Abstract
Gel mobility shift assays with His-tagged BldD isolated from Escherichia coli have illustrated that BldD is capable of specifically recognizing its own promoter region. DNase I and hydroxyl radical footprinting assays have served to delimit the BldD binding site, revealing that BldD recognizes and binds to a site just upstream from, and overlapping with, the −10 region of the promoter. How BldD binds to its promoter and the effect this binding has on the expression of BldD are discussed.
The life cycle of the multicellular bacterium Streptomyces coelicolor is complex by prokaryotic standards, progressing through a series of structurally and metabolically differentiated states. Morphological differentiation begins with a spore, which germinates through the outgrowth of vegetative hyphae and then produces aerial mycelium, upon which mature spores will develop (reviewed in references 2 and 5). This morphological differentiation has been found to be dependent on two main classes of genes: the bld genes, which are required for the erection of aerial hyphae (1, 7), and the whi genes, which are needed for the formation of mature spores (3). Physiological differentiation involves a shift from primary metabolism to secondary metabolism, where antibiotic production is observed. The developmental and metabolic cycles are not completely exclusive; the timing of aerial mycelium formation and that of antibiotic production coincide, suggesting that they may share regulatory elements. In support of this, it has been found that mutations in many of the bld genes not only result in the failure to form aerial hyphae but also adversely affect the production of secondary metabolites.
A number of the bld genes have been cloned, and of these, bldA, bldK, bldB, and bldD have been sequenced and at least partially characterized. It appears that while the bld genes have similar phenotypic characteristics, they play very different roles in the regulation of differentiation. bldA was found to encode a leucyl-tRNA, which is responsible for recognizing the rare UUA codon in mRNA (6), and bldK comprises a gene cluster coding for an oligopeptide permease (8). bldB and bldD encode small proteins possessing helix-turn-helix motifs, characteristic of DNA-binding proteins, and as such may function as transcription regulators (4, 9). To gain an understanding of how the bldD gene exerts its regulatory effects, the BldD protein was overexpressed and purified from Escherichia coli and then examined for its ability to bind DNA.
Overexpression and purification of BldD.
The bldD coding region was PCR amplified from chromosomal DNA of S. coelicolor J1501 and was cloned downstream of the six-His tag in the plasmid pQE9 (Qiagen) to yield pQE9BldD+. E. coli JM109 was transformed with pQE9BldD+, ampicillin-resistant transformants were selected, and the integrity of the recombinant plasmids was confirmed by DNA sequencing. Induction of the BldD fusion protein with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) resulted in the production of 50% soluble and 50% insoluble protein. The soluble protein fraction was then purified per the manufacturer’s recommendations (Qiagen).
DNA binding by BldD.
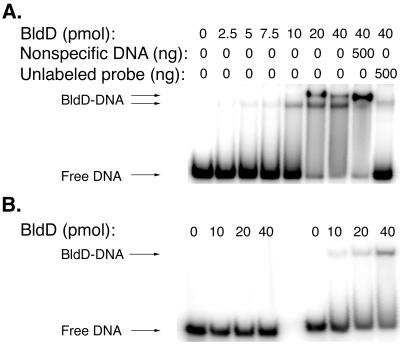
Previous analyses comparing mutant and wild-type bldD transcription had suggested that functional BldD may negatively regulate its own expression (4), so the bldD promoter region was selected as a target for the examination of BldD DNA-binding activity. A 257-bp probe (MAE11-12) (Table 1) was amplified by PCR, end labeled, and used to assay the binding of BldD at 30°C in a buffer consisting of 10 mM Tris-HCl (pH 7.8), 150 mM NaCl, 2 mM dithiothreitol, 1 μg of poly(dI-dC), and 10% glycerol. Two retarded fragments were seen when the DNA-protein complexes were run out on an 8% polyacrylamide gel (data not shown). The smaller of the two complexes was formed at low BldD concentrations and was followed by the appearance of a higher-molecular-weight complex as the amount of BldD was increased. The two retarded fragments were present in approximately equivalent amounts when maximal binding was attained, suggesting that there may be some equilibration between the two forms. Furthermore, the absence of a complete shift to the higher-molecular-weight complex suggested a single site for BldD binding on this 257-bp fragment. Attempts were then made to delimit the site recognized and bound by BldD, first using a 100-bp fragment (MAE1-4) and then using a 52-bp probe (MAE1-15) and a 77-bp probe (MAE16-4) (Table 1), overlapping by 22 bp, obtained by subdividing the 100-bp fragment. It was found that BldD was capable of binding to MAE16-4 (Fig. 1A) as strongly as to the original MAE11-12 fragment; however, there was absolutely no shift seen when the shorter MAE1-15 fragment was used as the binding target (Fig. 1B). Competition gel shift assays were also conducted, in which the addition of excess unlabeled, nonspecific DNA (BKL41-MAE5) (Table 1) from within the bldD coding sequence did not abolish binding by BldD and in which the addition of a 500-fold excess of unlabeled probe completely abolished BldD binding to the labeled fragments (Fig. 1A).
TABLE 1.
Oligonucleotides used for PCR amplification
| Strand and oligo- nucleotide | Sequence and positiona |
|---|---|
| Template | |
| BKL41 | 5′ (+460)CGCCGTCATCTACGACC(+477) |
| MAE1 | 5′ (−60)GGAAGAGTCGGTGCGGA(−44) |
| MAE6 | 5′ CGCGGATCC(+69)TCCAGCGAATACGCCAAAC(+87) |
| MAE11 | 5′ (−166)CGGTAGCAGGCTCACAG(−150) |
| MAE16 | 5′ (−31)ACGCAGCAGAGTAACGCTGCGTA(−9) |
| MAE18 | 5′ (−14)TGCGTAACCTCACAGTGAGTTACCAG(+12) |
| MAE36 | 5′ (−3)ACAGTGAGTTACCAGCCG(+15) |
| MAE46 | 5′ (−31)ACGCAGCAGCGTAACGCTGCG(−11) |
| MAE47 | 5′ (−31)ACGCAGCAGAGTTACGCTGCG(−11) |
| Nontemplate | |
| MAE4 | 5′ TCTAGA(+40)GCGGCAGGCTGTGTTGTC(+23) |
| MAE5 | 5′ GGTAAGCTT(+569)TCAGAGCTCGTCGTGGGAC(+551) |
| MAE12 | 5′ (+91)GAGCTGTTTGGCGTATTCG(+73) |
| MAE15 | 5′ (−9)TACGCAGCGTTACTCTGCTGCGT(−31) |
| MAE35 | 5′ (+2)ACTGTGAGGTTACGCAGC(−16) |
Oligonucleotide primer sequences and nucleotide locations (given in parentheses), relative to the bldD transcription start site. Nonhomologous extensions of the oligonucleotides are underlined.
FIG. 1.
Gel mobility shift assays of BldD binding to a 77-bp region (MAE16-4) of the bldD promoter. One to two nanograms of DNA end labeled with γ-32P was incubated with different amounts of BldD (0 to 40 pmol). The ability of BldD to shift its own promoter at increasing protein concentrations is seen in panel A. Specificity of binding is also illustrated in panel A by the addition of ∼500 ng of unlabeled, nonspecific competitor DNA (BKL41-MAE5) to the penultimate lane and ∼500 ng of unlabeled, specific competitor DNA (MAE16-4) to the last lane. Panel B illustrates the binding of BldD to MAE1-15 (left) and MAE18-4 (right) (1 to 2 ng of each probe) with increasing amounts of BldD (0 to 40 pmol).
The region contained within MAE16-4 was then further analyzed, and it was found that while the 52-bp MAE1-15 was not recognized by BldD (Fig. 1B), the addition of 9 bases to its 3′ end to yield MAE1-35 (Table 1; Fig. 2C) resulted in complete shifting (data not shown), indicating that these 9 bases are important for BldD binding. The region extending from MAE18-4 (Table 1; Fig. 2C), which included these 9 bases, showed defined but lower-affinity binding (Fig. 1B), suggesting that these 9 bases are required but are not sufficient for complete BldD binding. Gel shift experiments carried out with a fragment from MAE36-4 (Table 1) showed no shift (data not shown).
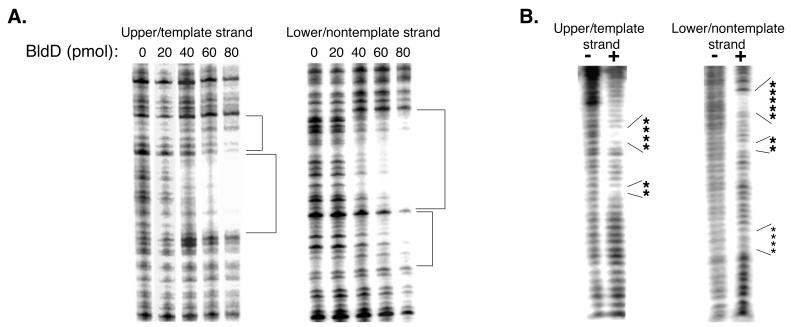
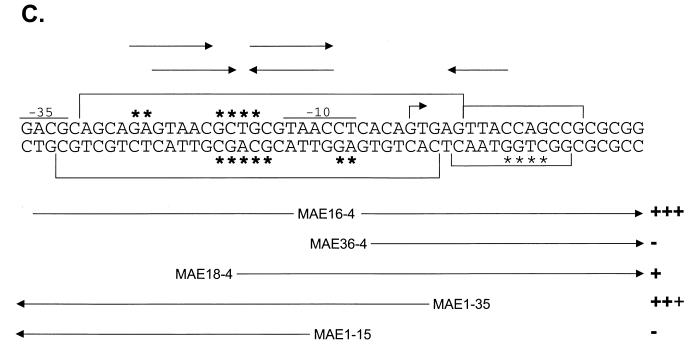
FIG. 2.
Localization of the BldD binding site. (A) DNase I footprints of the bldD promoter region, with different concentrations of BldD (0 to 80 pmol). Areas of strong protection are shown with large brackets, while weakly protected areas are indicated with small brackets. (B) Hydroxyl radical footprinting of the bldD promoter in the absence (−) or presence (+) of BldD (80 pmol). Large asterisks indicate strong protection by BldD, while small asterisks show sites of weak protection. (C) Summary of DNase I protection, hydroxyl radical protection, and gel mobility shift experiments. Areas protected from DNase I and the hydroxyl radical are as indicated in panels A and B. Fragments used for gel shift assays are outlined at the bottom, along with their relative abilities to shift DNA (+++, strong binding [100% bound]; +++, 90% bound; +, 20% bound; −, no binding). Potential BldD binding sites are indicated above the sequence, with the top set of arrows illustrating a direct repeat and the bottom set indicating an inverted repeat. Downstream of the transcription start site is a sequence that corresponds to a half site of the inverted repeat.
DNase I and hydroxyl radical footprinting experiments were carried out to more precisely localize the site of BldD interaction with its promoter. DNase I footprinting was conducted by using the 257-bp bldD promoter fragment (MAE11-12), which had been specifically labeled on either its upper or its lower strand. The reaction conditions were essentially the same as for the gel shift assays; however, the binding buffer was modified to include MgCl2 and CaCl2, and the salt concentration was changed to 50 mM KCl. In the presence of BldD, a single area spanning a 32-base segment of the DNA was found to be strongly protected from DNase I cleavage on both strands (Fig. 2A and C), and this area was found to encompass the bldD −10 promoter region and the transcription start site. A region of much weaker protection was also observed just downstream from the strongly protected area (Fig. 2A and C) on both strands.
A finer analysis of the BldD binding site with hydroxyl radical footprinting was then undertaken (12). The upper and lower strands of the 100-bp promoter fragment (MAE1-4; Table 1) were independently end labeled and were incubated in binding buffer, both alone and with BldD, in the absence of glycerol. Three microliters each of 0.5 mM Fe(EDTA)2−, 10 mM Na-ascorbate, and 0.3% H2O2 was added to each reaction mixture, and reactions were allowed to proceed for 2 min before being stopped with 5 μl of 0.1 M thiourea plus 3 μl of 100% glycerol. The resulting products were then applied to an 8% native polyacrylamide gel to separate any unbound probe from that bound by BldD, thereby reducing the background that would result from cleavage of the unbound fraction. The cleavage products were excised and purified per reference 10 and then electrophoresed on a 6% denaturing polyacrylamide gel. BldD was determined to afford strong protection to bases at positions −16 to −13 and −23 to −24 relative to the transcription start point (tsp). On the lower strand, BldD was seen to confer strong protection at positions −16 to −12 and −5 to −4 and much weaker protection at positions +9 to +12 (Fig. 2B and C). Taken together, the DNase I and hydroxyl radical footprinting results seem to suggest a site for BldD binding centered just upstream from the −10 region of the promoter. Examination of the sequence flanking this site has revealed an inverted repeat and a direct repeat (Fig. 2C) which overlap significantly and either of which may be the site recognized and bound by BldD. When the repeats were segmented and the gel shift assays were conducted, no binding was seen to the left half of the repeat, represented by MAE1-15 (Fig. 1B and 2C), while there was a defined single shifted band seen with MAE18-4 (Fig. 1B), which contained the entire right half of the repeat (Fig. 2C). This suggests that BldD binds preferentially to the right half of the repeat and then, after binding to the right, is capable of recognizing and binding to the left.
One possible interpretation of the presence of two shifted bands in the gel shift assay when both half sites are present is that the first shift represents binding to the higher-affinity right half site, and then a conformational change permits binding to the left half, which is seen as the second shift. Alternatively, the first shift may represent binding to the entire repeat, and upon the addition of more protein, some higher-order structure may be seen due to protein-protein interactions between excess BldD and BldD already complexed to the DNA. Finally, it appears that there is a single half site present just downstream from the bldD tsp (Fig. 2C), and it is possible that binding of BldD to the full inverted repeat may allow weak binding of BldD at this half site, either through protein-protein interactions or through a change in the DNA conformation. This would explain the weakly protected region observed immediately downstream from the tsp in the DNase I and hydroxyl radical footprints. Gel shift assays done by using a fragment with only this downstream region as the potential binding site (MAE36-4) showed no binding by BldD, which is not surprising if binding at the full repeat is required for stabilization of the weak binding downstream.
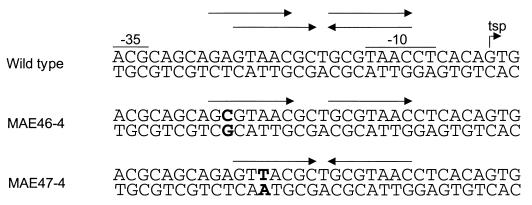
Attempts were made to clarify whether the inverted repeat or the direct repeat was the preferred binding motif by using mutagenic oligonucleotides. Perfect direct and indirect repeats were created (Fig. 3) and then subjected to gel shift assays; however, no difference from the results obtained with the wild-type fragment could be detected (data not shown). This suggests that subtle changes in the repeats cannot distinguish between binding to one or the other, although other factors suggest that it is the inverted repeat rather than the direct repeat that BldD recognizes. The pattern of protection observed with the hydroxyl radical footprinting showed that binding by BldD occurred symmetrically on both strands, supporting recognition of the inverted repeat. Prior analyses of the BldD sequence had revealed the presence of a C-terminal helix-turn-helix domain weakly similar to that of the LysR-like family of DNA-binding proteins (4). These LysR-like proteins recognize and bind to a consensus sequence containing a T-N11-A motif, usually within a 15-bp partially dyadic sequence (reviewed in reference 11). This would also be true for the BldD binding site if BldD bound to the imperfect inverted repeat, within which was contained a T-N11-A motif. It is possible that other genes controlled by BldD may possess sequences and/or spacings different from that of the bldD promoter, and the determination of a consensus binding sequence for BldD awaits their identification.
FIG. 3.
Mutagenesis of the bldD promoter region. The promoter region of the wild-type bldD gene is shown, together with portions of the fragments containing mutations within this region. MAE46-4 possesses a perfect direct repeat, and MAE47-4 has a perfect inverted repeat. The mutagenized bases are indicated in bold type.
Acknowledgments
This work was supported by the Alberta Heritage Foundation for Medical Research and the Natural Sciences and Engineering Research Council of Canada.
We thank Karen Anthony for her helpful advice regarding the overexpression and purification of BldD and for her gift of E. coli JM109, Gerald Stemke for his gift of pQE9, and Cyril Kay for his suggestions on how to maintain BldD in a soluble form. We are also very grateful for helpful discussions with Laura Frost, Linda Reha-Krantz, and Mark Glover and for critical reading of the manuscript by Laura Frost.
REFERENCES
- 1.Champness W C. New loci required for Streptomyces coelicolor morphological and physiological differentiation. J Bacteriol. 1988;170:1168–1174. doi: 10.1128/jb.170.3.1168-1174.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chater K F. Genetics of differentiation in Streptomyces. Annu Rev Microbiol. 1993;47:685–713. doi: 10.1146/annurev.mi.47.100193.003345. [DOI] [PubMed] [Google Scholar]
- 3.Chater K F. Taking a genetic scalpel to the Streptomyces colony. Microbiology. 1998;144:1465–1478. doi: 10.1099/00221287-144-6-1465. [DOI] [PubMed] [Google Scholar]
- 4.Elliot M, Damji F, Passantino R, Chater K, Leskiw B. The bldD gene of Streptomyces coelicolor A3(2): a regulatory gene involved in morphogenesis and antibiotic production. J Bacteriol. 1998;180:1549–1555. doi: 10.1128/jb.180.6.1549-1555.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kelemen G H, Buttner M J. Initiation of aerial mycelium formation in Streptomyces. Curr Opin Microbiol. 1998;1:656–662. doi: 10.1016/s1369-5274(98)80111-2. [DOI] [PubMed] [Google Scholar]
- 6.Lawlor E J, Baylis H A, Chater K F. Pleiotropic morphological and antibiotic deficiencies result from mutations in a gene encoding a tRNA-like product in Streptomyces coelicolor A3(2) Genes Dev. 1987;1:1305–1310. doi: 10.1101/gad.1.10.1305. [DOI] [PubMed] [Google Scholar]
- 7.Merrick M J. A morphological and genetic mapping study of bald colony mutants of Streptomyces coelicolor. J Gen Microbiol. 1976;96:299–315. doi: 10.1099/00221287-96-2-299. [DOI] [PubMed] [Google Scholar]
- 8.Nodwell J R, McGovern K, Losick R. An oligopeptide permease responsible for the import of an extracellular signal governing aerial mycelium formation in Streptomyces coelicolor. Mol Microbiol. 1996;22:881–893. doi: 10.1046/j.1365-2958.1996.01540.x. [DOI] [PubMed] [Google Scholar]
- 9.Pope M K, Green B, Westpheling J. The bldB gene encodes a small protein required for morphogenesis, antibiotic production, and catabolite control in Streptomyces coelicolor. J Bacteriol. 1998;180:1556–1562. doi: 10.1128/jb.180.6.1556-1562.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 11.Schell M A. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 12.Tullius T D, Dombroski B A. Hydroxyl radical “footprinting”: high-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein. Proc Natl Acad Sci USA. 1986;83:5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]