Abstract
Helicobacter pylori colonizes the human stomach and can cause gastroduodenal disease. Flagellar motility is regarded as a major factor in the colonizing ability of H. pylori. The functional roles of flagellar structural proteins other than FlaA, FlaB, and FlgE are not well understood. The fliD operon of H. pylori consists of flaG, fliD, and fliS genes, in the order stated, under the control of a ς28-dependent promoter. In an effort to elucidate the function of the FliD protein, a hook-associated protein 2 homologue, in flagellar morphogenesis and motility, the fliD gene (2,058 bp) was cloned and isogenic mutants were constructed by disruption of the fliD gene with a kanamycin resistance cassette and electroporation-mediated allelic-exchange mutagenesis. In the fliD mutant, morphologically abnormal flagellar appendages in which very little filament elongation was apparent were observed. The fliD mutant strain was completely nonmotile, indicating that these abnormal flagella were functionally defective. Furthermore, the isogenic fliD mutant of H. pylori SS1, a mouse-adapted strain, was not able to colonize the gastric mucosae of host mice. These results suggest that H. pylori FliD is an essential element in the assembly of the functional flagella that are required for colonization of the gastric mucosa.
Helicobacter pylori is a gram-negative, microaerophilic bacterium which colonizes the gastric antrum of the human stomach. Since Marshall and Warren first isolated and cultured this bacterium from biopsy specimens (25), extensive studies of its biology have been carried out and its complete genome sequence has been published (37). Epidemiological studies have consistently demonstrated that H. pylori is a causative agent of active chronic gastritis and peptic ulcers and that H. pylori is a primary risk factor for the development of intestinal type gastric adenocarcinoma (6, 15). More recently, this organism was also associated with mucosa-associated lymphoid tissue (MALT) and with B-cell MALT lymphomas (31). However, the actual mechanism by which gastroduodenal diseases develop in response to H. pylori infection remains unknown.
The putative pathogenic factors of H. pylori are categorized as colonization, persistence, and disease-inducing factors. Colonization in the host is a prerequisite of bacterial infection and subsequent pathogenesis. Motility by unipolar flagella, urease production, and adhesion to the gastric epithelial cells are all required for H. pylori colonization. A bacterial flagellum consists of three distinct parts connected in series: a basal body in the membrane, a short curved rod called the hook, and a long helical filament. The filament and the hook are each formed by regular assembly of a single species of protein subunit. Proteins called hook-associated proteins (HAPs) are required for joining the filament to the hook and for capping the distal tip of the filament. HAP1 and HAP3 are thought to function as structural adapters between the hook and the filament (13). HAP2 is thought to function as a capping structure at the distal end of the filament, which enables flagellin monomers to assemble into filaments (13, 14).
H. pylori is a motile, flagellated organism that probably penetrates the mucus layer by means of spiralling movements associated with its unique shape. The motility of H. pylori is provided by two to six polar, sheathed flagella, the filaments of which consist of two flagellin types, encoded by the genes flaA and flaB (35). The majority of the filament is composed of FlaA, while FlaB is found only in the base (18). The flagellar filament is linked to the basal body by means of the hook, which is a polymer of the FlgE hook protein (30). In vitro experiments with isogenic H. pylori strains with mutated flaA and flaB flagellin genes have shown that both flagellins are required for full motility (16). Colonization experiments with spontaneous nonmotile mutants, as well as with isogenic flaA, flaB, and flaA flaB mutants in the gnotobiotic piglet model of H. pylori infection, have demonstrated that the establishment of persistent infection requires full motility and the presence of both flagellins (4).
Full motility is an essential virulence factor of H. pylori and a potential target for therapy and vaccine development.
We describe here the molecular characterization of the H. pylori fliD gene, which encodes a 76-kDa HAP-2 homologue, and the effects of a mutation in the fliD gene on the assembly of flagellar filaments and on motility. Infection of mice with a fliD mutant of H. pylori SS1, a mouse-adapted strain, demonstrated that the FliD protein is necessary for colonization. The H. pylori fliD gene is interesting in that it is a structural gene which appears to play a role in genetic regulation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
H. pylori KCTC0217BP (Korean Collection for Type Cultures) was isolated from a Korean patient suffering from duodenal ulceration. H. pylori SS1 (Sydney strain) was kindly provided by A. Lee (University of New South Wales, Sydney, Australia). H. pylori strains were grown on selective blood agar plates containing 22 g of Columbia blood agar base (Difco)/liter, 20 g of tryptic soy agar (Difco)/liter, 7% (vol/vol) sheep blood, 10 μg of vancomycin (Sigma)/ml, 300 U of colistin (Sigma)/ml, and 2.5 μg of amphotericin B (Sigma)/ml. Plates were incubated at 37°C for 48 to 72 h under 10% CO2.
Expression of recombinant FliD protein.
The fliD gene was amplified by PCR with the oligonucleotide primers fliD/NNdeI and fliD/CBamHI (Table 1) according to standard PCR protocols. The amplified DNA fragment of 2,076 bp was digested with restriction enzymes NdeI and BamHI and then ligated into NdeI/BamHI-digested pET-15b (Novagen). The resulting plasmid was transformed into Escherichia coli BL21(DE3). The recombinant FliD protein was expressed and purified by using the pET His tag system according to the manufacturer’s instructions.
TABLE 1.
Oligonucleotides used for PCR amplification
| H. pylori gene | Primer | Sequencea | Position | Strand |
|---|---|---|---|---|
| fliD | fliD/NNdeI | ggaattccatATGGCAATAGGTTCATT | 853–869 | + |
| fliD/CBamHI | cgggatccTTAATTCTTTTTGGCCGC | 2893–2910 | − | |
| fliDseq#2 | GATATTTTTAGCCAAGTGG | 1216–1234 | + | |
| fliDseq#4 | TTCTTATAGCGTGCATACGG | 2457–2476 | + | |
| fliDseq#5 | TCTAAAGCCTGCTGTATCGC | 1621–1640 | − | |
| ureAb | UA5 | TTGATGCTCCACTACGCTGG | 2689–2708 | + |
| UA3 | GGGGTATGCACGGTTACGAG | 2935–2954 | − | |
| UALP | CACCCCAAAAGAGTTAGATA | 2667–2686 | + | |
| UARP | ATGTCTAAGCGTTTACCGAA | 3142–3161 | − |
Antibody preparation.
Polyclonal antibody against the outer membrane proteins (OMP) of H. pylori was produced in a rabbit. The animal was immunized with 140 μg of Sarkosyl-insoluble OMP prepared from H. pylori KCTC0217BP and boosted twice after 1 and 4 weeks, and blood was collected 10 days after the last injection. This anti-OMP antibody preparation was employed for genomic DNA library screening and Western blot analysis. Polyclonal antibody against H. pylori FliD was produced in a similar manner with 200 μg of purified recombinant FliD protein.
Electroporation of H. pylori.
H. pylori KCTC0217BP cells were harvested from blood agar plates, washed in a 15% glycerol–9% sucrose solution, and suspended in a final volume of 50 μl of glycerol-sucrose solution at 4°C (1010 bacteria per ml). Supercoiled plasmids (700 ng) containing the targeted gene that had been disrupted with a kanamycin resistance cassette were added to the cells. After a 1-min incubation on ice, the cells and DNA were transferred to a prechilled electroporation cuvette (0.2-cm electrode gap) and placed in a Gene Pulser apparatus (Bio-Rad). Pulses were applied with the electronic variables set to 25 F, 1.25 kV, and 200 Ω, giving a time constant of 4.6 ms. After electroporation, bacteria were grown on nonselective plates for 48 h to allow expression of antibiotic resistance and then transferred onto plates containing kanamycin (20 μg/ml) and grown for up to 6 days.
RNA isolation and Northern blot analysis.
Total RNA was extracted with Trizol reagent (Gibco BRL) from H. pylori KCTC0217BP and the fliD mutant of this strain grown for 48 h. Ten micrograms of RNA was resolved on 1% denaturing formaldehyde-agarose gels and transferred to a Hybond-N membrane (Amersham) by capillary blotting. The PCR product, amplified by primers flaA-5 (5′-GGAATTCCATATGGCTTTTCAGGTCAA-3′) and flaA-3 (5′-GCTCTAGACTAAGTTAAAAGCCTTAAG-3′), with genomic DNA of H. pylori KCTC0217BP as a template, was radiolabelled with the Megaprime DNA-labelling system (Amersham) and used as a probe. Hybridization was performed in ExpressHyb hybridization solution (Clontech) at 68°C for 18 h. The filter was washed at room temperature with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.05% sodium dodecyl sulfate (SDS) for 45 min and at 50°C for 30 min with 0.1× SSC–0.1% SDS. After autoradiography, RNA expression was quantified by densitometric analysis with a ScanJet 4c scanner (Hewlett-Packard) with SigmaGel, version 1.0, software.
Electron microscopy.
Bacteria were harvested from blood agar plates and gently resuspended in phosphate-buffered saline. For negative staining, a Formvar carbon-coated grid was floated on a drop of the sample suspension for 5 min. Excess sample was withdrawn by touching the edge of the grid to a cut of Whatman no. 1 filter paper. The grids were then floated onto a drop of 1% phosphotungstate (pH 6.8) for 1 min. The grids were examined with a JEOL JEM-1200EX or JEM-1010 transmission electron microscope.
Motility testing.
Motility was initially judged by viewing the movement of bacteria grown in liquid medium with an Olympus BX50 phase-contrast microscope. We also performed stab agar and single-colony motility tests to evaluate bacterial motility. Motility plates containing brucella broth (Difco) and 0.4% Bacto Agar were supplemented with 10% horse serum and antibiotics as described above. For the stab agar test, plates were inoculated by placing small slices of blood agar plates, densely grown with the strain to be tested, on the surface of the motility plates with the lawn side of the slice facing upward. Plates were incubated at 37°C for 4 days under microaerophilic conditions, and motility was assessed by examining the swarming halo. To examine single-colony motility, bacterial cells were harvested in brucella broth and diluted to about 101 to 102 cells/ml with motility medium. This bacterial suspension was poured onto the plates and incubated for up to 5 days. Single-colony morphology was examined with a phase-contrast microscope.
Animal experiments.
Specific pathogen-free, 6-week-old, female, C57BL/6 mice (Charles River) were inoculated with the bacterial suspension. The animals were dosed three times in a 2-day period with 0.4 ml of bacterial suspension (approximately 108 cells) by using a blunt-ended needle. Mice were sacrificed 10 days after the last inoculation. The stomach was excised, cut along the lesser curvature, and rinsed in saline to remove the contents. Half of the stomach was placed in 0.5 ml of brain heart infusion broth (Difco) and disrupted with a pellet pestle (Kontes Scientific Glassware), and the suspension was plated onto selective blood agar plates. The level of colonization was determined numerically by counting the viable bacteria thus recovered. Bacterial counts were expressed as CFU per gram of gastric tissue. The other half of the stomach was treated as previously described (9) and used for PCR amplification of the H. pylori ureA gene (21) to detect the presence of the bacteria. A nested PCR method was applied by using the ureA-specific oligonucleotide primers (UALP and UARP for primary PCR, UA5 and UA3 for nested PCR) listed in Table 1.
Nucleotide sequence accession number.
The nucleotide sequences of the fliD operon have been deposited in GenBank under the accession no. U82981.
RESULTS
Library screening and identification of clones expressing a 76-kDa antigenic protein.
A genomic expression library was constructed in Lambda ZapII (Stratagene) with HaeIII-digested genomic DNA of H. pylori KCTC0217BP and screened with anti-H. pylori OMP antibody. Approximately 1.5 × 105 plaques were screened, and 11 positive plaques were selected. Each clone was transformed into a pBluescript SK(−) phagemid by in vivo excision (33). When the protein profile of each clone was analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting, nine clones were found to express an H. pylori-specific protein with an estimated molecular mass of 76 kDa. A protein band of the same size was detected in H. pylori OMP as well as in whole-cell lysates with antiH. pylori OMP antibody (data not shown). Plasmid DNA was isolated from each clone expressing the 76-kDa protein and digested with EcoRI; DNA inserts of 3.4 and 5 kb were observed, and a plasmid containing a 3.4-kb insert was selected for further study.
Nucleotide and amino acid sequence analysis of the fliD gene.
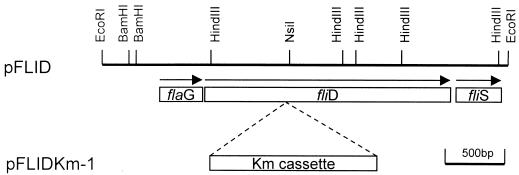
A restriction map of the selected clone, which was designated pFLID, was constructed (Fig. 1). The entire 3,390-bp nucleotide sequence of the pFLID insert was determined. The sequenced DNA fragment contained three open reading frames (ORFs), which appeared to comprise a single operon. The products of these ORFs showed significant homology with the bacterial FlaG, FliD, and FliS proteins, as demonstrated by an amino acid sequence homology search with the BLAST network service at the National Center for Biotechnology Information (1). The organization of these ORFs is shown in Fig. 1. A ς28-dependent promoter (5′-TtAA-N15-GCCGATAt-3′ in the H. pylori −35/−10 region, consistent with the E. coli promoter sequence 5′-TAAA-N15-GCCGATAA-3′ [nucleotides in the H. pylori sequence differing from those in the E. coli sequence are in lowercase]) was found upstream of the operon, and a Shine-Dalgarno ribosome binding site was found upstream of each ORF. Two transcription start points, nucleotide A at positions 417 and 419, were identified by primer extension experiments (data not shown). The fliD operon had the same gene organization in H. pylori KCTC0217BP and 26695, the complete genome sequences of which have been published previously (37). The amino acid sequences of FlaG and FliS from these two H. pylori strains showed significant homology, with 94% identity and 97% similarity for FlaG and identical sequences for FliS. However, FliD from these two strains showed size differences, having 685 amino acids in H. pylori KCTC0217BP and 674 amino acids in H. pylori 26695. The amino acid sequence divergence mainly occurred in the C-terminal region, while the first 654 residues from the N terminus showed 96% identity and 97% similarity. To rule out the possibility that this difference was due to misreading the fliD DNA sequences, we repeated the DNA sequencing and confirmed that the nucleotide sequences presented here are correct.
FIG. 1.
Restriction maps of the recombinant plasmids pFLID and pFLIDKm-1. The locations of three ORFs (flaG, fliD, and fliS genes) are indicated by arrows. pFLIDKm-1 was constructed by inserting a kanamycin resistance cassette into the NsiI restriction site of pFLID. Only the inserts of the plasmids are depicted.
Based on the calculated molecular masses of the proteins encoded by each ORF, the 76-kDa protein which reacted with anti-H. pylori OMP antibody appeared to be encoded by the second ORF. To confirm this, the N-terminal amino acid sequence of the partially purified 76-kDa protein was determined. The amino acid sequence thus obtained, AIGSLSSLGLGSKVL, matched residues 2 to 16 of the deduced amino acid sequence encoded by the second ORF perfectly, confirming that this ORF, designated the fliD gene, encodes a 76-kDa protein which is a HAP2 homologue of H. pylori. When the amino acid sequence of H. pylori FliD was aligned with HAP2 sequences from other bacteria, such as Salmonella typhimurium (12), Pseudomonas aeruginosa (2), Xenorhabdus nematophilus (7), Bacillus subtilis (3), and Vibrio parahaemolyticus (26), sequence similarities were predominantly apparent in the N- and C-terminal regions. The only observed modification in the N-terminal sequence of H. pylori FliD protein was the absence of the first methionine residue. The N-terminal amino acid sequence of FliD did not contain any conventional signal sequences for export, indicating that the FliD is not transported via the primary cellular pathway for protein export. H. pylori FliD, like the other axial components of the flagellum, contained no cysteine residues. The proline content of H. pylori FliD was 1.75 mol%; there were 12 proline residues in the entire sequence, and these residues were not evenly distributed throughout the sequence but rather seemed to be clustered. There were no proline residues in the last 109 amino acids from the C terminus. In this region, we observed a repeated motif of hydrophobic amino acids at intervals of seven residues, starting from isoleucine at position 613 to alanine at position 680. Such hydrophobic heptad repeats have also been reported in other bacterial HAP2 proteins (3, 12, 26) as structural elements for quaternary interactions in the flagellar axial structures. In contrast, the N-terminal region of FliD lacked such hydrophobic heptad repeats and contained proline residues.
Construction of an isogenic fliD mutant by allelic-exchange mutagenesis.
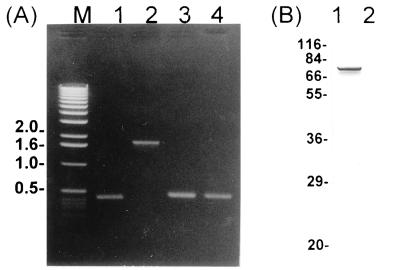
Bacterial HAP2 plays a role in flagellar morphogenesis as a flagellar capping protein, facilitating polymerization of the flagellin monomer at the tips of filaments (14). In order to examine the function of FliD in H. pylori, an isogenic mutant of H. pylori KCTC0217BP which would result in the null expression of FliD was constructed by the following method. A 1.4-kb SmaI restriction fragment of pILL600 (5) containing a gene encoding resistance to kanamycin (aph3′-III) was cloned into the unique NsiI site situated in the middle of the fliD gene after treatment of linearized pFLID with mung bean nuclease. The resulting plasmid was called pFLIDKm-1 (Fig. 1). H. pylori KCTC0217BP was transformed with pFLIDKm-1 by electroporation, and two independent transformants were obtained. Mutant strains were defined as descendants of a single colony of kanamycin-resistant H. pylori, and the genotypes of the mutants were characterized by PCR (Fig. 2A) with oligonucleotide primers specific for sequences upstream and downstream of the site of disruption (fliDseq#2 and fliDseq#5; Table 1). In mutant strains, a 1.8-kb DNA fragment containing the 1.4-kb kanamycin cassette was amplified, whereas a 0.4-kb DNA fragment was amplified from the wild type. Specific integration was also verified by nucleotide sequencing the 1.8-kb PCR fragment. When PCR was performed with primers (fliDseq#4 and fliD/CBamHI; Table 1) annealing downstream of the disruption site, identical 0.5-kb DNA fragments were amplified from both wild-type and mutant strains (Fig. 2A). A double-crossover event had taken place in all the strains tested, leading to replacement of the intact allele by the allele disrupted with the kanamycin resistance cassette. The expression of the FliD protein in the isogenic mutant was analyzed by Western blotting with antiH. pylori FliD antibody; these Western blots confirmed the disappearance of the 76-kDa FliD protein from the fliD mutant strain (Fig. 2B). All the mutant strains grew well and were not significantly affected in viability or growth characteristics.
FIG. 2.
Characterization of H. pylori KCTC0217BP wild type and isogenic fliD mutant by PCR (A) and Western blotting (B). (A) Lanes 1 and 2, DNA amplified by PCR with primers (fliDseq#2 and fliDseq#5) annealing up- and downstream of the kanamycin cassette insertion site in wild-type and mutant strains, respectively; lanes 3 and 4, DNA amplified by PCR with primers (fliDseq#4 and fliD/CBamHI) annealing downstream of the insertion site in wild-type and mutant strains, respectively; lane M, DNA size markers (in kilobases). (B) Western blotting of whole-cell lysates of H. pylori KCTC0217BP wild type (lane 1) and isogenic fliD mutant (lane 2). The blot was developed with anti-recombinant FliD antibody. Molecular mass markers (in kilodaltons) are shown on the left. Expression of the 76-kDa FliD protein was observed only in the wild-type strain.
Electron microscopy of H. pylori fliD mutant.
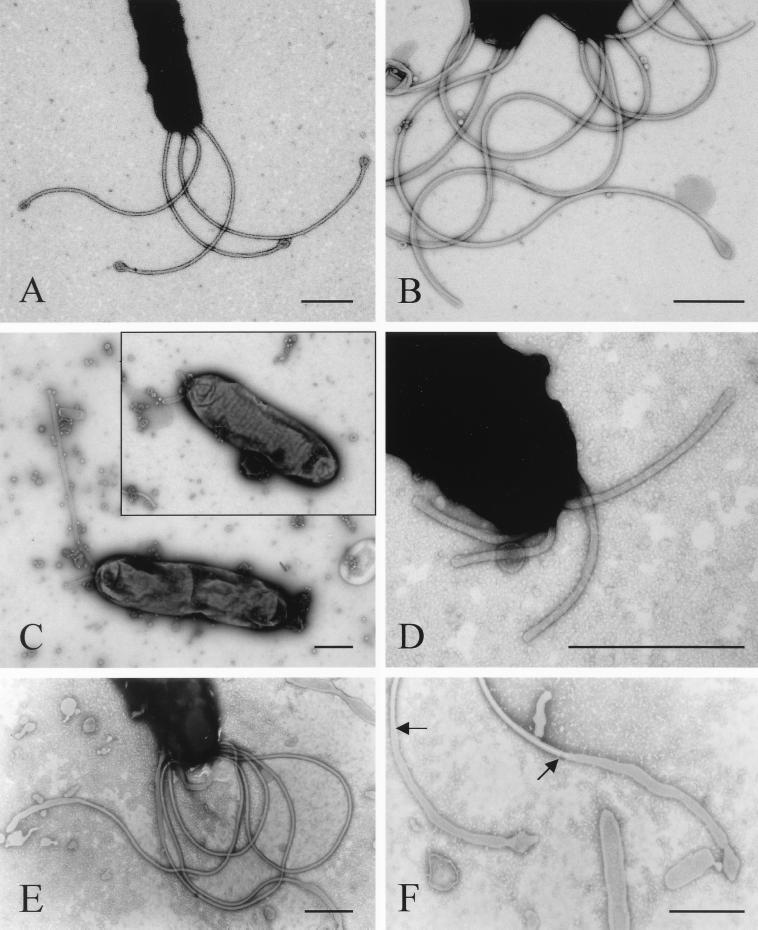
Flagellar morphology was examined by transmission electron microscopy after negative staining (Fig. 3). In wild-type strains, typical sheathed H. pylori flagella were observed. Inside the sheaths, the filament was elongated to the end of the tip and typical terminal bulbs were often observed (Fig. 3A and B). In mutant strains, truncated sheathed flagellum-like appendages were observed, but the numbers and sizes of these structures were highly variable (Fig. 3C and D). In most cases, it was difficult to observe elongated filaments inside the appendages, which looked like extensions of empty sheaths, even under higher magnification. Occasionally, more-typical sheathed flagella were observed in the fliD mutant, but filaments were not fully extended even in these structures, and empty flagellar sheaths were visible at the end of the flagella (Fig. 3E and F). Terminal bulbs were not observed in any fliD mutant strains. These findings demonstrate the requirement for FliD in the morphogenesis of flagella and, more specifically, in the elongation of the flagellar filament.
FIG. 3.
Electron microscopy of H. pylori KCTC0217BP wild type and isogenic fliD mutant, negatively stained with potassium phosphotungstate (pH 6.8). In wild-type cells, full-length sheathed flagella are observed (A). Within the wild-type flagellar sheath, filaments are elongated to the tips of the flagella and terminal bulbs are observed (B). In the fliD mutant, truncated flagella, variable in number and size, are observed (C), and terminal bulbs are not apparent. Filament elongation is not observed in the fliD mutant even under higher magnification (compare panels B and D). Typical sheathed flagella with truncated filaments are occasionally seen in the fliD mutant strains (E). The end points of the truncated filaments are indicated by arrows (F). Bars, 500 nm.
Effect of fliD gene mutation on flagellin gene expression.
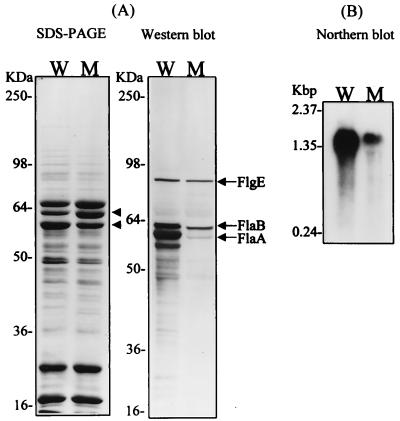
To further characterize the biochemical differences in flagellar structure between wild-type and fliD mutant H. pylori, whole-cell lysates were analyzed by SDS-PAGE and Western blotting with an anti-Fla polyclonal antibody (32), which was raised against purified flagellar filaments. Only minor differences in protein profiles, at around 60 kDa, between wild-type and fliD mutant strains were observed (Fig. 4A). Western blotting demonstrated that FlaA and FlaB expression in the fliD mutant was reduced to 7 and 57% of wild-type levels, respectively, whereas hook protein (FlgE) expression levels in the wild type and fliD mutants were identical. flaA mutant strains possess a unipolar tuft of truncated sheathed flagella (16), similar to the flagellar morphology observed in the fliD mutant strains. Thus, the flagellar morphology of the fliD mutant strains may have been due to reduced expression of FlaA resulting from fliD gene disruption. Given that both the flaA (23) and fliD genes are under the control of a ς28-dependent promoter, whereas the flaB and flgE genes are controlled by a ς54-regulated promoter (30, 35), we thought that transcriptional regulation might be involved in the reduced expression of FlaA. To test this prediction, total RNA was analyzed by Northern blotting with an flaA probe (Fig. 4B). The transcription level of the flaA gene in the fliD mutant was reduced to 20% of the wild-type level. In contrast, transcription of the ureA gene, which is constitutively expressed and presumably not related to flagellar biogenesis, was not affected (data not shown). These results indicated that the loss of FliD expression resulted in down-regulation of flaA gene expression at the transcriptional level. However, since FlaA expression was reduced to a greater degree than flaA transcription, another control mechanism, such as secretion of unpolymerized FlaA, appears to be involved in the regulation of FlaA levels.
FIG. 4.
Flagellin gene expression in wild-type and fliD mutant H. pylori strains. (A) Equal amounts of whole-cell lysates (20 μg of total protein) from wild-type (lane W) and fliD mutant (lane M) H. pylori KCTC0217BP were separated by SDS-PAGE. Two protein bands of around 60 kDa, showing different levels of expression, are indicated by arrowheads. The same gel was also analyzed by Western blotting with polyclonal antibody anti-Fla. Note the significant reduction of FlaA in the fliD mutant. The positions of the hook (FlgE) and major (FlaA) and minor (FlaB) flagellin proteins are indicated on the right. (B) Northern blotting demonstrated that flaA RNA levels were also reduced in the fliD mutant.
Functional analysis by motility testing.
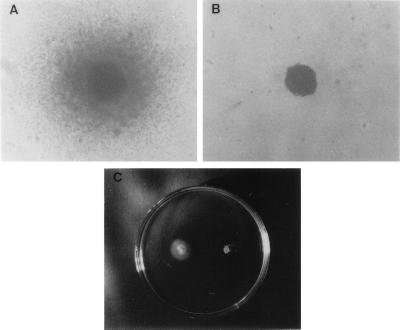
Wild-type H. pylori and fliD mutants, growing exponentially in liquid media, were observed under the phase-contrast microscope. Motile bacteria were all members of wild-type strains, not of the isogenic mutant strain. Since colony morphology on motility plates is a reliable indicator of the motility phenotype, single-colony motility and stab agar motility tests were performed to compare the motilities of wild-type and mutant H. pylori strains (Fig. 5). In the single-colony test, motility was assessed with the phase-contrast microscope by comparing the swarming halos surrounding single colonies. The wild-type strain formed colonies with the large diffuse spreading halo typical of motile bacteria (Fig. 5A). The fliD mutant strain produced small and sharply delineated colonies, a morphology typical of nonmotile bacteria (Fig. 5B). Likewise, in the stab agar test, swarming halo formation was observed only with the wild-type strain and the mutant strain showed no apparent motility (Fig. 5C).
FIG. 5.
Motility testing of H. pylori KCTC0217BP wild-type and isogenic fliD mutant strains. (A and B) Single-colony motilities of wild-type and fliD mutant strains, respectively. The wild-type strain formed diffuse colonies with large swarming halos, whereas the fliD mutant formed dense colonies. (C) Stab agar test of wild type (left) and fliD mutant (right).
Functional analysis by infectivity testing.
To determine whether the fliD mutant bacteria retained the ability to colonize the mucus layer of the stomach, an infection test using the H. pylori-mouse model was performed. H. pylori SS1, which is able to colonize mouse gastric mucosae and reach high infection levels (22), was used to assess the role of the fliD gene product in H. pylori colonization. We constructed an isogenic mutant of fliD in an H. pylori SS1 background by the method described above. Both wild-type and fliD mutant SS1 strains were administered orally to 6-week-old, female C57BL/6 mice. Ten days after oral inoculation, mice were sacrificed and one-half of each stomach was homogenized and plated on selective blood agar plates in order to culture colonizing H. pylori. We recovered 2.23 × 105 cells per gram of gastric tissue from the mice inoculated orally with wild-type H. pylori SS1 but none from those inoculated with the mutant (Table 2). The remaining half of the stomach tissue was used for PCR analysis in order to detect even very low levels of H. pylori in the gastric mucosa. The ureA gene was amplified only from the stomach tissues of mice inoculated with wild-type H. pylori SS1 (Table 2). These results strongly suggested that FliD is absolutely required for H. pylori to colonize and establish infection in the mouse model.
TABLE 2.
Colonization in mice by wild-type and fliD mutant H. pylori SS1
| Inoculuma | No. of mice colonized/totalb
|
CFU/g of gastric tissuec ± SD (105) | |
|---|---|---|---|
| By culture | By PCR | ||
| H. pylori SS1 | 3/3 | 3/3 | 2.23 ± 0.12 |
| H. pylori SS1 fliD::Km | 0/3 | 0/3 | 0 |
| Control | 0/3 | 0/3 | 0 |
Each mouse (strain C57BL/6) received 108 CFU of H. pylori SS1 wild type or fliD mutant in 0.4 ml of brain heart infusion (BHI) broth, twice at 2-day intervals. Control mice received 0.4 ml of BHI broth alone.
H. pylori colonization in gastric tissue was assessed at 10 days postinoculation by culture and PCR.
Numbers of H. pylori CFU recovered following homogenization of gastric tissue samples. The results are geometric means for three mice per group.
DISCUSSION
Many bacteria are propelled by the rotation of semirigid helical filaments called flagella. The flagellar filaments are joined via proteins called HAPs to a universal joint (the hook) which is connected to a motor (the basal body) embedded in the cytoplasmic membrane. HAPs are important in the formation of flagella, even though these proteins are only present in small amounts (13). HAP1 and HAP3 are involved in joining the filament to the hook. HAP2, also known as the distal capping protein, localizes to the tip of the flagellar filament, where it serves to plug the tip of the growing flagellum and promote polymerization of flagellin subunits (10, 13). Different HAP mutant phenotypes have been observed due to differences between sheathed and unsheathed flagella. S. typhimurium mutants with defects in genes encoding HAP1, HAP2, or HAP3 are immotile and secrete unpolymerized filament protein into the growth medium (11). When HAP2 protein was added exogenously to the HAP2 mutant, flagellin polymerization occurred and complete flagella were constructed (14). In contrast, V. parahaemolyticus mutants defective for these genes show different phenotypes (26). Mutants with defects in capping protein (HAP2) are motile but slow in semisolid motility plates, and mutants with defects in the joining proteins (HAP1 and HAP3) are immotile but do not secrete unpolymerized flagellin molecules. These mutants produced nonfunctional, severely truncated filaments that were not attached to the cell body. It seems likely that the flagellar sheath is responsible for the differences in phenotype between S. typhimurium and V. parahaemolyticus HAP mutants. The sheath may trap secreted, unpolymerized flagellin and substitute for the capping protein.
In this study, we identified and characterized the fliD operon of the human pathogen H. pylori. This operon consists of flaG, fliD, and fliS gene homologues, in the order stated, downstream of a sequence which closely resembles the consensus for the so-called ς28-dependent promoters. fliD operons containing fliD, fliS, and fliT genes have been found in other bacteria, including E. coli, S. typhimurium, and B. subtilis (3, 12, 17). The fliD genes of these bacteria encode the filament cap protein, also called HAP2, which facilitates the polymerization of endogenous flagellin at the tips of growing flagellar filaments. However, there are conflicting reports regarding the roles played by the fliS and fliT genes in flagellar formation (17). The fliS gene has been implicated as a chaperone involved in the export of flagellin, while fliT apparently has no effect on flagellar formation (38). In P. aeruginosa, the gene arrangement of the fliD operon is different; the fliT gene seems to be absent from this operon, and instead there is a duplication of the fliS gene (2). The gene arrangement of the H. pylori fliD operon also shows a distinctive feature in that the flaG gene homologue, instead of the fliT gene, is present. There appears to be no fliT gene homologue in the published H. pylori genome (37).
One of the interesting sequence features of flagellar axial proteins is a series of hydrophobic heptad repeats observed in their N- and C-terminal regions. These heptad repeats are characteristic of α-helical coiled coils and are believed to be important in quaternary interactions between respective protein subunits within the flagellar axial structures. Interestingly, hydrophobic heptad repeats were observed only in the C-terminal region in H. pylori FliD, as in other bacterial HAP2 homologues. These properties of HAP2 may reflect its special location. The filament cap occupies a unique location among all the axial substructures, in that its distal face is exposed to the environment (or probably to flagellar sheaths in sheathed flagella), rather than to quaternary interactions with other proteins. It has been suggested that the C-terminal region of HAP2 may be important in quaternary interactions with flagellin or with HAP3 at an earlier stage of the assembly process and that the N-terminal region is more free to diverge, perhaps because it is at the distal face of the subunit (12).
To elucidate the function of the fliD gene product of H. pylori, which possesses a unipolar bundle of sheathed flagella, we constructed isogenic mutants and compared their flagellar structures, motilities, and infectivities in a mouse model with those of the wild type.
In the fliD mutants, which were completely nonmotile, expression of the FliD protein was completely abolished and their flagella were sheathed but severely truncated. Interestingly, disruption of the fliD gene significantly decreased the expression of FlaA, the major flagellin subunit, partly by transcriptional down-regulation of flaA. This indicates that FliD, itself a flagellar structural component, plays a role in genetic regulation. In the process of flagellar assembly, filament elongation starts only after the hook-filament junction proteins (HAP1 and HAP3) and the filament-capping protein (FliD) are successively added to the full-length hook. Defects in the filament-capping protein may thus cause the failure of subsequent filament elongation. In this context, it is advantageous for the H. pylori fliD mutant strains to control the wasteful expression of FlaA by blocking gene transcription.
The regulation of motility and chemotaxis in S. typhimurium has been well studied (24). In S. typhimurium, flagellar operons are divided into three classes with respect to transcriptional hierarchy, and the genes required for flagellar biosynthesis are sequentially expressed according to a hierarchical pathway (19). Class 2 operons are positively regulated by class 1 genes, and class 3 operons are controlled by the FliA-FlgM regulatory system (29); FilA is a sigma factor (ς28) specific for class 3, whereas FlgM is an anti-sigma factor which binds FliA to prevent its association with RNA polymerase core enzyme. The fliD operon of S. typhimurium, together with the genes for chemoreception and the flagellin gene, is a class 3 regulon, and the expression of these genes is under the control of a ς28-dependent promoter. fliD mutations in S. typhimurium, in contrast to those in H. pylori, cause excessive export of the flagellum-specific anti-sigma factor, FlgM (39), resulting in overexpression of flagellar class 3 operons, including the flagellin gene, fliC (20). Therefore, the regulatory mechanism underlying flagellar biogenesis and control of motility in H. pylori appears to differ from that in S. typhimurium. Further evidence of such a difference in regulation is that no anti-sigma factor (FlgM) homologue has been found in the H. pylori genome (37). Instead, a transcriptional activator, FlgR, which functions both as an activator of ς54-regulated genes such as basal body and hook genes and as a repressor of the ς28-regulated flaA flagellin gene, has been identified (34). Given that the fliD gene is also under the control of a ς28-dependent promoter, transcription of the fliD gene may be regulated by FlgR, but no experimental evidence is available to confirm this suggestion. Little is known about the flagellar regulon in H. pylori. To understand our present observations in the full context of the flagellar regulon will therefore require more information.
A remarkable structural feature of H. pylori flagella is the flagellar sheath. The effects of flagellin expression on flagellar morphology have been extensively studied (16). Empty flagellar sheaths are occasionally seen on H. pylori flaA and flaA flaB mutants, in which flagellar filaments are severely truncated or absent. Thus, generation of the flagellar sheath appears to be an active process independent of filament production. The fliD mutant also showed truncated sheathed flagella similar to those of H. pylori flaA mutants. However, the motilities of the fliD and the flaA mutant strains are different. The fliD mutant strains are nonmotile, while the flaA mutant strains are slightly motile. This result indicates that FliD expression is essential for the formation and function of H. pylori flagella.
Links between flagella and virulence have been observed previously. Colonization experiments with nonmotile mutants in the gnotobiotic piglet model of H. pylori infection have demonstrated that full motility is an essential virulence factor of H. pylori and a possible target for novel therapeutic substances. In other bacterial species, loss of the ability to produce flagella generally results in a less virulent organism. Nonflagellated isolates of Campylobacter jejuni (8), Bordetella avium (28), Bacillus thuringiensis (40), and Clostridium chauvoei (36) were all found to be less invasive or less virulent than the parental strains in their respective in vitro and in vivo models of pathogenesis.
Recently, FliD has been implicated as a virulence factor of Proteus mirabilis. A fliD mutant of P. mirabilis was shown to be defective in colonization of the urinary tract and attenuated in virulence in a mouse model of ascending urinary tract infection (27). FliD of P. aeruginosa participates in mucin-specific adhesion, which is the initial event in colonization by this organism of the airways of cystic fibrosis patients (2). In the present study, FliD of H. pylori was shown to be essential for colonization as a result of its essential role in flagellar construction and establishment of motility. Construction of the flagellum is a complex process in which many genes are known to be involved. Although some of these genes have been characterized in H. pylori, little is known about the basic mechanisms and factors involved in flagellar synthesis in this organism. Recently, the sequence of the entire genome of H. pylori was published (37). Characterization of further H. pylori genes corresponding to known flagellar biosynthesis genes by reverse genetic approaches, as shown in this study, will provide more insights into the complex flagellar apparatus and the motility which these structures confer on bacteria. This information may provide practical ways to prevent or treat H. pylori infection in the gastric mucosa.
ACKNOWLEDGMENTS
We thank A. Lee for providing H. pylori SS1, A. Labigne for the gift of plasmid pILL600, P. W. O’Toole for rabbit anti-Fla, and Jae-Hak Park and Kyung-Ku Ahn for expert electron microscopy. We also thank Ji-Hyun Lee for excellent technical assistance in animal experiments.
This work was supported by the Korea Green Cross Corporation.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Arora S K, Ritchings B W, Almira E C, Lory S, Ramphal R. The Pseudomonas aeruginosa flagellar cap protein, FliD, is responsible for mucin adhesion. Infect Immun. 1998;66:1000–1007. doi: 10.1128/iai.66.3.1000-1007.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen L, Helmann J D. The Bacillus subtilis ςD-dependent operon encoding the flagellar proteins FliD, FliS, and FliT. J Bacteriol. 1994;176:3093–3101. doi: 10.1128/jb.176.11.3093-3101.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eaton K A, Suerbaum S, Josenhans C, Krakowka S. Colonization of gnotobiotic piglet by Helicobacter pylori deficient in two flagellin genes. Infect Immun. 1996;64:2445–2448. doi: 10.1128/iai.64.7.2445-2448.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferrero R L, Cussac V, Courcoux P, Labigne A. Construction of isogenic urease-negative mutants of Helicobacter pylori by allelic exchange. J Bacteriol. 1992;174:4212–4217. doi: 10.1128/jb.174.13.4212-4217.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forman D, Webb P, Parsonnet J. Helicobacter pylori and gastric cancer. Lancet. 1994;343:243–244. [PubMed] [Google Scholar]
- 7.Givaudan A, Lanois A, Boemare N. Cloning and nucleotide sequence of a flagellin encoding genetic locus from Xenorhabdus nematophilus: phase variation leads to differential transcription of two flagellin genes (fliCD) Gene. 1996;183:243–253. doi: 10.1016/s0378-1119(96)00452-0. [DOI] [PubMed] [Google Scholar]
- 8.Grant C C, Konkel M E, Cieplak W, Tompkins L S. Role of flagella in adherence, internalization, and translocation of Campylobacter jejuni in nonpolarized and polarized epithelial cell cultures. Infect Immun. 1993;61:1764–1771. doi: 10.1128/iai.61.5.1764-1771.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammar M, Tyszkiewicz T, Wadstrom T, O’Toole P W. Rapid detection of Helicobacter pylori in gastric biopsy material by polymerase chain reaction. J Clin Microbiol. 1992;30:54–58. doi: 10.1128/jcm.30.1.54-58.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Homma M, Iino T. Locations of hook-associated proteins in flagellar structure of Salmonella typhimurium. J Bacteriol. 1985;162:183–189. doi: 10.1128/jb.162.1.183-189.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Homma M, Iino T. Excretion of unassembled hook-associated proteins by Salmonella typhimurium. J Bacteriol. 1985;164:1370–1372. doi: 10.1128/jb.164.3.1370-1372.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Homma M, DeRoiser D J, Macnab R M. Flagellar hook and hook-associated proteins of Salmonella typhimurium and their relationship to other axial components of the flagellum. J Mol Biol. 1990;213:819–832. doi: 10.1016/S0022-2836(05)80266-9. [DOI] [PubMed] [Google Scholar]
- 13.Ikeda T, Homma M, Iino T, Asakura S, Kamiya R. Localization and stoichiometry of hook-associated proteins within Salmonella typhimurium flagella. J Bacteriol. 1987;169:1168–1173. doi: 10.1128/jb.169.3.1168-1173.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikeda T, Yamaguchi S, Hotani H. Flagellar growth in a filament-less Salmonella fliD mutant supplemented with purified hook-associated protein 2. J Biochem. 1993;114:39–44. doi: 10.1093/oxfordjournals.jbchem.a124136. [DOI] [PubMed] [Google Scholar]
- 15.International Agency for Research on Cancer. Schistosomes, liver flukes and Helicobacter pylori. IARC Monogr Eval Carcinog Risks Hum. 1994;61:177–220. [PMC free article] [PubMed] [Google Scholar]
- 16.Josenhans C, Labigne A, Suerbaum S. Comparative ultrastructural and functional studies of Helicobacter pylori and Helicobacter mustelae flagellin mutants: both flagellin subunits, FlaA and FlaB, are necessary for full motility in Helicobacter species. J Bacteriol. 1995;177:3010–3020. doi: 10.1128/jb.177.11.3010-3020.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawagishi I, Muller V, Williams A W, Irikura V M, Macnab R M. Subdivision of flagellar region III of the Escherichia coli and Salmonella typhimurium chromosomes and identification of two additional flagellar genes. J Gen Microbiol. 1992;138:1051–1065. doi: 10.1099/00221287-138-6-1051. [DOI] [PubMed] [Google Scholar]
- 18.Kostrzynska M, Betts J D, Austin J W, Trust T J. Identification, characterization, and spatial localization of two flagellin species in Helicobacter pylori flagella. J Bacteriol. 1991;173:937–946. doi: 10.1128/jb.173.3.937-946.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kutsukake K, Iino T. Role of the FliA-FlgM regulatory system on the transcriptional control of the flagellar regulon and flagellar formation in Salmonella typhimurium. J Bacteriol. 1994;176:3598–3605. doi: 10.1128/jb.176.12.3598-3605.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kutsukake K, Ohya Y, Iino T. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J Bacteriol. 1990;172:741–747. doi: 10.1128/jb.172.2.741-747.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Labigne A, Cussac V, Courcoux P. Shuttle cloning and nucleotide sequences of Helicobacter pylori genes responsible for urease activity. J Bacteriol. 1991;173:1920–1931. doi: 10.1128/jb.173.6.1920-1931.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee A, O’Rourke J, Ungria M C, Robertson B, Daskalpoulos G, Dixon M F. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology. 1997;112:1386–1397. doi: 10.1016/s0016-5085(97)70155-0. [DOI] [PubMed] [Google Scholar]
- 23.Leying H, Suerbaum S, Gels G, Haas R. Cloning and genetic characterization of a Helicobacter pylori flagellin gene. Mol Microbiol. 1992;6:2863–2874. doi: 10.1111/j.1365-2958.1992.tb01466.x. [DOI] [PubMed] [Google Scholar]
- 24.Macnab R M. Flagella and motility. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 123–145. [Google Scholar]
- 25.Marshall B J, Warren J R. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;i:1311–1314. doi: 10.1016/s0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- 26.McCarter L L. Genetic and molecular characterization of the polar flagellum of Vibrio parahaemolyticus. J Bacteriol. 1995;177:1595–1609. doi: 10.1128/jb.177.6.1595-1609.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mobley H L T, Belas R, Lockatell V, Chippendale G, Trifillis A, Johnson D E, Warren J W. Construction of a flagellum-negative mutant of Proteus mirabilis: effect on internalization by human renal epithelial cells and virulence in a mouse model of ascending urinary tract infection. Infect Immun. 1996;64:5332–5340. doi: 10.1128/iai.64.12.5332-5340.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore K M, Jackwood M W, Brown T P, Dreesen D W. Bordetella avium hemagglutination and motility mutants: isolation, characterization, and pathogenicity. Avian Dis. 1994;38:50–58. [PubMed] [Google Scholar]
- 29.Ohnishi K, Kutsukake K, Suzuki H, Iino T. A novel transcriptional regulation mechanism in the flagellar regulon of Salmonella typhimurium: an anti-sigma factor inhibits the activity of the flagellum-specific sigma factor, ςF. Mol Microbiol. 1992;6:3149–3157. doi: 10.1111/j.1365-2958.1992.tb01771.x. [DOI] [PubMed] [Google Scholar]
- 30.O’Toole P W, Kostrzynska M, Trust T J. Non-motile mutants of Helicobacter pylori and Helicobacter mustelae defective in flagellar hook production. Mol Microbiol. 1994;14:691–703. doi: 10.1111/j.1365-2958.1994.tb01307.x. [DOI] [PubMed] [Google Scholar]
- 31.Parsonnet J, Hansen S, Rodriguez L, Gelb A B, Warnke R A, Jellum E, Orenteich N, Vogelman J H, Freiedman G D. Helicobacter pylori infection and gastric lymphoma. N Engl J Med. 1994;330:1267–1270. doi: 10.1056/NEJM199405053301803. [DOI] [PubMed] [Google Scholar]
- 32.Porvollik S, Noonan B, O’Toole P W. Molecular characterization of a flagellar export locus of Helicobacter pylori. Infect Immun. 1999;67:2060–2070. doi: 10.1128/iai.67.5.2060-2070.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Short J M, Fernandez J M, Sorge J A, Huse W D. Lambda ZAP: a bacteriophage lambda expression vector with in vivo excision properties. Nucleic Acids Res. 1988;15:7583–7600. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spohn G, Scarlato V. Motility of Helicobacter pylori is coordinately regulated by the transcriptional activator FlgR, an NtrC homolog. J Bacteriol. 1999;181:593–599. doi: 10.1128/jb.181.2.593-599.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suerbaum S, Josenhans C, Labigne A. Cloning and genetic characterization of the Helicobacter pylori and Helicobacter mustelae flaB flagellin genes and construction of H. pylori flaA- and flaB-negative mutants by electroporation-mediated allelic exchange. J Bacteriol. 1993;175:3278–3288. doi: 10.1128/jb.175.11.3278-3288.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamura Y, Kijima-Tanaka M, Aoki A, Ogikubo Y, Takahashi T. Reversible expression of motility and flagella in Clostridium chauvoei and their relationship to virulence. Microbiology. 1995;141:605–610. doi: 10.1099/13500872-141-3-605. [DOI] [PubMed] [Google Scholar]
- 37.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Goldek A, McKenney K, Fitzgerald L M, Lee N, Adams M D, Hickey E K, Berg D E, Gocayne J D, Utterback T R, Peterson J D, Kelley J M, Cotton M D, Weidman J M, Fujii C, Bowman C, Watthey L, Wallin E, Hayes W S, Borodovsky M, Karp P D, Smith H O, Fraser C M, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 38.Yokoseki T, Kutsukake K, Ohnishi K, Iino T. Functional analysis of the flagellar genes in the fliD operon of Salmonella typhimurium. Microbiology. 1995;141:1715–1722. doi: 10.1099/13500872-141-7-1715. [DOI] [PubMed] [Google Scholar]
- 39.Yokoseki T, Iino T, Kutsukake K. Negative regulation by FliD, FliS, and FliT of the export of the flagellum-specific anti-sigma factor, FlgM, in Salmonella typhimurium. J Bacteriol. 1996;178:899–901. doi: 10.1128/jb.178.3.899-901.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang M Y, Lovgren A, Landen R. Adhesion and cytotoxicity of Bacillus thuringiensis to cultured Spodoptera and Drosophila cells. J Invertebr Pathol. 1995;66:46–51. doi: 10.1006/jipa.1995.1059. [DOI] [PubMed] [Google Scholar]