Abstract
The O polysaccharide (OPS) of the lipopolysaccharide (LPS) of Pseudomonas syringae pv. atrofaciens IMV 7836 and some other strains that are classified in serogroup O1 was shown to be a novel linear α-d-rhamnan with the tetrasaccharide O repeat →3)-α-d-Rhap-(1→3)-α-d-Rhap-(1→2)-α-d-Rhap-(1→2)-α-d-Rhap-(1→ (chemotype 1A). The same α-d-rhamnan serves as the backbone in branched OPSs with lateral (α1→3)-linked d-Rhap, (β1→4)-linked d-GlcpNAc, and (α1→4)-linked d-Fucf residues (chemotypes 1B, 1C, and 1D, respectively). Strains of chemotype 1C demonstrated variations resulting in a decrease of the degree of substitution of the backbone 1A with the lateral d-GlcNAc residue (chemotype 1C-1A), which may be described as branched regular ↔ branched irregular → linear OPS structure alterations (1C↔1C-1A → 1A). Based on serological data, chemotype 1D was suggested to undergo a 1D ↔ 1D-1A alteration, whereas chemotype 1B showed no alteration. A number of OPS backbone-specific monoclonal antibodies (MAbs), Ps(1-2)a, Ps(1-2)a1, Ps1a, Ps1a1, and Ps1a2, as well as MAbs Ps1b, Ps1c, Ps1c1, Ps1d, Ps(1-2)d, and Ps(1-2)d1 specific to epitopes related to the lateral sugar substituents of the OPSs, were produced against P. syringae serogroup O1 strains. By using MAbs, some specific epitopes were inferred, serogroup O1 strains were serotyped in more detail, and thus, the serological classification scheme of P. syringae was improved. Screening with MAbs of about 800 strains representing all 56 known P. syringae pathovars showed that the strains classified in serogroup O1 were found among 15 pathovars and the strains with the linear OPSs of chemotype 1A were found among 9 of the 15 pathovars. A possible role for the LPS of P. syringae and related pseudomonads as a phylogenetic marker is discussed.
More than 50 infraspecies taxa, so-called pathovars, of Pseudomonas syringae have been described on the basis of their distinctive pathogenicity to one or more host plants (67). Known phenotypic and genomic characters of P. syringae strains yield much information on the homogeneity of pathovars and their relatedness but cannot define the pathovar status of most strains (9, 12, 18, 35, 38, 41, 53, 59). Some progress in classification of P. syringae and related phytopathogenic pseudomonads has been achieved by DNA-DNA hybridization and ribotyping that resulted in delineation of nine genomospecies (12–14, 21, 47, 48, 56). However, these genomospecies cannot be differentiated systematically by phenotypic tests, and therefore, new phenotypic characters are necessary for this purpose and for more accurate allocation of strains to P. syringae pathovars. We suggest that the chemical structure and immunological specificity of the lipopolysaccharides (LPSs) of P. syringae could be reliable characters of this sort. The suggestion is based on the unique chemical structure, molecular biology, and biochemistry of the LPS molecule (see Discussion) (4, 20, 40, 49, 50, 62).
The LPSs of most gram-negative bacteria, including pseudomonads, are composed of three independently synthesized moieties: lipid A, core oligosaccharide, and O polysaccharide (OPS), with the structural conservatism decreasing in the order lipid A > core >> OPS (20, 40). A cascade of strongly conjoining genetic and biochemical events are related to LPS synthesis, transport, polymerization, and folding (4, 49, 50, 62). Thus, any replacement, gain, or loss of a sugar substituent and any change of a glycosidic linkage within the LPS structure has to be preceded by profound changes within the LPS-encoding genes. Therefore, the chemotypes and, correspondingly, serotypes of P. syringae LPSs may be suggested as a conservative phenotypic character (phylogenetic marker) having a high taxonomic impact.
Strains of different pathovars of P. syringae produce LPSs with linear or branched OPSs having l-, d-, or both l- and d-rhamnose (Rha) residues in the backbone and different lateral substituents (24–26, 58, 68). A number of branched P. syringae OPSs of chemotypes 1B, 1C, and 1D have the backbone 1A, composed of oligosaccharide repeating units (O repeats) with four α-d-Rhap residues (the structures of the chemical O repeats are shown in Table 1). However, until recently, no linear OPS of chemotype 1A had been described. Some other P. syringae OPSs are linear, irregular branched, or regular branched, composed of an O repeat backbone with three α-d-Rha residues (chemotype 2A) and a lateral (α1→4)-linked d-fucose residue (chemotype 2D) (references 29 and 58 and our unpublished data).
TABLE 1.
Structures of linear and regular branched OPSs of P. syringae serogroups O1 and O2
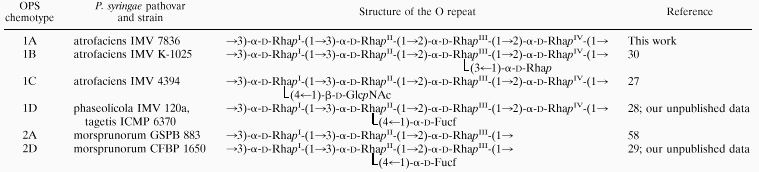 |
Immunochemical studies of P. syringae LPSs with known OPS structure by using monoclonal antibodies (MAbs) revealed a correlation between the OPS structure and the immunospecificity and allowed the inference of some group- and type-specific epitopes within OPSs (44–46). Strains with the backbone O repeats 1A and 2A were classified in serogroups O1 and O2, respectively, as a variety of serotypes (45, 46).
Recently, we described some peculiar immunological features of the LPS from P. syringae pv. atrofaciens IMV 7836 (46). In particular, this LPS (i) did not cross-react with any MAb specific to the lateral substituents of P. syringae OPSs, (ii) induced synthesis of antibodies that cross-reacted with branched OPSs having the backbone O repeat 1A and different lateral substituents, and (iii) induced production of MAbs which were specific to the homologous OPS only. Based on these findings, we suggested that this strain had a hitherto-unknown linear α-d-rhamnan OPS of chemotype 1A (Table 1). It was also suggested that in some P. syringae strains branched OPSs with the 1A backbone are irregular due to the presence of both linear and branched O repeats in various ratios.
Now we report the elucidation of the primary structure of the OPS of P. syringae pv. atrofaciens IMV 7836 and some other strains, which has confirmed our previous suggestion. We also report the results of serological and immunochemical studies of P. syringae LPSs with known OPS structure by the use of a panel of MAbs.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A total of around 800 P. syringae strains belonging to all 56 known pathovars (aceris, actinidiae, aesculi, amygdali, antirrhini, apii, aptata, atrofaciens, atropurpurea, avellanae [= Pseudomonas avellanae], berberidis, cannabina, ciccaronei, coriandricola, coronafaciens [= Pseudomonas coronafaciens], delphinii, dendropanacis, dysoxyli, eriobotryae, ficuserectae, garcae, glycinea [= Pseudomonas savastanoi pv. glycinea], helianthi, japonica, lachrymans, lapsa, maculicola, meliae, mellea, mori, morsprunorum, myricae, oryzae, panici, papulans, passiflorae, persicae, phaseolicola [= Pseudomonas savastanoi pv. phaseolicola], philadelphi, photiniae, pisi, porri, primulae, ribicola, savastanoi [= Pseudomonas savastanoi pv. savastanoi], sesami, striafaciens, syringae, tabaci, tagetis, theae, tomato, tremae, ulmi, viburni, and zizaniae), 82 P. syringae strains of unknown pathovars, and 59 strains of other species, including Pseudomonas (Pseudomonas aeruginosa, Pseudomonas cichorii, Pseudomonas fluorescens, Pseudomonas marginalis, and Pseudomonas viridiflava), Burkholderia (Burkholderia cepacia, Burkholderia gladioli, and Burkholderia glumae), Ralstonia solanacearum, Agrobacterium tumefaciens, and Xanthomonas campestris, were studied. The pathotype strains with all listed pathovars of P. syringae and related pseudomonads were screened. The bacteria were obtained from different collections of plant-pathogenic microorganisms (ATCC, CFBP, GSPB, ICMP, IMV, IPGR, ISPaVe, NCPPB, and PD). Despite the fact that not all of the pathovars listed above are valid (67), they are listed in a recent paper describing DNA relatedness among the pathovars of P. syringae and related pseudomonands (14) and therefore are used in this paper as well. A number of the strains used in this study have been characterized serologically and by some other phenotypic as well as genomic characters (2, 9, 21, 32, 35, 41, 42, 44–46, 54, 56, 61, 65). The bacteria were cultivated on solid potato dextrose agar (Difco Laboratories, Detroit, Mich.) at 20 to 22°C for 24 h.
Preparation of LPSs, OPSs, and Smith-degraded and synthetic polysaccharides.
For immunoassays, the LPSs were prepared as described previously (44, 46). For structural analysis, LPSs were produced in the preparative scale. The bacterial mass was harvested from solid medium with 0.5% NaCl solution containing 0.01% phenol, washed three times with the same solution, and separated by centrifugation. The washed cells were extracted with Tris-EDTA buffer (0.02 M Tris-HCl [pH 7.5 to 8.0], 0.0025 M EDTA, 3% NaCl, 0.1% NaN3) by stirring at 8,000 rpm for 4 h at room temperature. Insoluble material was removed by centrifugation, and the supernatant was intensively dialyzed against distilled water. The LPS and LPS-Opr complexes were precipitated by solid ammonium sulfate at 50% saturation. The precipitate was dissolved in 0.02 M Tris-HCl buffer (pH 8.0) containing Mg2+ and Ca2+. To deproteinize the LPSs, proteinase K was added (0.05 mg/ml) and the solution was dialyzed against several changes of the same buffer for 10 h. The deproteinized LPS was precipitated with ammonium sulfate. The precipitate was dissolved in a minimal volume of distilled water, dialyzed against distilled water, and lyophilized.
LPSs were degraded by hydrolysis with aqueous 2% acetic acid for 1.5 h at 100°C. High-molecular-mass OPSs were obtained by gel permeation chromatography of the water-soluble carbohydrate portion on a column (70 cm by 2.6 cm) of Sephadex G-50 with 0.05 M pyridinium acetate (pH 4.5) as the eluent and monitoring with a Knauer (Berlin, Germany) differential refractometer. Smith-degraded polysaccharides were prepared by oxidation of OPSs with 0.1 M NaIO4 for 36 h at room temperature in the dark followed by reduction with an excess of NaBH4, acidification with acetic acid, dialysis against distilled water, and lyophilization (15). A linear polysaccharide, 2As, composed on average of 10 trisaccharide O repeats (2A) (Table 1) was synthesized as described previously (60).
Chemical analyses.
Hydrolysis was performed with 2 M trifluoroacetic acid (120°C; 2 h), and d-rhamnose was identified by gas-liquid chromatography as alditol acetate (55) and acetylated (+)-2-octyl glycosides (33, 57) on a Hewlett-Packard 5880 instrument equipped with a DB-5 fused-silica capillary column with a temperature gradient of 160°C (1 min) to 250°C at 3°C/min. Methylation was performed with methyl iodide in dimethyl sulfoxide in the presence of solid sodium hydroxide (8); the methylated polysaccharide was recovered by extraction with ethyl acetate after dilution of the reaction mixture with water and hydrolyzed, and partially methylated alditol acetates were derived and analyzed by gas-liquid chromatography as described above.
NMR spectroscopy.
Samples were deuterium exchanged by lyophilization from 2H2O. One- and two-dimensional 1H and 13C nuclear magnetic resonance (NMR) spectra were recorded with a Bruker (Karlsruhe, Germany) DRX-500 spectrometer for solutions in 2H2O at 60°C. Bruker XWINNMR 1.2 software was used to acquire and process the NMR data. Chemical shifts are reported with internal acetone (δH, 2.225; δc, 31.45). A mixing time of 250 ms was used in a rotating-frame nuclear Overhauser effect (ROESY) experiment.
Production and characterization of MAbs.
P. syringae strains with linear and branched OPSs of LPSs were used as immunogens to produce MAbs in mice (Table 2). Immunization, generation of hybridomas, selection of particular cell lines, and defining of immunoglobulin classes and subclasses of MAbs were performed as previously described (44, 46). Initially, a number of hybridoma cell lines which synthesized MAbs reactive in enzyme-linked immunosorbent assay (ELISA) with the homologous antigen were produced. Based on the specificities and affinities of MAbs in different immunoassays with different antigens, including LPSs and OPSs with known structures of O repeats, a few suitable clones were selected which might vary slightly in affinity. After repeated recloning, most stable specific cell lines retaining the ability to produce MAbs were finally selected. Some cell lines with similar affinities and specificities were chosen based on the production of MAbs of different murine immunoglobulin classes and subclasses.
TABLE 2.
Murine MAbs against P. syringae LPSs with linear and branched OPSs
| MAb
|
Isotypeb | Produced against P. syringae pathovar and strain: | |
|---|---|---|---|
| Name | Former namea | ||
| Ps(1-2)a | Ps1a | IgG3 | atrofaciens IMV 7836 |
| Ps(1-2)a1 | IgG1 | helianthi CFBP 2149 | |
| Ps1a | Ps1a′ | IgG3 | atrofaciens IMV 7836 |
| Ps1a1 | IgG1 | avellanae ISPaVe C4 | |
| Ps1a2 | Ps1x | IgG1 | atrofaciens IMV 7836 |
| Ps1b | Ps1b | IgM | atrofaciens IMV K-1025 |
| Ps1c | Ps1c | IgM | atrofaciens NCPPB 2612 |
| Ps1c1 | IgG3 | atrofaciens NCPPB 2612 | |
| Ps1d | Ps1d | IgG3 | phaseolicola IMV 120a |
| Ps(1-2)d | IgG1 | phaseolicola IMV 120a | |
| Ps(1-2)d1 | IgM | tagetis ICMP 6370 | |
| Ps2a1 | IgG3 | morsprunorum GSPB 854 | |
| Ps2d | IgG2a | morsprunorum CFBP 1650 | |
As described in our previous report (4).
Ig, immunoglobulin.
MAbs (i.e., hybridomas) were designated as follows: Ps, P. syringae; 1, 2, (1-2), 3, and 4 indicate the O serogroup specificity [(1-2) indicates that the epitope is shared by serogroups O1 and O2]; a, a1, and a2 are epitopes localized within the OPS backbone; b, c, c1, d, and d1 are epitopes that include lateral sugar substituents (44, 46). The symbols ↓ and ↑ show weak and strong exposure of the epitope in ELISA. The designations of some MAbs produced earlier (46) were changed (Table 2).
Immunoassays.
Agglutination, ELISA, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and Western immunoblotting were performed basically as described previously (44, 46). Crude and proteinase K-digested LPSs, purified OPSs, Smith-degraded OPSs, and synthetic polysaccharide 2As were used to coat Nunc-Immuno MaxiSorp surface ELISA plates (Nunc, Roskilde, Denmark).
RESULTS
Structural studies of a linear O polysaccharide of chemotype 1A.
Sugar analysis of a chemotype 1A OPS from LPS of P. syringae pv. atrofaciens IMV 7836, including determination of the absolute configuration, revealed d-rhamnose as the only OPS component. Methylation analysis resulted in identification of equal amounts of 2,4-di-O-methylrhamnose and 3,4-di-O-methylrhamnose. Hence, OPS is linear, all rhamnose residues are in the pyranose form, and half of them are 3 substituted and half are 2 substituted.
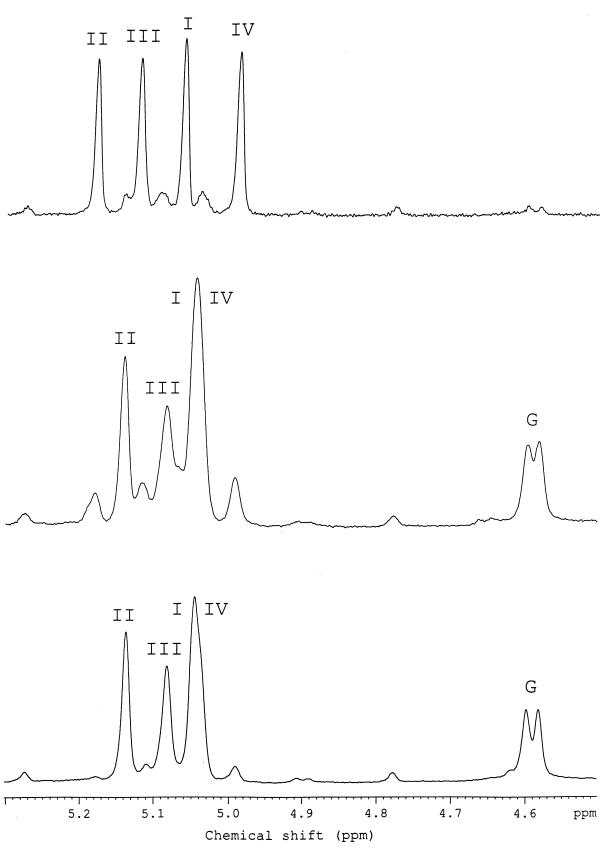
The 1H NMR spectrum of OPS (Fig. 1) contained, inter alia, signals for four anomeric protons at δ 4.96 to 5.17. Accordingly, the 13C NMR spectrum contained signals for four anomeric carbons at δ 102.1 to 103.4. Therefore, the O repeat of OPS contains four rhamnose residues, which were enumerated according to their sequence in the O repeat (RhaI to RhaIV; see below).
FIG. 1.
1H NMR spectrum of OPS of P. syringae pv. atrofaciens IMV 7836.
The 1H and 13C NMR spectra of the OPSs were assigned by using two-dimensional correlation spectroscopy and H-detected 1H,13C heteronuclear multiple-quantum coherence experiments, respectively (Table 3). The positions of the signals for H-5 at δ 3.72 to 3.87 and C-5 at δ 70.5 to 70.7 demonstrated that all rhamnose residues are α linked (compare the H-5 chemical shift δ 3.86 in α-Rhap but δ 3.39 in β-Rhap [22] and the C-5 chemical shift δ 70.0 in α-Rhap but δ 72.3 in β-Rhap [34]).
TABLE 3.
1H and 13C NMR data for the OPS of P. syringae pv. atrofaciens IMV 7836
| Sugar residue | Chemical shift (ppm) for:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H-1 | H-2 | H-3 | H-4 | H-5 | H-6 | C-1 | C-2 | C-3 | C-4 | C-5 | C-6 | |
| →3)-α-l-RhapI-(1→ | 4.96 | 4.15 | 3.83 | 3.56 | 3.76 | 1.27 | 103.1 | 71.1 | 79.1 | 72.5 | 70.6 | 17.9 |
| →3)-α-l-RhapII-(1→ | 5.03 | 4.12 | 3.90 | 3.57 | 3.87 | 1.27 | 103.2 | 71.2 | 79.1 | 72.8 | 70.4 | 17.9 |
| →2)-α-l-RhapIII-(1→ | 5.17 | 4.07 | 3.95 | 3.49 | 3.82 | 1.30 | 102.0 | 79.2 | 71.2 | 73.4 | 70.4 | 17.8 |
| →2)-α-l-RhapIV-(1→ | 5.11 | 4.08 | 3.89 | 3.48 | 3.72 | 1.30 | 101.9 | 79.2 | 71.1 | 73.5 | 70.5 | 17.8 |
Downfield displacements of the signals for C-3 of RhaI and RhaII and C-2 of RhaIII and RhaIV to δ 79.1 to 79.3, compared with their positions in the spectrum of nonsubstituted α-Rhap at δ 71.3 to 71.6 (34), demonstrated the substitution pattern of the rhamnose residues.
A two-dimensional ROESY experiment (the spectrum is not shown) revealed the following interresidue correlations between transglycosidic protons: RhaI H-1 and RhaII H-3, RhaII H-1 and RhaIII H-2, RhaIII H-1 and RhaIV H-2, RhaIV H-1 and RhaI H-3 at δ 5.03 and 3.83, 4.96 and 4.08, 5.11 and 4.07, and 5.17 and 3.90, respectively. In addition, the spectrum showed a strong cross peak, RhaII H-1 and RhaIII H-1, and weaker cross peaks, RhaIII H-1 and RhaIV H-1, RhaIII H-1 and RhaII H-5, and RhaIV H-1 and RhaIII H-5, all typical of (α1→2)-linked rhamnose disaccharides. Intraresidue H-1-H-2 cross peaks, but no H-1-H-3, H-5 cross peaks, were present for all rhamnose residues, thus confirming their α configuration.
Therefore, the ROESY data were in accord with the methylation and 13C NMR chemical shift data and finally confirmed that OPS of P. syringae pv. atrofaciens IMV 7836 is a linear OPS of chemotype 1A. The 1H NMR spectra of OPSs of P. syringae pv. glycinea GSPB 1991, P. syringae pv. helianthi CFBP 2149, and P. syringae pv. helianthi CFBP 1732 (NCPPB 2639) were identical to that of strain IMV 7836, and therefore, the OPSs have the same structure. It is worth noting that P. syringae pv. atrofaciens IMV 7836 was originally isolated and characterized serologically as having a chemotype 1C-1A OPS, indicating that chemotype 1C is unstable (see below).
Structural studies of irregular branched OPS of chemotype 1C-1A.
Three strains were selected for structural studies of OPSs, namely, P. syringae pv. glycinea CFBP 3192 (United States, 1990), NCPPB 1783 (CFBP 3211) (Yugoslavia, 1962), and NCPPB 3318 (CFBP 3215) (Italy, 1983). The OPSs of these strains were described by the serological formulae O1[(1-2)a,1c,1c1], O1[(1-2)a,(1-2)a1↓,1a,1a1,1c,1c1], and O1[(1-2)a,(1-2)a1↓,1a,1a1,1a2,1c], respectively, indicating the lack of strict regularity in the last two. The structural heterogeneity of the OPSs could be observed most clearly in the anomeric regions of the 1H NMR spectra, which are shown in Fig. 2, whereas less sensitive 13C NMR spectroscopy could not reliably reveal minor structures.
FIG. 2.
Resonance region of anomeric protons in the 1H NMR spectra of OPSs of P. syringae pv. glycinea NCPPB 3318 (top), NCPPB 1783 (middle), and CFBP 3192 (bottom). The Roman numerals refer to Rha residues enumerated as shown in Table 1; G, GlcNAc.
The 1H NMR spectrum of OPS of P. syringae pv. glycinea CFBP 3192 (Fig. 2, bottom) was practically identical to that of P. syringae pv. atrofaciens IMV 4394, which was reported to have a branched structure of chemotype 1C with the backbone 1A and the lateral (β1→4)-linked d-GlcNAc residue (Table 1) (27). The 1H NMR spectrum of OPS of P. syringae pv. glycinea NCPPB 1783 contained signals of the chemotype 1C O repeats as the major series, including those for H-1 of four Rha residues at δ 5.04 (2H), 5.08, and 5.14 (all broadened singlets) and one GlcNAc residue at δ 4.59 (d, J1,2 8 Hz) (Fig. 2, middle). The H-1 signals for four Rha residues of the chemotype 1A O repeats formed a minor series in this spectrum; the content of the minor O repeats could be estimated as 15% of the total. A smaller amount of the same minor series (<5%) could be detected in OPS of P. syringae pv. glycinea GSPB 3192 (Fig. 2, bottom). On the other hand, the spectrum of OPS of P. syringae pv. glycinea NCPPB 3318 contained signals for four Rha residues from the 1A O repeats as the major series in particular, signals for H-1 at δ 4.99, 5.06, 5.12, and 5.18 [Fig. 2, top]) and those from the 1C O repeats as the minor series, with the content of the latter estimated at about 10%.
Therefore, in addition to OPSs of “pure” chemotypes 1A and 1C that are characterized by the regular linear and regular branched O repeats, respectively, P. syringae strains can produce OPSs of a mixed (or transitional) chemotype, 1C-1A, which contain both types of O repeats in different ratios.
Characterization of MAbs and inferring of OPS-specific epitopes.
The MAbs selected for the current study are listed in Table 2. They had the following features in common: (i) they agglutinated heat-killed homologous bacterial cells, (ii) they reacted in ELISA with homologous purified OPSs and crude and deproteinized LPSs, and (iii) they revealed ladder-like profiles with smooth LPSs in Western immunoblotting. None of the polysaccharides that were released from LPSs by acidic hydrolysis reacted with MAb Pscor1, specific to the LPS core moiety (44), whereas each corresponding nondegraded LPS did. Specific epitopes recognized by these MAbs, except for epitope (1-2)a1, were sensitive to Smith degradation. A number of MAbs, which were designated by lowercase a, reacted with the homologous linear OPSs (Table 4), and hence, their specific epitopes are located within the backbone 1A (Table 1).
TABLE 4.
Immunochemical characterization of P. syringae LPSs with known OPS structures by using OPS-specific MAbs
| OPS chemotype | P. syringae pathovar and strain | Reactivity of proteinase K-digested LPSs in ELISA and Western immunoblotting with MAb:
|
|||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ps(1-2)a
|
Ps(1-2)a1
|
Ps1a
|
Ps1a1
|
Ps1a2
|
Ps1b
|
Ps1c
|
Ps1c1
|
Ps1d
|
Ps(1-2)d
|
Ps(1-2)d1
|
Ps2d
|
||||||||||||||
| Ea | Wb | E | W | E | W | E | W | E | W | E | W | E | W | E | W | E | W | E | W | E | W | E | W | ||
| 1A | atrofaciens IMV 7836 | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 1A | helianthi CFBP 2149 | + | + | + | + | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 1B | atrofaciens IMV K-1025 | + | + | + | + | + | + | − | − | − | − | + | + | − | − | − | − | − | − | − | − | − | − | − | − |
| 1C | atrofaciens IMV 4394 | + | − | − | − | − | − | − | − | − | − | − | − | + | + | + | ± | − | − | − | − | − | − | − | − |
| 1C-1Ac | glycinea NCPPB 1783 | + | + | ± | − | + | + | + | ± | − | − | − | − | + | + | + | ± | − | − | − | − | − | − | − | − |
| 1C-1Ad | glycinea NCPPB 3318 | + | + | + | + | + | + | + | + | + | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − |
| 1D | phaseolicola IMV 120a | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | + | + | + | − | − | − | − |
| 1D | tagetis ICMP 6370 | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | + | − | − | + | + | − | − |
| 1D-1Ae | phaseolicola GSPB 1552 | + | + | − | − | + | + | − | − | − | − | − | − | − | − | − | − | + | + | + | + | − | − | − | − |
| 2A | morsprunorum GSPB 883 | + | ± | + | ± | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 2D | morsprunorum CFBP 1650 | ± | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | + |
| 2D-2Af | morsprunorum GSPB 854 | + | ± | + | ± | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | + |
| 2ASg | ± | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
E, ELISA. Symbols: +, ±, and − correspond to optical densities at 492 nm of 1.5 to 2.0, 0.4 to 0.8, and 0 to 0.2, respectively.
W, Western blotting. Symbols: +, ±, and − correspond to strong, weak, and no reactivity, respectively.
The content of 1A linear O repeats is ∼15%.
The content of 1A linear O repeats is ∼90%.
The content of 1A linear O repeats is <10%.
The content of 2A linear O repeats is ∼60%.
A synthetic polysaccharide containing ∼10 2A O repeats.
MAbs to the backbone 1A also showed specific binding activity to heterologous branched LPSs and OPSs having an α-d-rhamnan backbone (Table 4) but not to those with l-rhamnan or mixed l- and d-rhamnan backbones (data not shown). Of these, MAb Ps(1-2)a demonstrated the widest spectrum of cross-reactivity. It reacted with LPSs and OPSs of both regular branched and mixed chemotypes 1B, 1C, 1D, 1A-1C, and 1A-1D in ELISA and, except for chemotypes 1C and 1D, in Western immunoblotting. MAb Ps(1-2)a also displayed a marked reactivity in ELISA with a synthetic polysaccharide, 2As (60), as well as with LPSs and OPSs of chemotype 2A and mixed chemotype 2A-2D; a weaker reactivity was observed with the crude LPS of chemotype 2D (Table 4). The crude LPSs of chemotypes 2A and 2A-2D reacted slightly in Western immunoblotting as well. These data showed that epitope (1-2)a occurs within both linear α-d-rhamnan backbones 1A and 2A containing four or three Rha residues in the O repeat. Therefore, ELISA with crude LPS extracts and MAb Ps(1-2)a may serve as a tool for identification of P. syringae serogroup O1 and O2 strains having an α-d-rhamnan OPS backbone. The wide reactivity and the lability towards Smith degradation suggested that MAb Ps(1-2)a recognizes a trisaccharide, α-d-Rhap-(1→2)-α-d-Rhap-(1→3)-α-d-Rhap, which corresponds to the fragments RhaIII-(1→2)-RhaIV-(1→3)-RhaI and RhaII-(1→2)-RhaIII-(1→3)-RhaI within the O repeats 1A and 2A, respectively. The lateral Fuc residue which is attached to RhaII almost completely masked epitope (1-2)a in chemotype 2D but only partially in chemotype 1D. Other lateral substituents, d-Rha and d-GlcNAc, affect the exposure of epitope (1-2)a in chemotypes 1B and 1C insignificantly.
MAb Ps(1-2)a1 cross-reacted in ELISA with LPSs and OPSs of chemotypes 1B, 1C-1A, 2A, and 2D-2A but not with those of chemotypes 1C, 1D, 1D-1A, and 2D (Table 4). It recognized neither regular branched chemotype 1D nor mixed chemotype 1D-1A OPS (e.g., in P. syringae pv. phaseolicola GSPB 1552). This could be accounted for by masking of the RhaII residue within epitope (1-2)a1 by the lateral d-Fuc residue in chemotype 1D and too low a content (<5%) of the 1A linear O repeats in chemotype 1D-1A. We could not chemically confirm the presence of the minor 1A linear O repeats in the OPS of strain P. syringae pv. phaseolicola GSPB 1552, but we inferred chemotype 1D-1A serologically (see below). On the other hand, epitope (1-2)a1 is strongly exposed by both chemotypes 2A and 2D-2A (Table 4). This is consistent with the previous findings that the content of the 2A linear O repeats in the mixed chemotype 2D-2A OPS is significant and could be demonstrated both serologically (46) and chemically (29). Epitope (1-2)a1 is only slightly exposed in ELISA, and not in Western immunoblotting, in OPSs of mixed chemotype 1C-1A with a low content of the 1A linear O repeats (e.g., in P. syringae pv. glycinea NCPPB 1783) and hence is also affected by the lateral d-GlcNAc residue attached to RhaI (chemotype 1C). Epitope (1-2)a1 is not influenced by the lateral d-Rha which is attached to RhaIII (chemotype 1B). Remarkably, unlike MAb Ps(1-2)a, MAb Ps(1-2)a1 did not react with the synthetic polysaccharide 2As (60) (Table 4) and a chemotype 2A OPS of Xanthomonas campestris pv. phaseoli var. fuscans GSPB 271 (unpublished data). This finding suggested that MAb Ps(1-2)a1 is specific to a region of the linkage between the OPS and the LPS core, which is suggested to be the same in all P. syringae O1 and O2 LPSs but different in X. campestris GSPB 271 LPS and absent from the synthetic polysaccharide 2AS. This region should be stable to periodate oxidation, since, unlike other backbone-located epitopes, epitope (1-2)a1 is resistant to Smith degradation.
MAb Ps1a cross-reacted both in ELISA and Western immunoblotting with LPSs and OPSs of chemotypes 1A and 1B, as well as those of mixed chemotypes 1C-1A and 1D-1A (Table 4). Unlike epitope (1-2)a1, the epitope 1a is exposed by OPS of chemotype 1D-1A with a low content of 1A linear O repeats. No LPS of regular branched chemotypes 1C and 1D reacted with MAb Ps1a in any immunoassay. MAb Ps1a is most useful for discrimination between strains with OPSs having the backbones 1A and 2A. It could be suggested that epitope 1a is associated with an O repeat fragment containing two consecutive 2-substituted Rha residues, RhaII-(1→2)-RhaIII-(1→2)-RhaIV, which is present in the backbone 1A but absent from the backbone 2A.
MAb Ps1a1 showed strong reactivity in ELISA with some LPSs and OPSs and a slight reactivity in Western immunoblotting with crude LPS of linear chemotype 1A (e.g., with those of the P. syringae pathovars atrofaciens IMV 7836 and helianthi NCPPB 2639 but not helianthi CFBP 2149). In addition, it reacted similarly with LPSs and OPSs of mixed chemotype 1C-1A with a high content of the linear O repeat 1A (e.g., P. syringae pv. glycinea NCPPB 1783 and NCPPB 3318) but did not react with those of chemotypes 1B, 1C, and 1D. Thus, epitope 1a1 is influenced by any lateral substituent. MAb Ps1a2 reacted in ELISA with LPSs, but not with OPSs, of chemotype 1A only. Hence, this epitope is completely masked by any lateral substituent. Like epitope 1a, epitopes 1a1 and 1a2 are not exposed by any OPSs with the backbone 2A, whether linear or branched, and are destroyed by Smith degradation. Remarkably, despite the fact that the 1A O repeats of OPSs of P. syringae pv. atrofaciens IMV 7836 and P. syringae pv. helianthi CFBP 2149 were shown to be chemically identical (see above), epitopes 1a1 and 1a2 are exposed only in the former (Table 4). These findings suggested that the exposure of epitopes 1a1 and 1a2 depends not only on the primary O repeat structure but also on the whole OPS conformation.
MAb Ps1b was produced against P. syringae pv. atrofaciens IMV K-1025 with an OPS of chemotype 1B (Table 4) (30). This MAb is specific to an epitope which includes the lateral (α1→3)-linked d-Rhap as the immunodominant sugar residue and has been characterized previously (46).
MAb Ps1c reacted strongly in any immunoassay with LPSs and OPSs of chemotypes 1C and 1C-1A independent of the content of 1A linear O repeats (Table 4). This finding confirmed the inference of epitope 1C as specific to the lateral (β1→4)-linked d-GlcpNAc residue (27, 46). The newly produced MAb Ps1c1 showed strong reactivity with OPS of chemotype 1C and weak reactivity with OPS of chemotype 1C-1A with a low content of 1A linear O repeats. Hence, like epitope 1c, epitope 1c1 is related to the lateral d-GlcNAc residue but, unlike epitope 1c, it alternates with epitopes (1-2)a1, 1a1, and 1a2. The reactivity of epitope 1c1 in Western immunoblotting was affected by treatment of LPS by proteinase K, suggesting that its exposure depends on the presence of LPS-associated proteins. These findings suggested that epitope 1c1 includes not only the lateral d-GlcNAc residue but also one or more d-Rha residues of the backbone 1A and is more influenced by the protein-dependent conformation of the OPS chain than epitope 1c.
MAbs Ps1d and Ps2d have been produced previously (46) and shown to be related to the lateral (α1→4)-linked d-Fucf residue in OPSs of chemotypes 1D (28) and 2D (29), respectively (Tables 1 and 4). The newly produced MAb Ps(1-2)d reacted strongly with LPSs of chemotypes 1D and 1D-1A. Epitope (1-2)d is coexposed with epitope 1d in all LPSs of chemotypes 1D and 1D-1A (Table 4) and with epitopes 2d and (1-2)d1 in some LPSs of chemotypes 2D, 2D-2A, and 1D (data not shown).
Another new Mab, Ps(1-2)d1, was produced against P. syringae pv. tagetis ICMP 6370, which belongs to serotype O1[(1-2)a↑,(1-2)d1,1d] and has OPS of chemotype 1D. It reacted with LPS of chemotype 1D in ELISA and Western immunoblotting. In the latter immunoassay, MAb Ps(1-2)d1 revealed ladder-like profiles of smooth LPS identical to those revealed by MAb Ps1d (data not shown), indicating that their epitopes are located within the same LPS molecules. Epitopes (1-2)d and (1-2)d1 strictly alternate in LPSs of strains of P. syringae pv. tagetis. Differences in the specificity of d-Fucf-containing epitopes may have the same nature as those of epitopes 1c and 1c1 (see above).
Western immunoblotting.
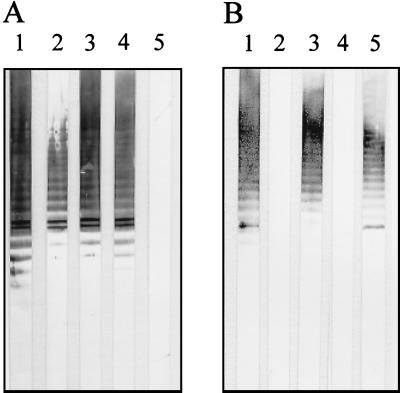
Proteinase K-digested LPSs from P. syringae pv. glycinea NCPPB 1783 and NCPPB 3318 having irregular OPSs of mixed chemotype 1C-1A with ∼15 and ∼90% 1C O repeats, respectively, were analyzed by Western immunoblotting with MAb Ps1c specific to the lateral (β1→4)-linked d-GlcpNAc and backbone-specific MAbs. As can be seen from Fig. 3, each of the LPSs showed practically identical ladder-like banding patterns with both types of MAbs. This finding suggested that in chemotype 1C-1A OPSs both O repeats 1C and 1A enter into the same polysaccharide chain.
FIG. 3.
Western immunoblot analysis of proteinase K-digested LPSs from P. syringae pv. glycinea NCPPB 3318 (A) and NCPPB 1783 (B) with MAbs Ps(1-2)a (lanes 1), Ps(1-2)a1 (lanes 2), Ps1a (lanes 3), Ps1a1 (lanes 4), and Ps1c (lanes 5).
OPS chemotypes of strains classified in serogroup O1 and their distribution in P. syringae pathovars.
The structure of the LPS core oligosaccharide of P. syringae pathovars and related phytopathogenic pseudomonads has not been elucidated yet. However, most of the bacteria listed in Materials and Methods cross-reacted with a panel of core-specific MAbs (44, 45), indicating structural similarities of their LPS core moieties. Since the core structure is known as a conservative part of LPS (19), this finding indicated a close relatedness of these bacteria. A serological screening of P. syringae strains with OPS-specific MAbs showed that strains with linear and branched OPSs having the backbone 1A (Table 1) emerged among pathovars actinidiae, aptata, atrofaciens, avellanae, glycinea, helianthi, japonica, panici, phaseolicola, philadelphi, pisi, primulae, tagetis, striafaciens, and syringae (Table 5 [only the most representative pathovars are shown]). No strain of other P. syringae pathovars, related pseudomonads, or other bacterial species tested cross-reacted with any serogroup O1-specific MAb, and therefore, they are not shown in Table 5.
TABLE 5.
Serotyping in ELISA of strains of different P. syringae pathovars classified in serogroup O1
| O1 serogroup serotypea | No. of strains of P. syringae pathovarb:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| actinidiae (12) | aptata (41) | atrofaciens (94) | avellanae (61) | glycinea (80) | helianthii (19) | pisi (31) | phaseolicola (71) | tagetis (5) | syringae (73) | |
| (1-2)a,(1-2)a11a,1a1,1a2 | 1 | 21 | 5 | 3 | 7 | |||||
| (1-2)a,(1-2)a11a | 2 | 1 | 20 | 12 | 1 | |||||
| (1-2)a,(1-2)a1,1a,1b | 45 | 17 | ||||||||
| (1-2)a↓,1c,1c1 | 2 | 19 | 2 | 5 | ||||||
| (1-2)a,1a,1c,1c1 | 35 | 22 | 7 | 27 | 16 | |||||
| (1-2)a,(1-2)a1,1a,1a1,1c,1c1 | 5 | 3 | 15 | |||||||
| (1-2)a,(1-2)a1,1a,1a1,1a2,1c↓ | 13 | |||||||||
| (1-2)a↓,(1-2)d,1d | 6 | 10 | 3 | 46 | ||||||
| (1-2)a,(1-2)d,1a,1d | 19 | 16 | ||||||||
| (1-2)a↑,(1-2)d1,1d | 5 | |||||||||
| Rough | 2 | 14 | 1 | 1 | 1 | 4 | ||||
Epitopes 1b, 1c, and 1d correspond to serogroups SYR1, APTPIS, and PHA, respectively; serotypes O1[(1-2)a,(1-2)a11a,1a1,1a2] and O1[(1-2)a,(1-2)a11a] correspond to serogroups HEL1 and HEL2 of the classification scheme of Saunier et al. (54).
Only the strains classified in serogroup O1 are shown. A limited number of strains from pathovars panici, philadelphi, primulae, and striafaciens were classified in serogroup O1, and therefore they are not shown. The total number of strains in each pathovar is shown in parentheses.
Different numbers of strains with the linear OPS of chemotype 1A (Table 1) were found within pathovars actinidiae, aptata, atrofaciens, avellanae, glycinea, helianthi, philadelphi, pisi, and syringae (Table 5). However, despite the fact that their OPSs were inferred to have the same chemotype, 1A, serologically they are not identical. Some strains of pathovars glycinea and helianthi were characterized by two serotypes, O1[(1-2)a,(1-2)a1,1a,1a1,1a2] and O1[(1-2)a,(1-2)a1,1a], whereas some strains of pathovars avellanae and syringae belonged to the former serotype only. Remarkably, when isolated, strains P. syringae pv. atrofaciens IMV 7836 and P. syringae pv. glycinea CFBP 3187, CFBP 3190, and CFBP 3360 exposed epitope 1c (54), characteristic of branched OPSs with the lateral d-GlcNAc residue (chemotypes 1C and 1C-1A). Later, during cultivation under laboratory conditions, they altered to the linear OPS chemotype 1A, as was also proved chemically for strain IMV 7836 (see above).
In contrast, OPS chemotype 1B (Table 1) showed no alteration. Strains of the corresponding serotype O1[(1-2)a,(1-2)a1,1a,1b] were found mainly in P. syringae pv. atrofaciens and to a lesser extent in pathovars japonica, striafaciens, and syringae. Since pathovars japonica and striafaciens are no longer valid (67), we suggest reclassifying the corresponding strains in pathovar atrofaciens.
OPSs of chemotype 1C and mixed chemotype 1C-1A (Table 1) are broadly spread among P. syringae pathovars classified in serogroup O1. They emerged to different extents in pathovars aptata, atrofaciens, glycinea, japonica, panici, pisi, and syringae (Table 5). A common feature of the corresponding strains is their tendency to alter the OPS chemotype from branched regular 1C to linear 1A via a cascade of transitional serotypes corresponding to mixed chemotype 1C-1A with different ratios of the O repeats 1C and 1A (Tables 1 and 4). The extent and completeness of the alteration varies from pathovar to pathovar. Thus, strains of pathovars aptata, atrofaciens, and pisi exhibited a restricted cascade of the transitional serotypes O1[(1-2)a↓,1c,1c1], O1[(1-2)a,1a,1c,1c1], and O1[(1-2)a,(1-2)a1↓,1a,1c,1c1], the most typical being serotype O1[(1-2)a,1a,1c,1c1]. Strains of P. syringae pv. glycinea demonstrated the widest spectrum of transitional serotypes, which can be described as follows: O1[(1-2)a↓,1c,1c1]↔O1[(1-2)a,1a↓,1a1↓, 1c,1c1]↔O1[(1-2)a,(1-2)a1↓,1a,1a1,1c,1c1]↔O1[(1-2)a,(1-2)a1, 1a,1a1,1a2,1c,1c1↓]↔O1[(1-2)a,(1-2)a1,1a,1a1,1a2,1c↓]→ O1[(1-2)a,(1-2)a1,1a] or O1[(1-2)a,(1-2)a1,1a,1a1,1a2]. However, most strains of P. syringae pv. glycinea belonged to serotypes O1[(1-2)a↓,1c,1c1] and O1[(1-2)a,(1-2)a1,1a,1a1,1a2,1c↓]. The serotype of some strains varied, depending on the growth conditions (data not shown), and therefore only the most typical transitional serotypes are shown in Table 5. P. syringae pv. glycinea CFBP 3187, CFBP 3190, and CFBP 3360, which had been originally described as exposing epitope 1c (chemotype 1C) (54), were shown later to convert under laboratory conditions into chemotype 1A (52a). We have found the culture conditions under which some OPS serotypes, including 1C and 1D, can be altered, and these data will be published elsewhere. Previously, based on the elucidation of a partial OPS structure, a variable content of the lateral d-GlcNAc residue has been reported in OPSs of a number of P. syringae pv. glycinea strains (3, 63). Together, these findings allowed the final conclusion that chemotype 1C is susceptible to 1C↔1C-1A→1A alterations under both natural and laboratory conditions.
OPS chemotype 1D and mixed chemotype 1D-1A (Table 1), as well as the corresponding two serotypes O1[(1-2)a↓,(1-2)d,1d] and O1[(1-2)a,(1-2)d,1a,1d], are typical of most strains of pathovar phaseolicola and some strains of pathovars actinidiae, atrofaciens, avellanae, helianthi, and syringae (Table 5). Under laboratory conditions, O1[(1-2)a↓,(1-2)d,1d]↔O1[(1-2)a,(1-2)d,1a,1d] serotype alterations have been observed only for pathovar phaseolicola. However, no strain of this pathovar belonged to serotype O1[(1-2)a,(1-2)a1,1a] (chemotype 1A). Most likely, the difference between the two serotypes is associated with the absence (chemotype 1D) or the presence (chemotype 1D-1A) of minor 1A linear O repeats in their OPSs (Tables 1, 4, and 5). The ability of chemotype 1D strains to lose the lateral α-d-Fucf residue from the OPS may account for the origin of chemotype 1A among strains of pathovars actinidiae, avellanae, and helianthi (see above).
Strains of a new stable serotype, O1[(1-2)a↑,(1-2)d1,1d], were described in P. syringae pv. tagetis (Table 4). In addition, P. syringae CFBP 4091, one of two strains of pathovar primulae tested, fell into the same serogroup (data not shown in Table 5). As in pathovar phaseolicola, the OPS of pathovar tegetis was of chemotype 1D (Table 1). The serological difference between strains of these two pathovars could be related to a minor modification of the OPS structure, which remains to be elucidated by additional chemical analyses.
To sum up, strains of P. syringae pathovars and other phytopathogenic pseudomonads within serogroup O1 can be characterized by a short or extended serotype, and it seems reasonable to characterize all collection strains by such a “serological passport.” Using a panel of MAbs specific to the LPS core, OPS backbone, and lateral substituents, certain strains can be classified in serologically homogeneous or heterogeneous pathovars. Together with the data on the host plant and the specific symptoms, the serotyping of freshly isolated strains may have a high practical impact. Therefore, MAbs against P. syringae LPSs may serve as tools in broad epidemiological and ecological studies of P. syringae pathovars and related bacteria.
DISCUSSION
Chemical studies of OPSs of LPSs of P. syringae pathovars and related pseudomonads have revealed a limited compositional and structural diversity (24–30, 58, 68). Using specific MAbs, a correlation between the molecular structure (chemotype) and the immunospecificity (serotype) of P. syringae OPSs has been demonstrated (24, 25, 44–46). These and the present data show that MAbs are a useful tool for inferring P. syringae OPS primary structures. In particular, the use of a panel of MAbs allows one (i) to identify strains with OPSs having an α-d-rhamnan backbone, (ii) to distinguish between OPSs of chemotypes 1A and 2A characterized by four or three α-d-Rha residues in the backbone O repeat, respectively, (iii) to determine linear, branched regular, and branched irregular OPS primary chemotypes, (iv) to estimate the degree of heterogeneity in branched irregular OPSs, and (v) to identify lateral substituents and the modes of their linkage in branched OPSs.
The data obtained also allowed a better understanding of the immunospecificity of P. syringae OPSs on the molecular level. Some backbone-associated epitopes depend strictly on the primary structure of the OPS O repeat. They are tightly colocalized or even overlapped within backbones 1A and 2A but seem to have different immunodominant rhamnose residues. Other epitopes may be conformation dependent or located in an intermediate region of the attachment of the OPS to the core of LPS. In branched OPSs, the immunodominant groups are the lateral sugar substituents. In OPSs of chemotypes 1B and 1C, these are (α1→3)-linked d-Rha and (β1→4)-linked d-GlcNAc, which define epitopes 1b and 1c (1c1), respectively (45, 46). Epitopes (1-2)d, (1-2)d1, 1d, and 2d in OPSs of basic chemotypes 1D and 2D are associated with the lateral (α1→4)-linked d-Fuc residue but seem to also include different neighboring rhamnose residues of the backbone that could be immunodominant for the particular epitopes. The possibility that some of these epitopes appeared as a result of postpolymerization modifications in LPS cannot be excluded.
Immunochemical studies of branched and linear OPSs by using MAbs demonstrated 1C (regular branched)→1C-1A (irregular branched)→1A (linear) chemotype alterations which correlated with the appearances of the backbone-located epitopes in the order (1-2)a→1a→1a1→(1-2)a1→1a2. Chemical studies showed that these serological variations are related to changes in the degree of substitution of the backbone 1A with the lateral d-GlcNAc residues. The same mechanism was proposed for 1D↔1D-1A chemotype alterations in branched OPSs with the lateral d-Fuc residues. However, the 1D↔1D-1A alterations were demonstrated only serologically and could not be confirmed chemically. More detailed chemical studies of serotype O1[(1-2)a,(1-2)d,1a,1d] OPS (suggested chemotype, 1D-1A) from P. syringae pv. phaseolicola GSPB 1552 are in progress. A similar phenomenon of nonstoichiometric glucosylation has been described for OPSs of some members of the family Enterobacteriaceae (17, 20, 43). It has been shown that the d-glucose residues are transferred to the OPS chain after polymerization and that the corresponding genes are located in the bacterial chromosome distant from the wbz (formerly rfb) cluster (16, 39, 43, 50).
A wide spectrum of P. syringae pathovars and some related phytopathogenic pseudomonads (14, 21, 67) from genomospecies 1 (pathovars aptata, atrofaciens, japonica, panici, and syringae), 2 (pathovars glycinea and phaseolicola), 7 (pathovars helianthi and tagetis), and 8 (pathovars avellanae and theae) were classified in serogroup O1. All of these bacteria revealed a cross-reactivity with a panel of core-specific MAbs, indicating a significant structural similarity of the LPS core moieties (44, 45). Furthermore, independent of the chemotype (1A, 1B, 1C, or 1D), all OPSs of this group possess identical backbone 1A and react with backbone-specific MAbs. Such relatedness of the LPS phenotypes of strains classified in serogroup O1 implies the relatedness of the corresponding LPS-encoding genes (20, 40, 49, 50). Even closer relatedness of these genes could be expected for the bacteria classified in each individual serotype within serogroup O1. Four basic serotypes, O1[(1-2)a,(1-2)a1,1a], O1[(1-2)a,(1-2)a1,1a,1b], O1[(1-2)a,1c,1c1], and O1[(1-2)a,(1-2)d,1d], were described within this serogroup, which correspond to OPS chemotypes 1A, 1B, 1C, and 1D, respectively. Independently of the pathovar and even the genomospecies (14), OPSs of chemotypes 1C and 1C-1A may undergo branched regular ↔ branched irregular → linear chemotype alterations. The OPS alteration to chemotype 1A may either reflect a natural mechanism of OPS diversification or be a case of “atavism” at the OPS structure level. Chemotype 1D undergoes only a branched regular → branched irregular alteration to chemotype 1D-1A. Chemotype 1B, which is characteristic mainly of pathovar atrofaciens (67), included in genomospecies 1 (14), did not show any alteration and thus may be considered most conservative.
A related homopolymer of α-d-rhamnose with a 2A trisaccharide O repeat (Table 1) also occurs widely in LPSs from different P. syringae pathovars; the corresponding strains are classified in serogroup O2. Remarkably, a linear OPS having the same structure of the chemical repeating unit (biological O repeats are unknown) has been found in LPSs from some strains of Burkholderia (formerly Pseudomonas) cepacia (7), Stenotrophomonas maltophilia (64), and X. campestris (30a). Moreover, it has also been described as a common, LPS-associated antigen called A-band polysaccharide (1, 31, 66) in P. aeruginosa strains of different serotypes, which are defined by another LPS-associated antigen, B-band polysaccharide or O-antigen (23, 36, 37). It is worth noting that in P. aeruginosa A-band and B-band polysaccharides are synthesized by two different pathways, one characteristic of homopolysaccharides (the Wzy [formerly Rfc]-independent pathway) and the other characteristic of heteropolysaccharides (the Wzy-dependent pathway) (5, 10, 52). A-band polysaccharide is assembled at the cytoplasmic face of the plasma membrane, and its polymerization is thought to occur by sequential sugar transfers by three α-d-rhamnosyl-transferases, WbpX, WbpY, and WbpZ, to a lipid intermediate (51). ATP-binding cassette transporter (or traffic ATPase) then translocates the polymerized polysaccharide across the plasma membrane prior to its ligation to core lipid A at the periplasmic space (52). The chromosomal genes wbpX, wbpY, and wbpZ are located in the A-band biosynthetic gene cluster (51). The genes encoding transferases for B-band LPS are localized in a different gene cluster. B-band O repeats (blocks) are synthesized on the cytoplasmic face of the inner membrane and then translocated by Wzx (formerly RfbX) to the periplasmic space, where they are polymerized by Wzy polymerase (the Wzy-dependent pathway). The B-band OPS length is modulated by Wzz (Rol, regulator of O-antigen length) (5, 6, 10). Recently, one gene, wbpL for transferase WbpL, that has been suggested to be required for initiation of both A-band and B-band LPS synthesis has been elucidated (6, 52).
Since B-band OPSs of P. aeruginosa (23) on the one hand and OPSs of P. syringae and related phytopathogenic pseudomonads (24–26, 68) on the other hand are structurally different, there is no reason to expect a close similarity in the genes encoding glycosyltransferases. In contrast, the A-band polysaccharide of P. aeruginosa is identical or similar to the OPS backbones of P. syringae strains classified in serogroups O2 and O1 with three and four α-d-Rha residues in the backbone O repeat (chemotypes 2A and 1A, respectively). Therefore, one can expect relatedness of the genes involved in biosynthesis of the corresponding LPSs in these two groups of bacteria. In future, it will be important to establish (i) by which basic pathway (Wzy independent or Wzy dependent) the synthesis of OPSs from serogroups O1 and O2 proceeds in P. syringae, (ii) whether there is any relatedness among the corresponding genes and enzymes, (iii) how close to each other the genes encoding assembly and transport of OPSs of P. syringae O1 and O2 are, and (iv) by which mechanism the lateral substitutions (Table 1) are transferred to the chemotype 1A and 2A OPS backbones.
The answers to these questions will shed light on the phylogenetic relatedness of the bacteria and on the origin of the A-band polysaccharide in P. aeruginosa and α-d-rhamnan-based OPS in P. syringae. For instance, could A-band polysaccharide be an ancient OPS which has been preserved during the evolution of P. aeruginosa strains and coexists now with diverse B-band polysaccharides as an example of molecular atavism? Could it be that OPS chemotype 1A in P. syringae, and later a number of branched OPS chemotypes 1B to 1D (Table 1), originated from OPS chemotype 2A as a result of vertical divergent evolution of LPS-encoding genes? A number of strains of P. syringae pathovars and some related phytopathogenic pseudomonads that have recently been delineated in different genomospecies (14) belong to serogroups O1 and O2 and synthesize OPSs of chemotypes shown in Table 1. If their LPS-encoding genes are related, the appearance of the corresponding phenotypes within different genomospecies could be a result of a horizontal transfer of the corresponding genes. Finally, it should be mentioned that the evolution of OPSs cannot be completely elucidated without an understanding of the molecular biology and biochemistry of the core moiety of LPS and its association with OPS.
ACKNOWLEDGMENTS
We thank Y. E. Tsvetkov (N. D. Zelinsky Institute of Organic Chemistry, Moscow, Russia) for the gift of a synthetic α-d-rhamnan. We also thank M. Jokela for excellent technical assistance.
This project was partly supported by a grant from the Tampere University Hospital Medical Research Fund.
REFERENCES
- 1.Arsenault T L, Hughes D W, MacLean D B, Szarek W A, Kropinski A M B, Lam J S. Structural studies on the polysaccharide portion of “A-band” lipopolysaccharide from a mutant (AK14O1) of Pseudomonas aeruginosa strain PAO1. Can J Chem. 1991;69:1273–1280. [Google Scholar]
- 2.Arseniejevic M, Obradovic A. A pathovar of Pseudomonas syringae causal agent of bacterial leaf spot and blight of pepper transplants. In: Rudolph K, Burr T J, Mansfield J W, Stead D, Vivian A, von Kietzell J, editors. Pseudomonas syringae pathovars and related pathogens. Dordrecht, The Netherlands: Kluwer Academic Press; 1997. pp. 61–66. [Google Scholar]
- 3.Barton-Willis P A, Wang M C, Staskawicz B, Keen N T. Structural studies on the O chain polysaccharides of lipopolysaccharides from Pseudomonas syringae pv. glycinea. Physiol Mol Plant Pathol. 1987;30:187–197. [Google Scholar]
- 4.Burrows L L, Charter D F, Lam J S. Molecular characterization of the Pseudomonas aeruginosa serotype O5 (PAO-1) B-band lipopolysaccharide gene cluster. Mol Microbiol. 1996;3:481–495. doi: 10.1046/j.1365-2958.1996.1351503.x. [DOI] [PubMed] [Google Scholar]
- 5.Burrows L L, Chow D, Lam J S. Pseudomonas aeruginosa B-band O-antigen chain length is modulated by Wzz (Rol) J Bacteriol. 1997;179:1482–1489. doi: 10.1128/jb.179.5.1482-1489.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burrows L L, Lam J S. Effect of wzx (rfbX) mutations on A-band and B-band lipopolysaccharide biosynthesis in Pseudomonas aeruginosa O5. J Bacteriol. 1999;181:973–980. doi: 10.1128/jb.181.3.973-980.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cerantola S, Montrozier H. Structural elucidation of two polysaccharides present in the lipopolysaccharide of a clinical isolate of Burkholderia cepacia. Eur J Biochem. 1997;146:360–366. doi: 10.1111/j.1432-1033.1997.00360.x. [DOI] [PubMed] [Google Scholar]
- 8.Ciucanu I, Kerek F. A simple and rapid method for the permethylation of carbohydrates. Carbohydr Res. 1984;131:209–217. [Google Scholar]
- 9.Clerc A, Manceau C, Nesme X. Comparison of randomly amplified polymorphic DNA with amplified fragment length polymorphism to assess genetic diversity and genetic relatedness within genospecies III of Pseudomonas syringae. Appl Environ Microbiol. 1998;64:1180–1187. doi: 10.1128/aem.64.4.1180-1187.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Kievit T R, Dasgupta T, Schweizer H, Lam J S. Molecular cloning and characterization of the rfc gene of Pseudomonas aeruginosa (serotype O5) Mol Microbiol. 1995;16:565–574. doi: 10.1111/j.1365-2958.1995.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 11.Denny T P. Phenotypic diversity in Pseudomonas syringae pv. tomato. J Gen Microbiol. 1988;134:1939–1948. doi: 10.1099/00221287-134-7-1949. [DOI] [PubMed] [Google Scholar]
- 12.Denny T P, Gilmour M N, Selander R J. Genetic diversity and relationships of two pathovars of Pseudomonas syringae. J Gen Microbiol. 1988;134:1949–1960. doi: 10.1099/00221287-134-7-1949. [DOI] [PubMed] [Google Scholar]
- 13.Gardan L, Bollet C, Abu Ghorrah M, Grimont P A D, Grimont F. DNA relatedness among pathovar strains of Pseudomonas syringae subsp. savastanoi Janse (1982) and proposal of Pseudomonas savastanoi sp. nov., 1990. Int J Syst Bacteriol. 1992;42:606–612. [Google Scholar]
- 14.Gardan L, Shafik H, Belouin S, Brosch R, Grimont F, Grimont P A D. DNA relatedness among the pathovars of Pseudomonas syringae and description of Pseudomonas tremae sp. nov. and Pseudomonas cannabina sp. nov. (ex. Sutic and Dowson 1959) Int J Syst Bacteriol. 1999;49:469–478. doi: 10.1099/00207713-49-2-469. [DOI] [PubMed] [Google Scholar]
- 15.Goldstein I J, Hay G W, Lewis B A, Smith F. Controlled degradation of polysaccharides by periodate oxidation, reduction, and hydrolysis. Methods Carbohydr Chem. 1965;5:361–370. [Google Scholar]
- 16.Helander I M, Moran A P, Mäkela P H. Separation of two lipopolysaccharide populations with different contents of O-antigen factor 122 in Salmonella enterica serovar Typhimurium. Mol Microbiol. 1992;6:2857–2862. doi: 10.1111/j.1365-2958.1992.tb01465.x. [DOI] [PubMed] [Google Scholar]
- 17.Hellerqist C G, Lindberg B, Svensson S, Holme T, Lindberg A A. Structural studies of the O-specific side chains of the cell wall lipopolysaccharides from Salmonella typhi and S. enteritidis. Acta Chem Scand. 1969;23:1588–1596. doi: 10.3891/acta.chem.scand.23-1588. [DOI] [PubMed] [Google Scholar]
- 18.Hendson M, Hildebrand D C, Huisman M N. Relatedness of Pseudomonas syringae pv. tomato, Pseudomonas syringae pv. maculicola, and Pseudomonas syringae pv. antirrini. J Appl Bacteriol. 1992;73:455–464. [Google Scholar]
- 19.Jann K, Jann B. Bacterial polysaccharide antigens. In: Sutherland I, editor. Surface carbohydrates of the procariote cells. New York, N.Y: Academic Press; 1977. pp. 247–287. [Google Scholar]
- 20.Jann K, Jann B. Structure and biosynthesis of O-antigen. In: Reitshel E T, editor. Handbook of endotoxin. 1. Chemistry of endotoxin. Amsterdam, The Netherlands: Elsevier Science Publishers; 1984. pp. 138–186. [Google Scholar]
- 21.Janse J D, Rossi P, Angelucci L, Scortichini M, Derks J H, Akkermans A D L, De Vrijer R, Psallidas P G. Reclassification of Pseudomonas syringae pv. avellanae as Pseudomonas avellanae (spec. nov.), the bacterium causing cancer of hazelnut (Corylus avellana L.) Syst Appl Microbiol. 1996;19:589–595. [Google Scholar]
- 22.Jansson P-E, Kenne L, Widmalm G. Computer-assisted structural analysis of polysaccharides with an extended version of CASPER using 1H- and 13C-NMR data. Carbohydr Res. 1989;188:169–191. doi: 10.1016/0008-6215(89)84069-8. [DOI] [PubMed] [Google Scholar]
- 23.Knirel Y A. Polysaccharide antigens of Pseudomonas aeruginosa. Crit Rev Microbiol. 1990;17:273–304. doi: 10.3109/10408419009105729. [DOI] [PubMed] [Google Scholar]
- 24.Knirel Y A, Ovod V V, Paramonov N A, Krohn K J. Structural heterogeneity in the O polysaccharide of Pseudomonas syringae pv. coriandricola GSPB 2028 (NCPPB 3780, W-43) Eur J Biochem. 1998;258:716–721. doi: 10.1046/j.1432-1327.1998.2580716.x. [DOI] [PubMed] [Google Scholar]
- 25.Knirel Y A, Ovod V V, Zdorovenko G M, Gvozdyak R I, Krohn K J. Structure of the O polysaccharide and immunochemical relationships between the lipopolysaccharides of Pseudomonas syringae pathovar tomato and pathovar maculicola. Eur J Biochem. 1998;258:657–661. doi: 10.1046/j.1432-1327.1998.2580657.x. [DOI] [PubMed] [Google Scholar]
- 26.Knirel Y A, Zdorovenko G M. Structures of O-polysaccharide chains of lipopolysaccharides as the basis for classification of Pseudomonas syringae and related strains. In: Rudolph K, Burr T J, Mansfield J W, Stead D, Vivian A, von Kietzell J, editors. Pseudomonas syringae pathovars and related pathogens. Dordrecht, The Netherlands: Kluwer Academic Press; 1997. pp. 475–480. [Google Scholar]
- 27.Knirel Y A, Zdorovenko G M, Dashunin V M, Yakovleva L M, Shashkov A S, Zakharova I Y, Gvozdyak R I, Kochetkov N K. Antigenic polysaccharides of bacteria. 15. Structure of the repeating unit of O-specific polysaccharide chain of Pseudomonas wieringae lipopolysaccharide. Bioorg Khim. 1986;12:1253–1262. [Google Scholar]
- 28.Knirel Y A, Zdorovenko G M, Shashkov A S, Gubanova N Y, Yakovleva L M, Gvozdyak R I. Antigenic polysaccharides of bacteria. 27. Structure of the O-specific polysaccharide chain of lipopolysaccharides from Pseudomonas syringae pv. atrofaciens 2399, phaseolicola 120a and Pseudomonas holci 8299, belonging to serogroup VI. Bioorg Khim. 1988;14:92–99. [PubMed] [Google Scholar]
- 29.Knirel Y A, Zdorovenko G M, Shashkov A S, Mamyan S S, Yakovleva L M, Solyanik L P, Zakharova I Y. Antigenic polysaccharides of bacteria. 26. Structure of the O-specific polysaccharide chains of lipopolysaccharides from Pseudomonas cerasi 467 and Pseudomonas syringae pv. syringae strains 218 and P-55, belonging to serogroups II and III. Bioorg Khim. 1988;14:82–91. [PubMed] [Google Scholar]
- 30.Knirel Y A, Zdorovenko G M, Yakovleva L M, Shashkov A S, Solyanik L P, Zakharova I Y. Antigenic polysaccharides of bacteria. 28. Structure of O-specific chain of lipopolysaccharide of Pseudomonas syringae pv. atrofaciens K-1025 and Pseudomonas holci 90a (serogroup II) Bioorg Khim. 1988;14:166–171. [PubMed] [Google Scholar]
- 30a.Knirel, Y. A., K. Rudolph, and V. Ovod. Unpublished data.
- 31.Kocharova N A, Knirel Y A, Kochetkov N K, Stanislavskii E S. Characterization of a rhamnan isolated from preparations of the lipopolysaccharides of Pseudomonas aeruginosa. Bioorg Khim. 1988;14:701–703. [PubMed] [Google Scholar]
- 32.Koike S T, Barak J D, Henderson D M, Gilbertson R L. Bacterial blight of leek: a new disease in California caused by Pseudomonas syringae. Plant Dis. 1999;83:165–170. doi: 10.1094/PDIS.1999.83.2.165. [DOI] [PubMed] [Google Scholar]
- 33.Leontein K, Lindberg B, Lönngren J. Assignment of absolute configuration of sugars by g.l.c. of their acetylated glycosides formed from chiral alcohols. Carbohydr Res. 1978;62:359–362. [Google Scholar]
- 34.Lipkind G M, Shashkov A S, Knirel Y A, Vinogradov E V, Kochetkov N K. A computer-assisted structural analysis of regular polysaccharides on the basis of 13C-n.m.r. data. Carbohydr Res. 1988;175:59–75. doi: 10.1016/0008-6215(88)80156-3. [DOI] [PubMed] [Google Scholar]
- 35.Little E, Gilbertson R L. Phenotypic and genotypic characters support placement of Pseudomonas syringae strains from tomato, celery, and cauliflower into distinct pathovars. In: Rudolph K, Burr T J, Mansfield J W, Stead D, Vivian A, von Kietzell J, editors. Pseudomonas syringae pathovars and related pathogens. Dordrecht, The Netherlands: Kluwer Academic Press; 1997. pp. 542–547. [Google Scholar]
- 36.Liu P V, Matsumoto H, Kusama H, Bergan T. Survey of heat-stable, major somatic antigens of Pseudomonas aeruginosa. Int J Syst Bacteriol. 1983;33:256–264. [Google Scholar]
- 37.Liu P V, Wang S P. Three new major somatic antigens of Pseudomonas aeruginosa. J Clin Microbiol. 1990;28:922–925. doi: 10.1128/jcm.28.5.922-925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Louws F G, Fulbright D W, Taylor Stephens C, De Bruin F J. Specific genomic fingerprints of phytopathogenic Xanthomonas and Pseudomonas pathovars and strains generated with repetitive sequences and PCR. Appl Environ Microbiol. 1994;60:2286–2295. doi: 10.1128/aem.60.7.2286-2295.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mäkelä P H, Stocker B A D. Genetics of lipopolysaccharide. In: Rietschel E T, editor. Handbook of endotoxin. 1. Chemistry of endotoxin. Amsterdam, The Netherlands: Elsevier Science Publisher; 1984. pp. 59–137. [Google Scholar]
- 40.Mamat U, Seydel U, Grimmecke D, Holst O, Rietschel E T. Lipopolysaccharides. In: Pinto B M, editor. Comprehensive natural products chemistry, vol. 3. Carbohydrates and their derivatives including tannins, cellulose, and related lignins. Amsterdam, The Netherlands: Elsevier; 1999. pp. 179–239. [Google Scholar]
- 41.Manceau C, Horvais A. Assessment of genetic diversity among strains of Pseudomonas syringae by PCR-restriction fragment length polymorphism analysis of rRNA operons with special emphasis on P. syringae pv. tomato. Appl Environ Microbiol. 1997;63:498–505. doi: 10.1128/aem.63.2.498-505.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maraite H, Weyns J. Pseudomonas syringae pv. aptata and pv. atrofaciens, specific pathovars or members of pv. syringae? In: Rudolph K, Burr T J, Mansfield J W, Stead D, Vivian A, von Kietzell J, editors. Pseudomonas syringae pathovars and related pathogens. Dordrecht, The Netherlands: Kluwer Academic Press; 1997. pp. 515–520. [Google Scholar]
- 43.Nikaido H, Nikaido K, Nakae T, Mäkela P H. Glucosylation of lipopolysaccharide in Salmonella: biosynthesis of O-antigen factor 122. 1. Over-all reaction. J Biol Chem. 1971;246:3902–3911. [PubMed] [Google Scholar]
- 44.Ovod V, Ashorn P, Yakovleva L, Krohn K. Classification of Pseudomonas syringae with monoclonal antibodies against the core and O-side chains of the lipopolysaccharide. Phytopathology. 1995;85:226–232. [Google Scholar]
- 45.Ovod V, Rudolph K, Krohn K. Serological classification of Pseudomonas syringae pathovars based on monoclonal antibodies towards the lipopolysaccharide O-chains. In: Rudolph K, Burr T J, Mansfield J W, Stead D, Vivian A, von Kietzell J, editors. Pseudomonas syringae pathovars and related pathogens. Dordrecht, The Netherlands: Kluwer Academic Press; 1997. pp. 526–531. [Google Scholar]
- 46.Ovod V, Rudolph K, Knirel Y, Krohn K. Immunochemical characterization of O polysaccharides composing the α-d-rhamnose backbone of lipopolysaccharide of Pseudomonas syringae and classification of bacteria into serogroups O1 and O2 with monoclonal antibodies. J Bacteriol. 1996;178:6459–6465. doi: 10.1128/jb.178.22.6459-6465.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palleroni N J, Ballard R W, Raiston E, Duodoroff M. Deoxyribonucleic acid homologies among some Pseudomonas species. J Bacteriol. 1972;110:1–11. doi: 10.1128/jb.110.1.1-11.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pecknold P C, Grogan R C. Deoxyribonucleic acid homology groups among phytopathogenic Pseudomonas species. Int J Syst Bacteriol. 1973;23:111–121. [Google Scholar]
- 49.Reeves P. Evolution of Salmonella O antigen variation by interspecific gene transfer on a large scale. Trends Genet. 1993;9:17–22. doi: 10.1016/0168-9525(93)90067-R. [DOI] [PubMed] [Google Scholar]
- 50.Reeves R R, Hobbs M, Valvano M A, Skurnik M, Whitfield C, Coplin D, Kido N, Klena J, Maskell D, Raetz C R H, Rick P D. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 1996;4:495–503. doi: 10.1016/s0966-842x(97)82912-5. [DOI] [PubMed] [Google Scholar]
- 51.Rocchetta H L, Burrows L L, Pacan J C, Lam J S. Three rhamnosyltransferases responsible for assembly of the A-band D-rhamnan polysaccharide in Pseudomonas aeruginosa: a fourth transferase, WbpL, is required for the initiation of both A-band and B-band lipopolysaccharide synthesis. Mol Microbiol. 1998;28:1103–1119. doi: 10.1046/j.1365-2958.1998.00871.x. [DOI] [PubMed] [Google Scholar]
- 52.Rocchetta H L, Lam J S. Identification and functional characterization of an ABC transport system involved in polysaccharide export of A-band lipopolysaccharide in Pseudomonas aeruginosa. J Bacteriol. 1997;179:4713–4724. doi: 10.1128/jb.179.15.4713-4724.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52a.Samson, R. Unpublished data.
- 53.Samson R, Shafik H, Benjama A, Gardan L. Description of the bacterium causing blight of leek as Pseudomonas syringae pathovar porri (pathovar nov.) Phytopathology. 1998;88:844–850. doi: 10.1094/PHYTO.1998.88.8.844. [DOI] [PubMed] [Google Scholar]
- 54.Saunier M, Malandrin L, Samson R. Distribution of Pseudomonas syringae pathovars into twenty-three O serogroups. Appl Environ Microbiol. 1996;62:2360–2374. doi: 10.1128/aem.62.7.2360-2374.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sawardeker J S, Sloneker J H, Jeanes A. Quantitative determination of monosaccharides as their alditol acetates by gas liquid chromatography. Anal Chem. 1965;37:1602–1603. [Google Scholar]
- 56.Scortichini M, Dettori M T, Marchesi U, Palombi M A, Rossi M P. Differentiation of Pseudomonas avellanae strains from Greece and Italy by rep-PCR genomic fingerprinting. J Phytopathol. 1998;146:417–420. [Google Scholar]
- 57.Shashkov A S, Senchenkova S N, Nazarenko E L, Zubkov V A, Gorshkova N M, Knirel Y A, Gorshkova R P. Structure of a phosphorylated polysaccharide from Shewanella putrefaciens strain S29. Carbohydr Res. 1997;303:333–338. doi: 10.1016/s0008-6215(97)00165-1. [DOI] [PubMed] [Google Scholar]
- 58.Smith A R W, Zamze S E, Munro S M, Carter K J, Hignett R C. Structure of the sidechain of lipopolysaccharide from Pseudomonas syringae pv. morsprunorum C28. Eur J Biochem. 1985;149:73–78. doi: 10.1111/j.1432-1033.1985.tb08895.x. [DOI] [PubMed] [Google Scholar]
- 59.Stead D E, Hennessy J, Elphistone J G, Wilson J K. Modern methods for classification of plant pathogenic bacteria including Pseudomonas syringae. In: Rudolph K, Burr T J, Mansfield J W, Stead D, Vivian A, von Kietzell J, editors. Pseudomonas syringae pathovars and related pathogens. Dordrecht, The Netherlands: Kluwer Academic Press; 1997. pp. 427–434. [Google Scholar]
- 60.Tsvetkov Y E, Backinowsky L V, Kochetkov N K. Synthesis of a common polysaccharide antigen of Pseudomonas aeruginosa as the 6-aminohexyl glycoside. Carbohydr Res. 1989;193:75–90. doi: 10.1016/0008-6215(89)85108-0. [DOI] [PubMed] [Google Scholar]
- 61.von Kietzel J, Rudolph K. Epiphitic occurrence of Pseudomonas syringae pv. atrofaciens. In: Rudolph K, Burr T J, Mansfield J W, Stead D, Vivian A, von Kietzell J, editors. Pseudomonas syringae pathovars and related pathogens. Dordrecht, The Netherlands: Kluwer Academic Press; 1997. pp. 29–34. [Google Scholar]
- 62.Whitfield C. Biosynthesis of lipopolysaccharide O antigens. Trends Microbiol. 1995;3:178–185. doi: 10.1016/s0966-842x(00)88917-9. [DOI] [PubMed] [Google Scholar]
- 63.Wingate V P M, Norman P M, Lamb C J. Analysis of the cell surface of Pseudomonas syringae pv. glycinea with monoclonal antibodies. Mol Plant-Microbe Interact. 1990;3:408–416. [Google Scholar]
- 64.Winn A M, Wilkinson S G. The O7 antigen of Stenotrophomonas maltophilia is a linear D-rhamnan with a trisaccharide repeating unit that is also present in polymers from some Pseudomonas and Burkholderia species. FEMS Microbiol Lett. 1998;166:57–61. doi: 10.1111/j.1574-6968.1998.tb13183.x. [DOI] [PubMed] [Google Scholar]
- 65.Yakovleva L, Pasichnik L, Porembskaya N, Zdorovenko G, Vassilev V. Serotypic variability of Pseudomonas syringae pv. tabaci strains. In: Rudolph K, Burr T J, Mansfield J W, Stead D, Vivian A, von Kietzell J, editors. Pseudomonas syringae pathovars and related pathogens. Dordrecht, The Netherlands: Kluwer Academic Press; 1997. pp. 297–299. [Google Scholar]
- 66.Yokota S, Kaya S, Sawada S, Kawamura T, Araki Y, Ito E. Characterization of a polysaccharide component of lipopolysaccharide from Pseudomonas aeruginosa HD 1908 (ATTC 27584) as d-rhamnan. Eur J Biochem. 1987;167:203–209. doi: 10.1111/j.1432-1033.1987.tb13324.x. [DOI] [PubMed] [Google Scholar]
- 67.Young J M, Saddler G S, Takikawa Y, DeBoer S H, Vauterin L, Gardan L, Gvozdyak R I, Stead D E. Names of plant pathogenic bacteria 1864–1995. ISPP Subcommittee on Taxonomy of Plant Pathogenic Bacteria. Rev Plant Pathol. 1996;75:721–763. [Google Scholar]
- 68.Zdorovenko E L, Ovod V, Shashkov A S, Kocharova N A, Knirel Y A, Krohn K. Structure of the O polysaccharide of the lipopolysaccharide of Pseudomonas syringae pv. garcae ICMP 8047. Biochemistry (Moscow) 1999;64:765–773. [PubMed] [Google Scholar]