Abstract
The Czc system of Ralstonia sp. strain CH34 mediates resistance to cobalt, zinc, and cadmium through ion efflux catalyzed by the CzcCB2A cation-proton antiporter. The CzcD protein is involved in the regulation of the Czc system. It is a membrane-bound protein with at least four transmembrane α-helices and is a member of a subfamily of the cation diffusion facilitator (CDF) protein family, which occurs in all three domains of life. The deletion of czcD in a Ralstonia sp. led to partially constitutive expression of the Czc system due to an increased transcription of the structural czcCBA genes, both in the absence and presence of inducers. The czcD deletion could be fully complemented in trans by CzcD and two other CDF proteins from Saccharomyces cerevisiae, ZRC1p and COT1p. All three proteins mediated a small but significant resistance to cobalt, zinc, and cadmium in Ralstonia, and this resistance was based on a reduced accumulation of the cations. Thus, CzcD appeared to repress the Czc system by an export of the inducing cations.
CzcD from Ralstonia sp. strain CH34 (formerly Alcaligenes eutrophus [1, 13]) and ZRC1p from Saccharomyces cerevisiae (6) were the first two published members of the cation diffusion facilitator (CDF) protein family (17, 23). The members of this family are all predicted to be membrane-bound proteins, mostly with six assumed transmembrane-spanning α-helices. The CDF proteins seem to interact with the divalent cations of zinc, cadmium, and cobalt. The transport of divalent heavy metal cations has been shown for COT1p from S. cerevisiae (2), for some mammalian ZnT proteins (21, 22), and for a CDF protein from Staphylococcus aureus (28).
On the other hand, CDF proteins were found to be involved in regulatory processes (5, 13): ZRC1p seems to regulate glutathione biosynthesis, and CzcD is involved in the regulation of a zinc, cobalt, and cadmium efflux system, the Czc system, which mediates resistance to these heavy metal cations in Ralstonia sp. strain CH34. The Czc system probably transports the toxic heavy metals across both membranes of the gram-negative bacterium (24). This transenvelope efflux is mediated by the CzcCB2A protein complex, a proton-cation antiporter (12). The czcCBA genes (16) are located on one of the two megaplasmids of strain CH34, plasmid pMOL30, and are flanked by genes encoding regulators, czcNI upstream and czcDRS downstream of czcCBA (13, 27). Transcription probably starts at four promoters, czcNp, czcIp, czcCp, and czcDp, and leads to a variety of transcripts in the czcNICBA region and to a tricistronic czcDRS message (4). In the current model of Czc system regulation (4), CzcN and CzcI may regulate the activity of a hypothetical extracellular function sigma factor while the two-component regulatory system made up of CzcR (response regulator) and CzcS (sensor histidine kinase) regulates the expression of CzcN.
CzcD is not essential for Czc system regulation but is needed to regulate the expression of a czcC::lacZ fusion when a constitutively expressed CzcCB2A efflux complex diminishes the cytoplasmic inducer concentration (13). In this publication, the function of CzcD in the Czc regulatory network is defined.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Tris-buffered mineral salts medium containing 2 g of sodium gluconate/liter was used to cultivate Ralstonia strains, all derivatives of the wild-type strain CH34(pMOL28, pMOL30) (9). Strain AE128(pMOL30) harbors megaplasmid pMOL30 with the czc determinant only, and strain AE104 is a plasmid-free, metal-sensitive derivative of strain CH34 (9). Strain DN175(pMOL30-9) has an insertion of a lacZ gene in the czcCBA operon between czcC and czcB (4). The czcD gene was cloned into the broad-host-range plasmid pVDZ′2 (3) under the control of the lac promoter which is constitutively expressed by Ralstonia (16), leading to plasmid pDNA176. Additionally, the S. cerevisiae genes ZRC1 and COT1 were PCR amplified and cloned into the same vector plasmid, leading to plasmids pDNA178 and pDNA177, respectively. Analytical-grade salts of CdCl2 · H2O, ZnCl2, and CoCl2 · 6H2O were used to prepare 1 M stock solutions, which were sterilized by filtration. Solid Tris-buffered media contained 20 g of agar/liter. Uptake experiments were performed by the filtration technique as described previously (19), but 10 mM Tris-HCl (pH 7.0) containing 10 mM MgCl2 was used to wash the cells on the filters. 109CdCl2 (1 Ci/g) and 65ZnCl2 (1.84 Ci/g) were from NEN (Brussels, Belgium), and 57CoCl2 (4,000 Ci/g) was from Amersham (Braunschweig, Germany).
Genetic techniques.
Standard molecular genetic techniques were used (10, 25). For conjugal gene transfer, overnight cultures of donor strain Escherichia coli S17/1 (26) and of the Ralstonia recipient strains grown at 30°C in complex medium were mixed (1:1) and plated onto nutrient broth agar. After overnight growth, the bacteria were suspended in saline (9 g of NaCl/liter), diluted, and plated onto selective media as previously described (11). The total RNA of the Ralstonia organisms was isolated as described previously (20). PCGENE (IntelliGenetics, Mountain View, Calif.) was used as the standard computer program for the analysis of DNA sequences.
Reporter gene fusions.
Fusion vectors pECD499 (lacZ fusions) and pECD500 (phoA fusions) (24) and E. coli CC118 were used (8). The specific activities of alkaline phosphatase (8) and β-galactosidase (13) were determined in triplicate as published previously (15). From each mean value, the negative control value (vector control without insert) was subtracted. The result was divided by the highest specific activity, which was 2.12 U/g (dry weight) for PhoA (fusion I-115) and 37.3 U/mg (dry weight) (control of a nonfused lacZ gene without leader) for LacZ, leading to the relative activities for each fusion point.
Construction of knockout mutations in czc genes (4).
To prevent polar effects mediated by the deletion of the czcD gene from megaplasmids pMOL30 containing czcNICBADRS (9) and pMOL30-9 harboring Φ(czcNIC-lacZ-czcBADRS) (4), czcD was exchanged for a small open reading frame encoding a polypeptide of 20 amino acids (aa). The first nine aa coded by this small open reading frame were identical with the nine amino-terminal amino acids of CzcD, and the last 9 aa were identical with the last carboxy-terminal amino acids of CzcD. Residues 10 and 11 were E and L, respectively, and were coded by the hexanucleotide recognition sequence CAATTG of the restriction endonuclease MunI. Thus, the 500 bp upstream of czcD were amplified by PCR, and this fragment ended with the 27-bp sequence coding for the first 9 aa of the respective gene product, followed by a MunI hexanucleotide. Secondly, the 500 bp downstream of czcD were amplified by PCR, and this fragment started with the MunI recognition sequence and the last 27 bp of the respective gene. Both fragments were fused by MunI restriction and ligation, cloned, verified by DNA sequencing, and finally cloned into pLO2 (7). The resulting plasmid was used for mutating Ralstonia organisms as described previously (4), leading to plasmid pMOL30-14 in strain DN182(pMOL30-14) (czcNICBA ΔczcD czcRS) and plasmid pMOL30-15 in strain DN183(pMOL30-15) [Φ(czcNIC-lacZ-czcBA) ΔczcD czcRS]. The mutant genotypes were verified by PCR and DNA sequencing.
Competitive RT-PCR (5).
DNase-treated total RNA was isolated from cells of Ralstonia sp. strains AE128(pMOL30) (9) and DN182(pMOL30-14) (ΔczcD) either without induction or after induction for 10 min with 300 μM Zn2+. One microgram of this total RNA was reverse transcribed with 100 U of Superscript II RT (Gibco BRL, Karlsruhe, Germany) and 50 pmol of random primer in a total volume of 20 μl. To determine the amount of czcCBA mRNA-specific cDNA for each strain and induction condition, different amounts of an internal DNA standard were added to 0.5 μl of the resulting cDNA solution, and the mixture was amplified with 100 μM concentrations of deoxynucleoside triphosphates, 10 pmol of primers (3′ antisense primer B, ATGCCACCGATTACCACCGTTGCGA, positions 7144 to 7120 [gbX98451] in czcA [positions 4092 to 7283], and 5′ sense primer A, ATTGGTTCATTCGTGCCCG, positions 6615 to 6633 in czcA] and 1 U of Taq polymerase (Qiagen, Hilden, Germany) in a total volume of 50 μl by the following PCR program: 2.30 min at 94°C, 1 min at 60°C, and 1 min at 72°C as the initial cycle, and a further 28 cycles of 1 min at 94°C, 1 min at 60°C, and 1 min at 72°C, with final extension for 5 min at 72°C. A 10-μl amount of each PCR product was analyzed on a 1.5% agarose gel stained with ethidium bromide. The relative amounts of czcCBA cDNA (529 bp) and internal DNA standard (237 bp) products were quantified after densitometric analysis with ScanPack 2.0 software (Biometra, Göttingen, Germany). For each lane, which represents cDNA with one of the internal DNA standards, the resulting spot intensities were first normalized for the lengths of the cDNA (529 and 237 bp, respectively, for czcCBA cDNA and the internal standard), and then the normalized czcCBA cDNA intensity was divided by the normalized density of the internal standard. For all lanes, the logarithm of this quotient was plotted against the logarithm of the amount of the internal standard used in the respective competition experiment. A linear regression was calculated for these points, and this line intercepts the x axis exactly at the point where the amount of the internal standard is identical with the amount of the czcCBA-specific cDNA in the reverse transcription (RT) probe. This value was used to calculate the czcCBA-specific cDNA per microgram of total RNA used.
Control assays were as follows: a complete assay without template, a complete assay with RNA template but without RT, and a complete assay with total RNA isolated from the plasmid-free Ralstonia strain AE104. All controls were negative.
For the construction of a specific internal standard, a 529-bp part of czcA was amplified with primers A and B. This fragment was purified and used as the template in a PCR experiment with a loop-out primer (ATTGGTTCATTCGTGCCCGGGCGGTGCTCAATGGTCTG) and primer B. The loop-out primer was identical in the first 19 nucleotides (underlined) to primer A, and the remaining 19 nucleotides (bold) were the base pairs in positions 6926 to 6944 (gbX98451) of czcA, which are located between the positions of primers A and B. Thus, a 237-bp PCR product which could be clearly differentiated from the 529-bp czcCBA cDNA product was amplified, but it had the same ends as the 529-bp fragment. The 237-bp PCR product was isolated, cleaned with QIAquick, quantified (GeneQuant; Pharmacia, Uppsala, Sweden), diluted, and used for competitive RT-PCR.
RESULTS
Topological analysis of CzcD.
The structure of the CzcD protein from Ralstonia was investigated by using translational lacZ and phoA fusions. Starting with the 5′ end, variously sized parts of the respective czcD gene were amplified and cloned into the fusion vectors pECD499 and pECD500 (24).
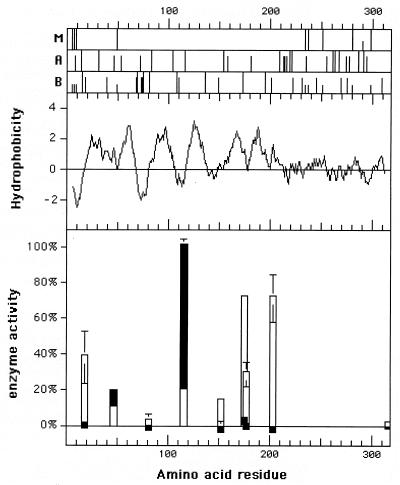
The amino terminus of CzcD is followed by four hydrophobic peaks (Fig. 1). Fusions between those peaks gave evidence for an alternation between periplasmic and cytoplasmic localizations of the fusion points (Fig. 1). Thus, the amino terminus of CzcD is followed by four transmembrane α-helices, I, II, III, and IV. The activity of the fusions plus the pattern of positively charged amino acid residues indicate a cytoplasmic localization of the N terminus and of the regions between helices II and III and downstream of helix IV. After these four spans (fusion position Y152), two more hydrophobic peaks occur. Although the fusions at position S203 after both peaks strongly indicate a cytoplasmic localization, two independent fusions between both peaks (T175 and W177) gave no evidence for a periplasmic localization (Fig. 1). LacZ and PhoA fusions at the carboxy terminus of CzcD had a very low specific activity. However, the absence of hydrophobic peaks downstream of S203 and the high LacZ activity of the S203 fusion could mean a cytoplasmic localization of the C terminus of CzcD.
FIG. 1.
Topological structure of CzcD. Parts of the czcD gene of Ralstonia were fused with the lacZ or phoA topological reporter gene. The czcD parts were from the 5′ end of the gene up to the points indicated by the positions of the bars. The relative activities of the reporter enzymes are indicated by overlapping white (LacZ) and black bars (PhoA), with error bars representing standard deviations. Above the hydrophobicity plot are the positions of putative metal-binding amino acid residues (in panel M, residues H [full-size bars] and C [half bars]), acidic amino acid residues (in panel A, residues D [two-thirds bars] and E [full-size bars]), and basic amino acid residues (in panel B, residues H [half bars], R [full-size bars], and K [two-thirds bars]).
Thus, both termini of CzcD are probably in the cytoplasm and the amino terminus is clearly followed by four transmembrane α-helices. The existence of the two subsequent transmembrane spans could not be proven; this may be the result of a limitation of the method used, or spans V and VI may be reversibly integrated into the membrane as part of the catalytic cycle or a regulatory event. However, CzcD should be an integral protein of the cytoplasmic membrane, with at least four and a maximum of six transmembrane α-helices.
A ΔczcD mutant is impaired in metal sensing.
Mutants with in-frame ΔczcD deletions were constructed from Ralstonia strains AE128(pMOL30) (czcNICBADRS) (9) and DN175(pMOL30-9) [Φ(czcNIC-lacZ-czcBADRS)] (4), leading to strains DN182(pMOL30-14) (czcNICBA ΔczcD czcRS) and DN183(pMOL30-15) [Φ(czcNIC-lacZ-czcBA) ΔczcD czcRS], respectively. As judged from the MICs, the resistances of the ΔczcD strains DN182 and DN183 to zinc were slightly lower than those of their isogenic CzcD+ strains, AE128 and DN175, respectively. In the presence of 7 mM Zn2+, both ΔczcD deletion strains produced only a few single colonies on solid medium while the wild-type strains displayed full growth (data not shown). There was no effect on the MICs of cobalt and cadmium and no significant difference in the induction of the β-galactosidase reporter gene at most metal cation concentrations when strains DN175 and DN183 were compared (data not shown). However, when the β-galactosidase activities of the ΔczcD strain and the wild-type strain were compared after induction by 300 μM Co2+, 100 μM Cd2+, or 10 μM Zn2+, the ΔczcD strain reached a higher expression level after 2 h (Table 1). In addition, the β-galactosidase activity in uninduced cells of the ΔczcD strain was twice as high as the activity in uninduced wild-type cells (Table 1).
TABLE 1.
Induction of czcCBA::lacZ in a ΔczcD mutant straina
| Inducerb | β-Galactosidase activity (mean ± SD) in bacterial strain
|
|
|---|---|---|
| DN175(pMOL30-9) | DN183(pMOL30-15) (ΔczcD) | |
| None | 120 ± 34 | 207 ± 36 |
| 10 μM Zn2+ | 256 ± 7 | 461 ± 42 |
| 300 μM Co2+ | 206 ± 15 | 293 ± 12 |
| 100 μM Cd2+ | 192 ± 32 | 341 ± 43 |
Means for two independent experiments for the determinations of the β-galactosidase activity are given in units per milligram (dry weight).
The cells were induced for 2 h with the respective heavy-metal cation.
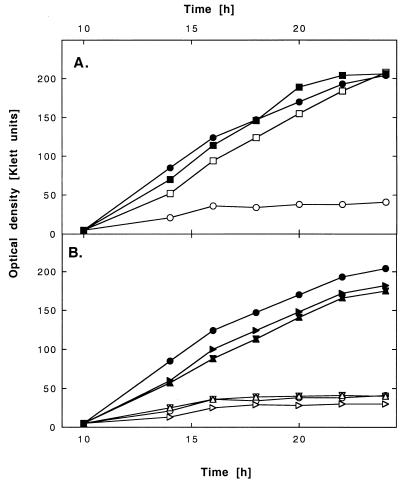
When strains AE128(pMOL30) and DN182(pMOL30-14) (ΔczcD) were precultivated in the absence of Zn2+ or in 300 μM Zn2+ and then transferred to a liquid medium containing 2.5 mM Zn2+, the ΔczcD deletion strain started to grow immediately in both cases but the wild-type strain grew only without a lag phase when it was preadapted in the presence of 300 μM Zn2+ (Fig. 2A). To examine this effect more closely, the concentration of czcCBA mRNA in both strains was judged by the amount of respective cDNA measured by competitive RT-PCR (Table 2). In both the deletion and wild-type strains, czcCBA was inducible, but the mRNA level in the deletion strain was 10-fold higher than that in the wild-type strain under noninduced and induced conditions (Table 2). Thus, CzcD represses czc induction either by inducer exclusion or by some kind of protein-protein interaction.
FIG. 2.
Effect of ΔczcD mutation on growth in presence of zinc. Cells of strain AE128(pMOL30) (● and ○), its ΔczcD mutant strain DN182(pMOL30-14) (■ and □), strain DN182 complemented in trans with pDNA176 containing czcD (▴ and ▵), pDNA178 containing ZRC1 (▾ and ▿), and pDNA177 containing COT1 (▸ and ▹) were cultivated for 48 h at 30°C in Tris-buffered mineral salts medium containing 2 g of sodium gluconate/liter as the carbon source. The cells were diluted in fresh medium containing 300 μM Zn2+ as the inducer (closed symbols) or no inducer (open symbols). Incubation was continued for 10 h, and then the cells were diluted to a cell density of up to 10 Klett units in fresh medium containing 2.5 mM Zn2+.
TABLE 2.
Concentration of czcCBA mRNA-originating cDNA depends on the presence of CzcDa
| Bacterial strain | Genotype | cDNA concn (ng/g of RNA) in:
|
Fold inductionc | |
|---|---|---|---|---|
| Uninduced cells | Induced cellsb | |||
| AE128(pMOL30)d | Wild type | 6, 16 | 762, 846 | 74 |
| DN182(pMOL30-14) | ΔczcD | 106, 268 | 4,950, 10,150 | 42 |
Two independent determinations are given.
The cells were induced for 10 min with 300 μM Zn2+ before the RNA was isolated.
Determined as the concentration in induced cells/concentration in uninduced cells.
Data from reference 4.
By comparing the mRNA data and the growth curves, it was found that a 15-fold overexpression of czcCBA in the ΔczcD deletion strain was sufficient for the cells to start growing immediately in the presence of 2.5 mM Zn2+. However, differences in the β-galactosidase activity were visible only in the presence of 10 μM Zn2+. This difference cannot be explained at the moment.
CzcD mediates a low-level metal ion resistance, probably by efflux.
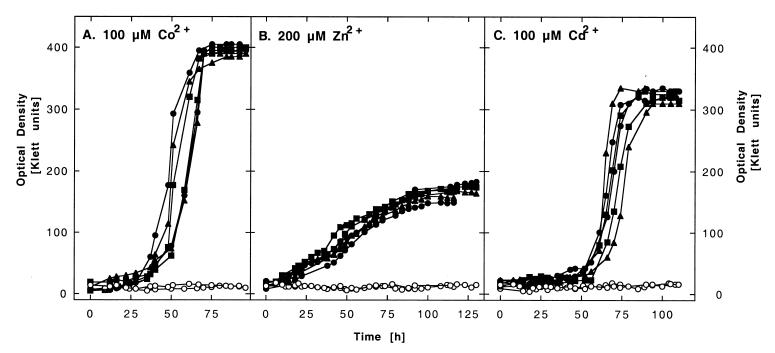
To determine if CzcD transports cations, the czcD gene was cloned into plasmid pVDZ′2 (3), leading to plasmid pDNA176. This plasmid was transferred to plasmid-free Ralstonia strain AE104. At most metal ion concentrations, no difference in metal ion resistance between AE104(pDNA176) and the negative control strain AE104(pVDZ′2) (data not shown) was observed. However, with 100 μM cobalt, 100 μM cadmium, or 200 μM zinc, the control strain AE104(pVDZ′2) was not able to grow, in contrast to AE104(pDNA176) (Fig. 3). Thus, the expression of CzcD mediates a small degree of metal ion resistance which is below the resistance level obtained when the protein CzcA is expressed alone (24). With the protection of CzcA, it took the cells 20 h to grow up to 300 Klett units in the presence of 200 μM Zn2+ (24); with CzcD, it took 125 h for the cells to reach 200 Klett units (Fig. 3). In both cases, the negative control strain did not grow.
FIG. 3.
CDF proteins mediate metal ion resistance. Ralstonia strain AE104 containing plasmids pVDZ′2 without an insert (○), pDNA176 with czcD (●), pDNA177 with COT1 (▴), or pDNA178 with ZRC1 (■) was cultivated in the presence of 100 μM Co2+ (A), 200 μM Zn2+ (B), or 100 μM Cd2+ (C), and the optical density over time was monitored. For each strain and metal ion, the results of two independent experiments are shown.
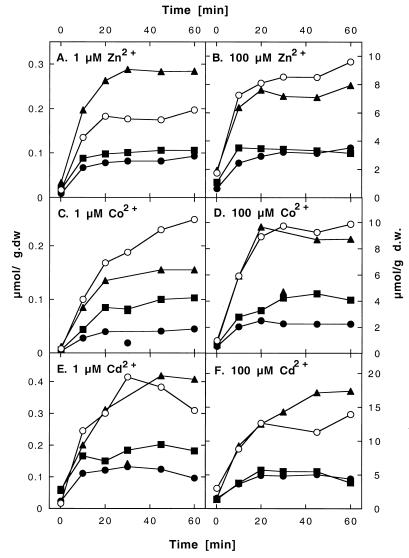
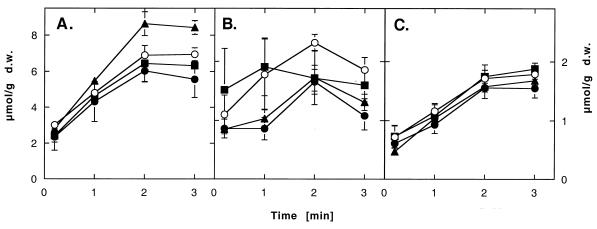
Resistance mediated by a membrane-integrated histidine-rich protein may be based on the binding of the metal by the histidine residues and/or metal ion efflux. Binding should increase the amount of metal ions in resistant cells compared to that in sensitive ones, while efflux should decrease the amount of cell-bound metal ions. When levels of cell-bound metal ions in cells with and without overexpressed CzcD were compared, resistant cells contained only 20 to 50% of the cobalt, zinc, or cadmium that the respective control cells contained (Fig. 4). There was no difference in the initial velocity of zinc or cadmium entry between cells of both strains (Fig. 5A and C). The uptake of cobalt in the first minutes was slow, with strong fluctuations between different experiments (Fig. 5B). Thus, CzcD mediates a reduced accumulation of Zn2+, Co2+, and Cd2+. Since the initial velocities of metal cation uptake with and without CzcD were identical, this reduced accumulation is probably based on the efflux of metal cations, at least in the cases of zinc and cadmium. Since CzcD is located in the cytoplasmic membrane, indicated by the fusion data, this efflux is across the cytoplasmic membrane.
FIG. 4.
Accumulation of heavy metal ions by Ralstonia strains expressing various CDF genes. Ralstonia strain AE104 containing plasmids pVDZ′2 without an insert (○), pDNA176 with czcD (●), pDNA177 with COT1 (▴), or pDNA178 with ZRC1 (■) was cultivated in Tris-buffered mineral salts medium containing 2 g of sodium gluconate/liter. The cells were harvested, washed, and suspended in 10 mM Tris-HCl buffer (pH 7.0) containing 2 g of sodium gluconate/liter. Radioactive metal isotopes at 1 or 100 μM were added, and incubation was continued with shaking at 30°C. Samples of 200 μl were removed, filtered (pore diameter, 0.45 μm), and washed twice on the filter with 2 ml of 10 mM Tris-HCl (pH 7.0) containing 10 mM MgCl2. Radioactivity was determined with a scintillation counter (Beckman, Munich, Germany), and the dry weight (dw) was determined from the optical density with a calibration curve.
FIG. 5.
Fast uptake of heavy metal ions by Ralstonia strains expressing various CDF genes. Ralstonia strain AE104 containing plasmids pVDZ′2 without an insert (○), pDNA176 with czcD (●), pDNA177 with COT1 (▴), or pDNA178 with ZRC1 (■) was cultivated in Tris-buffered mineral salts medium containing 2 g of sodium gluconate/liter. The cells were harvested, washed, and suspended in 10 mM Tris-HCl buffer (pH 7.0) containing 2 g of sodium gluconate/liter. The radioactive metal isotopes 65Zn2+ (A), 57Co2+ (B), and 109Cd2+ (C) at 100 μM were added, and metal uptake was measured as described for Fig. 4. d.w., dry weight.
ZRC1p and COT1p mediate a low-level metal ion resistance.
The genes of the two CDF proteins from yeast were cloned into plasmid pVDZ′2, leading to plasmids pDNA178 (containing ZRC1) and pDNA177 (containing COT1). Both plasmids mediated a degree of metal ion resistance comparable to that mediated by pDNA176 containing czcD (Fig. 3).
ZRC1p expression led to effects similar to CzcD expression concerning zinc and cadmium (Fig. 4 and 5), but ZRC1p was not as efficient as CzcD concerning the reduced accumulation of cobalt. If CzcD constitutes an efflux system for zinc, cobalt, and cadmium, then ZRC1p should function in the same manner. Although COT1p provided the same degree of metal resistance as ZRC1p and CzcD, the effect obtained by COT1p expression varied with the metal ion and its concentration (Fig. 4). With 100 μM Zn2+ or 1 μM Co2+, COT1p mediated a reduced accumulation, and it is probably an efflux system. In contrast, with 1 μM Zn2+, COT1p-containing cells accumulated more zinc than control cells, so COT1p should enhance the uptake and/or binding of zinc (Fig. 4A). Moreover, the uptake of 100 μM Zn2+ was faster in COT1p-containing cells than in CzcD- or ZRC1p-containing cells or control cells (Fig. 5A). Thus, the yeast CDF proteins work similarly to CzcD; however, the transport of cations across the cytoplasmic membrane may be in both directions, depending on the concentration.
Expression of ZRC1p and COT1p complements a czcD mutation.
To analyze the complementation of the ΔczcD mutation by czcD in trans, plasmid pDNA176 was transferred into DN182. The resulting transconjugant strain showed a lag phase of growth in the presence of 2.5 mM Zn2+ when the cells were not adapted by precultivation in 300 μM Zn2+ (Fig. 2B). Similarly to pDNA176, the ZRC1- and COT1-containing plasmids pDNA178 and pDNA177 were transferred into DN182(pMOL30-14) (ΔczcD). Both plasmids complemented the ΔczcD mutation as well as the czcD-containing plasmid (Fig. 2B). Thus, the two yeast CDF proteins were fully functional as participants of the Czc regulatory network.
DISCUSSION
In Ralstonia spp., CzcD and the two yeast CDF proteins ZRC1p and COT1p catalyze a reduced accumulation of heavy-metal ions, which is probably based on metal cation efflux. Since CzcD is located in the cytoplasmic membrane, this means there is an export of heavy-metal cations from the cytoplasm into the periplasm.
COT1p, which does not function well as an efflux system with 100 μM Co2+ in Ralstonia, nevertheless gives the same degree of resistance to 100 μM Co2+ as ZRC1p and CzcD do; therefore, the large histidine-rich domains of the protein might detoxify cobalt cations by binding them. The CDF proteins might function as a kind of heavy-metal buffer for the cell by performing the following: importing heavy metals when the magnesium transport system (18) is too slow to supply sufficient amounts of the trace elements cobalt or zinc (e.g., due to competitive inhibition by high magnesium concentrations), binding the heavy-metal cations, and exporting the heavy-metal cations when the cytoplasmic concentration becomes too high. Only when the capacity of this system is exhausted by too low or too high heavy-metal concentrations may additional energy sources have to be tapped to drive increased accumulation (e.g., by P-type or ATP-binding cassette uptake ATPases) or increased efflux (e.g., by P-type export ATPases or the Czc proton-cation antiporter).
CzcD not only protects the cell against toxic heavy metals, albeit at a lower level than CzcCB2A does, but also is involved in the regulation of expression of the CzcCB2A efflux system. Deletion of the czcD gene results in a higher czcCBA mRNA level in uninduced and induced cells, which is sufficient to produce enough CzcCB2A efflux complex for an initial protection against 2.5 mM Zn2+. As shown by primer extension and RT-PCR, the czcNICBA region is transcribed from three promoters, czcNp, czcIp, and czcCp (4). The two-component regulatory system CzcRS regulates only czcNp (4). CzcS, the sensor protein, may sense only cytoplasmic cations (14). Thus, since no other Czc protein is located in the cytoplasm except for CzcR, the presence of heavy metals in the cytoplasm should lead to transcription initiation from czcNp with a signal chain via CzcS and CzcR. Because deletion of the genes czcR or czcS does not abolish Czc system induction (4), other metal-sensing components are involved in Czc system regulation, and these may be located in the periplasm (e.g., CzcI) or the cytoplasmic membrane (e.g., CzcN).
ACKNOWLEDGMENTS
This work was supported by Forschungsmittel des Landes Sachsen-Anhalt, the Deutsche Forschungsgemeinschaft (Graduiertenkolleg Streß), and Fonds der Chemischen Industrie.
We thank Grit Schleuder for skillful technical assistance, Daniel van der Lelie for a COT1-containing plasmid, Oliver Lenz and Bärbel Friedrich for pLO2, and students of the radioisotope course of the Graduiertenkolleg Streß for helping with the uptake experiments. Also, we thank Joseph Lengeler for a very fruitful discussion.
REFERENCES
- 1.Brim H, Heyndrickx M, De Vos P, Wilmotte A, Springael D, Schlegel H G, Mergeay M. Amplified rDNA restriction analysis and further genotypic characterisation of metal-resistant soil bacteria and related facultative hydrogenotrophs. Syst Appl Microbiol. 1999;22:258–268. doi: 10.1016/S0723-2020(99)80073-3. [DOI] [PubMed] [Google Scholar]
- 2.Conklin D S, McMaster J A, Culbertson M R, Kung C. COT1, a gene involved in cobalt accumulation in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:3678–3688. doi: 10.1128/mcb.12.9.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deretic V, Chandrasekharappa S, Gill J F, Chatterjee D K, Chakrabarty A. A set of cassettes and improved vectors for genetic and biochemical characterization of Pseudomonas genes. Gene. 1987;57:61–72. doi: 10.1016/0378-1119(87)90177-6. [DOI] [PubMed] [Google Scholar]
- 4.Große C, Grass G, Anton A, Franke S, Navarrete Santos A, Lawley B, Brown N L, Nies D H. Transcriptional organization of the czc heavy-metal homoeostasis determinant from Alcaligenes eutrophus. J Bacteriol. 1999;181:2385–2393. doi: 10.1128/jb.181.8.2385-2393.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Inoue Y, Kobayashi S, Kimura A. Cloning and phenotypic characterization of a gene enhancing resistance against oxidative stress in Saccharomyces cerevisiae. J Ferment Bioeng. 1993;75:327–331. [Google Scholar]
- 6.Kamizomo A, Nishizawa M, Teranishi A, Murata K, Kimura A. Identification of a gene conferring resistance to zinc and cadmium ions in the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1989;219:161–167. doi: 10.1007/BF00261172. [DOI] [PubMed] [Google Scholar]
- 7.Lenz O, Schwartz E, Dernedde J, Eitinger M, Friedrich B. The Alcaligenes eutrophus H16 hoxX gene participates in hydrogenase regulation. J Bacteriol. 1994;176:4385–4393. doi: 10.1128/jb.176.14.4385-4393.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manoil C. Analysis of protein localization by use of gene fusions with complementary properties. J Bacteriol. 1990;172:1035–1042. doi: 10.1128/jb.172.2.1035-1042.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mergeay M, Nies D, Schlegel H G, Gerits J, Charles P, Van Gijsegem F. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J Bacteriol. 1985;162:328–334. doi: 10.1128/jb.162.1.328-334.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nies A, Nies D H, Silver S. Cloning and expression of plasmid genes encoding resistances to chromate and cobalt in Alcaligenes eutrophus. J Bacteriol. 1989;171:5065–5070. doi: 10.1128/jb.171.9.5065-5070.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nies D, Mergeay M, Friedrich B, Schlegel H G. Cloning of plasmid genes encoding resistance to cadmium, zinc, and cobalt in Alcaligenes eutrophus CH34. J Bacteriol. 1987;169:4865–4868. doi: 10.1128/jb.169.10.4865-4868.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nies D H. The cobalt, zinc, and cadmium efflux system CzcABC from Alcaligenes eutrophus functions as a cation-proton antiporter in Escherichia coli. J Bacteriol. 1995;177:2707–2712. doi: 10.1128/jb.177.10.2707-2712.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nies D H. CzcR and CzcD, gene products affecting regulation of resistance to cobalt, zinc, and cadmium (czc system) in Alcaligenes eutrophus. J Bacteriol. 1992;174:8102–8110. doi: 10.1128/jb.174.24.8102-8110.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nies D H, Brown N. Two-component systems in the regulation of heavy metal resistance. In: Silver S, Walden W, editors. Metal ions in gene regulation. London, England: Chapman & Hall; 1998. pp. 77–103. [Google Scholar]
- 15.Nies D H, Koch S, Wachi S, Peitzsch N, Saier M H., Jr CHR, a novel family of prokaryotic proton motive force-driven transporters probably containing chromate/sulfate antiporters. J Bacteriol. 1998;180:5799–5802. doi: 10.1128/jb.180.21.5799-5802.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nies D H, Nies A, Chu L, Silver S. Expression and nucleotide sequence of a plasmid-determined divalent cation efflux system from Alcaligenes eutrophus. Proc Natl Acad Sci USA. 1989;86:7351–7355. doi: 10.1073/pnas.86.19.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nies D H, Silver S. Ion efflux systems involved in bacterial metal resistances. J Ind Microbiol. 1995;14:186–199. doi: 10.1007/BF01569902. [DOI] [PubMed] [Google Scholar]
- 18.Nies D H, Silver S. Metal ion uptake by a plasmid-free metal-sensitive Alcaligenes eutrophus strain. J Bacteriol. 1989;171:4073–4075. doi: 10.1128/jb.171.7.4073-4075.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nies D H, Silver S. Plasmid-determined inducible efflux is responsible for resistance to cadmium, zinc, and cobalt in Alcaligenes eutrophus. J Bacteriol. 1989;171:896–900. doi: 10.1128/jb.171.2.896-900.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oelmüller U, Krüger N, Steinbüchel A, Friedrich C G. Isolation of prokaryotic RNA and detection of specific mRNA with biotinylated probes. J Microbiol Methods. 1990;11:73–84. [Google Scholar]
- 21.Palmiter R D, Cole T B, Findley S D. ZnT-2, a mammalian protein that confers resistance to zinc by facilitating vesicular sequestration. EMBO J. 1996;15:1784–1791. [PMC free article] [PubMed] [Google Scholar]
- 22.Palmiter R D, Findley S D. Cloning and functional characterization of a mammalian zinc transporter that confers resistance to zinc. EMBO J. 1995;14:639–649. doi: 10.1002/j.1460-2075.1995.tb07042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paulsen I T, Saier M H., Jr A novel family of ubiquitous heavy metal ion transport proteins. J Membr Biol. 1997;156:99–103. doi: 10.1007/s002329900192. [DOI] [PubMed] [Google Scholar]
- 24.Rensing C, Pribyl T, Nies D H. New functions for the three subunits of the CzcCBA cation-proton antiporter. J Bacteriol. 1997;179:6871–6879. doi: 10.1128/jb.179.22.6871-6879.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 26.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 27.van der Lelie D, Schwuchow T, Schwidetzky U, Wuertz S, Baeyens W, Mergeay M, Nies D H. Two component regulatory system involved in transcriptional control of heavy metal homoeostasis in Alcaligenes eutrophus. Mol Microbiol. 1997;23:493–503. doi: 10.1046/j.1365-2958.1997.d01-1866.x. [DOI] [PubMed] [Google Scholar]
- 28.Xiong A, Jayaswal R K. Molecular characterization of a chromosomal determinant conferring resistance to zinc and cobalt ions in Staphylococcus aureus. J Bacteriol. 1998;180:4024–4029. doi: 10.1128/jb.180.16.4024-4029.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]