Abstract
The immune response mediated by Th17 cells is essential in the pathogenesis of periodontitis. Emerging evidence has demonstrated that lipopolysaccharide from Porphyromonas gingivalis (Pg-LPS) could promote Th17-cell differentiation directly, while the downstream signaling remains elusive. This study was aimed to explore the role of JMJD3 (a JmjC family histone demethylase) and signal transducers and activators of transcription 3 (STAT3) in Th17-cell differentiation triggered by Pg-LPS and clarify the interaction between them. We found that the expression of JMJD3 and STAT3 was significantly increased under Th17-polarizing conditions. Pg-LPS could promote Th17-cell differentiation from CD4+ T cells, with an increased expression of JMJD3 and STAT3 compared to the culture without Pg-LPS. The coimmunoprecipitation results showed that the interactions of JMJD3 and STAT3, STAT3 and retinoid-related orphan nuclear receptor γt (RORγt) were enhanced following Pg-LPS stimulation during Th17-cell differentiation. Further blocking assays were performed and the results showed that inhibition of STAT3 or JMJD3 both suppressed the Th17-cell differentiation, JMJD3 inhibitor could reduce the expression of STAT3 and p-STAT3, while JMJD3 expression was not affected when STAT3 was inhibited. Taken together, this study found that JMJD3 could promote Pg-LPS induced Th17-cell differentiation by modulating the STAT3-RORc signaling pathway.
Keywords: P. gingivalis, lipopolysaccharide, Th17 cell, JMJD3, STAT3
Introduction
It is widely accepted that periodontitis is a microbial-related chronic inflammatory disease that induces the loss of the periodontal ligament and alveolar bone (Könönen et al., 2019). A skewed host immune and inflammatory response initiated by periodontal pathogens is considered to be the main cause for the initiation, establishment and progression of periodontal inflammation and tissue breakdown (Cekici et al., 2014). In this regard, CD4+ T helper (Th) cells, especially Th17 cells, are critically important in the pathogenesis of periodontitis. Exaggerated Th17 responses are proven to promote inflammatory bone loss and tissue damage in periodontitis (Adibrad et al., 2012; Eskan et al., 2012; Moutsopoulos et al., 2014). Notably, Th17 cells, originating from CD4+ T cells, are induced by retinoid-related orphan nuclear receptor γt (RORγt, encoding gene Rorc) (Ivanov et al., 2006). However, the underlying mechanisms that regulate Th17-cell differentiation in the periodontal inflammatory context remain undefined.
Porphyromonas gingivalis (P. gingivalis) is strongly correlated with advanced periodontal lesions (Lunar Silva and Cascales, 2021). Lipopolysaccharide (LPS), one of the most important virulence factors of P. gingivalis, is believed to induce a strong immune reaction by interacting with toll-like receptors (TLRs), which triggers the expression of proinflammatory cytokines and activates the NF-κB pathway (Yang et al., 2015; Heinbockel et al., 2018). These interactions stimulate the differentiation of CD4+ subtypes (particularly Th1 and Th17) (Candelli et al., 2021). Our previous study demonstrated that LPS from P. gingivalis (Pg-LPS) could promote Th17-cell differentiation directly by upregulating TLR2 expression on the T cell membrane (Zhang et al., 2019a). Therefore, the downstream signaling of TLRs in regulating Th17-cell differentiation induced by Pg-LPS needs to be further investigated.
Signal transducers and activators of transcription 3 (STAT3), a component of the IL-6-activated acute phase response factor complex (Heinrich et al., 1998), has been proven to be activated downstream of the TLR signaling pathway and participates in the regulation of inflammatory responses (Park et al., 2013; Lyu et al., 2016). Although STAT3 has been implicated to be closely associated with the differentiation of Th cells (Villarino et al., 2015), we lack insight into whether and how STAT3 regulates Th17-cell differentiation via TLRs. Genome-wide studies have illustrated clear correlations between epigenetic modifications and T cell differentiation (Wei et al., 2009; Zhang et al., 2012; Russ et al., 2014).
Recently, Lamere et al. (2017) found that H3K27me3 was demethylated during the early activation of CD4+ T cells. ChIP-seq analysis revealed reduced levels of H3K27me3 in the promoter regions of JAK2 and STAT3. JMJD3, a lysine K27 demethylase, is reported to have an important role in the activation of Th17-related cytokines (Wang et al., 2012). Another study demonstrated that JMJD3 shares direct targets with RORγt and STAT3 (Ciofani et al., 2012), suggesting a potential role of JMJD3 in regulating STAT3 and Th17-cell differentiation. Thus, in this preliminary study, we focused on the potential contribution of JMJD3 and STAT3 to Th17-cell differentiation triggered by Pg-LPS and further explored the interaction between JMJD3 and STAT3.
Materials and Methods
CD4+ T cell purification and in vitro differentiation
CD4+ T cells were magnetically separated from the splenocytes of 60 C57BL/6J mice (6- to 8-week-old, female) according to the manufacturer's instructions (130-104-454; Miltenyi Biotech, Germany). This study was approved by the Institutional Animal Care and Use Committee (IACUC), Sun Yat-sen University (SYSU-IACUC-2022-B0003). After sorting, CD4+ T cells were cultured in RPMI medium and stimulated with plate-bound anti-CD3 (5 μg/mL) and soluble anti-CD28 (2 μg/mL) for 5 days under the following conditions (Zhao et al., 2018): 2 ng/mL TGF-β, 30 ng/mL IL-6, 10 μg/mL anti-IFN-γ, 10 μg/mL anti-IL-4, 20 ng/mL IL-23, and 10 ng/mL IL-1β (Biolegend). The medium was replaced on the third day.
Th17 cells stimulated with Pg-LPS
Then, 1 μg/mL Pg-LPS (tlrl-Pglps; InvivoGen) was added to the CD4+ T cells under Th17-polarizing conditions. The cultures were incubated for 5 days for RNA analysis, FACS analysis, western blotting, and enzyme-linked immunosorbent assay (ELISA).
Coimmunoprecipitation
The cultured cells were solubilized in IP buffer. The protein lysate was incubated with 2 μg anti-STAT3 (Cell Signaling Technology) or anti-IgG (Beyotime, China) at 4°C overnight. Then, the pretreated Protein A/G Magnetic Beads (MCE) were incubated with 100 μg of the lysate proteins overnight at 4°C. The precipitates were washed with lysis buffer five times and then used for the immunoblotting assay using anti-JMJD3, anti-STAT3, or anti-RORγt antibody. For the anti-RORγt antibody, the horseradish peroxidase-conjugated anti-rabbit IgG light-chain (Abbkine, China) was used as the secondary antibody to avoid interference from IgG heavy chain.
STAT3 or JMJD3 blocking assay
The STAT3 inhibitor Stattic (MCE) or JMJD3 inhibitor GSK-J4 (Tocris) was added to the Th17-polarizing cultures with or without Pg-LPS. The cultures were further incubated in a humidified atmosphere of 5% CO2 at 37°C for 5 days for RNA analysis, FACS analysis, western blotting and ELISAs.
Cell viability assay
The effect of Stattic or GSK-J4 on CD4+ T cell viability was assessed by the cell counting kit-8 (CCK-8) assay (Beyotime) according to the manufacturer's instructions.
Flow cytometric analysis
CD4+ T cells were restimulated with 50 ng/mL phorbol-12-myristate-13-acetate and 500 ng/mL ionomycin in the presence of 10 μg/mL Brefeldin A (Sigma-Aldrich, Germany) for 5 h. After permeabilization, cells were stained with FluoresceinIsothiocyanate-conjugated anti-CD4 antibodies and anti-IL-17-antigen-presenting cell (APC) antibodies (BioLegend) for 20 min at room temperature. All flow cytometry analyses were performed on a FACSCalibur (BD Biosciences), and the data were analyzed by FlowJo software.
Enzyme-linked immunosorbent assay
The levels of IL-17A protein in the culture supernatants were measured by an ELISA kit (Wuhan Huamei Biotechnology, China) according to the manufacturer's instructions. The optical density values were measured by GEN5 at 450 nm.
Quantitative real-time PCR
RNA transcripts were quantified by real-time PCR as described (Zhang et al., 2021). The specific primer sequences are described in Table 1 (designed and synthesized by Tianyi Huiyuan Biotechnology, China).
Table 1.
Primers for Quantitative Polymerase Chain Reaction Analysis
| Forward primer (5′–3′) | Reverse primer (3′–5′) | |
|---|---|---|
| Jmjd3 | CACCGGACCCCAAGAAC | CTGTGGATGTTACCCGCAT |
| Stat3 | GAGAGCAGAAGGGAGCAA | CTCACAGAGTGGGGCAA |
| Il-17a | TTCACTTTCAGGGTCGAGA | GGGGTTTCTTAGGGGTCA |
| Rorc | GAACTTGGGGAACCAGAAC | TGGCATGTCTCTCGGAA |
| Gapdh | GGATGCTGCCCTTACCC | GTTCACACCGACCTTCACC |
Gapdh, glyceraldehyde-3-phosphate dehydrogenase; Il-17a, interleukin 17A; Jmjd3, Jumonji domain-containing protein 3; Rorc, RAR-related orphan receptor C; Stat3, signal transducer and activator of transcription 3.
Western blotting
Cells were lysed in RIPA buffer containing 1% protease and phosphatase inhibitor (Beyotime) on ice for 30 min. The lysates were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by immunoblotting with specific antibodies against JMJD3 (1:1000; Novus Biologicals), p-STAT3 (1:1000; Cell Signaling Technology), STAT3 (1:1000; Cell Signaling Technology), IL-17A (1:500; Abcam), and GAPDH (1:1000; Cell Signaling Technology).
Statistical analysis
Data were analyzed by GraphPad 7.0 software. Student's t test or the Mann–Whitney U test were used for appropriate comparisons between two groups. One-way analysis of variance was performed to determine the significance among multiple groups. A value of p < 0.05 was considered statistically significant. All experiments were repeated more than three times unless otherwise noted.
Results
Pg-LPS promotes the expression of JMJD3 and STAT3 in Th17-cell differentiation
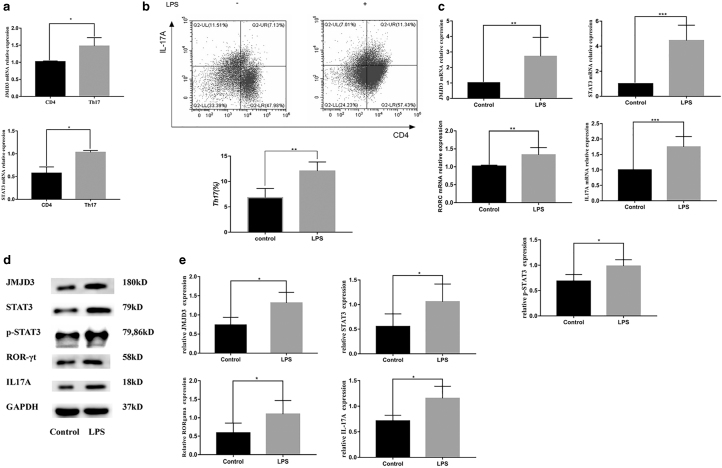
To determine the expression of JMJD3 and STAT3 during Th17-cell differentiation, purified CD4+ T cells were stimulated with specific antibodies for 5 days. Quantitative PCR results showed that the mRNA expression of JMJD3 and STAT3 was significantly increased under Th17-polarizing conditions (Fig. 1a). The Th17-cell group was indicated as the control group in the subsequent experiments.
FIG. 1.
Pg-LPS promotes the expression of JMJD3 and STAT3 in Th17-cell differentiation. (a) The mRNA expression of Jmjd3 and Stat3 was significantly increased in freshly isolated CD4+ T cells and activated CD4+ T cells under Th17-cell conditions. (b) After stimulation with Pg-LPS, CD4+ IL-17A+ T cells were identified by flow cytometry. The Th17-cell differentiation ratio was upregulated after Pg-LPS treatment. (c, d, e) The expression of RORC, IL-17A, JMJD3, and STAT3 was significantly upregulated with Pg-LPS at both the mRNA (c) and protein (d, e) levels. Data are presented as the mean ± SEM of at least three independent experiments. Significant changes are marked with *p < 0.05, **p < 0.01, and ***p < 0.001. Jmjd3, Jumonji domain-containing protein 3; Pg-LPS, LPS from Porphyromonas gingivalis; Rorc, RAR-related orphan receptor C; Stat3, signal transducer and activator of transcription 3.
After stimulation with Pg-LPS, CD4+IL-17A+ cells were determined by flow cytometry, and the results showed that the Th17-cell differentiation ratio was upregulated after Pg-LPS treatment compared to that of the control group without LPS (Fig. 1b). Meanwhile, the expression of RORγt and IL-17A was also significantly upregulated with Pg-LPS at both the mRNA and protein levels (Fig. 1d, e), suggesting that Pg-LPS could promote Th17-cell differentiation from CD4+ T cells in vitro.
Simultaneously, increased mRNA and protein expression levels of JMJD3 and STAT3 were observed in Th17-cell differentiation after Pg-LPS stimulation compared to the culture without Pg-LPS (Fig. 1c–e), suggesting that Pg-LPS could promote the expression of JMJD3 and STAT3 in Th17-cell differentiation.
STAT3 is correlated with JMJD3 in Pg-LPS-stimulated Th17-cell differentiation
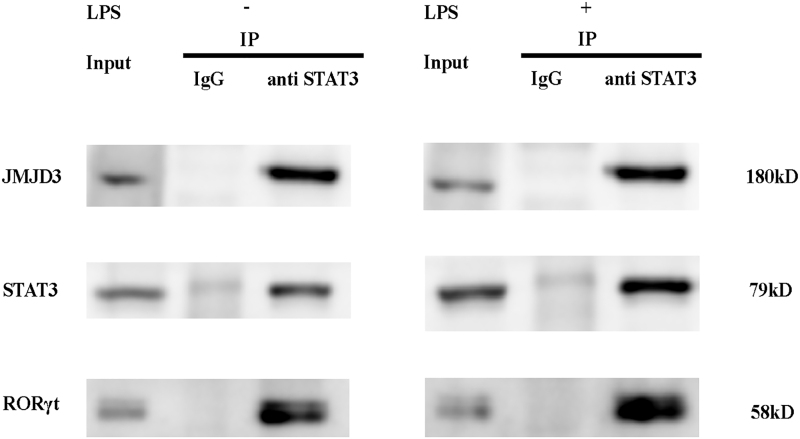
To explore the interaction between JMJD3 and STAT3, coimmunoprecipitation assays were performed. The results showed that JMJD3 directly interacted with STAT3 during Th17-cell differentiation with or without Pg-LPS stimulation (Fig. 2). And RORγt was identified in STAT3 immune complexes suggesting the interaction between STAT3 and RORγt. Furthermore, the interactions between JMJD3 and STAT3, STAT3 and RORγt were enhanced after Pg-LPS stimulation. Thus, we can infer that JMJD3 plays an important role during Pg-LPS-stimulated Th17-cell differentiation through its interaction with STAT3.
FIG. 2.
STAT3 interacts with JMJD3 and RORγt in Th17 cells. The associations of JMJD3 and STAT3, STAT3 and RORγt were enhanced after Pg-LPS stimulation. Total protein extracts were prepared from primary CD4+ T cells cultured for 5 days under Th17-polarizing conditions untreated or stimulated with Pg-LPS and then subjected to IP with anti-STAT3 antibody. The immunocomplexes were separated by SDS-PAGE and analyzed by western blot using indicated antibodies. Five percent of the total protein extracts were loaded as positive control (Input). Meanwhile, total protein extracts were immunoprecipitated with an antibody against rabbit IgG isotype as negative control (IgG). IP, immunoprecipitation; RORγt, retinoid-related orphan nuclear receptor γt; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
STAT3 regulates Th17-cell differentiation induced by Pg-LPS without affecting JMJD3
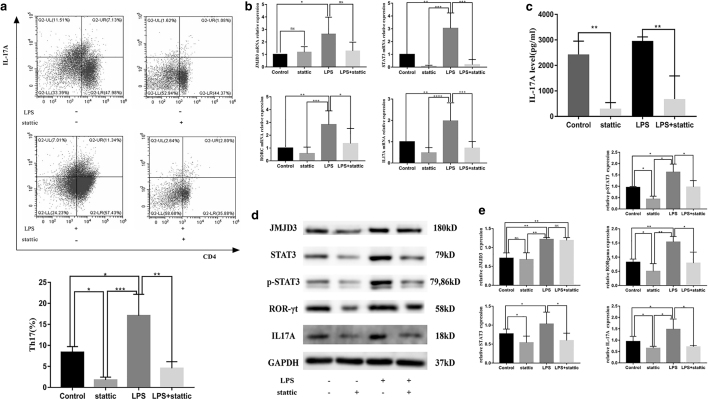
To determine whether STAT3 is involved in Pg-LPS-stimulated Th17-cell differentiation, a STAT3 blocking assay was performed. A concentration of 200 nM statttic was adopted in the blocking assay according to the CCK-8 assay (Supplementary Fig. S1).
The results showed that statttic could efficiently inhibit STAT3 expression at the mRNA and protein levels (p < 0.05) (Fig. 3b, d, e). Furthermore, the effects of STAT3 blockade on the proportion of Th17 cells differentiated from CD4+ T cells were investigated. The frequency of Th17 cells was significantly decreased after STAT3 blockade in both the control and LPS groups (p < 0.05) (Fig. 3a). The mRNA and protein expression of RORc and IL-17A were drastically decreased in the stattic-treated groups (p < 0.05) (Fig. 3b, d, e). The ELISA results also showed that stattic significantly reduced the concentration of supernatant IL-17A in both the control and LPS groups (p < 0.01) (Fig. 3c). However, there was no significant change in JMJD3 expression at either the mRNA or protein level (p > 0.05) (Fig. 3b, d, e), indicating that STAT3 can regulate Th17-cell differentiation triggered by Pg-LPS without affecting JMJD3.
FIG. 3.
STAT3 regulates Th17-cell differentiation induced by Pg-LPS without affecting JMJD3. Purified CD4+ T cells were treated with the STAT3 inhibitor stattic (200 nM), Pg-LPS (1 μg/mL), or stattic and Pg-LPS and cultured under the same conditions as the Th17 differentiation system. The cultures were further incubated for 5 days for RNA analysis, FACS analysis, western blotting, and ELISAs. (a) The frequency of Th17 cells was significantly decreased after STAT3 blockade in both the control and LPS groups. (b) The mRNA expression of Jmjd3, Stat3, Rorc, and Il-17a in the control, Stattic, LPS, and LPS+Stattic groups. (c) Stattic significantly reduced the concentration of supernatant IL-17A in both the control and LPS groups. (d, e) The protein levels of JMJD3, STAT3, RORγt, and IL-17A in the control, Stattic, LPS, and LPS+Stattic groups. Data are presented as the mean ± SEM of four independent experiments. Significant changes are marked with *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. ELISA, enzyme-linked immunosorbent assay; ns, no significant difference.
JMJD3 regulates Th17-cell differentiation under Pg-LPS through STAT3
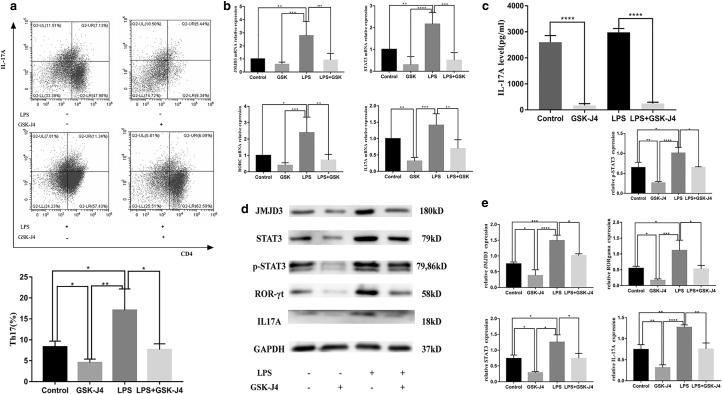
To further investigate the interaction between JMJD3 and STAT3 during Th17-cell differentiation by Pg-LPS stimulation, a JMJD3 blocking assay was also conducted. GSK-J4 (80 nM) was used in the blocking assay according to the CCK-8 assay (Supplementary Fig. S2). As shown in Figure 4a, LPS-induced Th17-cell differentiation was completely eliminated with additional GSK-J4 compared with the control group. This was further supported by quantitative real-time PCR and western blot analysis, which indicated that the levels of RORc and IL-17A were also profoundly reduced with GSK-J4 treatment (Fig. 4b, d, e).
FIG. 4.
JMJD3 regulates Th17-cell differentiation induced by Pg-LPS through STAT3. Purified CD4+ T cells were treated with the JMJD3 inhibitor GSK-J4 (80 nM), Pg-LPS (1 μg/mL), or GSK-J4 and Pg-LPS and cultured under the same conditions as the Th17 differentiation system. The cultures were further incubated for 5 days for RNA analysis, FACS analysis, western blotting, and ELISAs. (a) The frequency of Th17 cells was significantly decreased after JMJD3 blockade in both the control and LPS groups. (b) The mRNA expression of Jmjd3, Stat3, Rorc, and Il-17a in the control, GSK-J4, LPS, and LPS+ GSK-J4 groups. (c) GSK-J4 significantly reduced the concentration of supernatant IL-17A in both the control and LPS groups. (d, e) The protein levels of JMJD3, STAT3, RORγt, and IL-17A in the control, GSK-J4, LPS, and LPS+ GSK-J4 groups. Data are presented as the mean ± SEM of four independent experiments. Significant changes are marked with *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Consistently, cytokine levels in the supernatants as determined by ELISA showed that the amount of IL-17A production was much lower in Th17-polarizing conditions with GSK-J4 addition in both the control and LPS groups (Fig. 4c). Moreover, treatment with GSK-J4 dramatically suppressed the phosphorylated and total STAT3 levels, as shown at the protein and RNA levels (Fig. 4b, d, e). Thus, GSK-J4 is likely to inhibit Th17-cell differentiation by blocking the effect of LPS on JMJD3 and further inhibiting STAT3 expression. Taken together, our results demonstrate that JMJD3 regulates Th17-cell differentiation by affecting the expression and phosphorylation of STAT3.
Discussion
In addition to specific cytokines, the microbiota also plays important roles in inducing Th17-cell differentiation. Increasing evidence indicates that APCs, triggered by microbial infection, can lead to the differentiation of CD4+ T cells by presenting antigens and releasing proinflammatory cytokines (Boccasavia et al., 2021; Yin et al., 2021). In the periodontal microenvironment, P. gingivalis and Aggregatibacter actinomycetemcomitans have been reported to induce IL-17 production by activating CD14+ monocytes (Cheng et al., 2016). Generally, the recognition of inflammatory signals via pattern recognition receptors by APCs can activate the intracellular pathways involved in CD4+ T cell differentiation, especially TLRs (Mills, 2011; Li et al., 2018; Dias et al., 2019). LPS is widely recognized as an agonist of TLRs (Zeng et al., 2020; Ciesielska et al., 2021), which can induce the differentiation of Th17 cells (Wilson et al., 2009; McAleer et al., 2010; Park et al., 2015).
Interestingly, our previous study showed that Pg-LPS could promote Th17-cell differentiation directly by upregulating TLR2 expression on T cells in the absence of APC in vitro (Zhang et al., 2019a). Data from the present study further confirmed that under Th17-polarizing conditions, the expression of JMJD3 in CD4+ T cells was induced by Pg-LPS and further promoted Th17-cell differentiation by modulating the STAT3-RORγt signaling pathway. Taken together, periodontal pathogens can participate in the differentiation of Th17 cells through two different mechanisms. On the one hand, they induce the production of Th17 cells by providing antigens to APCs; on the other hand, they directly induce the production of Th17-related transcription factors, which leads to the differentiation of Th17 cells.
Moreover, it is well known that LPS can induce STAT3 activation (Chen et al., 2009). Upon the binding of LPS ligand to its receptor, intracellular activation of JAK2 is initiated, resulting in downstream STAT3 phosphorylation (Zeinalzadeh et al., 2021). After activation, phosphorylated STAT3 translocates into the nucleus (Pencik et al., 2016; Kitamura et al., 2017; Zheng et al., 2019) and acts as a transcription factor to promote the expression of downstream target genes, including Rorc and Il-17a (Heim, 1996; Yu et al., 2009; Johnson et al., 2018). ChIP-seq and RNA-seq studies revealed that STAT3 plays a critical role in the Th17 transcriptional program by binding to Th17-related gene loci (Durant et al., 2010). More recently, RORc was also reported to contain direct targets of STAT3 on Th17 cells (Sallusto, 2016; Tripathi et al., 2017; Wu et al., 2017; August, 2018). Consistent with previous studies, our work confirmed that STAT3 could bind to RORγt directly under Th17-polarizing conditions, showing the critical role of STAT3 in Th17-cell differentiation.
Generally, the expression of JMJD3 is low under normal conditions, but a variety of cellular stresses can induce the expression of JMJD3 (Zhang et al., 2019b). De Santa et al. (2009) reported that JMJD3 can be induced upon LPS stimulation, mediating inflammation-related genes in peripheral macrophages. Here, we confirmed that Pg-LPS induced JMJD3 expression along with elevated expression of STAT3, p-STAT3, and RORγt, and further enhanced their interactions, suggesting STAT3 and RORγt might be the targets of JMJD3.
A previous study has demonstrated that JMJD3 is involved in H3K27me3 modification at Rorc and Th17-related genes (Liu et al., 2015). The study of LaMere et al. (2017) confirmed that STAT3 promoter was evidently demethylated after CD4+ T cell activation. However, further ChIP-seq found no JMJD3 peak surrounding the STAT3 locus, and Hi-C did not reveal any distal JMJD3 sites interacting with the promoter. Two JMJD3 peaks surrounding the JAK2 locus were identified, indicating that JMJD3 may activate the phosphorylation of STAT3 by regulating the expression of JAK2 during CD4+ T cell differentiation, which further proved the complicated interaction between JMJD3 and STAT3 (LaMere et al., 2017).
An increasing number of studies have suggested that JMJD3 can interact with coactivators and activate the transcription of target genes as a transcription factor independent of its demethylase activity (Salminen et al., 2014; Zhang et al., 2019b; Ding et al., 2021). Actually, JMJD3 plays an important role in modulating cell-specific pro-inflammatory and anti-inflammatory immune responses, which may also be mediated by H3K27-independent mechanisms (Burchfield et al., 2015). Since there is still a lack of direct evidence to support the role of JMJD3 in regulating the expression of STAT3 by altering the enrichment of H3K27me3 at promoter, the H3K27 demethylation-independent JMJD3-mediated STAT3 gene activation in Th17-cell differentiation process cannot be excluded, therefore further research is required.
It has been reported that the regulation of JMJD3 is highly gene- and context-specific (Burchfield et al., 2015). STAT3 has been proven to bind to the Jmjd3 promoter in human glioma stem cells (Sherry-Lynes et al., 2017), which is consistent with published genomic data from murine embryonic stem cells (Kidder et al., 2008). The work of Przanowski et al. (2014) also indicates that JMJD3 cooperates with STAT1 and STAT3 and acts as their novel target to drive the expression of inflammatory genes in microglia. However, in the present study, JMJD3 and STAT3 blocking assays indicated that JMJD3 had a regulatory effect on STAT3 in the Th17-cell differentiation process with Pg-LPS stimulation, but STAT3 had no regulatory effect on JMJD3.
Consistent with our results, LaMere et al. (2017) reported that the expression of STAT3 and its phosphorylation in GSK-J4-treated cells or in JMJD3 knockdown cells were both impaired during CD4+ T cell activation. Therefore, different cell sources and disease states may be important reasons for the discrete function of STAT3 on JMJD3 in different experimental settings. Thus, further studies are required to elucidate the exact mechanism by which JMJD3 regulates STAT3 in the context of Th17 cells.
Conclusion
In this study, we demonstrated that JMJD3 was induced in CD4+ T cells stimulated with Pg-LPS under Th17-cell-polarizing conditions in vitro and further mediated STAT3 to enhance the expression of the key transcription factor RORγt, resulting in promoted Th17-cell differentiation. Nevertheless, the detailed mechanism still needs to be further investigated, as the effect of JMJD3 on STAT3 is complicated and not completely understood.
Supplementary Material
Authors' Contributions
L.G. and C.Z. conceptualized and designed the original idea. D.H. performed the experiments, acquired the figures, and revised the first draft. D.H., C.Z., and P.W. performed the literature search and analysis and finished the draft. X.L., L.G., and C.Z. provided valuable feedback and approved the final draft.
Authorship Confirmation Statement
All authors have read and approved the final article, and agree to submit it for consideration for publication in your journal.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon reasonable request.
Disclosure Statement
No competing financial interests exist.
Funding Information
This study was supported by National Natural Science Foundation of China (Grant Nos. 81870770, 82170959).
Supplementary Material
References
- Adibrad, M., Deyhimi, P., Ganjalikhani Hakemi, M., Behfarnia, P., Shahabuei, M., and Rafiee, L. (2012). Signs of the presence of Th17 cells in chronic periodontal disease. J Periodontal Res 47, 525–531. [DOI] [PubMed] [Google Scholar]
- August, A. (2018). Who regulates whom: ZNF341 is an additional player in the STAT3/T(H)17 song. Sci Immunol 3, eaat9779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccasavia, V.L., Bovolenta, E.R., Villanueva, A., Borroto, A., Oeste, C.L., van Santen, H.M., et al. (2021). Antigen presentation between T cells drives Th17 polarization under conditions of limiting antigen. Cell Rep 34, 108861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchfield, J.S., Li, Q., Wang, H.Y., and Wang, R.F. (2015). JMJD3 as an epigenetic regulator in development and disease. Int J Biochem Cell Biol 67, 148–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candelli, M., Franza, L., Pignataro, G., Ojetti, V., Covino, M., Piccioni, A., et al. (2021). Interaction between lipopolysaccharide and gut microbiota in inflammatory bowel diseases. Int J Mol Sci 22, 6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cekici, A., Kantarci, A., Hasturk, H., and Van Dyke, T.E. (2014). Inflammatory and immune pathways in the pathogenesis of periodontal disease. Periodontol 2000 64, 57–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L.P., Cai, M., Zhang, Q.H., Li, Z.L., Qian, Y.Y., Bai, H.W., et al. (2009). Activation of interleukin-6/STAT3 in rat cholangiocyte proliferation induced by lipopolysaccharide. Dig Dis Sci 54, 547–554. [DOI] [PubMed] [Google Scholar]
- Cheng, W.C., van Asten, S.D., Burns, L.A., Evans, H.G., Walter, G.J., Hashim, A., et al. (2016). Periodontitis-associated pathogens P. gingivalis and A. actinomycetemcomitans activate human CD14(+) monocytes leading to enhanced Th17/IL-17 responses. Eur J Immunol 46, 2211–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciesielska, A., Matyjek, M., and Kwiatkowska, K. (2021). TLR4 and CD14 trafficking and its influence on LPS-induced pro-inflammatory signaling. Cell Mol Life Sci 78, 1233–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofani, M., Madar, A., Galan, C., Sellars, M., Mace, K., Pauli, F., et al. (2012). A validated regulatory network for Th17 cell specification. Cell 151, 289–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa, F., Narang, V., Yap, Z.H., Tusi, B.K., Burgold, T., Austenaa, L., et al. (2009). Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. EMBO J 28, 3341–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias, A.S.O., Sacramento, P.M., Lopes, L.M., Sales, M.C., Castro, C., Araújo, A., et al. (2019). TLR-2 and TLR-4 agonists favor expansion of CD4(+) T cell subsets implicated in the severity of neuromyelitis optica spectrum disorders. Mult Scler Relat Disord 34, 66–76. [DOI] [PubMed] [Google Scholar]
- Ding, Y., Yao, Y., Gong, X., Zhuo, Q., Chen, J., Tian, M., et al. (2021). JMJD3: a critical epigenetic regulator in stem cell fate. Cell Commun Signal 19, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durant, L., Watford, W.T., Ramos, H.L., Laurence, A., Vahedi, G., Wei, L., et al. (2010). Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity 32, 605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskan, M.A., Jotwani, R., Abe, T., Chmelar, J., Lim, J.H., Liang, S., et al. (2012). The leukocyte integrin antagonist Del-1 inhibits IL-17-mediated inflammatory bone loss. Nat Immunol 13, 465–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim, M.H. (1996). The Jak-STAT pathway: specific signal transduction from the cell membrane to the nucleus. Eur J Clin Invest 26, 1–12. [DOI] [PubMed] [Google Scholar]
- Heinbockel, L., Weindl, G., Martinez-de-Tejada, G., Correa, W., Sanchez-Gomez, S., Bárcena-Varela, S., et al. (2018). Inhibition of lipopolysaccharide- and lipoprotein-induced inflammation by antitoxin peptide Pep19-2.5. Front Immunol 9, 1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich, P.C., Behrmann, I., Müller-Newen, G., Schaper, F., and Graeve L. (1998). Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem J 334, 297–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov, II., McKenzie, B.S., Zhou, L., Tadokoro, C.E., Lepelley, A., Lafaille, J.J., et al. (2006). The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126, 1121–1133. [DOI] [PubMed] [Google Scholar]
- Johnson, D.E., O'Keefe, R.A., and Grandis, J.R. (2018). Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol 15, 234–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidder, B.L., Yang, J., and Palmer, S. (2008). Stat3 and c-Myc genome-wide promoter occupancy in embryonic stem cells. PLoS One 3, e3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura, H., Ohno, Y., Toyoshima, Y., Ohtake, J., Homma, S., Kawamura, H., et al. (2017). Interleukin-6/STAT3 signaling as a promising target to improve the efficacy of cancer immunotherapy. Cancer Sci 108, 1947–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Könönen, E., Gursoy, M., and Gursoy, U.K. (2019). Periodontitis: a multifaceted disease of tooth-supporting tissues. J Clin Med 8, 1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMere, S.A., Thompson, R.C., Meng, X., Komori, H.K., Mark, A., and Salomon, D.R. (2017). H3K27 Methylation dynamics during CD4 T cell activation: regulation of JAK/STAT and IL12RB2 expression by JMJD3. J Immunol 199, 3158–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, R., Wang, J., Zhu, F., Li, R., Liu, B., Xu, W., et al. (2018). HMGB1 regulates T helper 2 and T helper17 cell differentiation both directly and indirectly in asthmatic mice. Mol Immunol 97, 45–55. [DOI] [PubMed] [Google Scholar]
- Liu, Z., Cao, W., Xu, L., Chen, X., Zhan, Y., Yang, Q., et al. (2015). The histone H3 lysine-27 demethylase Jmjd3 plays a critical role in specific regulation of Th17 cell differentiation. J Mol Cell Biol 7, 505–516. [DOI] [PubMed] [Google Scholar]
- Lunar Silva, I., and Cascales, E. (2021). Molecular strategies underlying Porphyromonas gingivalis virulence. J Mol Biol 433, 166836. [DOI] [PubMed] [Google Scholar]
- Lyu, J.H., Huang, B., Park, D.W., and Baek, S.H. (2016). Regulation of PHLDA1 expression by JAK2-ERK1/2-STAT3 signaling pathway. J Cell Biochem 117, 483–490. [DOI] [PubMed] [Google Scholar]
- McAleer, J.P., Liu, B., Li, Z., Ngoi, S.M., Dai, J., Oft, M., et al. (2010). Potent intestinal Th17 priming through peripheral lipopolysaccharide-based immunization. J Leukoc Biol 88, 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills, K.H. (2011). TLR-dependent T cell activation in autoimmunity. Nat Rev Immunol 11, 807–822. [DOI] [PubMed] [Google Scholar]
- Moutsopoulos, N.M., Konkel, J., Sarmadi, M., Eskan, M.A., Wild, T., Dutzan, N., et al. (2014). Defective neutrophil recruitment in leukocyte adhesion deficiency type I disease causes local IL-17-driven inflammatory bone loss. Sci Transl Med 6, 229ra240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, D.W., Lyu, J.H., Kim, J.S., Chin, H., Bae, Y.S., and Baek, S.H. (2013). Role of JAK2-STAT3 in TLR2-mediated tissue factor expression. J Cell Biochem 114, 1315–1321. [DOI] [PubMed] [Google Scholar]
- Park, J.H., Jeong, S.Y., Choi, A.J., and Kim, S.J. (2015). Lipopolysaccharide directly stimulates Th17 differentiation in vitro modulating phosphorylation of RelB and NF-κB1. Immunol Lett 165, 10–19. [DOI] [PubMed] [Google Scholar]
- Pencik, J., Pham, H.T., Schmoellerl, J., Javaheri, T., Schlederer, M., Culig, Z., et al. (2016). JAK-STAT signaling in cancer: from cytokines to non-coding genome. Cytokine 87, 26–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przanowski, P., Dabrowski, M., Ellert-Miklaszewska, A., Kloss, M., Mieczkowski, J., Kaza, B., et al. (2014). The signal transducers Stat1 and Stat3 and their novel target Jmjd3 drive the expression of inflammatory genes in microglia. J Mol Med (Berl) 92, 239–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ, B.E., Olshanksy, M., Smallwood, H.S., Li, J., Denton, A.E., Prier, J.E., et al. (2014). Distinct epigenetic signatures delineate transcriptional programs during virus-specific CD8(+) T cell differentiation. Immunity 41, 853–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto, F. (2016). Heterogeneity of human CD4(+) T cells against microbes. Annu Rev Immunol 34, 317–334. [DOI] [PubMed] [Google Scholar]
- Salminen, A., Kaarniranta, K., Hiltunen, M., and Kauppinen, A. (2014). Histone demethylase Jumonji D3 (JMJD3/KDM6B) at the nexus of epigenetic regulation of inflammation and the aging process. J Mol Med (Berl) 92, 1035–1043. [DOI] [PubMed] [Google Scholar]
- Sherry-Lynes, M.M., Sengupta, S., Kulkarni, S., and Cochran, B.H. (2017). Regulation of the JMJD3 (KDM6B) histone demethylase in glioblastoma stem cells by STAT3. PLoS One 12, e0174775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi, S.K., Chen, Z., Larjo, A., Kanduri, K., Nousiainen, K., Äijo, T., et al. (2017). Genome-wide analysis of STAT3-mediated transcription during early human Th17 cell differentiation. Cell Rep 19, 1888–1901. [DOI] [PubMed] [Google Scholar]
- Villarino, A.V., Kanno, Y., Ferdinand, J.R., and O'Shea, J.J. (2015). Mechanisms of Jak/STAT signaling in immunity and disease. J Immunol 194, 21–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Zhang, Y., Yang, X.O., Nurieva, R.I., Chang, S.H., Ojeda, S.S., et al. (2012). Transcription of Il17 and Il17f is controlled by conserved noncoding sequence 2. Immunity 36, 23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, G., Wei, L., Zhu, J., Zang, C., Hu-Li, J., Yao, Z., et al. (2009). Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 30, 155–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, R.H., Whitehead, G.S., Nakano, H., Free, M.E., Kolls, J.K., and Cook, D.N. (2009). Allergic sensitization through the airway primes Th17-dependent neutrophilia and airway hyperresponsiveness. Am J Respir Crit Care Med 180, 720–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X., Shou, Q., Chen, C., Cai, H., Zhang, J., Tang, S., et al. (2017). An herbal formula attenuates collagen-induced arthritis via inhibition of JAK2-STAT3 signaling and regulation of Th17 cells in mice. Oncotarget 8, 44242–44254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J., Zhao, Y., and Shao, F. (2015). Non-canonical activation of inflammatory caspases by cytosolic LPS in innate immunity. Curr Opin Immunol 32, 78–83. [DOI] [PubMed] [Google Scholar]
- Yin, X., Chen, S., and Eisenbarth, S.C. (2021). Dendritic cell regulation of T helper cells. Annu Rev Immunol 39, 759–790. [DOI] [PubMed] [Google Scholar]
- Yu, H., Pardoll, D., and Jove, R. (2009). STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer 9, 798–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeinalzadeh, E., Valerievich Yumashev, A., Rahman, H.S., Marofi, F., Shomali, N., Kafil, H.S., et al. (2021). The role of Janus Kinase/STAT3 pathway in hematologic malignancies with an emphasis on epigenetics. Front Genet 12, 703883. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zeng, X.Z., Zhang, Y.Y., Yang, Q., Wang, S., Zou, B.H., Tan, Y.H., et al. (2020). Artesunate attenuates LPS-induced osteoclastogenesis by suppressing TLR4/TRAF6 and PLCγ1-Ca(2+)-NFATc1 signaling pathway. Acta Pharmacol Sin 41, 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C., Xu, C., Gao, L., Li, X., and Zhao, C. (2021). Porphyromonas gingivalis lipopolysaccharide promotes T-hel per17 cell differentiation by upregulating Delta-like ligand 4 expression on CD14(+) monocytes. PeerJ 9, e11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J.A., Mortazavi, A., Williams, B.A., Wold, B.J., and Rothenberg, E.V. (2012). Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell 149, 467–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Gao, L., Xu, C., Li, X., Wang, P., Zhang, C., et al. (2019a). Porphyromonas gingivalis lipopolysaccharide promotes T- helper 17 cell differentiation from human CD4(+) naive T cells via toll-like receptor-2 in vitro. Arch Oral Biol 107, 104483. [DOI] [PubMed] [Google Scholar]
- Zhang, X., Liu, L., Yuan, X., Wei, Y., and Wei, X. (2019b). JMJD3 in the regulation of human diseases. Protein Cell 10, 864–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, M., Tan, Y., Peng, Q., Huang, C., Guo, Y., Liang, G., et al. (2018). IL-6/STAT3 pathway induced deficiency of RFX1 contributes to Th17-dependent autoimmune diseases via epigenetic regulation. Nat Commun 9, 583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Y., Ming, P., Zhu, C., Si, Y., Xu, S., Chen, A., et al. (2019). Hepatitis B virus X protein-induced SH2 domain-containing 5 (SH2D5) expression promotes hepatoma cell growth via an SH2D5-transketolase interaction. J Biol Chem 294, 4815–4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon reasonable request.