Abstract
Nucleic acids are important molecules of life and have recently emerged as important functional materials to synthesize, organize and assemble inorganic nanoparticles for various technological applications. In this study, we have systematically investigated the four basic nucleotides of DNA as precursors to form fluorescent nucleotide derived biodots (N-dots) with unique singlet oxygen generation properties by one-pot hydrothermal synthesis. It has been discovered for the first time that the nitrogenous base adenine accounts for the bright fluorescence, while the sugar and phosphate groups of the nucleotide endow the N-dots with good photo-stability. Among the N-dots synthesized in this study, adenosine triphosphate (ATP)-dots were found to exhibit the highest fluorescence quantum yield (QY) of 13.9%, whereas adenosine diphosphate (ADP)-dots exhibited the best photo-stability maintaining 97.6% photoluminescence intensity after continuous UV excitation for 30 min. Overall, deoxyadenosine monophosphate (dAMP)-dots display both high fluorescence QY (12.4%) and good photo-stability (91.9%). Most critically, dAMP-dots show the highest singlet oxygen generation with a remarkable singlet oxygen (1O2) quantum yield of 1.20 surpassing the 1O2 quantum yield of the conventional photosensitizer Rose Bengal (0.75). Further cellular experiments reveal that dAMP-dots possess excellent cellular uptake ability for successful fluorescent labeling with the ability to kill >60% HeLa cancer cells under white light treatment within 10 minutes. Additionally, N-dots possess excellent stability against both UV irradiation and DNase enzymatic action. These results demonstrate the unique physiochemical properties of N-dots, including an ultra-small size for cellular uptake, tunable photoluminescence for bioimaging, excellent aqueous solubility, high chemical stability and photo-stability as well as excellent singlet oxygen quantum yield with inherent biocompatibility for photodynamic therapy, which are important factors contributing to the promising theranostic applications in future personalized nanomedicine.
We report a new way to engineer nanomaterials from biomolecules (N-dots) with bright fluorescence and high singlet oxygen generation towards theranostic applications.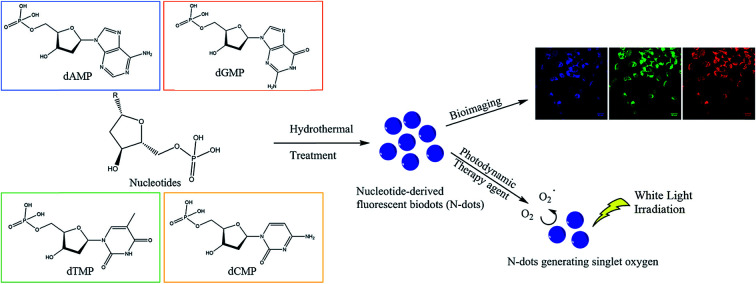
Introduction
DNA is a long chain polymer made from basic building blocks called nucleotides. It carries important genetic information of life and can self-assemble into various unique structures such as supercoils, G-quadruplexes and origami.1–4 Recently, the complementary base pairing properties of DNA have been exploited for nanoscale assembly of inorganic nanoparticles (NPs) such as gold-, silver- and metal oxide-NPs into functional hybrid materials for a wide range of technological applications.5,6 In addition to the assembly function, the natural doping of heteroatoms, such as nitrogen, oxygen and phosphorous, in DNA molecules also makes them a viable choice of precursor for the hydrothermal synthesis of fluorescent biodots, an emerging class of carbon-based nanomaterials.7–10 Due to the use of biomolecular precursors, the as-synthesized fluorescent biodots are often endowed with inherent biocompatibility, good aqueous solubility, chemical stability and inertness11,12 suitable for a wide range of biomedical applications.13–16 For example, fluorescent biodots derived from long chain DNAs have been developed for metal ion sensing (e.g., Hg2+, Ag+, and Fe2+), bioimaging and drug delivery.11,12,17–19 However, all these studies do not investigate the contribution of the four basic units of DNA (i.e., deoxyadenosine, deoxyguanosine, deoxythymidine and deoxycytidine nucleotides) to the formation of the resultant biodots, which are important to tailor the biodot properties for specific biomedical applications. Herein, we have conducted a systematic study to uncover the rational design of nucleotide-derived biodots (N-dots) with further insight into individual nucleotide contributions to the unique physiochemical properties and structure of the resultant N-dots.
Besides fluorescence properties, biodots can be endowed with additional functions such as antimicrobial and biofilm removal functions.9 To the best of our knowledge, the potential of using fluorescent nanodots derived from biomolecules with intrinsic singlet oxygen generation properties for photodynamic therapy (PDT) has not been reported. PDT is a non-invasive treatment for cancer with minimal side effects, negligible drug resistance and low cytotoxicity.20 It is based on the generation of highly reactive singlet oxygen through the interactions of photo-sensitizers, light and oxygen to result in cell death and tissue destruction.21–24 However, the widespread application of current organic-based PDT agents is mostly hindered by their poor aqueous solubility, photo-stability and biocompatibility.20 In this study, we have tackled these challenges by carefully investigating the contribution of functional groups and molecular structures of nucleotides to singlet oxygen generation for effective PDT treatment. In particular, our study involved the careful selection of nucleotide precursors for the hydrothermal synthesis of functional N-dots with desirable properties for simultaneous fluorescence imaging and photodynamic therapy.
In summary, we have presented the successful development of bright fluorescent nucleotide-derived nanodots as potential PDT agents with high singlet oxygen generation, excellent aqueous solubility and good biocompatibility. It was found that the presence of a photoluminescent center formed within the N-dots has provided them with excellent stability against both UV irradiation and DNase enzymatic action. Since typical photosensitizers with high singlet oxygen generation have poor fluorescence quantum yield, while other molecules with brighter fluorescence usually give low or negligible ROS generation, this work provides a plausible remedy to this dilemma and opens up a new way to engineer nanomaterials with multiple functionalities towards theranostic applications in future nanomedicine.
Experimental
Reagents and chemicals
Adenine, guanine, cytosine, thymine, adenosine, 2′-deoxyadenosine 5′-monophosphate, 2′-deoxyguanosine 5′-monophosphate, 2′-deoxycytidine 5′-monophosphate, 2′-deoxythymidine 5′-monophosphate, adenosine 5′-diphosphate, deoxyadenosine 5′-triphosphate, deoxyribonucleic acid sodium salt from salmon testes (approximately 2000 bp), 9,10-anthracenediyl-bis(methylene)dimalonic acid (ABDA) and 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) were purchased from Sigma-Aldrich. Deionized (DI) water with a specific resistance of 18.2 MΩ was used.
Materials characterization
The photoluminescence (PL) intensity was measured using a Tecan Infinite M200 Multimode Microplate Reader. The fluorescence images of the N-dot solution were obtained under the illumination of a High Performance 2UV™ Transilluminator. UV-vis absorption spectra were recorded with a Shimadzu UV-2450 UV-visible Spectrophotometer. X-ray Photoelectron Spectroscopy (XPS) was performed with Theta Probe XPS equipment. Dynamic Light Scattering (DLS) measurement was performed using a Malvern Zetasizer Nano Instrument. High-Resolution Transmission Electron Microscopy (HRTEM) images of N-dots were recorded using a Philips CM300 FEGTEM. Fourier Transform Infrared spectroscopy (FTIR) was performed using a PerkinElmer Fourier Transform Infrared Spectrometer. Time-resolved photoluminescence was measured using an Edinburgh Laser Flash Photolysis Spectrometer (LP920-KS). The N-dot sample was excited with a flash lamp pumped Q-switched Nd:YAG laser operating at the third harmonic wavelength of 374 nm (Ekspla NT 341 A). The photoluminescence decay at 450 nm was fitted with a bi-exponential function to obtain the lifetimes.
Synthesis of nucleotide-derived biodots (N-dots)
N-dots were hydrothermally synthesized in one step. Briefly, 100 mg of nucleotide precursors were dissolved in 10 mL DI water and heated to 60 °C until fully dissolved. This solution was then transferred to a Teflon-lined autoclave and heated at 180 °C for 12 h. After the autoclave completely cooled down, the reaction product was centrifuged at 13 000 rpm for 30 min and the supernatant was further dialyzed against DI water for 24 h.
Fluorescence quantum yield (QY) measurement
The fluorescence QY of N-dots was determined using a comparative method. The integrated photoluminescence (PL) intensity was plotted against the optical absorption value of N-dots (at an optimized excitation wavelength), with quinine sulfate as a reference. The QY was calculated using the equation below: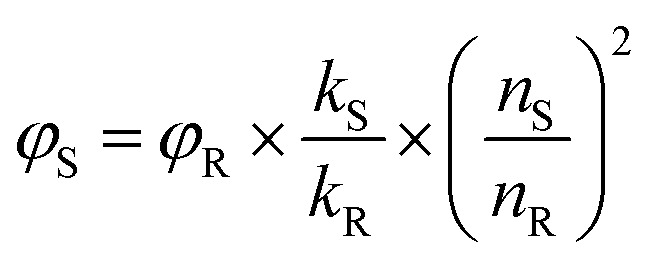 where φ is the quantum yield, k is the gradient(slope) of the lines obtained from the plot of integrated PL intensity (at an optimized excitation wavelength) against absorbance measured (at the optimized wavelength), n is the refractive index of the solvent, and the subscript S refers to the N-dot sample and R refers to the reference. The fluorescence QY of the reference dye, quinine sulfate dissolved in 0.1 M sulfuric acid, H2SO4 (n = 1.33), is 0.54. The N-dots were dissolved in DI water (n = 1.33). The absorption of the N-dots was maintained below 0.1 to prevent the re-absorption effect.
where φ is the quantum yield, k is the gradient(slope) of the lines obtained from the plot of integrated PL intensity (at an optimized excitation wavelength) against absorbance measured (at the optimized wavelength), n is the refractive index of the solvent, and the subscript S refers to the N-dot sample and R refers to the reference. The fluorescence QY of the reference dye, quinine sulfate dissolved in 0.1 M sulfuric acid, H2SO4 (n = 1.33), is 0.54. The N-dots were dissolved in DI water (n = 1.33). The absorption of the N-dots was maintained below 0.1 to prevent the re-absorption effect.
Detection of singlet oxygen generation and 1O2 quantum yield measurement
The N-dots were diluted until the absorbance level was below 0.2 for the wavelength range from 320–420 nm. The ABDA solution was used as a trapping agent for singlet oxygen in the measurement. The N-dots were first mixed with 10 μL of a 10 mM ABDA solution and topped up to 2 mL using DI water of the same dilution factor. The absorption of the prepared solution was measured before light irradiation and at every 10 minute interval up to a total of 60 minutes using a 150 mW white light source. The singlet oxygen generation rate was obtained by subtracting the absorbance value of the N-dots from that of the overall contribution of both the N-dots and ABDA. The percentage change in the absorbance peak value was then calculated by comparing each 10 minute interval. A baseline scan was conducted using 2 mL of DI water for all measurements.
Rose Bengal, an aqueous soluble photo-sensitizer, was used as the standard for the calculation of the 1O2 quantum yield. The 1O2 quantum yield was calculated using the equation below: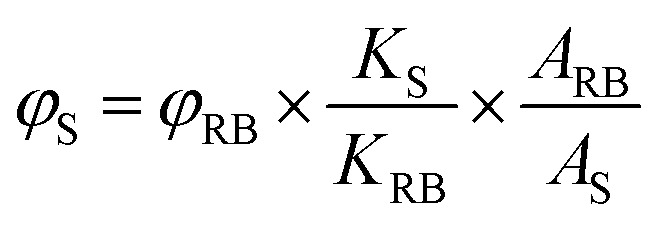 where φ is the quantum yield, K is the gradient of the decay rate constants of ABDA, A is the light absorbed which is calculated from the integration of the absorption bands in the wavelength range from 400 to 800 nm, and the subscript S refers to the N-dot sample and RB refers to the reference standard Rose Bengal. The 1O2 quantum yield of the reference compound, Rose Bengal, is 0.75 when dissolved in DI water.
where φ is the quantum yield, K is the gradient of the decay rate constants of ABDA, A is the light absorbed which is calculated from the integration of the absorption bands in the wavelength range from 400 to 800 nm, and the subscript S refers to the N-dot sample and RB refers to the reference standard Rose Bengal. The 1O2 quantum yield of the reference compound, Rose Bengal, is 0.75 when dissolved in DI water.
DNase enzymatic digestion of N-dots
The degradation of N-dots by DNase was evaluated by adding 240 μL of each of these N-dots into a mixture containing 100 μL of magnesium sulfate (12 mg mL−1), 400 μL of DNase dissolved in 0.85% sodium chloride (∼400 to 500 K units per mL), and 200 μL of sodium acetate buffer (pH 5.0, 136 mg mL−1) and topping up to 2 mL with deionized water. After 30 minutes' incubation at 25 °C, the photoluminescence intensity and absorbance of the resulting mixture solution were measured and subsequently compared to those of a series of control samples prepared in the absence of the N-dots and/or DNase.
Cytotoxicity test and photodynamic therapy (PDT)
The cell viability of N-dots was investigated using the MTT assay. A HeLa cell culture was maintained in high glucose DMEM medium (Invitrogen) with 1% penicillin–streptomycin (37 °C, 5% CO2) and 10% fetal bovine serum. HeLa cells were first seeded into 96-well plates (CoStar, IL, USA) at a density of 1 × 104 per well in 200 μL of media and incubated for 24 h. Various concentrations of N-dots were added to the cells and incubated for another 24 h. Subsequently, 20 μL of 5 mg mL−1 MTT solution was added to each well and further incubated for 4 h. Afterwards, the supernatant was aspirated and replaced with 200 μL DMSO leading to the formation of a violet solution. Cell viability was then determined by measuring the absorbance of the solution at 570 nm using a Tecan microplate reader.
To investigate the PDT effect, the cells with the N-dots were irradiated with 150 mW white light for 10 min and incubated for 24 h. Then, the MTT assay was conducted to calculate the viability of the cells after PDT treatment.
Cellular fluorescence imaging
To study the cellular uptake of N-dots, the cells were first incubated with the N-dot samples (∼1 mg mL−1) at 37 °C for 4 h. The cells were then washed three times with PBS buffer and the fluorescence images were obtained by confocal laser scanning microscopy (CLSM) (Fluoview 1000, Olympus, Japan) under 405, 488 or 504 nm excitation.
Results and discussion
Synthesis of photoluminescent nucleotide-derived biodots (N-dots)
The four types of nucleotides, which represent the basic building blocks of DNA, namely 2′-deoxyadenosine 5′-monophosphate (dAMP), 2′-deoxyguanosine 5′-monophosphate (dGMP), 2′-deoxythymidine 5′-monophosphate (dTMP) and 2′-deoxycytidine 5′-monophosphate (dCMP) were chosen as precursors to synthesize fluorescent biodots via a one-step hydrothermal route (Fig. S1a–d†). These nucleotide-derived biodots (N-dots) were subsequently investigated for bioimaging and PDT applications (Scheme 1).
Scheme 1. Synthesis of fluorescent biodots from four types of nucleotides for bioimaging and photodynamic therapy of cancer cells.
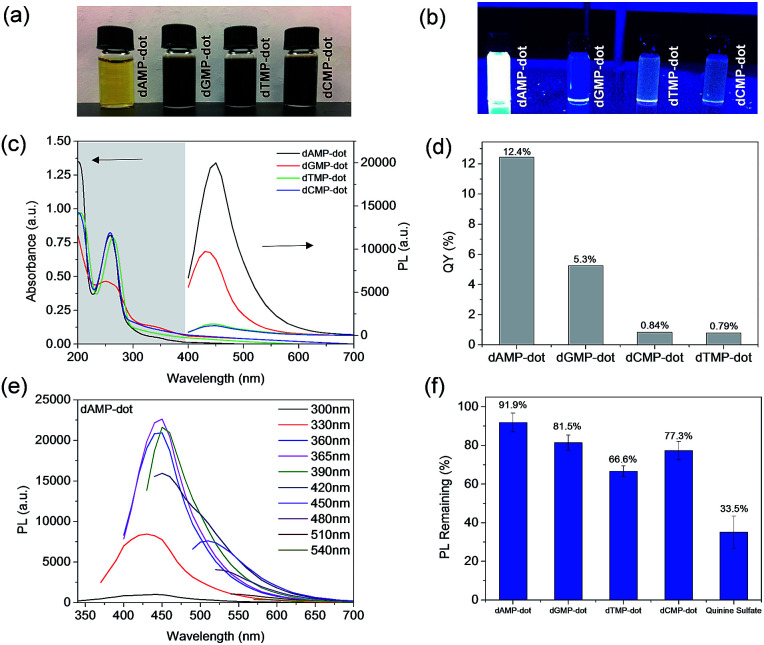
As shown in Fig. 1a, dAMP-dots appeared as a yellow solution while the other N-dot samples were dark-brown solutions under white light. Under UV irradiation, dAMP-dots exhibited the highest brightness among the four dots as shown in the color photograph in Fig. 1b. Indeed, dAMP-dots displayed the highest intensity in photoluminescent (PL) measurements when excited at 365 nm, which is almost two-fold higher than that of the second brightest dGMP-dots (Fig. 1c, right axis). We postulate that the difference in PL properties of the four N-dots may arise from the various nitrogenous bases in each nucleotide precursor which lead to contrasting structures of the resultant N-dots. The UV-vis absorption spectra of these four N-dots show three absorption peaks at ∼200 nm, 260 nm and 340 nm, respectively (Fig. 1c, left axis). The peaks at ∼200 nm and 260 nm are originally present in the four nucleotides (Fig. S2a†).25 This suggests that N-dots retained the heterocyclic rings of purines and pyrimidines from the nucleotide precursors. The absorption band at 260 nm is attributed to the π–π* transition as a result of the delocalized π electrons from the heterocyclic rings of purines and pyrimidines.26 Additionally, the appearance of a shoulder peak at 340 nm represents the n–π* transition which indicates the successful formation of N-dots.
Fig. 1. Photographs showing N-dots under (a) white light and (b) UV irradiation at 365 nm. (c) UV-vis absorption spectra of N-dots and photoluminescence (PL) intensity of the N-dots excited at 365 nm. (d) Fluorescence quantum yield comparison of respective N-dots. (e) Excitation-dependent PL spectra of dAMP-dots. (f) Photo-stability of the N-dots versus quinine sulfate.
On the other hand, three of the four N-dots including dAMP-dots, dTMP-dots and dCMP-dots exhibit maximum PL emission at 450 nm with 365 nm excitation, except the dGMP-dots which show an emission peak at 430 nm (Fig. 1e and S2b–d†). Furthermore, it is observed that the emission peak of each N-dot red-shifts as the excitation wavelength increases; meanwhile, the emission peak intensity decreases in a similar trend to the excitation dependent PL emission of biodots reported previously.11,27 This excitation dependent PL behavior could be attributed to the varying domain sizes as well as the differing sites of emission on the surface of N-dots.28 For a fair comparison, the fluorescence quantum yields (QY) of the dAMP (12.4%), dGMP (5.3%), dTMP (0.84%) and dCMP-dots (0.79%) were calculated, respectively (Fig. 1d). These data indicate that double-ring purines such as dAMP and dGMP give rise to N-dots with a much higher fluorescence QY than single-ring pyrimidines, suggesting that the fluorescence QY of N-dots is mainly determined by the bases in the nucleotides.
The photo-stability of each N-dot was also studied and compared to that of a standard organic dye, quinine sulfate. Unlike quinine sulfate which only retains ∼35% PL intensity after 30 min UV irradiation, the remaining PL of the N-dots is greater than ∼65% (Fig. 1f). The dAMP-dots which conserved over 90% PL intensity upon UV irradiation were found to be the most photo-stable ones among all the N-dots tested under the same experimental conditions. This result indicates that the dAMP-dots are less susceptible to high oxidation potential from the UV generated OH radicals.
Investigation of structure–optical properties of N-dots and their derivatives
Among the four types of N-dots, dAMP-dots were identified to be the most photo-stable (91.9% conserved PL intensity) biodots with the highest fluorescence QY of 12.4%. Since only the nitrogenous base differs in each nucleotide precursor, it is postulated that the molecular structure of the adenine base might play a crucial role in the formation of the photoluminescent center during the hydrothermal synthesis process. Hence, a more detailed investigation was conducted on compounds containing the adenine nucleobase such as adenine, adenosine, adenosine diphosphate (ADP) and adenosine triphosphate (ATP) which have comparable chemical structures to dAMP (Fig. S1e–h†) to better understand the roles of sugar and phosphate groups. The N-dots derived from adenine containing nucleotides are termed A-dots in this work.
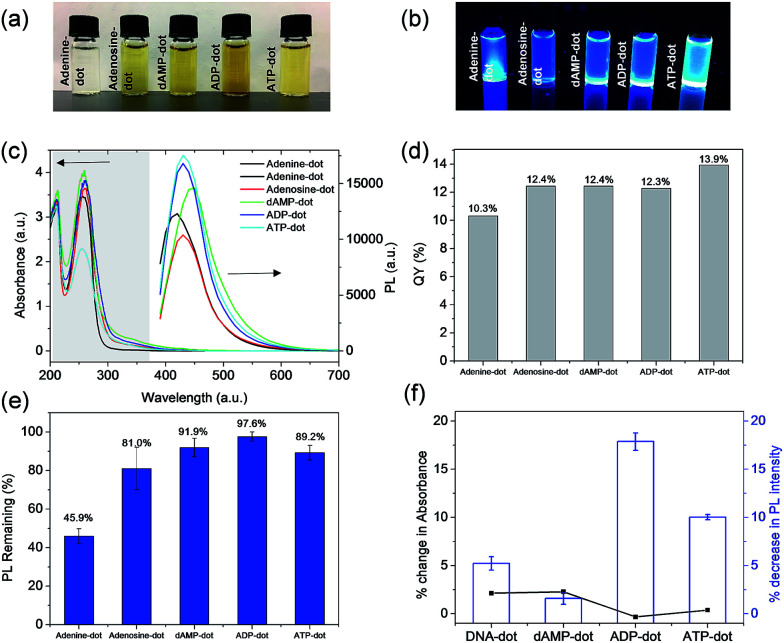
As shown in Fig. 2a, most solutions are yellowish in color under white light except adenine-dots, which appear to be colorless. Under UV irradiation, all A-dots exhibited bright blue emission (Fig. 2b) among which the ATP-dots displayed the brightest fluorescence. Correspondingly, the PL intensity of these A-dots was measured, and a similar trend was observed in which the ATP-dots have the highest PL intensity followed by ADP, dAMP, adenine and adenosine-dots (Fig. 2c). This further confirms that the high PL intensity of A-dots could be mainly attributed to the presence of the adenine base, while the sugar group also has a significant effect. It also suggests that the additional phosphate groups present in the ADP and ATP-dots could endow the A-dots with trapping states accompanied by different series of energy levels, which allows the electronic transitions among bonding (σ and π), antibonding (σ* and π*) and non-bonding (n) orbitals,29,30 leading to the enhanced PL intensity as compared to the dAMP-dots. Similar to those of other N-dots, UV-vis absorption spectra of most A-dots also show three absorption peaks at ∼200 nm, 260 nm and 340 nm, respectively, except the adenine-dots with no shoulder peak observed (Fig. 2c). Likewise, the peaks at ∼200 nm and 260 nm are originally present in the precursors (Fig. S3a†). The A-dots also exhibit excitation-dependent PL similar to that of the other N-dots where the emission peak intensity red-shifts and decreases as the excitation wavelength increases (Fig. S3b–d†). The fluorescence QYs of the adenine (10.3%), adenosine (12.4%), dAMP (12.4%), ADP (12.3%) and ATP-dots (13.9%) were calculated accordingly (Fig. 2d). The fluorescence QYs of the N-dots are higher than those of the dots derived from nucleic acids and comparable to those of many of the carbon dots derived from other precursors (see Table S2† for comparison).
Fig. 2. Photographs of A-dots under (a) white light and (b) UV irradiation at 365 nm excitation. (c) UV-vis absorption spectra of A-dots and PL intensity of the A-dots at 365 nm excitation. (d) Fluorescence quantum yield and (e) photo-stability of the A-dots. (f) % Change in PL of dAMP, ADP and ATP-dots in the presence of DNase enzyme (inset shows percentage % change in absorbance).
As shown in Fig. 2e, ADP-dots exhibit the highest photo-stability with ∼97% of PL intensity conserved upon continuous UV irradiation. It is also notable that A-dots derived from the phosphate group containing precursors displayed better resistance to photo-bleaching whereas the phosphate-free adenine and adenosine-dots have poorer photo-stabilities of 45.9% and 81.8%, respectively. This result suggests that the phosphate group is necessary to establish a stable chemical structure within the N-dots, which then makes the N-dots less susceptible towards the high oxidation potential from OH radicals generated by UV exposure. In comparison, ADP-dots exhibit better photo-stability than ATP-dots, which could be due to the different biological functions of these two precursors. Under physiological conditions, ATP molecules being in a higher energy state take part in cellular processes such as respiration to produce more stable ADP. Hence, the N-dots may have inherited their biological properties from these biomolecular precursors.
Moreover, the biological stabilities of dAMP, ADP and ATP-derived dots in the presence of DNase were also investigated. Overall, these N-dots demonstrated good bio-stability with less than 3% change in absorbance and their respective PL intensities were also highly conserved (Fig. 2f). This result indicates that these N-dots are minimally affected by DNase degradation which usually targets the phosphodiester bond. The presence of a photoluminescent center formed within the N-dots may have endowed the N-dots with excellent stability against both UV irradiation and DNase enzymatic action.
Singlet oxygen (1O2) generation performance of N-dots
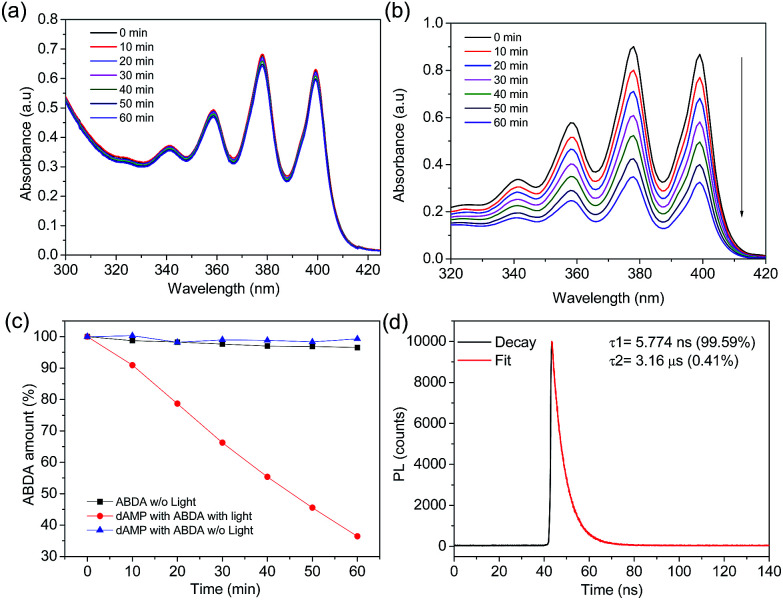
The singlet oxygen productivity of N-dots was measured using an anthracene derivative, ABDA, as a chemical probe for the detection of singlet oxygen (1O2). In the absorption spectrum, ABDA displays four characteristic bands at 342, 359, 378 and 400 nm (Fig. S4b†). Under 150 mW white light irradiation, the absorption spectrum of the ABDA solution in the presence of each N-dot is recorded for a duration of 60 min at 10 min intervals. ABDA is capable of trapping singlet oxygen specifically which in turn undergoes a pseudo first order reaction to form an endoperoxide product (Fig. S4a†) which leads to a decrease in absorbance of their four characteristic bands in 60 min.31 The amount of ABDA consumed corresponds to the amount of singlet oxygen produced.
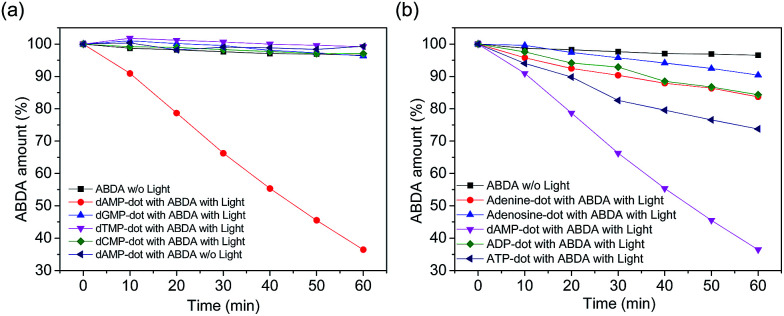
The absorbance of ABDA in the presence of N-dots at 400 nm was plotted against the illumination duration to determine the efficiency of 1O2 generation (Fig. 3). The 1O2 generation efficiency was estimated by the decay rate of ABDA.31 Our results show that dAMP-dots have the highest singlet oxygen generation ability as compared to other N-dots tested under the same conditions.
Fig. 3. Time-dependent decay profile of ABDA as a singlet oxygen indicator in the presence of (a) different N-dots and (b) A-dots under white light irradiation (150 mW). ABDA decay in the absence of light (w/o light) is used as a reference (black square line).
In the presence of dAMP-dots, the characteristic bands of ABDA decreased steadily in the span of 60 minutes (Fig. 4b) which demonstrates the singlet oxygen generation ability of dAMP-dots (Fig. 4c). Since the characteristic absorbance bands of ABDA in the presence of dAMP-dots barely change under dark conditions (Fig. 4a), it can be concluded that the singlet oxygen was generated by photosensitizing rather than other molecular interactions between ABDA and dAMP-dots. The process of radiation relaxation is relatively fast, as suggested by the fluorescence lifetime study of the dAMP-dots dissolved in water (Fig. 4d). Fitting of the photoluminescence decay profile of dAMP-dots indicates components of 5.774 ns (99.59%) and 3.16 μs (0.41%), which yields an averaged lifetime (τAV) of 18.71 ns.
Fig. 4. (a) Time dependent UV-vis absorption spectrum of the ABDA solution in the presence of dAMP-dots without light. (b) Time dependent UV-vis absorption spectra of ABDA solution in the presence of dAMP-dots with light. The arrow indicates the direction of time increase. (c) Time course decay of ABDA solution in the presence of dAMP-derived biodots with light (circle line) and without light (triangle line) and ABDA solution in the absence of light (square line). (d) Fluorescence decay profile of 374 nm laser excited dAMP-dots in aqueous solution.
The 1O2 quantum yield of dAMP-dots was calculated to be 1.20 with respect to a reference photosensitizer, Rose Bengal (Fig. S5a–f†). The calculated value was much greater than that of most of the conventional photo-sensitizers such as Rose Bengal (0.75), riboflavin (0.54),32 porphyrins (0.44–0.85)33 and Ce6 (∼0.63).34 Furthermore, this value was among the highest for the carbon dots with intrinsic PDT effects (see Table S1† for comparison). This demonstrates the high potential of N-dots to be used as a water-soluble biocompatible photosensitizer for simultaneous fluorescence imaging and photodynamic therapy.
Formation mechanism of dAMP-dots and characterization
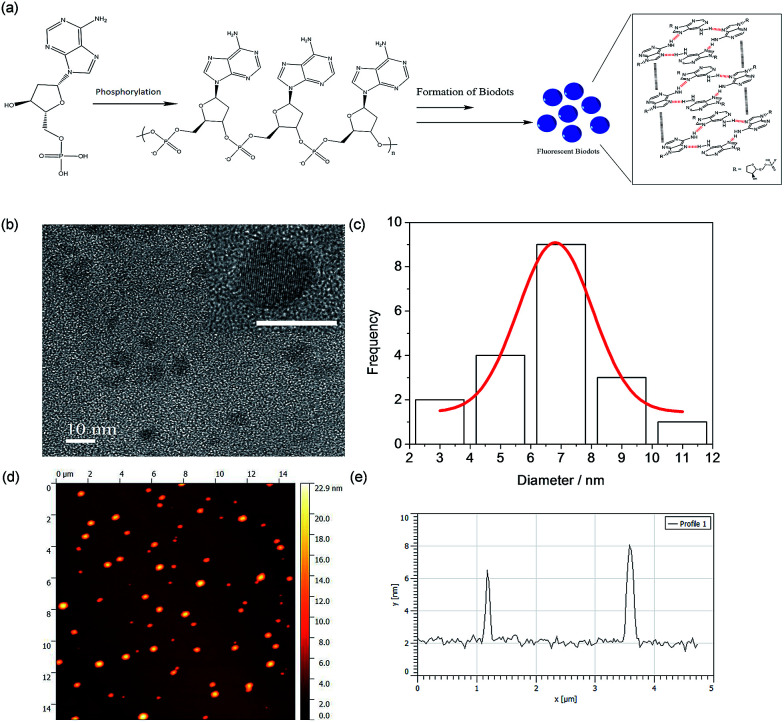
To better understand the formation mechanism of N-dots, surface functional groups of the best performance dAMP-dots were analyzed by X-ray Photoelectron Spectroscopy (XPS) as shown in Fig. S6.† The deconvolution of the P 2s scan of dAMP-dots shows that the PO− bond was retained. From the O 1s scan, only the –C–O–C– bond is present while the C–OH bond is absent. This result suggested that the nucleotide precursor may have undergone polymerization via phosphorylation.
The morphology of the dAMP-dots was also investigated using Transmission Electron Microscopy (TEM) and Atomic Force Microscopy (AFM). From the TEM image in Fig. 5b, it can be observed that the dAMP-dots exhibit good mono-dispersity with an average size of 7 ± 1 nm (Fig. 5c). The HRTEM image shows clear lattice fringes with an interplanar spacing of 0.22 nm, corresponding to the (100) plane of graphitic carbon. The X-ray diffraction (XRD) pattern displays two diffraction peaks centered at 2θ = 25° and 42°, which are attributed to the graphitic carbon lattice spacing of (002) and (100) planes, respectively, further confirming the crystalline structure of the dAMP-dots (Fig. S7†). The AFM measurement indicates that the nanodots are about 4–6 nm in height (Fig. 5e).
Fig. 5. (a) Proposed formation mechanism of dAMP-dots. (b) TEM image of dAMP-dots; inset shows the HRTEM image with a clear lattice (scale bar represents 10 nm). (c) Size distribution of dAMP-dots. (d) AFM image of dAMP-dots on mica. (e) Height profile of dAMP-dots.
Based on these characterization results, we propose a possible mechanism in Fig. 5a. The precursor, dAMP, first undergoes polymerization through phosphorylation where the phosphate groups (PO4−) and the hydroxyl groups (OH) on the sugar base form an –O–P–O– bond. Subsequently, through self-assembly, a photoluminescent center is formed via stacking of the aromatic nucleobases. Thereafter, the negatively charged phosphate groups on the backbone are located outside the sp2 carbon-like center and wrap the center to form an N-dot.11,17
Cytotoxicity study and applications of dAMP-dots for cellular imaging and photodynamic therapy (PDT)
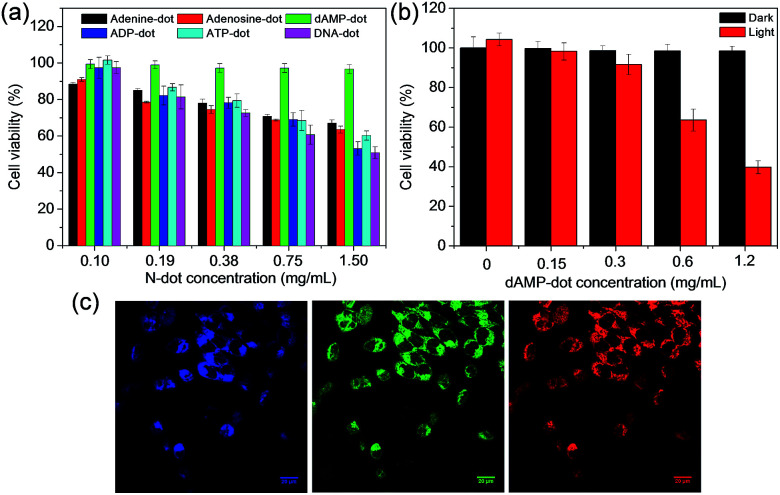
After the investigation on the optical and structural properties of all N-dots, in vitro studies were conducted to further evaluate the intracellular uptake capability of dAMP-dots. A good imaging agent should possess both biocompatibility and cellular uptake efficiency. Thus, the cytotoxicity test was first carried out on dAMP-dots using HeLa cancer cells as a model cell line. After 24 h incubation, dAMP-dot treated HeLa cells exhibit >95% cell viability even at a high concentration of 1.5 mg mL−1 (Fig. 6a). The usage of a natural biomolecular precursor, dAMP, in the absence of toxic solvents and harsh treatment processes may have contributed to the good biocompatibility and low cytotoxicity of dAMP-dots. We have carried out zeta potential measurement for dAMP-dots. The results show that the dAMP-dots are positively charged (16.5 ± 1.8 mV), which allows quick initial interaction with the negatively charged cell membrane, thus enabling efficient uptake. Subsequently, the intracellular uptake of dAMP-dots was evaluated by Confocal Laser Scanning Microscopy (CLSM) (Fig. 6b). The confocal fluorescence images in Fig. 6c indicate successful intracellular uptake of the dAMP-dots, displaying bright blue, green and red fluorescence under different excitations due to the excitation dependent PL properties of the biodots (Fig. 6c). These results show that the dAMP-dots are able to penetrate the cell membrane and enter the cells for efficient fluorescent labeling. Furthermore, confocal fluorescence images were also obtained for ADP and ATP-dots (Fig. S8†). The dAMP-dot staining gives the brightest fluorescence image followed by the ADP-dots and lastly the ATP-dots. Interestingly, ADP-dots and ATP-dots do not show equal efficiency in the intracellular uptake. This could be due to the presence of additional phosphate groups which may have suppressed their intracellular uptake ability.
Fig. 6. (a) MTT cell proliferation assay with the A-dots. (b) Cellular toxicity of dAMP-dots with (red bars) or without (black bars) white light irradiation for 10 min at various concentrations. (c) Confocal fluorescence images of HeLa cells stained with dAMP-dots under 405 nm, 488 nm and 564 nm excitation, giving rise to different emission colors, i.e., blue, green and red (from left to right).
As the dAMP-dots are able to generate the highest level of singlet oxygen, the application of dAMP-dots for photodynamic therapy (PDT) was studied in HeLa cells. In this study, increasing concentrations of dAMP-dots (from 0 to 0.62 mg mL−1) were added to the cancer cells and irradiated with 150 mW white light for 10 minutes, followed by 24 h incubation. It can be observed that at 0.62 mg mL−1, dAMP-derived dots were able to induce significant cytotoxicity upon 10 min of white light treatment, resulting in ∼60% decrease in cell viability (Fig. 6b). On the other hand, no dark toxicity (cells retain more than 95% viability) was observed in the absence of white light. Additionally, the confocal fluorescence images of the dAMP-dot treated and light illuminated HeLa cells were also obtained to further evaluate the PDT effect of the dAMP-dots. It can be observed that the light-treated HeLa cells display a different morphology with a rounded cell shape indicating cell shrinkage as compared to the non-treated HeLa cells that are flattened and spread out (Fig. S9†). This could be an early sign of cell apoptosis due to the singlet oxygen generated from the dAMP-dots. Thus, it revealed good therapeutic effects of dAMP-dots as a potential PDT agent in generating singlet oxygen to kill cancer cells.
Conclusions
In summary, we have conducted a systematic study to unravel the nucleotide base effects of DNA on the synthesis of N-dots with intrinsic singlet oxygen generation ability. Among all the N-dots, the nitrogenous base adenine-containing precursors lead to the formation of A-dots with the highest photoluminescent (PL) intensity. The presence of phosphate groups in the A-dots could help to enhance the PL intensity and photo-stability of the resultant N-dots. Comprehensive optical and structural characterization revealed that the dAMP-dots possess the beneficial characteristics of a theranostic agent for both bioimaging and therapeutic applications. In particular, the dAMP-dots displayed good cellular uptake capability and biocompatibility which are inherited from their biomolecular precursor. Furthermore, the dual-functional dAMP-dots also exhibit a high 1O2 quantum yield of 1.20 surpassing the performance of most conventional photosensitizers. As such, they can be used as non-invasive photodynamic therapeutic agents which are capable of inducing cell death upon white light irradiation. The study herein provides new insights into the rational design of biodots with multiple functionalities leading to the development of efficient theranostic agents for simultaneous cellular tracking and therapeutic treatment.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was financially supported by the Institute of Materials Research and Engineering, Agency for Science, Technology and Research (A*STAR), Singapore, under the Biomimetic and Biomedical Program exploratory fund IMRE/16-1P1401.
Electronic supplementary information (ESI) available. See DOI: 10.1039/c9na00058e
Notes and references
- Seeman N. C. Annu. Rev. Biochem. 2010;79:65–87. doi: 10.1146/annurev-biochem-060308-102244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S. Prakash V. Halder S. Chakraborty K. Krishnan Y. Nat. Nanotechnol. 2015;10:645. doi: 10.1038/nnano.2015.130. [DOI] [PubMed] [Google Scholar]
- Leung K. Chakraborty K. Saminathan A. Krishnan Y. Nat. Nanotechnol. 2019;14:176–183. doi: 10.1038/s41565-018-0318-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty K. Veetil A. T. Jaffrey S. R. Krishnan Y. Annu. Rev. Biochem. 2016;85:349–373. doi: 10.1146/annurev-biochem-060815-014244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta A. Medintz I. L. Nanoscale. 2016;8:9037–9095. doi: 10.1039/C5NR08465B. [DOI] [PubMed] [Google Scholar]
- Zhou L. Ren J. Qu X. Mater. Today. 2017;20:179–190. doi: 10.1016/j.mattod.2016.09.012. [DOI] [Google Scholar]
- Xu H. V. Zheng X. T. Mok B. Y. L. Ibrahim S. A. Yu Y. Tan Y. N. J. Mol. Eng. Mater. 2016;04:1640003. doi: 10.1142/S2251237316400037. [DOI] [Google Scholar]
- Zheng X. T., Xu H. V. and Tan Y. N., in Advances in Bioinspired and Biomedical Materials Volume 2, American Chemical Society, 2017, ch. 7, vol. 1253, pp. 123–152 [Google Scholar]
- Xu H. V. Zheng X. T. Wang C. Zhao Y. Tan Y. N. ACS Appl. Nano Mater. 2018;1:2062–2068. doi: 10.1021/acsanm.8b00465. [DOI] [Google Scholar]
- Xu H. V. Zheng X. T. Zhao Y. Tan Y. N. ACS Appl. Mater. Interfaces. 2018;10:19881–19888. doi: 10.1021/acsami.8b04864. [DOI] [PubMed] [Google Scholar]
- Guo C. X. Xie J. Wang B. Zheng X. Yang H. B. Li C. M. Sci. Rep. 2013;3:2957. doi: 10.1038/srep02957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song T. Zhu X. Zhou S. Yang G. Gan W. Yuan Q. Appl. Surf. Sci. 2015;347:505–513. doi: 10.1016/j.apsusc.2015.04.143. [DOI] [Google Scholar]
- Tripathi K. M. Tyagi A. Ashfaq M. Gupta R. K. RSC Adv. 2016;6:29545–29553. doi: 10.1039/C5RA27432J. [DOI] [Google Scholar]
- Tyagi A. Tripathi K. M. Singh N. Choudhary S. Gupta R. K. RSC Adv. 2016;6:72423–72432. doi: 10.1039/C6RA10488F. [DOI] [Google Scholar]
- Devi S. Gupta R. K. Paul A. K. Kumar V. Sachdev A. Gopinath P. Tyagi S. RSC Adv. 2018;8:32684–32694. doi: 10.1039/C8RA06460A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi S. Gupta R. K. Paul A. K. Tyagi S. Mater. Res. Express. 2018;6:025605. doi: 10.1088/2053-1591/aaf03c. [DOI] [Google Scholar]
- Li Q. H. Zhang L. Bai J. M. Liu Z. C. Liang R. P. Qiu J. D. Biosens. Bioelectron. 2015;74:886–894. doi: 10.1016/j.bios.2015.07.018. [DOI] [PubMed] [Google Scholar]
- Cheng G. Zhang W. Zhou Y. Ge Q. Huang C. Anal. Methods. 2015;7:6274–6279. doi: 10.1039/C5AY01151E. [DOI] [Google Scholar]
- Ding H. Du F. Liu P. Chen Z. Shen J. ACS Appl. Mater. Interfaces. 2015;7:6889–6897. doi: 10.1021/acsami.5b00628. [DOI] [PubMed] [Google Scholar]
- Ge J. Lan M. Zhou B. Liu W. Guo L. Wang H. Jia Q. Niu G. Huang X. Zhou H. Meng X. Wang P. Lee C. S. Zhang W. Han X. Nat. Commun. 2014;5:4596. doi: 10.1038/ncomms5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreaden E. C. Alkilany A. M. Huang X. Murphy C. J. El-Sayed M. A. Chem. Soc. Rev. 2012;41:2740–2779. doi: 10.1039/C1CS15237H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z. Hsu Y.-C. Li L.-B. Wang L.-W. Song X.-D. Yow C. M. N. Lei X. Musani A. I. Luo R.-C. Day B. J. J. Innovative Opt. Health Sci. 2015;08:1530002. doi: 10.1142/S1793545815300025. [DOI] [Google Scholar]
- DeRosa M. C. Crutchley R. J. Coord. Chem. Rev. 2002;233:351–371. doi: 10.1016/S0010-8545(02)00034-6. [DOI] [Google Scholar]
- Choi Y. Kim S. Choi M.-H. Ryoo S.-R. Park J. Min D.-H. Kim B.-S. Adv. Funct. Mater. 2014;24:5781–5789. doi: 10.1002/adfm.201400961. [DOI] [Google Scholar]
- Baev A. A., Absorption Spectra of Minor Bases: their Nucleosides, Nucleotides, and Selected Oligoribonucleotides, Springer, 2013 [Google Scholar]
- Hughes M. E. Brandin E. Golovchenko J. A. Nano Lett. 2007;7:1191–1194. doi: 10.1021/nl062906u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X. T. Ananthanarayanan A. Luo K. Q. Chen P. Small. 2015;11:1620–1636. doi: 10.1002/smll.201402648. [DOI] [PubMed] [Google Scholar]
- Lim S. Y. Shen W. Gao Z. Chem. Soc. Rev. 2015;44:362–381. doi: 10.1039/C4CS00269E. [DOI] [PubMed] [Google Scholar]
- Yuan Y. H. Liu Z. X. Li R. S. Zou H. Y. Lin M. Liu H. Huang C. Z. Nanoscale. 2016;8:6770–6776. doi: 10.1039/C6NR00402D. [DOI] [PubMed] [Google Scholar]
- Shang J. Ma L. Li J. Ai W. Yu T. Gurzadyan G. G. Sci. Rep. 2012;2:792. doi: 10.1038/srep00792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y. Geng J. Ong E. Y. Chellappan V. Tan Y. N. Adv. Healthcare Mater. 2016;5:2528–2535. doi: 10.1002/adhm.201600312. [DOI] [PubMed] [Google Scholar]
- Regensburger J. Knak A. Maisch T. Landthaler M. Baumler W. Exp. Dermatol. 2012;21:135–139. doi: 10.1111/j.1600-0625.2011.01414.x. [DOI] [PubMed] [Google Scholar]
- Mathai S. Smith T. A. Ghiggino K. P. Photochem. Photobiol. Sci. 2007;6:995–1002. doi: 10.1039/B705853E. [DOI] [PubMed] [Google Scholar]
- Mojzisova H. Bonneau S. Maillard P. Berg K. Brault D. Photochem. Photobiol. Sci. 2009;8:778–787. doi: 10.1039/B822269J. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.