Abstract
The genes for dihydropteroate synthase of Mycobacterium tuberculosis and Mycobacterium leprae were isolated by hybridization with probes amplified from the genomic DNA libraries. DNA sequencing revealed an open reading frame of 840 bp encoding a protein of 280 amino acids for M. tuberculosis dihydropteroate synthase and an open reading frame of 852 bp encoding a protein of 284 amino acids for M. leprae dihydropteroate synthase. The dihydropteroate synthases were expressed under control of the T5 promoter in a dihydropteroate synthase-deficient strain of Escherichia coli. Using three chromatography steps, we purified both M. tuberculosis and M. leprae dihydropteroate synthases to >98% homogeneity. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis revealed molecular masses of 29 kDa for M. tuberculosis dihydropteroate synthase and 30 kDa for M. leprae dihydropteroate synthase. Gel filtration of both enzymes showed a molecular mass of ca. 60 kDa, indicating that the native enzymes exist as dimers of two identical subunits. Steady-state kinetic parameters for dihydropteroate synthases from both M. tuberculosis and M. leprae were determined. Representative sulfonamides and dapsone were potent inhibitors of the mycobacterial dihydropteroate synthases, but the antimycobacterial agent p-aminosalicylate, a putative dihydropteroate synthase inhibitor, was a poor inhibitor of the enzymes.
Tuberculosis (TB) and leprosy remain major public health problems in many regions of the world. The resurgence of Mycobacterium tuberculosis, the etiological agent for TB, has been especially worrisome because of the high risk of TB infection among human immunodeficiency virus (HIV)-positive populations (21, 40). Further, coinciding with frequent TB-HIV coinfection is the emergence of virulent multidrug-resistant TB which is refractory to standard anti-TB agents (24). Likewise, a major problem of leprosy treatment has been the growing resistance of Mycobacterium leprae to dapsone, a mainstay therapy for more than two decades. The emerging resistance has created an urgent need for new therapeutics and targets to combat the spread of drug-resistant mycobacteria.
A successful approach to selective antimicrobial chemotherapy has been to exploit the inhibition of targets unique and vital to the pathogen. Central to this approach has been the folate biosynthesis pathway, which generates folate cofactors essential for continued DNA and RNA synthesis (6). Unlike mammals, which utilize exogenous sources of folates, many prokaryotes and protozoa must synthesize these essential cofactors de novo. Dihydropteroate synthase (DHPS; EC 2.5.1.15) is one of several crucial enzymes in the de novo biosynthesis of folate cofactors that have been important targets for antimicrobial agents.
Dihydropteroate synthase catalyzes the condensation of p-aminobenzoic acid (pABA) and 6-hydroxymethyl-7,8-dihydropterin pyrophosphate (HPOPP) to form 7,8-dihydropteroate (31). The latter is an essential precursor of the folate cofactor, tetrahydrofolate. DHPS is the target for important antimicrobial agents such sulfonamides and dapsone, which are competive inhibitors with respect to pABA (2).
The genes coding for DHPS from a number of microorganisms have been cloned and sequenced (11, 16, 19, 25, 32, 35, 38). The DHPSs of Escherichia coli (11), Pneumocystis carinii (3, 37), Plasmodium falciparum (35), and Neisseria meningitidis (12) were successfully expressed in heterologous systems, with two DHPS structures of Staphylococcus aureus (14) and E. coli (1) solved to date. While this work was in progress, the DNA sequences for M. leprae and M. tuberculosis DHPSs were deposited in public databases. The DNA sequences of mycobacterial DHPS have the following EMBL accession numbers: M. leprae (locus MLCB2548), AL023093; M. tuberculosis (locus MTCY7H7B), Z95557; and M. tuberculosis H37Rv (locus MTBH37Rv), AL123456.
Nevertheless, little information is available on the DHPS in mycobacteria, largely because sufficient amounts of enzyme have not been available for study. In the present work, we describe the isolation, cloning, and expression in E. coli of DHPSs from M. tuberculosis and M. leprae. The availability of large amounts of these enzymes should facilitate studies on directed molecular approaches toward the design of potential second-generation antimicrobial agents.
MATERIALS AND METHODS
Materials.
Restriction endonucleases and other DNA-modifying enzymes were obtained from New England Biolabs and Gibco-Life Technologies. The plasmid and DNA purification columns were from Qiagen. The Random Primed DNA labeling kit was from Boehringer Mannheim (Mannheim, Germany). [α-32P]dCTP (3,000 Ci/mmol) and [carboxyl-14C]pABA (58 Ci/mol) were from Amersham and Moravek Biochemicals, respectively. The substrate HPOPP was a gift from Carmen J. Allegra, National Cancer Institute, National Institutes of Health, Bethesda, Md. Reverse-phase C18 Bakerbond SPE columns were from J. T. Baker. DEAE-Sepharose, DyeMatrix Gel Green A, and hydroxylapatite (Bio-Gel HTP) were purchased from Pharmacia, Amicon, and Bio-Rad, respectively. Dapsone, sulfamethoxazole, sulfamethoxypyridazine, and p-aminosalicylate (PAS) were obtained from Sigma. Oligonucleotides were synthesized in the BioService Unit, BIOTEC Center, National Science and Technology Development Agency, Thailand, and the Biomolecular Resource Center, University of California at San Francisco. Other chemicals and reagents were of the highest purity commercially available.
Bacterial strains and plasmids.
M. tuberculosis H37Rv was cultivated at the Department of Microbiology, Faculty of Medicine, Siriraj Hospital, Bangkok, Thailand. The M. leprae genomic DNA library constructed in pYUB18 cosmid (4) was a gift from William R. Jacobs, Jr., Howard Hughes Medical Institute, Albert Einstein College of Medicine, New York, N.Y. The DHPS-deficient E. coli strain C600ΔfolP::Kmr, used for the expression of M. tuberculosis and M. leprae DHPSs, was provided by Gote Swedberg, Uppsala University, Uppsala, Sweden (12). E. coli DH5α (Life Technologies) was used as a common host strain for plasmid-mediated transformations and general manipulation of recombinant plasmids. Plasmid pBluescript KS+ was from Stratagene. The expression vector pKOS007-90 was a gift from Kosan Bioscience (Burlingame, Calif.).
Preparation of probes for genomic screening.
Two degenerate primers, DHPS-1 (5′ GTCGAATTCGA(CT)TC(GATC)TT(CT)TC(GATC)GA(CT)GG) and DHPS-2 (5′ GACGGATCCGA(CT)TC(GATC)CC(GATC)CC(GAT)AT(GA)TC), encoding the sequences DSFSDG and DIGGES, respectively, were used in a PCR with either M. tuberculosis genomic DNA or the M. leprae cosmid library as a template. A separate 125-bp DNA fragment was amplified from each DNA template. EcoRI and BamHI restriction sites (underlined) were introduced in the primers to facilitate cloning, and each amplified fragment was cloned into pBluescript KS+ for DNA sequence analysis. The deduced amino acid sequences were compared to DHPS sequences reported for other organisms.
Cloning of mycobacterial DHPS.
The cloned 125-bp DNA fragments were 32P labeled and used as probes. A Southern blot of BamHI-digested M. tuberculosis genomic DNA was screened, the hybridizing region of the gel was excised, and the extracted DNA was used for the construction of a minilibrary in pBluescript KS+, which was then screened for DHPS clones. For M. leprae, the genomic cosmid library was screened for the desired clones. Cosmids which hybridized to the probes were analyzed by restriction analysis, and the desired DNA fragments were subcloned into pBluescript KS+. Full-length DHPS clones were verified by DNA sequence analysis.
Construction of expression clones.
pKOS007-90, an expression vector utilizing the T5 promoter (15), was modified by inserting the synthetic adapters DHPS-3 (5′ TATGGCGGCCGCATCGATGGTACCCGGGGATCCGAGCTCGTCGA CA) and DHPS-4 (5′ AGCTTGTCGACGAGCTCGGATCCCCGGGTACCATCGATGCGGCCGCCA) containing NotI-ClaI-KpnI-SmaI-BamHI-SacI-SalI between the NdeI and HindIII sites to facilitate subcloning. The resulting plasmid, pKOS007-90PL, was then used for construction of the DHPS expression clones. The complete sequences of the M. tuberculosis and M. leprae DHPS genes were amplified from the pBluescript KS+ clones by using primer pairs DHPS-5 (5′ CAGGAATTCCATATGAGTCCGGCGCCCGTGC)–DHPS-6 (5′ GACGGATCCGCTGCCCGCCCACTCG) for M. tuberculosis DHPS and DHPS-7 (5′ GGAATTCCATATGAGTTTGGCGCCAGTGC)–DHPS-8 (GACGGATCCATTCGGTCAGCCATCACA) for M. leprae DHPS. The NdeI and BamHI restriction sites introduced at the 5′ ends of the sense and antisense primers (underlined) allow cloning of mycobacterial DHPS genes into the corresponding sites of pKOS007-90PL. The resulting clones, pKOS-TBDHPS and pKOS-LPDHPS, were transformed into E. coli C600ΔfolP::Kmr.
Expression of mycobacterial DHPSs.
pKOS-TBDHPS- and pKOS-LPDHPS/pREP4-GroES (8)-transformed E. coli C600ΔfolP::Kmr cells were grown on Luria-Bertani agar plates supplemented with kanamycin (40 μg/ml) and ampicillin (100 μg/ml). A fresh overnight culture from a single colony (0.2% inoculum) was used to inoculate each plate. The culture was then grown at 37°C with vigorous shaking. When the optical density at 600 nm of the cultures reached ∼0.7 to 0.8, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM. The culture was allowed to grow for an additional 24 h at 37°C for pKOS-TBDHPS and 24 h at 25°C for pKOS-LPDHPS before harvesting by centrifugation at 10,000 × g for 15 min at 4°C. The cell pellets obtained after centrifugation were resuspended in 20 mM Tris-HCl (pH 7.5)–1 mM EDTA–1 mM dithiothreitol (DTT)–20% glycerol (buffer A) containing leupeptin (10 μg/ml), phenylmethylsulfonyl fluoride (20 μg/ml), trypsin inhibitor (50 μg/ml), and 1 mM benzamidine-HCl. The cells were disrupted by two passages through a French pressure cell at 15,000 lb/in2, and the extracts were centrifuged at 30,000 × g for 30 min at 4°C. The clear supernatant was used for DHPS assays and purification. Protein concentration was determined as described elsewhere (27).
Purification of mycobacterial DHPSs.
All buffers contained 20% glycerol, and the entire purification process was carried out at 4°C. The crude supernatant (∼18 ml) was applied to a 1- by 8-cm column of DEAE-Sepharose preequilibrated with buffer A containing 50 mM NaCl. The column was washed with 60 ml of equilibration buffer followed by a 60-ml linear gradient to 0.5 M NaCl at a flow rate of 1 ml/min. Fractions with DHPS activity were pooled (∼30 ml), and the sample was diluted with buffer A to reduce the NaCl concentration to <100 mM. The pooled sample was then circulated at a flow rate of 0.5 ml/min through a DyeMatrix Gel Green A column (1.5 by 5 cm) preequilibrated with buffer A containing 100 mM NaCl. The column was washed with 10 mM sodium phosphate buffer (pH 7.0)–1 mM DTT–20% glycerol (buffer B) containing 1 mM EDTA and 100 mM NaCl until protein was undetectable in the effluent. Then a linear gradient of 0.1 to 1.0 M NaCl in buffer B was applied. Fractions with DHPS activity were pooled (∼30 ml) and diluted with buffer B to reduce the NaCl concentration to <400 mM. The sample was then loaded onto a Bio-Gel HTP column (1 by 7 cm) preequilibrated with buffer B containing 0.1 mM EDTA and 400 mM NaCl. The column was washed with 50 ml of equilibration buffer, and a linear gradient of 10 to 400 mM sodium phosphate buffer (pH 7.0) was applied. The active DHPS was eluted as a sharp peak at approximately 100 mM sodium phosphate buffer (pH 7.0). Active fractions were pooled and concentrated, and aliquots were fast frozen in liquid nitrogen and stored at −80°C.
Phylogenetic tree.
The dendrogram of approximate sequence relationships was generated by using the Pileup program of the Wisconsin Package (version 9.1; Genetics Computer Group, Madison, Wis.). Similarity scores are used to create a clustering order based on a strategy called UPGMA (unweighted pair-group method using arithmetic averages), the results of which are represented by the dendrogram.
Enzyme assay.
DHPS activity was determined by monitoring the amount of [14C]dihydropteroate produced from the substrate, [14C]pABA, as described elsewhere (22, 28) except that substrate and product were separated on 3-ml reverse-phase C18 Bakerbond SPE columns. Reaction mixtures (50 μl) contained 50 mM Tris-HCl (pH 8.3), 5 mM MgCl2, 5 mM DTT, 100 μg of bovine serum albumin per ml, 10 μM HPOPP, 5 μM [14C]pABA (58 Ci/mol), and enzyme (∼5 mU). Unless specified, the reaction was initiated with enzyme. Control reactions contained all the reagents except enzyme. After incubation at 37°C for 10 min, reaction mixtures were quenched by immersing the reaction tubes in boiling water for 2 min and centrifuged at 10,000 × g for 30 min. An aliquot of the clear supernatant (40 μl) was applied to a C18 Bakerbond SPE column activated with 5 ml of acetonitrile and equilibrated with 5 ml of 10 mM sodium phosphate (pH 7.0). The column was washed with 5 ml of the same buffer to remove the unreacted [14C]pABA, and [14C]dihydropteroate was eluted from the column with 1 ml of acetonitrile. The eluate (∼1 ml) was mixed with 9 ml of scintillation cocktail (Bio-Safe II), and radioactivity was counted on a Beckman LS 3801 scintillation counter. One unit of DHPS was defined as the amount of enzyme required to produce 1 nmol of dihydropteroate per min at 37°C.
Kinetics and inhibition studies.
Steady-state kinetic parameters were obtained by determination of DHPS activity in the presence of various concentrations of [14C]pABA (0.2 to 5.0 μM) or HPOPP (0.5 to 8.0 μM) while the concentration of the other substrate, HPOPP or pABA, was held at a constant concentration of 10 or 0.5 μM, respectively. Kinetic parameters were calculated by using a nonlinear least-squares fit of the data to the Michaelis-Menten equation. Data points were obtained from two independent experiments and were fit to equation III-5 for competitive inhibition (29).
Nucleotide sequence accession numbers.
The nucleotide sequences of DHPS genes of M. tuberculosis and M. leprae reported in this paper have been submitted to GenBank and assigned accession no. AF117617 and AF117618, respectively.
RESULTS
Cloning and nucleotide sequence of DHPSs from M. tuberculosis and M. leprae.
Similar strategies were used for cloning the DHPS genes of M. tuberculosis and M. leprae. A homologous 125-bp gene fragment was amplified from the corresponding genomic DNA of each organism, using degenerate primers designed to encode two motifs (DSFSDG and DIGGES) which are highly conserved in bacterial DHPSs (11, 19, 25, 32). Characterization of both 125-bp fragments revealed significant sequence homology to other bacterial DHPSs. We labeled the fragments with 32P and used them as probes to screen for the full-length genes. Southern blot analysis of BamHI-digested genomic DNA from M. tuberculosis showed hybridization at 2.7 kb, and a minilibrary was prepared by cloning size-selected DNA into pBluescript KS+. This library was screened with the M. tuberculosis homologous probe, and a clone, pKS-TBDHPS, containing a 2.7-kb fragment was obtained. For M. leprae, screening the cosmid library yielded a positive cosmid. Southern blot analysis of a BamHI digest of this cosmid showed a strongly hybridizing 2.4-kb fragment, which was subcloned into pBluescript KS+ to yield pKS-LPDHPS. Sequence analysis of the 2,718-bp DNA insert from pKS-TBDHPS revealed an open reading frame of 840 bp encoding a 280-amino-acid DHPS. Likewise, sequence analysis of the 2,412-bp insert from pKS-LPDHPS revealed an open reading frame of 852 bp encoding a 284-amino-acid DHPS. It is noteworthy that GTG, which codes for Val, was an initiation codon for the DHPS genes of both M. tuberculosis and M. leprae. While this work was in progress, Cole et al. reported the complete genome sequence of M. tuberculosis H37Rv (9), of which the sequence of folP (SPTREMBL 006274) was completely identical to the sequence AF117617 reported in this paper.
Comparison with DHPS sequences from other organisms.
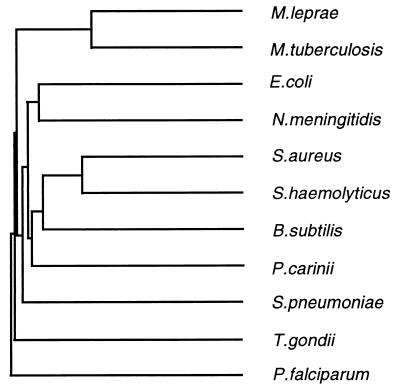
Alignment of the predicted M. tuberculosis and M. leprae DHPS amino acid sequences revealed that the two proteins were highly homologous, with 78% identical amino acid residues (data not shown). The mycobacterial DHPS sequences showed moderate homology to other known bacterial DHPS amino acid sequences (35 to 39% identity) and to the DHPS domains of polyproteins from certain eukaryotes (26 to 37% identity to sequences from P. carinii, Toxoplasma gondii, and P. falciparum). Figure 1 is a dendrogram showing approximate relationships among the mycobacterial DHPS sequences and a selection of those thus far reported from other organisms.
FIG. 1.
Dendrogram of approximate sequence relationships among DHPSs from M. tuberculosis (this work), M. leprae (this work), E. coli (11), S. aureus (14), S. haemolyticus (16), Bacillus subtilis (32), S. pneumoniae (19), and N. meningitidis (25) and DHPS domains of polyproteins from P. carinii (38), P. falciparum (5, 35), and T. gondii (23). (Failure to match accepted phylogenetic branching order can be seen in certain cases, in particular where a species with an expected history of relatively rapid evolutionary change is involved, such as P. falciparum.)
Expression of mycobacterial DHPSs.
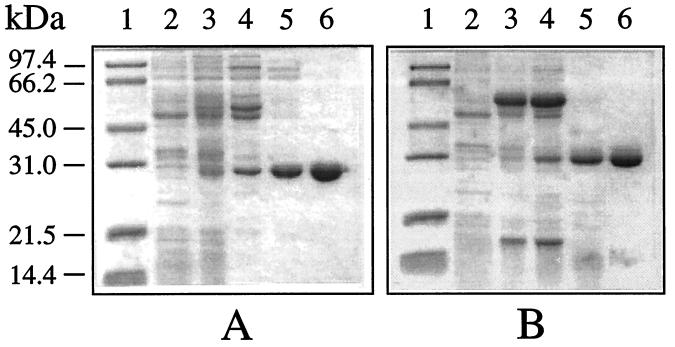
Initial attempts to express M. tuberculosis and M. leprae DHPSs in E. coli were complicated by the presence of host DHPS with a molecular mass indistinguishable from that for the mycobacterial enzymes. Therefore, the DHPS-deficient E. coli strain C600ΔfolP::Kmr (12) was used as the host for the expression system. Two sets of primers (DHPS-5–DHPS-6 and DHPS-7–DHPS-8) were designed to facilitate cloning of the DHPS sequence between NdeI-BamHI sites of the expression plasmid pKOS007-90PL. The resulting recombinant plasmids harboring M. tuberculosis DHPS (pKOS-TBDHPS) and M. leprae DHPS (pKOS-LPDHPS) were transformed into E. coli C600ΔfolP::Kmr and used to express DHPS under control of the T5 promoter. IPTG induction at 37°C for 24 h resulted in expression of M. tuberculosis DHPS as a soluble protein. The expressed product could be visualized as a thin protein band with a molecular mass of ∼29 kDa (Fig. 2A, lane 3). The expressed M. tuberculosis DHPS was estimated to be ca. 5% of the total soluble protein in the crude extract, with a specific activity of 19 nmol/min/mg of protein.
FIG. 2.
SDS-PAGE analysis of expression and purification of M. tuberculosis (A) and M. leprae (B) DHPSs. Lanes: 1, molecular size markers (masses are shown at the left); 2, host cell extract as negative control; 3, crude extract; 4, DEAE-Sepharose pool; 5, DyeMatrix Gel Green A pool; 6, Bio-Gel HTP pool.
Under the same induction conditions as for M. tuberculosis DHPS (37°C for 24 h), M. leprae DHPS was poorly expressed and formed inactive inclusion bodies. Since a lower induction temperature (25°C) improved the solubility of the expressed enzyme (data not shown), the expression of M. leprae DHPS was performed at 25°C for 24 h. The yield of soluble M. leprae DHPS was further improved by the presence of the chaperonins GroEL and GroES. The plasmid encoding these two proteins was cotransformed with pKOS-LPDHPS into E. coli C600ΔfolP::Kmr, and this system yielded a specific activity of 4 nmol/min/mg of protein, which was about sixfold higher than the specific activity obtained in the absence of chaperonins (data not shown). Even with these improvements in expression, the ∼30-kDa DHPS band was difficult to visualize by Coomassie staining after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 2, lane 3B). The intense band of molecular mass ∼60 kDa represents the coexpressed chaperonins (Fig. 2B, lanes 3 and 4).
Purification and characterization.
The DHPSs of M. tuberculosis and M. leprae were purified by passage through three consecutive chromatographic columns. Both enzymes were purified three- to fourfold with about an 80% yield after passage through the first chromatographic column, DEAE-Sepharose (Table 1). At this stage of purification, Coomassie-stained bands corresponding to the predicted sizes of M. tuberculosis DHPS (∼29 kDa) and M. leprae DHPS (∼30 kDa) could be visualized after SDS-PAGE (Fig. 2A, lane 4) (see also Fig. 4B, lane 4). The next purification step, on a DyeMatrix Green A column, resulted in complete separation of the coexpressed chaperonins from M. leprae DHPS (Fig. 2B, lane 5). This step resulted in a ∼50% loss of total activity and a five- to eightfold increase in specific activity (Table 1). The final chromatographic step, using Bio-Gel HTP, resulted in >98% pure M. tuberculosis DHPS (Fig. 2A, lane 6), with an overall ∼32-fold purification and ∼30% yield (Table 1). A 44-fold purification and 24% yield were obtained for M. leprae DHPS, although SDS-PAGE revealed some minor low-molecular-weight protein impurities (Fig. 2B, lane 6). The overall yield of the purified mycobacterial DHPSs was estimated to be 3 to 4 mg/liter of E. coli culture.
TABLE 1.
Purification of recombinant DHPSs of M. tuberculosis and M. lepraea
| Step | Protein (mg) | DHPS activity
|
Fold purification | Yield (%) | |
|---|---|---|---|---|---|
| Total activity (nmol/min) | Sp act (nmol/min/mg) | ||||
| Crude extractb | 88 (195) | 1,632 (786) | 19 (4) | 1 (1) | 100 (100) |
| DEAE-Sepharose | 17 (53) | 1,293 (634) | 75 (12) | 4 (3) | 79 (81) |
| DyeMatrix Green A | 2.0 (3.0) | 653 (306) | 327 (102) | 18 (26) | 40 (39) |
| Bio-Gel HTP | 0.8 (1.1) | 477 (191) | 596 (174) | 32 (44) | 29 (24) |
Numbers in parentheses are data for M. leprae DHPS.
From 250-ml E. coli culture.
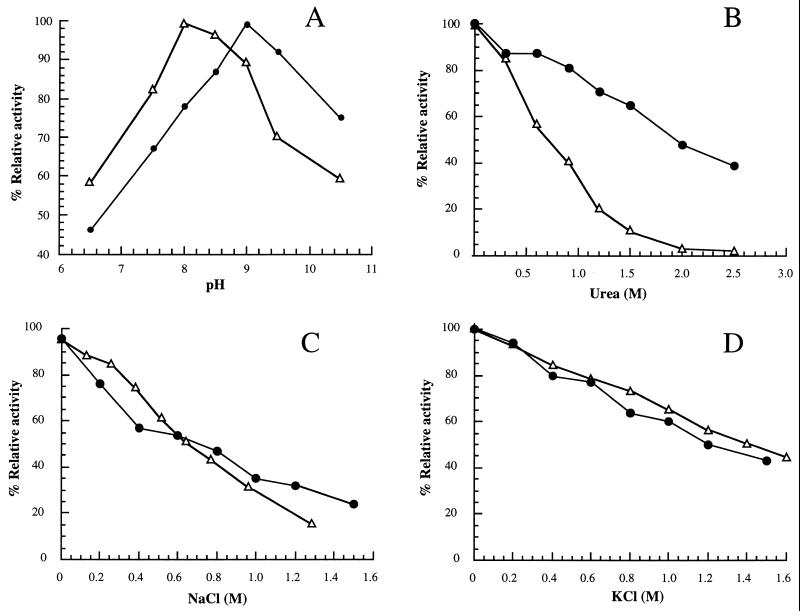
FIG. 4.
Optimal pH and effects of urea and salts. Purified recombinant DHPSs of M. tuberculosis (●) and M. leprae (▵) were tested for optimal pH (A) and effects of urea (B), NaCl (C), and KCl (D).
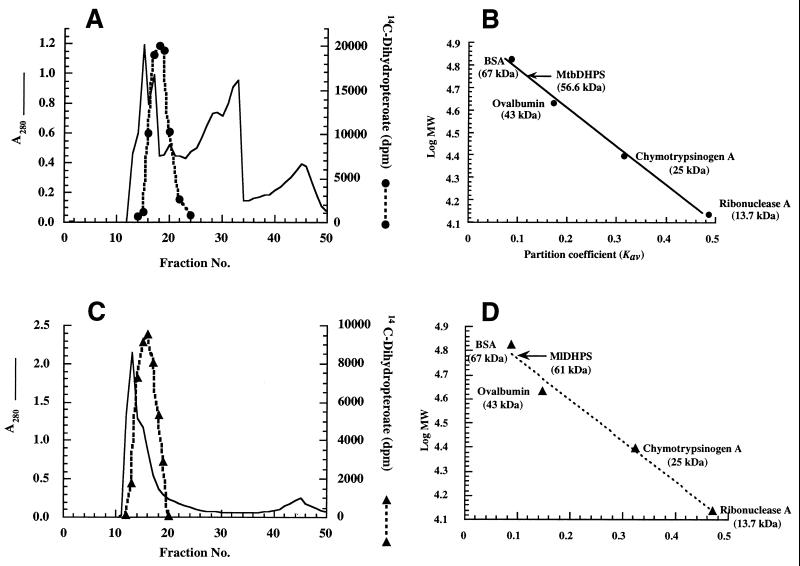
The molecular and kinetic properties of M. tuberculosis and M. leprae DHPSs were investigated. Figures 3A and C show the Sephadex G-100 purification profiles of DHPSs of M. tuberculosis and M. leprae, respectively. The apparent molecular masses calculated from gel filtration data were ∼56 kDa for M. tuberculosis DHPS (Fig. 3B) and ∼61 kDa for M. leprae DHPS (Fig. 3D). These values are twice the molecular masses determined by SDS-PAGE (Fig. 2), suggesting that the enzymes are dimers of identical subunits as reported for the DHPSs of E. coli (34), S. aureus (14), Streptococcus pneumoniae (19), and T. gondii (23). The pIs calculated from the deduced amino acid sequences of DHPSs of M. tuberculosis and M. leprae were 4.92 and 5.42, respectively. The optimal pHs for the activity of M. tuberculosis and M. leprae DHPSs were 9.0 and 8.0, respectively (Fig. 4A). The enzymes from both sources were inactivated 50% or more by 0.6 to 2 M NaCl, KCl, and urea (Fig. 4B to D). The DHPSs of M. tuberculosis and M. leprae were not stable, with 10 to 30% loss of activity upon storage at −80°C for 1 month in 0.1 M sodium phosphate buffer (pH 7.0) containing 20% glycerol. Other storage conditions have not been assessed.
FIG. 3.
Gel filtration chromatography of mycobacterial DHPSs. Partially purified recombinant DHPSs of M. tuberculosis (MtbDHPS) (A) and M. leprae (MlDHPS) (C) were loaded onto Sephadex G-100 columns (2.8 by 47 cm) and eluted at a flow rate of 0.5 ml/min with 20 mM Tris HCl (pH 7.5)–1 mM EDTA–1 mM DTT. Protein concentration and DHPS activity were monitored by A280 and DHPS assay, respectively. The native molecular masses of M. tuberculosis DHPS (B) and M. leprae DHPS (D) were estimated to be 56.6 and 61 kDa, respectively, from the plots of Kav versus log molecular mass (MW). BSA, bovine serum albumin.
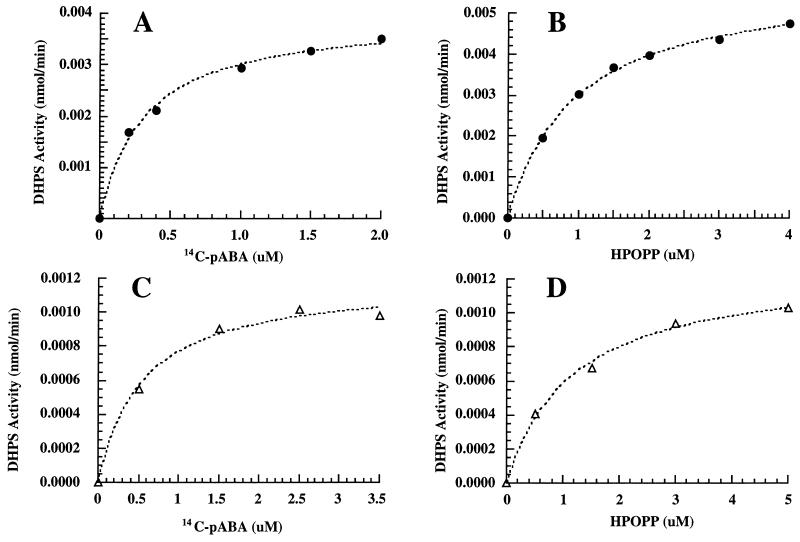
The kinetic parameters for M. tuberculosis and M. leprae DHPSs for the substrates pABA and HPOPP were determined. Figure 5 illustrates the typical kinetics of M. tuberculosis and M. leprae DHPSs in the presence of various concentrations of substrates pABA and HPOPP. The kcat for M. tuberculosis DHPS was 35 ± 3 min−1, while that for M. leprae DHPS was 10.6 ± 0.04 min−1. The Kms of M. tuberculosis DHPS for pABA and HPOPP were 0.37 ± 0.08 and 1.03 ± 0.07 μM, respectively. The Kms of M. leprae DHPS for pABA and HPOPP were 0.6 ± 0.1 μM and 1.2 ± 0.3 μM, respectively.
FIG. 5.
Steady-state kinetics of the purified recombinant DHPSs of M. tuberculosis (●) and M. leprae (▵), determined by assaying DHPS activities in the presence of various concentrations of [14C]pABA and HPOPP. The Kms for [14C]pABA (A and C) and HPOPP (B and D) were determined as described in Materials and Methods.
We assessed the inhibitory effects of a sulfone (dapsone), two sulfonamides (sulfamethoxazole and sulfamethoxypyridazine), and PAS on the purified enzymes. Dapsone, sulfamethoxazole, and sulfamethoxypyridazine were potent inhibitors of both M. tuberculosis and M. leprae DHPSs, with Kis in the range of 12 to 32 nM, while PAS was a much less potent inhibitor, with Ki values of ∼1 μM for both enzymes (Table 2). Dapsone has been reported to be active against M. leprae and M. avium complex (13, 17). Dapsone and sulfamethoxazole are only moderately active against M. tuberculosis, as determined from MIC90s (MICs at which 90% of strains are inhibited) (Table 2) (13, 39). To facilitate comparison, the MICs reported for sulfamethoxazole and dapsone were calculated and found to be from >1,000 to 10,000 times higher than the Kis of the compounds, suggesting that the compounds may have difficulty in accessing the target. In contrast to the poor inhibition of DHPS, PAS has been reported to be highly active against the growth of M. tuberculosis (Table 2) (10).
TABLE 2.
Effects of sulfa and sulfa analogues on the activity and/or growth of M. tuberculosis and M. leprae DHPSs
DISCUSSION
The sulfonamides and sulfones are used alone or in combination with dihydrofolate reductase inhibitors for the treatment of certain microbial infections. The drugs act by inhibition of DHPS, which blocks de novo folate biosynthesis and results in a cessation of DNA synthesis. Attempts to study the M. tuberculosis and M. leprae enzymes have been difficult due to the slow growth of M. tuberculosis and the lack of an in vitro cultivation system for M. leprae. To circumvent these difficulties, we cloned the genes encoding DHPSs of M. tuberculosis and M. leprae from corresponding genomic DNA libraries, expressed them in E. coli, and then purified and characterized the enzymes.
The DHPSs of M. tuberculosis and M. leprae are highly homologous, with 219 of 280 (78%) identical residues (data not shown). The mycobacterial DHPSs showed strong homology to the enzymes from most bacterial sources but exhibited lower homology to those in protozoa. Similar to other mycobacterial DNA sequences, those encoding M. tuberculosis and M. leprae DHPS have high (60 to 67%) G+C contents.
Like other bacterial DHPSs thus far reported (11, 14, 16, 19, 25, 32), M. tuberculosis and M. leprae DHPSs are monofunctional. In contrast, DHPSs from eukaryotic organisms are on multifunctional polypeptides containing other enzymes of folate biosynthesis (5, 23, 35, 38). The observed subunit sizes of M. tuberculosis DHPS (∼29 kDa) and M. leprae DHPS (∼30 kDa) (Fig. 2) are approximately half the sizes of the native proteins, indicating that the enzymes are homodimers.
Inhibitors targeting DHPS are used for the treatment of mycobacterial infections; dapsone is used for the treatment of leprosy (30), and sulfadimethoxine and PAS are used to treat infections caused by M. avium and M. tuberculosis, respectively (18, 36). While sulfonamide and sulfone inhibition of DHPS is well documented, the mode of action of PAS remains controversial. The structural similarity between PAS and sulfonamides suggests that its general mode of action is through inhibition of biosynthesis of folate (20). PAS was initially thought to exert its action by blocking the biosynthesis of mycobactin, a lipid-soluble compound believed to be involved in iron chelation and transport (26, 33). However, evidence from subsequent studies supported the proposal that the compound presumably blocked the function of salicylate and not its conversion to mycobactin (7). As expected, dapsone, sulfamethoxazole, and sulfamethoxypyridazine were potent inhibitors of the recombinant DHPSs, with Kis in the low nanomolar range. In contrast, PAS was a relatively poor inhibitor, with a Ki of ∼1 μM (Table 2). However, as a growth inhibitor of M. tuberculosis, PAS was 25- to 90-fold more potent than the sulfonamides or sulfone. In the absence of compensatory factors (e.g., increased transport, accumulation), these results suggest that the primary mode of antimycobacterial action of PAS may not involve inhibition of DHPS.
ACKNOWLEDGMENTS
This work was supported by a Thailand National Science and Technology Development Agency Career Development Grant to W.S. and by USPHS grant AI 32784 to D.V.S.
We thank Ute Schellenberger for developing the DHPS assay.
REFERENCES
- 1.Achari A, Somers D O, Champness J N, Bryant P K, Rosemond J, Stammers D K. Crystal structure of the anti-bacterial sulfonamide drug target dihydropteroate synthase. Nat Struct Biol. 1997;4:490–497. doi: 10.1038/nsb0697-490. [DOI] [PubMed] [Google Scholar]
- 2.Alford R H, Wallace R J, editors. Antimycobacterial agents. 4th ed. Vol. 1. Philadelphia, Pa: Churchill Livingstone Inc.; 1995. [Google Scholar]
- 3.Ballantine S P, Volpe F, Delves C J. The hydroxymethyldihydropterin pyrophosphokinase domain of the multifunctional folic acid synthesis Fas protein of Pneumocystis carinii expressed as an independent enzyme in Escherichia coli: refolding and characterization of the recombinant enzyme. Protein Expr Purif. 1994;5:371–378. doi: 10.1006/prep.1994.1054. [DOI] [PubMed] [Google Scholar]
- 4.Belisle J T, Pascopella L, Imamine J M, Brennan P J, Jacobs W R., Jr Isolation and expression of a gene cluster responsible for biosynthesis of the glycopeptidolipid antigens of Mycobacterium avium. J Bacteriol. 1991;173:6991–6997. doi: 10.1128/jb.173.21.6991-6997.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brooks D R, Wang P, Read M, Watkins W M, Sims P F G, Hyde J E. Sequence variation of the hydroxymethyldihydropterin pyrophosphokinase: dihydropteroate synthase gene in lines of the human malaria parasite, Plasmodium falciparum, with differing resistance to sulfadoxine. Eur J Biochem. 1994;224:397–405. doi: 10.1111/j.1432-1033.1994.00397.x. [DOI] [PubMed] [Google Scholar]
- 6.Brown G M, Williamson J M. Biosynthesis of riboflavin, folic acid, thiamine and pantothenic acid. Adv Enzymol. 1982;53:345–381. doi: 10.1002/9780470122983.ch9. [DOI] [PubMed] [Google Scholar]
- 7.Brown K A, Ratledge C. The effect of p-aminosalicyclic acid on iron transport and assimilation in mycobacteria. Biochim Biophys Acta. 1975;385:207–220. doi: 10.1016/0304-4165(75)90349-9. [DOI] [PubMed] [Google Scholar]
- 8.Caspers P, Stieger M, Burn P. Overproduction of bacterial chaperones improves the solubility of recombinant protein tyrosine kinases in Escherichia coli. Cell Mol Biol. 1994;40:635–644. [PubMed] [Google Scholar]
- 9.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail M A, Rajandream M-A, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 10.Collins L A, Franzblau S G. Microplate alamar blue assay versus BACTEC 460 system for high-throughput screening of compounds against Mycobacterium tuberculosis and Mycobacterium avium. Antimicrob Agents Chemother. 1997;41:1004–1009. doi: 10.1128/aac.41.5.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dallas W S, Gowen J E, Ray P H, Cox M J, Dev I K. Cloning, sequencing, and enhanced expression of the dihydropteroate synthase gene of Escherichia coli MC4100. J Bacteriol. 1992;174:5961–5970. doi: 10.1128/jb.174.18.5961-5970.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fermer C, Swedberg G. Adaptation to sulfonamide resistance in Neisseria meningitidis may have required compensatory changes to retain enzyme function: kinetic analysis of dihydropteroate synthase from N. meningitidis expressed in a knockout mutant of Escherichia coli. J Bacteriol. 1997;179:831–837. doi: 10.1128/jb.179.3.831-837.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez A H, Berlin O G W, Bruckner D A. In-vitro activity of dapsone and two potentiators against Mycobacterium avium complex. J Antimicrob Chemother. 1989;24:19–22. doi: 10.1093/jac/24.1.19. [DOI] [PubMed] [Google Scholar]
- 14.Hampele I C, D’Arcy A, Dale G E, Kostrewa D, Nielsen J, Oefner C, Page M G, Schonfeld H J, Stuber D, Then R L. Structure and function of the dihydropteroate synthase from Staphylococcus aureus. J Mol Biol. 1997;268:21–30. doi: 10.1006/jmbi.1997.0944. [DOI] [PubMed] [Google Scholar]
- 15.Kealey J T, Liu L, Santi D V, Betlach M C, Barr P J. Production of a polyketide natural product in nonpolyketide-producing prokaryotic and eukaryotic hosts. Proc Natl Acad Sci USA. 1998;95:505–509. doi: 10.1073/pnas.95.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kellam P, Dallas W S, Ballantine S P, Delves C J. Functional cloning of the dihydropteroate synthase gene of Staphylococcus haemolyticus. FEMS Microbiol Lett. 1995;134:165–169. doi: 10.1111/j.1574-6968.1995.tb07932.x. [DOI] [PubMed] [Google Scholar]
- 17.Kerkering T M, Espinael-Ingroff A, Dalton H, Warren N. Program and abstracts of the 7th International Conference on AIDS. Florence, Italy: Istituto Superiore di Sanita; 1991. In-vitro susceptibility of dapsone against M. tuberculosis and strains of M. avium and M. intracellulare isolated from AIDS patients, abstr. W. A. 1024. [Google Scholar]
- 18.Lehmann J. Para-aminosalicylic acid in the treatment of tuberculosis. Lancet. 1946;i:15–16. doi: 10.1016/s0140-6736(46)91185-3. [DOI] [PubMed] [Google Scholar]
- 19.Lopez P, Espinosa M, Greenberg B, Lacks S A. Sulfonamide resistance in Streptococcus pneumoniae: DNA sequence of the gene encoding dihydropteroate synthase and characterization of the enzyme. J Bacteriol. 1987;169:4320–4326. doi: 10.1128/jb.169.9.4320-4326.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McClatchy J K. Antituberculosis drugs: mechanisms of action, drug resistance, susceptibility testing, and assays of activity in biological fluids. Baltimore, Md: Williams & Wilkins; 1980. [Google Scholar]
- 21.Nakajima H. Tuberculosis: a global emergency. World Health. 1993;4:3. [Google Scholar]
- 22.Okinaka O, Iwai K. A radioassay for dihydropteroate-synthesizing enzyme activity. Anal Biochem. 1969;31:174–182. doi: 10.1016/0003-2697(69)90255-3. [DOI] [PubMed] [Google Scholar]
- 23.Pashley T V, Volpe F, Pudney M, Hyde J E, Sims P F G, Delves C J. Isolation and molecular characterization of the bifunctional hydroxymethyldihydropterin pyrophosphokinase-dihydropteroate synthase gene from Toxoplasma gondii. Mol Biochem Parasitol. 1997;86:37–47. [PubMed] [Google Scholar]
- 24.Paul, N., and K. Arata. 1993. A deadly duo—TB and AIDS. World Health July-August:7–9.
- 25.Radstrom P, Fermer C, Kristiansen B E, Jenkins A, Skold O, Swedberg G. Transformational exchanges in the dihydropteroate synthase gene of Neisseria meningitidis: a novel mechanism for acquisition of sulfonamide resistance. J Bacteriol. 1992;174:6386–6393. doi: 10.1128/jb.174.20.6386-6393.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ratledge C, Brown K A. Inhibition of mycobactin formation in Mycobacterium smegmatis by p-aminosalicylate. A new proposal for the mode of action of p-aminosalicylate. Am Rev Respir Dis. 1972;106:774–776. doi: 10.1164/arrd.1972.106.5.774. [DOI] [PubMed] [Google Scholar]
- 27.Read S M, Northcote D H. Minimization of variation in the response to different proteins of the Coomassie blue G dye-binding assay for protein. Anal Biochem. 1981;116:53–64. doi: 10.1016/0003-2697(81)90321-3. [DOI] [PubMed] [Google Scholar]
- 28.Richey D P, Brown G M. The biosynthesis of folic acid. IX. Purification and properties of the enzymes required for the formation of dihydropteroic acid. J Biol Chem. 1969;244:1582–1592. [PubMed] [Google Scholar]
- 29.Segel I H. Enzyme kinetics: behavior and analysis of steady-state and rapid equilibrium enzyme systems. New York, N.Y: Wiley-Interscience; 1975. [Google Scholar]
- 30.Shepard C C, Tolentino J G, McRae D H. The therapeutic effect of 4.4′-diacetyldiaminodiphenylsulfone (DADDS) in leprosy. Am J Trop Med Hyg. 1968;17:192–201. doi: 10.4269/ajtmh.1968.17.192. [DOI] [PubMed] [Google Scholar]
- 31.Shiota T, Disraely M N, McCann M P. The enzymatic synthesis of folate-like compounds from hydroxymethyldihydropteridine pyrophosphate. J Biol Chem. 1964;239:2259–2266. [PubMed] [Google Scholar]
- 32.Slock J, Stahly D P, Han C-Y, Six E W, Crawford I P. An apparent Bacillus subtilis folic acid biosynthesis operon containing pab, an amphibolic trpG gene, a third gene required for synthesis of para-aminobenzoic acid and the dihydropteroate synthase gene. J Bacteriol. 1990;172:7211–7226. doi: 10.1128/jb.172.12.7211-7226.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snow G A. Mycobactins: iron-chelating growth factors from mycobacteria. Bacteriol Rev. 1970;34:99–125. doi: 10.1128/br.34.2.99-125.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Talarico T L, Dev I K, Dallas W S, Ferone R, Ray P H. Purification and partial characterization of 7,8-dihydropteroate synthase from Escherichia coli MC4100. J Bacteriol. 1991;173:7029–7032. doi: 10.1128/jb.173.21.7029-7032.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Triglia T, Cowman A F. Primary structure and expression of the dihydropteroate synthase gene of Plasmodium falciparum. Proc Natl Acad Sci USA. 1994;91:7149–7153. doi: 10.1073/pnas.91.15.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsukamura M. Bacteriostatic effects of sulfadimethoxine and kitasamycin on Mycobacterium avium-M. intracellulare complex. Kekkaku. 1983;58:247–250. [PubMed] [Google Scholar]
- 37.Volpe F, Ballantine S P, Delves C J. The multifunctional folic acid synthesis fas gene of Pneumocystis carinii encodes dihydroneopterin aldolase, hydroxymethyldihydropterin pyrophosphokinase and dihydropteroate synthase. Eur J Biochem. 1993;216:449–458. doi: 10.1111/j.1432-1033.1993.tb18163.x. [DOI] [PubMed] [Google Scholar]
- 38.Volpe F, Dryer M, Scaife J G, Darby G, Stammers D K, Delves C J. The multifunctional folic acid synthesis fas gene of Pneumocystis carinii appears to encode dihydropteroate synthase and hydroxymethyldihydropterin pyrophosphokinase. Gene. 1992;112:213–218. doi: 10.1016/0378-1119(92)90378-3. [DOI] [PubMed] [Google Scholar]
- 39.Wallace R J, Jr, Nash D R, Steele L C, Steingrube V. Susceptibility testing of slowly growing mycobacteria by a microdilution MIC method with 7H9 broth. J Clin Microbiol. 1986;24:976–981. doi: 10.1128/jcm.24.6.976-981.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.World Health Organization. The World Health Report 1996—fighting disease, fostering development. World Health Forum. 1997;18:1–8. [PubMed] [Google Scholar]
- 41.Yajko D M, Madej J J, Lancaster M V, Sanders C A, Cawthon V L, Gee B, Babst A, Hadley W K. Colorimetric method for determining MICs of antimicrobial agents for Mycobacterium tuberculosis. J Clin Microbiol. 1995;33:2324–2327. doi: 10.1128/jcm.33.9.2324-2327.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]