Abstract
Inherited retinal dystrophies are caused by mutations in more than 250 genes, each of them carrying several types of mutations that can lead to different clinical phenotypes. Mutations in Retinitis Pigmentosa GTPase-Regulator (RPGR) cause X-linked Retinitis pigmentosa (RP). A nucleotide substitution in intron 9 of RPGR causes the increase of an alternatively spliced isoform of the mature mRNA, bearing exon 9a (E9a). This introduces a stop codon, leading to truncation of the protein. Aiming at restoring impaired gene expression, we developed an antisense RNA-based therapeutic approach for the skipping of RPGR E9a. We designed a set of specific U1 antisense snRNAs (U1_asRNAs) and tested their efficacy in vitro, upon transient cotransfection with RPGR minigene reporter systems in HEK-293T, 661W, and PC-12 cell lines.
We thus identified three chimeric U1_asRNAs that efficiently mediate E9a skipping, correcting the genetic defect. Unexpectedly, the U1–5′antisense construct, which exhibited the highest exon-skipping efficiency in PC-12 cells, induced E9a inclusion in HEK-293T and 661W cells, indicating caution in the choice of preclinical model systems when testing RNA splicing-correcting therapies. Our data provide a proof of principle for the application of U1_snRNA exon skipping-based approach to correct splicing defects in RPGR.
Keywords: alternative splicing, U1 snRNA, exon skipping, Retinitis Pigmentosa GTPase-regulator (RPGR), RNA Therapeutics
Introduction
In the past several decades, genetic studies have significantly advanced our understanding of inherited retinal dystrophies (IRDs).
Retinitis pigmentosa (RP, MIM# 268000) is the most common form of IRD with a prevalence of ∼1 in 3,000 individuals [1]. Although clinical symptoms of RP may be similar among patients, the genetic pattern of inheritance is complex. Indeed, RP is a heterogeneous disease associated with sequence variants in more than 71 genes, resulting in different forms of inheritance: autosomal dominant RP (23 genes), autosomal recessive (43 genes), and X-linked RP (XLRP) (5 genes) [2] (RetNet, see https://sph.uth.edu/RetNet/sum-dis.htm#A-genes).
Mutations in the Retinitis Pigmentosa GTPase Regulator gene (RPGR; MIM# 312610) are responsible for 70%–80% of XLRP cases [3–6]. RPGR is located on chromosomal region Xp21.1 and spans about 172 kb. RPGR mRNA is found in at least 12 alternatively spliced isoforms and is widely expressed in various tissues (eg, kidney, brain, retina, lung, and testis), as is its protein.
RPGR is essential for the survival of retinal cells, and in humans it is expressed by both rod and cone photoreceptors [7]. Outside the retina, RPGR has also been detected at the transition zone of primary and motile cilia of airway epithelial cells and centrosomes/basal bodies of cultured cells [7,8]. Several studies have contributed to defining the ciliary localization of RPGR and its interacting proteins in the retina [9]. Some of the alternatively spliced isoforms are retina specific [10]. Indeed, the two major subsets of transcripts identified in the retina contain exon ORF15 (RPGRORF15) or exon 19 (RPGR1–19) [11,12]. Nevertheless, the mechanism of action of RPGR in photoreceptors is still not well understood. Splicing of RPGR is precisely regulated in a tissue-dependent fashion and mutations in RPGR frequently interfere with the expression of alternative transcript isoforms [13–15].
In the retina, an RPGR splicing isoform has been described to include pseudoexon 9a (E9a). This exon, 136 bases long, is 418 base pairs (bp) downstream of the 5′ splice site of intron 9. Moreover, a c.1059 + 363G>A nucleotide substitution 55 bp upstream of E9a was reported in a patient with a mild RP phenotype [15]. This substitution affects E9a recognition by the splicing machinery, increasing, by a factor of ∼3.5, the levels of E9a-containing RPGR transcripts in cone photoreceptors [15]. The presence of E9a in the mature mRNA results in a premature stop codon and the truncation of the protein. Moreover, E9a RPGR has a peculiar expression pattern, being mainly present in cone photoreceptors rather than in rods. Specifically, this isoform is localized in the inner segment of cones, whereas RPGR is normally expressed in the connective cilium [15].
E9a belongs to a class of alternative exons naturally occurring in several genes, named “poison exons” that contain a premature termination codon [16]. Alternative splicing of poison exons is predicted to lead to nonsense-mediated decay (NMD). Poison exons are proposed to play an important role in tissue-specific gene expression regulation and in development. Mutations in the sequences responsible for the splicing of these exons often result in diseases, by altering a finely tuned regulatory process [16].
Nothing is known about the specific regulatory role of poison exon E9a in RPGR. However, we performed a comparative analysis of RPGR gene sequence across animal species. By a bioinformatic analysis of reference genomes, we could determine that, despite the conservation of the general exon–intron architecture of the RPGR gene and of the size of its exons, E9a is only present in human and chimpanzee, and highly conserved (data not shown). Therefore, its putative regulatory role must have evolved recently in primates.
Both human proteins, RPGRORF15 and RPGREx1–19, are involved in cilia regulation signaling pathways, but their role is unclear.
The RPGRORF15 is a protein with 1,152 amino acids and has a Glu-Gly-rich region in the C-terminal domain, while the protein RPGREx1–19 contains 815 amino acids with an isoprenylation motif at the C-terminus. Both isoforms share exon 1–14 and this suggests that they have some common function. Indeed, RPGR isoforms may compete with each other for the availability of endogenous binding partners. Therefore, maintaining the optimal RPGREx1–19/RPGRORF15 ratio has a crucial role in optimal cilia growth and ciliary trafficking regulation [16]. However, as reported in the literature by Moreno-Leon et al., [17,18], both isoforms, RPGREx1–19 and RPGRORF15, are regulated by independent mechanisms and both of them have different functional properties, different cellular and tissue localization, and different levels of expression during retinal development and maturation [18].
AAV-based delivery of RPGRORF15 is in advanced clinical development: a clinical trial investigated a gene therapy approach for X-linked RP, through an AAV8 vector-delivered codon-optimized human RPGRORF15 coding sequence (CDS). The initial results of this study were successful and showed an amelioration of the patients' visual field [19]. However, as mutations in RPGR may impact several RPGR splicing isoforms at the same time, future gene therapy strategies will need to implement ways to restore all RPGR isoforms, in the correct relative amounts. In this regard, universal gene addition or cell-based therapies, highly promising techniques [20–22], may not be the optimal choice to treat RPGR mutations.
We propose, in this study, an exon-skipping strategy, which, besides taking advantage of the endogenous processes guaranteeing a balance among the different RPGR isoforms, would have the additional advantage of being functional only in the cell types in which the target pre-mRNA is expressed.
Intensive research in the field of splicing modulation has led to the development of successful exon-skipping approaches by engineered U1 small nuclear RNA (U1 snRNA) able to mask the mutated sites, thus modulating the aberrant alternative splicing [23,24]. U1 snRNA recognizes the 5′ splice site and mediates the first step in spliceosome assembly [25].
Several examples of successful applications of U1 snRNA in therapeutic exon skipping are described in the literature [26–29]. U1 snRNA has been shown to be a useful vector for the stable expression of antisense molecules [23,29]. The U1 snRNA expression cassettes are small (about 600 bp) and they work efficiently both in in vitro and in vivo systems and can be delivered as part of lentiviral [30] or AAV vectors [26,28,29].
In this study, we aimed at developing an exon-skipping U1 antisense snRNA (U1_asRNA) to restore the levels of E9a in RPGR bearing the c.1059 + 363G>A deep intronic nucleotide substitution. We designed two single U1_asRNA molecules, U1_3′ and U1_5′, and a double-target U1_3′5′ carrying two distinct antisense sequences targeted against two splicing-regulating regions. This latter strategy has already been used to increase exon skipping efficiency [26–28]. We tested whether the three chimeric constructs were able to modulate E9a skipping in HEK-293T, 661W, and PC-12 cell lines, by cotransfecting them with a RPGR minigene reporter system, recapitulating RPGR mRNA expression observed in patients (MINI mut) and unaffected control individuals (MINI wt).
Our results demonstrate that a new therapeutic strategy based on U1_snRNA molecules can efficiently be used to correct the splicing of RPGR transcripts. Additionally, the finding that U1_5′asRNA induces E9a skipping in PC-12 cells and inclusion in HEK-293T and 661W cells, calls for caution in the choice of the preclinical model systems used to test U1_asRNAs.
Materials and Methods
The experiments described do not require IRB approval.
Computational analysis of RPGR exon 9a (E9a) splicing
Splice sites were explored by analyzing the RPGR genomic sequences from exon 9 to 10 with the NNSPLICE 0.9 program [31] (https://omictools.com/nnsplice-tool). The resulting scores for the different splice sites were reported (Table 1). The branch point analysis was conducted on intron 9 using the Human Splicing Finder (www.umd.be/HSF/) [32]. The binding of splicing regulatory proteins was investigated by using ESEFinder 2.0 (http://krainer01.cshl.edu/tools/ESE2/) [33] and SpliceAid site predictions [34] (www.introni.it/splicing.html).
Table 1.
Splice Site Scores of Retinitis Pigmentosa GTPase-Regulator Sequence from Exon 9 to 10.
| Splice site | Score | Sequence 5′- 3′ |
|---|---|---|
| Acceptor E9 (3′ splice site) | 0.97 | ttcattatttttgcattttAGatatcggccttatgtatact |
| Donor E9 (5′ splice site) | 0.99 | taaattgGTaagggc |
| Acceptor E9a (3′ splice site) | 0.00 | ataaacaaagcgttttggcAGggcacggtggctcactcctg |
| Donor E9a (5′ splice site) | 0.93 | taagccagGTatgatg |
| Acceptor E10 (3′ splice site) | 0.83 | ttatgtggatttatgctgcAGgttgcttgtggtggatgtca |
| Donor E10 (5′ splice site) | 0.19 | agagaggGTacaatt |
According to NNSPLICE 0.9, higher scores indicate stronger splice sites. A perfect match to the splice site consensus sequence has score 1.
Construction of RPGR reporter minigenes
To generate minigene constructs, we cloned genomic sequences spanning a region from intron 8 to 10 of the RPGR gene into the mammalian expression vector pcDNA3 (Life Technologies, Carlsbad, CA). RPGR wild-type minigene construct was generated (Fig. 2A) by amplification from a pool of human female genomic DNAs (Promega, Milan, Italy) using the following primers:
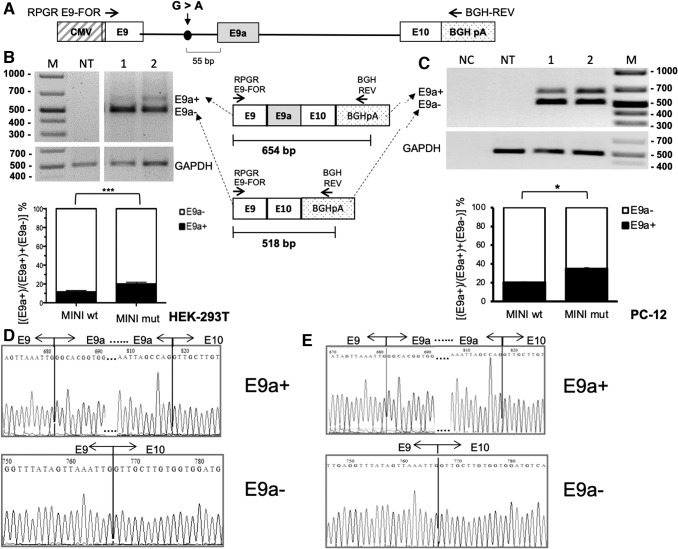
FIG. 2.
Setting up a minigene reporter system of RPGR E9–E10 splicing and E9a inclusion-inducing mutations. (A) Schematic representation of the RPGR minigene construct (not in scale). The position of the mutation is indicated by a dot, while horizontal arrows represent primers for RT-PCR analysis. The endogenous RPGR mRNA is not amplified by this primer pair, as the reverse primer anneals to the reporter-specific portion of the RNA transcribed from the BGH polyA cassette. The alternative splicing of exon 9a leads to the production of two different splice isoforms (E9a+ and E9a−). As depicted in the schematic diagram below, the presence of E9a in the reporter RPGR transcript yields a 528-bp-long amplicon (E9a+), while the amplification of the reporter RPGR transcript devoid of E9a produces a 392-bp-long amplicon (E9a−). (B) RT-PCR analysis of RPGR mRNA levels on RNA from HEK-293T cells transfected with wild-type minigene (MINI wt, lane 1) and mutant minigene (MINI mut, lane 2). The analysis has been performed in triplicate, and one representative gel is shown. The histogram represents the densitometric analysis of the bands relative to the two different isoforms (E9a+ and E9a−) normalized to GAPDH (P < 0.001). Data are shown as mean ± SD (n = 3). (c) RT-PCR on RNA from transfected PC-12 cells. Lane 1: wild-type minigene (MINI wt), Lane 2: mutant minigene (MINI mut). The histogram represents the densitometric analysis of the bands relative to the two different isoforms (E9a+ and E9a−) normalized to GAPDH (P < 0.05). Data are shown as mean ± SD (n = 3). (D) and (E) E9a+ and E9a− bands from lanes 2, of the gels in (B) and (C), respectively, were eluted and sequenced. Chromatograms show the correct fusion between exons 9 and 9a, and exons 9a and 10, in the E9a+ band, and exons 9 and 10 in the E9a− band.
RPGR-For: 5′-ctaggtacccacagagaccatagagagtg-3′ and
RPGR-Rev: 5′-ctactcgagaagtttgttagcactcaactctaa-3′
PCR amplification was performed in a final volume of 50 μL, using the Cloned Pfu DNA polymerase (Agilent Technology, Santa Clara, CA), according to the manufacturer's protocol.
The PCR products and the pcDNA3 vector (Life Technologies) were cut with endonucleases KpnI and XhoI (New England Biolabs) and purified using the QiAquick Gel Extraction Kit according to the manufacturer's protocol (Qiagen, Hilden, Germany). The digested fragments were cloned into the pcDNA3 vector using T4 DNA ligase (New England Biolabs), according to the manufacturer's protocol.
The generated RPGR minigene wild-type (MINI wt) was used as the template for mutant RPGR minigene production (MINI mut). To introduce the c.1059 + 363G >A mutation, we used the Quick-change II XL Site-directed Mutagenesis Kit (Agilent Technologies), according to the manufacturer's protocol. The PCR amplification was performed in a final volume of 50 μL in a reaction mixture containing the RPGR wild-type minigene, as template, and the following specific primers: mut_RPGR-For: 5′-gctgaattaaatgttaaactctcaaatcctgcacaacag-3′ and mut_RPGR-Rev: 5′-ctgttgtgcaggatttgagagtttaacatttaattcagc-3′. The sequence of both wild-type and mutant minigenes and the exact point mutation position into the RPGR mutant minigene were verified by sequencing (BMR Genomics, Padova, Italy).
Cloning of U1 expression constructs
Four chimeric U1_snRNA constructs were obtained by ligation of two different inverse polymerase chain reaction (PCR) products. The first containing the U1 promoter, and the second containing the U1 sequence plus the chimeric sequence, as described in Denti et al. [35]. To generate the U1_5′, U1_3′ and U1_Scramble constructs, a first PCR was performed on the human U1 snRNA gene using the forward primer U1cas-up For: 5′-ctagctagcggtaaggaccagcttctttg-3′ and three different reverse primers: U1_5′RPGR Rev: 5′-caaaaattagccaggtatgatggcatgagatcttgggcctctgc-3′, U1_3′RPGR Rev: 5′-cgttttggcagggcacggtggatgagatcttgggcctctgc-3′, or U1_Scramble RPGR Rev: 5′-tcaattattccgcgagacgcagcatgagatcttgggcctctgc-3′.
All PCR assays were carried out in 50 μL final volume in a reaction mixture containing 1X of Cloned Pfu DNA polymerase (Agilent Technology), according to the manufacturer's protocol. Each of the three PCR fragments produced was then ligated with a PCR product from the human U1 snRNA gene using the U1-univ For primer: 5′-ggcaggggagataccatgatc-3′ and U1Cas-down Rev primer: 5′-ctagctagcggttagcgtacagtctac-3′.
To generate the U1_5′3 ′ constructs, a PCR product, obtained using primers U1Cas-up For and U1 5′RPGR Rev, was ligated with another PCR product, generated using primers U1 3′RPGR-II For: 5′-ccaccgtgccctgccaaaacgggcaggggagataccatgatc-3′ and U1Cas-down Rev. Ligation was performed in a final volume of 20 μL by incubation at 16°C for 3 h, according to the manufacturer's protocol (T4 DNA ligase; New England Biolabs, NEB, Ipswich, MA).
All the ligation products were subsequently amplified by PCR using U1cas-up ForNheI and U1cas-down RevNheI primers. The PCR reaction was performed according to the manufacturer's protocol of Cloned Pfu DNA polymerase (Agilent Technology).
After amplification and purification, the resulting fragments and 10 μg of the pAAV-2.1CMVeGFP vector were digested with NheI restriction enzyme (New England Biolabs, NEB) in 50 μL final volume. The PCR products were then cloned in the forward orientation into a pAAV-2.1CMVeGFP plasmid at the NheI restriction site upstream of the CMV promoter [36]. Ligation was performed as described above, using T4 DNA ligase, (New England Biolabs, NEB). We confirmed the exact sequence and the orientation of the inserts within the pAAV-2.1CMVeGFP vector by automated DNA sequencing (BMR Genomics), using the AAV Rev primer (5′-ccatatatgggctatgaataatg-3′).
Binding energy (ΔG°37) prediction
RNAfold algorithms were used to predict secondary structures and the minimum free energy (ΔG°37, kcal/mol) for the RPGR pre-mRNA [37,38] (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi). The DuplexFold, from the package RNAstructure 3.5 [39] (http://rna.urmc.rochester.edu/RNAstructureWeb/), was used to calculate the binding energy (ΔG°37overall, kcal/mol) for each RPGR mRNA–Chimeric RNA interaction.
Cell culture and transfection conditions
Wild-type and mutant RPGR minigenes (MINI wt, MINI mut) and chimeric U1_snRNA constructs (U1_3′, U1_5′, U1_3′5 ′, and U1_Scramble) were transiently cotransfected into Human embryonic kidney (HEK-293T), rat Pheochromocytoma (PC-12), and mouse retinal tumor (661W) (ATCC, American Type Culture Collection, Manassas, VA) cell lines. HEK-293T and 661W cells were maintained in Dulbecco's modified Eagle's medium (DMEM) with red phenol supplemented with 10% fetal bovine serum (FBS), 2 mM Glutamine, 100 U/μL Penicillin/Streptomycin, and were grown in a humidified incubator at 37°C and 5% CO2. Lipofectamine 2000 (Life-Technologies) was used for cotransfection with RPGR minigenes and chimeric U1 snRNAs, according to the manufacturer's instructions.
Rat PC-12 cells (ATCC entry CRL-1721) were grown at 37°C (5% CO2) in supplemented DMEM with 4.5% glucose (Lonza, Visp, Switzerland) supplemented with 10% FBS (Gibco, Grand Island, NY), 5% horse serum (Gibco), 1 mM glutamine (Gibco), and 1 mM Penicillin/Streptomycin (Gibco). Cells were seeded in T-75 cm2 flasks (Corning, NY) coated with 50 ng/mL poly-D-lysine hydrobromide (Sigma-Aldrich, St. Louis, MO), to achieve 70% confluence.
The same amount of each U1_asRNA plasmids (200 ng) was used either alone (U1_3′, U1_5′, U1_3′5 ′ or U1_Scramble) or in combination [U1_3′ (100 ng) + U1_5′ (100 ng)]. For each cotransfection experiment, the same amount of MINI mut or MINI wt was used (400 ng).
Transfection of PC-12 cells with RPGR minigenes and chimeric U1 snRNAs was performed using the Neon-Transfection System MPK5000 (Life-Technologies) under the following conditions: three 10 ms long pulses each one of 1,500 V as described in Covello et al. [40]. HEK-293T (4.0 × 104 cells/well) and PC-12 (3.0 × 105 cells/well) cells were plated onto 24-multiwell plates. An amount of 400 ng of each RPGR minigenes (MINI wt or MINI mut) plus 200 ng of each different chimeric U1 snRNAs was used for cotransfection of both HEK-293T and PC-12 cells unless specifically mentioned. After 48 h of cotransfection, cells were trypsinized and subsequently washed once with phosphate-buffered saline (PBS). Pellets were collected and stored at −80°C for RNA analysis.
RNA extraction and semiquantitative RT-PCR analyses
Total RNA was extracted from HEK-293T, PC-12, and 661W cells, using TRIzol Reagent (Life Technologies) according to the manufacturer's instructions. RNA was treated with DNase (TURBO DNA-free Kit; Life Technologies) and concentrations checked by NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, NC). Five hundred nanograms of RNA were reverse transcribed in cDNA using random hexamer primers and the RevertAid First Strand cDNA Synthesis Kit, according to the manufacturer's protocols (Thermo Scientific, Waltham, MA).
In semiquantitative RT-PCR, it is important to select the appropriate number of cycles so that the amplification product is clearly visible on an agarose gel and can be quantified, and that amplification is in the exponential range and has not reached a plateau yet. The optimal number of cycles has to be in the same range for the specific RNA of interest and the control, so that both can be measured on the same gel.
We optimized the protocol and defined the final amplification cycles by performing a gradient PCR [41,42]. The condition of 35 cycles were chosen because none of the RNAs analyzed reached a plateau at the end of the amplification protocol: each PCR product (amplicons of interest and control GAPDH gene) was in the exponential phase of amplification. Interestingly, similar semiquantitative RT-PCR protocols with 35 cycles have been reported for use with splicing reporter minigene systems by other authors [43,44].
For all semiquantitative RT-PCR analyses, we applied the following amplification protocol, a 1 min denaturation at 94°C, 35 amplification cycles (30 s at 94°C, 30 s at 58°C, and 1 min at 72°C), and a final extension for 7 min at 72°C. PCR reactions were carried out in 25 μL final volume in a reaction mixture containing 50 ng of diluted cDNA and 1 U Taq DNA polymerase (Applied Biosystems by Life Technologies), according to the manufacturer's protocols.
The endogenous RPGR expression levels in both cell lines, HEK-293T and PC-12, were detected by semiquantitative RT-PCR assay, by using two pairs of specific primers:
human RPGR: hRPGR_E7_For: 5′-caatcacagaacaccccaga-3′; hRPGR_E10_Rev: 5′-tgacatccaccacaagcaacc-3′
rat RPGR: rRPGR_E7_For: 5′ tgatcaatcacagatctcc-3′; rRPGR_E10_Rev: 5′- tgacatccaccacaggcaatc-3′.
These primers discriminate between the two alternative endogenous RPGR transcripts (E9a+ and E9a-) producing two different amplicons: 390 bp for the isoform without E9a (E9a-) and 526 bp for the isoform with E9a (E9a+).
To analyze the transcripts deriving from wild-type and mutant minigene reporters, the semiquantitative RT-PCR assay of cDNAs from HEK-293T, PC-12, and 661W cells was performed using primers: RPGRE9-For; 5′-cggccttatgtatacttttgg-3′ and BGH-Rev; 5′-tagaaggcacagtcgagg-3′ that anneal to the RPGR exons 9 and BGH plasmid region, respectively.
The pair of primers produce an amplicon of 518 bp for the isoform without E9a (E9a-) and an amplicon of 654 bp for the isoform with E9a (E9a+). An amplicon of 496 bp from GAPDH region was amplified using primers GAPDH For; 5′-tgacctcaactacatggtctaca-3′ and GAPDH Rev; 5′-cttcccattctcggccttg-3′ and was used as an internal control (housekeeping gene). Densitometric analyses were carried out with the Image Lab 2.0 software (Bio-Rad, Hercules, CA). After background correction, band intensities were normalized to the GAPDH levels. The assay was performed in triplicate and the mean ± SD was calculated.
RT-PCR assay to evaluate U1_asRNA expression in cotransfected cells
The expression levels and the integrity of the U1 snRNA chimeric constructs transfected in HEK-293T and PC-12 cells were analyzed through semiquantitative RT-PCR. The primer U1 + 130_Rev; 5′-agcacatccggagtgcaatg-3′, which anneals to the body of the U1 snRNA molecule [35], was used in combination with a specific primer for the engineered antisense modified U1 tail: U1-RPGR3′_For: 5′-ccaccgtgccctgccaaaacg-3′; U1-RPGR5′_For: 5′-gccatcatacctggctaatttttg-3′ or U1-Scramble_For: 5′-gctgcgtctcgcggaataattga-3′. To analyze the expression level of double antisense U1_3′5′ construct, the U1 + 130 Rev and U1-RPGR5′_For primers were used. PCR reactions were performed as described above for semiquantitative RT-PCR with an annealing temperature of 54°C. The U1_asRNA amplification products had a size of ∼100 bp.
RNA immunoprecipitation assay
HEK-293T cells were grown in 150-mm culture dishes (1.8 × 107 cells/dish) at 37°C in 5% CO2 atmosphere. After 24 h, the cells were cotransfected with U1 snRNA constructs (U1_3′, U1_5′ and U1_Scramble) (c.a. 15 μg) and RPGR mutant minigene (MINI mut) (c.a. 28 μg) using the Lipofectamine 2000 method according to the manufacturer's protocol (Life-Technologies). Forty-eight hours after cotransfection, the culture medium was removed, and ice-cold PBS was added to the cells. The culture dishes were placed on ice and irradiated once with 150 mJ/cm2 at 350 UV for 1 min using Ultraviolet Crosslinker, UVP CL-1000L Model: 365nm UV (Thermo Fisher Scientific, Illikirch Cedex, France).
Cells were harvested with a cell scraper, centrifuged briefly to remove PBS, and then nuclear protein fraction was obtained from HEK-293T cells by a modified version of the protocol described by Dignam and collaborators [45]. Briefly, the pellet was harvested, resuspended into 5X Cytoplasmic Extract (CE) buffer with NP-40 (10 mM HEPES pH 7.0, 10 mM KCl, 1.5 mM MgCl2, 0.1 M EDTA, 0.6% (v/v) NP-40, 1 mM DTT, 100 U/mL-1 RNase out (Life-Technologies), 2 U/mL Turbo Dnase (Ambion; Life-Technologies), and proteinase inhibitor cocktail (Sigma-Aldrich ), adjusted to pH 7.6. After centrifugation (1,200 rpm for 10 min at 4°C), cells were resuspended in CE buffer without NP-40.
Cells were centrifuged at 4°C (3,000 rpm for 5 min), and an equal volume of Nuclear Extract (NE) buffer (20 mM HEPES pH7.9, 400 mM NaCl, 1 mM EDTA, 1X Proteinase inhibitor cocktail (Sigma-Aldrich), 100 U/mL-1 RNase out (Life-Technologies), 2 U/mL Turbo DNase (Ambion; Life-Technologies), and 25% (v/v) glycerol, adjusted to pH 8.0) was added to this pellet and was kept on ice for 10 min. After homogenization, the cell suspension was centrifuged for 5 min at 14,000 rpm at 4°C. The final protein concentration in the NE extracts was determined using the colorimetric Pierce BCA Protein Assay Kit (Thermo Scientific). Pellets were stored at −80°C.
Immunoprecipitation was carried out from the nuclear fraction by Dynabeads Protein A (Life Technologies) with the Anti-U1A antibody (U1-70K) (Abcam, Cambridge, United Kingdom) and the Normal Rabbit IgG (Millipore, Darmstadt, Germany) as a negative control. Five microliters of both antibodies (anti-U1-70k and IgG) were used with 200 μL of the nuclear extract, and the Rip-Chip assay was performed as described by Keene et al. [46] that was optimized to minimize inappropriate interaction.
RNAs were extracted from immunoprecipitation fractions using TRIzol reagent, according to the manufacturer's instructions (TRIzol Reagent; Life Technologies) and were treated with DNase (the TURBO DNA-free Kit; Life Technologies). RNA was used to perform U1_snRNA chimeric screening, according to the procedure reported in the section RT-PCR assay to evaluate U1_asRNA expression in cotransfected cells. Ten microliters of PCR products were analyzed on 2.5% agarose gel. The amplicons were about 100 bp long, as predicted.
Statistical analyses
Semiquantitative RT-PCR densitometric data were expressed as mean ± SD (n = 3) and were compared using two-way ANOVA, followed by Bonferroni's comparisons test, for multiple comparisons test, and unpaired two-tailed Student's t-test, for two group comparison (GraphPad Software, San Diego, CA. www.graphpad.com). Statistical significance is denoted with asterisks (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
Results
Computational analysis of splicing-relevant sequences in RPGR exon 9a (E9a)
To understand how to modulate splicing of E9a, we undertook a bioinformatics analysis of sequences essential for the recognition of the alternative exon. We also investigated the effect of the g.26652G>A nucleotide substitution on these sequences.
The first analysis carried out by using NNSPLICE 0.9 program [31] (https://omictools.com/nnsplice-too l), showed that E9a has a weak consensus sequence at its 3′ splice site, indicating that a Splicing Enhancer might assist E9a splicing, whereas its 5′ splice site is similar to the consensus sequence (Table 1). Additionally, we analyzed whether the nucleotide substitution might create a new branch site with a stronger consensus. However, using the branch point analysis bioinformatic program “Human Splicing Finder” [32] (www.umd.be/HSF/), we found no consensus change in the branch sites pattern of intron 9 (data not shown). A secondary structure prediction (Supplementary Fig. S1A) indicates that the c.1059 + 363G>A intronic nucleotide substitution destabilizes a 13-bp-long stem loop in the intron, while not affecting the accessibility of the E9a acceptor site.
Identification of a defined Exon Splicing Enhancer (ESE) sequence [47] on E9a would provide an optional target site for the design of chimeric U1_snRNAs able to mediate E9a skipping. A first scan for consensus binding sites of four known splicing regulator proteins (SF2/ASF, SC35, SRp40, and SRp55) was performed with ESEFinder [33] and identified several potential binding sites for these proteins in E9a sequence (Supplementary Fig. S1B). However, a subsequent analysis of E9a was performed with SpliceAid [34] (www.introni.it/splicing.html), which searches consensus binding sites of 71 positive and negative regulators of splicing. This analysis did not show the presence of a clear and defined ESE, since both splice-enhancing and splice-silencing RNPs are predicted to bind to the same regions (Supplementary Fig. S1C).
Our bioinformatic analyses suggested that E9a does not have any evident ESEs. Accordingly, we decided to target E9a splice sites. Indeed, masking these sites should result in E9a skipping as a consequence of the splicing machinery failure to recognize these critical cis-acting sequences.
To test the hypothesis that mutation c.1059 + 363G>A affects a splicing regulatory sequence, we also performed a comparative analysis of the mutant and wild-type intronic region sequences upstream of E9a, through ESEFinder (Supplementary Fig. S1B) and SpliceAid (Supplementary Fig. S1C).
The first program indicated that mutation c.1059 + 363G>A abolishes a binding site for SC35 protein in the mutant RPGR pre-mRNA, 54 to 46 nucleotides upstream of the 3′ss (Supplementary Fig. S1B), while the second showed that the mutation introduces a new binding site for Sam68 and Sam68-like SLM-2 proteins in the mutant RPGR pre-mRNA, 58 to 52 nucleotides upstream of E9a 3′ss (Supplementary Fig. S1C). The loss of SC35 binding, the gain of Sam68/SLM-2 binding, or both actions at the same time, might explain the mechanism underlying the pathological effect of the mutation.
Analysis of exon 9a (E9a) presence in RPGR mRNA in cells in culture.
We aimed at analyzing E9a presence in RPGR mRNA in three independent cell models, to add robustness to the analysis. Moreover, the chosen cell lines had to be easy to grow, to maintain, and to transfect. Finally, the cell models should recapitulate as much as possible the repertoire of auxiliary splicing factors typical of photoreceptors and other neurons. Alternative splicing is indeed a mechanism increasing gene-expression diversity, which is widely used by neurons, and exon-skipping therapeutical strategies should be tested taking the tissue-specific peculiarities of this mechanism into account [24].
To our knowledge there is presently no cell line able to recapitulate photoreceptor characteristics in vitro. High levels of expression of both RPGR mRNA and RPGR protein have been reported in the human adrenal gland medulla (www.proteinatlas.org/ENSG00000156313-RPGR) that has a neural crest origin as do other neuronal types. We, therefore, chose to use two different adrenomedullary cell lines: HEK-293T and PC-12.
HEK-293T (fetal human embryonic kidney cells) [48] derive from HEK-293 that, despite their name, have presumably been established from a human (female) embryonic adrenal precursor cell [49]. HEK-293T cells are hypotriploid and carry three copies of the X chromosome.
PC-12 cells derive from a pheochromocytoma in a (male) rat adrenal medulla [50] and grow in culture as undifferentiated neuroblasts. PC-12 are described to have 40 chromosomes (38 autosomes plus XY) and therefore to be hypodiploid. They are a commonly employed model system for studies of neuronal development and function and are relatively easy to passage, culture, and transfect [40].
The use of PC-12 cells of rat origin, additionally, allows to enquire whether the therapeutical strategy could be tested in rodent models of the disease, in the future.
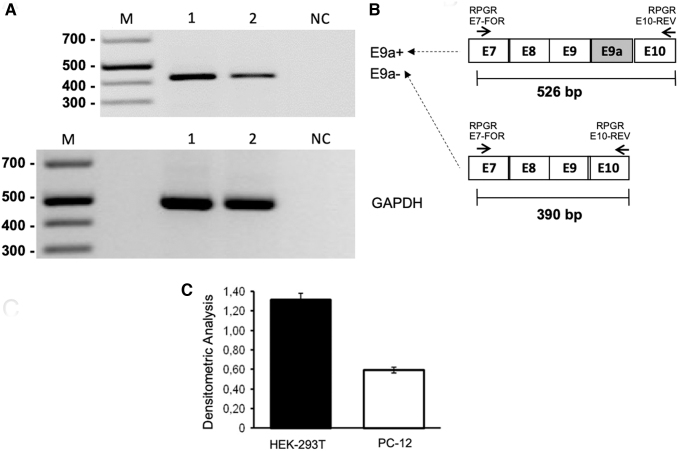
By semiquantitative RT-PCR performed with primers annealing to exons 7 and 10 of RPGR mRNA, we evaluated the level of endogenous RPGR expression in HEK-293T and PC-12. We showed that, in both cell lines, endogenous RPGR mRNA was detectable in the form of E9a−, while the E9a+ isoform was not present (Fig. 1). RPGR mRNA was more abundant in HEK-293T than in PC-12 cells.
FIG. 1.
Analysis of exon 9a inclusion in RPGR mRNA in HEK-293T and PC-12 cells in culture. (A) Semiquantitative RT-PCR analysis was performed with primers annealing to exon 7 (forward) and exon 10 (reverse) of human and rat RPGR mRNA, respectively. Lane 1: HEK-293T cells, Lane 2: PC-12 cells. As depicted in the schematic diagram to the right, (B) the presence of E9a would yield a 324-bp-long amplicon (E9a+), while the amplification of RPGR transcript devoid of E9a produces an E9a-118-bp-long amplicon. GAPDH was used as a housekeeping gene. One representative gel of three is shown. Only E9a− bands were detected. Their intensities were measured by densitometric analysis and reported in the histogram. (C) Values are represented as mean ± SD (n = 3).
Reporter minigenes recapitulate RPGR exon 9a (E9a) alternative splicing.
To study E9a skipping, in vitro, we constructed reporter systems by cloning the RPGR genomic region containing exon 9, the entire intron 9 and exon 10 in the expression vector pCDNA3. We produced both the RPGR wild-type reporter (MINI wt) and the mutant one (MINI mut) bearing the c.1059 + 363G>A mutation located 55 bp upstream of the E9a 3′splice site (Fig. 2A). We expect the RPGR mutant reporter to generate higher levels of transcripts containing E9a compared with the wt reporter, as observed in RP patients [15].
To test if the reporter constructs were able to recapitulate alternative splicing of E9a in presence or absence of the c.1059 + 363G>A nucleotide substitution, we transiently transfected either MINI wt or MINI mut in HEK-293T and PC-12 cells. By semiquantitative RT-PCR, using primers targeting RPGR exon 9 and the BGHpA region (Fig. 2A), we observed that in both cell lines the nucleotide substitution was able to increase E9a levels of about two-fold compared with the wild-type (Fig. 2B, C). PCR products were analyzed to confirm the exact sequence of the two mRNA RPGR isoforms, with and without E9a, in HEK-293T and PC-12 cells transfected with MINI mut reporters (Fig. 2D, E, respectively).
Design of antisense U1_snRNAs to induce exon skipping of RPGR exon9a (E9a)
To induce skipping of E9a in the context of our RPGR minigene reporters, we designed and generated three chimeric constructs in the backbone of the U1_snRNA.
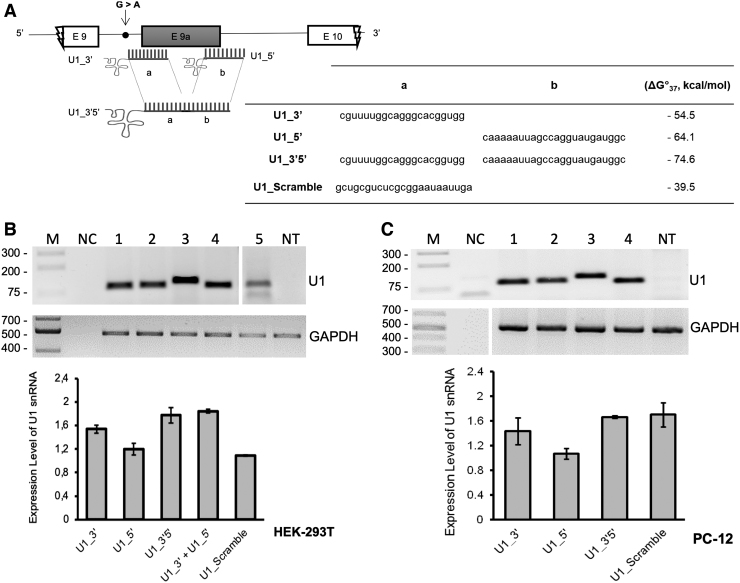
The nucleotide sequence required for the recognition of the 5′ splice site (positions 3–10 at the 5′-end of U1 snRNA) was substituted with an antisense sequence complementary either to the 3′ splice site (U1_3′), 5′ splice site (U1_5′), or both splice sites simultaneously (U1_3′5′) (Fig. 3A). This latter construct was created because, according to literature, there is a higher efficiency of exon skipping by employing double-target U1 asRNAs [26–28]. As a negative control, we additionally cloned an U1_Scramble construct, bearing a 23-nt-long sequence (Fig. 3A) with no targets in mammalian cells.
FIG. 3.
Validation of chimeric antisense U1 snRNA construct expression. (A) Schematic representation of chimeric U1 snRNAs with splicing correction activity on the minigene model of RPGR (not in scale). The table summarizes the target regions of the different constructs, the scramble sequence (5′-to- 3′ direction), and the values of the hybridization energy of Chimeric U1_snRNAs/RPGR pre-mRNA (ΔG°37, Kcal/mol), predicted by using DuplexFold tool. (B, C) Expression of chimeric antisense U1 constructs in HEK-293T (B) and PC-12 (C) was assayed by RT-PCR, using primers U1 + 130 Rev and the appropriate U1-RPGRFor (see Materials and Methods section). One representative gel of three is shown in both (B) and (C). The histogram shows the densitometric analysis as mean ± SD (n = 3).
Since the different antisense constructs, U1_3′, U1_5′, U1_3′5′, and U1_Scramble, considerably extended the length of the U1_snRNA, we checked both integrity and expression levels of U1 antisense RNAs after cotransfection in HEK-293T and PC-12 cells. By performing semiquantitative RT-PCR as described in Materials and Methods, we observed the presence of specific amplification products, confirming that all designed molecules were present and expressed at comparable levels (Fig. 3B, C). As expected, the sizes of the amplicons (around 100 bp) slightly varied, depending on the antisense sequence introduced, and on the different forward primers used.
Antisense U1_snRNAs restore RPGR transcript expression patterns
To assess the feasibility of E9a skipping, we carried out cotransfection experiments by using the U1_3′, U1_5′, U1_3′5′, U1_3′ + U1_5′, and U1_Scramble constructs in combination with the RPGR MINI mut minigenes, in HEK293 cells.
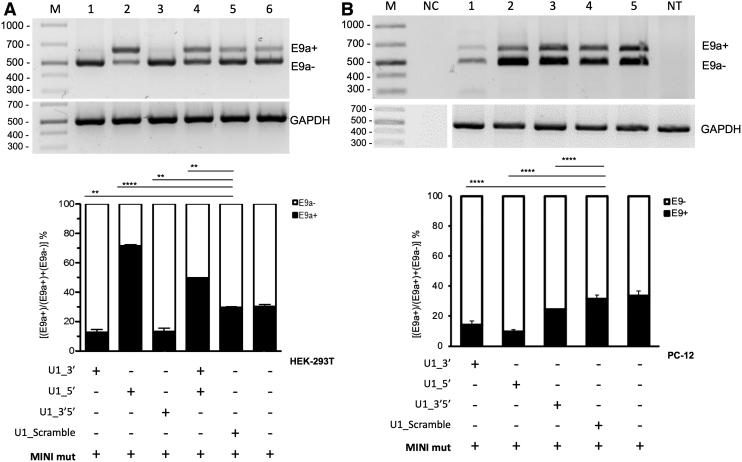
When using MINI mut, we observed a reduction of E9a (E9a+) mRNAs of about 50% using U1_3′ or U1_3′5′ in HEK-293T cells, when compared with the transfection of the minigene alone. Surprisingly, the U1_5′ led to a 130% increase in E9a+ transcripts (Fig. 4A).
FIG. 4.
Chimeric antisense U1 snRNA-induced exon 9a skipping in HEK-293T and PC-12 cell lines transfected with RPGR mutant minigene. (A) Semiquantitative RT-PCR of RNA from HEK-293T cells transfected with RPGR mutant minigene (MIN mut) alone or in combination with chimeric U1_snRNA plasmids. E9a reduction using U1_3′ or U1_3′5′ is significant (P < 0.01) compared with U1_Scramble. (B) Semiquantitative RT-PCR of RNA from PC-12 cells transfected with RPGR mutant minigene (MINI mut) alone or in combination with chimeric U1 snRNAs. E9a reduction using any of the three U1 constructs is significant (P < 0.001) compared with U1_Scramble. One representative gel of three is shown in both (A) and (B). Densitometric analysis of E9a+ and E9a− amplicons, from three independent experiments, is shown for both cell lines. GAPDH is used as an internal control. Data are shown as mean ± SD (n = 3). Statistical analysis was performed with a global statistical test and is reported in Supplementary Table S1 (P-value: **P < 0.01; ***P < 0.001).
Additionally, when both constructs U1_3′ and U1_5′ were cotransfected, E9a (E9a+) inclusion increased in mature mRNA, suggesting that U1_5′ has a dominant effect on U1_3′. The fact that the total amount of U1_asRNA was maintained (transfecting only half of the amount of U1_5′ compared with when it was transfected alone) explains why exon inclusion was 50% lower than that obtained by transfecting U1_5′ alone.
The U1_Scramble control did not cause any significant alteration in the RPGR expression, as expected. Overall, our results indicate that the U1_3′ and U1_3′5′ constructs (Fig. 4A) induce the exon skipping correction for c.1059 + 363G>A nucleotide substitution.
Statistical analysis of these data was performed by a global statistical test (two-way ANOVA) and detailed data are reported in Supplementary Table S1.
All the E9a+ and E9a- bands in Fig. 4A were eluted and sequenced, confirming the correct joining of exons 9 and 10, or exons 9, 9a and 10, respectively. In particular, sequencing of bands in lanes 2 and 4 confirmed that the lower band consists in the transcript devoid of E9a, and the upper band results from the correct joining of E9a in between exon 9 and exon 10 (Supplementary Fig. S2).
We then selected one of our best-performing chimeric U1 snRNA, U1_3′, and tested it at decreasing concentrations (200, 66, 20 ng) over RPGR MINI mut transfected HEK-293T cells, to assess if its effect was dose dependent. Semiquantitative RT-PCR assay clearly showed that the U1_Scramble does not affect E9a levels at the different tested doses, on the contrary U1_3′ also showed measurable E9a skipping at lower doses (Supplementary Fig. S3).
To test the exon skipping effects in a different cellular environment, we analyzed total RNA isolated from PC-12 cells transfected with the RPGR MINI mut alone or in combination with our U1_snRNAs (Fig. 4B). All three designed U1_asRNAs (U1_3′, U1_5′, and U1_3′5′) induced E9a skipping, although at different levels. Indeed, the constructs' ability to reduce the inclusion of the E9a depended on the targeted sequence and ranged from ∼70% reduction for U1_5′ to ∼20% reduction for U1_3′5′ compared with the MINI mut alone. The U1_Scramble did not influence RPGR E9a splicing, as expected (Fig. 4B).
Sequence analysis of both E9a+ and E9a- PCR isoform products was performed on all bands shown in Fig. 4B and showed restoration of the reading frames after treatment with U1_snRNA constructs and the presence of the exact junction between exon 9 and exon 10 in the products correlated to skipping. Supplementary Fig. S4 reports the chromatograms relative to lane 2 of Fig. 4B.
Thus, interestingly, we observed that the efficiency of our constructs is not only dependent on the target sequence but also on the cell type. In contrast to the results obtained in human HEK-293T cells (Fig. 4A), U1_5′ led to efficient E9a skipping in rat PC-12 cells (Fig. 4B).
To analyze whether the difference in the outcome of U1_5′ transfection (exon skipping or exon inclusion) might be due to the origin of the cell line (human or rodent), we repeated the experiment in 661W cells, a cell line derived from the retinal tumor of a mouse expressing the SV40 T antigen [51,52]. In this cell line, the nucleotide substitution in MINI mut reporter increases E9a levels of about 1.4-fold compared with MINI wt (Supplementary Fig. S5A). We observed a reduction of E9a (E9a+) mRNAs of about 15% using U1_3′ or U1_3′5′ in 661W cells, when compared with the transfection of the minigene alone (Supplementary Fig. S5B). Similar to HEK293T cells, in 661W cells, the U1_5′ led to a 160% increase in E9a+ transcripts (Supplementary Fig. S5B).
The U1_asRNAs also recognize the sequence of wild-type RPGR pre-mRNA. However, semiquantitative RT-PCR analysis carried out using the RPGR MINI wt with the U1_snRNA constructs (U1_3′, U1_5′, and U1_3′5′) did not show any significant variation on endogenous RPGR mRNA expression levels for both HEK-293T and PC-12 cell lines, compared with the nontreated transfected RPGR wild-type (p = n.s.; n = 3) (Supplementary Figs. S6A and B, respectively).
Chimeric antisense U1 asRNAs are able to form snRNP complexes
We next performed an RNA ImmunoPrecipitation (RIP) assay [46] to test whether the U1_asRNA constructs are able to bind U1-70K protein, upon cotransfection of HEK-293T cells with the RPGR MINI mut and U1_3′, U1_5′ or U1_Scramble constructs. U1-70K protein behaves as a trans-acting factor that, by interacting and binding to U1 snRNA in the first step of the spliceosome assembly, mediates the pre-mRNA splicing (Fig. 5B) [28]. The ability to bind to U1-70K not only is a prerequisite for the assembly into a functional snRNP, but also a check over the correct folding of the chimeric RNA, as U1-70K binds to loop I in U1 snRNA (Fig. 5B). It is assumed that chimeric antisense snRNAs correctly assembled in an snRNP are also protected from degradation and therefore more stable and more effective.
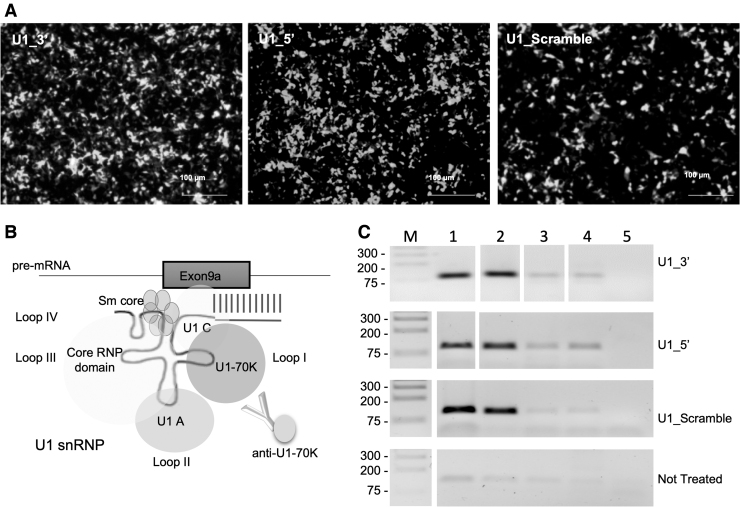
FIG. 5.
Chimeric U1_5′ and U1_3′ assemble with U1-70k ribonucleoprotein in transfected HEK-293T cells. (A) Representative images of HEK-293T cells 48 h after cotransfection with the RPGR mutant minigene and the different chimeric U1_asRNA constructs (Zeiss Axio Observer Z1 microscope; Scale bar: 100 μm). (B) Schematic representation of the U1 snRNP particle (not in scale). (C) Semiquantitative RT-PCR of the immunoprecipitated RNA using anti U1-70k antibodies was performed with a U1 + 130 primer and specific U1 primers, as described in Materials and Methods. NE: Nuclear extracts; IPP: NE sample + U1-70K antibody; IgG: NE sample + IgGs; Input: NE sample + Beads; Ctr: No Template PCR control.
Nuclear extracts were prepared from transfected cells (Fig. 5A) and immunoprecipitated with an anti U1-70 K antibody. Immunoprecipitated RNP complexes were then dissociated into RNA and protein. The RNA was isolated and checked for integrity and length by RT-PCR. As expected, PCR products were detected in Nuclear Extracts (NE) and enriched in the samples immunoprecipitated with anti-U1-70K (Fig. 5C). PCR products of U1_snRNAs were barely detectable in the sample treated with normal rabbit IgG conjugated to beads, Input, and no-template control (Fig. 5C). These results indicate that U1_3′, U1_5′, and U1_Scramble RNA constructs are able to assemble into a functional U1 snRNP.
Discussion
Most eukaryotic genes contain pseudoexons, sequences that look like perfect exons but are ignored by the splicing machinery based on rules that are yet to be fully understood. In recent years, research has highlighted roles for pseudoexons not only as regulators of the splicing process but also as possible causes of human diseases [53].
Here, we focused on an intronic mutation that affects pseudoexon splicing but does not alter the consensus sequence of the pseudoexon splice sites. This type of mutation acts mainly by deleting or creating intronic splicing silencers or intronic splicing enhancers or changing the strength of the consensus sequence of the branch point [54–57]. There are two advantages in targeting splicing mutations instead of mutations leading to missense or frameshift. First, since these mutations influence splicing, the achievement of correction of the altered splicing pattern is sufficient to obtain a therapeutic benefit. Indeed, if constitutive exons will not be skipped, functional analysis of the rescued proteins is not needed. Second, if the mutations affect splicing by creating new cis-acting sequences, the disease can be quickly addressed by identifying and masking those sequences with antisense oligonucleotide approaches. For these reasons, we selected the intronic nucleotide substitution c.1059 + 363G>A of the RPGR gene [15] as an optimal target for applying splicing-correction approaches in the retina.
Neidhardt et al. [15] reported that the nucleotide substitution c.1059 + 363G>A (which they called g.26652G>A) in the RPGR gene increases the levels of the alternatively spliced E9a nonsense pseudoexon. Our bioinformatic analyses were not conclusive about whether this nucleotide substitution causes an increase of the alternatively spliced E9a RPGR mRNA through the generation or removal of cis-acting sequences. We excluded that the mutation introduced a new branch point. ESEFinder indicated that mutation c.1059 + 363G>A abolishes a binding site for SC35 protein in the mutant RPGR pre-mRNA, while SpliceAid showed that the mutation introduces a new binding site for Sam68 and Sam68-like SLM-2 proteins. Both effects could explain the gain of E9a inclusion.
SC35 [58] (now known as SRSF2) is a member of the serine/arginine (SR)-rich family of pre-mRNA splicing factors, which constitute part of the spliceosome. It contains an RNA recognition motif for binding RNA and a domain rich in serines and arginines (RS domain) for binding other proteins. It interacts with other spliceosomal components to form a bridge between the 5′- and 3′-splice site-binding components, U1 snRNP and U2AF, and has been observed to affect both alternative 5′ and 3′ splice site selection [59]. Therefore, c.1059 + 363G>A mutation-induced loss of SC35 binding has the potential to change the splicing pattern of RPGR intron 9, although the hypothesis of a direct involvement of SC35 would require experimental validation.
Sam68 has roles in signaling, transcription, and alternative splicing, and undergoes multiple post-translational modifications that finely modulate its subcellular localization, interaction with signaling proteins, affinity for target RNAs, and function. A possible role for Sam68 in splicing was initially suggested by its binding to an intronic regulatory element located between the polypyrimidine tract and the 3′ splice site of the β-tropomyosin pre-mRNA [60]. Sam68 binds to U2AF65, a component of the U2 snRNP, and its phosphorylation by ERK1/2 was proposed to fine-tune occupancy of the 3′ splice site by U2AF65 [61].
While Sam68 is ubiquitously expressed in neuronal and non-neuronal cells, its close homologs SLM1 and SLM2 exhibit a remarkable, mutually exclusive expression in neuronal cell types [62–65]. SLM2 was shown to interact with splicing factors and to modulate splicing of reporter minigenes [66] and to be responsible for a hierarchical mechanism for establishing cell type-specific expression of neuronal splicing regulators [67].
In the context of the RPGR c.1059 + 363G>A mutation, Sam68 and/or SML2 could function as intronic splicing enhancers, by attracting U2AF65/35 to the weak 3′ss of E9a, thus favoring exon inclusion.
In the absence of a clear indication for an Exonic or Intronic Splicing Enhancer, we decided to direct chimeric U1_snRNAs toward either splice sites of E9a (U1_5′, U1_3′) or both splice sites at the same time (U1_3′5′), aiming at reducing the splice site recognition in the presence of the G > A nucleotide alteration. U1_snRNAs typically recognize the 5′ splice site by the binding of their 5′ end to the −3/+ 6 exon/intron consensus sequence. This binding needs to be reversible since U1 snRNA has to detach from the 5′ splice site so that the splicing reaction can proceed further. To increase the specificity and affinity of U1 to its target site, we increased the length of the region recognized by our chimeric U1_asRNAs to 21, 24, or 45 nucleotides.
Using RNAfold, it was possible to predict the secondary structure and the ΔG°37 free energy of both wild-type and mutant RPGR pre-mRNA sequences. Therefore, the Chimeric U1_snRNAs were designed to take into consideration the folding energy (data not shown) and the binding energy between the U1_snRNA Chimeras and RPGR pre-mRNA target regions (Fig. 3A).
Comparing the average of binding Gibbs' free energy (ΔG°37, −7.6 to −14.2 kcal/mol) of endogenous U1_snRNA with that of our chimeric U1_snRNAs, we observed that the ΔG°37 (−54.5 to −74.6 kcal/mol) values of our U1_snRNA were lower than those of the endogenous U1_snRNA (Fig. 3A). Therefore, the computational analysis confirmed the efficient binding of our chimeric U1_snRNAs to the RPGR pre-mRNA target sequence, suggesting that they could play a role, in vitro, in E9a exon-skipping.
To recapitulate the wild-type and mutant conditions in vitro, we generated minigene reporter systems [68,69]. We designed a wild-type minigene (MINI wt), and one carrying the c.1059 + 363G>A mutation (MINI mut) (Fig. 2A). These constructs were transfected either in Human Embryonic Kidney (HEK-293T) cells or rat pheochromocytoma (PC-12) cells. Both cells express the endogenous RPGR mRNA exclusively without the E9a pseudoexon (Fig. 1).
Increased levels of E9a transcripts were detected upon MINI mut transfection compared with cells in which the MINI wt was transfected, recapitulating the splicing pattern of patients' cells affected by the c.1059 + 363G >A mutation [15]. Similar results were obtained when transfecting the MINI wt and MINI mut reporter constructs in 661W cells.
Neidhardt et al. [15] have estimated that in human retinas from healthy individuals ∼4% of RPGR transcripts bear E9a (Fig. 1C in [15]).
In our hands, upon transfection of our wild-type RPGR minigene, E9a is included in 12% of RPGR minigene-derived transcripts in HEK-293T, in 18% of said transcripts in PC12 cells (Fig. 2B, C) and in 23% of transcripts in 661W cells (Supplementary Fig. S5A).
These observations are in line with the general understanding that alternative splicing is not a black and white process and small amounts of nonsense alternative (pseudo-)exons (also termed “poison exons”) are often spliced in gene transcripts, possibly as part of a finely tuned post-transcriptional regulation of gene expression [16].
Neidhardt et al. [15] describe a mutation in the RPGR gene (c.1059 + 363G>A), which correlates with X-linked Retinitis Pigmentosa and significantly increases E9a inclusion in RPGR transcripts. When comparing RPGR transcripts in blood cells from the affected individual to those of his two unaffected brothers, Neidhardt et al. report a 3-times higher level of e-containing RPGR transcript.
Therefore, our mutant minigene system recapitulates the molecular defect observed in the patient (ie, the increase of E9a-containing transcripts). In fact, upon transfection of an RPGR minigene bearing mutation g.26652 G > A (MINI mut) E9a is included in 20% of RPGR minigene-derived transcripts in HEK-293T (Fig. 2B), in 30% of said transcripts in PC12 cells (Fig. 2C) and in 33% of transcripts in 661 W cells (Supplementary Fig. S5A).
By testing U1_asRNAs against the RPGR minigene system, we observed that a chimeric U1_asRNA directed toward the 3′ splice site (U1_3′) in HEK-293T, PC-12, and 661W cells was able to achieve a significant E9a exon skipping (Fig. 4 and Supplementary Fig. S5B).
Interestingly, we observed a different effect of the U1_5′ construct activity. We designed this construct with a 24 bp long antisense sequence to have a stable heteroduplex conformation and to promote masking of the splice site, in line with results reported in the literature [70].
The U1_snRNA directed toward the 5′ splice site unexpectedly caused a significant increase in E9a levels when tested in the HEK-293T (Fig. 4A) and 661W (Supplementary Fig. S5B). Indeed, sequence analysis performed on the E9a+ PCR product (Supplementary Fig. S2) confirmed that the fragment corresponded to the sequence containing E9, E9a, and E10 perfectly joined.
On the contrary, the U1_5′ was as capable as the U1_3′ of skipping E9a in PC-12 cells, restoring the normal expression of the alternatively spliced RPGR mRNA (Fig. 4B). These findings bring into question why this approach does not work in all cell types in the same way. Additionally, it has to be noted that U1_5′ does not induce E9a inclusion in MINI wt-transfected HEK-293T cells (Supplementary Fig. S5A).
Overall, it seems that c.1059 + 363G>A mutation facilitates E9a inclusion and the U1_5′ in HEK-293T and 661W cells acts over the mutated minigene similarly to adapted U1 snRNAs, which are typically designed with shorter complementarity (around 10 bp) [71–73]. It was demonstrated, indeed, that when complementarity between U1_snRNA and 5′splice site is extended to 11 nt, 5′ splice site recognition is increased [74]. Notably, RIP experiments were performed to check the capability of our U1_asRNA chimeras to bind the U1-70K protein, proving that they can form stable and functional snRNPs (Fig. 5).
We also analyzed the effect of the double antisense construct U1_3′5′. This construct led to splicing correction in HEK-293T and 661W at a comparable level of that induced by U1_3′ (Fig. 4A and Supplementary Fig. S5B), while in PC-12 U1_3′5′ was less efficient than U1_3′ (Fig. 4B).
Interestingly, while evaluating the combined effect of U1_3′ and U1_5′ antisense constructs, we observed an increase of E9a levels in between those observed with each of the two chimeric RNA (Fig. 4A and Supplementary Fig. S5B). This result suggests that U1_5′ exon-inclusion effect overtakes U1_3′ exons-skipping effect. The observed levels of E9a inclusion were 50% lower than that obtained by transfecting U1_5′ alone, in line with the reduced amounts of U1_5′construct transfected.
Our study provides evidence that both our chimeric constructs U1_3′ and U1_3′5′, tested in HEK-293T, PC-12, and 661W cell lines, are efficient at inducing exon skipping of RPGR exon 9 in the presence of nucleotide substitution c.1059 + 363G> A. However, one of our constructs, U1_5′, induces exon skipping or exon inclusion, depending on the cellular background.
The observations of Fig. 5, together with our bioinformatic analysis might shed some light on the unexpected exon-including effect of U1_5′. In fact, in HEK-293T cells, both U1_5′ and U1_3′ assemble in a functional U1 snRNP. However, the participation of this “adapted” U1 snRNP in a functional spliceosome depends on whether the defined binding site of the U1 snRNP (the target sequence) is correctly positioned with respect to a branch point, a 3′ss, and several binding sites of auxiliary splicing factors (proteins).
The chimeric U1_5′ happens to be directed against a binding site, which is also the 5′ss of E9a and has therefore the potential to act as an adapted U1 snRNA. On the contrary, U1_3′ is attracted to the E9a acceptor splice site (3′ss) by virtue of its sequence, complementary to the 21 nucleotides surrounding the acceptor site itself. Here, it will act according to two possible scenarios: either U1_3′ interferes with the binding of U2AF to E9a 3′ss, therefore inducing skipping of E9a, or it also acts as an adapted U1 snRNA, defining a new donor site (5′ss), which would then splice to the canonical intron 9 3′ss (acceptor site).
If coupled with a subsequent splicing step between the canonical 5′ss and this newly defined 3′ss (“3′ recursive splicing” [75]), this latter scenario also results in the skipping of E9a. In this context, the c.1059 + 363G>A mutation might facilitate the role of the “adapted” U1_5′, either by abolishing a binding site for SC35, which might be needed to bridge the “canonical” 5′ss and 3′ss of intron 9, or by creating a new binding site for Sam68 and/or SLM2, which might increase the strength of the otherwise very weak 3′ss of E9a. Of note, the U1_5′ exon-inclusion effect is not observed on the wild-type reporter minigene transfected in HEK-293T cells (Supplementary Fig. S6).
Unfortunately, we are not able to explain why the same exon-including effect of U1_5′ is not observed in PC12 cells. This cell-type-specific effect might have to do with the peculiarity of the pool of auxiliary splicing factors present in each cell type.
Taken together, these results suggest that a new therapeutic strategy based on U1 snRNA molecules could efficiently be used to restore the physiological RPGR gene splicing, but call for caution in the choice of the model system used to study its efficiency.
The results presented here constitute proof-of-concept data that should be followed by further preclinical trials in patient-derived cells and in animal models, to test efficacy in correcting the genetic defect, with more emphasis on the correction of the genotype–phenotype pattern. However, unfortunately, and to our best knowledge, no patient-derived cell model of RPGR E9a inclusion is available at the moment that could be used for further studies, nor any animal model.
RPGR mutation c.1059 + 363G> A has only been described in one patient by Neidhardt et al. in 2007 [15] and no other E9a inclusion-inducing mutation has been reported so far. Therefore, patient-derived induced pluripotent stem cell (iPSC) models of the mutation have not been generated.
For the purpose of obtaining a cell model to be used for proof-of-concept studies, ZFN or CRISPR-Cas9 technologies could be used to introduce the RPGR c.1059 + 363G G > A mutation into commercially available control human iPSCs. These cells could be then differentiated into retinal cells, to validate the therapeutic approach in relevant human cell types [76]. However, methods for retinal differentiation based on two-dimensional (2D) cell culture have been so far unable to generate all structural components, such as the inner and outer segments or the spatial information for photoreceptor cells, making it difficult to fully recapitulate retinal diseases in cell culture [77,78].
In the last 10 years, significant progress has been made in achieving three-dimensional (3D) retinal differentiation from iPSCs [79,80]. As an example, Deng et al. generated iPSCs from three patients with frameshift mutation in the RPGR gene and differentiated these cells into retinal pigmented epithelium (RPE) cells and 3D retinae, demonstrating significant defects in photoreceptors and cilia, and therefore that the model recapitulates the disease in vitro [81]. However, protocols for 3D retinal differentiation from iPSCs are still rather laborious and long.
E9a is not conserved in rodents, so it would be impossible to generate a mouse model of X-linked Retinitis Pigmentosa by appropriately introducing a point mutation in mouse RPGR intron 9, similarly to what has been done to model Spinal Muscular Atrophy [82]. However, the general exon–intron architecture of the RPGR gene and the sequence and size of its “canonical” exons are conserved.
With the aim of generating a disease model to be used to preclinically validate the therapeutical approach proposed here, a “humanized” mouse model could be generated, by using the endogenous mouse RPGR gene and homologous recombination to replace mouse RPGR intron 9 (2,756 bp and not containing E9a) with human RPGR intron 9 (2,105 bp and containing E9a).
The “humanization” of mice to recapitulate the splicing pattern of human genes has been extensively employed for the generation of animal models of several diseases and has been recently reviewed [83]. For example, using standard homologous recombination and mouse ES cell techniques, a mouse model of Familial Dysautonomia (FD) was generated, which carries the complete human IKBKAP locus with the FD IVS20 + 6T>C splice mutation [84].
More recently, programmable nucleases, such as ZFN, TALEN, and CRISPR-associated (Cas) nucleases, have been used to generate targeted double-strand breaks (DSBs) in the genome. These DSBs would then constitute the sites for the targeted insertion of the human gene portion through Homology Directed Repair (HDR). For instance, Baker et al. reported replacing the mouse cancer gene KMT2D with the human gene, using just one Cas9 cleavage at the genome target site [85].
Another outcome of our work is that the choice of U1 antisense chimeras directed against the 3′ss might avoid the unwanted effect of exon inclusion observed here with the U1 antisense chimeras directed against the 5′ss, and might therefore be advisable when choosing the target sequences of U1-vectored exon-skipping strategies. Unfortunately, we do not have access to similar observations having been made by other research groups, possibly due to the tendency of the scientific community not to share unpredicted (so-called “negative”) results [86]. Therefore, it is difficult at this point to generalize our finding as a universal indication.
For several decades, RNA therapeutics to regulate gene expression have been developed toward clinical use at a steady pace. In recent years the field has witnessed a welcomed acceleration and there are now 11 approved RNA therapies based on antisense oligonucleotides, aptamers, and small interfering RNAs, with many others in the pipeline both in academia and industry [87,88]. The development of these therapeutic approaches, all based on oligonucleotides, has been initially hampered by the poor stability and high toxicity of these synthetic compounds [87].
Developments in oligonucleotide chemistry have improved the drug properties and reduced their cost, while the main hurdle for their application to a wider range of disorders remains delivery to target tissues [88].
Efforts are being made to develop methods, such as conjugates or nanoparticles, to deliver nucleic acid-based therapeutics across biological barriers and to specific tissues [87,88].
In this context, chimeric U1 constructs have been developed as an alternative way to vector exon skipping-inducing antisense sequences to the target cells [23]. Upon the introduction of the corresponding DNA sequence in the cells, the chimeric RNAs are transcribed in the nucleus, thus avoiding toxicity and resulting in chimeric U1 molecules whose stability is favored by their secondary structure and their assembly in U1 snRNP particles.
The U1 promoter/U1_asRNA cassette is relatively small (ca. 600 bp) and can easily be accommodated in the limited capacity of Adeno-Associated Viral (AAV) vectors. Based on our previous experience in a mouse model of Duchenne Muscular Dystrophy, exon-skipping U1asRNAs vectored by AAV show a good distribution and are effective and stable [26,29]. In recent years AAV has become the pre-eminent vector for gene therapy, and is particularly well suited for delivery to the nervous system [89]. Compared with the administration of synthetic antisense oligonucleotides, AAV-vectored antisense U1 RNAs would have the advantages of long-term efficacy and lower toxicity, while presenting challenges similar to those of gene therapies in regulatory approvals.
On the condition that a preclinical proof-of-concept study is successful, the described therapeutic approach described in this study, could be translated into clinical trials through subretinal injection [89–91] that is performed by administering the therapeutic molecule between the photoreceptor cell layer and the RPE.
Supplementary Material
Acknowledgments
The authors would like to acknowledge Annalisa Rossi (Laboratory of Transcriptional Networks, Department CIBIO, University of Trento) for the helpful suggestions with regard to RIP assay protocol.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
This work was funded by the Italian Ministry of Health (Project GR-2008-1136933), by Department CIBIO, University of Trento (Grant number 40201033) to M.A.D., and supported by COST Actions-BM1207 Networking toward clinical application of antisense-mediated exon skipping and by COST Action CA17103 Delivery of Antisense RNA Therapeutics.
Supplementary Material
References
- 1. Ferrari S, Di Iorio E, Barbaro V, Ponzin D, Sorrentino FS, and Parmeggiani F. (2011). Retinitis Pigmentosa: genes and Disease Mechanisms. Curr Genomics 12:238–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Daiger SP, Sullivan LS and Bowne SJ. (2013). Genes and mutations causing retinitis pigmentosa. Clin Genet 84:132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Meindl A, Dry K, Herrmann K, Manson E, Ciccodicola A, Edgar A, Carvalho MRS, Achatz H, Hellebrand H, et al. (1996). A gene (RPGR) with homology to the RCC1 guanine nucleotide exchange factor is mutated in X-linked retinitis pigmentosa (RP3). Nat Genet 13:35–42. [DOI] [PubMed] [Google Scholar]
- 4. Roepman R, Bauer D, Rosenberg T, Van Duijnhoven G, Van De Vosse E, Platzer M, Rosenthal A, Ropers HH, Cremers FPM and Berger W. (1996). Identification of a gene disrupted by a microdeletion in a patient with X-linked retinitis pigmentosa (XLRP). Hum Mol Genet 5:827–833. [DOI] [PubMed] [Google Scholar]
- 5. Roepman R, Van Duijnhoven G, Rosenberg T, APinckers JLG, Bleeker-Wagemakers LM, Bergen AAB, Post J, Beck A, Reinhardt R, et al. (1996). Positional cloning of the gene for X-linked retinitis pigmentosa 3: homology with the guanine-nucleotide-exchange factor RCC1. Hum Mol Genet 5:1035–1041. [DOI] [PubMed] [Google Scholar]
- 6. Petrs-Silva H and Linden R. (2013). Advances in gene therapy technologies to treat retinitis pigmentosa. Clin Ophthalmol 8:127–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hong DH, Pawlyk B, Sokolov M, Strissel KJ, Yang J, Tulloch B, Wright AF, Arshavsky VY and Li T. (2003). RPGR isoforms in photoreceptor connecting cilia and the transitional zone of motile cilia. Invest Ophthalmol Vis Sci 44:2413–2421. [DOI] [PubMed] [Google Scholar]
- 8. Khanna H, Hurd TW, Lillo C, Shu X, Parapuram SK, He S, Akimoto M, Wright AF, Margolis B, et al. (2005). RPGR-ORF15, which is mutated in retinitis pigmentosa, associates with SMC1, SMC3, and microtubule transport proteins. J Biol Chem 280:3580–33587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Khanna H. (2018). More than meets the eye: current understanding of RPGR function. Adv Exp Med Biol 1074:521–538. [DOI] [PubMed] [Google Scholar]
- 10. Kirschner R, Rosenberg T, Schultz-Heienbrok R, Lenzner S, Feil S, Roepman R, FCremers PMM, Ropers HH and Berger W. (1999). RPGR transcription studies in mouse and human tissues reveal a retina-specific isoform that is disrupted in a patient with X-linked retinitis pigmentosa. Hum Mol Genet 8:1571–1578. [DOI] [PubMed] [Google Scholar]
- 11. Vervoort R, Lennon A, Bird AC, Tulloch B, Axton R, Miano MG, Meindl A, Meitinger T, Ciccodicola A and Wright AF. (2000). Mutational hot spot within a new RPGR exon in X-linked retinitis pigmentosa. Nat Genet 25:462–466. [DOI] [PubMed] [Google Scholar]
- 12. He S, Parapuram SK, Hurd TW, Behnam B, Margolis B, Swaroop A and Khanna H. (2008). Retinitis Pigmentosa GTPase Regulator (RPGR) protein isoforms in mammalian retina: insights into X-linked Retinitis Pigmentosa and associated ciliopathies. Vision Res 48:366–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Buskin A, Zhu L, Chichagova V, Basu B, Mozaffari-Jovin S, Dolan D, Droop A, Collin J, Bronstein R, et al. (2018). Disrupted alternative splicing for genes implicated in splicing and ciliogenesis causes PRPF31 retinitis pigmentosa. Nat Commun 9:4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schmid F, Glaus E, Cremers FPM, Kloeckener-Gruissem B, Berger W and Neidhardt J. (2010). Mutation-and tissue-specific alterations of RPGR transcripts. Invest Ophthalmol Vis Sci 51:1628–1635. [DOI] [PubMed] [Google Scholar]
- 15. Neidhardt J, Glaus E, Barthelmes D, Zeitz C, Fleischhauer J and Berger W. (2007). Identification and characterization of a novel RPGR isoform in human retina. Hum Mutat 28:797–807. [DOI] [PubMed] [Google Scholar]
- 16. Carvill GL and Mefford HC. (2020). Poison exons in neurodevelopment and disease. Curr Opin Genet Dev 65:98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moreno-Leon L, West EL, O'Hara-Wright M, Li L, Nair R, He J, Anand M, Sahu B, Chavali VRM, et al. (2021). RPGR isoform imbalance causes ciliary defects due to exon ORF15 mutations in X-linked retinitis pigmentosa (XLRP). Hum Mol Genet 29:3706–3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang Q, Giacalone JC, Searby C, Stone EM, Tucker BA and Sheffield VC. (2019). Disruption of RPGR protein interaction network is the common feature of RPGR missense variations that cause XLRP. Proc Natl Acad Sci USA 116:1353–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cehajic-Kapetanovic J, Xue K, Martinez-Fernandez de la Camara C, Nanda A, Davies A, Wood LJ, Salvetti AP, Fischer MD, Aylward JW, et al. (2020). Initial results from a first-in-human gene therapy trial on X-linked retinitis pigmentosa caused by mutations in RPGR. Nat Med 26:354–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Beltran WA, Cideciyan AV, Lewin AS, Iwabe S, Khanna H, Sumaroka A, Chiodo VA, Fajardo DS, Román AJ, et al. (2012). Gene therapy rescues photoreceptor blindness in dogs and paves the way for treating human X-linked retinitis pigmentosa. Proc Natl Acad Sci USA 109:2132–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sahel JA, Marazova K and Audo I. (2015). Clinical characteristics and current therapies for inherited retinal degenerations. Cold Spring Harb Perspect Med 5:1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu Z, Hiriyanna S, Qian H, Mookherjee S, Campos MM, Gao C, Fariss R, Sieving PA, Li T, et al. (2015). A long-term efficacy study of gene replacement therapy for RPGR-associated retinal degeneration. Hum Mol Genet 24:3956–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martone J, De Angelis FG and Bozzoni I. (2012). U1 snRNA as an effective vector for stable expression of antisense molecules and for the inhibition of the splicing reaction. Methods Mol Biol 867:239–257. [DOI] [PubMed] [Google Scholar]
- 24. Siva K, Covello G and Denti MA. (2014). Exon-skipping antisense oligonucleotides to correct missplicing in neurogenetic diseases. Nucleic Acid Ther 24:69–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guiro J and O'Reilly D. (2015). Insights into the U1 small nuclear ribonucleoprotein complex superfamily. Wiley Interdiscip Rev RNA 6:79–92. [DOI] [PubMed] [Google Scholar]
- 26. Denti MA, Incitti T, Sthandier O, Nicoletti C, De Angelis FG, Rizzuto E, Auricchio A, Musarò A and Bozzoni I. (2008). Long-term benefit of adeno-associated virus/antisense-mediated exon skipping in dystrophic mice. Hum Gene Ther 19:601–608. [DOI] [PubMed] [Google Scholar]
- 27. Ferri L, Covello G, Caciotti A, Guerrini R, Denti MA and Morrone. A (2016). Double-target Antisense U1snRNAs Correct Mis-splicing Due to c.639 + 861C>T and c.639 + 919G>A GLA deep intronic mutations. Mol Ther Nucleic Acids 5:e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Incitti T, De Angelis FG, Cazzella V, Sthandier O, Pinnarò C, Legnini I and Bozzoni I. (2010). Exon skipping and duchenne muscular dystrophy therapy: selection of the most active U1 snRNA antisense able to induce dystrophin exon 51 skipping. Mol Ther 18:1675–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Denti MA, Rosa A, D'Antona G, Sthandier O, De Angelis FG, Nicoletti C, Allocca M, Pansarasa O, Parente V, et al. (2006). Body-wide gene therapy of Duchenne muscular dystrophy in the mdx mouse model. Proc Natl Acad Sci USA 103:3758–3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hartmann L, Neveling K, Borkens S, Schneider H, Freund M, Grassman E, Theiss S, Wawer A, Burdach S, et al. (2010). Correct mRNA processing at a mutant TT splice donor in FANCC ameliorates the clinical phenotype in patients and is enhanced by delivery of suppressor U1 snRNAs. Am J Hum Genet 87:480–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reese MG, Eeckman FH, Kulp D and Haussler D. (1997). Improved splice site detection in Genie. J Comput Biol 4:311–323. [DOI] [PubMed] [Google Scholar]
- 32. Desmet F-O, Hamroun D, Lalande M, Collod-Béroud G, Claustres M and Béroud C. (2009). Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 37:e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cartegni L, Wang J, Zhu Z, Zhang MQ and Krainer AR. (2003). ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acid Res 31:3568–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Piva F, Giulietti M, Nocchi L and Principato G. (2009). SpliceAid: a database of experimental RNA target motifs bound by splicing proteins in humans. Bioinformatics 25:1211–1213. [DOI] [PubMed] [Google Scholar]
- 35. Denti MA, Rosa A, D'Antona G, Sthandier O, De Angelis FG, Nicoletti C, Allocca M, Pansarasa O, Parente V, et al. (2006). Chimeric adeno-associated virus/antisense U1 small nuclear RNA effectively rescues dystrophin synthesis and muscle function by local treatment of mdx mice. Hum Gene Ther 17:565–574. [DOI] [PubMed] [Google Scholar]
- 36. Auricchio A, Kobinger G, Anand V, Hildinger M, O'Connor E, Maguire AM, Wilson JM and Bennett J. (2001). Exchange of surface proteins impacts on viral vector cellular specificity and transduction characteristics: the retina as a model. Hum Mol Genet 10:3075–3081. [DOI] [PubMed] [Google Scholar]
- 37. Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M and Turner DH (2004). Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci USA 101:7287–7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lorenz R, Bernhart SH, Höner zu Siederdissen C, Tafer H, Flamm C, Stadler PF and Hofacker. IL (2011). ViennaRNA Package 2.0. Algorithms Mol Biol 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mathews DH, Burkard ME, Freier SM, Wyatt JR and Turner DH. (1999). Predicting oligonucleotide affinity to nucleic acid targets. RNA 5:1458–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Covello G, Siva K, Adami V and Denti MA. (2014). An electroporation protocol for efficient DNA transfection in PC12 cells. Cytotechnology 66:543–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fraile-Bethencourt E, Valenzuela-Palomo A, Díez-Gómez B, Caloca MJ, Gómez-Barrero S and Velasco. EA (2019). Minigene Splicing Assays Identify 12 Spliceogenic Variants of BRCA2 Exons 14 and 15. Front Genet 10:503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Garanto A, Duijkers L and Collin RW. (2015). Species-dependent splice recognition of a cryptic exon resulting from a recurrent intronic CEP290 mutation that causes congenital blindness. Int J Mol Sci 16:5285–5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Marone M, Mozzetti S, De Ritis D, et al. (2001). Semiquantitative RT-PCR analysis to assess the expression levels of multiple transcripts from the same sample. Biol Proced Online 3:19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Meadus WJ. (2003). A semi-quantitative RT-PCR method to measure the in vivo effect of dietary conjugated linoleic acid on porcine muscle PPAR gene expression. Biol Proced Online 5:20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dignam JD, Lebovitz RM and Roeder RG. (1983). Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11:1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Keene JD, Komisarow JM and Friedersdorf MB. (2006). RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc 1:302–307. [DOI] [PubMed] [Google Scholar]
- 47. Zhu J, Mayeda A and Krainer AR. (2001). Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol Cell 8:1351–1361. [DOI] [PubMed] [Google Scholar]
- 48. DuBridge RB, Tang P, Hsia HC, Leong PM, Miller JH and Calos MP. (1987). Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol Cell Biol 7:379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shaw G, Morse S, Ararat M and Graham FL. (2002). Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J 16:869–871. [DOI] [PubMed] [Google Scholar]
- 50. Greene LA and Tischler AS. (1976). Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc Natl Acad Sci USA 73:2424–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tan E, Ding XQ, Saadi A, Agarwal N, Naash MI and Al-Ubaidi MR. (2004). Expression of cone-photoreceptor-specific antigens in a cell line derived from retinal tumors in transgenic mice. Invest Ophthalmol Vis Sci 45:764–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wheway G, Nazlamova L, Turner D and Cross S (2019). 661W photoreceptor cell line as a cell model for studying retinal ciliopathies. Front Genet 10:308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dhir A and Buratti E. (2010). Alternative splicing: role of pseudoexons in human disease and potential therapeutic strategies: minireview. FEBS J 277:841–855. [DOI] [PubMed] [Google Scholar]
- 54. Cooper TA, Wan L and Dreyfuss G. (2009). RNA and Disease. Cell 136:777–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lewandowska MA. (2013). The missing puzzle piece: splicing mutations. Int J Clin Exp Pathol 6:2675–2682. [PMC free article] [PubMed] [Google Scholar]
- 56. Tazi J, Bakkour N and Stamm S. (2009). Alternative splicing and disease. Biochim Biophys Acta 1792:14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cooper TA. (2005). Use of minigene systems to dissect alternative splicing elements. Methods 37:331–340. [DOI] [PubMed] [Google Scholar]
- 58. Fu XD and Maniatis T. (1992). The 35-kDa mammalian splicing factor SC35 mediates specific interactions between U1 and U2 small nuclear ribonucleoprotein particles at the 3’ splice site. Proc Natl Acad Sci U S A 89:1725–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fu XD, Mayeda A, Maniatis T and Krainer AR. (1992). General splicing factors SF2 and SC35 have equivalent activities in vitro, and both affect alternative 5’ and 3’ splice site selection. Proc Natl Acad Sci USA 89:11224–11228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Grossman JS, Meyer MI, Wang YC, Mulligan GJ, Kobayashi R and Helfman DM. (1998). The use of antibodies to the polypyrimidine tract binding protein (PTB) to analyze the protein components that assemble on alternatively spliced pre-mRNAs that use distant branch points. RNA 4:613–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tisserant A and König H. (2008). Signal-regulated Pre-mRNA occupancy by the general splicing factor U2AF. PLoS One 3:e1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Di Fruscio M, Chen T and Richard S. (1999). Characterization of Sam68-like mammalian proteins SLM-1 and SLM-2: SLM-1 is a Src substrate during mitosis. PNAS 96:2710–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Venables JP, Vernet C, Chew SL, Elliott DJ, Cowmeadow RB, Wu J, Cooke HJ, Artzt K and Eperon IC. (1999). T-STAR/ETOILE: a novel relative of SAM68 that interacts with an RNA-binding protein implicated in spermatogenesis. Hum Mol Genet 8:959–969. [DOI] [PubMed] [Google Scholar]
- 64. Iijima T, Wu K, Witte H, Hanno-Iijima Y, Glatter T, Richard S and Scheiffele P. (2011). SAM68 regulates neuronal activity-dependent alternative splicing of neurexin-1. Cell 147:1601–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Iijima T, Iijima Y, Witte H and Scheiffele P. (2014). Neuronal cell type-specific alternative splicing is regulated by the KH domain protein SLM1. J Cell Biol 204:331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Stoss O, Olbrich M, Hartmann AM, Konig H, Memmott J, Andreadis A and Stamm S. (2001). The STAR/GSG family protein rSLM-2 regulates the selection of alternative splice sites. J Biol Chem 276:8665–8673. [DOI] [PubMed] [Google Scholar]
- 67. Traunmüller L, Bornmann C and Scheiffele P. (2014). Alternative splicing coupled nonsense-mediated decay generates neuronal cell type-specific expression of SLM proteins. J Neurosci 34:16755–16761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gaildrat P, Killian A, Martins A, Tournier I, Frébourg T and Tosi M. (2010). Use of splicing reporter minigene assay to evaluate the effect on splicing of unclassified genetic variants. Methods Mol Biol 653:249–257. [DOI] [PubMed] [Google Scholar]
- 69. Kar A, Fushimi K, Zhou X, Ray P, Shi C, Chen X, Liu Z, Chen S and Wu JY. (2011). RNA Helicase p68 (DDX5) Regulates tau Exon 10 Splicing by Modulating a Stem-Loop Structure at the 5’ Splice Site. Mol Cell Biol 31:1812–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. De Angelis FG, Sthandier O, Berarducci B, Toso S, Galluzzi G, Ricci E, Cossu G and Bozzoni I. (2002). Chimeric snRNA molecules carrying antisense sequences against the splice junctions of exon 51 of the dystrophin pre-mRNA induce exon skipping and restoration of a dystrophin synthesis in Δ48–50 DMD cells. Proc Natl Acad Sci USA 99:9456–9461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Pinotti M, Rizzotto L, Balestra D, Lewandowska MA, Cavallari N, Marchetti G, Bernardi F and Pagani F. (2008). U1-snRNA mediated rescue of mRNA processing in severe factor VII deficiency. Blood 111:2681–2684. [DOI] [PubMed] [Google Scholar]
- 72. Glaus E, Schmid F, Da Costa R, Berger W and Neidhardt J. (2011). Gene therapeutic approach using mutation-adapted U1 snRNA to correct a RPGR Splice defect in patient-derived cells. Mol Ther 19:936–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sánchez-Alcudia R, Pérez B, Pérez-Cerdá C, Ugarte M and Desviat LR. (2011). Overexpression of adapted U1snRNA in patients' cells to correct a 5′ splice site mutation in propionic acidemia. Mol Genet Metab 102:134–138. [DOI] [PubMed] [Google Scholar]
- 74. Freund M, Hicks MJ, Konermann C, Otte M, Hertel KJ and Schaal H. (2005). Extended base pair complementarity between U1 snRNA and the 5′ splice site does not inhibit splicing in higher eukaryotes, but rather increases 5′ splice site recognition. Nucleic Acids Res 33:5112–5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gazzoli I, Pulyakhina I, Verwey NE, Ariyurek Y, Laros JF, PA ‘t Hoen and Aartsma-Rus A. (2016). Non-sequential and multi-step splicing of the dystrophin transcript. RNA Biol 13:290–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Jin Z-B, Okamoto S, Osakada F, Homma K, Assawachananont J, Hirami Y, Iwata T and Takahashi. M (2011). Modeling retinal degeneration using patient-specific induced pluripotent stem cells. PLoS One 6:e17084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ikeda H, Osakada F, Watanabe K, Mizuseki K, Haraguchi T, Miyoshi H, Kamiya D, Honda Y, Sasai N and Yoshimura N. (2005). Generation of Rx(+)/Pax(6+) neural retinal precursors from embryonic stem cells. Proc Natl Acad Sci USA 102:11331–11336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Osakada F, Jin ZB, Hirami Y, Ikeda H, Danjyo T, Watanabe K, Sasai Y and Takahashi M. (2009). In vitro differentiation of retinal cells from human pluripotent stem cells by small-molecule induction. J Cell Sci 122:3169–3179. [DOI] [PubMed] [Google Scholar]
- 79. Eiraku M, Takata N, Ishibashi H, Kawada M, Sakakura E, Okuda S, Sekiguchi K, Adachi T and Sasai Y. (2011). Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 472:51–56. [DOI] [PubMed] [Google Scholar]
- 80. Zhong X, Gutierrez C, Xue T, Hampton C, Vergara MN, Cao L-H, Peters A, Park TS, Zambidis ET and Meyer. JS (2014). Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat Commun 5:4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Deng WL, Gao ML, Lei XL, Lv JN, Zhao H, He KW, Xia XX, Li LY, Chen YC, Li YP, Pan D, Xue T and Jin ZB. (2018). Gene correction reverses ciliopathy and photoreceptor loss in iPSC-derived retinal organoids from retinitis pigmentosa patients. Stem Cell Reports 10:1267–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Gladman JT, Bebee TW, Edwards C, Wang X, Sahenk Z, Rich MM and Chandler DS. (2010). A humanized SMN gene containing the SMN2 nucleotide alteration in exon 7 mimics SMN2 splicing and the SMA disease phenotype. Hum Mol Genet 19:4239–4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Aartsma-Rus A and van Putten M. (2020). The use of genetically humanized animal models for personalized medicine approaches. Dis Model Mech 13:dmm041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hims MM, Shetty RS, Pickel J, Mull J, Leyne M, Liu L, Gusella JF and Slaugenhaupt SA. (2007). A humanized IKBKAP transgenic mouse models a tissue-specific human splicing defect. Genomics 90:389–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Baker O, Tsurkan S, Fu J, Klink B, Rump A, Obst M, Kranz A, Schröck E, Anastassiadis K and Stewart AF. (2017). The contribution of homology arms to nuclease-assisted genome engineering. Nucleic Acids Res 45:8105–8115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Eilers W and Denti MA. (2021). Delivery of antisense RNA therapeutics: turning negative results into a positive development. Nucleic Acid Ther 31:183–184. [DOI] [PubMed] [Google Scholar]
- 87. Godfrey C, Desviat LR, Smedsrød B, Piétri-Rouxel F, Denti MA, Disterer P, Lorain S, Nogales-Gadea G, Sardone V, Anwar R, El Andaloussi S, Lehto T, Khoo B, Brolin C, van Roon-Mom WM, Goyenvalle A, Aartsma-Rus A and Arechavala-Gomeza V. (2017). Delivery is key: lessons learnt from developing splice-switching antisense therapies. EMBO Mol Med 9:545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Hammond SM, Aartsma-Rus A, Alves S, Borgos SE, Buijsen RAM, Collin RWJ, Covello G, Denti MA, Desviat LR, Echevarría L, Foged C, Gaina G, Garanto A, Goyenvalle AT, Guzowska M, Holodnuka I, Jones DR, Krause S, Lehto T, Montolio M, Van Roon-Mom W and Arechavala-Gomeza V. (2021). Delivery of oligonucleotide-based therapeutics: challenges and opportunities. EMBO Mol Med 13:e13243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Peters CW, Maguire CA and Hanlon KS. (2021). Delivering AAV to the central nervous and sensory systems. Trends Pharmacol Sci 42:461–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Peng Y, Tang L and Zhou Y. (2017). Subretinal injection: a review on the novel route of therapeutic delivery for vitreoretinal diseases. Ophthalmic Res 58:217–226. [DOI] [PubMed] [Google Scholar]
- 91. Ghazi NG, Abboud EB, Nowilaty SR, Alkuraya H, Alhommadi A, Cai H, Hou R, Deng WT, Boye SL, et al. (2016). Treatment of retinitis pigmentosa due to MERTK mutations by ocular subretinal injection of adeno-associated virus gene vector: results of a phase I trial. Hum Genet 135:327–343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.