model was created by Biocytogen. Homologous regions 4.1 kb upstream of Ep300 exon 6 and 5.1 kb downstream of exon 6 were subcloned from a BAC clone from C57BL/6J mouse genomic BAC library. A LoxP site was inserted upstream of exon 6. Downstream of exon 6 the following sequences were inserted: an FRT-flanked Neo-resistance positive-selection cassette; a mLoxp sequence; a reverse-orientation cDNA minigene containing Ep300 exon 6, Znf384-V5-HA, and polyA tail; a LoxP sequence; and a mLoxp sequence. After linearization, the targeting vector was transfected into C57BL/6 embryonic stem cells by electroporation. ES cells were selected by G418 treatment and screened by PCR and Southern blotting. Seven positive clones were identified by Southern blotting with a 5′ probe, a 3′ probe, and a 5′ Neo probe. Ten positive clones were injected into Balb/c blastocysts and implanted into pseudopregnant females. Chimeric mice were crossed with C57BL/6 mice to obtain F1 mice carrying the recombinant allele containing the floxed Ep300::Znf384-V5-HA allele and the neomycin selection cassette. Mice were bred with Flipase-expressing mice to remove the neomycin selection cassette. Genotype was determined by extracting DNA from tail snip and performing PCR using KAPA2G Fast Genotyping Mix (Roche) and primers XWY LoxP-F/ XWY LoxP-R (Supplementary Table S13). To generate conditional Ep300::Znf384-V5-HA fusion mice, mice homozygous for the floxed alleles of Ep300::Znf384-V5-HA were crossed with Vav-Cre mice (37). Animals that became moribund were euthanized, and blood, bone marrow, and spleen were analyzed using morphology, flow cytometry, and histopathologic analysis.
To confirm recombination, DNA was extracted from peripheral blood using QIAamp DNA Blood Mini Kit (Qiagen) and PCR was performed using Phusion High-Fidelity DNA polymerase (New England BioLabs) and the following primer pairs: EZ_Fus_F1/EZ_Fus_R1 (Rxn 1), XWY LoxP-F/EZ_Fus_R1 (Rxn 2), and XWY LoxP-F/XWY LoxP-R (Rxn 3).
Mouse Flow Cytometry Analysis.
Postmortem immunophenotyping of bone marrow and spleen cells from transplanted mice was performed on the GFP-positive or GFP, RFP-double positive cells to determine the lineage of disease in engrafted samples using the following panel of antibodies: B220-BV605 (BioLegend, 103244, clone RA3–6B2, 1:20); Gr1-PerCP-Cy5.5 (BD Biosciences, 552093, clone RB6–8C5, 1:50); TER119-V500 (BD Biosciences, 562120, clone TER-119, 1:50); CD3-APC (BD Biosciences, 553066, clone 145–2C11, 1:50); Mac1-Alexa700 (BD Biosciences, 557960, clone M1/70, 1:50); CD41-BUV396 (BD Bioscience, 564056, MWReg30, 1:100); and CD19-APC-Cy7 (BD Biosciences, 552854, clone 1D3, 1:50).
Peripheral blood collected by retroorbital bleed and immunophenotyping of Ep300::Znf384-V5-HA conditional mice was performed as described above. Flow analysis of the HSPCs compartments was performed using the following antibodies: CD4-BV605 (BD Biosciences, 740336, RM4–4, 1:100); CD8-BV605 (BD Biosciences, 563152, 53–6.7, 1:100); CD19-BV605 (BD Biosciences, 563148, 1D3, 1:100); CD3-BV605 (BioLegend, 100237, 17A3, 1:100); TER119-BV605 (BD Biosciences, 747740, TER119, 1:100); B220-BV605 (BioLegend, 103244, RA3–6B2, 1:100); Gr1-BV605 (BioLegend, 108440, RB6–8C5, 1:100); FLT3-PE (BD Biosciences, 553842, A2F10.1, 1:100); Sca1-PerCP-Cy5.5 (eBioscience, 45–5981–82, D7, 1:100); CD150-PE-Cy7 (BioLegend, 115914, TC15–12F12.2, 1:100); CD48-Alexa700 (BioLegend, 103426, HM48–1, 1:100); ckit-APC-eF780 (eBioscience, 47–1171–82, 2B8, 1:100); CD34-FITC (BD Biosciences, 553733, RAM34, 1:100).
Postmortem flow analysis of bone marrow and spleen cells from NSG-SGM3 transplanted with human CD34 cells was performed using a panel of multilineage markers [CD34 APC-Cy7 (BioLegend, 343513, Clone 581, 1:50), CD38 PE-Cy7 (BD Biosciences, 335790, clone HB7, 1:50), CD45 Alexa700 (Invitrogen, MHCD4529, HI30, 1:50), CD19 BV605 (BD Biosciences, 562653, SJ25C1, 1:50), CD33 BV711 (BioLegend, 366624, clone P67.6, 1:50), CD10 BV421 (BD Biosciences, 562902, clone HI10a, 1:50)] was performed on the GFP-positive population to determine the lineage of disease in engrafted samples.
Experimental Human Modeling
Human Hematopoietic In Vitro Assays.
CD34 cells were cultured in X-VIVO 10 (Lonza), supplemented with 1% BSA (Sigma), recombinant human SCF (100 ng/mL), FLT-3 ligand (100 ng/mL), thrombopoietin (TPO; 50 ng/mL), and IL7 (10 ng/mL) (PeproTech) for 24 hours prior to transduction. Cells and viral supernatants (pCL20c MSCV-ires-GFP lentivirus expressing the wild-type or fusion genes of interest) were added to 96-well non-tissue culture–treated plates and maintained at 37°C for 48 hours. CD34, GFP double-positive cells were fluorescence-activated cell sorted directly into complete methylcellulose (H4034, STEMCELL Technologies) that included β-Mercaptoethanol, recombinant human SCF, IL3, erythropoietin (EPO), granulocyte colony-stimulating factor (G-CSF), and GM-CSF with the addition of recombinant human FLT-3 ligand (20 ng/mL), and IL6 (50 ng/mL; PeproTech). One-hundred cells were plated in duplicate and colonies were allowed to differentiate for 10 days before counting and morphologic assessment of hematopoietic colonies. Flow cytometry analysis using a panel of multilineage markers [CD34 APC-Cy7 (BioLegend, 343513, clone 581, 1:50), CD38 PE-Cy7 (BD Biosciences, 335790, clone HB7, 1:50), CD45 Alexa700 (Invitrogen, MHCD4529, HI30, 1:50), CD19 BV605 (BD Biosciences, 562653, SJ25C1, 1:50), CD33 BV711 (BioLegend, 366624, clone P67.6, 1:50), CD10 BV421 (BD Biosciences, 562902, clone HI10a, 1:50)] was performed on the GFP-positive cells to determine colony immunophenotype.
For the MS-5 single-cell differentiation assay, Cells were fluorescence-activated cell sorted for HSC, MPP, CMP, MEP, or GMP populations using the following antibodies: CD45RA FITC (BD Biosciences, 561882, clone HI100, 1:50), CD90 PE (BD Biosciences, 561970, clone 5E10, 1:50), CD34 APC-Cy7 (BioLegend, 343513, Clone 581, 1:50), CD38 PE-Cy7 (BD Biosciences, 335790, clone HB7, 1:50), CD49f BV605 (BD Biosciences, 740416, clone GoH3, 1:50), CD71 BV711 (BD Biosciences, 563767, clone M-A712, 1:50), CD7 V450 (BD Biosciences, 642916, clone M-T701, 1:50), CD10 V421 (BD Biosciences, 562902, clone HI10a, 1:50), CD135 A647 (BD Biosciences, 563494, clone 4G8, 1:50). HSC, MPP, CMP, MEP and GMP populations were prestimulated in X-VIVO 10, 1% BSA supplemented with recombinant human SCF (100 ng/mL), FLT-3 ligand (100 ng/mL), TPO (50 ng/mL), and IL7 (10 ng/mL; PeproTech) for 4 hours before adding lentivirus. Viral supernatants (pCL20c MSCV-ires-GFP lentivirus expressing the wild-type or ZNF384 fusion genes of interest) were added and GFP-positive cells were isolated by FACS 72 hours after transduction.
Ninety-six–well flat-bottom plates (Nunc, Thermo Fisher) were coated with 0.2% gelatin prior to seeding 1.5 × 103 MS-5 cells (Creative Bioarray) in MyeloCult media (H5100, STEMCELL Technologies) supplemented with hydrocortisone (1 µmol/L/mL). Cells were allowed to adhere for 48 hours prior to cell sorting. Before cell sorting, media were changed to StemPro-34 SFM with nutrients (Life Technologies) supplemented with penicillin, streptomycin, l-glutamine, lipids (STEMCELL Technologies), and the following recombinant human cytokines: SCF (100 ng/mL), FLT-3 ligand (20 ng/mL), TPO (100 ng/mL), EPO (3 ng/mL), IL6 (50 ng/mL), IL3 (10 ng/mL), IL11 (50 ng/mL), GM-CSF (20 ng/mL), IL2 (10 ng/mL) and IL7 (20 ng/mL; PeproTech). GFP-positive cells were fluorescence-activated cell sorted into each well. One week after sorting, fresh media were added to the wells. Two weeks after sorting, the contents of each well were harvested and analyzed by flow cytometry. The following antibodies were used to assess lineage: CD41 APC (BioLegend, 303709, clone HIP8, 1:50), CD235a PE (BioLegend, 349106, clone HI264, 1:50), CD45 APC-H7 (BD Biosciences, 560178, clone 2D1, 1:50), CD11b PE-Cy7 (BioLegend, 301322, clone ICRF44, 1:50), CD15 BV605 (BD Biosciences, 562980, clone W6D3, 1:50), and CD33 BV711 (BioLegend, 366624, clone P67.6, 1:50). Immunophenotype was determined when there were at least 10 GFP-positive cells.
Human Hematopoietic Cell In Vivo Assays.
CD34-positive cells were prestimulated in X-VIVO 10 supplemented with 1% BSA, and recombinant human SCF (100 ng/mL), FLT-3 ligand (100 ng/mL), TPO (50 ng/mL), and IL7 (10 ng/mL; PeproTech) for 24 hours before adding lentivirus. Cells and viral supernatants (pCL20c MSCV-ires-GFP lentivirus expressing the wild-type or fusion genes of interest) were added to non-tissue culture–treated plates and maintained at 37°C. Twenty-four hours after transduction, cells were collected and 3.0 × 104 cells were transplanted through intrafemoral injections into sub-lethally (250 Rad) irradiated 8- to 12-week-old NSG or NSG-SGM3 mice (Jackson Laboratory). A portion of cells were retained in liquid culture and the percent of GFP-positive cells was determined by flow cytometry 72 hours posttransduction. Animals that became moribund were euthanized, and blood, bone marrow, and spleen were analyzed for evidence of leukemia, using morphology, flow cytometry, and histopathologic analysis. For secondary transplantation, sub-lethally irradiated (250 Rad) 8- to 12-week-old NSG-SGM3 mice were injected intrafemorally with 6.0 × 105 unsorted cells or 5.0 × 104 fluorescence-activated cell sorted; CD34, GFP double-positive, bone marrow cells from the primary leukemic mice.
Immunoblotting.
Cryopreserved patient samples or cell lines were lysed in RIPA buffer (Sigma) and 30 μg of protein was loaded and run on 4%–12% NuPage Bis-Tris gels (Life Technologies) at 160 V for 1 hour. Blots were probed with anti-ZNF384 (Abcam, Ab176689, 1:1,000).
Preclinical Drug Studies
In Vivo FLT3 Inhibitor Therapy.
The xenograft model of human TCF3::ZNF384 B/myeloid MPAL (SJMPAL046472) was established using diagnostic bone marrow (Supplementary Table S2; ref. 7). Primary leukemia cells (1 × 106 cells per mouse) were transplanted via tail vein injection into sub-lethally irradiated (250 Rad) NSG-SGM3 mice (Jackson Laboratory). Bone marrow harvested from engrafted mice was transduced with lentivirus expressing YFP and luciferase to enable bioluminescent imaging using a Xenogen IVIS-200 system and Living Image software (Caliper Life Sciences; ref. 52) in subsequent transplants. Engraftment was monitored weekly by bioluminescent imaging and flow cytometric analysis of human CD45+, CD33+, and CD19+ leukemic blasts in the peripheral blood. Gilteritinib (SJ001009805–01, MedChem Express, HY-12432A, lot no. 29622) dosing was determined by a performing a plasma pharmacokinetic profile on 12-week-old NSG-SGM3 mice using a qualified LC/MS-MS assay. For efficacy studies, mice were randomized to receive vehicle or treatment when bioluminescent flux reached 2 × 109 p/s at week 2 posttransplant. Gilteritinib was administered by oral gavage at 20 mg/kg/day in 0.5% methylcellulose (type 400 cPs) at 1 mg/mL. Disease burden was assessed every 7 days using bioluminescent imaging and flow cytometric measurement of human CD45, CD33+, and CD19+ blasts in the peripheral blood. Treatment ended at week 5 posttransplant due to vehicle-treated mice reaching a humane endpoint. Spleen and bone marrow were harvested at time of sacrifice.
Transcriptomic/Genomic/Epigenomic Analyses
RNA-seq and Analysis.
RNA was isolated from GFP+ ARF-null pre-B cells (indicating expression of CL20-MSCV-ires-GFP containing wild-type or fusion inserts) or flow sorted tumor cells using the RNeasy Mini Kit or AllPrep DNA/RNA Mini Kits (Qiagen). The integrity of RNA was analyzed by Bioanalyzer (Agilent) and quantitated using RiboGreen (Thermo Fisher Scientific). Libraries were prepared from total RNA with the TruSeq Stranded Total RNA Library Prep Kit (Illumina). Libraries were quantified using the Quant-iT PicoGreen dsDNA assay (Life Technologies), Kapa Library Quantification Kit (Kapa Biosystems,) or low pass sequencing on a MiSeq Nano v2 (Illumina). One-hundred cycle paired-end sequencing was performed on an Illumina HiSeq 2500, HiSeq 4000, or NovaSeq 6000. Sequencing was paired end and performed using total RNA and stranded RNA-seq. Paired-end reads from RNA-seq were aligned to mm10 reference genome and BAM files constructed. HTSeq (ref. 53; version 0.6.1p1) was used to generate gene-level counts and estimate FPKM based on GENCODE (ref. 54; vM9). Voom (55) was used for gene differential expression analysis after trimmed mean normalization (TMM).
Major hematopoietic cell types in the mouse models were investigated using xCell, a tool that performs cell type enrichment analysis from gene expression data for 64 immune and stroma cell types (36). Relative enrichment scores (Z-score) were displayed in the heatmap. Read counts from RNA-seq data were imported to DESeq2 R package for differential gene expression analysis. To perform gene-set enrichment analysis (GSEA), all the genes were ranked according to the fold-change and significance from differential analysis. GSEA was performed using mSigDB C2 genes and curated gene sets from in-house analyses (Supplementary Table S14).
For analysis of xenograft tumors (n = 6) with limiting material, the low input RNA library preparation kit (NuGen Ovation V2) was used with Kapa Hyper Kit (KK8504), and sequencing was performed with paired end setting. Paired-end reads were aligned to hg38 reference genome using STAR (v2.7) and gene quantification was estimated using RSEM (v1.3.1) with nonstranded setting. Enrichment analysis of gene sets from 14 B-ALL subtypes was performed using single-sample GSEA (10, 56). Relative enrichment score (Z-score) was displayed in the heatmap.
Analysis of ZNF384 Exon 8 Usage.
PDX cells from SJBALL004031_X18–4052 (primary engrafted NSG-SGM3 PDX) and SJMPAL046472_X2-PAVRCX-063S5–1411 (secondary engrafted NSG-SGM3 PDX) and JIH-5 cells (7) were thawed and stained with the following human-specific antibodies (all from BD Biosciences, catalog number in parentheses): anti-CD45-FITC (345808), anti-CD19-BV605 (562653) and anti-CD33 PE-Cy7 (333946). Cells were then sorted in two or more of the following CD45+ tumor populations (PT): PT1: CD33+CD19+; PT2: CD33−CD19−; PT3: CD33−CD19+; PT4: CD33−CD19−. Sorted cells were collected, resuspended in RLT buffer (Qiagen, catalog no. 79216) and processed for DNA and RNA extraction by the AllPrep DNA/RNA Mini Kit (Qiagen, catalog no. 80204). RNA samples were sequenced by total stranded RNA-seq (SJMPAL046472_X2-PAVRCX-063S5–1411_PT1 and SJMPAL047180_C2-JIH5_PT1) or low input RNA-seq (SJBALL004031_X16–4052: PT1, PT2, PT3 and PT4; SJMPAL046472_X3-PAVRCX-063S5–1411_PT2 and SJMPAL047180_C4-JIH5_PT3). After sequencing, quality control was conducted using FastQC (version 0.11.5). Sequencing reads were then mapped to the GRCh37 human genome reference by STAR (version 2.4.2a). Mixture of Isoforms (MISO; ref. 57) Bayesian Inference model (version 0.5.4) was used for quantification and comparison of alternative splicing events. The MISO algorithm counts the numbers of reads that are common to both isoforms and the reads that are exclusive to one isoform or the other, to estimate the percent spliced-in (PSI) values in a given sample. A built-in utility in MISO, sashimi_plot, was used for visualizing raw RNA-seq read densities along exons and junctions.
scRNA-seq and Analysis.
Mouse bone marrow cells were isolated into cold PBS supplemented with 2% BSA to generate single-cell suspensions. Red blood cells (RBC) were removed using red blood cell lysis buffer (BioLegend), resuspended in PBS/2% BSA, and filtered through a 40-μm cell strainer. An aliquot of 1 × 106 cells was removed as bulk bone marrow. Lineage depletion was performed on the remaining cells using the mouse Lineage Cell Depletion Kit (Miltenyi Biotec) according to the manufacturer's recommendations. Lineage-negative cells were then stained with streptavidin-APC (BioLegend, 405207, 1:100). Live (DAPI-negative) cells were purified by FACS from the bulk marrow samples and live, lineage-negative (APC negative) cells were purified from the lineage-negative samples. Cells were washed and counted. scRNA-seq libraries were generated using Chromium 10X Next GEM Single Cell 3′ v3.1 (10X Genomics) by combining 5,000 bulk bone marrow cells and 5,000 enriched bone marrow lineage-negative cells for each sample. Final library quality was assessed using an Agilent Bioanalyzer High Sensitivity Chip (Agilent Technologies). Samples were sequenced using the Illumina NovaSeq with 28 (barcode+UMI) + 91(read) setting, with a median depth of over 50,000 reads per cell.
Single-cell data generated using the 10X Genomics 3′ NEXT GEM gene expression kit (v3.1) was aligned and quantified using Cell Ranger (v5.0.0) against mouse genome GRCm38 (refdata-gex-mm10–2020-A). Cells with fewer than 200 or higher than 6,000 features, or mitochondria content higher than 5% were removed. The feature counts data of the selected cells from one wild-type and two samples expressing EP300::Znf384-V5-HA transcript were merged together using canonical correlation analysis (58) from Seurat (59). A total of 21,705 cells were used for further analysis. Clusters of cells were identified using Seurat, partitioned into five major superclusters using Monocle3 (60), and characterized on the basis of gene expression of major hematopoietic cell types. Differential expression between wild-type and EP300::Znf384-V5-HA was performed for the five major partitions separately using Seurat.
Whole-Exome Sequencing and Analysis.
DNA was extracted from 2–5 × 106 sorted tumor cells using the AllPrep DNA/RNA Mini Kit (Qiagen). Libraries were constructed, checked for quality control, and sequenced as previously described (61). Briefly, construction utilized DNA tagmentation (fragmentation and adapter attachment) performed with the reagent provided in the Illumina Nextera rapid exome kit and was performed using the Caliper Biosciences Sciclone G3 (PerkinElmer). First-round PCR (ten cycles) was performed using Illumina Nextera kit reagents and clean-up steps using AMPure XP beads (Beckman Coulter Genomics/Agencourt). Target capture utilized Illumina Nextera Rapid Capture Expanded Exome and supplied hybridization and associated reagents. The prehybridization pool size was 12 samples, and second-round PCR (ten cycles) was performed with Nextera kit reagents.
Whole-exome sequencing was performed with 100 bp paired-end setting. The paired-end sequencing reads were mapped to mouse genome mm10 using BWA mem (version 0.7.17). Read sorting based on chromosome position and PCR duplication marking was performed using Picard (http://broadinstitute.github.io/picard/, version 2.9.4). Then the reads were realigned around potential indel regions by GATK (version 3.7) IndelRealigner module. Sequencing depth and coverage were assessed on the basis of coding regions (∼34 Mb) defined by RefSeq genes, with median value of 82% of 100× coding exon coverage. UnifiedGenotyper (within GATK v3.7) module was applied to call SNVs and Indels from leukemia and paired samples. Variant annotation against RefSeq gene was done using ANNOVAR (62).
ChIP-seq.
ARF-null pre-B cells were fluorescence-activated cell sorted for live, GFP-positive cells (indicating expression of CL20-MSCV-ires-GFP containing wild-type or fusion inserts) and used for crosslinking. ChIP assays were carried out as described previously (63). Briefly, 2 × 107 cells were incubated for 10 minutes in 1% formaldehyde in PBS at room temperature, quenched by the addition of 1/10 volume of 2.5 mol/L glycine. Cells were then washed three times with cold PBS-containing proteinase inhibitors and lysed on ice for 10 minutes in lysis buffer (50 mmol/L HEPES, pH 7.9, 140 mmol/L NaCl, 1 mmol/L EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100). Chromatin was washed twice in washing buffer (10 mmol/L Tris-HCl, pH 8, 200 mmol/L NaCl, 1 mmol/L EDTA, 0.5 mmol/L EGTA) and then twice in shearing buffer (0.1% SDS, 10 mmol/L Tris-HCl, pH 8, 1 mmol/L EDTA) before resuspension in 1 mL of shearing buffer. Chromatin was sonicated in AFA fiber milliTUBEs (Covaris) using a Covaris E210 sonicator (5% duty cycle, 200 cycles per burst, 105 Watts, water level 6) at 4°C for 15 minutes. Sheared chromatin was centrifuged for 10 minutes at 13,200 × g at 4°C, and the supernatant was mixed with an equal amount of ChIP dilution buffer (0.1% SDS, 30 mmol/L Tris-HCl, pH 8, 1 mmol/L EDTA, 300 mmol/L NaCl, 2% Triton X-100) before ChIP experiments. Immunoprecipitation was initially performed with an antibody to HA (Abcam, Ab9110) and a normal rabbit IgG control (Santa Cruz Biotechnology, Sc2027) using 2 μg of antibody per ChIP. To prepare ChIP-seq libraries, 10 ng of ChIP DNA was end repaired and adaptor ligation was performed using the NEB Next ChIP-Seq Library Prep Reagent Set for Illumina (New England BioLabs). Libraries were purified after 14 rounds of PCR amplification with Q5 DNA Hot-Start polymerase (New England BioLabs). Each ChIP-seq library underwent 50-cycle single-end sequencing using TruSeq SBS kit v3 on an Illumina HiSeq 2000. After confirming synonymy between anti-ZNF384 and anti-HA, anti-HA was used for the remaining experiments. Experiments were performed with three biological replicates.
ChIPmentation Sequencing.
ARF-null pre-B cells were fluorescence-activated cell sorted for live, GFP-positive cells (indicating expression of CL20-MSCV-ires-GFP containing wild-type or fusion inserts) and used immediately for crosslinking. One to two million cells were fixed in 150 µL PBS with 1% PFA (Pierce) for 10 minutes before quenching with 0.125 mol/L glycine for 5 minutes. Cells were washed twice with 150 µL cold PBS supplement with protease inhibitors (Pepstatin, leupeptin, PMSF, and sodium butyrate) and pellets were snap frozen before storing at −80°C. Subsequent work was performed on ice and cool buffers and solutions were used unless otherwise specified. The pellet was lysed in 130 µL sonication buffer (10 mmol/L Tris-HCl pH 8.0, 2 mmol/L EDTA pH 8.0, 0.25% SDS, and protease inhibitors) and sonicated with a Covaris E210 sonicator (2% duty cycle, 200 cycles per burst, 105 Watts, water level 6) for 9 minutes in an AFA fiber microTUBE (Covaris) until the size of most fragments was in the range of 200–700 base pairs. Lysates were centrifuged at 14,000 × g for 10 minutes at 4°C, and the supernatant containing the sonicated chromatin was transferred to a new tube.
The lysate was then diluted in RIPA buffer (final concentration: 10 mmol/L Tris-HCl pH 8.0, 1 mmol/L EDTA pH 8.0, 140 mmol/L NaCl, 1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate, and protease inhibitors) to a volume of 200 µL per immunoprecipitation. The histone 3 lysine 27 acetyl (H3K27Ac) antibody (1 µL/IP, Cell Signaling Technology, catalog no. 8173S) was added to the chromatin and rotated overnight at 4°C. For each immunoprecipitation, 10 µL Protein G Dynabeads (Thermo Fisher) were prepared by washing twice and resuspending in PBS supplemented with 0.1% BSA. The beads rotated overnight at 4°C to block to surface with BSA. The next morning, the beads were added to antibody/chromatin mixture and rotated 2 hours at 4°C. For control libraries, an immunoprecipitation with 2.5 µL of a nonspecific IgG rabbit antibody (Santa Cruz Biotechnology, Sc2027) was used. Beads were washed subsequently with RIPA-LS (10 mmol/L Tris-HCl pH 8.0, 1 mmol/L EDTA pH 8.0, 140 mmol/L NaCl, 1% Triton X-100, 0.1% SDS and 0.1% sodium deoxycholate; twice), RIPA-HS (10 mmol/L Tris-HCl pH 8.0, 1 mmol/L EDTA pH 8.0, 500 mmol/L NaCl, 1% Triton X-100, 0.1% SDS and 0.1% DOC; twice), RIPA-LiCl (10 mmol/L Tris-HCl pH 8.0, 1 mmol/L EDTA pH 8.0, 250 mmol/L LiCl, 0.5% sodium deoxycholate and 0.5% NP40; twice), and 10 mmol/L Tris-HCL pH 8.0 (once). Following the wash with Tris, the beads and Tris were transferred to a new tube and the supernatant was discarded.
Beads were then resuspended in 25 µL of the tagmentation reaction mix containing 5 µL 5X Tagmentation buffer, 19 µL nuclease free water, and 1 µL Tagment DNA Enzyme from the Nextera DNA Sample Prep Kit (Illumina) and incubated at 37 °C for 5 minutes in a thermocycler. The reaction was stopped by adding 150 µL of RIPA-LS and incubating on ice for 5 minutes. The beads were washed with RIPA-LS (twice) and twice with cold Tris-HCl pH 8.0, 1 mmol/L EDTA. Beads were then resuspended in 48 µL ChIP elution buffer (10 mmol/L Tris-HCl pH 8.0, 5 mmol/L EDTA, 300 mmol/L NaCl, and 0.4% SDS) containing 2 µL of Proteinase K (NEB) and incubated at 55°C for 1 hour followed by 65°C for 8 hours. The beads were magnetized and supernatant was transferred to a new tube. DNA was purified with Qiagen MinElute Kit and eluted in 22 µL Qiagen buffer EB.
To estimate the optimum number of enrichment cycles, 2 µL of DNA was added to a 10 µL qPCR reaction containing 0.15 µmol/L primers, 5 µL Kapa HiFi HotStart ReadyMix (Kapa Biosystems), 0.2 µL KAPA SYBR Rox Low 50X (Kapa Biosystems), and 0.1 µL SYBR 100X (Thermo Fisher); using the following program: 72°C for 5 minutes, 98°C for 30 seconds, 25 cycles of 98°C for 10 seconds, 63°C for 30 seconds, and 72°C for 30 seconds, and a final elongation at 72°C for 1 minute. Kapa HiFi HotStart ReadyMix was incubated at 98°C for 30 seconds before preparation of all PCR reactions (qPCR and final enrichment PCR), to activate the hot-start enzyme. Library was amplified in a 50 µL reaction using 20 µL of DNA, 0.75 µmol/L primers, and 25 µL Kapa HiFi HotStart ReadyMix. The number of cycles was determined by using the quantitation cycle (Cq) value determined by the qPCR reaction and adding 1 (N = Cq+1). Libraries were purified using AMPureXP beads at a beads-to-sample ratio of 1.8:1, followed by a size selection with AMPureXP beads to recover libraries with a fragment length of 200–400 base pairs.
Input libraries were made by reserving 20 µL of input chromatin for reverse crosslinking and Qiagen MinElute as described above. The input DNA was tagmented at 55°C for 5 minutes in 5 µL reactions containing 2.5 µL DNA, 1 µL 1:10 Tagment DNA enzyme, and Tagmentation buffer. The DNA was cleaned up using the Qiagen MinElute Kit and the PCR was performed as described above with 12 cycles. A detailed protocol can be found at https://www.medical-epigenomics.org/papers/schmidl2015/ (version 1.14).
Library preparation was performed using custom Nextera primers as described (64). The libraries were sequenced by the Hartwell Center using the Illumina HiSeq3000/4000 platform and the 25 base pair, paired-end configuration.
ChIP-seq Analysis.
ChIP-seq data from anti-ZNF384, anti-HA and anti-H3K27ac were analyzed using ENCODE pipeline (https://github.com/ENCODE-DCC/chip-seq-pipeline2). Briefly, raw sequencing reads were mapped to mouse genome mm10 with BWA (ref. 65; version 0.7.13-r1126). Duplicated reads were marked with Picard and only uniquely mapped reads extracted by Samtools (66) were retained for analysis. Each read was extended to estimated fragment size by SPP (67) (version 1.1) and bigwig files were generated with the track normalized to 15 million unique mapped reads. Each sample was reviewed to confirm clear peaks by IGV (ref. 68; 3.0.beta) and consistency between anti-HA samples and anti-ZNF384 samples. Anti-HA produced stronger peaks than anti-ZNF384 and nonspecific binding was not observed, therefore anti-HA was used for all ChIP-seq experiments.
For differential binding analysis, peaks were called with MACS2 (ref. 69; version 2.1.1). Differential binding analysis was performed using DiffBind R package and peaks with P < 0.01 were defined as differential peaks. Differential peaks between TCF3::ZNF384 versus ZNF384 were used to generate a heatmap and average profile for HA signal at the peak regions. The same methods were used to compare differential histone modifications, H3K4me1, H3K18Ac, and H3K27Ac; and TCF3::ZNF384 isoforms, with exon 8 versus without exon 8. qPCR (ΔΔCt method) was employed to validate ChIP-seq results.
Differential binding sites were annotated to genes if their promoter (transcription start site ± 2kb) overlapped the binding sites and GSEA was performed using differentially expressed genes from RNA-Seq analysis by their rank of log2 fold change. Enriched gene sets were obtained through GREAT (ref. 43; version 4.0.4) using 1,139 peaks identified as showing increased binding by TCF3::ZNF384 compared with wild-type ZNF384 and 1,128 peaks identified as showing increased binding by TCF3::ZNF384 with exon 8 compared to without exon 8. ClueGO (version 2.5.8; ref. 70) was further applied on the genes within 10kb of the 1,139 peaks to identify pathways with increasing regulation by ZNF384 fusion proteins.
For differential peak analysis, genomic sequences in fasta format were extracted from the mouse genome based on the coordinates of peaks. Discriminative mode of MEME-ChIP (http://meme-suite.org/tools/meme-chip; ref. 71) was used for motif analysis. To perform motif enrichment analysis in “Fusion up” peaks, the primary sequences were set to be fasta sequences for “Fusion up” peaks and the control sequences were set to be fasta sequences for “Fusion down” peaks. Similarly, for motif enrichment analysis in “Fusion down” peaks, fasta sequences for “Fusion down” peaks and “Fusion up” peaks were set to be primary and control sequences, respectively (72).
HiChIP Library Construction.
ARF-null pre-B cells were fluorescence-activated cell sorted for live, GFP-positive cells (indicating expression of CL20-MSCV-ires-GFP containing wild-type or fusion inserts) and we used 1.1 × 107 fresh cells per sample. All H3K27ac HiChIP experiments were performed using the Arima-HiC+ kit (Arima Genomics A101020) according precisely to the manufacturer's protocols using the cell culture crosslinking protocol [Arima-HiC+ document numbers A160430 v00 (HiChIP) and A160169 v00 (library preparation)]. We used the H3K27ac antibody from Active Motif (am91194). Uniquely barcoded HiChIP libraries were pooled and sequenced to an average depth of 300 million reads on an Illumina HiSeq instrument.
HiChIP Data Analysis.
Paired-end reads of 100 bp were trimmed for adapters using cutadapt (version 1.9, paired-end mode, default parameter with “-m 25 -O 6”; ref. 73) and then processed by HiC-Pro (version 2.11.4; ref. 74) using mouse genome mm10 (GRCm38 from GENCODE) and Arima fragment file (Ligation sites GATC, GANTC). Bowtie2–2.2.4, samtools-1.2, R-3.4.0, Python-2.7.12 were configured for HiC-Pro. allValidPairs files from HiC-Pro pipeline were then used to generate bigwig files for IGV inspection (make_viewpoints.py from HiC-Pro utils). All samples (TAF15::ZNF384, TCF3::ZNF384, wild-type) have two biological replicates with good depth (209M to 282M pairs) and comparable metrics with the published data (GEO id: GSE80820; ref. 75), such as valid interaction rate (78.98% to 92.79% compared with GEO 78.65% to 80.53%). Each sample called ∼20k loops by FitHiChIP (76) while only about ∼300 loops have been called as differential loops (by FitHiChIP, calling with ChIP-seq data). However, review of chromatin interaction profiles does not identify any strongly correlated with H3K27ac ChIP-seq differential peaks.
Proteomic Analysis
RIME.
RIME experiments were performed and analyzed as previously described (77). HEK293T cells were cultured in DMEM supplemented with 10% FBS; 1× penicillin, streptomycin and l-glutamine (Gibco). Cells (8 × 106) were transfected with 30 μg of pCL20c MSCV-ires-GFP plasmids (empty vector, ZNF384-HA, HA-TAF15::ZNF384, or TCF3::ZNF384-HA) using Fugene HD (Promega) according to the manufacturer's recommendations. After culturing for 48 hours, media were removed and cells were crosslinked with PBS containing 1% formaldehyde (Thermo Fisher) for 10 minutes at room temperature. Crosslinking was quenched by adding glycine to a final concentration of 0.125 mol/L for 5 minutes. Cells were washed with ice-cold PBS containing proteinase inhibitor cocktail (Sigma-Aldrich) four times and frozen on dry ice. Beads were prepared by using 50 µL of Dynabeads Protein G (Invitrogen) and 5 μg rabbit HA antibody (Abcam, Ab9110) for each sample. Beads were washed twice in PBS containing 5 mg/mL BSA before bringing them up in PBS/BSA with 1:50 antibody. Beads and antibody rotated overnight at 4°C. Cell pellets were resuspended in LB1 buffer [50 mmol/L HEPES-KOH (pH 7.5), 140 mmol/L NaCl, 1 mmol/L EDTA, 10% glycerol, 0.5% Igepal CA-630, and 0.25% Triton X-100] containing proteinase inhibitor cocktail, followed by rotation mixing for 10 minutes at 4°C. Nuclei were pelleted and resuspended in LB2 buffer [10 mmol/L Tris-HCL (pH 8.0), 200 mmol/L NaCl, 1 mmol/L EDTA and 0.5 mmol/L EGTA] containing proteinase inhibitor cocktail and rotated at 4°C for 5 minutes. The samples were resuspended in LB3 buffer [10 mmol/L Tris-HCl (pH 8), 100 mmol/L NaCl, 1 mmol/L EDTA, 0.5 mmol/L EGTA, 0.1% sodium deoxycholate and 0.5% N-lauroylsarcosine] containing proteinase inhibitor cocktail. Chromatin was sheared by sonication with a Covaris E210 sonicator (2% duty cycle, 200 cycles per burst, 105 Watts, water level 6) for 9 minutes in an AFA fiber microTUBE (Covaris) until the size of most fragments were in the range of 100–1,000 bp. Lysates were centrifuged at 14,000 × g for 10 minutes at 4°C, and the supernatant containing the sonicated chromatin was transferred to a new tube. Beads were washed three times in PBS/BSA and resuspended in the chromatin lysate. The bead-bound antibody and chromatin were rotated overnight at 4°C. Beads were then washed 10 times with 1 mL ice-cold RIPA buffer and twice with freshly made 500 µL 100 mmol/L ammonium bicarbonate. Supernatant was removed and chromatin-bound beads were frozen on dry ice and stored at −80°C.
The beads were subjected to overnight digestion at 37°C by adding trypsin at a final concentration of 15 ng/µL (Pierce), as previously described (77, 78). The next day, a second digestion step was performed by adding trypsin for 4 hours. Then, the tryptic peptides were acidified with 5% formic acid and were purified using the Ultra-Micro C18 Spin Columns (Harvard Apparatus) according to manufacturer's instructions. Samples were dried with vacuum concentrator and resuspended in 15 µL 0.1% formic acid prior to mass spectrometry analysis. Purified peptides were analyzed on a Dionex Ultimate 3000 UHPLC system coupled with the Q-Exactive (Thermo Scientific) mass spectrometer. The full scans were performed in the Orbitrap in the range of 400 to 1600 m/z at 70k resolution. For MS2, the top 10 most intense precursors were selected at resolution 17,5k. A 2.0 Th isolation window was used for isolation and the HCD collision energy was set at 28%. The collected HCD tandem mass spectra were processed with the SequestHT search engine in Proteome Discoverer 2.4 against a human uniprot database containing over 20,000 entries along with custom databases containing the sequences of FO (Supplementary Table S15). The SequestHT for the HCD spectra included the following parameters: Precursor Mass Tolerance 20 ppm, Fragment Mass Tolerance 0.02 kDa, number of maximum missed cleavages sites 2 and Dynamic Modifications were Oxidation of M (+15.995Da) and Deamidation of N/Q (+0.984Da). The confidence level for peptide identifications was estimated with the Percolator node using decoy database search and peptides were filtered for q-value < 0.01. Any proteins that appeared in any of the three empty vector control RIME experiments were excluded. FO-specific interactors were considered as those occurring in at least six of eight independent replicates but not occurring in any of the three ZNF384-HA replicates.
Coimmunoprecipitation to Validate RIME Interactions.
HEK293T cells were cultured in DMEM supplemented with 10% FBS; 1× penicillin, streptomycin, and l-glutamine (Gibco). Cells (2.4 × 107) were transfected with 90 μg of pCL20c MSCV-ires-GFP plasmids (empty vector, ZNF384-HA, HA-TAF15::ZNF384, or TCF3::ZNF384-HA) using Fugene HD (Promega) according to manufacturer's recommendation. After culturing for 48 hours, media were removed and cells were washed twice with ice-cold PBS-containing proteinase inhibitor cocktail (Sigma-Aldrich) and scraped from plates. Cells were washed with cold Buffer A (20 mmol/L Tris-HCL (pH 7.5), 20 mmol/L NaCl, 0.2 mmol/L EDTA) containing proteinase inhibitor cocktail (PI) and resuspended in Buffer A + PI for 10 minutes on ice. One volume of Buffer A with 0.04% Igepal CA-630 + PI was added to each sample and they incubated on ice for 10 minutes. Nuclei were pelleted at 1,200 × g for 7 minutes and washed twice with Buffer A + PI. Nuclei were resuspended in Buffer B (20 mmol/L Tris-HCl (pH 7.5), 150 mmol/L NaCl, 0.2 mmol/L EDTA, 1% Igepal CA-630) + PI and sonicated with the Diagenode Bioruptor 300 at low intensity for cycles of 30 seconds on/30 seconds off until nuclei were lysed. Nuclear lysate was centrifuged at 20,000 × g for 30 minutes at 4°C, moved to tubes with prepared anti-HA Dynabeads Protein G (Invitrogen; as described in RIME method) or saved as input (20 µL), and rotated overnight at 4°C. Beads were then washed three times with 1 mL ice-cold RIPA buffer and twice with freshly made 500 µL 100 mmol/L ammonium bicarbonate. Supernatant was removed and chromatin-bound beads were frozen on dry ice and stored at −80°C.
Beads were resuspended in 50 µL NuPage LDS Sample Buffer, NuPage Reducing Agent, and water (Novex) and incubated at 70°C for 10 minutes. Beads were separated using a magnetic stand and lysates were loaded into a NuPage 4%–12% Bis-Tris Mini Gel using MOPS SDS Running Buffer. Protein was transferred using the iBlot 2 and membrane was blocked for 1 hour in 10% milk. The blot was incubated overnight at 4°C with 1:50 mouse MED23 antibody (BD Biosciences, 550429), washed with TBS-Tween, incubated for 1 hour with 1:5,000 stabilized peroxidase conjugated goat anti-mouse (Thermo Scientific, 32430), washed with TBS-Tween, and developed with Clarity ECL Substrate (Bio-Rad). The membrane was stripped using Restore PLUS Western Blot Stripping Buffer (Thermo Scientific) according to the manufacturer's recommendations. The membrane was blocked for 1 hour in 10% milk, incubated overnight at 4°C with 1:5,000 rabbit HA antibody (Abcam, Ab9110), washed with TBS-Tween, incubated for 1 hour with 1:5,000 stabilized peroxidase conjugated goat anti-rabbit (Thermo Scientific, 32460), washed with TBS-Tween, and developed with Clarity ECL Substrate (Bio-Rad).
Statistical Analysis
Data analyses were performed using Prism version 8.4.3 (GraphPad). Data are presented as means ± SD. Significance was determined using Student t test or two-way ANOVA Sidak multiple comparison test. Kaplan–Meier analysis and the Mantel–Cox log-rank test were used for survival data. The P values are listed in the figures or figure legends.
Resource Availability
Lead Contact.
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Charles Mullighan (Charles.mullighan@stjude.org).
Materials Availability.
The mouse line and plasmids generated in this study are available upon request from the lead contact.
Data and Code Availability.
ChIP-seq (GSE181499), RNA-seq (GSE181499), scRNA-seq (GSE181499), and HiChIP (GSE181499) data have been deposited at GEO and WES data have been deposited at Sequence Read Archive under BioProject ID PRJNA752404. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD028481. All are publicly available as of the date of publication. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Authors’ Disclosures
I. Iacobucci reports other support from Amgen and other support from Mission Bio outside the submitted work. Y. Chang reports grants from NIH during the conduct of the study. C.G. Mullighan reports personal fees from Illumina, personal fees from Amgen, grants from Pfizer, and grants from AbbVie outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
K.M. Dickerson: Conceptualization, data curation, formal analysis, validation, investigation, methodology, writing–original draft. C. Qu: Software, formal analysis, investigation, visualization. Q. Gao: Formal analysis, visualization. I. Iacobucci: Data analysis and manuscript editing. Z. Gu: Formal analysis. H. Yoshihara: Investigation. E.A. Backhaus: Investigation. Y. Chang: Investigation. L.J. Janke: Investigation. B. Xu: Formal analysis. G. Wu: Formal analysis. E.K. Papachristou: Formal analysis, investigation, methodology. C.S. D'Santos: Formal analysis, investigation, methodology. K.G. Roberts: Data curation. C.G. Mullighan: Conceptualization, resources, formal analysis, supervision, funding acquisition, investigation, writing–original draft, project administration, writing–review and editing.
Supplementary Material
Acknowledgments
We thank the Animal Resource Center, Genome Sequencing Facility, Center for Applied Bioinformatics, Center for Proteomics and Metabolomics, and Flow Cytometry and Cell Sorting core facilities of St Jude Children's Research Hospital. We thank Pankaj Ghate, Chun Shik Park and Ashley Hill for technical assistance. This work was supported by the American Lebanese Syrian Associated Charities of St. Jude Children's Research Hospital; The St. Jude Children's Research Hospital Chromatin Collaborative; NCI R35 Outstanding Investigator Award CA197695 (to C.G. Mullighan), the Henry Schueler 41&9 Foundation (to C.G. Mullighan).
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
References
- 1. Hunger SP, Mullighan CG. Acute lymphoblastic leukemia in children. N Engl J Med 2015;373:1541–52. [DOI] [PubMed] [Google Scholar]
- 2. Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016;127:2391–405. [DOI] [PubMed] [Google Scholar]
- 3. Kurzer JH, Weinberg OK. Acute leukemias of ambiguous lineage: clarification on lineage specificity. Surg Pathol Clin 2019;12:687–97. [DOI] [PubMed] [Google Scholar]
- 4. Gerr H, Zimmermann M, Schrappe M, Dworzak M, Ludwig WD, Bradtke J, et al. Acute leukaemias of ambiguous lineage in children: characterization, prognosis and therapy recommendations. Br J Haematol 2010;149:84–92. [DOI] [PubMed] [Google Scholar]
- 5. Rubnitz JE, Onciu M, Pounds S, Shurtleff S, Cao X, Raimondi SC, et al. Acute mixed lineage leukemia in children: the experience of St Jude Children's Research Hospital. Blood 2009;113:5083–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Matutes E, Pickl WF, Van't Veer M, Morilla R, Swansbury J, Strobl H, et al. Mixed-phenotype acute leukemia: clinical and laboratory features and outcome in 100 patients defined according to the WHO 2008 classification. Blood 2011;117:3163–71. [DOI] [PubMed] [Google Scholar]
- 7. Alexander TB, Gu Z, Iacobucci I, Dickerson K, Choi JK, Xu B, et al. The genetic basis and cell of origin of mixed phenotype acute leukaemia. Nature 2018;562:373–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Montefiori LE, Bendig S, Gu Z, Chen X, Polonen P, Ma X, et al. Enhancer hijacking drives oncogenic BCL11B expression in lineage-ambiguous stem cell leukemia. Cancer Discov 2021;11:2846–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li JF, Dai YT, Lilljebjorn H, Shen SH, Cui BW, Bai L, et al. Transcriptional landscape of B cell precursor acute lymphoblastic leukemia based on an international study of 1,223 cases. Proc Natl Acad Sci U S A 2018;115:E11711–E20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gu Z, Churchman ML, Roberts KG, Moore I, Zhou X, Nakitandwe J, et al. PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat Genet 2019;51:296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gu Z, Churchman M, Roberts K, Li Y, Liu Y, Harvey RC, et al. Genomic analyses identify recurrent MEF2D fusions in acute lymphoblastic leukaemia. Nat Commun 2016;7:13331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yasuda T, Tsuzuki S, Kawazu M, Hayakawa F, Kojima S, Ueno T, et al. Recurrent DUX4 fusions in B cell acute lymphoblastic leukemia of adolescents and young adults. Nat Genet 2016;48:569–74. [DOI] [PubMed] [Google Scholar]
- 13. Shago M, Abla O, Hitzler J, Weitzman S, Abdelhaleem M. Frequency and outcome of pediatric acute lymphoblastic leukemia with ZNF384 gene rearrangements including a novel translocation resulting in an ARID1B/ZNF384 gene fusion. Pediatr Blood Cancer 2016;63:1915–21. [DOI] [PubMed] [Google Scholar]
- 14. Qian M, Zhang H, Kham SK, Liu S, Jiang C, Zhao X, et al. Whole-transcriptome sequencing identifies a distinct subtype of acute lymphoblastic leukemia with predominant genomic abnormalities of EP300 and CREBBP. Genome Res 2017;27:185–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zaliova M, Kotrova M, Bresolin S, Stuchly J, Stary J, Hrusak O, et al. ETV6/RUNX1-like acute lymphoblastic leukemia: a novel B-cell precursor leukemia subtype associated with the CD27/CD44 immunophenotype. Genes Chromosomes Cancer 2017;56:608–16. [DOI] [PubMed] [Google Scholar]
- 16. Marincevic-Zuniga Y, Dahlberg J, Nilsson S, Raine A, Nystedt S, Lindqvist CM, et al. Transcriptome sequencing in pediatric acute lymphoblastic leukemia identifies fusion genes associated with distinct DNA methylation profiles. J Hematol Oncol 2017;10:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McClure BJ, Heatley SL, Kok CH, Sadras T, An J, Hughes TP, et al. Pre-B acute lymphoblastic leukaemia recurrent fusion, EP300-ZNF384, is associated with a distinct gene expression. Br J Cancer 2018;118:1000–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hirabayashi S, Ohki K, Nakabayashi K, Ichikawa H, Momozawa Y, Okamura K, et al. ZNF384-related fusion genes define a subgroup of childhood B-cell precursor acute lymphoblastic leukemia with a characteristic immunotype. Haematologica 2017;102:118–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Torrungruang K, Alvarez M, Shah R, Onyia JE, Rhodes SJ, Bidwell JP. DNA binding and gene activation properties of the Nmp4 nuclear matrix transcription factors. J Biol Chem 2002;277:16153–9. [DOI] [PubMed] [Google Scholar]
- 20. Yang S, Adaway M, Du J, Huang S, Sun J, Bidwell JP, et al. NMP4 regulates the innate immune response to influenza A virus infection. Mucosal Immunol 2021;14:209–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thunyakitpisal P, Alvarez M, Tokunaga K, Onyia JE, Hock J, Ohashi N, et al. Cloning and functional analysis of a family of nuclear matrix transcription factors (NP/NMP4) that regulate type I collagen expression in osteoblasts. J Bone Miner Res 2001;16:10–23. [DOI] [PubMed] [Google Scholar]
- 22. Nakamoto T, Yamagata T, Sakai R, Ogawa S, Honda H, Ueno H, et al. CIZ, a zinc finger protein that interacts with p130(cas) and activates the expression of matrix metalloproteinases. Mol Cell Biol 2000;20:1649–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nakamoto T, Izu Y, Kawasaki M, Notomi T, Hayata T, Noda M, et al. Mice deficient in CIZ/NMP4 develop an attenuated form of K/BxN-serum induced arthritis. J Cell Biochem 2016;117:970–7. [DOI] [PubMed] [Google Scholar]
- 24. He L, Fan X, Li Y, Chen M, Cui B, Chen G, et al. Overexpression of zinc finger protein 384 (ZNF 384), a poor prognostic predictor, promotes cell growth by upregulating the expression of Cyclin D1 in Hepatocellular carcinoma. Cell Death Dis 2019;10:444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Martini A, La Starza R, Janssen H, Bilhou-Nabera C, Corveleyn A, Somers R, et al. Recurrent rearrangement of the Ewing's sarcoma gene, EWSR1, or its homologue, TAF15, with the transcription factor CIZ/NMP4 in acute leukemia. Cancer Res 2002;62:5408–12. [PubMed] [Google Scholar]
- 26. Corveleyn A, Janssen H, Martini A, Somers R, Cools J, Marynen P. Cellular transformation of NIH3T3 fibroblasts by CIZ/NMP4 fusions. J Cell Biochem 2005;94:1112–25. [DOI] [PubMed] [Google Scholar]
- 27. Liu YF, Wang BY, Zhang WN, Huang JY, Li BS, Zhang M, et al. Genomic profiling of adult and pediatric B-cell acute lymphoblastic leukemia. EBioMedicine 2016;8:173–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yaguchi A, Ishibashi T, Terada K, Ueno-Yokohata H, Saito Y, Fujimura J, et al. EP300-ZNF384 fusion gene product up-regulates GATA3 gene expression and induces hematopoietic stem cell gene expression signature in B-cell precursor acute lymphoblastic leukemia cells. Int J Hematol 2017;106:269–81. [DOI] [PubMed] [Google Scholar]
- 29. Yamamoto H, Hayakawa F, Yasuda T, Odaira K, Minamikawa Y, Tange N, et al. ZNF384-fusion proteins have high affinity for the transcriptional coactivator EP300 and aberrant transcriptional activities. FEBS Lett 2019;593:2151–61. [DOI] [PubMed] [Google Scholar]
- 30. Paietta E, Roberts KG, Wang V, Gu Z, Buck GAN, Pei D, et al. Molecular classification improves risk assessment in adult BCR-ABL1-negative B-ALL. Blood 2021;138:948–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Roberts KG, Gu Z, Payne-Turner D, McCastlain K, Harvey RC, Chen IM, et al. High frequency and poor outcome of philadelphia chromosome-like acute lymphoblastic leukemia in adults. J Clin Oncol 2017;35:394–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Roberts KG, Li Y, Payne-Turner D, Harvey RC, Yang YL, Pei D, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med 2014;371:1005–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Waanders E, Gu Z, Dobson SM, Antic Z, Crawford JC, Ma X, et al. Mutational landscape and patterns of clonal evolution in relapsed pediatric acute lymphoblastic leukemia. Blood Cancer Discov 2020;1:96–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Signer RA, Montecino-Rodriguez E, Witte ON, Dorshkind K. Immature B-cell progenitors survive oncogenic stress and efficiently initiate Ph+ B-acute lymphoblastic leukemia. Blood 2010;116:2522–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Williams RT, Roussel MF, Sherr CJ. Arf gene loss enhances oncogenicity and limits imatinib response in mouse models of Bcr-Abl-induced acute lymphoblastic leukemia. Proc Natl Acad Sci U S A 2006;103:6688–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol 2017;18:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, Speck NA. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature 2009;457:887–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tusi BK, Wolock SL, Weinreb C, Hwang Y, Hidalgo D, Zilionis R, et al. Population snapshots predict early haematopoietic and erythroid hierarchies. Nature 2018;555:54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Izzo F, Lee SC, Poran A, Chaligne R, Gaiti F, Gross B, et al. DNA methylation disruption reshapes the hematopoietic differentiation landscape. Nat Genet 2020;52:378–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Heng TS, Painter MW, Immunological Genome Project Consortium. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol 2008;9:1091–4. [DOI] [PubMed] [Google Scholar]
- 41. Notta F, Zandi S, Takayama N, Dobson S, Gan OI, Wilson G, et al. Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science 2016;351:aab2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Billerbeck E, Barry WT, Mu K, Dorner M, Rice CM, Ploss A. Development of human CD4+FoxP3+ regulatory T cells in human stem cell factor-, granulocyte-macrophage colony-stimulating factor-, and interleukin-3-expressing NOD-SCID IL2Rgamma(null) humanized mice. Blood 2011;117:3076–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 2010;28:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Griffith M, Griffith OL, Krysiak K, Skidmore ZL, Christopher MJ, Klco JM, et al. Comprehensive genomic analysis reveals FLT3 activation and a therapeutic strategy for a patient with relapsed adult B-lymphoblastic leukemia. Exp Hematol 2016;44:603–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Piragyte I, Clapes T, Polyzou A, Klein Geltink RI, Lefkopoulos S, Yin N, et al. A metabolic interplay coordinated by HLX regulates myeloid differentiation and AML through partly overlapping pathways. Nat Commun 2018;9:3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bhagwat AS, Roe JS, Mok BYL, Hohmann AF, Shi J, Vakoc CR. BET bromodomain inhibition releases the mediator complex from select cis-regulatory elements. Cell Rep 2016;15:519–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen X, Zhao J, Gu C, Cui Y, Dai Y, Song G, et al. Med23 serves as a gatekeeper of the myeloid potential of hematopoietic stem cells. Nat Commun 2018;9:3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Borggrefe T, Yue X. Interactions between subunits of the Mediator complex with gene-specific transcription factors. Semin Cell Dev Biol 2011;22:759–68. [DOI] [PubMed] [Google Scholar]
- 49. Perl AE, Martinelli G, Cortes JE, Neubauer A, Berman E, Paolini S, et al. Gilteritinib or chemotherapy for relapsed or refractory FLT3-mutated AML. N Engl J Med 2019;381:1728–40. [DOI] [PubMed] [Google Scholar]
- 50. Whitlock CA, Witte ON. Long-term culture of murine bone marrow precursors of B lymphocytes. Methods Enzymol 1987;150:275–86. [DOI] [PubMed] [Google Scholar]
- 51. Williams RT, den Besten W, Sherr CJ. Cytokine-dependent imatinib resistance in mouse BCR-ABL+, Arf-null lymphoblastic leukemia. Genes Dev 2007;21:2283–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Boulos N, Mulder HL, Calabrese CR, Morrison JB, Rehg JE, Relling MV, et al. Chemotherapeutic agents circumvent emergence of dasatinib-resistant BCR-ABL kinase mutations in a precise mouse model of Philadelphia chromosome-positive acute lymphoblastic leukemia. Blood 2011;117:3585–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31:166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 2012;22:1760–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Law CW, Chen Y, Shi W, Smyth GK. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 2014;15:R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Unni AM, Lockwood WW, Zejnullahu K, Lee-Lin SQ, Varmus H. Evidence that synthetic lethality underlies the mutual exclusivity of oncogenic KRAS and EGFR mutations in lung adenocarcinoma. Elife 2015;4:e06907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods 2010;7:1009–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36:411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM III, et al. Comprehensive integration of single-cell data. Cell 2019;177:1888–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 2014;32:381–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Iacobucci I, Wen J, Meggendorfer M, Choi JK, Shi L, Pounds SB, et al. Genomic subtyping and therapeutic targeting of acute erythroleukemia. Nat Genet 2019;51:694–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang J, McCastlain K, Yoshihara H, Xu B, Chang Y, Churchman ML, et al. Deregulation of DUX4 and ERG in acute lymphoblastic leukemia. Nat Genet 2016;48:1481–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 2013;10:1213–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol 2008;26:1351–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol 2011;29:24–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008;9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009;25:1091–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 2009;37:W202–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mohammed H, Taylor C, Brown GD, Papachristou EK, Carroll JS, D'Santos CS. Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes. Nat Protoc 2016;11:316–26. [DOI] [PubMed] [Google Scholar]
- 73. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal 2011;17:10–2. [Google Scholar]
- 74. Servant N, Varoquaux N, Lajoie BR, Viara E, Chen CJ, Vert JP, et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol 2015;16:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mumbach MR, Rubin AJ, Flynn RA, Dai C, Khavari PA, Greenleaf WJ, et al. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat Methods 2016;13:919–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bhattacharyya S, Chandra V, Vijayanand P, Ay F. Identification of significant chromatin contacts from HiChIP data by FitHiChIP. Nat Commun 2019;10:4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Papachristou EK, Kishore K, Holding AN, Harvey K, Roumeliotis TI, Chilamakuri CSR, et al. A quantitative mass spectrometry-based approach to monitor the dynamics of endogenous chromatin-associated protein complexes. Nat Commun 2018;9:2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Glont SE, Papachristou EK, Sawle A, Holmes KA, Carroll JS, Siersbaek R. Identification of ChIP-seq and RIME grade antibodies for Estrogen Receptor alpha. PLoS One 2019;14:e0215340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP-seq (GSE181499), RNA-seq (GSE181499), scRNA-seq (GSE181499), and HiChIP (GSE181499) data have been deposited at GEO and WES data have been deposited at Sequence Read Archive under BioProject ID PRJNA752404. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD028481. All are publicly available as of the date of publication. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.