Abstract
We demonstrate that IS1397, a putative mobile genetic element discovered in natural isolates of Escherichia coli, is active for transposition into the chromosome of E. coli K-12 and inserts specifically into palindromic units, also called repetitive extragenic palindromes, the basic element of bacterial interspersed mosaic elements (BIMEs), which are found in intergenic regions of enterobacteria closely related to E. coli and Salmonella. We could not detect transposition onto a plasmid carrying BIMEs. This unprecedented specificity of insertion into a well-characterized chromosomal intergenic repeated element and its evolutionary implications are discussed.
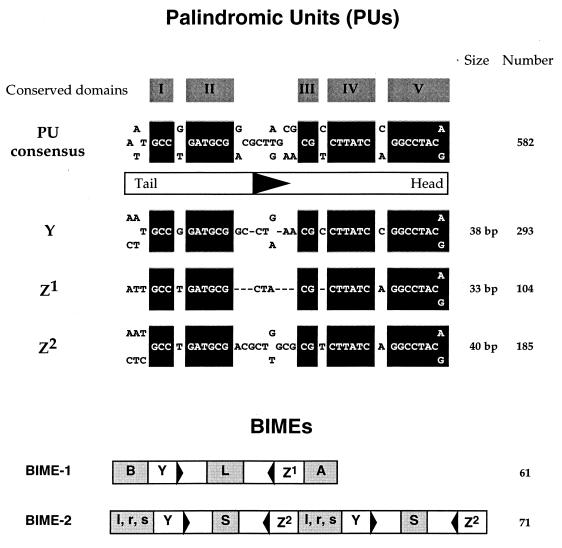
Bacterial interspersed mosaic elements (BIMEs) are repeated structures found on the chromosomes of Escherichia coli and other enterobacteria (3, 13). They are positioned in intergenic regions, between convergent operons, or between genes belonging to the same operon and are composed of several motifs assembled with a precise organization, which is summarized in Fig. 1. The basic motif is a palindromic unit (PU), also known as a repetitive extragenic palindromic (REP) sequence (11, 17, 26), which is an imperfect palindromic sequence (Fig. 1) that can confer a stem-loop secondary structure to mRNA. Two PU types, Z and Y, which differ at positions 7 and 32 of their sequence (respectively T/A and G/C), have been defined. The Z family can be divided in two classes (Z1 and Z2) according to their size (top of Fig. 1). PUs can be found singly or in clusters, in which they alternate in orientation and type. In this case, they are separated by other conserved motifs (extra PU motifs) to form BIMEs (bottom of Fig. 1). Two BIME families have been described previously (13). In BIME-1, one Z1 and one Y PU are placed head to head, are separated by an L motif, and are flanked by an A motif on the Z1 side and a B motif on the Y side. The BIME-2 family structure involves a basic element consisting of one Z2 and one Y placed head to head, separated by an l, r, or s motif. This basic element can be repeated (up to six times), and each repeat is separated from the following one by an S motif. The chromosome of E. coli K-12 contains 61 BIME-1, 71 BIME-2, and 49 occurrences of other associations between PUs and extra PU motifs which are different from the ones mentioned (hereafter referred to as atypical BIMEs).
FIG. 1.
BIME organization. PUs (11, 17) and BIMEs (3, 13) have been described previously. Important features are summarized here. (Top) Consensus sequences for PUs (upper part) and for the three PU types, Y, Z1, and Z2 (lower part). Domains conserved between the three types are boxed in black. Nucleotides G between domains I and II as well as C between domains IV and V are found in Y, while the same positions are occupied respectively by T and A in Z. These sequences are globally palindromic, with asymmetry elements which allow orientation of the structure, which is drawn under the PU consensus, from tail to head. A black triangle indicates PU orientation. The right-hand column shows the number of occurrences on the E. coli chromosome (1). (Bottom) BIMEs are composed of PU repeats in which both PU types (Z and Y) and orientation alternate (13). Members of BIME-1 are typically composed of one Y and one Z1 in head-to-head orientations, placed between A and B and separated by L. Members of BIME-2 are repeats of Y and Z2 alternating in opposite orientations, separated by S (between heads) and/or by either an s, l, or r motif (between tails).
IS1397 is a putative insertion sequence related to members of the IS3 family. This sequence was discovered during the study of intergenic regions in several isolates of E. coli (2). These regions had been chosen because they contained typical BIMEs. In the three cases analyzed, IS1397 was found to be inserted in a PU. In a second step, cloning and analysis of chromosomal DNA fragments from EPEC25 and ECOR49, two strains hosting numerous copies of the IS, confirmed this observation and supported the hypothesis of a target site specificity for IS1397 insertion into PUs. In this study, we investigated IS1397 in K-12, the laboratory strain of E. coli which normally does not contain IS1397. We developed a genetic tool which allowed us to select for transposition events from a donor plasmid carrying a genetically tagged version of IS1397. Our results show that IS1397 is a fully active insertion sequence for transposition into the chromosome of E. coli K-12 and complete the demonstration of the high specificity of insertion into extragenic PUs. This case represents the most striking example of sequence specificity for IS insertion.
MATERIALS AND METHODS
Media and standard procedures.
Luria-Bertani (LB) medium was used for growth of all E. coli strains. Kanamycin and ampicillin were used at 25 and 50 μg/ml, respectively. Restriction enzymes and DNA-modifying enzymes were purchased from New England Biolabs or Boehringer Mannheim and used as recommended. Oligonucleotides were purchased from Genset. Plasmid DNA manipulations were carried out by standard procedures (24). PCR was performed with the Amersham PCR kit as recommended and with an MJ Research Inc. PTC-100 apparatus.
Plasmids.
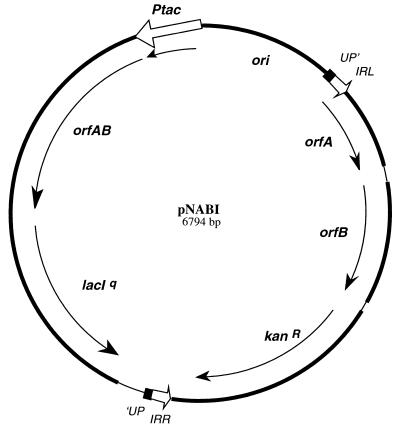
pNABI (Fig. 2) is a composite P15A-based plasmid carrying a modified version of IS1397 in which a kanamycin resistance cassette has been inserted between orfB and the right-end inverted repeat (IRR). This IS is flanked by the same sequences (an interrupted PU sequence with a 4-bp duplication) that were found in the mtlA to mtlD region of EPEC25, a natural enteropathogenic isolate of E. coli in which IS1397 had been described originally. This module was inserted between the XbaI (bp 1424) and Tth111-1 (bp 3698) sites of plasmid pACYC184 (8). The second important component of pNABI is an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible orfAB cassette composed of a Ptac promoter, a ribosome binding site, an open reading frame encoding the expected OrfAB fusion protein (22), and lacIq. This module was inserted between the NcoI (bp 3944) and XmnI (bp 635) sites of plasmid pACYC184 (8). The structure of pNABI is as follows: bp 1 to 795, P15A origin of replication from pACYC184 (bp 635 to 1429) (8); bp 796 to 798, linker (CTT); bp 799 to 837, EPEC25 mtlA to mtlD intergenic region, interrupted PU (2); bp 838 to 887, IS1397 IRL (bp 1 to 51) (2); bp 888 to 1409, IS1397 orfA coding sequence (bp 52 to 573) (2); bp 1410 to 1513, IS1397 orfA-orfB intergenic region (bp 574 to 677) (2); bp 1514 to 2236, IS1397 orfB coding sequence (bp 678 to 1400) (2); bp 2237 to 2261, pUC18 multiple cloning site (MCS) (bp 424 to 452) (33); bp 2262 to 2282, pUC19 MCS (bp 399 to 419) (33); bp 2283 to 3549, pUC4K kanamycin resistance cassette (BamHI fragment, bp 408 to 1674) (28); bp 3550 to 3554, linker TCTAG; bp 3555 to 3586, IS1397 IRR (bp 1401 to 1432) (2); bp 3587 to 3606, EPEC25 mtlA to mtlD intergenic region, interrupted PU (2); bp 3607 to 3631, pUC18 MCS (bp 430 to 454) (33); bp 3632 to 3879, end of the chloramphenicol resistance gene from pACYC184 (bp 3703 to 3950) (8); bp 3880 to 5048, lacIq PCR fragment generated with plasmid CIMER (27) as a template and the two oligonucleotides C6TCTAGACCATGGTCACTGC3GCT3CCAGTCG3 and C6TCTAGAGCTAGCACCATCGAATGGTGCA4CCT3CGCGG as primers—this fragment was cut with XbaI and introduced into the NheI site artificially introduced downstream of the orfAB gene; and bp 5049 to 6394, assembly of two PCR fragments generated with, as a template, a pUC19 recombinant plasmid in which a HindIII EPEC25 chromosomal DNA fragment overlapping the mtlA-to-mtlD intergenic region contains IS1397 (2). The first fragment, containing orfA, was made with the following two primers: G6ATCCAAGGACCATAGATTATGA3CATTCAT3GAAGTA4CTTGCCGC and G6ACGCGTGCTAGCTCCTGGCGCCT7CCAGAAGATGCTCCTGCATGGC. The second fragment, containing orfB, was made with the following two primers: GAACATCTTCTGGA7GGCGCCTGGAGCAGGTGA6CGA3GTCATCC and CAGT3CAGCTAGCCGGCT5GATACTC. These fragments were cloned separately and recombined by being cut with KasI before their insertion into pNABI. This created an artificial in-frame fusion between orfA and orfB in which the wild-type palindromic sequence characteristic of frameshift windows (22), --- - --- -> <-- -- -- -- CTG GAA AAA AAG CGC CAG GAG CTG GAG AAA AAA CGA AAG TCA TCC AGA GCC TGA GGT L E K K R Q E L E K K R K S S R A * has been replaced by the following, which creates orfAB, an in-frame fusion between orfA and orfB due to the deletion of one nucleotide and the disruption of the palindrome (underlined): CTG GAA AAA AAG GCG CCT GGA GCA GGT GAA AAA ACG AAA GTC ATC CAG AGC CTG AGG T L E K K A P G A G E K T K V I Q S L R. The structure of pNABI continues as follows: bp 6395 to 6413, ribosome binding site from pMAL-p2 bp (New England Biolabs) (bp 1513 to 1527); and bp 6414 to 6794, Ptac promoter from pDR540 (Pharmacia) (bp 1 to 389) (23).
FIG. 2.
Plasmid pNABI. pNABI is a composite low-copy-number plasmid composed of two modules. The first (clockwise) is IS1397, in which a kanamycin resistance gene has been inserted downstream of orfAB. In the second (counterclockwise), the expression of OrfAB is under the control of the Ptac promoter and is repressed by the product of lacI (IPTG inducible). Details are given in Materials and Methods.
pUP3 has been described previously (12). It consists of a DNA fragment with NcoI ends filled with Klenow polymerase and cloned into the SmaI site of pUC18. This fragment contains a 3-PU BIME-2 present in the E. coli malE-malF intergenic region. The structure of this region is malE→Y′→s←Z2 S Y→ malF→ (see the legend of Fig. 3 for explanations of the symbols). The BIME is flanked by SacI and BclI sites on one side and by a BamHI site on the other side, so that successive rounds of ligations between SacI-BclI and SacI-BamHI fragments created a series of recombinant plasmids with increasing numbers of PUs, up to 99 occurrences (33 tandem repeats of the same BIME-2 repeated in tandem). The largest plasmid, called pUP99, was digested with BamHI and EcoRI, and its insert was transferred into pTZ18 (Pharmacia), which contains an f1 origin of replication. Since the new plasmid, called pTZ99, contained sequences located between the EcoRI (bp 213) and PvuII (bp 415) sites of pTZ18 that were also present in pNABI, these were removed by ligating the large PvuII-RsaI fragment from pTZ99 to the large PvuII fragment from pTZ18, creating pTZ99Δ.
FIG. 3.
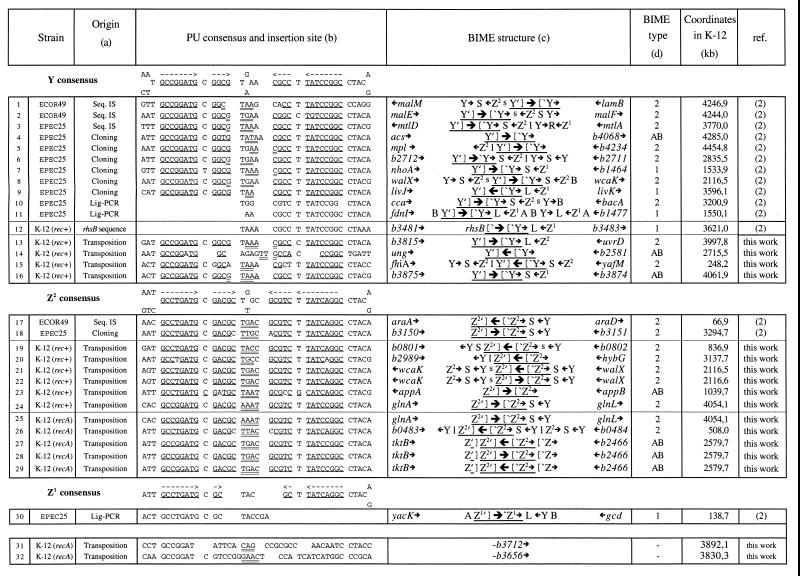
Insertion sites of IS1397. Natural or laboratory examples have been separated into four boxes according to the PU type: Y (insertions 1 to 15), Z2 (insertions 16 to 28), Z1 (insertion 29), and non-PU (insertions 30 and 31). The consensus sequences for Y, Z2, and Z1 are indicated on top of each box. (a) DNA sequencing of cloned PCR fragments from intergenic regions containing IS1397 (“Seq. IS”), of ligation-mediated PCR cloned fragments (“Lig-PCR”), and of clones from a representative library of EPEC25 chromosomal DNA (“cloning”) has been described previously (2). All other examples (“Transposition”) are new examples from the selection of transposition events in E. coli K-12. (b) The various insertion sites are indicated. Duplicated nucleotides on each side of the IS are doubly underlined. Pairing nucleotides from the stem of the palindrome are singly underlined and also labelled with arrows above consensus sequences for Y, Z2, and Z1. (c) Flanking genes are indicated, as well as the structure of the BIME in which IS1397 had transposed. Small arrows indicate, the orientation of genes or PUs. The various BIME accessory motifs (A, B, S, s, L, l, and r) (13) are included. The large arrow symbolizes IS1397 and its orientation within the interrupted PU, which is underlined. (d) The BIME type (BIME-1 or BIME-2) is shown. AB, atypical BIMEs which are either isolated PUs or combinations of Z and Y with accessory motifs that are different from BIME-1 or BIME-2.
Bacterial strains.
We used strains JM109 [recA1 endA1 gyrA96 thiA hsdR17 (rK− mK+) relA1 supE44 Δ(lac-proAB) (F′ traD proAB lacIqZΔM15)] (33), TG1 [supE hsdΔ5 thiA Δ(lac-proAB) (F′ proAB lacIqZΔM15)] (Promega), PL0 [F− Δ(lac-proAB) strA trp ara thiA Valr galE φ80dlac lacIΔ169) (25), JC10289 (a gift from A. J. Clark), PL1 [F− Δ(lac-proAB) strA trp ara thiA Valr galE φ80dlac lacIΔ169 recA [see below—recA transduction of strain PL0 by a P1 phage stock made on JC10289]), PL2 (PL0/pNABI), PL3 (PL1/pNABI), and P4 (JM109/pNABI/pTZ99Δ).
Selection of transposition events.
Independent clones of PL2 and PL3 strains were grown overnight at 37°C in liquid LB medium containing kanamycin, and a 500-μl volume of each culture was plated on an LB plate containing kanamycin and 10−3 M IPTG. After overnight incubation at 37°C, the plates were replicated onto LB plus kanamycin (without IPTG). After a 24-h incubation at 37°C, these plates were replicated on LB plus kanamycin plus 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) plates to check the LacI+ phenotype of IPTG-resistant colonies. Blue colonies (which had possibly lost the plasmid) were restreaked and grown in liquid medium for subsequent plasmid preparation by the minilysate alkaline lysis technique. These preparations were analyzed by agarose gel electrophoresis and used to transform strain PL0 to Kmr.
Chromosomal DNA extraction.
The centrifugation pellet from a 1-ml overnight culture was resuspended in 500 μl of TE (10 mM Tris-HCl [pH 8], 1 mM EDTA). Then 14 μl of 10% sodium dodecyl sulfate (SDS) and 13 μl of proteinase K (20 mg/ml in 100 mM Tris-HCl [pH 7.5]–100 mM CaCl2) were added, and incubation at 37°C was continued until lysis. Two phenol and one chloroform-isoamyl alcohol extractions were followed by ethanol precipitation. The DNA pellet was resuspended in 400 μl of TE.
Southern hybridization.
Chromosomal DNAs were digested with BglII (which does not cut inside IS1397), and 1 to 5 μg was loaded on a 1% agarose gel. After electrophoresis, DNA was transferred to a Hybond N+ membrane (Amersham) by using a Transvac TE80 vacuum blotter (Hoeffer Scientific Instruments) as follows: 0.25 N HCl for 15 min and 0.4 N NaOH for 1 h. After transfer, the membrane was neutralized for 15 mn in 1 M Tris-HCl (pH 7.5) and incubated for 1 h in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 2 h in hybridization buffer (6× SSC, 0.1% SDS, 0.05% skim milk powder) at 65°C. The heat-denatured probe was added, and hybridization was carried out overnight at 65°C. The probe was an internal EcoRI fragment from IS1397 32P labelled with the Promega nick-translation kit. The membrane was washed twice at room temperature with 2× SSC–0.1% SDS for 30 min and twice at 68°C in 0.2× SSC–0.1% SDS for 45 min. The membrane was wrapped in Saran Wrap and exposed on an autoradiographic film.
DNA cloning of IS1397 insertion sites.
BglII-digested chromosomal DNAs were ligated to pUC18 vector cut with BamHI and treated with phosphatase (pUC18 BamHI/BAP; Pharmacia Biotech), and E. coli TOP10 cells were transformed as recommended by the supplier (Invitrogen). For fragments which were too long to be cloned in pUC18, chromosomal DNAs were codigested with ApaLI, BglII, NcoI, and NdeI (none of which have restriction sites in the IS), and a 3′-A residue was added by using Taq polymerase activity (Invitrogen). DNAs were then ligated to the pGEM-T vector, and E. coli JM109 cells were transformed as recommended by the supplier (Promega). In both cases, ampicillin- and kanamycin-resistant clones were selected on LB-kanamycin-ampicillin plates.
DNA sequencing.
DNA sequencing was performed with Qiagen-purified plasmid DNAs (Qiagen spin minipreps or Qiagen midi preps as recommended) and with a Thermo Sequenase Cy5.5 dye terminator cycle-sequencing kit (Amersham Pharmacia Biotech). Sequencing reactions were run on an Amersham Pharmacia Biotech 4X4 automatic sequencer. The following primers were used for sequencing: lig-PCR-A (GCCGTAGAAATGATGCCTGC), complementary to codons 20 to 26 of orfA) (2) and Km seq out (CACGAGGCAGACCTCAGCGC), corresponding to a region located between the end of the Kmr gene and the IRR of IS139 (2), as found on plasmid pNABI (Fig. 2 and see below). Chromosomal regions flanking the IS were identified on the Colibri Web Server (8a).
RESULTS
Transposition events.
To study IS1397 transposition, pNABI was constructed. It is composed of two modules. The first contains IS1397 flanked by an interrupted PU and containing a kanamycin resistance gene inserted between orfB and IRR. The second contains an orfAB in-frame fusion under the control of the IPTG-inducible Ptac promoter. lacI was included in the construct to achieve repression of the toxic OrfAB protein in all E. coli strains. pNABI was used to transform PL0, a lacZY+ lacI strain. In the presence of IPTG, OrfAB is expressed, and this induction was found to be lethal to the cells. IPTG-resistant colonies could be isolated after overnight culture on LB-kanamycin-IPTG plates at 37°C (see Materials and Methods). These were found to arise from distinct events: (i) a mutation within orfAB resulting in a nontoxic protein (in this case, LacI is still expressed and the presence of this marker is revealed by a white phenotype after restreaking on LB–kanamycin–X-Gal plates without IPTG; (ii) a deletion encompassing orfAB and lacI; or (iii) transposition of the IS1397-Kmr module into the chromosome, with a loss of the pNABI donor plasmid. Due to the loss of lacI, these last two events would give blue colonies on kanamycin–X-Gal plates. To discriminate between them, the presence or absence of a plasmid carrying the kanamycin resistance gene was checked by analyzing minilysates by agarose gel electrophoresis or by analyzing for the ability to transform a kanamycin-sensitive strain into a resistant one.
Approximately 5 × 108 CFU from each of 26 separate liquid cultures of independent PL2 (rec+) or PL3 (rec) clones were plated on IPTG plates. Resistant colonies were obtained for 24 PL2 and 22 PL3 plates. Of these, 19 PL2 and 20 PL3 plates contained blue colonies when streaked on LB–X-Gal plates. Two LacI− colonies from each plate were checked for the absence of plasmid, either by analyzing plasmid minipreparations by agarose gel electrophoresis or by using the same minipreparations to transform PL0 to kanamycin resistance. When used in parallel, both tests always led to the same conclusion: approximately half of the clones tested had lost the plasmid, so that 11 PL2 and 7 PL3 clones could be examined for the presence of an IS1397-Kmr insertion in the chromosome. BglII digests of chromosomal DNA were analyzed by Southern hybridization with an internal IS1397 DNA fragment as a probe (results not shown). Under these conditions, all candidates showed a single band (ranging from approximately 7 to 15 kb) hybridizing to the probe. In all cases, the two colonies from the same plate proved to be identical, i.e., containing a plasmid or, when plasmid free, displaying a single IS-containing chromosomal BglII DNA fragment of identical size. We cloned these BglII fragments directly in pUC18. When this was not successful, we used the TA cloning procedure on DNA fragments obtained after chromosomal DNA was cut with a cocktail of enzymes which did not cut within IS1397-Kmr. This allowed us to analyze the new junctions between the IS and the chromosome. The results are presented in Fig. 3.
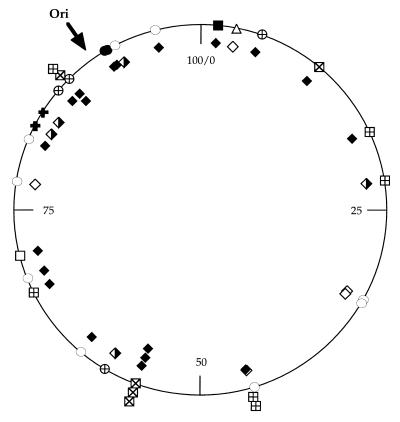
As we expected from our previous results, all insertions were found to have occurred within a PU, with two possible exceptions (see Discussion) in the recA context (Fig. 3, column b). In all instances, insertion took place in the loop, with a 3- to 4-bp duplication. This confirmed the tight IS1397 specificity of insertion into PUs. Insertion localizations seem to follow the PU distribution on the E. coli chromosomal map (1) (Fig. 4). However, some regions were overrepresented. (i) In walX-wcaK, three independent insertions were characterized. One (insertion 8) occurred in EPEC25 in the central Y PU, while two (insertions 21 and 22), selected in E. coli K-12, had occurred in the central Z2 PU with either IS1397 orientation. (ii) Two insertions were found between glnA and glnL, one (insertion 24) in E. coli K-12 rec+ and the other (insertion 25) in recA, both in the same Z2 PU in the same IS orientation. (iii) Three occurrences (insertions 27 to 29) were identified in E. coli K-12 recA at the same position within the Z2 PU, between b2466 and tktB, with the two possible IS1397 orientations.
FIG. 4.
Chromosomal locations of the various IS1397 insertions in E. coli. Circular map of E. coli and positions of the various occurrences of IS1397 insertions. The origin of replication is indicated by an arrow, and coordinates are indicated in minutes. Symbols for PUs are as follows: ○, Y (EPEC25); ●, Y (ECOR49); ⊕, Y (K-12 rec+); ✚, non-PU; ▵, Z1 (EPEC25); □, Z2 (EPEC25); ■, Z2 (ECOR49); ⊞, Z2 (K-12 rec+); ⊠, Z2 (K-12 rec). Inside the circle, symbols for BIMEs are as follows: ◊, BIME-1; ⧫, BIME-2; ◅, Atypical BIME.
Transposition into a plasmid-borne target.
pTZ99Δ and pNABI carry compatible replication origins and specify different antibiotic resistances, and so both plasmids can be maintained in the same cell. We analyzed whether the recombinant IS1397-Kmr borne by pNABI could integrate into PTZ99Δ. For this, minilysates were prepared from cultures of strain P4 (which contains both pNABI and pTZ99Δ) grown overnight in LB containing kanamycin and ampicillin. These preparations were used to transform PL0. All the Ampr Kmr colonies were found to contain both intact plasmids when checked by agarose gel electrophoresis. To avoid the problem of double transformants, we infected strain P4 with M13mp18 phage, and supernatants of overnight cultures in LB plus ampicillin and kanamycin were first filtered to eliminate parental cells and used to transform JM109 or TG1 for Ampr or for Ampr Kmr. This protocol takes advantage of the presence of an active M13 origin of replication on pTZ99Δ, which ensures an effective encapsidation by this bacteriophage. Despite an extensive search for colonies resistant to both antibiotics, we never observed recombinant plasmids harboring IS1397-Kmr integrated into pTZ99Δ. Hence, considering the titer of pseudo-viral particles able to confer Ampr (around 109/ml), we can estimate that the frequency of transposition into pTZ99Δ is less than 10−8. The method we used involved M13 encapsidation of an expected recombinant plasmid carrying IS1397-Kmr. One could argue that integration of IS1397-Kmr into pTZ99Δ would create a plasmid of excessive length or incompatible for encapsidation. This point was checked by using pTZ99, the progenitor of pTZ99Δ, which has two regions of homology to pNABI. In this case, M13 culture supernatants were able to transduce Ampr and Kmr in a single step. The plasmid content of the resistant colonies was analyzed and revealed either a large plasmid resulting from the recombination between pTZ99 and pNABI or two plasmids resulting from an intramolecular recombination within the latter (not shown). This rules out the possibility that a large plasmid was encapsidated in M13 or that IS1397 has a deleterious effect on encapsidation. It thus appears that transposition of IS1397 into a plasmid is truly a rare event (<10−8) which could not be detected by our method.
DISCUSSION
We discovered IS1397 when we studied the polymorphism of intergenic regions containing BIMEs among natural or laboratory strains of E. coli (2). Three independent experiments indicated that IS1397 was systematically associated with PUs. This paper extends these findings to the case of E. coli K-12, a strain which was shown to be free of IS1397, with the exception of the rhsB locus, which contains a truncated form of the IS (2). Our results demonstrate that IS1397 is a fully active insertion sequence, able to transpose from a plasmid to the chromosome of E. coli K-12.
The genetic procedure used to select for transposition events relies on the toxicity of OrfAB. Such a phenomenon has been described for IS1 (19) and is well documented for Tn5 (30, 34, 35). The reasons for the toxicity of the IS1397 transposase are currently being investigated.
The emergence of IPTG-resistant clones is the result of several events. Beside any alteration rendering OrfAB nontoxic for the cell, which still leaves an intact lacI on the plasmid and is easily detected, we observed lacI clones still harboring a plasmid. We hypothesized that in this case a deletion encompassing lacI and orfAB on pNABI had occurred. This was checked in 2 cases of 19 obtained by sequencing the regions of interest. The remaining cases, i.e., IPTG- and kanamycin-resistant clones having lost the plasmid, which represent roughly half of the lacI clones, were all caused by transposition events. This class is thus the result of two unrelated events: transposition and loss of the plasmid. The selection procedure we used did not allow us to dissociate them, so that the number of clones having undergone transposition is probably larger than what could be selected. We measured the frequency of spontaneous plasmid loss in the absence of selection pressure (kanamycin resistance). We could estimate this rate to be around 5 × 10−4 per generation. If transposition occurs first, the plasmid can be lost, since kanamycin resistance has moved into the chromosome. We observed a highly heterogeneous proportion (from 0 to nearly 100%) of lacI colonies in the different independent PL2 or PL3 clones. This could be explained by a different timing in transposition during preculture, with IPTG allowing us to select subsequently for plasmid loss. Not only is IPTG a selective agent for transposition events, but also it induces the expression of OrfAB, the putative transposase from IS1397. Overexpression of this protein could induce transposition. However, it does not seem that IS1397 moves to many chromosomal sites within the same cell, since we never observed on Southern blots any clone harboring more than one IS chromosomal location. We therefore believe that the result of our selection is the emergence of clones having first undergone transposition of the IS, which can take place anytime during the preculture, followed by the loss of the plasmid, which is no longer required to sustain kanamycin resistance, and then counterselection in the presence of IPTG because of the production of a toxic protein.
Transposition into the chromosome could be readily detected. The sequences flanking IS1397 are shown in Fig. 3. All but two cases (insertions 31 and 32) are characteristic PUs (column b) with a 4-bp duplication at the junction. This signature is a hallmark of transposition and proves that in E. coli K-12 transposition did not occur through recombination between a resident PU and the interrupted PU sequence (originating from mtlA to mtlD) flanking IS1397-Kmr. It will be interesting to examine whether the presence of such a truncated PU is necessary for transposition and target specificity. Insertion took place very precisely in the central part of PUs, particularly in Z2, where in almost all cases the four nucleotides of the loop were duplicated (the only exceptions are insertion 20, where only three residues were duplicated, and insertion 18, where the last nucleotides of the stem do not match, creating a 6-nucleotide central part). Insertion seems less precise for Y. The duplication always overlapped the central part of the palindrome but often included one residue belonging to the “stem.” There might be a slight preference for some central sequences. Half of the 32 examples are distributed among three sequences: TGAC was found six times and accounts for half of the Z2 examples. This sequence was found only in Z2 even though it is equally close to the Y consensus sequences, TGAA and TAAA, which were found four and six times, respectively, and account altogether for 62.5% of the Y examples. However, when we compared the distribution of PU or BIME types in E. coli to the distribution of the PUs or BIMEs found as targets for transposition, we did not observe a statistical difference, indicating that transposition occurs randomly among PUs and BIMEs (data not shown).
As mentioned, two cases (insertions 30 and 31) are not PUs, since they are both located within a coding sequence and are not palindromic but share the sequence GCCGGAT with the Y subtype. These two cases were observed in a recA context. However, from our results it does not seem that RecA is involved in target recognition. We computed the number of occurrences of the sequences GCCGGATG and GCCTGATG, which are part of the stem in the two main PU types in the chromosome of E. coli. Only 37% (454 of 1,241) of these sequences are found in PUs, indicating that these sequences are far less attractive targets for transposition when they are found alone than when they belong to a PU. PUs can therefore be considered the true target for IS1397 transposition.
IS1397 is to our knowledge the first example of an insertion sequence with such a striking transposition target consensus (for a review, see reference 10). For other IS or transposons, the consensus is very weak or totally undefined. Table 1 summarizes a few examples of recently identified target consensus sequences for different bacterial mobile genetic elements. One can imagine that IS1397, being targeted into PUs, will transpose systematically into intergenic regions, which is less detrimental to the host than random jumping, which can inactivate genes. Indeed, we observed that IS1397, like many other insertion sequences, had no polar effect on the expression of downstream genes when insertion took place in one orientation. For instance, in strain ECOR49, where IS1397 insertions have been characterized, IS1397 is inserted between malE and malF with the same orientation (Fig. 3, insertion 2). The strain is phenotypically Mal−, but the introduction of a plasmid expressing MalT, the activator of the maltose operons, reversed this phenotype, showing that ECOR49 lacks this protein and that the IS did not inactivate the expression of malF. In contrast, IS1397 is inserted between araA and araD in the opposite orientation (Fig. 3, insertion 17). The strain is Ara−, but revertants could be obtained and all contained deletions of the IS, showing that the IS had a polar effect in this case. IS1397 could thus propagate safely for the host, limiting the risk of abortive events to insertions in the wrong orientation into a BIME placed in front of an essential gene. A comparable “safe” strategy has been used by Tn7, which chose attTn7 as a specific site for insertion (9). However, such a strategy limits the possibility of spreading. As discussed below, Tn7 has developed an alternative strategy to solve this dilemma. IS1397, with the choice of PUs, automatically solves this problem in a simpler fashion because these sequences are widespread in the chromosome of E. coli (579 occurrences) and also in other enterobacteria. Another advantage of selecting a consensus sequence for integration is that intramolecular transposition (i.e., transposition of an IS within its own sequence) is prevented.
TABLE 1.
Examples of palindromic target consensus sites for IS insertions
| Insertion | Sequence | Reference |
|---|---|---|
| IS231A | GGGnCCC | 15 |
| Tn10 | GCTnAGC | 16 |
| IS50/TN5 | AGNTY RANCT | 14 |
| IS903 | TTTYAnnnnnnnnnnnTRAAA | 18 |
| IS3 | TAnGAAAnnTTCnTA | 20 |
| IS6 | AnGCAGTnnAAAnTGCnT | 20 |
| IS21 | GGAGSnGGCnYYYRnnGCCnSCTCC | 20 |
| IS30 | TAAAAAWGGCnRYCGCnWTTTTTA | 20 |
| IS1397 | MHTGCCKGATGCGRCG(C)TDRM(G)CGYCTTATCMGGCCTATR (PU) | This work |
Symbols: Y, C or T; R, A or G; n, A, G, C, or T; S, G or C; W, A or T; D, A, G, or T; B, G, C, or T; K, G or T; M, A or C. Complementary sequences of the palindromes are underlined.
Recognition of target sites by mobile genetic elements results from a variety of different mechanisms which are poorly understood. Homology between the target and the ends of the element has been proposed, for instance in the case of Tn3 (29). Some transposases have been shown to recognize their target directly (10). In other instances, the recognition occurs through an interaction with a DNA binding protein which is distinct from the transposase. The best-documented case is Tn7, which transposes either into a specific site, attTn7, or to nonspecific sites (9), with a preference for replicating DNAs in this case (31). Both mechanisms invoke an interaction between a specific protein and the target DNA sequence: transposon-encoded TnsD binds attTn7 and recruits TnsC, the transposition regulator. DNA-bound TnsD also interacts with the transposase TnsA+B, triggering its endonucleolytic and recombinogenic activities (4, 5). TnsE, another Tn7-encoded protein, can substitute for TnsD, leading to transposition into replicating plasmids or episomes. In contrast to transposition into the chromosome, no IS1397 transposition could be detected when the target was on pTZ99Δ. This multicopy plasmid, which contains 33 tandemly repeated BIMEs totaling 99 PUs, brings a substantial amount of PUs to the cell, probably more than the chromosome, which contains 579 PUs. This observation can be connected to the finding that PUs seem specific to chromosomes, since they were never detected on nonchromosomal genetic elements such as episomes, plasmids, or bacteriophages. An attractive hypothesis is that PUs deal with the organization of the chromosome by scaffolding complex structures which include proteins. Several BIME binding proteins have indeed been described. They all play a role in either DNA replication (e.g., DNA polymerase I [12]) or DNA folding: IHF binds the L motif of BIME-1 (7, 21), and DNA gyrase binds PUs (32). If PU-containing structures involved in nucleoid organization were the actual target for IS1397 transposition, one could explain why PUs are efficient targets on the chromosome but not on plasmids. This would imply that IS1397 is driven to its target by an interaction between OrfAB, its transposase, and proteins bound to PUs. Such an example of interaction between a transposase and a DNA binding protein has been described for Tn5 transposase, which binds topoisomerase I (34, 35). An alternative explanation to IS1397 transposase specificity would be that chromosomal PUs, bound to proteins, adopt a special conformation which renders them competent for a productive interaction with the transposase. Both hypotheses are currently being investigated.
ACKNOWLEDGMENTS
This work was supported in part by the Programme de Recherche Fondamentale en Microbiologie et Maladies Infectieuses et Parasitaires from Ministère Français de l’Education Nationale de la Recherche et de la Technologie.
REFERENCES
- 1.Bachellier, S., J.-M. Clément, and M. Hofnung. Short palindromic repetitive DNA elements in enterobacteria: a survey. Res. Microbiol., in press. [DOI] [PubMed]
- 2.Bachellier S, Clément J-M, Hofnung M, Gilson E. Bacterial interspersed mosaic elements (BIMEs) are a major source of sequence polymorphism in Escherichia coli intergenic regions including specific associations with a new insertion sequence. Genetics. 1997;145:551–562. doi: 10.1093/genetics/145.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachellier S, Saurin W, Perrin D, Hofnung M, Gilson E. Structural and functional diversity among bacterial interspersed mosaic elements (BIMEs) Mol Microbiol. 1994;12:61–70. doi: 10.1111/j.1365-2958.1994.tb00995.x. [DOI] [PubMed] [Google Scholar]
- 4.Bainton R, Gamas P, Craig N L. Tn7 transposition in vitro proceeds through an excised transposon intermediate generated by staggered breaks in DNA. Cell. 1991;65:805–816. doi: 10.1016/0092-8674(91)90388-f. [DOI] [PubMed] [Google Scholar]
- 5.Bainton R J, Kubo K M, Feng J N, Craig N L. Tn7 transposition: target DNA recognition is mediated by multiple Tn7-encoded proteins in a purified in vitro system. Cell. 1993;72:931–943. doi: 10.1016/0092-8674(93)90581-a. [DOI] [PubMed] [Google Scholar]
- 6.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 7.Boccard F, Prentki P. Specific interaction of IHF with RIBs, a class of bacterial repetitive DNA elements located at the 3′ end of transcription units. EMBO J. 1993;12:5019–5027. doi: 10.1002/j.1460-2075.1993.tb06195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a.Colibri Web Server. [Online.] http://www.pasteur.fr/Bio/Clibri.html. [4 May 1999, last date accessed.]
- 9.Craig N L. Transposon Tn7. Curr Top Microbiol Immunol. 1996;204:27–48. doi: 10.1007/978-3-642-79795-8_2. [DOI] [PubMed] [Google Scholar]
- 10.Craig N L. Target site selection in transposition. Annu Rev Biochem. 1997;66:437–474. doi: 10.1146/annurev.biochem.66.1.437. [DOI] [PubMed] [Google Scholar]
- 11.Gilson E, Clément J-M, Brutlag D, Hofnung M. A family of dispersed repetitive extragenic palindromic DNA sequences in E. coli. EMBO J. 1984;3:1417–1421. doi: 10.1002/j.1460-2075.1984.tb01986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilson E, Perrin D, Hofnung M. DNA polymerase I and a protein complex bind specifically to E. coli palindromic unit highly repetitive DNA: implications for bacterial chromosome organization. Nucleic Acids Res. 1990;18:3941–3952. doi: 10.1093/nar/18.13.3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilson E, Saurin W, Perrin D, Bachellier S, Hofnung M. Palindromic units are part of a new bacterial interspersed mosaic element (BIME) Nucleic Acids Res. 1991;19:1375–1383. doi: 10.1093/nar/19.7.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goryshin I Y, Miller J A, Kil Y V, Lanzov V A, Reznikoff W S. Tn5/IS50 target recognition. Proc Natl Acad Sci USA. 1998;95:10716–10721. doi: 10.1073/pnas.95.18.10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hallet B, Rezshazy R, Mahillon J, Delcour J. IS231A insertion specificity: consensus sequence and DNA bending at the target site. Mol Microbiol. 1994;14:131–139. doi: 10.1111/j.1365-2958.1994.tb01273.x. [DOI] [PubMed] [Google Scholar]
- 16.Halling S M, Kleckner N. A symmetrical six-base-pair target site sequence determines Tn10 insertion specificity. Cell. 1982;28:155–163. doi: 10.1016/0092-8674(82)90385-3. [DOI] [PubMed] [Google Scholar]
- 17.Higgins C F, Ferro-Luzzi Ames G, Barnes W M, Clément J-M, Hofnung M. A novel intercistronic regulatory element of prokaryotic operons. Nature. 1982;298:760–762. doi: 10.1038/298760a0. [DOI] [PubMed] [Google Scholar]
- 18.Hu W-H, Derbyshire K M. Target choice and orientation preference of the insertion sequence IS903. J Bacteriol. 1998;180:3039–3048. doi: 10.1128/jb.180.12.3039-3048.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lane D, Cavaillé J, Chandler M. Induction of the SOS response by IS1 transposase. J Mol Biol. 1994;242:339–350. doi: 10.1006/jmbi.1994.1585. [DOI] [PubMed] [Google Scholar]
- 20.Olasz F, Kiss J, Konig P, Buzas Z, Stalder R, Arber W. Target specificity of insertion element IS30. Mol Microbiol. 1998;28:691–704. doi: 10.1046/j.1365-2958.1998.00824.x. [DOI] [PubMed] [Google Scholar]
- 21.Oppenheim A B, Rudd K E, Mendelson I, Teff D. Integration host factor binds to a unique class of complex repetitive extragenic DNA sequences in Escherichia coli. Mol Microbiol. 1993;10:113–122. doi: 10.1111/j.1365-2958.1993.tb00908.x. [DOI] [PubMed] [Google Scholar]
- 22.Polard P, Prère M F, Chandler M, Fayet O. Programmed translational frameshifting and initiation at an AUU codon in gene expression of bacterial insertion sequence IS911. J Mol Biol. 1991;222:465–477. doi: 10.1016/0022-2836(91)90490-w. [DOI] [PubMed] [Google Scholar]
- 23.Russell D R, Bennett G N. Construction and analysis of in vivo activity of E. coli promoter hybrids and promoter mutants that alter the −35 to −10 spacing. Gene. 1985;20:231–243. doi: 10.1016/0378-1119(82)90042-7. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 25.Schmeissner U, Ganem D, Miller J H. Revised gene-protein map for the lacI gene-lac repressor system. J Mol Biol. 1977;117:572–575. doi: 10.1016/0022-2836(77)90058-4. [DOI] [PubMed] [Google Scholar]
- 26.Stern J M, Ferro-Luzzi Ames G, Smith N H, Robinson E C, Higgins C F. Repetitive extragenic palindromic sequences: a major component of the bacterial genome. Cell. 1984;37:1015–1026. doi: 10.1016/0092-8674(84)90436-7. [DOI] [PubMed] [Google Scholar]
- 27.Szmelcman S, Clément J-M, Jehanno M, Schwartz O, Montagnier L, Hofnung M. Export and one-step purification from Escherichia coli of a MalE-CD4 hybrid protein that neutralizes HIV in vitro. J Acquired Immune Defic Syndr. 1990;3:859–872. [PubMed] [Google Scholar]
- 28.Taylor L A, Rose R E. A correction in the nucleotide sequence of the Tn903 kanamycin resistance determinant in pUC4K. Nucleic Acids Res. 1988;16:358. doi: 10.1093/nar/16.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tu C P, Cohen S N. Translocation specificity of the Tn3 element: characterization of sites of multiple insertions. Cell. 1980;19:151–160. doi: 10.1016/0092-8674(80)90396-7. [DOI] [PubMed] [Google Scholar]
- 30.Weinreich M D, Yigit H, Reznikoff W S. Overexpression of the Tn5 transposase results in filamentation, aberrant nucleoid segregation, and cell death: analysis of Escherichia coli and transposase suppressor mutations. J Bacteriol. 1994;176:5494–5504. doi: 10.1128/jb.176.17.5494-5504.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolkow C A, DeBoy R T, Craig N L. Conjugating plasmids are preferred targets for Tn7. Genes Dev. 1996;10:2145–2157. doi: 10.1101/gad.10.17.2145. [DOI] [PubMed] [Google Scholar]
- 32.Yang Y, Ames G F-L. DNA gyrase binds to the family of prokaryotic repetitive extragenic palindromic sequences. Proc Natl Acad Sci USA. 1988;85:8850–8854. doi: 10.1073/pnas.85.23.8850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 34.Yigit H, Reznikoff W S. Examination of the Tn5 transposase overproduction phenotype in Escherichia coli and localization of a suppressor of transposase overproduction killing that is an allele of rpoH. J Bacteriol. 1997;179:1704–1713. doi: 10.1128/jb.179.5.1704-1713.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yigit H, Reznikoff W S. Escherichia coli topoisomerase I copurifies with Tn5 transposase, and Tn5 transposase inhibits topoisomerase I. J Bacteriol. 1999;181:3185–3192. doi: 10.1128/jb.181.10.3185-3192.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]