Abstract
To allow for the molecular analysis of halorespiration by the strictly anaerobic gram-positive bacterium Desulfitobacterium dehalogenans, halorespiration-deficient mutants were selected and characterized following insertional mutagenesis by the conjugative transposon Tn916. To facilitate rapid screening of transconjugants, a highly efficient method for the growth of single colonies on solidified medium has been developed. A streptomycin-resistant mutant of D. dehalogenans was isolated and mated with Enterococcus faecalis JH2-2 carrying Tn916. Insertion of one or two copies of Tn916 into the chromosome of D. dehalogenans was observed. From a total of 2,500 transconjugants, 24 halorespiration-deficient mutants were selected based upon their inability to use 3-chloro-4-hydroxyphenylacetic acid as an electron acceptor. Physiological characterization led to the definition of three phenotypic classes of mutants that differed in their ability to use the additional terminal electron acceptors nitrate and fumarate. The activities of hydrogenase and formate dehydrogenase were determined, and the transposon insertion sites in selected mutants representing the different classes were analyzed on the sequence level following amplification by inverse PCR. The results of the molecular characterization as well as the pleiotropic phenotypes of most mutants indicate that genes coding for common elements shared by the different respiratory chains present in the versatile D. dehalogenans have been disrupted.
Halogenated hydrocarbons are present in the environment in large quantities due to their past and present application in industry and agriculture, e.g., as solvents, pesticides, and preservatives, and to natural production, compromising environmental integrity and health (11, 12). The biodegradability of these compounds under different environmental conditions has been studied extensively (8). In particular, higher halogenated hydrocarbons can often be degraded solely under anaerobic conditions by reductive dehalogenation. In contrast to aerobic degradation processes, however, only limited knowledge exists on reductive dehalogenation at the molecular level (8, 16). Recently, a rapidly increasing number of anaerobic bacteria has been isolated that are able to couple the reductive dehalogenation to energy conservation and hence to growth (7, 14). These bacteria can use chloroalkenes, e.g., tetra- and trichloroethene, or chloroaromatic compounds, such as chlorophenols or 3-chlorobenzoate, as the terminal electron acceptor. Among these, the gram-positive genus Desulfitobacterium comprises a major group of isolates, including Desulfitobacterium dehalogenans, which is able to couple the reductive dechlorination of different ortho-chlorinated phenolic compounds to growth with lactate, pyruvate, formate, or hydrogen as the electron donor. In addition, pyruvate is also used for fermentative growth (24).
Isolation of these strains and their expected potential for application in in situ biodegradation of halo-organic pollutants also led to an increased interest in the molecular bases of this novel anaerobic respiratory pathway. To date, efforts have mainly focused on reductive dehalogenase as the key enzyme in halorespiration. The inducible ortho-chlorophenol-reductive dehalogenase was purified from D. dehalogenans and characterized at the biochemical and genetic level (25). A comparison with other chlorophenol- and tetrachloroethene-reductive dehalogenases shows that all enzymes are either membrane bound or membrane associated and contain Fe-S clusters and, with one exception, a corrinoid as redox centra (for a recent review, see reference 14). The presence of two iron-sulfur clusters as determined by electron paramagnetic resonance analysis was confirmed by the identification of one ferredoxin-like motif and one truncated iron-sulfur cluster binding motif in the deduced primary sequence. Similar results were obtained for the tetrachloroethene-reductive dehalogenase of the gram-negative Dehalospirillum multivorans (17). Furthermore, both reductive dehalogenases share a twin arginine (RR) leader sequence, which is cleaved off in the mature proteins. These RR leader sequences are thought to play a major role in the maturation and translocation of mainly periplasmic proteins binding different redox cofactors (3).
In order to enable a molecular characterization of additional components involved in halorespiratory electron transport, as well as in folding, targeting, and regulation of the reductive dehalogenase in D. dehalogenans, we tested the use of the conjugative transposon Tn916 for the isolation of halorespiration-deficient (HRD) mutants. Tn916 was the first conjugative transposon to be identified, and members of the Tn916-Tn1545 family have been found in or introduced into more than 50 different gram-positive and -negative species (5).
We report here the development of an efficient conjugation system for the integration of Tn916 in the chromosome of the newly described halorespiring D. dehalogenans. This allowed for the isolation of HRD mutants that were characterized by analyzing their respiratory and biochemical properties as well as by sequence analysis of the insertion sites.
MATERIALS AND METHODS
Materials.
3-Chloro-4-hydroxyphenylacetic acid (Cl-OHPA) was purchased from Sigma-Aldrich Chemie (Zwijndrecht, The Netherlands) and filtered prior to use. All gasses were obtained from Hoek Loos (Schiedam, The Netherlands). When appropriate, experiments were carried out in an anaerobic glove box (Coy Laboratories Products, Grass Lake, Mich.) under an atmosphere of 96% N2 and 4% H2. The oxygen concentration was kept low with the palladium catalyst RO-20 provided by BASF (Arnhem, The Netherlands).
Bacterial strains, plasmids, and culture conditions.
The bacterial strains used in this study are listed in Table 1. Strains of Escherichia coli were grown in Luria Bertani medium at 37°C (19). Enterococcus faecalis was cultivated at 30°C in M17 broth (Oxoid, Basingstoke, United Kingdom) supplemented with 0.5 g of glucose/liter. Strains of D. dehalogenans were routinely grown anaerobically (100% N2 gas phase) at 37°C in a phosphate-buffered medium (pH 7.5) with a low chloride concentration. The basal medium contained (in grams per liter of demineralized water): Na2HPO4 · 2H2O, 3.56; K2HPO4, 3.31; KH2PO4, 0.87; (NH4)2HPO4, 0.61; MgCl2 · 6H2O, 0.22; CaCl2 · 2 H2O, 0.03; resazurin, 0.0005. Prior to inoculation, the basal medium was supplemented with 0.04 g of Na2S/liter, 0.04 g of cysteine-HCl/liter, 1 g of yeast extract/liter, and trace elements and vitamin solution as recommended by the German Collection of Microorganisms. The electron donor and acceptor were added to the desired concentrations. If appropriate, the media were amended with ampicillin (100 μg/ml), streptomycin (2,000 μg/ml), and tetracycline (5 μg/ml). For the long-term storage of cultures of D. dehalogenans at −80°C, prior to freezing them, glycerol was added to a final concentration of 20% from an anoxic stock solution inside the glove box.
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant characteristicsa | Reference or source |
|---|---|---|
| E. faecalis JH2-2 | Rifr Fusr Tn916 (Tetr) | 15 |
| E. coli XL-1 Blue | recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 [F′ lacIqZM15 Tn10 (Tetr)] | Stratagene |
| D. dehalogenans | ||
| JW/IU-DC1 (DSM 9161) | Wild-type strain | DSMb |
| HSS1 | Strr | This study |
| HRD0 | Strr Tn916 (Tetr) | This study |
| HRD2 (class I) | Strr Tn916 (Tetr) HRD | This study |
| HRD1, -3 to -5, -7 to -24 (class II) | Strr Tn916 (Tetr) HRD Nar− | This study |
| HRD6 (class III) | Strr Tn916 (Tetr) HRD Nar− Ffr− | This study |
Rifr, rifampin resistant; Fusr, fusidic acid resistant; Tetr, tetracycline resistant; Strr, streptomycin resistant; HRD, no growth with Cl-OHPA plus formate or lactate; Nar−, no growth with nitrate plus formate or lactate; Ffr−, no growth with fumarate plus formate.
DSM, Deutsche Sammlung von Mikroorganismen.
The cloning vectors pUC18 and pUC19 were purchased from Amersham Pharmacia Biotech (Uppsala, Sweden), and the PCR product cloning vectors pGEM-T and pMON38201 were obtained from Promega (Madison, Wis.) and Monsanto (St. Louis, Mo.), respectively.
Plating of D. dehalogenans.
Basal medium was autoclaved in serum bottles in the presence of agar. After the addition of vitamins, trace elements, 0.1 g of yeast extract/liter, 20 mM electron donor and -acceptor, and, if necessary the appropriate antibiotics, the medium was reduced with 0.04 g of both Na2S and cysteine-HCl/liter. Plates were poured in the glove box and stored there for 1 day. Cells from the exponential growth phase were diluted appropriately into reduced basal medium and distributed on the solidified medium in aliquots of 100 μl with sterile glass beads. The plates were incubated at 37°C upside down under N2 in a gas-tight jar. Application of 0.8% BBL agar purified (Becton Dickinson, Meylan, France) resulted in an average efficiency of plating (EOP, equal to CFU/number of cells counted) of 0.85 ± 0.14, compared to 0.15 ± 0.05 in the case of agar of normal purity (Difco Laboratories, Detroit, Mich.). An agar concentration of 1% or higher resulted in a 2.7-fold decrease in efficiency. For all subsequent experiments, 0.8% BBL agar purified was routinely used for the plating of D. dehalogenans.
For further subcultivation, colonies were picked inside the glove box, resuspended in 0.5 ml of reduced basal medium, and transferred to 120-ml serum bottles with 20 ml of anaerobic medium containing the appropriate substrates and antibiotics.
Isolation of D. dehalogenans HSS1.
Spontaneous streptomycin-resistant mutants of D. dehalogenans were isolated following growth in liquid culture in the presence of 20 mM pyruvate and 10 μg of streptomycin/ml. In subsequent cultures, the concentration of streptomycin was raised stepwise to 150 μg/ml. D. dehalogenans HSS1 was finally isolated from a single colony grown on a plate containing 20 mM lactate, 20 mM Cl-OHPA, and 200 μg of streptomycin/ml.
Filter mating, selection of transconjugants, and screening for halorespiration deficiency.
For the conjugation experiments, exponentially growing cultures of E. faecalis JH2-2 and D. dehalogenans HSS1 were diluted into fresh medium containing the appropriate antibiotics and grown to an A660 of 0.4 to 0.5 and 0.2 to 0.3, respectively. E. faecalis JH2-2 was cultivated at 30 or 37°C under either aerobic or anaerobic conditions (optimized protocol, 37°C and anaerobic). Subsequently, the cultures were transferred to a glove box, washed twice, and resuspended in reduced basal medium. Donor and recipient cultures were mixed in different ratios and filtered on a 0.45-μm-pore-size HA filter (Millipore, Bedford, Mass.). The membrane was placed with the bacterium side up on an agar plate containing 20 mM pyruvate and incubated at 30 or 37°C in an anaerobic jar under N2 for 6 to 16 h (optimized protocol, 37°C for 16 h). The cells were resuspended from the membrane in reduced basal medium, and transconjugants were selected on plates containing 20 mM pyruvate, 2,000 μg of streptomycin/ml, and 5 μg of tetracycline/ml. To determine the influence of the mating procedure on the viability of D. dehalogenans HSS1, plates without tetracycline were inoculated in parallel. Screening for HRD mutants of D. dehalogenans HSS1 was performed by streaking colonies of transconjugants on plates containing 2,000 μg of streptomycin/ml, 5 μg of tetracycline/ml, and either 20 mM pyruvate or 20 mM lactate and 20 mM Cl-OHPA.
DNA isolation and manipulation.
Chromosomal DNA of D. dehalogenans was isolated as described previously (25).
Plasmid DNA was isolated from E. coli by using the alkaline lysis method, and standard DNA manipulations were performed according to established procedures (19) and manufacturers’ instructions. Enzymes were purchased from Life Technologies B.V. (Breda, The Netherlands, Boehringer Mannheim GmbH (Mannheim, Germany), or New England Biolabs (Beverly, Mass.). Oligonucleotides were obtained from Life Technologies Inc. Hybond-N+ nylon transfer membrane (Amersham Life Science, Little Chalfont, United Kingdom) was used for Southern blot analysis. Probes for hybridization experiments were labeled by nick translation in the presence of [α-32P]dATP (Amersham Pharmacia Biotech). As specific probe for the tetracycline resistance determinant tetM, a 4.2-kb HincII restriction fragment of the Tn919 tetM gene from pCI182 was used (13).
Characterization of Tn916 insertion sites.
Chromosomal fragments flanking the sites of Tn916 insertion were amplified by inverse PCR (22). The divergent primer pairs BG 285-BG 287 (BG 285, 5′-GAC CTT GAT AAA GTG TGA TAA GTC C-3′, nucleotides (nt) 62 to 38; BG 287, 5′-GGA GTT TTA GCT CAT GTT GAT GC-3′, nt 12141 to 12163) and BG 286-BG 288 (BG 286, 5′-CTC GAA AGC ACA TAG AAT AAG GC-3′, nt 17956 to 17978; BG 288, 5′-CCA CGC TTC CTA ATT CTG TAA TCG-3′, nt 12231 to 12208) were used to specifically amplify the upstream and downstream flanking fragments, respectively. The sequence of the primers was based on the nucleotide sequence of Tn916 (GenBank accession no. U09422) (9). Chromosomal DNA was digested with HindIII and ligated at a concentration of 0.5 ng/μl. Five nanograms of self-ligated DNA was used as the template in a 25-μl PCR mixture containing 2 ng of each primer; 2 mM MgCl2; 200 μM (each) dATP, dCTP, dGTP, and dTTP; and 1 U of Expand Long Template enzyme mixture (Boehringer Mannheim GmbH). The DNA was amplified with the GeneAmp PCR System 2400 (Perkin-Elmer Cetus, Norwalk, Conn.). After the mixture was preheated to 94°C for 2 min, 35 amplification cycles were performed, consisting of denaturation at 94°C for 20 s, primer annealing at 55°C for 30 s, and elongation at 68°C for 3 min. From cycle 6 onward, the elongation was extended by 20 s per cycle to increase the yield. A final extension of 7 min at 68°C was performed. The PCR products were purified from agarose gel by Gene Clean (Bio 101, La Jolla, Calif.) and cloned into E. coli with pGEM-T or pMON38201.
For the further elucidation of the D. dehalogenans hyd locus identified in HRD mutant 6 (HRD6), the divergent primer pair BG 345-BG 346 (BG 345, 5′-CCATTCGATACCATGAGACC-3′, nt 1235 to 1254; BG 346, 5′-GTACTAATGATTCGATACTGGG-3′, nt 1222 to 1201) was used for inverse PCR from PstI-digested chromosomal DNA of the parental strain. The reaction conditions were as described above, and the PCR product was cloned and sequenced. For additional information on the hyf-hyc locus (HRD4), a 2.5-kb EcoRI-BamHI fragment was isolated from a D. dehalogenans genomic library, cloned, and sequenced.
DNA sequencing.
DNA sequencing was performed with a Li-Cor (Lincoln, Nebr.) DNA sequencer 4000L. Plasmid DNA used for sequencing reactions was purified with the QIAprep Spin Miniprep kit (Qiagen GmbH, Hilden, Germany). Reactions were performed with the Thermo Sequenase fluorescent-labeled primer cycle sequencing kit (Amersham Pharmacia Biotech). Infrared-labeled universal sequencing primers were purchased from MWG Biotech (Ebersberg, Germany). Sequence similarity searches and alignments were performed with the BLAST 2.0 program (1) (National Center for Biotechnology Information, Bethesda, Md.) and the DNAstar package (DNASTAR Inc., Madison, Wis.), respectively.
Enzyme and protein assays.
Harvesting of cells and preparation of cell extracts by sonication under anoxic conditions were performed as described previously (25). Enzyme activities in cell extracts were determined spectrophotometrically at 30°C in rubber-stoppered N2-flushed cuvettes by following the oxidation or reduction of benzylviologen or methylviologen at 578 nm (ɛ578 = 9.2 mM−1 × cm−1 or 9.7 mM−1 × cm−1, respectively). Formate dehydrogenase and hydrogenase were assayed in 1 ml of 100 mM Tris-HCl (pH 8.0) containing 1 mM benzylviologen and 10 mM formate or 1 ml H2, respectively. Fumarate reductase activity was determined in 1 ml of 100 mM sodium phosphate buffer (pH 7.5) containing 5 mM titanium(III)citrate-reduced benzylviologen and 10 mM fumarate. For all assays, 1 U of enzyme activity corresponds to the amount of enzyme catalyzing the conversion of 1 μmol of substrate or 2 μmol of benzyl- or methylviologen per min. For the semiquantitative determination of formate-H2 lyase activity, 200 μl of cell extract was added to 1.8 ml of 100 mM Tris-HCl (pH 7.8) buffer containing 10 mM formate and incubated at 30°C. The formation of hydrogen was followed by gas chromatography. Protein was determined according to the method of Bradford, with bovine serum albumin as the standard (4).
Nucleotide sequence accession numbers.
The nucleotide sequences of the sites of Tn916 insertion in the different mutants have been deposited in the GenBank database under accession no. AF157637 (HRD2 upstream), AF157638 (HRD2 downstream), AF157642 (HRD22), AF176224 (HRD4-1), AF157639 (HRD4-2 upstream), AF157640 (HRD4-2 downstream), and AF157641 (HRD6).
RESULTS
Development of a Tn916-based transposition system for D. dehalogenans.
To allow for the genetic analysis of halorespiration by D. dehalogenans, a transposition system was designed and optimized based on the broad-host-range conjugative transposon Tn916 (5). Previously, it had been reported that D. dehalogenans showed a poor EOP on agar plates (24). However, application of strict anaerobic conditions, an optimized agar concentration (0.8%), and an appropriate source of agar (BBL agar purified) resulted in a highly efficient plating system (EOP, 0.85 ± 0.14). On plates containing 20 mM pyruvate, colonies that were translucent, white, and spherical with a diameter of 2 to 3 mm appeared after 3 days of incubation. Similar results were obtained with 20 mM pyruvate or lactate as the electron donor and nitrate, fumarate, or Cl-OHPA as the electron acceptor (data not shown).
To enable counterselection following conjugation, a streptomycin-resistant mutant, strain HSS1, was isolated and showed an EOP of 0.85 ± 0.1 on plates containing up to 4,000 μg of streptomycin/ml, while for the parental strain, D. dehalogenans DSM 9161, the EOP was below 10−8 on plates containing 50 μg of streptomycin/ml. No resistance to other antibiotics (tetracycline, chloroamphenicol, or rifampin) was observed in strain HSS1 (data not shown).
The conjugative transposon Tn916 was introduced into D. dehalogenans HSS1 by filter mating with the streptomycin-sensitive E. faecalis JH2-2, which carries a single copy of Tn916 on its chromosome and is widely used as a Tn916 donor in matings of gram-positive bacteria (5, 6). After 5 days of incubation on plates containing streptomycin and tetracycline, transconjugant colonies of D. dehalogenans developed. Subsequently, conjugal transfer was optimized by varying growth of the donor, mating conditions, and the donor/recipient ratio (Table 2). The highest conjugation frequencies were observed when the E. faecalis donor was grown at 37°C in the absence of oxygen and mated in a fourfold excess over the recipient D. dehalogenans HSS1 for 16 h at 37°C, resulting in 3 × 10−6 transconjugants per recipient.
TABLE 2.
Conjugation frequencies for mating of E. faecalis JH2-2 and D. dehalogenans HSS1
| Preincubation E. faecalis | Mating conditions | Donor/recipient ratio | Conjugation frequency (107)a |
|---|---|---|---|
| Aerobic, 37°C | 37°C, 16 h | 0.5 | 1.1 |
| Aerobic, 30°C | 30°C, 6 h | 10.0 | 4.0 |
| Aerobic, 37°C | 37°C, 6 h | 6.0 | 6.7 |
| Anaerobic, 37°C | 37°C, 16 h | 4.0 | 30.0 |
Conjugation frequencies were calculated as number of transconjugants per recipient.
The insertion of Tn916 in the chromosome of D. dehalogenans HSS1 was confirmed by Southern blot analysis with a tetM-specific probe and revealed many single copy insertions, occasional insertion of two copies, and an apparently random distribution (data not shown) (see below).
HRD mutants and their phenotypic classification.
A total of 2,500 tetracycline-resistant transconjugants obtained from several independent matings were analyzed for halorespiration deficiency. Transconjugants which showed growth on pyruvate but not on lactate–Cl-OHPA within 7 days were considered HRD. After rescreening, a total of 24 HRD mutants were isolated and subsequently subjected to a series of 20-ml batch incubations in the presence of 20 mM lactate or formate as the electron donor and fumarate, nitrate, or Cl-OHPA as the electron acceptor. As a control, fermentative growth with 20 mM pyruvate was followed. Furthermore, a transconjugant which had not lost the ability to grow upon halorespiration (HRD0) was used as a control strain. Deficiency in using a certain combination of electron donor and acceptor was defined by a lag phase increased more than twofold in growth compared to strain HRD0. This criterion was chosen because continuous incubation in the medium resulted in the growth of tetracycline-resistant revertants in which the transposon probably had moved from the original site of integration to another site. This was confirmed by Southern analysis with a tetM-specific probe, as the specific hybridizing signals progressively disappeared with successive cultivation under selective conditions (results not shown). Based on the results, the strains could be grouped into three major classes, which differed in their capacities to utilize additional electron acceptors (Table 1). The first class contained only one mutant, HRD2, that was found to be solely deficient in halorespiration with lactate or formate as the electron donor. Class II was the major group, comprising 22 HRD mutants which had lost the ability to grow by both halorespiration and nitrate respiration with lactate or formate as the electron donor. The third phenotypic group again included only a single mutant, strain HRD6, which was not only impaired in the use of Cl-OHPA and nitrate but also fumarate as the electron acceptor. The pleiotropic phenotypes of most mutants indicate the disruption of genes that encode common elements shared by the different respiratory chains present in this organism.
Genetic characterization of HRD mutants.
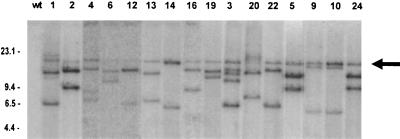
Chromosomal DNA was isolated from HRD transconjugants and digested with HindIII. Tn916 contains a single HindIII site, yielding left and right arms of 12.2 and 5.8 kb, respectively (9). The fragments were separated by agarose gel electrophoresis and transferred to membranes that were hybridized to a 4.2-kb HincII restriction fragment of the Tn919 tetM gene (Fig. 1). While no DNA fragments from the recipient D. dehalogenans strain HSS1 hybridized with the tetM probe, most of the mutant strains exhibited two hybridizing fragments, indicating chromosomal integration of a single copy of Tn916. Only HRD1, -3, and -4 showed more than two hybridizing bands, suggesting multiple transposon insertions. The difference in the sizes of the hybridizing fragments in the mutants indicates that the transposon inserted in an apparently random manner into the chromosome of D. dehalogenans. An additional 18-kb hybridizing fragment was found with different intensities in all mutants, suggesting the presence of a circular intermediate of Tn916 (10).
FIG. 1.
Hybridization of HindIII-digested chromosomal DNA from wild-type (wt) and tetracycline-resistant HRD mutants of D. dehalogenans with a probe for tetM (obtained from pCI182 [13]). The autoradiograph was digitally corrected for differences in DNA concentration. Lanes: 1, strain HSS1; 2 to 17, mutant strains HRD1, -2, -4, -6, -12, -13, -14, -16, -19, -3, -20, -22, -5, -9, -10, and -24, respectively. DNA size markers are in kilobase pairs. Fragments smaller than 4 kb were not found to hybridize with the tetM probe. The arrow indicates the possible 18-kb circular intermediate of Tn916.
Characterization of the Tn916 insertion sites in different HRD mutants.
Inverse PCR was used to specifically amplify the Tn916-flanking fragments from the chromosomal DNA of representative HRD mutants. The identity of relevant PCR products was verified by Southern blot analysis of HindIII-digested chromosomal DNA isolated from the different mutants. Fragments hybridizing with the different PCR products also hybridized strongly with a radiolabeled tetM probe, confirming that the primers used specifically yielded the amplification of Tn916-flanking fragments (data not shown).
Characterization of the Tn916 insertion sites indicated that 4 of the 24 HRD-mutants isolated carried the transposon inserted at the same site but in different orientations (HRD5-HRD24 and HRD9-HRD10). Interestingly, all these mutants arose from independent mating experiments. Similarly, Tn916 insertion sites were identical for HRD3, HRD20, and HRD22, with HRD20 carrying the transposon in the inverse orientation compared to those in HRD3 and HRD22.
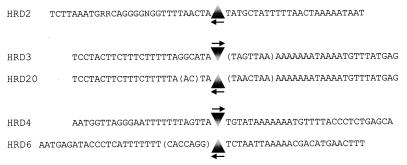
Nucleotide sequence analysis of the flanking chromosomal regions showed that the target sites are generally AT rich (Fig. 2). In two cases, the wild-type sequence of the insertion site was determined, revealing the presence of additional nucleotides after the insertion event, as shown for HRD3, HRD20, and HRD6. These short so-called coupling sequences are a common feature of Tn916 insertion sites and are a result of the integration event (5). The insertions were located in the vicinity (i.e., within potential promoter and terminator sequences) of open reading frames that encode regulatory proteins or enzymes of anaerobic respiratory pathways present in D. dehalogenans (Fig. 3).
FIG. 2.
Nucleotide sequences of the Tn916 insertion sites in different HRD mutants of D. dehalogenans. The triangles indicate the sites and orientations of Tn916 insertion (nt 1 → 18032). The bases given in parentheses for HRD3, -6, and -20 were not present in the wild-type sequence.
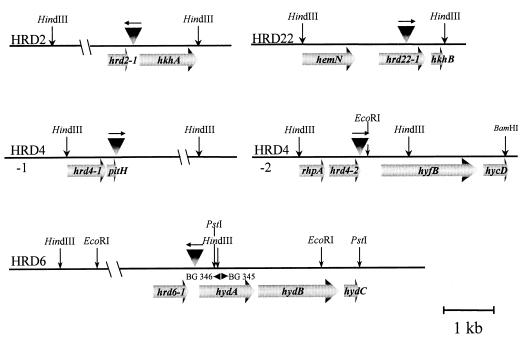
FIG. 3.
Physical map of the Tn916 insertion sites in different class I, II, and III HRD mutants of D. dehalogenans. The triangles indicate the sites and orientations of transposon integration. The horizontal arrows show identified open reading frames. Restriction sites and oligonucleotides are indicated by vertical arrows and by arrowheads, respectively.
Four representatives of class I, II, and III mutants were analyzed in detail. In the class I mutant, HRD2, Tn916 had inserted downstream of hrd2-1, which shows limited similarity (20% identity at the amino acid level) with cheX, encoding a chemotaxis-related protein in different strains of Treponema. It is located upstream of hkhA, revealing significant similarity (22% identity at the protein level) with a gene coding for a sensory transduction histidine kinase in Methanobacterium thermoautotrophicum (GenPept accession no. AAB84866).
In the class II mutant HRD22, the transposon inserted in the hrd22-1 gene, which could encode a 30-kDa protein of unknown function. This gene is followed by hkhB, which may encode a member of the two-component sensor histidine kinases (28% identity at the protein level with YwpD from Bacillus subtilis; GenPept accession no. CAB05945). One kilobase downstream of the transposon insertion site in HRD22 is the hemN gene, which shows strong similarity with genes coding for oxygen-independent coproporphyrinogen III oxidases that are involved in porphyrin biosynthesis (Fig. 3).
The class II mutant HRD4 contains two Tn916 insertions (Fig. 3). One copy of the transposon inserted in the pitH gene showing significant similarity with genes encoding low-affinity inorganic phosphate transporters (60% identity at the protein level with YkaB from B. subtilis, GenPept accession no. CAB13141). A second copy of Tn916 had inserted downstream of hrd4-2, coding for a hypothetical protein, and rhpA, exhibiting significant similarity with genes encoding putative regulatory proteins (44% identity at the protein level with the hypothetical protein YwhH from B. subtilis [GenPept accession no. CAB15775] and 25% with EbsC from E. faecalis [GenPept accession no. AAC36853]). The putative proteins encoded by the two genes identified downstream of the insertion, hyfB and hycD, show the highest similarity to the gene products of hyfB (31% identity) and hycD (25% identity), respectively, which are integral membrane subunits of two formate-hydrogen lyase complexes present in E. coli (2), and to the proteins encoded by genes from an unidentified Mycobacterium tuberculosis operon [GenBank accession no. Z74410).
In the class III mutant strain HRD6, Tn916 had inserted downstream of an open reading frame, hrd6-1, encoding a protein of unknown function, and in the presumed promoter region 200 bp upstream of a potential hydrogenase-encoding gene cluster (Fig. 3). The predicted gene products of hydA, hydB, and hydC show significant homology with the small, large, and B-type cytochrome subunits of membrane-bound periplasmic quinone-reactive hydrogenases of different bacteria.
Biochemical characterization of HRD mutants.
The results of both the physiological and the genetic characterizations of the isolated HRD mutants indicated the disruption of either structural or regulatory components shared by different respiratory chains present in D. dehalogenans. Key respiratory enzyme activities were determined for the class II mutants HRD4 and HRD22, as well as for HRD6 (class III), and compared to those of a transconjugant strain showing a wild-type phenotype with respect to its halorespiring ability (HRD0). The class I mutant HRD2 revealed a rather high degree of instability, making a more detailed biochemical characterization impossible. Formate dehydrogenase, hydrogenase, and fumarate reductase activities were determined in cell extracts prepared from cells grown by fermentation on 20 mM pyruvate. These experiments indicated that the class II mutant strain HRD4 had strongly reduced activity in both formate dehydrogenase (50 U/mg) and hydrogenase activity (60 U/mg) in contrast to the fumarate activity (1,910 U/mg), which was even higher than that found in the wild-type strain (520, 640, and 720 U/mg in HRD0 for the above-mentioned enzymes, respectively). Semiquantitative determination of H2 formation upon formate addition indicated no significant differences in formate-H2 lyase activity in HRD0 and HRD4. Similar results were obtained for the class III mutant, HRD6. Remarkably, the class II mutant HRD22 was affected only in formate dehydrogenase activity (40 U/mg). The results obtained from the activity measurements could be qualitatively confirmed by hydrogenase and formate dehydrogenase activity staining experiments. After polyacrylamide gel electrophoresis of cell extracts under nondenaturing conditions, the experiments were performed with benzylviologen as the electron acceptor and showed single bands for each enzyme, although some material did not enter the gel (data not shown).
DISCUSSION
Halorespiration is a recently discovered mode of anaerobic respiration carried out by a rather wide range of gram-positive and -negative bacteria. Nevertheless, our current knowledge of the architecture, bioenergetics, and control of this respiratory pathway at the molecular level is very limited and is restricted to the molecular characterization of reductive dehalogenases (17, 25). Here we describe the development of a plating, delivery, and screening system that allowed integration of the conjugative transposon Tn916 in the chromosome of the newly described o-chlorophenol-respiring gram-positive bacterium D. dehalogenans. Subsequently, HRD mutants that were genetically and physiologically characterized and found to be deficient in the biosynthesis of one or more of the components of the halorespiratory electron transfer chain were isolated.
Using E. faecalis JH2-2 as a donor in anaerobic filter matings, we were able to accomplish Tn916 integration into the chromosome of genetically marked D. dehalogenans HSS1 with a frequency of 3 × 10−6 transconjugants per recipient (0.8 × 10−6 per donor). This transposition frequency is within the range commonly observed for Tn916, which varies from <10−9 to >10−4 per donor (5). Based on their inability to use Cl-OHPA as an electron acceptor, we isolated a total of 24 HRD mutants. Based on subsequent physiological characterization, the mutants could be grouped in three major phenotypic classes. Only one mutant was deficient solely in halorespiration (class I). The rest of the mutants (22 class II and 1 class III mutants) were all impaired in both halorespiration and nitrate respiration. This suggests that the nitrate respiratory and halorespiratory chains share common components, the function of which has been affected by the insertion of the transposon.
Southern blot analysis revealed that Tn916 integrated in single copy, but occasionally two copies, into the chromosome of D. dehalogenans. Furthermore, the presence of a large, 18-kb Tn916-specific band suggests there is a circular intermediate of Tn916 (10) in virtually all mutants, indicating that the transposon is still mobile in D. dehalogenans (Fig. 1). Both the occurrence of multiple insertions and the remaining mobility of the transposon in transconjugants of D. dehalogenans might explain the relative instability of the mutants, which was most pronounced in the single class I mutant, HRD2.
The repeated isolation of identical integrants from independent conjugation experiments suggests saturation mutagenesis. Another possible explanation would be the preferential integration of Tn916 at specific sites, i.e., the occurrence of hot spots of mutation. Sequence analysis revealed that the insertion sites are very AT rich, ranging from 75 to 91% compared to an average AT content of the D. dehalogenans genome of 55% (24). In most cases, insertions were situated in the immediate vicinity of potential promoter and terminator sequences. This is a frequently observed feature of transposons belonging to the Tn916-Tn1545 family, and it was proposed that the special conformation of DNA carrying those sequences, like strong bending or supercoiling, could possibly be recognized by the integrase (18, 21).
The results of the genetic characterization of the Tn916 insertion site in HRD2 indicate that regulatory functions might be impaired in this class I mutant (Table 3). Considering that halorespiration is induced in D. dehalogenans by the presence of different o-chlorophenols (23), it is tempting to speculate that the hkhA gene product may be the o-chlorophenol-sensing part of a two-component regulatory system. Unfortunately, high instability made a more detailed physiological and biochemical investigation of this interesting mutant impossible.
TABLE 3.
Characterization and putative functions of selected HRD mutants
| Clone | Class | Additional phenotypea | Putative disruption | Putative function |
|---|---|---|---|---|
| HRD2 | I | Upstream hkhA (regulator) | Regulation, Cl-OHPA sensing | |
| HRD22 | II | Nar− | Downstream hemN (maturation), upstream hkhB (regulator) | Regulation or maturation |
| HRD4 | II | Nar− | Upstream hyf-hyc, downstream rhpA (regulator) | Formate-hydrogen lyase (respiration) |
| HRD6 | III | Nar−, Ffr− | Upstream hydABC | Uptake hydrogenase (respiration) |
All HRD mutants are deficient in growth with Cl-OHPA plus formate or lactate; Nar−, no growth with nitrate plus formate or lactate; Ffr−, no growth with fumarate plus formate.
In the class II mutant HRD22, the site of Tn916 integration was located 0.5 kb upstream of a second putative sensor histidine kinase-encoding gene, hkhB, and downstream of hemN, a gene encoding a protein involved in porphyrin biosynthesis. This suggests that regulatory functions involved in maturation of redox complexes might be disrupted in this mutant (Table 3).
The results from both molecular and biochemical analysis of different class II and III HRD mutants indicate an involvement of respiratory complexes in halorespiration in D. dehalogenans (Table 3). In HRD6, Tn916 inserted upstream of a hydrogenase-encoding operon. HRD4 carried two copies of the transposon, of which one had inserted in a gene encoding a putative phosphate transporter. However, the second copy was found upstream of genes encoding subunits of a putative formate-H2 lyase, suggesting that this insertion is responsible for the observed respiration-deficient phenotype. As HRD6 cannot utilize any of the tested electron acceptors, the presumed hyd gene product seems to play a central role in the different respiratory chains. The hydrogenase might play an essential role in electron transfer to the terminal reductases. For a long time, it has been assumed that the formate-H2 lyase complex (Hyc complex) is only involved in fermentation in the extensively studied facultative anaerobe E. coli (20). However, it was recently proposed that the formate-H2 lyase, which is likely to be encoded by the E. coli hyf operon, might be proton translocating and thereby involved in respiratory metabolism (2). According to this hypothesis, the hydrogen evolved by Hyf would be oxidized by an electron transport chain involving a menaquinone-reducing uptake hydrogenase and a terminal reductase (2). Such a respiratory chain may be impaired in HRD4 and HRD6, suggesting that it may indeed be an essential part of both the halo- and nitrate respiratory chains in D. dehalogenans. In both mutants, hydrogenase and formate dehydrogenase activities were severely affected, suggesting coordinated regulation of the different enzyme complexes within the proposed respiratory chains. Inactivation of the hydrogenase thus also affects formate dehydrogenase activity (which might be part of the formate-hydrogen lyase complex), and vice versa. In the latter case, the lack of hydrogen formation in a formate-hydrogen lyase-deficient mutant might affect the induction of the hydrogenase.
A priori, it was expected that the HRD mutants obtained would be impaired in the key enzyme o-chlorophenol-reductive dehalogenase (CprA), additional structural components of the halorespiratory chain, and enzymes involved in processing and targeting of respiratory complexes or regulatory functions. However, the characterization of the 24 HRD mutants isolated revealed that none of the first type was obtained. A mutation in cprA is not expected to be lethal, as the organism is rather versatile in its ability to use alternative electron acceptors. The possibility of multiple copies of cprA can also be ruled out based on results obtained by Southern blot analysis (25). The detailed analysis of the isolated mutants on the physiological, biochemical, and genetic levels reported here complements our earlier identification of the cpr operon, provides insight into the complexity of halorespiration, and indicates that the halorespiratory chain and other electron transport chains are integrated.
ACKNOWLEDGMENTS
This work was partly supported by grants from the Studienstiftung des Deutschen Volkes and the Chinese Academy of Sciences and contract BIO4-98-0303 of the European Union.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J H, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews S C, Berks B C, McClay J, Ambler A, Quail M A, Golby P, Guest J R. A 12 cistron Escherichia coli operon (hyf) encoding a putative proton translocating formate hydrogenlyase system. Microbiology. 1997;143:3633–3647. doi: 10.1099/00221287-143-11-3633. [DOI] [PubMed] [Google Scholar]
- 3.Berks B C. A common export pathway for proteins binding complex redox cofactors? Mol Microbiol. 1996;22:393–404. doi: 10.1046/j.1365-2958.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantification of microgram quantities of proteins utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Clewell D B, Flannagan S E, Jaworski D D. Unconstrained bacterial promiscuity: The Tn916-Tn1545 family of conjugative transposons. Trends Microbiol. 1995;3:225–236. doi: 10.1016/s0966-842x(00)88930-1. [DOI] [PubMed] [Google Scholar]
- 6.de Vos W M, Kleerebezem M, Kuipers O P. Expression systems for industrial Gram-positive bacteria with low guanine and cytosine content. Curr Opin Biotechnol. 1997;8:547–553. doi: 10.1016/s0958-1669(97)80027-4. [DOI] [PubMed] [Google Scholar]
- 7.El Fantroussi S, Naveau H, Agathos S N. Anaerobic dechlorinating bacteria. Biotechnol Prog. 1998;14:167–188. doi: 10.1021/bp980011k. [DOI] [PubMed] [Google Scholar]
- 8.Fetzner S. Bacterial dehalogenation. Appl Microbiol Biotechnol. 1998;50:633–657. doi: 10.1007/s002530051346. [DOI] [PubMed] [Google Scholar]
- 9.Flannagan S E, Zitzow L A, Su Y A, Clewell D B. Nucleotide sequence of the 18-kb conjugative transposon Tn916 from Enterococcus faecalis. Plasmid. 1994;32:350–354. doi: 10.1006/plas.1994.1077. [DOI] [PubMed] [Google Scholar]
- 10.Gawron-Burke C, Clewell D B. Regeneration of insertionally inactivated streptococcal DNA fragments after excision of transposon Tn916 in Escherichia coli: strategy for targeting and cloning of genes from gram-positive bacteria. J Bacteriol. 1984;159:214–221. doi: 10.1128/jb.159.1.214-221.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gribble G W. Naturally occurring organohalogen compounds—a comprehensive survey. Fortschr Chem Org Naturst. 1996;68:1–423. [PubMed] [Google Scholar]
- 12.Hileman B. Concerns broaden over chlorine and chlorinated hydrocarbons. Chem Eng News. 1993;71:11–20. [Google Scholar]
- 13.Hill C, Venema G, Daly C, Fitzgerald G F. Cloning and characterization of the tetracycline resistance determinant of and several promoters from within the conjugative transposon Tn919. Appl Environ Microbiol. 1988;54:1230–1236. doi: 10.1128/aem.54.5.1230-1236.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holliger C, Wohlfarth G, Diekert G. Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol Rev. 1998;22:383–398. [Google Scholar]
- 15.Jacob A E, Hobbs S J. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J Bacteriol. 1974;117:360–372. doi: 10.1128/jb.117.2.360-372.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohn W W, Tiedje J M. Microbial reductive dehalogenation. Microbiol Rev. 1992;56:482–507. doi: 10.1128/mr.56.3.482-507.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neumann A, Wohlfarth G, Diekert G. Tetrachloroethene dehalogenase from Dehalospirillum multivorans: cloning, sequencing of the encoding genes, and expression of the pceA gene in Escherichia coli. J Bacteriol. 1998;180:4140–4145. doi: 10.1128/jb.180.16.4140-4145.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Renault P, Nogrette J F, Galleron N, Godon J J, Ehrlich S D. Specificity of insertion of Tn1545 transposon family in Lactococcus lactis subsp. lactis. Dev Biol Stand. 1995;85:535–541. [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 20.Sawers G. The hydrogenases and formate dehydrogenases of Escherichia coli. Antonie Leeuwenhoek. 1994;66:57–88. doi: 10.1007/BF00871633. [DOI] [PubMed] [Google Scholar]
- 21.Scott J R, Bringel F, Marra D, Van Alstine G, Rudy C K. Conjugative transposition of Tn916: preferred targets and evidence for conjugative transfer of a single strand and for a double-stranded circular intermediate. Mol Microbiol. 1994;11:1099–1108. doi: 10.1111/j.1365-2958.1994.tb00386.x. [DOI] [PubMed] [Google Scholar]
- 22.Triglia T, Peterson M G, Kemp D J. A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 1988;16:8186. doi: 10.1093/nar/16.16.8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Utkin I, Dalton D D, Wiegel J. Specificity of reductive dehalogenation of substituted ortho-chlorophenols by Desulfitobacterium dehalogenans JW/IU-DC1. Appl Environ Microbiol. 1995;61:346–351. doi: 10.1128/aem.61.1.346-351.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Utkin I, Woese C, Wiegel J. Isolation and characterization of Desulfitobacterium dehalogenans gen. nov., sp. nov., an anaerobic bacterium which reductively dechlorinates chlorophenolic compounds. Int J Syst Bacteriol. 1994;44:612–619. doi: 10.1099/00207713-44-4-612. [DOI] [PubMed] [Google Scholar]
- 25.van de Pas B A, Smidt H, Hagen W R, van der Oost J, Schraa G, Stams A J M, de Vos W M. Purification and molecular characterization of ortho-chlorophenol reductive dehalogenase, a key enzyme of halorespiration in Desulfitobacterium dehalogenans. J Biol Chem. 1999;274:20287–20292. doi: 10.1074/jbc.274.29.20287. [DOI] [PubMed] [Google Scholar]