Abstract
The β-lactamase inhibitor clavulanic acid is formed by condensation of a pyruvate-derived C3 unit with a molecule of arginine. A gene (pyc, for pyruvate converting) located upstream of the bls gene in the clavulanic acid gene cluster of Streptomyces clavuligerus encodes a 582-amino-acid protein with domains recognizing pyruvate and thiamine pyrophosphate that shows 29.9% identity to acetohydroxyacid synthases. Amplification of the pyc gene resulted in an earlier onset and higher production of clavulanic acid. Replacement of the pyc gene with the aph gene did not cause isoleucine-valine auxotrophy in the mutant. The pyc replacement mutant did not produce clavulanic acid in starch-asparagine (SA) or in Trypticase soy broth (TSB) complex medium, suggesting that the pyc gene product is involved in the conversion of pyruvate into the C3 unit of clavulanic acid. However, the β-lactamase inhibitor was still formed at the same level as in the wild-type strain in defined medium containing d-glycerol, glutamic acid, and proline (GSPG medium) as confirmed by high-pressure liquid chromatography and paper chromatography. The production of clavulanic acid by the replacement mutant was dependent on addition of glycerol to the medium, and glycerol-free GSPG medium did not support clavulanic acid biosynthesis, suggesting that an alternative gene product catalyzes the incorporation of glycerol into clavulanic acid in the absence of the Pyc protein. The pyc replacement mutant overproduces cephamycin.
Clavulanic acid, a clinically used β-lactamase inhibitor, is synthesized by condensation of arginine (25, 29) with a C3 unit derived from pyruvate or glycerate. Incorporation of labelled pyruvate or glycerate into clavulanic acid has been reported (9, 28), but the nature of the intermediate substrate that binds to the amino group of arginine remains uncertain. Gutman et al. (12) proposed an intact incorporation of β-hydroxypropionate into the C3 unit based on the unchanged 3H/14C ratio in clavulanic acid following incorporation of double-labelled [2-3H, 2-14C]hydroxypropionate, which suggests that no tritium is lost from the C-2 methylene group during incorporation of the C3 unit. The oxidation levels of the carbon atoms of β-hydroxypropionate are the same as those in clavulanic acid.
Townsend and Ho showed that glycerate is a precursor of the C3 unit (28), but the use of glycerate as the intermediate precursor of the C3 unit would require removal of the hydroxyl group at carbon 2 of the glycerate, which is not present in the β-lactam ring of clavulanic acid. Feeding experiments using racemic [1,2-13C, 2,3,3,-2H] glycerate showed that no incorporation of the hydrogen at C-2 of glycerate into the C-6 of clavulanic acid occurred (23).
Recently Thirkettle et al. (27) proposed that pyruvate (and not glycerate) is the most likely primary metabolic source of the three β-lactam carbons of clavulanic acid. However, the reactions required to convert pyruvate to the C3 unit remain unknown. The utilization of pyruvate as primary precursor does not exclude the involvement of β-hydroxypropionate as the final intermediate in the condensation reaction.
The genes for clavulanic acid biosynthesis are clustered together with the genes for cephamycin C biosynthesis, forming the so-called β-lactam supercluster (14, 30). Several genes of the cluster (cas2, bls, pah, car, and claR) have been assigned specific biosynthesis roles (1, 2, 17, 19, 22), but the genes encoding the formation of the C3 precursor and the initial condensation step that results in formation of the N-carboxyethylarginine intermediate (10) remain to be identified. Since the genes involved in precursor formation are sometimes associated with β-lactam biosynthesis clusters (18), it is of great interest to characterize the genes involved in formation and/or condensation of the C3 unit.
We have identified a gene, pyc, that encodes a protein ressembling acetohydroxyacid synthases and appears to be involved in the conversion of pyruvate into the C3 unit of clavulanic acid. Inactivation of this gene leads to the inability to produce clavulanic acid in starch-asparagine medium but not in GSPG (glycerol, glutamic acid, proline) medium, suggesting that an alternative pathway bypasses the conversion of pyruvate into the C3 unit.
MATERIALS AND METHODS
Bacterial strains, plasmids, and DNA procedures.
Streptomyces clavuligerus ATCC 27064 and the disrupted mutants obtained in this work were grown in GSPG medium (24), SA (starch-asparagine) medium (19), and TSB (Trypticase soy broth) liquid cultures as described previously (24). GSP medium is GSPG medium without glycerol; SAG medium is SA medium supplemented with 15 g of glycerol per liter. All fermentation experiments were repeated twice, using triplicate flasks. Data are the means ± standard deviation (SD) for duplicate bioassays in triplicate flasks. Defined LAT minimal medium (15) was used to test the auxotrophy of the disrupted mutants.
Plasmids pBSSK(+) and pBSKS(+) were obtained from Stratagene. Plasmid pHZ1351, used for disruption experiments, was provided by Z. Deng (Wuham, China). Streptomyces-Escherichia coli bifunctional vector pULVK99 (6) was used to express the pyc gene in high copy number. DNA manipulations, restriction endonuclease digestions, ligations, and E. coli transformations were performed according to standard procedures (26).
Southern blot hybridization.
For Southern hybridizations, 10 μg of digested DNA of each strain was used. The probes were labelled with a Dig-High Prime kit (Boehringer Mannheim), and 0.2 μg of denatured probe was used in the hybridizations, using the conditions suggested by the manufacturer.
Gene disruption and gene replacement.
Two different strategies were followed. (i) The 2.1-kb EcoRI-BglII insert present in plasmid pBSKS2.1 was digested with BstEII to linearize the plasmid. The BstEII site is located 1,534 nucleotides (nt) from the 5′ end and 215 nt from the 3′ end of open reading frame 2 (ORF2). The BstEII cohesive ends were filled, and a 1.3-kb DNA fragment containing the aph gene was inserted into the gap, with the orientation of the aph gene opposite that of ORF2; the DNA was then circularized. This construction was subcloned as a XhoI-SpeI 3.4-kb DNA fragment into BamHI-digested pHZ1351 (containing an unstable replication origin [3]), which was transformed into S. clavuligerus. Using standard procedures (21), we found that 50% of the transformants were kanamycin resistant and thioestrepton sensitive after two steps of sporulation in antibiotic-free medium.
(ii) Alternatively, two oligonucleotides (O1 [5′TTGGATCCGGTTTCGCCGGGGTGTT3′, corresponding to nt 597–616 of ORF2] and O2 [5′TTGGATCCACCAGGTCATCGACTCCAT3′, corresponding to nt 1183 to 1205 of ORF2]) were used to amplify by PCR a BamHI-ended 4.5-kb DNA fragment containing the pBSKS(+) vector, 616 nt of the 5′ end of ORF2, and 566 nt of the 3′ end of ORF2. A 1.3-kb BamHI DNA fragment containing the aph gene was ligated to give a circular construction. Afterwards, a 2.8-kb SpeI-XhoI DNA fragment containing the aph gene flanked by the 5′ and 3′ ends of ORF2 was subcloned into pHZ1351 and used to transform S. clavuligerus. After one step of sporulation in antibiotic-free medium, 97% of the transformants were found to be kanamycin resistant and thiostrepton sensitive, indicating that chromosomal integration by recombination had occurred.
Clavulanic acid, cephamycin, and alanyl-clavam production.
Clavulanic acid was quantified by bioassay using Klebsiella pneumoniae ATCC 29665 as described previously (24). The presence of clavulanic acid in the broth was confirmed by derivatization with imidazole as described by Bird et al. (4) and analysis by high-pressure liquid chromatography (HPLC) using the conditions indicated by Foulstone and Reading (11); under those conditions, clavulanic acid eluted with a retention time of 5.5 min. Additionally, clavulanic acid was identified by paper chromatography using Whatman chromatography paper (Whatman International Ltd.). The samples were developed in acetonitrile–n-propanol–H2O (1:1:1), and the clavulanic acid spot was identified by bioautography using K. pneumoniae. Under these conditions, pure clavulanic acid has an Rf of 0.67. Cephamycin C and alanyl-clavam were quantified by standard bioassay using E. coli ESS 22–31 and Bacillus strain ATCC 27860, respectively, as described previously (22, 25). Growth was estimated as cell DNA content, using salmon sperm DNA as the control as described by Burton (5).
Sequence analysis.
Sequence analysis was done with the Editseq and Geneplot programs (DNAStar). The FASTA3 program was used to search the Swiss Protein database, and the Prosite program was used to look for specific motifs in the proteins. Alignment of proteins was done with the Clustal V program (DNAStar).
Nucleotide sequence accession number.
The nucleotide sequence of pyc has been deposited in the EMBL database under accession no. AJ238211.
RESULTS
Cloning and sequencing of ORF2.
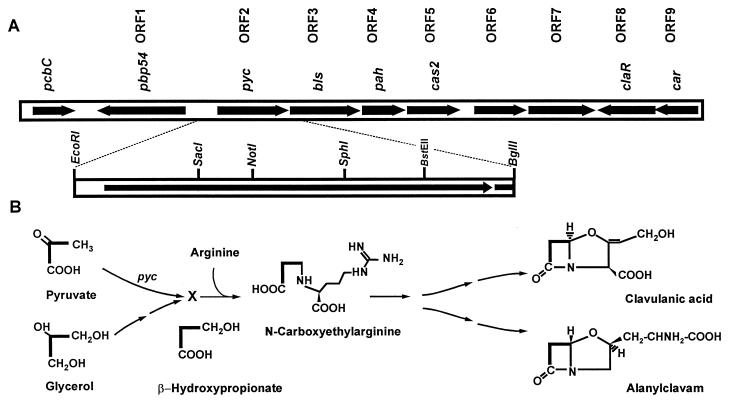
Using DNA from λ phage Φ-AR3, which contains a 15-kb insert of S. clavuligerus DNA of the region downstream of the pcbC gene (21), a 2.1-kb EcoRI-BglII fragment, located downstream of the pbp54 gene (ORF1 in Fig. 1; also named pcbR [20]), was subcloned into vectors pBSSK(+) and pBSKS(+) and sequenced in both orientations. A complete ORF (ORF2; nt 145 to 1893) located 315 nt upstream of pbp54 and an incomplete ORF (ORF3) downstream of ORF2 were found in the DNA fragment. ORF3 corresponds to the gene bls described by Bachmann et al. (2), encoding the β-lactam synthetase. ORF2 has 1,758 nt and a G+C content of 68.8% (Fig. 1). The presence of the pbp54 gene upstream and in the opposite direction to ORF2 and the positive expression of ORF2 in plasmid pVKBE2.1, a pVK99-derived plasmid containing a 2.1-kb EcoRI-BglII fragment carrying ORF2, indicates that a promoter is present in the 143 nt upstream of ORF2.
FIG. 1.
(A) Clavulanic acid gene cluster of S. clavuligerus. The restriction map of the region corresponding to ORF2 (pyc) has been amplified. (B) Proposed pathway for conversion of pyruvate to the C3 unit of clavulanic acid. Pyruvate is converted to an intermediate, X, by the pyc gene product. X may correspond to β-hydroxypropionate.
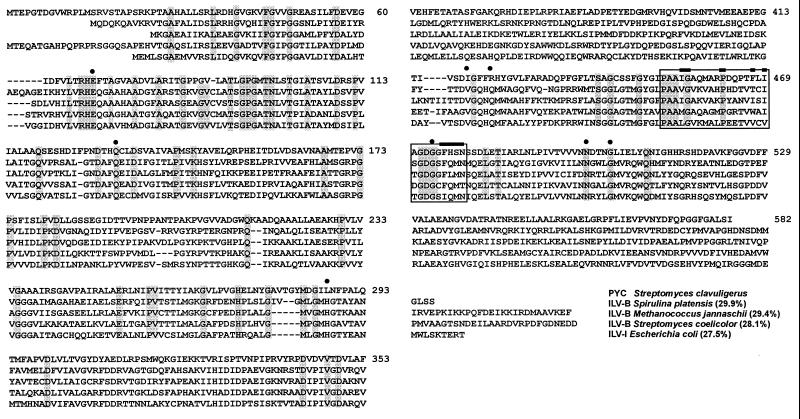
The amino acid sequence encoded by ORF2 has a significant similarity to acetohydroxyacid synthases (Fig. 2), with identical amino acids conserved in specific motifs along the sequence. The highest degree of similarity was with the ILVA-B protein (the acetolactate synthase large subunit) of Spirulina platensis (29.9% amino acid identity), but similar identities were found with the homologous proteins of Streptomyces coelicolor (28.1%), Methanococcus jannaschii (29.4%), and E. coli (27.5%). Acetohydroxyacid synthases activate two pyruvate units forming acetolactic acid. Five of the eight amino acids that have been described to be conserved in the active center of acetohydroxyacid synthases (7) are present in the protein encoded by ORF2 (amino acids 69, 132, 472, 499, and 503 in Fig. 2). Also, a conserved motif characteristic of enzymes that require thiamine pyrophosphate (13), such as acetohydroxyacid synthases, is found at amino acid positions 452 to 478, which suggests that ORF2 encodes a protein that requires thiamine pyrophosphate and binds pyruvate. However, the protein is not an acetohydroxyacid synthase for primary metabolism (see below); the similarity between ILVA-B proteins is usually higher (i.e., the ILVA-B proteins of S. coelicolor and M. jannaschii are 43.2% identical) than that observed with the ORF2-encoded protein.
FIG. 2.
Comparison of the amino acid sequence encoded by pyc with sequences of the large subunit of acetohydroxyacid synthases of Spirulina platensis (P27868), M. jannaschii (Q57725), S. coelicolor (E1390257), and E. coli (P00893). Shaded boxes contain identical amino acids. Amino acids which form part of the active center of acetohydroxyacid synthase (7) are indicated by dots. The box extending from amino acids 452 to 478 corresponds to the conserved thiamine pyrophosphate binding motif, with the consensus amino acids in the motif indicated with bars.
Disruption of the gene encoded by ORF2 by insertion or replacement does not lead to isoleucine-valine auxotrophy.
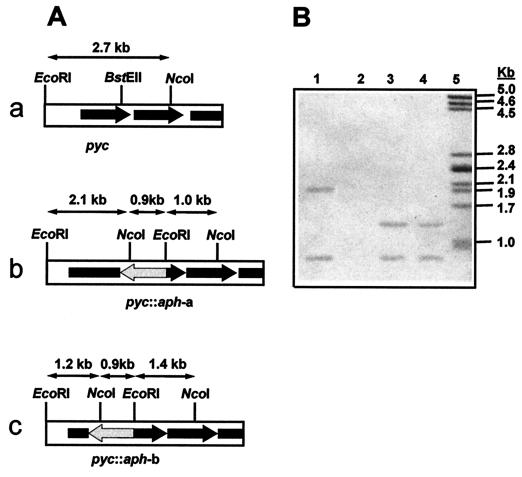
To elucidate the role of the protein encoded by ORF2 in cephamycin C and/or clavulanic acid biosynthesis, ORF2 was disrupted. Two types of disrupted mutants were obtained: (i) S. clavuligerus ORF2::aph-a, which has a 1.3-kb aph cassette inserted at position 1534 of the gene; and (ii) S. clavuligerus ORF2::aph-b, which contains the 1.3-kb insertion cassette in place of nt 760 to 1328 of ORF2.
Both disrupted mutants gave hybridizing bands of the expected size with the aph probe (Fig. 3A). S. clavuligerus ORF2::aph-a gave bands of 0.9 and 2.1 kb (lane 1), and S. clavuligerus ORF2::aph-b showed bands of 0.9 and 1.4 kb (Fig. 3B, lanes 3 and 4); the wild-type strain gave no hybridization (lane 2). With the ORF2 probe, the S. clavuligerus ORF2::aph-a mutant gave bands of 2.1 and 1.0 kb, and the S. clavuligerus ORF2::aph-b strain showed hybridization bands of 1.2 and 1.4 kb; the wild-type strain gave a single 2.7-kb hybridization band (not shown).
FIG. 3.
(A) Scheme of the DNA region around the pyc gene in wild-type S. clavuligerus 27064 (a), disrupted mutant S. clavuligerus pyc::aph-a (b), and replacement mutant S. clavuligerus pyc::aph-b (c). (B) Hybridization of EcoRI-NcoI-digested DNA with the 1.3-kb BamHI probe containing the aph gene. Lanes: 1, S. clavuligerus pyc::aph-a; 2, S. clavuligerus 27064, 3 and 4, S. clavuligerus pyc::aph-b; 5, size marker (λ phage digested with PstI).
Acetohydroxyacid synthases catalyze the first step of the isoleucine-valine pathway. The two disrupted mutants, S. clavuligerus ORF2::aph-a and S. clavuligerus ORF2::aph-b, were tested for isoleucine and valine requirement in LAT minimal medium with and without 50 μg of each of those amino acids per ml. Both disrupted mutants were prototrophs and grew well in the absence of amino acid supplementation. Growth did not improve in isoleucine-valine-supplemented plates. However the presence of a pyruvate-recognizing active center in the deduced protein indicated that ORF2 encodes a pyruvate-converting enzyme, and the gene has been named pyc.
Replacement of pyc blocks clavulanic acid production except in GSPG medium.
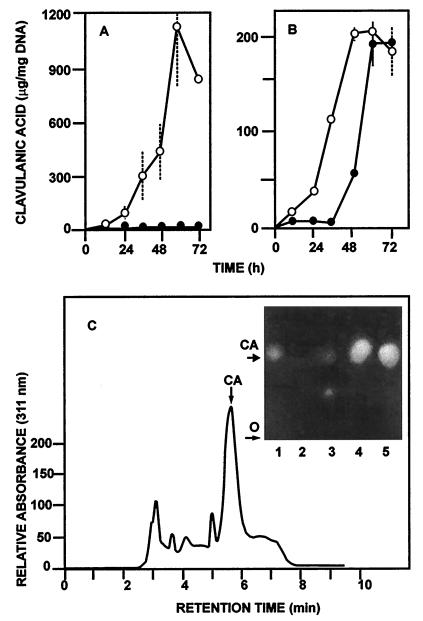
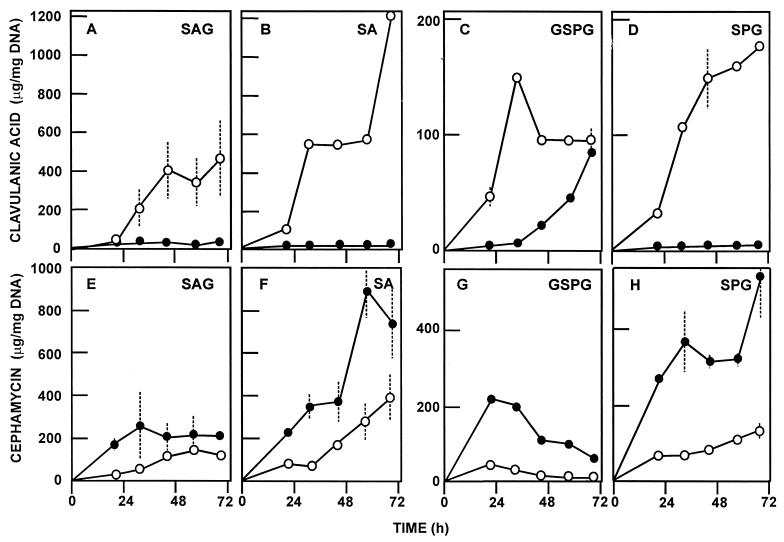
Clavulanic acid production by both strains was tested in solid MEY and Trypticase soy agar media. S. clavuligerus pyc::aph-a (containing the aph gene inserted in the 3′ region of pyc) produced the same levels of clavulanic acid as the wild-type strain. This suggests that the 72 amino acids which are lacking in this mutant are not essential for clavulanic acid biosynthesis. However, the replacement mutant S. clavuligerus pyc::aph-b did not produce clavulanic acid in either of these media. To further study the effect of the pyc deletion on the production of clavulanic acid, cephamycin, and alanyl-clavam, S. clavuligerus pyc::aph-b was grown in liquid cultures in either defined GSPG or SA medium or complex TSB medium. As expected, the replacement mutant did not produce clavulanic acid in either TSB medium (not shown) or SA medium (Fig. 4A). However, clavulanic acid was produced by this mutant in GSPG medium (see below); the onset of clavulanic acid biosynthesis was delayed 24 h in relation to the wild-type strain (Fig. 4B), but the level of the β-lactamase inhibitor at 72 h was up to 95% of that seen in the control S. clavuligerus strain. It seems that glycerol induces an alternative enzyme system for its conversion into the C3 unit of clavulanic acid.
FIG. 4.
Production of clavulanic acid by wild-type S. clavuligerus 27064 (○) and replacement mutant S. clavuligerus pyc::aph-b (●) in SA (A) and GSPG (B) media. (C) HPLC elution profile of the broth corresponding to 48 h of S. clavuligerus pyc::aph-b growth in GSPG medium shown in panel B. The arrow indicates the peak that coeluted with a sample of pure clavulanic acid (CA). (Inset) Bioautography of a paper chromatography of culture broths. Lanes: 1, pure clavulanic acid (Rf 0.67); 2 and 3, S. clavuligerus pyc::aph-b grown in SA medium; 4, S. clavuligerus 27064 grown in GSPG medium; 5, S. clavuligerus pyc::aph-b grown in GSPG medium. SD are given as discontinuous bars for the wild-type strain and as solid bars for the S. clavuligerus pyc::aph-b mutant.
To confirm that the antibiotic produced was clavulanic acid, several tests were performed: (i) bioassays were done with K. pneumoniae as the sensitive strain in the presence and absence of penicillin G, (ii) samples were subjected to paper chromatography and bioassayed with pure clavulanic acid as the control, and (iii) clavulanic acid was identified in the supernatants by HPLC chromatography. The results showed that the bioactivity was due to a β-lactamase inhibitory substance (since the inhibition zone decreased considerably in the absence of penicillin G in the bioassay) with an Rf of 0.67, identical to that found in broths of the wild-type strain and to that of the pure clavulanic acid (Fig. 4C, inset). A peak eluting with a retention time of 5.5 min that cochromatographed with pure clavulanic acid was found by HPLC in the GSPG-grown cultures of the S. clavuligerus pyc::aph-b mutant (Fig. 4C).
Alanyl-clavam, detected by bioassay and confirmed by methionine reversion, was produced by the wild-type strain in GSPG medium. The production of alanyl-clavam by the S. clavuligerus pyc::aph-b mutant was in the range of 45 to 66% of that of the control at different times of the culture.
Clavulanic acid production by the pyc-deleted mutant is dependent on the presence of glycerol.
The production of clavulanic acid in GSPG medium suggested the existence of a different gene involved in the formation of the C3 unit in the disrupted mutant. Therefore, clavulanic acid formation was tested in GSP medium (containing glutamate, sucrose, and proline) compared with GSPG medium, (which contains glycerol [15 g/liter]) as well as in SA medium compared with SAG medium (SA medium supplemented with glycerol [15 g/liter]). acid-specific production are lower than in SPG medium, which reflects an increase of growth due to the presence of glycerol in GSPG. However, as shown in Fig. 5C and D, clavulanic acid production by the pyc replacement mutant is dependent on the presence of glycerol in GSPG medium. The absence of glycerol in the medium dramatically decreased also the production of alanyl-clavam by both strains (to less than 2% of the control condition), suggesting that the alternative pathway that converts glycerol into the C3 unit may be directly related to clavam biosynthesis. Therefore, it seems that an alternative gene to pyc encodes an enzyme able to form the C3 unit from d-glycerol instead of pyruvate in GSPG medium.
FIG. 5.
Production of clavulanic acid (A to D) and cephamycin C (E to H) by S. clavuligerus 27064 (○) or replacement mutant S. clavuligerus pyc::aph-b (●) in the indicated culture media. SD are given as discontinuous bars.
Amplification of the pyc gene results in an earlier onset and higher production of clavulanic acid.
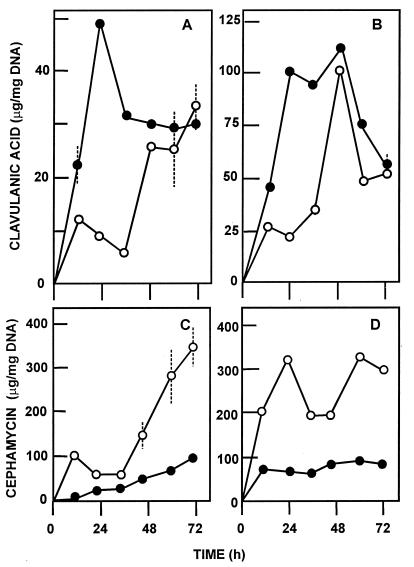
In parallel, the effect of pyc gene dosage on antibiotic production was tested by using multicopy plasmid pVKEB2.1 (9.9 kb). The higher pyc gene dosage in S. clavuligerus[pVKEB2.1] results in an earlier formation of clavulanic acid and higher levels of this compound than in the control strain, S. clavuligerus[pULVK99] (Fig. 6).
FIG. 6.
Effect of pyc gene amplification on clavulanic acid and cephamycin biosynthesis. Clavulanic acid (A and B) and cephamycin C (C and D) biosynthesis by S. clavuligerus [pULVK99] (○) and S. clavuligerus[pVKBE2.1] (●) in GSPG medium (A and C) or SA medium (B and D). SD are given as discontinuous bars.
The pyc-replacement mutant overproduces cephamycin.
Another interesting finding is the observation that the pyc replacement mutant is an overproducer of cephamycin in three different media, GSPG, SA (Fig. 5), and the soy meal-based TSB (not shown). In parallel, the amplification of pyc in S. clavuligerus[pVKBE2.1] resulted in lower production of cephamycin (Fig. 6C and D). The increase in cephamycin biosynthesis by the pyc deletion mutant might be explained by sparing pyruvate for use in the lysine (α-aminoadipic acid) pathway. Lysine is formed through the dihydrodipicolinic intermediate by condensation of pyruvate with the four-carbon aspartic acid semialdehyde. Alternatively, the Pyc protein may have a repressor effect on cephamycin biosynthesis that is lacking in the pyc replacement mutant. This hypothesis is being further investigated to establish if the transcripts (or enzymes) for the cephamycin gene cluster are enhanced in the pyc-disrupted mutant.
DISCUSSION
The cloned pyc gene shows 29.9% identity in specific motifs to acetohydroxyacid synthases, enzymes that condense two pyruvate units to form acetolactic acid. An S. clavuligerus strain disrupted in the pyc gene is prototrophic and does not require isoleucine or valine to grow, thus ruling out the possibility that the pyc gene encodes a functional acetohydroxyacid synthase. The conserved amino acids in pyruvate-binding motifs suggest, however, that pyruvate is a substrate of this enzyme.
The pyc gene product is required for clavulanic acid biosynthesis in the standard SA medium; this medium has been optimized for clavulanic acid production with very high yields of the β-lactamase inhibitor. We propose that the pyc gene is involved in the conversion of pyruvate to a hydroxyacid, presumably β-hydroxypropionate (Fig. 1). Genes for precursor formation are sometimes clustered with antibiotic biosynthesis genes (18). The pyc gene of the clavulanic acid cluster appears to play a role in formation of the C3 unit, just as the lysine-6-aminotransferase encoded by the lat gene of the cephamycin cluster is involved in formation of the α-aminoadipic acid precursor of cephamycin (8, 16).
An interesting finding is the observation that S. clavuligerus lacking the pyc gene is able to produce clavulanic acid in GSPG, a defined medium containing glycerol in addition to proline and glutamate (24). When glycerol was removed from this medium, clavulanic acid production by the replacement deletion mutant dropped to barely detectable levels. These results suggest that there is a gene(s) that encodes an enzyme able to convert glycerol into the C3 precursor independently from the pyc gene product. This alternative enzyme system is possibly repressed in starch- or soy meal-supplemented SA (Fig. 5A and B) or TSB medium. This result explains some of the previous confusion about the incorporation of C3 precursors into clavulanic acid. Incorporation of glycerol (9, 28), propionate (9), β-hydroxypropionate (12), pyruvate (23, 28), and d- and l-glycerate (28) has been reported. The reported incorporation of labelled precursors must have been dependent on the biosynthetic route for the C3 unit used in each growth medium.
Our results indicate that there are at least two different pathways involved in formation of C3 unit; the first one, encoded by the pyc gene, appears to use pyruvate as the substrate and is catalyzed by the acetohydroxyacid synthase-like pyc gene product. These observations support the recent proposal of Pitlik and Townsend (23) that pyruvate is the real precursor of the C3 unit. Interestingly, in defined medium containing glycerol, this three-carbon alcohol provides a good level of clavulanic acid production, suggesting that it is converted to the C3 unit. The parallel gene might be involved in the formation of the C3 unit for clavam biosynthesis since in the absence of glycerol, the formation of alanyl-clavam is drastically reduced in the wild-type strain and the pyc replacement mutant.
ACKNOWLEDGMENTS
This research was supported by grants from the CICYT, Madrid (BIO-96-0827), and from Antibióticos, S.A. (León), Spain. R. Pérez-Redondo received a fellowship from the University of León.
We thank Sonia Campoy and Luis M. Lorenzana for help in measuring clavulanic acid by HPLC and paper chromatography.
REFERENCES
- 1.Aidoo K A, Wong A, Alexander D C, Rittamer R A R, Jensen S E. Cloning, sequencing and disruption of a gene from S. clavuligerus involved in clavulanic acid biosynthesis. Gene. 1994;147:41–46. doi: 10.1016/0378-1119(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 2.Bachmann B O, Li R, Townsend C A. β-Lactam synthetase: a new biosynthetic enzyme. Proc Natl Acad Sci USA. 1998;95:9082–9086. doi: 10.1073/pnas.95.16.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bao K, Zhou X, Kieser T, Deng Z. Abstracts of ISBA’97. 1997. pHZ1351, a broad host-range plasmid vector useful for gene cloning and for gene replacement in Streptomyces hygroscopicus KMP3, abstr. 4P15. Beijing, People’s Republic of China. [Google Scholar]
- 4.Bird A E, Bellis J M, Gasson B C. Spectrophotometric assay of clavulanic acid by reaction with imidazole. Analyst. 1982;107:1241–1245. [Google Scholar]
- 5.Burton K. A study of the conditions and mechanism of the diphenylamine reaction for the colorimetric estimation of deoxyribonucleic acid. Biochem J. 1956;62:315–323. doi: 10.1042/bj0620315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chary V K, Fuente J L, Liras P, Martín J F. amy as a reporter gene for promoter activity in Nocardia lactamdurans: comparison of promoters of the cephamycin cluster. Appl Environ Microbiol. 1997;63:2977–2982. doi: 10.1128/aem.63.8.2977-2982.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chipman D, Barak Z, Schloss J V. Biosynthesis of 2-aceto-2-hydroxyacids:acetolactate synthases and acetohydroxyacid synthases. Biochim Biophys Acta. 1998;1385:401–419. doi: 10.1016/s0167-4838(98)00083-1. [DOI] [PubMed] [Google Scholar]
- 8.Coque J J R, Liras P, Laíz L, Martín J F. A gene encoding lysine-6-aminotransferase, which forms the β-lactam precursor α-aminoadipic acid, is located in the cluster of cephamycin biosynthetic genes in Nocardia lactamdurans. J Bacteriol. 1991;173:6258–6264. doi: 10.1128/jb.173.19.6258-6264.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elson S W, Oliver R S. Studies on the biosynthesis of clavulanic acid. I. Incorporation of 13C-labelled precursors. J Antibiot. 1978;31:586–592. doi: 10.7164/antibiotics.31.586. [DOI] [PubMed] [Google Scholar]
- 10.Elson S W, Baggaley K H, Fulston M, Nicholson N H, Tyler J W, Edwards J, Holms H, Hamilton I, Mousdale D M. Two novel arginine derivatives from a mutant of Streptomyces clavuligerus. J Chem Soc Chem Commun. 1993;1993:1211–1212. [Google Scholar]
- 11.Foulstone M, Reading C. Assay of amoxicillin and clavulanic acid, the components of aumgmentin, in biological fluids with high-performance liquid chromatography. Antimicrob Agents Chemother. 1982;178:6310–6318. doi: 10.1128/aac.22.5.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutman A L, Ribon V, Boltanski A. Incorporation of β-hydroxypropionate into the β-lactam residue of clavulanic acid. J Chem Soc Chem Commun. 1985;1985:1627–1629. [Google Scholar]
- 13.Hawkins C F, Borges A, Perham R N. A common structural motif in thiamine pyrophosphate-binding enzymes. FEBS Lett. 1989;255:77–82. doi: 10.1016/0014-5793(89)81064-6. [DOI] [PubMed] [Google Scholar]
- 14.Hodgson J E, Fosberry A P, Rawlinson N S, Ross H N M, Neal R J, Arnell J C, Earl A J, Lawlor E J. Clavulanic acid biosynthesis in Streptomyces clavuligerus: gene cloning and characterization. Gene. 1995;166:49–55. doi: 10.1016/0378-1119(95)00560-9. [DOI] [PubMed] [Google Scholar]
- 15.Madduri K, Stuttard S, Vining L C. Lysine catabolism in Streptomyces spp. is primarily through cadaverine: β-lactam producers also make α-aminoadipate. J Bacteriol. 1989;17:1299–1302. doi: 10.1128/jb.171.1.299-302.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madduri K, Stuttard S, Vining L C. Cloning and location in Streptomyces spp. of a gene governing lysine-6-aminotransferase, an enzyme initiating β-lactam biosynthesis in Streptomyces spp. J Bacteriol. 1991;173:985–988. doi: 10.1128/jb.173.3.985-988.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marsh E N, Chang Dah-Tsyr M, Townsend C A. Two isoenzymes of clavaminate synthase central to clavulanic acid formation: cloning and sequencing of both genes from Streptomyces clavuligerus. Biochemistry. 1992;31:12648–12657. doi: 10.1021/bi00165a015. [DOI] [PubMed] [Google Scholar]
- 18.Martín J F. New aspects of gene and enzymes for β-lactam antibiotic biosynthesis. Appl Microbiol Biotechnol. 1998;50:1–15. doi: 10.1007/s002530051249. [DOI] [PubMed] [Google Scholar]
- 19.Paradkar A S, Jensen S E. Functional analysis of the gene encoding the clavaminate synthase 2 isoenzyme involved in clavulanic acid biosynthesis in Streptomyces clavuligerus. J Bacteriol. 1995;177:1307–1314. doi: 10.1128/jb.177.5.1307-1314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paradkar A S, Kwanema A, Aidoo K A, Wong A, Jensen S E. Molecular analysis of a β-lactam resistance gene encoded within the cephamycin gene cluster of Streptomyces clavuligerus. J Bacteriol. 1996;178:6266–6274. doi: 10.1128/jb.178.21.6266-6274.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pérez-Llarena F J, Liras P, Rodríguez-García A, Martín J F. A regulatory gene (ccaR) required for cephamycin and clavulanic acid production in Streptomyces clavuligerus: amplification results in overproduction of both β-lactam compounds. J Bacteriol. 1997;179:2053–2059. doi: 10.1128/jb.179.6.2053-2059.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pérez-Redondo R, Rodríguez-García A, Martín J F, Liras P. The claR gene of Streptomyces clavuligerus, encoding a LysR-type regulatory protein controlling clavulanic acid biosynthesis, is linked to the clavulanate-9-aldehyde reductase (car) gene. Gene. 1998;211:311–321. doi: 10.1016/s0378-1119(98)00106-1. [DOI] [PubMed] [Google Scholar]
- 23.Pitlik J, Townsend C A. The fate of [2,3,3,-2H3, 1,2-13C2]-d,l-glycerate in clavulanic acid biosynthesis. Chem Commun. 1997;1997:225–226. [Google Scholar]
- 24.Romero J, Liras P, Martín J F. Dissociation of cephamycin and clavulanic acid biosynthesis in Streptomyces clavuligerus. Appl Microbiol Biotechnol. 1984;20:318–325. [Google Scholar]
- 25.Romero J, Liras P, Martín J F. Utilization of ornithine and arginine as specific precursors of clavulanic acid. Appl Environ Microbiol. 1986;52:892–897. doi: 10.1128/aem.52.4.892-897.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Thirkettle J E, Baldwin J E, Edwards J, Griffin J P, Schofield C J. The origin of the β-lactam carbons of clavulanic acid. Chem Commun. 1997;1997:1025–1026. [Google Scholar]
- 28.Townsend C A, Ho M. Biosynthesis of clavulanic acid: origin of the C3 unit. J Am Chem Soc. 1985;107:1066–1068. [Google Scholar]
- 29.Valentine B B, Bailey C R, Doherty A, Morris J, Elson S W, Baggaley K H, Nicholson N H. Evidence that arginine is a later metabolic intermediate than ornithine in the biosynthesis of clavulanic acid by Streptomyces clavuligerus. J Chem Soc Chem Commun. 1993;1993:1210–1212. [Google Scholar]
- 30.Ward J M, Hodgson J E. The biosynthetic genes for clavulanic acid and cephamycin production occur as a “super-cluster” in three Streptomyces. FEMS Microbiol Lett. 1993;110:239–242. doi: 10.1111/j.1574-6968.1993.tb06326.x. [DOI] [PubMed] [Google Scholar]