ABSTRACT
In vitro fertilization and embryo transfer (IVF-ET) is one of the effective methods to treat female infertility. Poor endometrial receptivity (ER) is an important factor leading to embryo implantation dysfunction, which can reduce pregnancy rate of IVF-ET. The mice model with embryo implantation dysfunction in vivo and attachment model of trophoblast (JAR) spheroids in vitro were constructed. The levels of lncRNA NEAT1, HOXA10, CTCF and markers of ER were detected. The cell proliferation was measured. The interaction between lncRNA NEAT1 and CTCF, HOXA10 promoter and CTCF were confirmed. LncRNA NEAT1 and HOXA10 levels in infertile patients and mice model with embryo implantation dysfunction were increased. In vitro experiments showed that down-regulation of lncRNA NEAT1 improved EECs proliferation and ER marker expressions. LncRNA NEAT1 could bind to CTCF, and CTCF could bind to HOXA10 promoter and down-regulate HOXA10 gene expression by regulating histone modification level. The lncRNA NEAT1/CTCF/HOXA10 signaling pathway regulated EECs proliferation and ER establishment in vitro and in vivo. Our study suggested that lncRNA NEAT1 could up-regulate HOXA10 promoter activity and its expression by combining with CTCF, thus improving EECs proliferation and ER establishment, and ultimately facilitating embryo implantation.
KEYWORDS: Endometrial receptivity, lncRNA NEAT1, HOXA10, CTCF
Introduction
As a common gynecological disease, female infertility seriously harms the physical and mental health of women, and its prevalence rate increases year by year. It has become a global medical and sociological problem that needs to be solved urgently [1]. The pathogenic factors of female infertility are mainly the pathological changes related to the reproductive system (uterus, fallopian tube and ovary). In addition, the abnormal immune function and social/psychological/spiritual factors also have obvious influence on infertility [2–4]. At present, in vitro fertilization and embryo transfer (IVF-ET) is the core technology of human assisted reproductive technology (ART), which is one of the effective technologies in the treatment of infertility [5]. Following IVF treatment the number of transferred embryos has increased, however, the clinical pregnancy rate was between 40% and 60%. It has been found that embryo implantation dysfunction is one of the main reasons leading to low pregnancy rate, and the success of embryo implantation depends on endometrial receptivity (ER) [6,7]. Therefore, improving ER is the key to improve the success rate of IVF-ET.
Long non-coding RNA (lncRNA) regulation on endometrium receptivity by acting in competing endogenous RNA theory has been explored [8,9]. LncRNA nuclear paraspeckle assembly transcript 1 (NEAT1) is transcribed by RNA polymerase II, and has been proven to be involved in various pathophysiological processes such as inflammation and tumor [10,11]. Previous studies have shown that the expression of lncRNA NEAT1 is significantly increased in the placenta with intrauterine growth retardation (IUGR), suggesting that lncRNA NEAT1 may be closely related to trophoblast cell function and placental dysfunction [12]. A recent study using genome-wide analysis found that lncRNA NEAT1 level in endometrial tissues of patients with recurrent implantation failure (RIF) was significantly up-regulated from that of normal controls [8]. The effect of lncRNA NEAT1 on ER has not been confirmed by studies, so its specific mechanism is worth exploring.
Homeobox genes (HOX) are a class of transcriptional regulatory genes that control embryonic development and cell differentiation. HOXA10 gene is mainly expressed in the endometrium, and plays an important role in the proliferation and differentiation of endometrium stromal cells, the establishment of ER, the formation of pinopodes and microvilli, embryo implantation and development, etc. CTCF (CCCTC-binding factor), a protein containing 11 zinc finger structures, is well known as a transcription regulatory factor [13,14]. Previous studies have reported that in breast cancer cells, CTCF can bind to the promoter region of HOXA10 gene and maintain the methylation state of H3K27, thus inhibiting the activity of HOXA10 promoter and leading to gene silencing [15].
It is worth noting that the interaction between CTCF and lncRNA metastasis associated in lung denocarcinoma transcript 1 (MALAT1) has been explored by Akihiro, and lncRNA MALAT1 is known as NEAT2, which emphasizes the importance of the interaction between lncRNA and CTCF protein [16]. Bioinformatics software (RNainter) predicted that there was a binding site between CTCF protein and lncRNA NEAT1. We speculated that lncRNA NEAT1 might regulate HOXA10 promoter activity and gene expression through CTCF, thus affecting the establishment of ER.
Materials and methods
Study subjects
A schematic diagram for summarizing the methodology was shown in the Supplementary figure 1. Serum samples were collected from healthy controls (n = 20), early pregnant women (n = 20) and infertile women (n = 20) at the age of 25–35. Total samples were obtained from Henan Provincial People’s Hospital between 2018 and 2019. Healthy controls: normal menstrual cycle, excluding other gynecological abnormalities. Infertility patients: failure to achieve a clinical pregnancy after 12 months, excluding pathological salpingemphraxis. Pregnant women: confirmed by ultrasound diagnosis at 6–10 gestational weeks. LncRNA NEAT1 and HOXA10 mRNA levels in the serum were detected. All procedures were conducted in accordance with ethical guidelines of the institutional and national research committees and the World Medical Association Declaration of Helsinki. This study was approved by the Medical Ethics Committee of Henan Provincial People’s Hospital, and all participants provided written informed consent. All patients had signed informed consent prior to the study.
Animals and treatments
Female C57BL/6 mice (8–12 weeks old) were purchased from Charles River (Beijing, China), and were kept in light/dark cycle for 12 h at 22–25°C for free feeding. They were provided with sufficient food and water and allowed to adapt environment for 1 week before formal experiments. All experimental procedures were approved by the Committee on Animal Care of Henan Provincial People’s Hospital.
The mice model with embryo implantation dysfunction was established as previously described [17]. In brief, female mice were mated with fertile male mice to induce pregnancy after intraperitoneal injection of pregnant mare serum gonadotropin (PMSG, 10 IU) and human chorionic gonadotophin (HCG, 10 IU). Each mouse was injected subcutaneously with mifepristone (RU486) dissolved in sesame oil solution (4 mg/kg) at 9 AM on day 4 of gestation. The mice were sacrificed by cervical dislocation on the day 11, and their uterine tissues were separated. Then, lncRNA NEAT1 level in mice model with embryo implantation dysfunction was detected.
To confirm the role of lncRNA NEAT1, the mice were transfected with shRNA or sh-NEAT1 at 9 AM on day 3, the day before modeling. Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) and shRNA or sh-NEAT1 (100 μmol/L) were mixed according to the instructions. After the 20 min of the incubation, they were injected into the uterus of mice. On the day 11 of gestation, the mice were sacrificed by cervical dislocation, and their uterine tissues were separated.
Cell lines and cell culture
The Ishikawa cell line (endometrial epithelial cells, EECs), JAR cell line (human choriocarcinoma cells) and HEK293T cells were obtained from the American Type Culture Collection (ATCC) and Shanghai Cell Bank of the Chinese Academy of Sciences. Ishikawa and JAR cells were cultured in fresh RPMI 1640 medium (Gibco, Thermo Fisher Scientific) containing 10% fetal bovine serum (FBS; Gibco), penicillin and streptomycin antibiotics (100 U/mL) at an environment of 37°C and 5% CO2 [18]. HEK293T cells were cultured in DMEM high glucose medium containing 10% (v/v) FBS and 1% penicillin/streptomycin (Thermo Fisher Scientific). The cells were cultured at 37°C with 5% CO2.
Cell transfection
The small interfering RNAs (siRNA) targeting lncRNA NEAT1 (si-NEAT1) or CTCF (si-CTCF) was designed and synthesized by ApplyBio (Henan, China). The full-length lncRNA NEAT1 or CTCF cDNA were PCR amplified and cloned into the pcDNA3.1 vector to generate pcDNA-NEAT1 or pcDNA-CTCF constructs. Corresponding controls (pcDNA) were transfected into Ishikawa cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Ishikawa cells were seeded in a 24-well plate and transfected for 48 h. After that, quantitative real-time polymerase chain reaction (qRT-PCR) was used to examine the transfection efficiency.
The attachment of JAR spheroids to endometrial epithelial cell monolayers
The in vitro attachment model of trophoblast spheroids constructed by JAR cells and Ishikawa cells was used to study endometrial adhesion and ER, which has been widely recognized [19]. Endometrial epithelial cells were pretreated with 10−9 M 17β-estradiolum (E2), and incubated in the RPMI 1640 medium for 3 days. JAR spheroids were prepared according to the reported standard procedure [20], and then transferred to pretreated monolayer Ishikawa cells (50 spheroids/well). After co-culture for 1 hour, the cells were centrifuged at 80–90 rpm for 4 min, and the cells were gently washed twice with PBS. The number of spheroids was calculated by light microscope to calculate the attachment rate. The experiment was repeated at least three times.
RNA extraction and quantitative real time-PCR
Total RNA from uterine tissues and Ishikawa cells was isolated using the Trizol reagent (Vigorous Biotech), and then reverse transcribed into cDNA using cDNA Synthesis Kit (Takara). QRT-PCR assay was used to validate lncRNA NEAT1 level, HOXA10 and CTCF mRNA levels, which were performed on the ABI 7500 Fast Real-Time PCR System. QRT-PCR reaction was in 20 μL reaction volumes containing cDNA, primers, and SYBR Green Real-time PCR Master Mix (Roche). The 2−ΔΔCt method was used to access the relative RNA expression levels. GAPDH was used as an internal reference for lncRNA NEAT1, CTCF and HOXA10.
Primer sequences for qPCR were provided:
LncRNA NEAT1 Forward: 5’-AGTTAGCGACAGGGAGGGATGC-3’;
Reverse: 5’-TGTCCCCTGAAGCCCTGAGCTA-3’;
LncRNA NEAT1 (mouse) Forward: 5’-GGCACAAGTTTCACAGGCCTACATGGG-3’;
Reverse: 5’-GCCAGAGCTGTCCGCCCAGCGAAG-3’;
HOXA10 Forward: 5’-ACACTGGAGCTGGAGAAGGA-3’;
Reverse: 5’-TCACTTGTCTGTCCGTGAGG-3’;
HOXA10 (mouse) Forward: 5’-AGGTGGACGCTGCGGCTAATCTCTA-3’;
Reverse: 5’-GCCCCTTCCGAGAGCAGCAAAG-3’;
CTCF Forward: 5’-CAGTGGAGAATTGGTTCGGCA-3’;
Reverse: 5’-CTGGCGTAATCGCACATGGA-3’.
Western blotting
The protein expressions of HOXA10, CTCF and markers of ER were analyzed by using Western blot analysis. Cells were rinsed in phosphate buffer, and then dissociated by using RIPA lysis buffer. Protein lysates were generated after centrifugation for 20 min at 12,000 × g. The concentrations of protein extracts were determined using the Bradford method (Bio-Rad). The equal amount of protein was electrophoresed on SDS-PAGE, and then transferred to polyvinylidene difluoride (PVDF) membranes (Millipore). PVDF membranes were blocked in 5% nonfat milk for at room temperature, and then incubated with anti-HOXA10 (0.2–0.6 µg/mL, ab191470, Abcam), anti-CTCF (1:5000, ab188408, Abcam), anti-Integrin αV (1:5000, ab179475, Abcam), anti-Integrin β3 (1:1000, ab119992, Abcam), anti-LIF (3–5 µg/mL, ab138002, Abcam) and anti-Osteopontin (1:500, ab166709, Abcam) overnight at 4°C. Next, all membranes were incubated with secondary antibody at room temperature for 1 h. Immunoblots were visualized by Enhanced Chemiluminescence Kit (Beyotime, Shanghai, China), and then recorded the chemiluminescent signals of protein bands. GAPDH was used as an internal control.
Hematoxylin and eosin staining
The uterine tissues of the mice in each group were removed and then rinsed with pre-cooled PBS. After fixation with 4% paraformaldehyde for 48 h, routine paraffin embedding and section were performed. Hematoxylin and Eosin (HE) Staining Kit (Beyotime, Shanghai, China) was used for HE staining. The pathological changes of uterine tissues were observed with a CKX41 phase-contrast microscope (Olympus, Tokyo, Japan).
Cell proliferation assay
A Cell Counting Kit-8 (CCK-8) (Boster, China) was used to evaluate the proliferative activity following the manufacturer’s instructions. 96-well plates were utilized to immediately seed cells at a density of 3x103 cells per well. To examine cell viability, 10 μL of CCK8 solution was added into each well and incubated at 37°C for 2 h. The absorbance at 450 nm was measured with a microplate reader.
Fluorescence in situ hybridization (FISH)
FISH probe with lncRNA NEAT1 was purchased from Guangzhou RiboBio Co., Ltd. The slides were dewaxed and rehydrated in a conventional way and treated in water bath at 100°C for 20 min. After digestion with protease for 10 min and denaturation at 85°C, hybridization was carried out overnight at 37°C under dark conditions. Finally, nucleation was stained with DAPI and fluorescence reaction occurred. The reaction results were observed under microscope.
Clone formation assays
For the clone formation assay, 1x103 cells per well were seeded in six-well plates. The culture medium should be replaced every 2–3 days. After 2 weeks of incubation, the surviving colonies were washed and stained with crystal violet. After the images were taken under an inverted microscope (Olympus), the colonies containing >50 cells were counted.
RNA immunoprecipitation and RNA pull-down assays
The interaction between lncRNA NEAT1 and CTCF protein was detected by RNA immunoprecipitation (RIP) and RNA pull-down experiments. RIP assay was performed by the use of a Millipore EZ-Magna RIP RNA-Binding Protein Immunoprecipitation kit (Millipore) in line with the manufacturer’s recommendations. Briefly, Ishikawa cells transfected with pc-DNA CRNDE were incubated in RIP buffer for 30 min. Then they were incubated with magnetic beads conjugated to the CTCF antibody for 6 h at 4°C. Finally, lncRNA NEAT1 level in the coprecipitation was detected through qPCR. RNA pull-down assay was performed using the Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher). Ishikawa cells were transfected with biotinylated lncRNA NEAT1 for 48 h. After the cells were lysed with lysis buffer, the lysates were incubated with M-280 streptavidin magnetic beads (Invitrogen). The bound RNAs were purified by adding TRIzol (Invitrogen), input and the pull-down beads for further Western blot analysis.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assay was performed using the EZ-Magna ChIP™ Chromatin Immunoprecipitation Kit (Millipore). Ishikawa cells in the immunoprecipitation reaction were treated with 1% formaldehyde at room temperature for 10 min, and crosslinking was stopped by using PBS solution. Immunoprecipitation was performed by incubating with antibodies against CTCF or H3K4me3 (ab1012, Abcam), H3K27me3 (ab6002, Abcam) and immunoglobulin G (IgG) and Dynabeads Protein A/G magnetic beads overnight. Co-precipitated DNA was quantified using the Qiagen PCR Cleanup Kit.
Luciferase reporter assays
The HEK293T cells were seeded in 12-well plates and co-transfected with pcDNA or pcDNA-CTCF, and the pGL4 luciferase vector (Promega, E4481) fused with the HOXA10 promoter. After transfection for 48 h, the firefly luciferase activities in each well were calculated by a Firefly Luciferase Reporter Gene Assay Kit (Beyotime) following the manufacturer’s instructions.
Statistical analysis
All cell experiments in this study had at least three independent biological replicates for quantitative analysis. SPSS 16.0 (SPSS Inc, Chicago, USA) was employed for statistical analysis. Data are presented as mean ± SD. Our in vivo data has been proven to be normality distributed after assessment with the Kolmogorov-Smirnov test. Differences between two groups were analyzed by Student’s t-test, and differences between multiple groups were analyzed by ANOVA and Post Hoc test (Tukey test). P values less than 0.05 were considered significant.
Results
Expression of lncRNA NEAT1 in infertile patients and in mice model with embryo implantation dysfunction
To measure the levels of lncRNA NEAT1 and HOXA10 mRNA in the serum, we collected venous blood from healthy non-pregnant women, early pregnant women and infertile women. The results of qRT-PCR showed that lncRNA NEAT1 level was lower in early pregnant patients and higher in the infertile patients, while HOXA10 mRNA level was higher in the early pregnant patients and lower in the infertile patients, when compared with healthy non-pregnant patients (Figure 1a,b). Next, we constructed a mice model with embryo implantation dysfunction. After the uterine horns of mice were isolated, we found that the number of implanted embryos in the RU486 group was significantly reduced (Figure 1c). Moreover, the pathological changes of uterine tissues in RU486 group were as follows: endometrial shedding, a large number of inflammatory cells and so on (Figure 1d). ISH assays showed that lncRNA NEAT1 was expressed in the functional layer of endometrium (Supplementary figure 2). Compared with the control group, the level of lncRNA NEAT1 was higher in the RU486 group, while HOXA10 mRNA level and protein expression were lower (Figure 1e,f). These results suggest that lncRNA NEAT1 and HOXA10 are closely related to embryo implantation.
Figure 1.
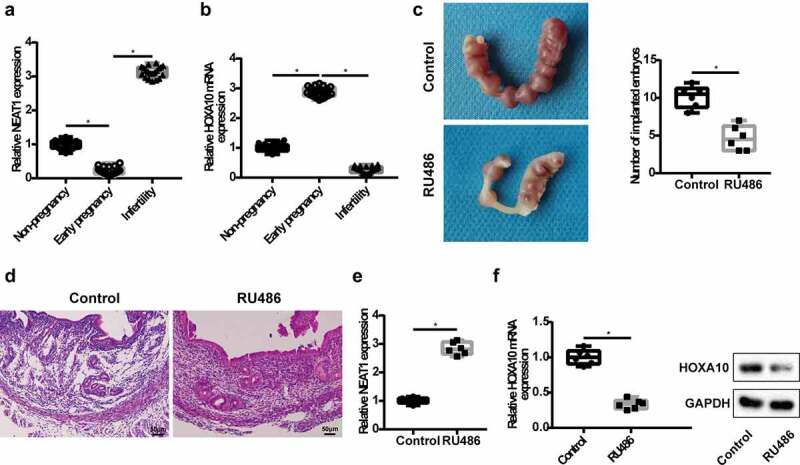
Expression of lncRNA NEAT1 in infertile patients and in mice model with embryo implantation dysfunction. Venous blood samples were collected from healthy non-pregnant women (n = 20), early pregnant women (n = 20) and infertile women (n = 20). QRT-PCR detected lncRNA NEAT1 (a) and HOXA10 mRNA (b) level in the serum of patients. *P< 0.05 vs. non-pregnancy group or early pregnancy group. A mice model with embryo implantation dysfunction was constructed, mice were divided into control group and RU486 group (n = 6/group). (c) The number of implanted embryos was counted. (d) The pathological changes of uterine tissues were detected by HE staining. QRT-PCR detected lncRNA NEAT1 (e) and HOXA10 mRNA level (f) in the uterine tissues. (F) Western blot was used to detect HOXA10 protein expression in the uterine tissues. *P < 0.05 vs. control.
LncRNA NEAT1 regulates endometrial epithelial cell proliferation and ER establishment
Ishikawa cell line is a class of highly differentiated endometrial adenocarcinoma cells, which has similar characteristics of normal EECs. Ishikawa cells were divided into control, pcDNA, pcDNA-NEAT1, si-control, and si-NEAT1, and the expression of lncRNA NEAT1 was detected by qRT-PCR. LncRNA NEAT1 level was significantly up-regulated after cell transfection with pcDNA-NEAT1, and down-regulated after cell transfection with si-NEAT1 (Figure 2a). CCK8 and clone formation assay results showed that, compared with the control group, overexpression of lncRNA NEAT1 reduced the proliferation of Ishikawa cells, while interference with lncRNA NEAT1 increased the proliferation of Ishikawa cells (Figure 2b,c). Based on this, we infer that down-regulation of lncRNA NEAT1 level can improve the proliferation of EECs.
Figure 2.
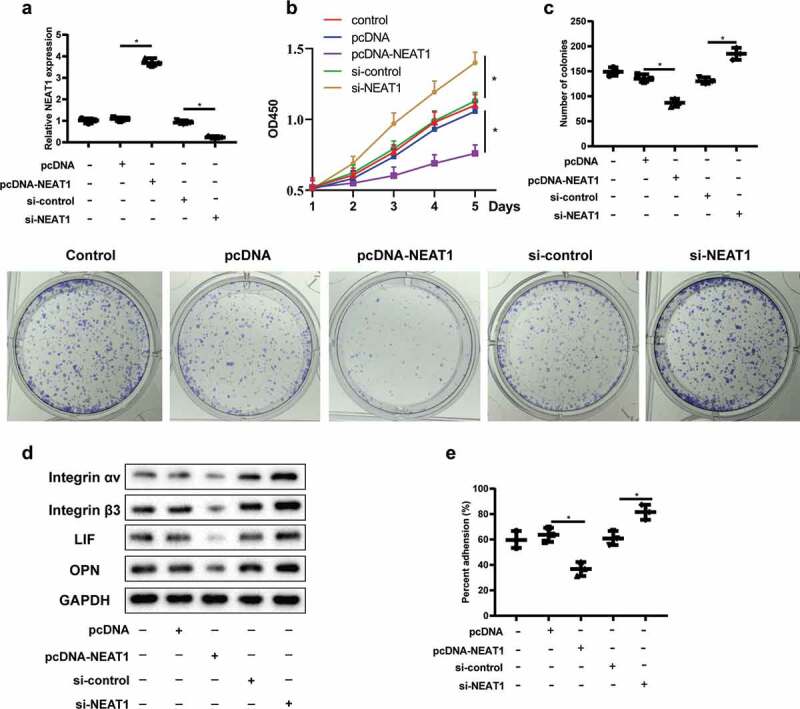
LncRNA NEAT1 regulates endometrial epithelial cell proliferation and ER establishment. Ishikawa cells were divided into control, pcDNA, pcDNA-NEAT1, si-control, and si-NEAT1. (a) The level of lncRNA NEAT1 was detected by qRT-PCR. CCK8 (b) and clone formation assay (c) detected the proliferation of Ishikawa cells in different groups. (d) Integrin αVβ3, leukemia inhibitor factor (LIF) and osteopontin (OPN) expressions were detected by Western blot assay. 2D in vitro model was established to simulate the interaction between embryos and endometrium. (e) The attachment rate of JAR cells to Ishikawa cells was analyzed. *P < 0.05 vs. pcDNA or si-control.
Integrin αVβ3, leukemia inhibitor factor (LIF) and osteopontin (OPN) were specifically expressed during endometrial implantation window, and were used as markers of ER [21,22]. The results showed that overexpression of lncRNA NEAT1 could decrease the protein expression of Integrin αVβ3, LIF and OPN, while interference with lncRNA NEAT1could increase the protein expressions of the above factors (Figure 2d). JAR cell lines originated from human choriocarcinoma cells and have the characteristics of trophoblastic cells, which can secrete placental hormones and differentiate into syncytiotrophoblast cells in vitro. Based on the fact that JAR cells can be attached to Ishikawa cells, 2D in vitro model was established to simulate the interaction between embryos and endometrium [23]. The attachment rate of JAR cells to Ishikawa cells was declined after Ishikawa cells transfection of pcDNA-NEAT1, while interference with lncRNA NEAT1 enhanced the attachment rate (Figure 2e). These results suggest that lncRNA NEAT1 can regulate the establishment of ER.
The mechanism of lncRNA NEAT1 regulating HOXA10 expression
Bioinformatics software (RNAINTER) was used to predict the presence of binding sites between CTCF and lncRNA NEAT1. RIP and RNA pull-down experiments were further used to prove that lncRNA NEAT1 could interact with CTCF protein (Figure 3a,b). Furthermore, studies have shown that CTCF can bind to the promoter region of HOXA10 gene [15]. In this study, ChIP assay was used to verify the binding of CTCF with HOXA10 promoter (Figure 3c). Furthermore, studies have shown that the methylation level in the promoter region of HOXA10 is related to the formation of pinocytosis and the establishment of ER [24]. Ishikawa cells were divided into si-control group and si-CTCF group, and then we analyzed the level of histone modification in the HOXA10 binding region using ChIP-PCR. The results showed that CTCF and histone H3 Lys27 trimethylation (H3K27Me3) levels were significantly reduced after interference with CTCF, while histone H3 Lys4 trimethylation (H3K4Me3) level was not significantly changed (Figure 3d). To investigate the effect of CTCF on HOXA10 expression, Ishikawa cells were transfected with pcDNA or pcDNA-CTCF. We found that CTCF mRNA and protein levels were up-regulated after overexpression of CTCF (Figure 3e). In addition, the activity of HOXA10 promoter was significantly reduced, HOXA10 mRNA and protein levels were down-regulated (figure 3f,g). The above indicates that CTCF can bind to the promoter of HOXA10 gene, maintain the methylation state of H3K27, inhibit the activity of the promoter, and eventually lead to the silencing of HOXA10 gene.
Figure 3.

The mechanism of lncRNA NEAT1 regulating HOXA10 expression. RIP (a) and RNA pull-down experiments (b) were used to prove that lncRNA NEAT1 could interact with CTCF protein. (c) ChIP assay was used to verify the binding of CTCF with HOXA10 promoter. Ishikawa cells were divided into si-control group and si-CTCF group. (d) The level of histone modification in the HOXA10 binding region was detected by using ChIP-PCR. *P < 0.05 vs. si-control. Ishikawa cells were transfected with pcDNA or pcDNA-CTCF. (e) The levels of CTCF mRNA and protein were detected by qRT-PCR and Western blot. (f) The activity of HOXA10 promoter was detected by luciferase reporter assay. (g) QRT-PCR and Western blot detected HOXA10 mRNA and protein levels. *P < 0.05 vs. pcDNA.
LncRNA NEAT1 regulates HOXA10 expression through CTCF and affects endometrial epithelial cell proliferation and ER establishment
In order to verify the influence of the signaling pathway on the proliferation and tolerance establishment of endometrial epithelial cells, Ishikawa cells were divided into pcDNA, pcDNA-NEAT1, pcDNA-NEAT1+ si-control, pcDNA-NEAT1+ si-CTCF, and pcDNA-NEAT1+ si-CTCF+si-HOXA10. QRT-PCR was used to detect the level of NEAT1, and the results showed that overexpression of NEAT1 could up-regulate its expression level, but interference with CTCF and HOXA10 did not affect its expression level (Figure 4a). Overexpression of lncRNA NEAT1 up-regulated the protein expression of CTCF, but does not affected its mRNA level, and interference with HOXA10 had no effect on the expression of CTCF (Figure 4b). In addition, overexpression of lncRNA NEAT1 could down-regulate HOXA10 mRNA level and protein expression, while interference with CTCF reversed this effect, resulting in up-regulation of HOXA10 expression (Figure 4c). These results confirm that lncRNA NEAT1 regulates HOXA10 expression through CTCF.
Figure 4.
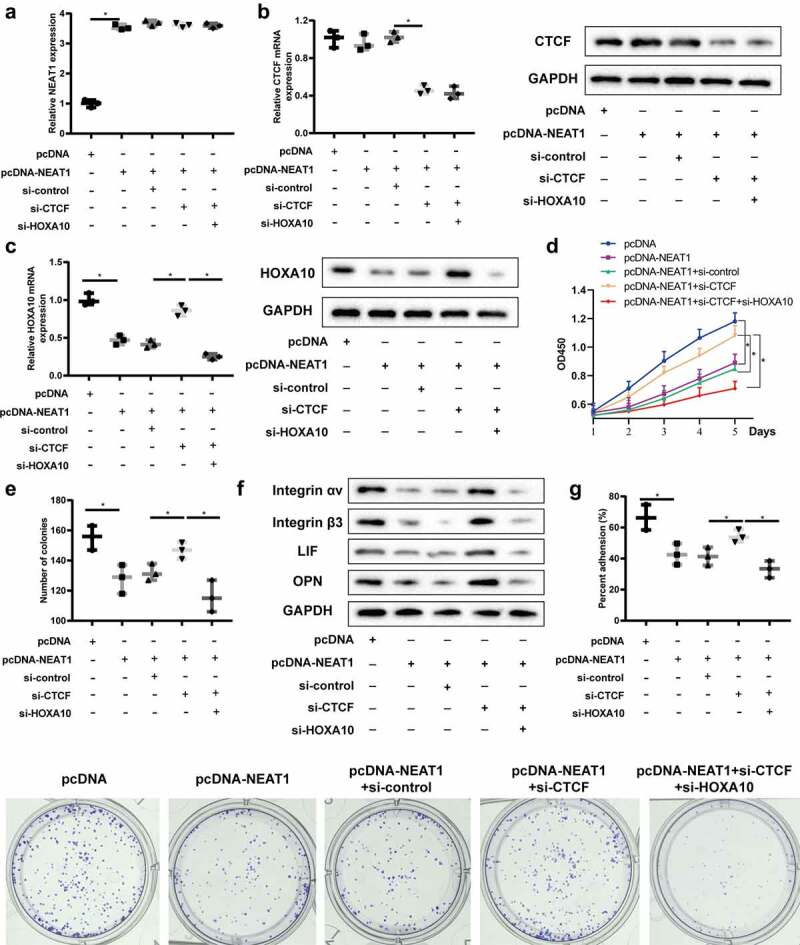
LncRNA NEAT1 regulates HOXA10 expression through CTCF and affects endometrial epithelial cell proliferation and ER establishment. Ishikawa cells were divided into pcDNA, pcDNA-NEAT1, pcDNA-NEAT1+ si-control, pcDNA-NEAT1+ si-CTCF, and pcDNA-NEAT1+ si-CTCF+si-HOXA10. (a) QRT-PCR was used to detect the level of lncRNA NEAT1. QRT-PCR and Western blot detected CTCF (b) and HOXA10 (c) mRNA and protein levels. CCK8 (d) and clone formation assay (e) detected the proliferation of Ishikawa cells in different groups. (f) Integrin αVβ3, leukemia inhibitor factor (LIF) and osteopontin (OPN) expressions were detected by Western blot assay. (g) The attachment rate of JAR cells to Ishikawa cells was analyzed. *P < 0.05 vs. pcDNA or pcDNA-NEAT1+ si-control or pcDNA-NEAT1+ si-CTCF.
CCK8 and clone formation assay results showed that overexpression of lncRNA NEAT1 reduced the proliferation of Ishikawa cells, while interference with CTCF improved the proliferation of Ishikawa cells (Figure 4d,e). What is more, down-regulation of HOXA10 could reduce the proliferation ability of Ishikawa cells (Figure 4d,e). We also found that overexpression of lncRNA NEAT1 could decrease the protein expression of Integrin αVβ3, LIF and OPN, while interference with CTCF reversed this effect, and down-regulation of HOXA10 could increase the protein expressions of the above factors (figure 4f). As shown in Figure 4g, the attachment rate of JAR cells to Ishikawa cells was declined after Ishikawa cells transfection of pcDNA-NEAT1, while interference with CTCF enhanced the attachment rate, and silenced HOXA10 reversed this effect. Based on this, we inferred that lncRNA NEAT1/CTCF/HOXA10 axis could affect endometrial epithelial cell proliferation and ER establishment.
LncRNA NEAT1 can enhance the ER establishment and embryo implantation in vivo
To confirm whether lncRNA NEAT1 could mediate ER establishment and embryo implantation in vivo, a mice model with embryo implantation dysfunction was established, and lncRNA NEAT1 was overexpressed after injecting sh-NEAT1 into the uterus of mice (Figure 5c). After the uterine horns of mice were isolated, we found that the number of implanted embryos was significantly increased in the sh-NEAT1 group (Figure 5a). We observed an increased number of endometrial epithelial cells, smaller glandular lacuna, and uniform size of glands (Figure 5b). Compared with the control group, the protein expression of CTCF was down-regulated in the sh-NEAT1 group, while Integrin αVβ3, LIF and OPN protein expressions were up-regulated (Figure 5d). In addition, interfering lncRNA NEAT1 could significantly increase HOXA10 mRNA level. These results suggest that down-regulation of lncRNA NEAT1 could enhance the establishment of ER and ultimately facilitate mouse embryo implantation. We also showed a schematic diagram for summarizing these results in Supplementary Figure 3.
Figure 5.
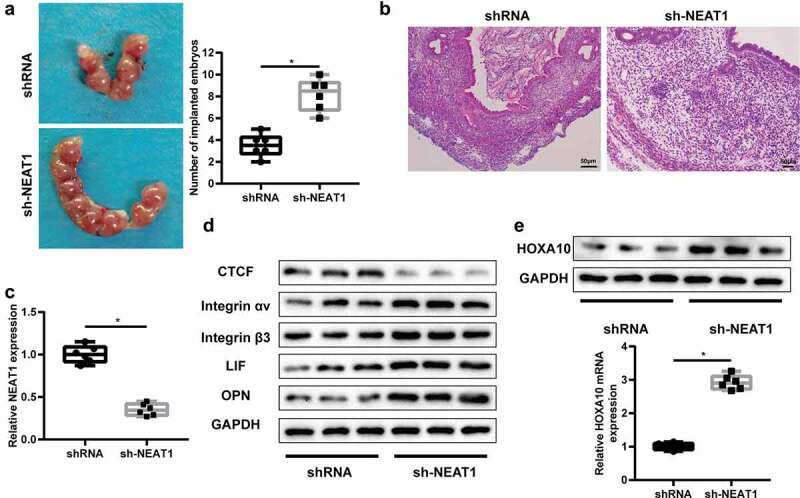
LncRNA NEAT1 can enhance the ER establishment and embryo implantation in vivo. A mice model with embryo implantation dysfunction was established. After injecting shRNA and sh-NEAT1 into the uterus, mice were divided into shRNA group and sh-NEAT1 group (n = 6/group). (a) The number of implanted embryos was counted. (b) The pathological changes of uterine tissues were detected by HE staining. (c) QRT-PCR detected lncRNA NEAT1 level in the uterine tissues. (d) Integrin αVβ3, leukemia inhibitor factor (LIF) and osteopontin (OPN) expressions were detected by Western blot assay. (e) QRT-PCR and Western blot was used to detect HOXA10 expression in the uterine tissues. *P < 0.05 vs. shRNA.
Discussion
As we all known, receptive endometrium is one of the core factors for successful embryo implantation. During the last 30 years, the success rate of embryo transfer has been significantly increased, while the pregnancy rate following IVF-ET is still rather low, suggesting the importance of the success of embryo implantation in this technology, which is closely associated with the endometrial receptivity. The functions of lncRNAs during oocyte maturation and early embryo development have been studied in depth, but their roles in embryo implantation are rarely reported. Multiple differentially expressed lncRNAs were found in endometrial tissues of patients with RIF and women with successful pregnancy after ET at the implantation window, indicating that lncRNAs can affect ER and embryo implantation through various pathways including immune response, metabolic process and cell cycle regulation [8,9,25,26]. A recent study found that lncRNA NEAT1 may regulate endometrial receptivity by acting on miRNA through the construction of ceRNA regulatory network, but the specific mechanism has not been confirmed [8]. This study was the first to investigate the effect of lncRNA NEAT1 on endometrial receptivity and its mechanism. Hence, we investigated the role of NEAT1 and its potential regulation mechanism in affecting endometrial receptivity in the present study. We first found that lncRNA NEAT1 expression was increased in infertility patients and mice model with embryo implantation dysfunction. In addition, down-regulation of lncRNA NEAT1 was found to improve the proliferation ability of EECs and the establishment of ER in vitro. However, we believe that the current sample size is small, and it is necessary to increase the peripheral blood sample size.
The important functions of homeobox genes are to regulate embryonic development and endometrial receptivity, and to determine the directed differentiation and proliferation of cells [27,28]. Previous studies have shown that HOXA10 expression is abnormal in endometriosis, which is related to the methylation state of the promoter region [29,30]. In addition, the increased methylation level in the promoter region of HOXA10 is not conducive to the formation of Pinocytosis and the establishment of ER [24]. Therefore, inhibiting methylation level can up-regulate the expression level of HOXA10, thus improving ER [31]. We demonstrated for the first time that CTCF regulated histone modification of HOXA10 promoter in EECs, thereby down-regulating HOXA10 gene expression. Furthermore, in vitro experiments confirmed that lncRNA NEAT1 could interact with CTCF protein, and then CTCF could affect the proliferation of EECs. The effect of lncRNA NEAT1/CTCF/HOXA10 signaling pathway on the proliferation of EECs and establishment of ER was verified in vivo.
ER refers to the ability of the endometrium to accept the blastocyst and allow the blastocyst to induce a series of changes in the endometrium stroma during a specific period of the menstrual cycle, thus enabling the embryo to be implanted. This specific period, known as the implantation window, usually occurs between 7 and 9 days after ovulation [32]. Integrin αVβ3 is a major cell surface receptor involved in the adhesion reaction between early embryo and endometrium, which promotes embryo implantation and regulates embryo growth [33]. LIF is a secreted glycoprotein, which as a multifunctional cytokine participates in follicle development, affects ER, and regulates embryo implantation and growth [34]. OPN, a class of phosphorylated glycoproteins, is the ligand of integrin αVβ3. The combination of OPN and integrin αVβ3 can mediate the adhesion of the blastocyst to the endometrium, which is essential for the establishment of ER [35]. In this study, we found that interference or overexpression of lncRNA NEAT1, CTCF and HOXA10 could affect the expression of the above markers. In a previous study, lncRNA NEAT1 has been proven to be required for corpus luteum formation and the establishment of pregnancy in mice, which seems to be contradictory with our conclusion [36]. Although the importance of the corpus luteum in pregnancy should be acknowledged, the factors that affect endometrial receptivity are very complex, which may lead to the different results.
In conclusion, the expression of lncRNA NEAT1 was increased in infertility patients, and our results suggested that down-regulation of lncRNA NEAT1 could inhibit the protein expression of CTCF, further improve the activity of HOXA10 promoter and its gene expression level, thus enhancing the proliferation of EECs and receptivity establishment of ER, and ultimately facilitating the implantation of mice embryos. This study provides new ideas for improving the success rate of IVF-ET and may improve the development of infertility in the future.
Supplementary Material
Funding Statement
Medical Science and Technology Research project (Joint construction) of Henan Province of China (Grant No. 2018020391).
Availability of data and material
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15384101.2022.2075198
References
- [1].Esfandyari S, Chugh RM, H-s P, et al. Mesenchymal stem cells as a bio organ for treatment of female infertility. Cells. 2020;9(10):2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Jobling P, O’Hara K, Hua S.. Female reproductive tract pain: targets, challenges, and outcomes. Front Pharmacol. 2014;5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Crain DA, Janssen SJ, Edwards TM, et al. Female reproductive disorders: the roles of endocrine-disrupting compounds and developmental timing. Fertil Steril. 2008;90(4):911–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Y-x Z, S-r C, P-p S, et al. Using mesenchymal stem cells to treat female infertility: an update on female reproductive diseases. Stem Cells Int. 2019;2019. DOI: 10.1155/2019/9071720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhao C, Wei Z, Yang J, et al. Characterization of the vaginal microbiome in women with infertility and its potential correlation with hormone stimulation during in vitro fertilization surgery. Msystems. 2020;5(4). DOI: 10.1128/mSystems.00450-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Frantz S, Parinaud J, Kret M, et al. Decrease in pregnancy rate after endometrial scratch in women undergoing a first or second in vitro fertilization. A multicenter randomized controlled trial. Hum Reprod. 2019;34(1):92–99. [DOI] [PubMed] [Google Scholar]
- [7].Gardner DK, Lane M, Stevens J, et al. Blastocyst score affects implantation and pregnancy outcome: towards a single blastocyst transfer. Fertil Steril. 2019;112(4):E81–E84. [DOI] [PubMed] [Google Scholar]
- [8].Xu H, Zhou M, Cao Y, et al. Genome-wide analysis of long noncoding RNAs, microRNAs, and mRNAs forming a competing endogenous RNA network in repeated implantation failure. Gene. 2019;720:144056. [DOI] [PubMed] [Google Scholar]
- [9].Feng C, Shen J-M, Lv -P-P, et al. Construction of implantation failure related lncRNA-mRNA network and identification of lncRNA biomarkers for predicting endometrial receptivity. Int J Biol Sci. 2018;14(10):1361–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhang P, Cao L, Zhou R, et al. The lncRNA Neat1 promotes activation of inflammasomes in macrophages. Nat Commun. 2019;571(7766):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhang M, Weng W, Zhang Q, et al. The lncRNA NEAT1 activates Wnt/beta-catenin signaling and promotes colorectal cancer progression via interacting with DDX5. J Hematol Oncol. 2018;11(1):113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gremlich S, Damnon F, Reymondin D, et al. The long non-coding RNA NEAT1 is increased in IUGR placentas, leading to potential new hypotheses of IUGR origin/development. Placenta. 2014;35(1):44–49. [DOI] [PubMed] [Google Scholar]
- [13].Roy AR, Ahmed A, DiStefano PV, et al. The transcriptional regulator CCCTC-binding factor limits oxidative stress in endothelial cells. J Biol Chem. 2018;293(22):8449–8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].s K, Yu N-K, Shim K-W, et al. Remote memory and cortical synaptic plasticity require neuronal CCCTC-Binding factor (CTCF). J Neurosci. 2018;38(22):5042–5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mustafa M, Lee J-Y, Kim MH. CTCF negatively regulates HOXA10 expression in breast cancer cells. Biochem Biophys Res Commun. 2015;467(4):828–834. [DOI] [PubMed] [Google Scholar]
- [16].Fujimoto A, Furuta M, Totoki Y, et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet. 2016;48(5):500–+. [DOI] [PubMed] [Google Scholar]
- [17].Ran H, Kong S, Zhang S, et al. Nuclear Shp2 directs normal embryo implantation via facilitating the ERa tyrosine phosphorylation by the Src kinase. Proc Natl Acad Sci U S A. 2017;114(18):4816–4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Guoa C, Zhaoa W. Bao. X-f: lncRNA NNT-AS1 affect progesterone resistance by regulating miR-542-3p/survivin axis in endometrial cancer. Clini Surg Res Commun. 2019;2(3):25–35. [Google Scholar]
- [19].Ullah K, Rahman TU, Pan H-T, et al. Serum estradiol levels in controlled ovarian stimulation directly affect the endometrium. J Mol Endocrinol. 2017;59(2):105–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hohn HP, Linke M, Denker HW. Adhesion of trophoblast to uterine epithelium as related to the state of trophoblast differentiation: in vitro studies using cell lines. Mol Reprod Dev. 2000;57(2):135–145. [DOI] [PubMed] [Google Scholar]
- [21].Toktay E, Selli J, Gurbuz MA, et al. Effects of soy isoflavonoids (genistein and daidzein) on endometrial receptivity. Iran J Basic Med Sci. 2020;23(12):1603–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zarrin Y, Bakhteyari A, Nikpour P, et al. A study on the presence of osteopontin and alpha3beta1 integrin in the endometrium of diabetic rats at the time of embryo implantation. J Reprod Infertil. 2020;21(2):87–93. [PMC free article] [PubMed] [Google Scholar]
- [23].H-WR L, Y-X L, T-T L, et al. Effect of ulipristal acetate and mifepristone at emergency contraception dose on the embryo-endometrial attachment using an in vitro human trophoblastic spheroid and endometrial cell co-culture model. Hum Reprod. 2017;32(12):2414–2422. [DOI] [PubMed] [Google Scholar]
- [24].Li F, Zhang M, Zhang Y, et al. GnRH analogues may increase endometrial Hoxa10 promoter methylation and affect endometrial receptivity. Mol Med Rep. 2015;11(1):509–514. [DOI] [PubMed] [Google Scholar]
- [25].Fan LJ, Han HJ, Guan J, et al. Aberrantly expressed long noncoding RNAs in recurrent implantation failure: a microarray related study. Systems Biology in Reproductive Medicine. 2017;63(4):269–278. [DOI] [PubMed] [Google Scholar]
- [26].Li D, Jiang W, Jiang Y, et al. Preliminary functional inquiry of lncRNA ENST00000433673 in embryo implantation using bioinformatics analysis. Syst Biol Reprod Med. 2019;65(2):164–173. [DOI] [PubMed] [Google Scholar]
- [27].Du H, Taylor HS. The Role of Hox Genes in Female Reproductive Tract Development, Adult Function, and Fertility. Cold Spring Harb Perspect Med. 2016;6(1):a023002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Xu B, Geerts D, Bu Z, et al. Regulation of endometrial receptivity by the highly expressed HOXA9, HOXA11 and HOXD10 HOX-class homeobox genes. Hum Reprod. 2014;29(4):781–790. [DOI] [PubMed] [Google Scholar]
- [29].Andersson KL, Bussani C, Fambrini M, et al. DNA methylation of HOXA10 in eutopic and ectopic endometrium. Hum Reprod. 2014;29(9):1906–1911. [DOI] [PubMed] [Google Scholar]
- [30].Wu Y, Halverson G, Basir Z, et al. Aberrant methylation at HOXA10 may be responsible for its aberrant expression in the endometrium of patients with endometriosis. Am J Obstet Gynecol. 2005;193(2):371–380. [DOI] [PubMed] [Google Scholar]
- [31].Wang L, Tan YJ, Wang M, et al. DNA methylation inhibitor 5-Aza-2 ‘-Deoxycytidine modulates endometrial receptivity through upregulating HOXA10 expression. Reprod Sci. 2019;26(6):839–846. [DOI] [PubMed] [Google Scholar]
- [32].Lessey BA, Young SL. What exactly is endometrial receptivity? Fertil Steril. 2019;111(4):611–617. [DOI] [PubMed] [Google Scholar]
- [33].Zhang D, Wei J, Wang J, et al. Difucosylated oligosaccharide Lewis Y is contained within integrin alpha v beta 3 on RL95-2 cells and required for endometrial receptivity. Fertil Steril. 2011;95(4):1446–U1296. [DOI] [PubMed] [Google Scholar]
- [34].Seli E, Kayisli UA, Cakmak H, et al. Removal of hydrosalpinges increases endometrial leukaemia inhibitory factor (LIF) expression at the time of the implantation window. Hum Reprod. 2005;20(11):3012–3017. [DOI] [PubMed] [Google Scholar]
- [35].Casals G, Ordi J, Creus M, et al. Expression pattern of osteopontin and alpha v beta 3 integrin during the implantation window in infertile patients with early stages of endometriosis. Hum Reprod. 2012;27(3):805–813. [DOI] [PubMed] [Google Scholar]
- [36].Nakagawa S, Shimada M, Yanaka K, et al. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development. 2014;141(23):4618–4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
The data that support the findings of this study are available from the corresponding author upon reasonable request.


