Abstract
IntI1 integrase is a member of the prokaryotic DNA integrase superfamily. It is responsible for mobility of antibiotic resistance cassettes found in integrons. IntI1 protein, as well as IntI1-COOH, a truncated form containing its carboxy-terminal domain, has been purified. Electrophoretic mobility shift assays were carried out to study the ability of IntI1 to bind the integrase primary target sites attI and aadA1 attC. When using double-stranded DNA as a substrate, we observed IntI1 binding to attI but not to attC. IntI1-COOH did not bind either attI or attC, indicating that the N-terminal domain of IntI1 was required for binding to double-stranded attI. On the other hand, when we used single-stranded (ss) DNA substrates, IntI1 bound strongly and specifically to ss attC DNA. Binding was strand specific, since only the bottom DNA strand was bound. Protein IntI1-COOH bound ss attC as well as did the complete integrase, indicating that the ability of the protein to bind ss aadA1 attC was contained in the region between amino acids 109 and 337 of IntI1. Binding to ss attI DNA by the integrase, but not by IntI1-COOH, was also observed and was specific for the attI bottom strand, indicating similar capabilities of IntI1 for binding attI DNA in either double-stranded or ss conformation. Footprinting analysis showed that IntI1 protected at least 40 bases of aadA1 attC against DNase I attack. The protected sequence contained two of the four previously proposed IntI1 DNA binding sites, including the crossover site. Preferential ssDNA binding can be a significant activity of IntI1 integrase, which suggests the utilization of extruded cruciforms in the reaction mechanisms leading to cassette excision and integration.
Since the introduction of antibiotics in the treatment of human infectious diseases, antibiotic resistance has spread quickly and dramatically among bacteria. Such explosive dissemination is a consequence of the mobility of the antibiotic-resistance-encoding genes, which are usually found in plasmids, transposons, and integrons (12, 27). Integrons are a class of site-specific recombination elements which consist of two conserved sequence regions (5′ and 3′ conserved regions), flanking a central variable region (Fig. 1), in which antibiotic resistance genes are found organized as resistance cassettes (14). The recombinative activity of integrons resides in an enzyme, the integrase, belonging to the broad family of phage integrases (1, 6). The integrase gene is part of the integron 5′ conserved region (19, 20). The integrase acts mainly at two sites in DNA. The attI site is close to the integrase gene. It signals the boundary of the integron 5′ conserved region (15). The attC sites flank each of the inserted resistance cassettes in the central variable region of the integron (13). While integron attI sites are well conserved in type I integrons, attC sites, also called 59-bp elements (13) and recombination hot spots (20), are very heterogeneous in sequence and length. In the case of class I integrons, recombination can occur across any pair of att sites, but each att site is used at widely different frequencies (7, 15, 19). attI and attC can be considered IntI1 primary sites. Besides, IntI1 can act on secondary sites. The secondary sites consist of a degenerate pentanucleotide with the sequence GWTMW (8). Integration of gene cassettes into these secondary sites in naturally occurring plasmids in vivo has been described elsewhere (22, 25). Therefore, IntI1 seems to be quite flexible in the sequence that it requires to carry out its site-specific recombination. IntI1-mediated recombination across attI and attC sites results in mobility of preexisting resistance cassettes among integrons. Cassettes can be excised from integrons and be reintegrated by attC-attI or attC-attC site-specific recombination (3, 14). Recombination at secondary sites may be important in the recruitment of chromosomal genes into cassettes, allowing the flux of chromosomal genes into mobile genetic elements (8, 9).
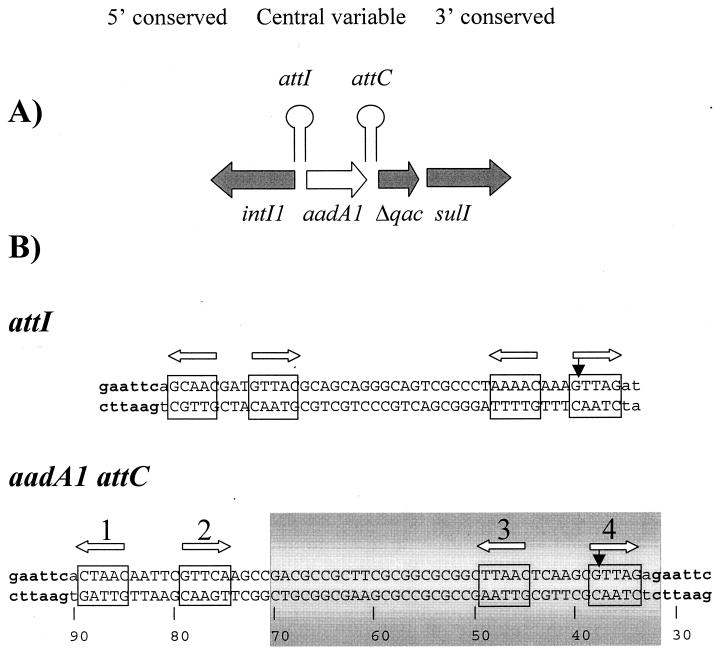
FIG. 1.
(A) Simplified diagram of the type I integron in Tn21 to illustrate integron organization and location of the att sites. (B) Nucleotide sequence of the IntI1 primary sites (attI and attC) in plasmids pSU18R2 and pSU18R3. The DNA sequences show the GTTAG crossover site at the right (3′) end. This orientation defines the top and bottom nomenclature for the ssDNAs. Filled vertical arrows indicate the crossover sites, and open horizontal arrows point to the four putative pentanucleotide components (boxed and numbered) of each att site. The numbers below the bottom strand of the aadA1 attC site start at the beginning of the reverse M13 priming site of the pSU18 vector sequence and correspond to numbers in Fig. 6. The shaded area indicates the region protected by IntI1 from DNase I attack.
Evidence from in vivo experiments indicates that IntI1 primary sites may be seen as a composite of four single sites organized as two inverted pairs at each att end and a central sequence with some degree of secondary structure (7, 26). The single sites could be the basic unit for integrase-DNA interaction. Some authors have identified this single site with a degenerate 7-bp site, GTTRRRY, also called a core site (26). GWTMW pentanucleotides have also been proposed as the single integrase binding sites (7, 9). The purification and DNA binding properties of IntI1 have been recently reported (4, 10, 11). The results available indicated that the integrase bound efficiently to the attI site in vitro but that no binding to attC was observed. Regarding the integrase binding sites, in one study (4), binding to attI was in a region quite separate from the known integrase crossover site. In another study (11), four clear-cut binding sites were reported, two of them, I and II, flanking the crossover site while the other two, III and IV, were located three and five helix turns, respectively, to the left of the crossover. Site III could be the same strong binding site described in reference 4. Furthermore, sites I, II, and III overlap three of the pentanucleotides proposed to be part of IntI1 primary sites. In spite of these reports, we are still far from understanding the biochemical details of IntI1-mediated site-specific reaction, and even some basic aspects such as IntI1 interaction with attC are obscure. In this work, we used an alternative approach to purify IntI as well as a truncated form containing the IntI1 carboxy-terminal domain (IntI1-COOH) and used these proteins to study the binding to Int primary sites in both double-stranded (ds) and single-stranded (ss) forms.
Purification of native IntI1 and IntI1-COOH.
A DNA fragment containing a complete intI1 gene was amplified by PCR from plasmid pSU2056 (19) by using the primers PETInt (CCCCATATGAAAACCGCCACTGCGCC) and universal M13, digested with NdeI and BamHI, and cloned into the same sites of the pET3a plasmid (23) as described in reference 24. In the resulting plasmid, called pET3aInt, the IntI1 open reading frame was expressed from the T7 promoter (28). The in vivo activity of the integrase from plasmid pET3aInt was checked in a conduction assay as described in reference 19. Plasmid pSU18R2, which contained the site aadA1 attC, formed site-specific cointegrates with plasmid R388 (5) at a frequency of 10−2, indicating that the integrase produced by the plasmid pET3aInt was active in vivo. Overexpression of the IntI1 protein from pET3aInt was assayed in five different Escherichia coli BL21 derivatives, although we obtained overexpression only in strain C41 (21). Following IPTG (isopropyl-β-d-thiogalactopyranoside) induction, an overexpressed band of approximately 38 kDa (calculated size, 38,357 Da) was visualized on sodium dodecyl sulfate (SDS) gels. Most of the IntI1 protein produced in strain C41 grown at 37°C was present in the form of insoluble inclusion bodies. These inclusion bodies were used as a source of IntI1 protein to obtain IntI1-specific rabbit antiserum as described in reference 16. Soluble IntI1 protein could be obtained by growing and inducing the cultures at 25°C (21, 28). E. coli C41 cells containing plasmid pET3aInt were grown in 50 ml of Luria-Bertani medium (containing ampicillin [100 μg ml−1]) with shaking at 25°C to an optical density of 0.4 at 600 nm. Expression of the IntI1 protein was induced by addition of IPTG to 1.0 mM for 4 h at the same temperature (28). Cells were harvested by centrifugation and resuspended in 5 ml of 50 mM Tris (pH 7.5)–5 mM EDTA–5% sucrose–0.05 mg of lysozyme ml−1. After 20 min at room temperature, the cells were pelleted and resuspended in 5 ml of high-salt lysis buffer (50 mM Tris [pH 7.5], 500 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol [DTT], 0.25% Triton X-100). After a 15-min incubation at 4°C, the cells were disrupted by sonication. The cell lysates were centrifuged at 18,000 × g for 20 min at 4°C. By this method, the majority of the integrase produced was recovered in soluble form; however, it did not bind to any of the commonly used ion-exchange media (phosphocellulose, carboxymethyl cellulose, SP-Sepharose, DEAE-cellulose, or Mono Q-Sepharose [Amersham-Pharmacia Biotech]) when assayed with different buffers and pH conditions. When the lysis of the cells was done in low-salt lysis buffer containing 50 mM NaCl, most of the IntI1 protein was found in the pellet after a low-speed centrifugation step. Thus, we decided to purify the IntI1 protein from this pellet. The pellet was washed with 2.5 ml of lysis buffer containing 1% Triton X-100 for 15 min at 4°C, centrifuged as before, and washed again for 15 min at 4°C with 2.5 ml of lysis buffer containing 2 M NaCl. The washed pellet was resuspended in 2.5 ml of 50 mM Tris (pH 7.5)–1 mM EDTA–1 mM DTT–1 M NaCl–10 mM CHAPS (3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate) and centrifuged at 100,000 × g for 15 min at 4°C to eliminate insoluble material. The IntI1 protein thus purified was more than 80% homogeneous as judged by SDS-polyacrylamide gel electrophoresis. Only the major protein band showed a reaction with polyclonal antibody against the IntI1 integrase in Western blot assays (Fig. 2).
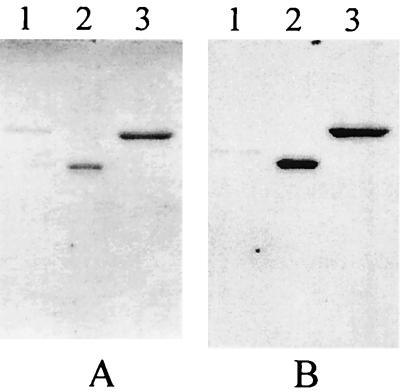
FIG. 2.
Western blot analysis of the IntI1 protein purified fractions. (A) Coomassie blue-stained polyacrylamide-SDS gel. (B) Western blot of a similar gel immunoprobed with the anti-IntI1 antibody. Lanes 1, control strain extract; lanes 2, purified IntI1-COOH protein; lanes 3, purified IntI1 protein.
To obtain a truncated form of IntI1 similar to the C170 carboxy-terminal domain of λ integrase, which contains most of the protein catalytic functions (29), we used a structure-based alignment to locate the equivalent to λ integrase helix F in IntI1 (18). This α-helix stretch is at the beginning of the C170 carboxy-terminal domain of λ integrase. In this way, we selected the primer PETCOOH (CCCCATATGCCGTCGCGGCGCTTGCC) and used it along with the universal M13 primer to amplify by PCR an 862-bp DNA fragment from plasmid pSU2056 corresponding to the carboxy-terminal domain of IntI1. This PCR fragment was cloned as described above into the expression vector pET3a (23) to yield the plasmid pET3aCOOH. The cloned region lacked 107 amino acids of the amino-terminal half of IntI1 and presumably contained all the structurally conserved elements present in the carboxy-terminal domain of members of the Int family (1, 6). The IntI1-COOH protein was overexpressed in the strain BL21(DE3) derivative C41 (21). E. coli C41(pET3aCOOH) cells were grown at 37°C in 250 ml of Luria-Bertani medium (containing ampicillin [100 μg ml−1]) to an optical density of 0.4 at 600 nm and induced with IPTG as described above. A protein with an apparent molecular mass of 26 kDa on SDS gels was overexpressed (predicted size, 25,493 Da). Cultures were lysed as described above. After sonication, the cell debris was removed and the supernatants were centrifuged at 100,000 × g for 15 min at 4°C. The clarified supernatant was applied to a phosphocellulose P11 (Whatman) column equilibrated in lysis buffer without Triton X-100. The protein bound to the phosphocellulose was washed with the same buffer containing 0.2 M NaCl. A linear gradient of NaCl (0.3 to 0.5M) in the same buffer was applied to the column. The fractions containing the IntI1-COOH protein were pooled and concentrated 50 times by ultrafiltration with Centricon 10 filters, before being applied to a high-resolution gel filtration column (Superdex 75-HR; Amersham-Pharmacia Biotech). The fractions containing the IntI1-COOH protein were again pooled and stored. The purified Int-COOH polypeptide showed a size of 46 kDa in the gel filtration column, suggesting that in solution it behaved as a dimer. Only the major protein band in the preparation, corresponding to the IntI1-COOH protein, reacted with anti-Int antibody in Western blots (Fig. 2).
IntI1 protein binds specifically to ss att sites.
To determine the DNA binding specificity of purified IntI1, we studied its behavior by gel mobility shift assays with two different DNA fragments representing the two different IntI1 primary sites: a fragment containing aadA1 attC from In2 and the other containing the attI site. As a source of aadA1 attC DNA, we constructed plasmid pSU18R2 by cloning a PCR product obtained from plasmid pSU2068 (19) into the pSU18 vector (2) by using primers R1 (GGGAATTCTCTAACAATTCGTTCAAGCCGACGCC) and R2 (GGGAATTCTCTAACGCTTGAGTTAAGCCGCGCC). Similarly, we obtained plasmid pSU18R3 as a source of attI by cloning a PCR product from the incW plasmid R388 (5), by using the primers RHS31 (GGGAATTCAGCAACGATGTTACGCA) and RHS32 (GGGCATGCCTAACTTTGTTTTAGGG). The DNA sequence of interest in plasmids pSU18R2 and pSU18R3 is shown in Fig. 1. dsDNA fragments for binding assays were generated by PCR with the plasmids pSU18R2 and pSU18R3 as templates and the oligonucleotides M13 and rM13 as primers. The PCR products were labeled with [α-32P]dCTP (Amersham) and separated in an agarose gel, and excised bands were eluted with the QiaQuick gel extraction kit (Qiagen). Labeled purified DNA fragments (1 pmol) were incubated with either IntI1, IntI1-COOH, or control extracts for 15 min at 30°C in a 20-μl final volume containing 50 mM Tris (pH 7.5), 100 mM NaCl, 1 mM CHAPS, 0.2 mM EDTA, 5% glycerol, 1 mM DTT, 1.5 μg of poly(dI-dC) DNA, and 0.7 μg of bovine serum albumin. After this incubation period, the binding reaction mixtures were electrophoresed at room temperature in 5% polyacrylamide gels in 0.5× Tris-borate-EDTA buffer. When the purified IntI1 protein was added to the 112-bp DNA fragment containing attI, the mobility of the fragment was specifically lowered, indicating IntI1 binding to this sequence (Fig. 3). On the other hand, no retardation was observed with the 167-bp aadA1 attC fragment. The purified IntI1-COOH protein was unable to bind to either attI or attC dsDNA (Fig. 3). IntI1 binding to attI but not to attC was expected from previous publications (4, 11), although it was surprising, taking into account that IntI1 from the pET3aInt plasmid exhibited a high in vivo recombination activity at attC. Trying to find an explanation for this apparent paradox, we thought that IntI1 could bind to DNA forms different from the standard double helix. To investigate this possibility, we decided to perform gel retardation assays with ssDNA corresponding to aadA1 attC. To obtain ssDNA fragments, an asymmetric PCR in which the primer for the desired strand was 10 times more concentrated than the other was used. After amplification, two products were observed on agarose gels. One of them showed the mobility of the dsDNA product. The other was the ssDNA strand as corroborated by susceptibility to S1 nuclease, resistance to restriction endonucleases, hybridization in native Southern blots (17), and DNA sequencing. ssDNA fragments were radiolabeled at the 5′ end with T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP. Labeled purified ssDNA was used in band shift assays in the conditions described above. We labeled and purified each of the two strands of attC and incubated them with IntI1. As shown in the left panel of Fig. 4, IntI1 specifically retarded the bottom strand of aadA1 attC. This retardation was also observed with IntI1-COOH but not when control extracts were tested. To confirm that the protein responsible for this band shift was really IntI1, Int proteins were synthesized by using an in vitro transcription and translation reticulocyte system (TNT T7 reticulocyte lysate system [Promega]), in the presence of [35S]methionine. In vitro-synthesized IntI1 exhibited the same site-specific ssDNA binding activity as the protein overproduced in E. coli C41 (data not shown). As the IntI1 protein produced in the reticulocyte lysate is free of other E. coli proteins, we can conclude that IntI1 was responsible for the binding to the aadA1 attC bottom strand. Furthermore, since the IntI1-COOH protein also exhibited the same ssDNA binding activity, it can be deduced that the ability of IntI1 to bind attC ssDNA resided in its C-terminal domain. To demonstrate the specificity of IntI1 binding to ssDNA, competition experiments were carried out with an excess of nonlabeled identical DNA (Fig. 5). This resulted in partial inhibition of the IntI1 binding. A more-than-40-fold excess of competitor DNA was necessary to completely displace the labeled ssDNA from the complexes (Fig. 5, lanes 3 to 6). Addition of an equimolar or a twofold excess of the nonlabeled opposite single strand also diminished the formation of specific retardation bands, due to the production of the dsDNA form by annealing of the two complementary ssDNAs (Fig. 5, lanes 7 and 8). On the other hand, binding to attC was not abolished when unrelated competitor DNA was added (Fig. 5, lanes 9 and 10). These results support the idea that IntI1 binds to ssDNA in a sequence-specific manner. Furthermore, the addition of polyclonal anti-IntI1 antibody to the binding reaction supershifted the IntI1-DNA complex, and a large proportion of the radioactivity remained in the well of the gel, presumably because the complex of DNA with the integrase and the antibody could not enter the gel matrix (Fig. 5, lanes 11 and 12). This effect was not observed with preimmune serum (Fig. 5, lane 13). It is interesting to note that the antibody did not inhibit IntI1 DNA-binding function.
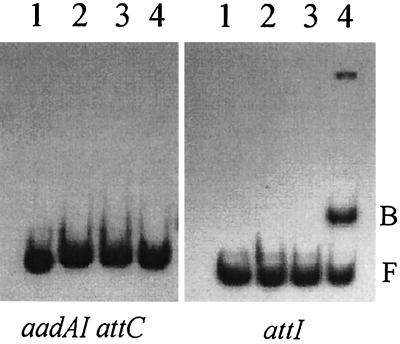
FIG. 3.
Gel mobility shift assays with dsDNA fragments containing aadA1 attC (left) or the attI site (right). Lanes 1, control dsDNA; lanes 2, dsDNA plus E. coli C41 control extract; lanes 3, dsDNA plus pure IntI1-COOH; lanes 4, dsDNA plus purified IntI1. F, free dsDNA; B, DNA-protein complex.
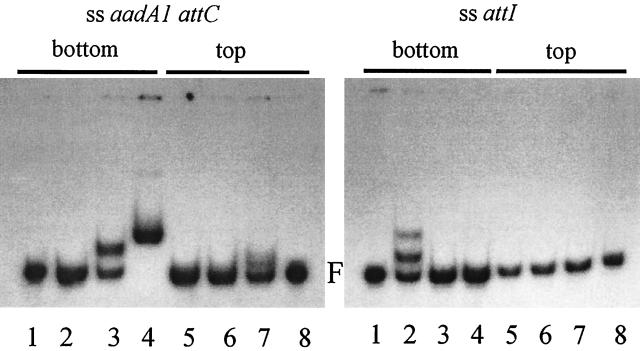
FIG. 4.
Gel retardation assays with ssDNA fragments containing either the top or the bottom strands of the aadA1 attC site (left) or the attI site (right). The substrate DNA used is indicated at the top of the figure. Protein extracts in each lane are as follows: left panel, lane 1, control DNA; lane 2, E. coli C41 control extract; lane 3, IntI1-COOH; lane 4, IntI1; lane 5 E. coli C41 control extract; lane 6, IntI1-COOH; lane 7, IntI1; lane 8, control DNA; right panel, lane 1, DNA alone; lane 2, purified IntI1; lane 3, IntI1-COOH; lane 4, E. coli C41 control extract; lane 5, IntI1; lane 6, IntI1-COOH; lane 7, E. coli C41 control extract; lane 8, control DNA. F, free DNA.
FIG. 5.
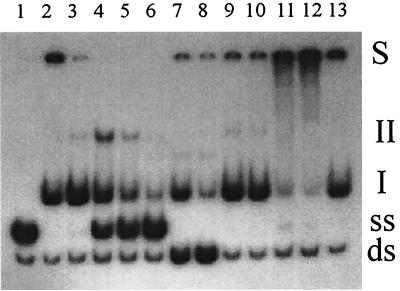
Competition of IntI1 binding to the bottom strand of the aadA1 attC site and supershift experiments with anti-IntI1 antibody. Lane 1, control DNA; lane 2, IntI1 control binding reaction; lanes 3 to 6, competition by the unlabeled attC bottom strand; addition of 2-, 10-, 20-, and 40-fold excess unlabeled bottom-strand aadA1 attC, respectively; lanes 7 and 8, competition by attC top strand; addition of an identical (lane 7) or twofold (lane 8) excess amount of the top attC aadA1 strand; lanes 9 and 10, heterologous DNA competition; addition of an identical (lane 9) or twofold (lane 10) excess amount of the unrelated competitor (an oligonucleotide mix); lanes 11 to 13, supershift assays with specific antisera; lane 11, incubation of IntI1 with polyclonal anti-IntI1 antibody before binding reaction; lane 12, incubation with polyclonal anti-IntI1 antibody after binding reaction; lane 13, incubation with preimmune serum after binding reaction. ds, dsDNA; ss, ssDNA; I and II, different protein-DNA complexes.
In addition to aadA1 attC, IntI1 binding to a different attC site, aadB attC from the plasmid pRAY (25), was also analyzed. Purified IntI1 was able to bind to the bottom strand of aadB attC but not to the ds form (data not shown).
IntI1 could also recognize and bind to the attI site in ss form (Fig. 4, right), but the proportion of the total DNA retarded was less than that for ss aadA1 attC. IntI1 binding to ss attI showed the same strand specificity observed for the attC site. Only the attI bottom strand was significantly bound to the integrase. This binding was not observed with IntI1-COOH. From these results, we can reasonably conclude that IntI1 bound both ds and ss attI but that only ss attC was efficiently bound by IntI1 in vitro. The binding to both ss primary sites was specific for the bottom DNA strand. This strand specificity is difficult to interpret; however, it could be related to the peculiar asymmetry of the IntI1 primary sites which show the crossover position being near the 3′ end. In previously published works (4, 10, 11), the DNA used for the binding experiments was linearly ds. In these conditions, IntI1 binds only to attI. The use of ss substrates allowed us to detect binding to attC, an activity that was necessary to explain IntI1 activity. Our results combine well with in vivo observations, suggesting that IntI1 may bind to attI and attC in different ways. It could be that attI is recognized and bound by IntI1 in ds form but that attC is more easily used if it is denatured than if it is present in ds form. Binding of enzymes in the integrase family to ssDNA has been described previously. Both the Flp and Cre proteins are capable of binding ssDNA. However, in the case of Flp, the sequence of the ssDNA binding site was different from that of the ds Flp recognition target, probably suggesting that this capability is unrelated to the site-specific recombination (30). In the case of IntI1, the ssDNA binding site included the recombination crossover point, indicating a relationship with the recombining activity and probably revealing some structural requirement of the target DNAs as discussed below.
Footprinting analysis of IntI1-aadA1 binding.
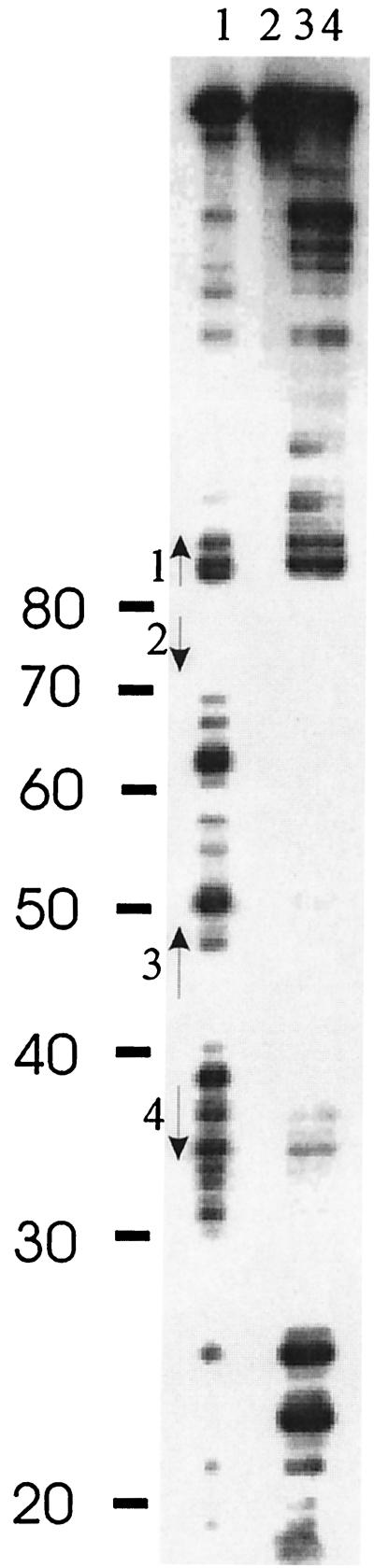
DNase I protection experiments were used to localize the precise IntI1 binding sites on the attC ssDNA fragment. Two picomoles of 5′-end-labeled ssDNA aliquots was incubated with 0.1 and 0.5 μg of purified IntI1, in a buffer containing 50 mM Tris (pH 7.5), 100 mM NaCl, 5 mM MgCl2, 1 mM CaCl2, 5% glycerol, 1 mM DTT, 0.2 mM EDTA, and 1.5 μg of poly(dI-dC) DNA in a 20-μl final volume, for 15 to 20 min at 30°C. Then, 10−1 DNase I Kunitz units was added to each binding reaction mixture and incubated for 15 min at 30°C. The resulting products were separated on 6% polyacrylamide sequencing gels and visualized by autoradiography (Fig. 6). About 40 bases, including most of the attC sequence, were protected. Some faint bands were observed, probably indicating sites of incomplete protection, and some new DNase I-hypersensitive sites appeared close to the 5′ end of the protected fragment. Protection was essentially complete between nucleotides 31 and 70 (Fig. 1 and 6), possibly indicating that the DNA was so covered by protein that DNase I had no access to it. The protected region included two of the four putative single IntI1 binding sites proposed previously (7, 26), considered either pentanucleotides (GWTWM) or heptanucleotides (GTTRRRY). The two sites in the 3′ end of attC (pentanucleotides 3 and 4 [Fig. 1]) were completely protected. Pentanucleotide 1 was out of the protected region, and the condition of pentanucleotide 2 is unknown due to the the lack of DNase I-hypersensitive sites in this region. If we accept the structural similarities between attI and attC sites, we can try to compare these results with the data available for IntI1 binding to attI (4, 11). Gravel et al. showed that three single sites defined as GTTR were found in the part of attI protected by IntI1 (11). These sites are equivalent to pentanucleotides 2, 3, and 4 of aadA1 attC. Collis et al. (4) showed protection of a single site, GTTACGC, corresponding to pentanucleotide 2. Taking this data into account, we could assume that pentanucleotide 2 of attC is also protected, even when we cannot draw this conclusion from our results.
FIG. 6.
DNase I footprinting analysis of the aadA1 attC bottom strand. The ssDNA was labeled at the 5′ end, incubated with purified IntI1 protein, subjected to limited cleavage by DNase I, and electrophoresed on a 6% sequencing gel. Lane 1, DNase I cleavage pattern of the DNA alone; lane 2, binding reaction without DNase I treatment; lane 3, DNase I cleavage pattern of the DNA incubated with 0.1 μg of IntI1; lane 4, DNase I cleavage pattern of the DNA incubated with 0.5 μg of IntI1. Nucleotide numbering starts from the 5′ end of the labeled fragment, as indicated in Fig. 1. Locations of the pentanucleotides previously proposed as IntI1 binding sites are shown by vertical arrows and numbered as in Fig. 1.
An interesting property of IntI1 primary sites is that they are large palindromic sequences, and they could be denatured or extruded as cruciforms from supercoiled DNA molecules. The ssDNA could equally fold back as hairpins in our in vitro binding assays. This property could justify the IntI1 capability of binding ssDNA. Furthermore, processes that stabilize cruciforms or involve transient denaturation of DNA, such as replication or transcription, could be favorable for IntI1-mediated reactions, especially at sites with poor sequence symmetry (9). More speculative interpretations of the observed ssDNA binding ability are possible, especially in the context of the already-proposed theory of an RNA origin for the cassettes. In this scenario, IntI1 should be able to bind to RNA or to ssDNA as a preliminary step for integration. Specific binding of Int to ssDNA, or to RNA, could also be important in regulation of integron gene expression.
Finally, we should mention that, in spite of the availability of IntI1 purified protein, we have been unable to reproduce any biochemical activity of IntI1 related to recombination. We have assayed for DNA nicking and topoisomerase activities of IntI1 with negative results. This suggests that the Int-DNA complexes described so far seem to be incomplete. Accordingly, we should wait until some activity is reproduced in vitro before drawing conclusions on the molecular mechanism of IntI1-mediated recombination.
Acknowledgments
This work was supported by a grant to J.M.G.L. from the Spanish “Fondo de Investigación Sanitaria.”
We are grateful to J. E. Walker for the E. coli BL21 derivatives and to Don B. Clewell for critical reading of the manuscript.
REFERENCES
- 1.Argos P, Landy A, Abremski K, Egan J B, Haggard-Ljungquist E, Hoess R H, Kahn M L, Kalionis B, Narayana S V, Pierson III L S, Stenberg N, Leong J M. The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 1986;5:433–440. doi: 10.1002/j.1460-2075.1986.tb04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartolomé B, Jubete Y, Martínez E, de la Cruz F. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene. 1991;102:75–78. doi: 10.1016/0378-1119(91)90541-i. [DOI] [PubMed] [Google Scholar]
- 3.Collis C M, Grammaticopoulos G, Briton J, Stokes H W, Hall R M. Site-specific insertion of gene cassettes into integrons. Mol Microbiol. 1993;9:41–52. doi: 10.1111/j.1365-2958.1993.tb01667.x. [DOI] [PubMed] [Google Scholar]
- 4.Collis C M, Kim M J, Stokes H W, Hall R M. Binding of the purified integron DNA integrase IntI1 to integron- and cassette-associated recombination sites. Mol Microbiol. 1998;29:477–490. doi: 10.1046/j.1365-2958.1998.00936.x. [DOI] [PubMed] [Google Scholar]
- 5.Datta N, Hedges R W. Trimethoprim resistance conferred by W plasmids in Enterobacteriaceae. J Gen Microbiol. 1972;72:349–355. doi: 10.1099/00221287-72-2-349. [DOI] [PubMed] [Google Scholar]
- 6.Esposito D, Scocca J J. The integrase family of tyrosine recombinases: evolution of a conserved active site domain. Nucleic Acids Res. 1997;25:3605–3614. doi: 10.1093/nar/25.18.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Francia M V, Avila P, de la Cruz F, García Lobo J M. A hot spot in plasmid F for site-specific recombination mediated by Tn21 integron integrase. J Bacteriol. 1997;179:4419–4425. doi: 10.1128/jb.179.13.4419-4425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Francia M V, de la Cruz F, García Lobo J M. Secondary-sites for integration mediated by the Tn21 integrase. Mol Microbiol. 1993;10:823–828. doi: 10.1111/j.1365-2958.1993.tb00952.x. [DOI] [PubMed] [Google Scholar]
- 9.Francia M V, García Lobo J M. Gene integration in the Escherichia coli chromosome mediated by Tn21 integrase (Int21) J Bacteriol. 1996;178:894–898. doi: 10.1128/jb.178.3.894-898.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gravel A, Messier N, Roy P H. Point mutations in the integron integrase IntI1 that affect recombination and/or substrate recognition. J Bacteriol. 1998;180:5437–5442. doi: 10.1128/jb.180.20.5437-5442.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gravel A, Fournier B, Roy P H. DNA complexes obtained with the integron integrase IntI1 at the attI site. Nucleic Acids Res. 1998;26:4347–4355. doi: 10.1093/nar/26.19.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grinsted J, de la Cruz F, Schmitt R. The Tn21 subgroup of bacterial transposable elements. Plasmid. 1990;24:163–189. doi: 10.1016/0147-619x(90)90001-s. [DOI] [PubMed] [Google Scholar]
- 13.Hall R M, Brookes D E, Stokes H W. Site-specific insertion of genes into integrons: role of the 59-base element and determination of the recombination cross-over point. Mol Microbiol. 1991;5:1941–1959. doi: 10.1111/j.1365-2958.1991.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 14.Hall R M, Collis C M. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol Microbiol. 1995;15:593–600. doi: 10.1111/j.1365-2958.1995.tb02368.x. [DOI] [PubMed] [Google Scholar]
- 15.Hansson K, Skold O, Sundstrom L. Non-palindromic attI sites of integrons are capable of site-specific recombination with one another and with secondary targets. Mol Microbiol. 1997;26:441–453. doi: 10.1046/j.1365-2958.1997.5401964.x. [DOI] [PubMed] [Google Scholar]
- 16.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 17.Hill S A, Stahl M M, Stahl F W. Single-strand DNA intermediates in phage lambda’s Red recombination pathway. Proc Natl Acad Sci USA. 1997;94:2951–2956. doi: 10.1073/pnas.94.7.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon H J, Tirumalai R, Landy A, Ellenberger T. Flexibility in DNA recombination: structure of the lambda integrase catalytic core. Science. 1997;276:126–131. doi: 10.1126/science.276.5309.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martínez E, de la Cruz F. Genetic elements involved in Tn21 site-specific integration, a novel mechanism for the dissemination of antibiotic resistance genes. EMBO J. 1990;9:1275–1281. doi: 10.1002/j.1460-2075.1990.tb08236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martínez E, de la Cruz F. Transposon Tn21 encodes a RecA-independent site-specific integration system. Mol Gen Genet. 1988;211:320–325. doi: 10.1007/BF00330610. [DOI] [PubMed] [Google Scholar]
- 21.Miroux B, Walker J E. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 22.Recchia G D, Hall R M. Plasmid evolution by acquisition of mobile gene cassettes: plasmid pIE723 contains the aadB gene cassette precisely inserted at a secondary site in the incQ plasmid RSF1010. Mol Microbiol. 1995;15:179–187. doi: 10.1111/j.1365-2958.1995.tb02232.x. [DOI] [PubMed] [Google Scholar]
- 23.Rosenberg A H, Lade B N, Chui D S, Lin S W, Dunn J J, Studier F W. Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene. 1987;56:125–135. doi: 10.1016/0378-1119(87)90165-x. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Segal H, Elisha B G. Identification and characterization of an aadB gene cassette at a secondary site in a plasmid from Acinetobacter. FEMS Microbiol Lett. 1997;153:321–326. doi: 10.1111/j.1574-6968.1997.tb12591.x. [DOI] [PubMed] [Google Scholar]
- 26.Stokes H W, O’Gorman D B, Recchia G D, Parsekhian M, Hall R M. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol Microbiol. 1997;26:731–745. doi: 10.1046/j.1365-2958.1997.6091980.x. [DOI] [PubMed] [Google Scholar]
- 27.Stokes H W, Hall R M. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol Microbiol. 1989;3:1669–1683. doi: 10.1111/j.1365-2958.1989.tb00153.x. [DOI] [PubMed] [Google Scholar]
- 28.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 29.Tirumalai R S, Healey E, Landy A. The catalytic domain of λ site-specific recombinase. Proc Natl Acad Sci USA. 1997;94:6104–6109. doi: 10.1073/pnas.94.12.6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu X D, Sadowski P D. Selection of novel, specific single-stranded DNA sequences by Flp, a duplex-specific DNA binding protein. Nucleic Acids Res. 1998;26:1329–1336. doi: 10.1093/nar/26.5.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]