Abstract
Obesity is a problem of modern health care that causes the occurrence of many concomitant diseases: arterial hypertension, diabetes mellitus, non-alcoholic fatty liver disease, and cardiovascular diseases. New strategies for the treatment and prevention of obesity are being developed that are based on using probiotics for modulation of the gut microbiota. Our study aimed to evaluate the bacterial composition of the gut of obese patients before and after two weeks of lactic acid bacteria (Lactobacillus acidophilus, Lactiplantibacillus plantarum, Limosilactobacillus fermentum, and Lactobacillus delbrueckii) intake. The results obtained showed an increase in the number of members of the phylum Actinobacteriota in the group taking nutritional supplements, while the number of phylum Bacteroidota decreased in comparison with the control group. There has also been an increase in potentially beneficial groups: Bifidobacterium, Blautia, Eubacterium, Anaerostipes, Lactococcus, Lachnospiraceae ND3007, Streptococcus, Escherichia-Shigella, and Lachnoclostridium. Along with this, a decrease in the genera was demonstrated: Faecalibacterium, Pseudobutyrivibrio, Subdoligranulum, Faecalibacterium, Clostridium sensu stricto 1 and 2, Catenibacterium, Megasphaera, Phascolarctobacterium, and the Oscillospiraceae NK4A214 group, which contribute to the development of various metabolic disorders. Modulation of the gut microbiota by lactic acid bacteria may be one of the ways to treat obesity.
Keywords: microbiome, obesity, lactic acid bacteria, 16S rRNA, sequencing
1. Introduction
The gut microbiome strongly contributes to the overall health of the host including the risk of developing obesity. Although our knowledge about the composition of the human microbiome is evolving quite rapidly, there are many questions about the metabolic mechanisms and functionality of the human microbiome. It is known that probiotics have a positive effect in the context of various diseases, particularly those associated with gastroenterology [1]. A deeper understanding of the effect of probiotics on the composition of the microbiome should help in the treatment and prevention of various diseases.
The prevalence of obesity worldwide continues to rise [2]. Even though obesity is a metabolically complex and multifactorial disease, aberrations in the composition of the gut microbiome make a great contribution to the formation of this pathology [3,4]. There are many studies describing dysbiosis in people with obesity, characterized by less diversity [5,6,7]. Dysbiosis likely contributes to the development of obesity and metabolic complications through various mechanisms including changes in the regulation of energy, gut hormones, pro-inflammatory mechanisms, and others [8].
Since weight loss drugs often have specific toxic side effects, it is necessary to look for more moderate and effective substitutes. Some research suggests that the gut microbiota is directly linked to obesity and taking probiotics may have a positive impact on weight loss [9,10].
Lactic acid bacteria are often used as probiotics. Many species of lactic acid bacteria can boost nutrient absorption, improve the functioning of the gastrointestinal tract, lower serum cholesterol levels, prevent obesity, inhibit the growth of putrefactive bacteria in the intestine, and improve immune function [11,12,13,14].
Lactobacillus acidophilus is one of the main commercially important species of lactic acid bacteria used as a probiotic [15]. L. acidophilus promotes nutrient absorption, has a beneficial effect on digestive functions, and prevents diarrhea [16].
There is growing evidence that L. plantarum may benefit human health [17]. For example, there is evidence that L. plantarum can lower cholesterol levels in adults with hypercholesterolemia, potentially reducing the risk of coronary heart disease [18]. Additionally, L. plantarum often exhibits the broadest ability to inhibit pathogen growth among the Lactobacilli species [19,20].
L. fermentum is one of the most promising probiotics. It is often used as a typical reference species in comparative investigations with different probiotics due to its health benefits [21]. There are studies that describe the potential beneficial role of L. fermentum in various diseases such as intestinal inflammation [22], respiratory infections [23], and hepatic injury [24].
Despite the reported beneficial properties of these lactic acid bacteria species, supporting studies are needed to further understand the role of these probiotics in modulating gut microbial populations.
It is possible that restoring the gut microbiome to a healthy state could improve the conditions associated with obesity. However, the role of probiotics in the prevention and treatment of obesity has not yet been fully explored.
The purpose of this work was to study the effect of probiotic intake on the composition of the gut microbiome in obese people.
2. Materials and Methods
2.1. Experiment Design
At the beginning of the study, each patient completed a questionnaire that included questions about age, nutrition, physical activity, a history of infectious and non-infectious pathologies, taking medications including hormonal and antibacterial drugs (for the last 6 months), and there were clarifications on anthropometric indicators (height; weight for calculating body mass index (BMI)) and waist circumference (OT). Based on the results of the survey, eight patients were selected, who were subsequently included in the group with diagnosed obesity.
At the beginning of the study, each of the eight patients had a fecal sampling, approximately 20 ± 5 g. Then, each patient consumed 55 g of the supplement, which consisted of lactic acid bacteria, with food for two weeks. During this period, patients did not change their usual diet in any way. At the end of the 2-week supplementation, approximately 20 ± 5 g of feces were again collected from each patient for microbiome profiling.
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Ethics Board of Voronezh State University (Protocol N42-03, 7 March 2022). Before the beginning of the study, all patients gave their written consent to the use of anonymized personal data for scientific purposes.
2.2. Dietary Supplement of Lactic Acid Bacteria
Probiotic bacteria were obtained from LLC “White Lily” (Voronezh, Russia). After purification, the bacterial cultures received the following numbers: Lactobacillus delbrueckii subsp. bulgaricus VSUET15; Lactiplantibacillus plantarum VSUET13; Lactobacillus acidophilus VSUET12; and Limosilactobacillus fermentum VSUET14 for further placement in the Microbiological Museum of the Voronezh State University of Engineering Technologies.
For subsequent work, the bacteria were added to 100 mL of sterilized skimmed milk. To improve the fermentation process in the mixture of bacteria, Streptococcus thermophilus was added. The activation of microbial cells of the concentrate occurred due to mixing and further 4 h exposure at a constant temperature of 37 ± 1 °C, cyclically stirring (shaking) for a uniform distribution of bacterial cells in the mixture. Then, the produced concentrate was immediately added to pasteurized (92 ± 2 °C, exposure 2–8 min) and chilled (37–42 °C) skimmed milk with constant stirring. The density of microorganisms in the finished product was adjusted to 108 CFU/mL. Subsequently, the cultures were kept at a temperature of −u °C for no more than three days. The viable cells of lactic acid bacteria were accounted on MRS agar.
2.3. 16S rRNA Gene Sequencing
A commercial Quick-DNA Fungal/Bacterial Microprep Kit (Zymo Research; Irvine, CA, USA) was used to obtain a DNA product from a biomaterial.
Analysis of the bacterial diversity of the intestinal microbiome by sequencing based on the Ion Torrent PGM platform was carried out for the V3 hypervariable region of the 16S rRNA gene.
For further PCR analysis, two universal primers 337F and 518R (the sequence of primers is presented in Table 1) and a commercial set of the 5X Screen Mix-HS Master Mix Kit (Eurogen, Moscow, Russia) were used.
Table 1.
The primers used in the study.
| Name of the Primer | Primer Sequence |
|---|---|
| 337F | 5′-GACTCCTACGGGAGGCWGCAG-3′ |
| 518R | 5′-GTATTACCGCGGCTGCTGG-3′ |
The PCR protocol is presented in Table 2.
Table 2.
The PCR protocol for amplification of the V3 hypervariable region of the 16S rRNA gene.
| Stage Name | Temperature | Time | The Number of Cycles |
|---|---|---|---|
| General denaturation | 94 °C | 4 min | 1 |
| Denaturation | 94 °C | 30 s | 37 |
| Primer annealing | 53 °C | 30 s | |
| Elongation | 72 °C | 30 s | |
| Final elongation |
72 °C | 5 min | 1 |
We used AMPureXP magnetic beads to purify PCR-derived amplicons (Beckman Coulter, Brea, CA, USA) for further preparation of the sequencing libraries. This is a necessary step for preparing high-quality sequencing libraries.
For the preparation of sequencing libraries, a commercial NEBNext Fast DNA Kit (New England Biolabs, Ipswich, MA, USA) was used in the work according to the method provided by the manufacturer. In further work, the resulting library was barcoded with a commercial NEXTflex Kit (Ion Torrent; 64 adapters; PerkinElmer, Inc., Waltham, MA, USA). Then, another cleaning of the finished libraries was carried out with AMPureXP magnetic particles (Beckman Coulter, Brea, CA, USA). The Ion Torrent PGM platform was used for sequencing. Libraries were loaded onto an Ion 318™ Chip v2 BC using the standard protocols of the Ion PGM Hi-Q View OT2 Kit for further enrichment using emulsion PCR and the Ion PGM Hi-Q View Sequencing Kit for post-processing and loading libraries onto the chip as well as for running the instrument itself (ThermoFisher Scientific, Madison, WI, USA).
2.4. Statistical Analysis
The obtained analysis results were processed using the R programming language in the R Studio environment (V SEARCH v software.2.8.2; version 1.1.414 ©2009-2018 RStudio Inc., RStudio PBC, Boston, MA, USA).
At the initial stage, BAM files were generated for each sample, which were later modified into the FastQ format based on the fileexporterwin plugin.
To exclude low-quality reads from the subsequent analysis, we used the maximum expected error threshold, all reads for which values of less than 1.0 were eliminated (DADA2 package). In addition, the reads were truncated to an optimized total size, followed by demultiplexing. After all the manipulations, the reads were dereplicated and, as a result, all identical reads were combined into unique sequences. Then, based on the UNOISE2 algorithm, we formed operational taxonomic units (OTUs).
Generic identification was carried out using version 132 of the SILVA database (https://www.arb-silva.de/, accessed on 14 March 2022). The taxonomy was determined relative to the peak limit of identity with variant amplicon sequences corresponding to 100%. All raw sequences obtained from the analyses for each sample are available in the BioProject repository (Bio Project:PRJNA862929).
Statistical analysis of the sequencing data was carried out on the basis of GraphPad Prism 9 software (GraphPad, San Diego, CA, USA). The statistical significance of differences in the microbiome profiles of patients was achieved using the Mann–Whitney U test. The average abundance of individual taxa was expressed as the mean ± standard error of the mean (SEM). The intra-sample alpha diversity score was expressed using Shannon’s test.
3. Results
In total, 128,390 unique reads were obtained during sequencing, corresponding to 103 bacterial genera (Table S1).
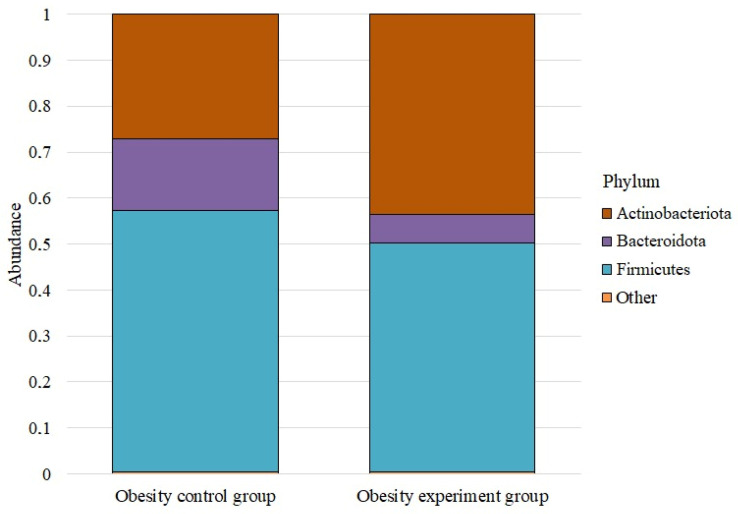
We analyzed the microbiome composition at the phylum level for the study groups (Figure 1).
Figure 1.
The average abundance of phyla in the studied groups.
The phylum Firmicutes dominated in both cases. Before taking lactic acid bacteria, its abundance was 0.569 ± 0.072; after taking it, it decreased to 0.498 ± 0.048. The phylum Actinobacteriota was the next most numerous both before and after taking the lactic acid bacteria supplement. In the control group, its abundance was 0.272 ± 0.025, while in the experimental group, it was 0.436 ± 0.036. The abundance of the phylum Bacteroidota before taking the supplement was 0.155 ± 0.078; after, it was—0.061 ± 0.020. Proteobacteria were next, with an abundance of 0.003 ± 0.001 and 0.005 ± 0.001, respectively. The number of Verrucomicrobiota before taking lactic acid bacteria was 0.0008 ± 0.001; after, it was — 0.0002 ± 0.001. The Desulfobacterota phylum was the least numerous; in both groups, its abundance was less than 0.005.
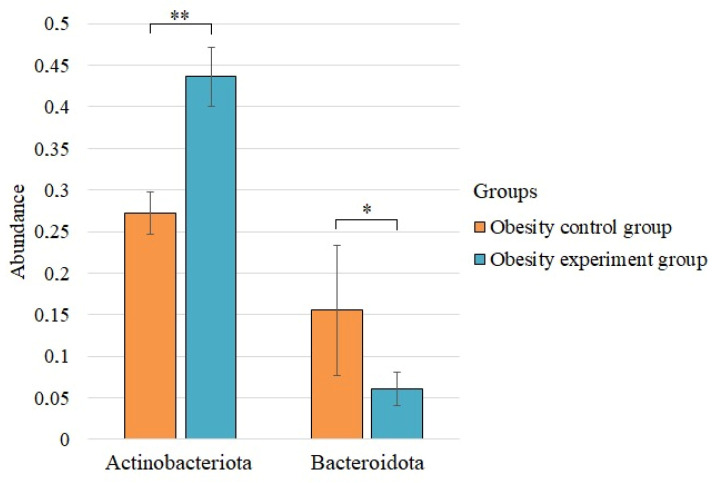
We found statistically significant differences before and after the intake of lactic acid bacteria for the two phyla (Figure 2).
Figure 2.
The statistically significant differences in the phylum composition of the control group of obesity and the group of patients with obesity after taking lactic acid bacteria. * p < 0.05; ** p < 0.01.
The abundance of the phylum Actinobacteriota increased significantly after supplementation with lactic acid bacteria, while the abundance of Bacteroidota decreased.
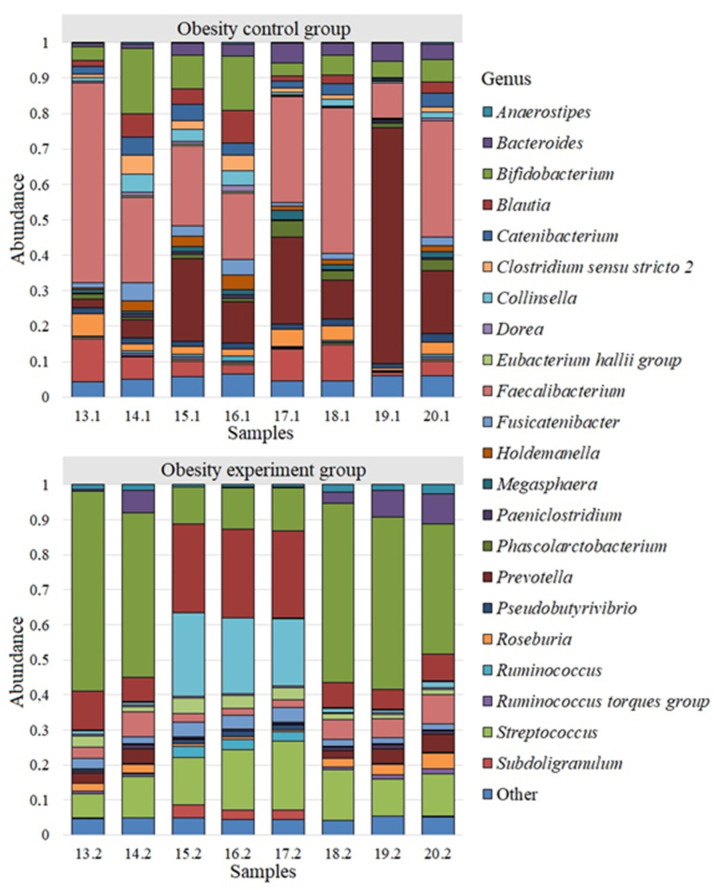
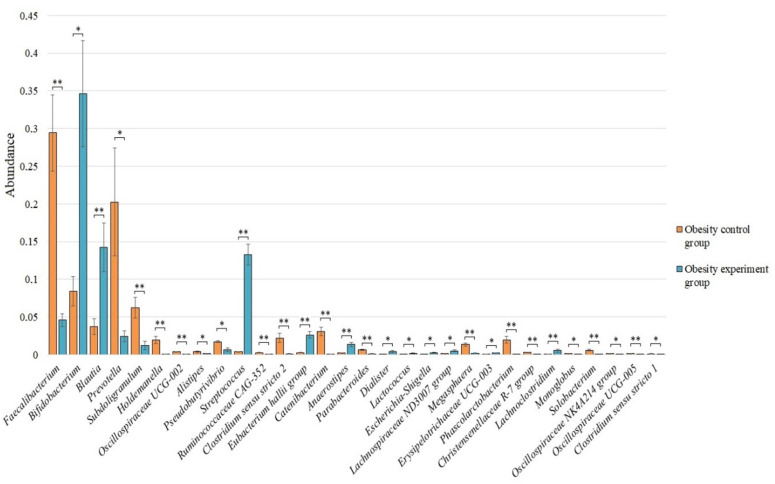
Bacterial genera whose abundance was less than 0.005 were grouped into “Other Genus”. We analyzed the differences among all identified genera of bacteria for each study group. Twenty-two of the most common bacterial genera were identified for the group of obese patients participating in a study evaluating the effects of a nutritional supplement containing lactic acid bacteria on the gut microbiome composition (Figure 3).
Figure 3.
The microbiome of obesity patients before and after the intake of lactic acid bacteria.
We detected a different microbiome landscape in the group of patients with obesity. Faecalibacterium with a population of 0.294 ± 0.05 was also the most common genus. The next largest genera were Prevotella—0.203 ± 0.072; Bifidobacterium—0.084 ± 0.019; Subdoligranulum—0.062 ± 0.014; Blautia—0.037 ± 0.011; Bacteroides—0.035 ± 0.006; Roseburia—0.032 ± 0.006; Catenibacterium—0.031 ± 0.005; Fusicatenibacter—0.024 ± 0.006; Collinsella—0.022 ± 0.006; Clostridium sensu stricto 2—0.022 ± 0.006; Phascolarctobacterium—0.020 ± 0.005; Holdemanella—0.019 ± 0.005; Pseudobutyrivibrio—0.017 ± 0.001; Megasphaera—0.013 ± 0.002. The average abundance of other genus in the group of patients with obesity was less than 0.001.
We observed a microbiome landscape dominated by the genus Bifidobacterium—0.346 ± 0.070 after a two-week intake of lactic acid bacteria in obese patients. The next largest genera were Blautia—0.142 ± 0.033; Streptococcus—0.132 ± 0.014; Collinsella—0.088 ± 0.038; Faecalibacterium—0.046 ± 0.009; Bacteroides—0.034 ± 0.013; Fusicatenibacter—0.028 ± 0.004; Eubacterium hallii group—0.026 ± 0.004; Prevotella—0.024 ± 0.007; Roseburia—0.021 ± 0.005; Anaerostipes—0.014 ± 0.003; Subdoligranulum—0.012 ± 0.006; Ruminococcus—0.011 ± 0.005. The number of other bacterial genera was less than 0.010.
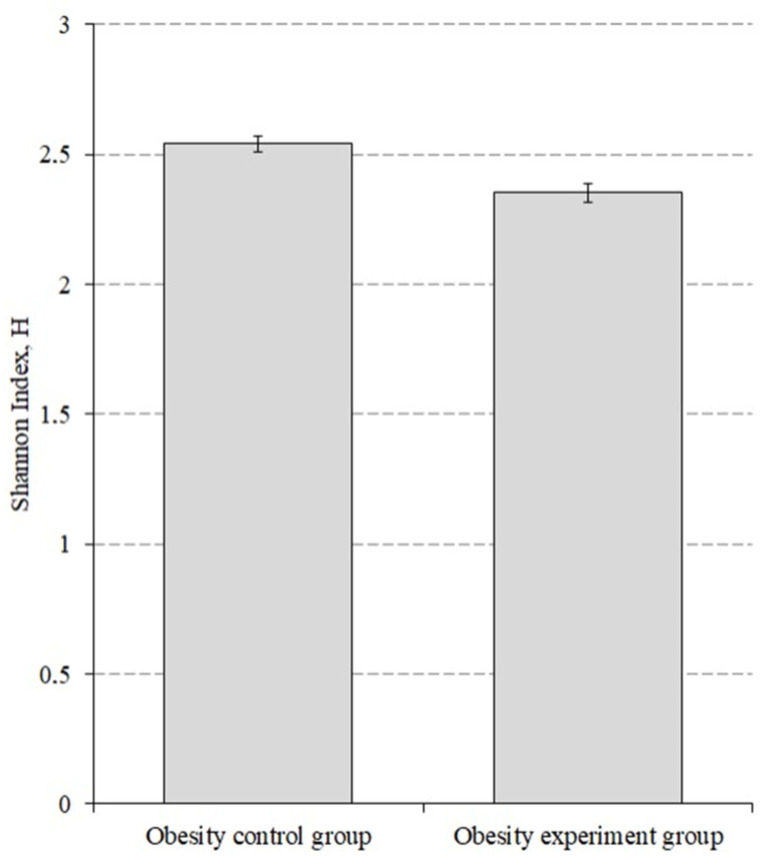
We also evaluated the alpha diversity before and after lactic acid bacteriaintake by obesity group of patients (Figure 4).
Figure 4.
The alpha diversity index for the obesity patients before and after the intake of lactic acid bacteria. p < 0.001.
The alpha diversity index before taking the lactic acid bacteria was 2.540 ± 0.031, while after taking it decreased to 2.351 ± 0.035 (p < 0.001).
Statistically significant differences in the microbiome composition before and after the intake of lactic acid bacteria by an obesity group of patients were identified for 30 bacterial genera (Figure 5).
Figure 5.
The statistically significant differences in the content of the genera of bacteria among the obesity control group and the obesity group of patients after the intake of lactic acid bacteria. * p < 0.05; ** p < 0.01.
After taking lactic acid bacteria, we observed a statistically significant increase in the genus Bifidobacterium—0.346 ± 0.070 vs. 0.084 ± 0.019 (p < 0.05), Blautia—0.142 ± 0.033 vs. 0.037 ± 0.011 (p < 0.01), Streptococcus—0.132 ± 0.014 vs. 0.004 ± 0.000 (p < 0.01), Eubacterium hallii group—0.026 ± 0.005 vs. 0.003 ± 0.001 (p < 0.01), Anaerostipes—0.013 ± 0.003 vs. 0.002 ± 0.001 (p < 0.01), Dialister—0.004 ± 0.001 vs. 0.000 ± 0.000 (p < 0.05), Lactococcus—0.002 ± 0.001 vs. 0.000 ± 0.000 (p < 0.05), Escherichia-Shigella—0.003 ± 0.001 vs. 0.000 ± 0.000 (p < 0.05), Lachnospiraceae ND3007 group—0.005 ± 0.001 vs. 0.001 ± 0.000 (p < 0.05), Erysipelotrichaceae UCG-003—0.002 ± 0.000 vs. 0.001 ± 0.000 (p < 0.05), and Lachnoclostridium—0.005 ± 0.002 vs. 0.001 ± 0.000 compared with the pre-supplement microbiome control obesity group, respectively.
At the same time, we observed a decrease in the number for the genus Faecalibacterium—0.046 ± 0.009 vs. 0.294 ± 0.051 (p < 0.01), Prevotella—0.024 ± 0.007 vs. 0.203 ± 0.072 (p < 0.05), Subdoligranulum—0.012 ± 0.006 vs. 0.062 ± 0.014 (p < 0.01), Holdemanella—0.000 ± 0.000 vs. 0.019 ± 0.005 (p < 0.01), Oscillospiraceae UCG-002—0.001 ± 0.000 vs. 0.004 ± 0.000 (p < 0.01), Alistipes—0.001 ± 0.000 vs. 0.004 ± 0.001 (p < 0.05), Pseudobutyrivibrio—0.006 ± 0.002 vs. 0.017 ± 0.001 (p < 0.05), Ruminococcaceae CAG-352—0.001 ± 0.000 vs. 0.003 ± 0.000 (p < 0.01), Clostridium sensu stricto 2—0.001 ± 0.000 vs. 0.022 ± 0.006 (p < 0.01), Catenibacterium—0.000 ± 0.000 vs. 0.031 ± 0.005 (p < 0.01), Parabacteroides—0.001 ± 0.000 vs. 0.007 ± 0.001 (p < 0.01), Megasphaera—0.002 ± 0.001 vs. 0.013 ± 0.002 (p < 0.01), Phascolarctobacterium—0.000 ± 0.000 vs. 0.020 ± 0.005 (p < 0.01), Christensenellaceae R-7 group—0.000 ± 0.000 vs. 0.003 ± 0.000 (p < 0.01), Monoglobus—0.000 ± 0.000 vs. 0.001 ± 0.000 (p < 0.05), Solobacterium—0.000 ± 0.000 vs. 0.006 ± 0.001 (p < 0.01), Oscillospiraceae NK4A214 group—0.000 ± 0.000 vs. 0.001 ± 0.000 (p < 0.05), Oscillospiraceae UCG-005—0.000 ± 0.000 vs. 0.001 ± 0.000 (p < 0.01), and Clostridium sensu stricto 1—0.000 ± 0.000 vs. 0.001 ± 0.000 (p < 0.05) after taking the lactic acid bacteria compared to the obesity control, respectively.
4. Discussion
4.1. Changes in Microbiome Composition at the Phylum Level
The phylum Actinobacteriota, whose increase we observed after the ingestion of lactic acid bacteria, includes members that can have a beneficial effect on intestinal health. It is also known that there is an inverse correlation of the number of this phylum with indicators characteristic of irritable bowel syndrome [25,26].
After 14 days of lactic acid bacteria intake, the number of the Bacteroidota phyla decreased. Members of this type are the main colonizers of the gastrointestinal tract. Many of these have the ability to produce butyrate, which is involved in maintaining gut health [27]. However, it is also known that an increase in individual members of Bacteroidota is associated with the development of obesity and inflammatory bowel disease [28,29].
4.2. Changes in the Relative Abundance of Beneficial Bacteria
In our study, we observed changes in the abundance of some potentially beneficial bacteria, particularly the genera Bifidobacterium, Blautia, Eubacterium hallii group, Anaerostipes, Lactococcus, and Lachnospiraceae ND3007 group. In contrast, the number of representatives of some other beneficial genus (e.g., Subdoligranulum, Holdemanella, Parabacteroides) decreased after probiotics in obese patients.
Members of the genus Bifidobacterium are commensal microorganisms found in the human gastrointestinal tract and are traditionally considered beneficial to human health [30]. A study by Cerano C. Da Silva et al. showed that Bifidobacterium species were involved in the reduction of body weight and fat mass in Sprague-Dawley rats fed a high-fat diet [31]. This suggests that Bifidobacterium spp. potentially have anti-obesity properties.
Members of the genus Blautia are well-known as butyrate-producing bacteria. This metabolite may explain the beneficial role of these bacteria in glucose metabolism and inflammation associated with obesity [32,33].
Eubacterium hallii group is a butyrate-producing bacterium [34]. Unlike many other bacteria (e.g., Roseburia) that produce butyrate from monosaccharides, E. hallii can produce butyrate from lactate acetate in a low pH environment [35]. In a study by Shanthadevi Udayappan et al., the oral administration of active E. hallii improved the insulin sensitivity in severely resistant db/db mice insulin. In addition, E. hallii has been shown to contribute to changes in SCFA production and bile acid composition. The ability to exert these effects suggests that E. hallii is capable of beneficially affecting insulin sensitivity [32].
Anaerostipes is the most efficient consumer of lactate in the human colon [36]. It was previously shown that some bacteria of this genus are able to reduce glucose levels [37], and can also promote the production of propionate through inositol or phytate, thereby reducing the risk of metabolic diseases [38]. These data point to the potential health benefits of bacteria of the genus Anaerostipes.
The genus Lactococcus is a species of lactic acid bacteria that produce lactic acid as a result of the fermentation of carbohydrates [39]. Many species of Lactococcus are widely used as starter bacteria in the production of cheese and other fermented dairy products. However, there have been several studies showing the possibility of the presence of strains of Lactococcus in the flora of the gastrointestinal tract of humans or animals [40,41]. They are not pathogenic to humans, especially Lactococcus lactis and Lactococcus garvieae, which are able to produce antimicrobial agents [42]. Some species of bacteria of the genus Lactococcus including Lactococcus lactis and Lactococcus petauri have probiotic properties [43,44]. All of this suggests that the intake of probiotics based on lactic acid bacteria against the background of obesity contributes to an increase in the number of beneficial bacteria of the genus Lactococcus.
Members of the Lachnospiraceae ND3007 group are putative producers of short-chain fatty acids (SCFAs) [45]. It has been shown that with a significant and inverse correlation, the Lachnospiraceae ND3007 group has been associated with a decrease in the blood glucose levels in rats with diabetes [33].
It is known that bacteria of the genus Subdoligranulum are SCFA producers and are conditionally positive for the body [46]. It has also been previously noted that the genus Subdoligranulum is positively associated with overall metabolic health including lower body mass index and fat mass in adult humans [47].
The genus Holdemanella is considered to be a potential health contributor [48]. A positive correlation of the bacterial genus Holdemanella with the consumption of fermented dairy products, carbohydrates, and fiber was previously reported [49].
Members of the genus Parabacteroides are saccharolytic bacteria and produce major fermentation end products such as acetic acid and succinic acid. According to numerous studies, the relative abundance of Parabacteroides has been negatively associated with BMI [50,51,52,53] In addition, according to studies, some species such as Parabacteroides goldsteinii and Parabacteroides distasonis are considered as promising probiotics that may alleviate obesity and metabolic dysfunctions associated with obesity [53,54]. However, in our study, we observed a decrease in the abundance of this genus after the intake of lactic acid bacteria.
4.3. Changes in the Number of Pathogenic and Opportunistic Bacteria after Taking Probiotics
As a result of taking probiotics, we observed a decrease in some pathogenic bacteria such as Clostridium sensu stricto 2 and Solobacterium. Furthermore, an increase in the relative abundance of the potentially pathogenic genus Streptococcus was observed.
Previously, the bacterial genus Clostridium sensu stricto 2 was identified in human feces and clinical specimens such as blood, peritoneal and pleural fluid as well as lung biopsies from lung infections [55]. Members of this genus are associated with the development of infectious diseases: gas gangrene as well as mixed infections and metritis [56]. Thus, the data obtained indicate that this genus of bacteria has a high pathogenicity, which contributes to the development of infectious diseases.
Solobacterium is a Gram-positive obligate anaerobic genus in the family Erysipelotrichidae with one known species. This genus is part of the salivary microbiome, but is also found in small amounts in the colon [57,58], which refers to conditionally pathogenic microorganisms. Numerous studies have revealed a positive relationship between the increase in the abundance of this genus and the development of obesity [59,60]
Most Streptococcus species are considered pathogenic, but Streptococcus thermophilus, for example, is a probiotic [61]. S. thermophilus was in the composition of the final product containing lactic acid bacteria (see Materials and Methods). S. thermophilus is known to ferment lactose and sucrose, and can also metabolize fructose monosaccharide [62]. High fructose levels may be due to low numbers of these beneficial bacteria [63].
4.4. Changes in the Number of Bacteria Associated with Obesity
In our study, we found an increase in obesity-associated bacteria such as Dialister and Lachnoclostridium. Furthermore, some bacteria with a role associated with obesity decreased including Faecalibacterium, Alistipes, Prevotella, Catenibacterium, Megasphaera, Phascolarctobacterium, Christensenellaceae R-7 group, Monoglobus, and the Oscillospiraceae NK4A214 group.
A study by Yanrong Lv et al. showed that the abundance of Dialister bacteria was lower in the overweight group compared to the lean group [64]. In our study, the intake of lactic acid bacteria led to an increase in the number of these bacteria in obese people. It is possible that bacteria of the genus Dialister play some role in the development of obesity, but more detailed studies of these bacteria are needed to understand the exact mechanisms.
In a study by Ana Nogal et al., increased levels of Lachnoclostridium bacteria led to lower levels of circulating acetate, which ultimately led to increased visceral fat. However, the study only looked at women and also only measured the changes in acetate, not all SCFAs [65]. We found that the relative abundance of members of the genus Lachnoclostridium increased in obese patients after taking probiotics. This suggests that more research is needed on the role of Lachnoclostridium bacteria in obesity.
Studies using the analysis of the association of individual microbial genera (according to 16S rRNA gene sequencing) with obesity or type 2 diabetes mellitus showed that the reduced activity of Faecalibacterium, Alistipes is significant in relation to obesity [66]. This is consistent with the results obtained in our study for these genera of bacteria.
The genus Alistipes is known to be anaerobic bacteria found mainly in the microbiota of the gastrointestinal tract of healthy humans [67]. According to the results of studies, a marked decrease in the abundance of the genus Alistipes was observed in people with obesity compared with people with normal weight [66]. Additionally, the results were revealed in a study by Duan et al. (2021), where significant differences in the microbiota between people diagnosed with obesity and the control group, and the number of bacteria of the genus Alistipes significantly decreased [68]. In our study, a trend toward a decrease in the number of this genus of bacteria was obtained after two weeks of supplementation containing lactic acid bacteria. The differences between the obtained results and the literature data indicate an incomplete study of this genus of bacteria in relation to obesity.
Prevotella also decreased after two weeks of dietary lactic acid bacteria supplementation, and these results are consistent with the results of a study by Duan et al. (2021), where a significant difference was demonstrated between the obese and control group [68]. Hu H.J. et al. also reported an increase in the abundance of the genus Prevotella in people with obesity [69]. It has been established that bacteria of this genus are able to separate insoluble plant fiber and ferment soluble carbohydrates to form SCFAs [70]. The revealed ability of the genus Prevotella may be useful on a low-calorie diet, but in turn can lead to obesity on a fairly high-calorie diet. The results of studies demonstrate the association of the abundance of the Prevotella enterotype with a long-term diet that included a high intake of soluble carbohydrates and simple sugars and a low intake of proteins, amino acids, and saturated fats [71,72].
The study by Martínez-Cuesta et al. (2021) noted that bacteria of the genus Catenibacterium were significantly higher in obese participants, while the bacterial genus Alistipes was reduced [73].
Megasphaera is a genus of Gram-negative, non-motile, coccoid, opportunistic bacteria [74]. It is a part of the resident microflora of the human colon. This genus produces SCFAs: butyric, isobutyric, valeric, caproic, isovaleric, and isocaproic. This bacterium can use lactate and produce beneficial butyrate [75]. However, the pathway of its formation is associated with the production of ammonia, which can harm the host [76]. A direct correlation was found between the abundance of the genus and indicators characteristic of obesity [51,77,78].
Phascolarctobacterium is a relatively numerous genus of bacteria in the human gastrointestinal tract. A study in obese rats showed that an increase in Phascolarctobacterium was positively correlated with an increase in body weight, fat mass, plasma leptin, triglycerides, and glucose tolerance [79]. It is also associated with unfavorable glycemic metabolism in a healthy young population [80]. In addition, according to some data, this genus can be considered as a biomarker for type 2 diabetes mellitus [81].
Most members of the Christensenellaceae R-7 group genus are saccharolytic, and they can ferment carbohydrates with the accumulation of a mixture of SCFAs [82]. Studies have also confirmed a negative correlation between the abundance of the Christensenellaceae R-7 group and body weight and visceral fat content [82,83,84]. Although Christensenellaceae R-7 is believed to be reduced in patients suffering from intestinal diseases, we noted a reduction in this genus with a probiotic mixture of lactic acid bacteria.
The genus Monoglobus is one of the main specialized taxa involved in the fermentation of pectin and mannan in the human colon [85,86]. There are few studies describing the role of the genus Monoglobus in the effect of metabolic disorders. It is known to be inversely correlated with BMI as well as with the development of ulcerative colitis [87,88].
According to the literature data, the abundance of the Oscillospiraceae NK4A214 group genus has a direct correlation with obesity, regardless of the cohort studied, and therefore is a contender for one of the prognostic genera for obesity [89].
4.5. Changes in the Abundance of Bacteria Whose Role Is Ambiguous or Unknown
The role of some bacteria, the number of which changed in our study, is still poorly understood.
Escherichia and Shigella are two closely related genera that share bioenergetic mechanisms that allow them to fill a specific niche in the gut microbiome ecosystem [90]. The genus Escherichia-Shigella includes four Escherichia species, four Shigella species, and several lineages currently unclassified as “Escherichia cryptic clades” [91]. All of them have different functions, some species exhibit pathogenic properties, while others are described as beneficial to humans [92,93,94]. In our study, a limitation of the approach is that the type of assay used combines different species of bacteria of the genus Escherichia-Shigella with a wide range of functions. Thus, this does not allow us to draw clear conclusions about the role played by the increase in the relative abundance of bacteria of the genus Escherichia-Shigella in the case of obesity after the ingestion of lactic acid bacteria.
In our study, a reduction in bacteria belonging to the genus Oscillospiraceae UCG-002 was demonstrated after two weeks of lactic acid supplementation. Bacteria of the genus Oscillospiraceae UCG-002 are known to significantly contribute to aspartate degradation and glycine degradation [95].
The bacterial genus Ruminococcaceae CAG-352 also showed a decrease after two weeks of lactic acid bacteria supplementation. The genus Ruminococcaceae CAG-352 is known to be Gram-positive bacteria [96]. We did not find evidence of an association of this genus of bacteria with obesity in the literature.
For the genus Oscillospiraceae UCG-005, we observed a decrease in abundance after lactic acid bacteria supplementation, but the relationship of this genus with any metabolic pathologies in humans is poorly understood. Further research is needed to shed light on the contribution of this genus to gut health.
The contribution of Clostridium sensu stricto 1 to human gut health is poorly understood. There are data that have found a direct correlation of Clostridium sensu stricto 1 with acute pancreatitis in rats [97] and with necrotic enteritis [98]. It is also thought that an abundance of Clostridium sensu stricto 1 may be responsible for the visceral hypersensitivity seen in irritable bowel syndrome [99]. Further studies are needed to suggest what role this bacterial cluster plays in the human gut. We found a reduction of this kind after the intake of lactic acid bacteria in obese patients.
Little is known about the role of bacteria of the genus Erysipelotrichaceae UCG-003, so it is difficult to say exactly what role members of this genus play.
5. Conclusions
The studied mixture of lactic acid bacteria, consisting of strains of L. acidophilus VSUET12, L. plantarum VSUET13, L. fermentum VSUET14, and Lactobacillus delbrueckii subsp bulgaricus VSUET15, which was taken by obese people, had a significant effect on changing the gut microbiome landscape. According to the results of the study, we observed at the phylum level, a decrease in Bacteroidota and an increase in Actinobacteriota, both of which belong to the main phylum that form the core of the microbiome. At the generic level, a 14-day intake of the mixture of lactic acid bacteria led to the enrichment of the potentially beneficial groups Bifidobacterium, Blautia, E. halli group, Anaerostipes, Lactococcus, and Lachnospiraceae ND3007. We also observed an increase in the genera Streptococcus, Escherichia-Shigella, and Lachnoclostridium, which contributed to gut health and is highly member-dependent.
We also found a decrease in the genera Faecalibacterium, Pseudobutyrivibrio, Subdoligranulum, Faecalibacterium, Clostridium sensu stricto 1 and 2, Catenibacterium, Megasphaera, Phascolarctobacterium, Oscillospiraceae NK4A214 group, which can be associated with the development of various metabolic disorders as well as inflammatory bowel diseases. At the same time, the abundance of beneficial genera such as Subdoligranulum, Holdemanella, Alistipes, Parabacteroides, Christensenellaceae R-7, Monoglobus, and Solobacterium also decreased. The decrease in the genera Prevotella, Oscillospiraceae UCG-002, and UCG-005, Ruminococcaceae CAG-352 cannot be interpreted unambiguously. The effect of some of them on the host organism depends on the specific species and strain, while others are poorly understood.
Thus, based on the data obtained, we cannot unambiguously conclude on the beneficial effect of the mixture of the studied lactic acid bacteria on the gut of obese people. Positive microbiome changes were observed against the background of the intake, but along with these, there were changes in the gut microbiota that could not be interpreted as positive.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/nu14163384/s1, Table S1: The abundance of microorganisms for the samples.
Author Contributions
Conceptualization, M.S. and V.P.; Methodology, M.S. and E.P., Software, M.G.; Validation, M.G. and I.B.; Formal analysis, M.G., Y.S. and I.B.; Investigation, I.B. and Y.S.; Resources, V.P.; Data curation, M.G. and Y.S.; Writing—original draft preparation, M.G., Y.S., I.B. and P.C.; Writing—review and editing, M.S. and V.P.; Visualization, M.G. and I.B.; Supervision, M.S. and V.P.; Project administration, V.P.; Funding acquisition, V.P. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Ethics Board of Voronezh State University (Protocol N42-03, 7 March 2022).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Sequencing data are available in the NCBI BioProject database (BioProject: PRJNA862929).
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was supported by the Ministry of Science and Higher Education of the Russian Federation in the framework of the national project “Science” (project FZGW-2020-0001, unique number of the register of State tasks 075001X39782002).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Versalovic J. The Human Microbiome and Probiotics: Implications for Pediatrics. Ann. Nutr. Metab. 2013;63((Suppl. 2)):42–52. doi: 10.1159/000354899. [DOI] [PubMed] [Google Scholar]
- 2.Blüher M. Obesity: Global Epidemiology and Pathogenesis. Nat. Rev. Endocrinol. 2019;15:288–298. doi: 10.1038/s41574-019-0176-8. [DOI] [PubMed] [Google Scholar]
- 3.Schütz F., Figueiredo-Braga M., Barata P., Cruz-Martins N. Obesity and Gut Microbiome: Review of Potential Role of Probiotics. Porto Biomed. J. 2021;6:e111. doi: 10.1097/j.pbj.0000000000000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tseng C.H., Wu C.Y. The Gut Microbiome in Obesity. J. Formos. Med. Assoc. 2019;118:S3–S9. doi: 10.1016/j.jfma.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 5.Manichanh C., Rigottier-Gois L., Bonnaud E., Gloux K., Pelletier E., Frangeul L., Nalin R., Jarrin C., Chardon P., Marteau P., et al. Reduced Diversity of Faecal Microbiota in Crohn’s Disease Revealed by a Metagenomic Approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turnbaugh P.J., Hamady M., Yatsunenko T., Cantarel B.L., Duncan A., Ley R.E., Sogin M.L., Jones W.J., Roe B.A., Affourtit J.P., et al. A Core Gut Microbiome in Obese and Lean Twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lambeth S.M., Carson T., Lowe J., Ramaraj T., Leff J.W., Luo L., Bell C.J., Shah V.O. Composition, Diversity and Abundance of Gut Microbiome in Prediabetes and Type 2 Diabetes. J. Diabetes Obes. 2015;2:1. doi: 10.15436/2376-0949.15.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baothman O.A., Zamzami M.A., Taher I., Abubaker J., Abu-Farha M. The Role of Gut Microbiota in the Development of Obesity and Diabetes. Lipids Health Dis. 2016;15:108. doi: 10.1186/s12944-016-0278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen Y.-L., Zhang L.-Q., Yang Y., Yin B.-C., Ye B.-C., Zhou Y. Advances in the Role and Mechanism of Lactic Acid Bacteria in Treating Obesity. Food Bioeng. 2022;1:101–115. doi: 10.1002/fbe2.12002. [DOI] [Google Scholar]
- 10.Wei B., Peng Z., Xiao M., Huang T., Zheng W., Xie M., Xiong T. Three Lactic Acid Bacteria with Anti-Obesity Properties: In Vitro Screening and Probiotic Assessment. Food Biosci. 2022;47:101724. doi: 10.1016/j.fbio.2022.101724. [DOI] [Google Scholar]
- 11.Seo M.K., Park E.J., Ko S.Y., Choi E.W., Kim S. Therapeutic Effects of Kefir Grain Lactobacillus-Derived Extracellular Vesicles in Mice with 2,4,6-Trinitrobenzene Sulfonic Acid-Induced Inflammatory Bowel Disease. J. Dairy Sci. 2018;101:8662–8671. doi: 10.3168/jds.2018-15014. [DOI] [PubMed] [Google Scholar]
- 12.Shoukat S. Potential Anti-Carcinogenic Effect of Probiotic and Lactic Acid Bacteria in Detoxification of Benzo[a]Pyrene: A Review. Trends Food Sci. Technol. 2020;99:450–459. doi: 10.1016/j.tifs.2020.02.029. [DOI] [Google Scholar]
- 13.Thiennimitr P., Yasom S., Tunapong W., Chunchai T., Wanchai K., Pongchaidecha A., Lungkaphin A., Sirilun S., Chaiyasut C., Chattipakorn N., et al. Lactobacillus Paracasei HII01, Xylooligosaccharides, and Synbiotics Reduce Gut Disturbance in Obese Rats. Nutrition. 2018;54:40–47. doi: 10.1016/j.nut.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 14.Wu X.Z., Wen Z.G., Hua J.L. Effects of Dietary Inclusion of Lactobacillus and Inulin on Growth Performance, Gut Microbiota, Nutrient Utilization, and Immune Parameters in Broilers. Poult. Sci. 2019;98:4656–4663. doi: 10.3382/ps/pez166. [DOI] [PubMed] [Google Scholar]
- 15.Bull M., Plummer S., Marchesi J., Mahenthiralingam E. The Life History of Lactobacillus Acidophilus as a Probiotic: A Tale of Revisionary Taxonomy, Misidentification and Commercial Success. FEMS Microbiol. Lett. 2013;349:77–87. doi: 10.1111/1574-6968.12293. [DOI] [PubMed] [Google Scholar]
- 16.Allen S.J., Martinez E.G., Gregorio G.V., Dans L.F. Probiotics for Treating Acute Infectious Diarrhoea. Cochrane Database Syst. Rev. 2010;2010:CD003048. doi: 10.1002/14651858.CD003048.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toshimitsu T., Gotou A., Furuichi K., Hachimura S., Asami Y. Effects of 12-Wk Lactobacillus Plantarum OLL2712 Treatment on Glucose Metabolism and Chronic Inflammation in Prediabetic Individuals: A Single-Arm Pilot Study. Nutrition. 2019;58:175–180. doi: 10.1016/j.nut.2018.07.116. [DOI] [PubMed] [Google Scholar]
- 18.Costabile A., Buttarazzi I., Kolida S., Quercia S., Baldini J., Swann J.R., Brigidi P., Gibson G.R. An in Vivo Assessment of the Cholesterol-Lowering Efficacy of Lactobacillus Plantarum ECGC 13110402 in Normal to Mildly Hypercholesterolaemic Adults. PLoS ONE. 2017;12:e0187964. doi: 10.1371/journal.pone.0187964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arena M.P., Capozzi V., Russo P., Drider D., Spano G., Fiocco D. Immunobiosis and Probiosis: Antimicrobial Activity of Lactic Acid Bacteria with a Focus on Their Antiviral and Antifungal Properties. Appl. Microbiol. Biotechnol. 2018;102:9949–9958. doi: 10.1007/s00253-018-9403-9. [DOI] [PubMed] [Google Scholar]
- 20.Davoodabadi A., Soltan Dallal M.M., Rahimi Foroushani A., Douraghi M., Sharifi Yazdi M., Amin Harati F. Antibacterial Activity of Lactobacillus Spp. Isolated from the Feces of Healthy Infants against Enteropathogenic Bacteria. Anaerobe. 2015;34:53–58. doi: 10.1016/j.anaerobe.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 21.Salvetti E., Torriani S., Felis G.E. The Genus Lactobacillus: A Taxonomic Update. Probiotics Antimicrob. Proteins. 2012;4:217–226. doi: 10.1007/s12602-012-9117-8. [DOI] [PubMed] [Google Scholar]
- 22.Rodríguez-Nogales A., Algieri F., Garrido-Mesa J., Vezza T., Utrilla M.P., Chueca N., Garcia F., Olivares M., Rodríguez-Cabezas M.E., Gálvez J. Differential Intestinal Anti-Inflammatory Effects of Lactobacillus Fermentum and Lactobacillus Salivarius in DSS Mouse Colitis: Impact on MicroRNAs Expression and Microbiota Composition. Mol. Nutr. Food Res. 2017;61:1700144. doi: 10.1002/mnfr.201700144. [DOI] [PubMed] [Google Scholar]
- 23.Maldonado J., Cañabate F., Sempere L., Vela F., Sánchez A.R., Narbona E., López-Huertas E., Geerlings A., Valero A.D., Olivares M., et al. Human Milk Probiotic Lactobacillus Fermentum CECT5716 Reduces the Incidence of Gastrointestinal and Upper Respiratory Tract Infections in Infants. J. Pediatr. Gastroenterol. Nutr. 2012;54:55–61. doi: 10.1097/MPG.0b013e3182333f18. [DOI] [PubMed] [Google Scholar]
- 24.Nardone G., Compare D., Liguori E., di Mauro V., Rocco A., Barone M., Napoli A., Lapi D., Iovene M.R., Colantuoni A. Protective Effects of Lactobacillus Paracasei F19 in a Rat Model of Oxidative and Metabolic Hepatic Injury. Am. J. Physiol. Gastrointest Liver Physiol. 2010;299:G669–G679. doi: 10.1152/ajpgi.00188.2010. [DOI] [PubMed] [Google Scholar]
- 25.Krogius-Kurikka L., Lyra A., Malinen E., Aarnikunnas J., Tuimala J., Paulin L., Mäkivuokko H., Kajander K., Palva A. Microbial Community Analysis Reveals High Level Phylogenetic Alterations in the Overall Gastrointestinal Microbiota of Diarrhoea-Predominant Irritable Bowel Syndrome Sufferers. BMC Gastroenterol. 2009;9:95. doi: 10.1186/1471-230X-9-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salonen A., de Vos W.M., Palva A. Gastrointestinal Microbiota in Irritable Bowel Syndrome: Present State and Perspectives. Microbiology. 2010;156:3205–3215. doi: 10.1099/mic.0.043257-0. [DOI] [PubMed] [Google Scholar]
- 27.Shao J., Li Z., Gao Y., Zhao K., Lin M., Li Y., Wang S., Liu Y., Chen L. Construction of a “Bacteria-Metabolites” Co-Expression Network to Clarify the Anti-Ulcerative Colitis Effect of Flavonoids of Sophora Flavescens Aiton by Regulating the “Host-Microbe” Interaction. Front. Pharmacol. 2021;12:710052. doi: 10.3389/fphar.2021.710052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stojanov S., Berlec A., Štrukelj B. The Influence of Probiotics on the Firmicutes/Bacteroidetes Ratio in the Treatment of Obesity and Inflammatory Bowel Disease. Microorganisms. 2020;8:1715. doi: 10.3390/microorganisms8111715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwon H., Chae S.H., Jung H.J., Shin H.M., Ban O.H., Yang J., Kim J.H., Jeong J.E., Jeon H.M., Kang Y.W., et al. The Effect of Probiotics Supplementation in Postoperative Cancer Patients: A Prospective Pilot Study. Ann. Surg. Treat. Res. 2021;101:281–290. doi: 10.4174/astr.2021.101.5.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salazar N., Neyrinck A.M., Bindels L.B., Druart C., Ruas-Madiedo P., Cani P.D., de los Reyes-Gavilán C.G., Delzenne N.M. Functional Effects of EPS-Producing Bifidobacterium Administration on Energy Metabolic Alterations of Diet-Induced Obese Mice. Front. Microbiol. 2019;10:1809. doi: 10.3389/fmicb.2019.01809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.An H.M., Park S.Y., Lee D.K., Kim J.R., Cha M.K., Lee S.W., Lim H.T., Kim K.J., Ha N.J. Antiobesity and Lipid-Lowering Effects of Bifidobacterium Spp. in High Fat Diet-Induced Obese Rats. Lipids Health Dis. 2011;10:116. doi: 10.1186/1476-511X-10-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Udayappan S., Manneras-Holm L., Chaplin-Scott A., Belzer C., Herrema H., Dallinga-Thie G.M., Duncan S.H., Stroes E.S.G., Groen A.K., Flint H.J., et al. Oral Treatment with Eubacterium Hallii Improves Insulin Sensitivity in Db/Db Mice. NPJ Biofilms Microbiomes. 2016;2:16009. doi: 10.1038/npjbiofilms.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Q., Xiao X., Li M., Yu M., Ping F., Zheng J., Wang T., Wang X. Vildagliptin Increases Butyrate-Producing Bacteria in the Gut of Diabetic Rats. PLoS ONE. 2017;12:e0184735. doi: 10.1371/journal.pone.0184735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Louis P., Young P., Holtrop G., Flint H.J. Diversity of Human Colonic Butyrate-Producing Bacteria Revealed by Analysis of the Butyryl-CoA:Acetate CoA-Transferase Gene. Environ. Microbiol. 2010;12:304–314. doi: 10.1111/j.1462-2920.2009.02066.x. [DOI] [PubMed] [Google Scholar]
- 35.Duncan S.H., Louis P., Flint H.J. Lactate-Utilizing Bacteria, Isolated from Human Feces, That Produce Butyrate as a Major Fermentation Product. Appl. Environ. Microbiol. 2004;70:5810–5817. doi: 10.1128/AEM.70.10.5810-5817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muñoz-Tamayo R., Laroche B., Walter É., Doré J., Duncan S.H., Flint H.J., Leclerc M. Kinetic Modelling of Lactate Utilization and Butyrate Production by Key Human Colonic Bacterial Species. FEMS Microbiol. Ecol. 2011;76:615–624. doi: 10.1111/j.1574-6941.2011.01085.x. [DOI] [PubMed] [Google Scholar]
- 37.Bui T.P.N., Mannerås-Holm L., Puschmann R., Wu H., Troise A.D., Nijsse B., Boeren S., Bäckhed F., Fiedler D., deVos W.M. Conversion of Dietary Inositol into Propionate and Acetate by Commensal Anaerostipes Associates with Host Health. Nat. Commun. 2021;12:4798. doi: 10.1038/s41467-021-25081-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ioannou G.N. The Role of Cholesterol in the Pathogenesis of NASH. Trends Endocrinol. Metab. 2016;27:84–95. doi: 10.1016/j.tem.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 39.Rattanachaikunsopon P., Phumkhachorn P. Lactic Acid Bacteria: Their Antimicrobial Compounds and Their Uses in Food Production. Sch. Res. Libr. Ann. Biol. Res. 2010;1:218–228. [Google Scholar]
- 40.Gruzza M., Duval-Iflah Y., Ducluzeau R. Colonization of the Digestive Tract of Germ-Free Mice by Genetically Engineered Strains of Lactococcus Lactis: Study of Recombinant DNA Stability. Microb. Releases Viruses Bact. Fungi. 1992;1:165–171. [PubMed] [Google Scholar]
- 41.Klijn N., Weerkamp A.H., de Vos W.M. Genetic Marking of Lactococcus Lactis Shows Its Survival in the Human Gastrointestinal Tract. Appl. Environ. Microbiol. 1995;61:2771. doi: 10.1128/aem.61.7.2771-2774.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyburgh C.M., Bragg R.R., Boucher C.E. Lactococcus Garvieae: An Emerging Bacterial Pathogen of Fish. Dis. Aquat. Organ. 2017;123:67–79. doi: 10.3354/dao03083. [DOI] [PubMed] [Google Scholar]
- 43.Yerlikaya O. Probiotic Potential and Biochemical and Technological Properties of Lactococcus Lactis Ssp. Lactis Strains Isolated from Raw Milk and Kefir Grains. J. Dairy Sci. 2019;102:124–134. doi: 10.3168/jds.2018-14983. [DOI] [PubMed] [Google Scholar]
- 44.Li O., Zhang H., Wang W., Liang Y., Chen W., Ud Din A., Li L., Zhou Y. Complete Genome Sequence and Probiotic Properties of Lactococcus Petauri LZys1 Isolated from Healthy Human Gut. J. Med. Microbiol. 2021;70:001397. doi: 10.1099/jmm.0.001397. [DOI] [PubMed] [Google Scholar]
- 45.Nishiwaki H., Hamaguchi T., Ito M., Ishida T., Maeda T., Kashihara K., Tsuboi Y., Ueyama J., Shimamura T., Mori H., et al. Short-Chain Fatty Acid-Producing Gut Microbiota Is Decreased in Parkinson’s Disease but Not in Rapid-Eye-Movement Sleep Behavior Disorder. mSystems. 2020;5:e00797-20. doi: 10.1128/mSystems.00797-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cani P.D., de Hase E.M., van Hul M. Gut Microbiota and Host Metabolism: From Proof of Concept to Therapeutic Intervention. Microorganisms. 2021;9:1302. doi: 10.3390/microorganisms9061302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Hul M., le Roy T., Prifti E., Dao M.C., Paquot A., Zucker J.D., Delzenne N.M., Muccioli G., Clément K., Cani P.D. From Correlation to Causality: The Case of Subdoligranulum. Gut Microbes. 2020;12:1849998. doi: 10.1080/19490976.2020.1849998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang C., Liang D., Li X., Liu J., Fan M., Jing M., Wang Y., Zhang Y., Fang Y., Li D. Characteristics of Gut Microbial Profiles of Offshore Workers and Its Associations with Diet. Front. Nutr. 2022;9:1183. doi: 10.3389/fnut.2022.904927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma Y., Zhu L., Ma Z., Gao Z., Wei Y., Shen Y., Li L., Liu X., Ren M. Distinguishing Feature of Gut Microbiota in Tibetan Highland Coronary Artery Disease Patients and Its Link with Diet. Sci. Rep. 2021;11:18486. doi: 10.1038/s41598-021-98075-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang S., Wu P., Tian Y., Liu B., Huang L., Liu Z., Lin N., Xu N., Ruan Y., Zhang Z., et al. Gut Microbiota Serves a Predictable Outcome of Short-Term Low-Carbohydrate Diet (LCD) Intervention for Patients with Obesity. Microbiol. Spectr. 2021;9:e00223-21. doi: 10.1128/Spectrum.00223-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palmas V., Pisanu S., Madau V., Casula E., Deledda A., Cusano R., Uva P., Vascellari S., Loviselli A., Manzin A., et al. Gut Microbiota Markers Associated with Obesity and Overweight in Italian Adults. Sci. Rep. 2021;11:5532. doi: 10.1038/s41598-021-84928-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zeng Q., Li D., He Y., Li Y., Yang Z., Zhao X., Liu Y., Wang Y., Sun J., Feng X., et al. Discrepant Gut Microbiota Markers for the Classification of Obesity-Related Metabolic Abnormalities. Sci. Rep. 2019;9:13424. doi: 10.1038/s41598-019-49462-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang K., Liao M., Zhou N., Bao L., Ma K., Zheng Z., Wang Y., Liu C., Wang W., Wang J., et al. Parabacteroides Distasonis Alleviates Obesity and Metabolic Dysfunctions via Production of Succinate and Secondary Bile Acids. Cell Rep. 2019;26:222–235. doi: 10.1016/j.celrep.2018.12.028. [DOI] [PubMed] [Google Scholar]
- 54.Wu T.R., Lin C.S., Chang C.J., Lin T.L., Martel J., Ko Y.F., Ojcius D.M., Lu C.C., Young J.D., Lai H.C. Gut Commensal Parabacteroides Goldsteinii Plays a Predominant Role in the Anti-Obesity Effects of Polysaccharides Isolated from Hirsutella Sinensis. Gut. 2019;68:248–262. doi: 10.1136/gutjnl-2017-315458. [DOI] [PubMed] [Google Scholar]
- 55.Lawson P.A., Rainey F.A. Proposal to Restrict the Genus Clostridium Prazmowski to Clostridium Butyricum and Related Species. Int. J. Syst. Evol. Microbiol. 2016;66:1009–1016. doi: 10.1099/ijsem.0.000824. [DOI] [PubMed] [Google Scholar]
- 56.Biström M., Moisander-Jylhä A.M., Heinikainen S., Pelkola K., Raunio-Saarnisto M. Isolation of Clostridium Limosum from an Outbreak of Metritis in Farmed Mink. Acta Vet. Scand. 2016;58:49. doi: 10.1186/s13028-016-0230-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barrak I., Stájer A., Gajdács M., Urbán E. Small, but Smelly: The Importance of Solobacterium Moorei in Halitosis and Other Human Infections. Heliyon. 2020;6:e05371. doi: 10.1016/j.heliyon.2020.e05371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alauzet C., Aujoulat F., Lozniewski A., Brahim S., Domenjod C., Enault C., Lavigne J.P., Marchandin H. A New Look at the Genus Solobacterium: A Retrospective Analysis of Twenty-Seven Cases of Infection Involving S. Moorei and a Review of Sequence Databases and the Literature. Microorganisms. 2021;9:1229. doi: 10.3390/microorganisms9061229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu Y., Chi X., Zhang Q., Chen F., Deng X. Characterization of the Salivary Microbiome in People with Obesity. PeerJ. 2018;2018:e4458. doi: 10.7717/peerj.4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reimer R.A., Delzenne N.M. Dieting for Success: What Baseline Gut Microbiota Can Tell You About Your Chances of Losing Weight. Gastroenterology. 2021;160:1933–1935. doi: 10.1053/j.gastro.2021.02.021. [DOI] [PubMed] [Google Scholar]
- 61.Schots P.C., Jansen K.M., Mrazek J., Pedersen A.M., Olsen R.L., Larsen T.S. Obesity-Induced Alterations in the Gut Microbiome in Female Mice Fed a High-Fat Diet Are Antagonized by Dietary Supplementation with a Novel, Wax Ester-Rich, Marine Oil. Nutr. Res. 2020;83:94–107. doi: 10.1016/j.nutres.2020.09.002. [DOI] [PubMed] [Google Scholar]
- 62.Hutkins R.W., Morris H.A. Carbohydrate Metabolism by Streptococcus Thermophilus: A Review. J. Food Prot. 1987;50:876–884. doi: 10.4315/0362-028X-50.10.876. [DOI] [PubMed] [Google Scholar]
- 63.Jones R.B., Alderete T.L., Kim J.S., Millstein J., Gilliland F.D., Goran M.I. High Intake of Dietary Fructose in Overweight/Obese Teenagers Associated with Depletion of Eubacterium and Streptococcus in Gut Microbiome. Gut Microbes. 2019;10:712–719. doi: 10.1080/19490976.2019.1592420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lv Y., Qin X., Jia H., Chen S., Sun W., Wang X. The Association between Gut Microbiota Composition and BMI in Chinese Male College Students, as Analysed by next-Generation Sequencing. Br. J. Nutr. 2019;122:986–995. doi: 10.1017/S0007114519001909. [DOI] [PubMed] [Google Scholar]
- 65.Nogal A., Louca P., Zhang X., Wells P.M., Steves C.J., Spector T.D., Falchi M., Valdes A.M., Menni C. Circulating Levels of the Short-Chain Fatty Acid Acetate Mediate the Effect of the Gut Microbiome on Visceral Fat. Front. Microbiol. 2021;12:1943. doi: 10.3389/fmicb.2021.711359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thingholm L.B., Rühlemann M.C., Koch M., Fuqua B., Laucke G., Boehm R., Bang C., Franzosa E.A., Hübenthal M., Rahnavard A., et al. Obese Individuals with and without Type 2 Diabetes Show Different Gut Microbial Functional Capacity and Composition. Cell Host Microbe. 2019;26:252–264. doi: 10.1016/j.chom.2019.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shkoporov A.N., Chaplin A.V., Khokhlova E.V., Shcherbakova V.A., Motuzova O.V., Bozhenko V.K., Kafarskaia L.I., Efimov B.A. Alistipes Inops Sp. Nov. and Coprobacter Secundus Sp. Nov., Isolated from Human Faeces. Int. J. Syst. Evol. Microbiol. 2015;65:4580–4588. doi: 10.1099/ijsem.0.000617. [DOI] [PubMed] [Google Scholar]
- 68.Duan M., Wang Y., Zhang Q., Zou R., Guo M., Zheng H. Characteristics of Gut Microbiota in People with Obesity. PLoS ONE. 2021;16:e0255446. doi: 10.1371/journal.pone.0255446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu H.J., Park S.G., Jang H.B., Choi M.G., Park K.H., Kang J.H., Park S.I., Lee H.J., Cho S.H. Obesity Alters the Microbial Community Profile in Korean Adolescents. PLoS ONE. 2015;10:e0134333. doi: 10.1371/journal.pone.0134333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Flint H.J., Bayer E.A., Rincon M.T., Lamed R., White B.A. Polysaccharide Utilization by Gut Bacteria: Potential for New Insights from Genomic Analysis. Nat. Rev. Microbiol. 2008;6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- 71.Wu G.D., Chen J., Hoffmann C., Bittinger K., Chen Y.Y., Keilbaugh S.A., Bewtra M., Knights D., Walters W.A., Knight R., et al. Linking Long-Term Dietary Patterns with Gut Microbial Enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Durbán A., Abellán J.J., Latorre A., Moya A. Effect of Dietary Carbohydrate Restriction on an Obesity-Related Prevotella-Dominated Human Fecal Microbiota. Metagenomics. 2013;2:235722. doi: 10.4303/mg/235722. [DOI] [Google Scholar]
- 73.Martínez-Cuesta M.C., del Campo R., Garriga-García M., Peláez C., Requena T. Taxonomic Characterization and Short-Chain Fatty Acids Production of the Obese Microbiota. Front. Cell. Infect. Microbiol. 2021;11:516. doi: 10.3389/fcimb.2021.598093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bordigoni A., Lo C.I., Yimagou E.K., Diop K., Nicaise B., Raoult D., Desnues C., Fenollar F. Megasphaera Vaginalis Sp. Nov. and Anaerococcus vaginimassiliensis sp. nov., New Bacteria Isolated from Vagina of French Woman with Bacterial Vaginosis. New Microbes New Infect. 2020;37:100706. doi: 10.1016/j.nmni.2020.100706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Feng W., Ao H., Peng C. Gut Microbiota, Short-Chain Fatty Acids, and Herbal Medicines. Front. Pharmacol. 2018;9:1354. doi: 10.3389/fphar.2018.01354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Anand S., Kaur H., Mande S.S. Comparative In Silico Analysis of Butyrate Production Pathways in Gut Commensals and Pathogens. Front. Microbiol. 2016;7:1945. doi: 10.3389/fmicb.2016.01945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Deng K., Shuai M., Zhang Z., Jiang Z., Fu Y., Shen L., Zheng J.-S., Chen Y. Temporal Relationship among Adiposity, Gut Microbiota, and Insulin Resistance in a Longitudinal Human Cohort. BMC Med. 2022;20:171. doi: 10.1186/s12916-022-02376-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kaplan R.C., Wang Z., Usyk M., Sotres-Alvarez D., Daviglus M.L., Schneiderman N., Talavera G.A., Gellman M.D., Thyagarajan B., Moon J.Y., et al. Gut Microbiome Composition in the Hispanic Community Health Study/Study of Latinos Is Shaped by Geographic Relocation, Environmental Factors, and Obesity. Genome Biol. 2019;20:219. doi: 10.1186/s13059-019-1831-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Muñiz Pedrogo D.A., Jensen M.D., van Dyke C.T., Murray J.A., Woods J.A., Chen J., Kashyap P.C., Nehra V. Gut Microbial Carbohydrate Metabolism Hinders Weight Loss in Overweight Adults Undergoing Lifestyle Intervention with a Volumetric Diet. Mayo Clin. Proc. 2018;93:1104–1110. doi: 10.1016/j.mayocp.2018.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu F., Li P., Chen M., Luo Y., Prabhakar M., Zheng H., He Y., Qi Q., Long H., Zhang Y., et al. Fructooligosaccharide (FOS) and Galactooligosaccharide (GOS) Increase Bifidobacterium but Reduce Butyrate Producing Bacteria with Adverse Glycemic Metabolism in Healthy Young Population. Sci. Rep. 2017;7:11789. doi: 10.1038/s41598-017-10722-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang T.Y., Zhang X.Q., Chen A.L., Zhang J., Lv B.H., Ma M.H., Lian J., Wu Y.X., Zhou Y.T., Ma C.C., et al. A Comparative Study of Microbial Community and Functions of Type 2 Diabetes Mellitus Patients with Obesity and Healthy People. Appl. Microbiol. Biotechnol. 2020;104:7143–7153. doi: 10.1007/s00253-020-10689-7. [DOI] [PubMed] [Google Scholar]
- 82.Tavella T., Rampelli S., Guidarelli G., Bazzocchi A., Gasperini C., Pujos-Guillot E., Comte B., Barone M., Biagi E., Candela M., et al. Elevated Gut Microbiome Abundance of Christensenellaceae, Porphyromonadaceae and Rikenellaceae Is Associated with Reduced Visceral Adipose Tissue and Healthier Metabolic Profile in Italian Elderly. Gut Microbes. 2021;13:1880221. doi: 10.1080/19490976.2021.1880221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Alcazar M., Escribano J., Ferré N., Closa-Monasterolo R., Selma-Royo M., Feliu A., Castillejo G., Luque V., Closa-Monasterolo R., Escribano J., et al. Gut Microbiota Is Associated with Metabolic Health in Children with Obesity. Clin. Nutr. 2022;41:1680–1688. doi: 10.1016/j.clnu.2022.06.007. [DOI] [PubMed] [Google Scholar]
- 84.Wang X., Zhu L., Li X., Wang X., Hao R., Li J. Effects of High Fructose Corn Syrup on Intestinal Microbiota Structure and Obesity in Mice. NPJ Sci. Food. 2022;6:17. doi: 10.1038/s41538-022-00133-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim C.C., Healey G.R., Kelly W.J., Patchett M.L., Jordens Z., Tannock G.W., Sims I.M., Bell T.J., Hedderley D., Henrissat B., et al. Genomic Insights from Monoglobus Pectinilyticus: A Pectin-Degrading Specialist Bacterium in the Human Colon. ISME J. 2019;13:1437–1456. doi: 10.1038/s41396-019-0363-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sauvaitre T., Etienne-Mesmin L., Sivignon A., Mosoni P., Courtin C.M., van de Wiele T., Blanquet-Diot S. Tripartite Relationship between Gut Microbiota, Intestinal Mucus and Dietary Fibers: Towards Preventive Strategies against Enteric Infections. FEMS Microbiol. Rev. 2021;45:fuaa052. doi: 10.1093/femsre/fuaa052. [DOI] [PubMed] [Google Scholar]
- 87.Ahrens A.P., Culpepper T., Saldivar B., Anton S., Stoll S., Handberg E.M., Xu K., Pepine C., Triplett E.W., Aggarwal M. A Six-Day, Lifestyle-Based Immersion Program Mitigates Cardiovascular Risk Factors and Induces Shifts in Gut Microbiota, Specifically Lachnospiraceae, Ruminococcaceae, Faecalibacterium Prausnitzii: A Pilot Study. Nutrients. 2021;13:3459. doi: 10.3390/nu13103459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kozhakhmetov S., Babenko D., Kozhakhmetova S., Tuyakova A., Nurgaziyev M., Nurgozhina A., Muhanbetganov N., Chulenbayeva L., Sergazy S., Gulyayev A., et al. Gut Modulation of Dysbiosis Induced by Dextran Sulfate Sodium. Food Biosci. 2021;42:101167. doi: 10.1016/j.fbio.2021.101167. [DOI] [Google Scholar]
- 89.Yu Z., Yu X.-F., Zhao X., Ren P.-G. Greater Alteration of Gut Microbiota Occurs in Childhood Obesity than Adulthood Obesity. bioRxiv. 2022:492992. doi: 10.1101/2022.05.22.492992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Johnson J.R. Shigella and Escherichia Coli at the Crossroads: Machiavellian Masqueraders or Taxonomic Treachery? J. Med. Microbiol. 2000;49:583–585. doi: 10.1099/0022-1317-49-7-583. [DOI] [PubMed] [Google Scholar]
- 91.van der Putten B.C.L., Matamoros S., Schultsz C., Arcilla M.S., Bootsma M.C.J., van Genderen P.J., Goorhuis A., Grobusch M., van Hattem J.M., de Jong M.D., et al. Genomic Evidence for Revising the Escherichia Genus and Description of Escherichia ruysiae sp. nov. bioRxiv. 2019:781724. doi: 10.1101/781724. [DOI] [Google Scholar]
- 92.Palleroni N.J., Genus I. Bergey’s Manual of Systematic Bacteriology, The Proteobacteria, Part B the Gammaproteobacteria. 2nd ed. Volume 2. Springer; New York, NY, USA: 2005. Pseudomonas Migula 1894 (Nom. Cons., Opin. 5 of the Jud. Comm. 1952, 121) pp. 323–373. [Google Scholar]
- 93.Sims G.E., Kim S.H. Whole-Genome Phylogeny of Escherichia Coli/Shigella Group by Feature Frequency Profiles (FFPs) Proc. Natl. Acad. Sci. USA. 2011;108:8329–8334. doi: 10.1073/pnas.1105168108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gao Y.D., Zhao Y., Huang J. Metabolic Modeling of Common Escherichia Coli Strains in Human Gut Microbiome. BioMed Res. Int. 2014;2014:694967. doi: 10.1155/2014/694967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Atzeni A., Bastiaanssen T.F.S., Cryan J.F., Tinahones F.J., Vioque J., Corella D., Fitó M., Vidal J., Moreno-Indias I., Gómez-Pérez A.M., et al. Taxonomic and Functional Fecal Microbiota Signatures Associated With Insulin Resistance in Non-Diabetic Subjects With Overweight/Obesity Within the Frame of the PREDIMED-Plus Study. Front. Endocrinol. 2022;13:804455. doi: 10.3389/fendo.2022.804455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tomnikov N. Master’s Thesis. University of Turku; Turku, Finland: Jun 13, 2022. Optimization of Microbial DNA Extraction in 96-Format from Fecal Samples for Next Generation Sequencing. [Google Scholar]
- 97.Liu J., Luo M., Qin S., Li B., Huang L., Xia X. Significant Succession of Intestinal Bacterial Community and Function During the Initial 72 Hours of Acute Pancreatitis in Rats. Front. Cell. Infect. Microbiol. 2022;12:808991. doi: 10.3389/fcimb.2022.808991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang W.Y., Lee Y., Lu H., Chou C.H., Wang C. Analysis of Gut Microbiota and the Effect of Lauric Acid against Necrotic Enteritis in Clostridium Perfringens and Eimeria Side-by-Side Challenge Model. PLoS ONE. 2019;14:e0205784. doi: 10.1371/journal.pone.0205784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li Y.J., Li J., Dai C. The Role of Intestinal Microbiota and Mast Cell in a Rat Model of Visceral Hypersensitivity. J. Neurogastroenterol. Motil. 2020;26:529. doi: 10.5056/jnm20004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data are available in the NCBI BioProject database (BioProject: PRJNA862929).