Abstract
Background
Hepatitis C virus (HCV) screening is critical to HCV elimination efforts. Simplified diagnostics are required for low-resource settings and difficult-to-reach populations. This retrospective study assessed performance of rapid diagnostic tests (RDTs) for detection of HCV antibodies.
Methods
Two lots of 13 RDTs were evaluated at 3 laboratories using archived plasma samples from 4 countries (Nigeria, Georgia, Cambodia, and Belgium). HCV status was determined using 3 reference tests according to a composite algorithm. Sensitivity and specificity were evaluated in HIV-infected and HIV-uninfected populations. Operational characteristics were also assessed.
Results
In total, 1710 samples met inclusion criteria. In HIV-uninfected samples (n = 384), the majority of RDTs had sensitivity ≥98% in 1 or both lots and most RDTs had specificity ≥99%. In HIV-infected samples (n = 264), specificity remained high but sensitivity was markedly lower than in HIV-uninfected samples; only 1 RDT reached >95%. The majority of HIV-infected samples for which sensitivity was low did not have detectable HCV viral load/core antigen. Interreader variability, lot-to-lot variability, and rate of invalid runs were low for all RDTs (<2%).
Conclusions
HCV RDTs should be evaluated in the intended target population, as sensitivity can be impacted by population factors such as HIV status.
Clinical Trials Registration
Keywords: hepatitis C virus, in vitro diagnostics, rapid diagnostic test, low- and middle-income country, HCV screening, specificity, sensitivity
In this performance evaluation of 13 rapid diagnostic tests for hepatitis C virus antibody detection, sensitivity and specificity were high in HIV-uninfected plasma samples. In HIV-infected samples, specificity remained high, but sensitivity was markedly reduced.
In 2015, the number of people with chronic hepatitis C (HCV) infection worldwide was estimated at 71 million [1]. However, only around 20% of people with HCV are aware of their HCV status [1]. HCV screening is critical to the success of HCV elimination targets, but in low- and middle-income countries (LMICs), where standardized laboratory tests are expensive and often not covered by public health systems, screening of at-risk populations for HCV infection remains very limited [2]. The burden of HCV in LMICs is particularly high, representing over 70% of the global total [3]. As such, the World Health Organization (WHO) strategy to eliminate HCV has highlighted an urgent need for simplified diagnostic tests for use in low-resource settings, as well as for difficult-to-reach populations in high-income countries, such as people who inject drugs [4].
Screening for HCV is performed through the detection of HCV-specific antibodies. WHO guidelines recommend the use of a single quality-assured serological in vitro diagnostic test, either a laboratory-based immunoassay or a rapid diagnostic test (RDT) [5]. For many LMICs, where equipped laboratories and trained staff are limited, RDTs may be most appropriate, as they are quick and easy to perform without the need for laboratory equipment. RDTs have proved effective in other disease areas; for example, the wide availability of low-cost RDTs for the diagnosis of human immunodeficiency virus (HIV) has substantially increased access to testing, resulting in more than 600 million people being tested for HIV in LMICs from 2010 to 2014 [6].
The lack of quality-assured RDTs for HCV serology testing has been identified as an important barrier to large-scale access to HCV diagnosis [2]. While a number of HCV RDTs are commercially available, many do not have quality assurance status (eg, stringent regulatory authority approval or WHO prequalification [7]). Additionally, data on the quality and performance of many tests are limited, especially in LMICs. WHO recommendations on performance criteria for procurement of in vitro diagnostics for HCV, which also serve as guidance for WHO prequalification, recommend a sensitivity of ≥98% and a specificity of ≥97% for HCV serology RDTs in plasma or serum specimens [8]. Data on sensitivity and specificity of existing RDTs and RDTs in development can help to determine whether additional tests may be suitable for WHO prequalification, and results of independent performance evaluations can support countries in their choice to procure tests that meet international performance criteria.
Furthermore, some studies have noted a potential negative impact of HIV coinfection on the sensitivity of some HCV RDTs [9–11]. This may be due to the compromised immune system of people living with HIV limiting the production of anti-HCV antibodies; data on RDT performance by CD4 count (an indicator of immune status in HIV-positive people) have been identified as a research gap for HCV serology testing [5]. Given the high burden of HIV in LMICs, and the substantial proportion of people with HCV and HIV coinfection worldwide (approximately 2.3 million) [1], understanding the effect of HIV status on HCV RDT performance will be crucial to HCV elimination efforts.
The objective of this study was to evaluate the performance of a range of HCV RDTs using clinical samples collected from different geographic regions, as well as from HIV-infected individuals, in order to identify tests that could be used for HCV screening in LMICs or difficult-to-reach populations.
METHODS
Study Design
This was an observational, retrospective, multicenter laboratory evaluation of 13 HCV RDTs (NCT04033887; Table 1). Nine RDTs were on-market products, 1 RDT (HCV-only Ab Test; Biosynex SA) had its configuration adapted to only evaluate the HCV line (the on-market product configuration is a triplex test with additional lines for the detection of HIV antibodies and hepatitis B virus surface antigen; the evaluated version lacked the test lines for HIV and hepatitis B), and 3 RDTs were still in late-stage development at the time of the study (defined here as prototype).
Table 1.
HCV RDTs Included in the Study
| Manufacturer | Test Name | Country | Test Format |
|---|---|---|---|
| SD Biosensor | Standard Q HCV Ab | Korea | Lateral flow |
| Antron Laboratories | HCV Hepatitis Virus Antibody Test | Canada | Lateral flow |
| Beijing Wantal Biological Pharmacy Enterprise | HCV-Ab Rapid Test | China | Lateral flow |
| InTec | Rapid Anti-HCV Test | China | Lateral flow |
| Premier Medical Corporation | First Response HCV Card Test | India | Lateral flow |
| Arkray Healthcare | Signal HCV Version 3.0 | India | Flow through |
| J. Mitra & Co. | TRI DOT HCV | India | Flow through |
| Biosynex SA | Modified HCV-only Ab Test | France | Lateral flow |
| Abbott Diagnostics | SD Bioline HCV | United States | Lateral flow |
| OraSure | OraQuick HCV | United States | Lateral flow |
| BioLytical Laboratories | Prototype HCV Ab Test | Canada | Flow through |
| Chembio Diagnostic Systems | Prototype DPP HCV | United States | Lateral flow |
| Access Bio | Prototype Care Start HCV | United States | Lateral flow |
Abbreviations: AB, antibody; HCV, hepatitis C virus; RDT, rapid diagnostic test.
Tests were evaluated at 3 laboratories: the Nigerian Institute of Medical Research (Lagos, Nigeria), the Lugar Center at the National Center for Disease Control and Public Health (Tbilisi, Georgia), and the Institute of Tropical Medicine HIV/Sexually Transmitted Diseases (STD) Reference Laboratory (Antwerp, Belgium). Testing was performed on randomly selected locally archived frozen plasma samples from these 3 laboratories. Additionally, samples obtained from the Sihanouk Hospital Center of Hope (Phnom Penh, Cambodia) were tested at the Institute of Tropical Medicine HIV/STD Reference Laboratory; samples were frozen in Cambodia and remained frozen throughout transportation to Belgium. All sites received approval for the study from the respective institutional review boards. Testing was performed between September 2018 and March 2019.
All samples were ethylenediaminetetraacetic acid (EDTA)-treated plasma samples taken from people aged ≥18 years, with a minimum volume of 1.5 mL and known HIV status. Information on HCV and HIV treatment status of the sample donors was available. No further information on the characteristics of the sample donors were collected as part of this study. Samples were nonhemolytic, had <3 freeze-thaw cycles, and had been stored at or below −70°C. Samples were collected between 2008 and 2018 (92% collected between 2014 and 2018). Samples were excluded if generic consent for further use was missing. Prior to commencement of testing, each site prepared small aliquots from the master samples to eliminate the need for multiple freeze-thaw cycles.
HCV antibody status was determined using 3 reference tests, of which 2 were WHO prequalification approved enzyme immunoassays (EIA; Murex Anti-HCV version 4.0, DiaSorin S.A., and INNOTEST HCV Ab IV, Fujirebio Europe) and 1 was a line immunoassay (LIA; MP Diagnostics HCV blot 3.0, MP Biomedicals). A signal to cutoff ratio of ≥1 (based on the measured optical density) was used for the EIAs; interpretation of LIA results was performed according to manufacturer instructions. HCV antibody status of each sample was determined according to a composite algorithm incorporating the results of all 3 reference tests (Supplementary Table 1). A similar algorithm has previously been used in WHO prequalification evaluation protocols, although the WHO algorithm does not require LIA confirmation for samples testing negative on both EIAs [12].
Outcomes
The primary outcomes were point estimates of sensitivity and specificity with 95% confidence intervals of the 13 HCV RDTs in HIV-infected and -uninfected samples. Secondary outcomes included sensitivity and specificity of the 13 RDTs in the overall population (regardless of HIV status), interreader variability, lot-to-lot variability, and the rate of invalid runs. Exploratory outcomes included point estimates of sensitivity and specificity with 95% confidence intervals of the 13 RDTs in HIV-infected and -uninfected samples with active HCV infection measured by the presence of detectable HCV viral load (VL) or core antigen (cAg) in the sample. Analysis of test performance by CD4 count range (<200 cells/mm3 [severely immunocompromised], 200–500 cells/mm3 [immunocompromised], or >500 cells/mm3 [not immunocompromised]) in HIV-infected samples and by HCV genotype in HIV-uninfected and -infected samples was also performed.
RDT Performance Assessments
Each sample was tested on 2 independently produced lots of each RDT and each result was read and recorded by 3 independent readers. RDT results were interpreted according to the manufacturer’s instructions. Samples were scored as either positive (reactive), negative (nonreactive), or invalid on each RDT based on the concordance of at least 2 out of 3 reader results. For all samples that were scored invalid, a repeat test was performed once on the same lot.
Operators/readers of the RDTs were blinded to the results of the reference standard tests. The sequence in which samples were tested was varied for each RDT to avoid bias related to recognition patterns. Operators and reader sequences were also varied.
Statistical Analyses
For an average sensitivity of 85% and specificity of 80%, a minimum sample size of 400 for sensitivity analyses and 502 for specificity analyses was required to obtain point estimates with a precision of ± 5% and power of 80% to obtain a confidence interval with total width of 10% or less [13].
Point estimates were obtained, with 95% confidence intervals based on Wilson score method, for sensitivity and specificity. Interreader variability was assessed by Fleiss kappa coefficient (κ) (agreement was defined as concordance between 2/3 or 3/3 results for each RDT). Lot-to-lot variability was evaluated by assessing performance in each lot using final valid RDT outcomes (excluding repeatedly invalid results), and the rate of invalid runs was calculated as the ratio between runs marked as invalid and the total number.
RESULTS
Sample Characteristics
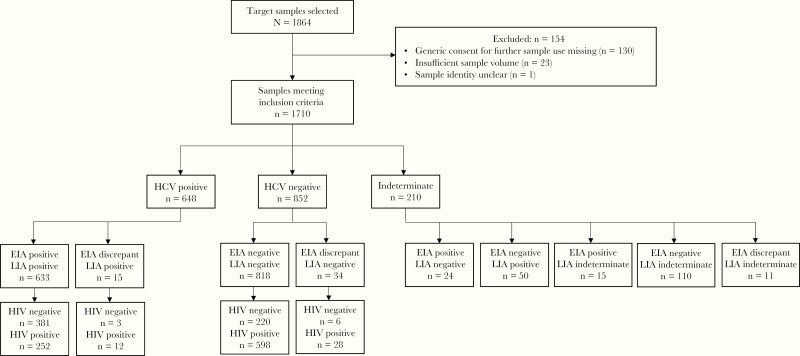
Of 1864 samples selected, 1710 met inclusion criteria. In total, 648 samples were HCV antibody positive, of which 264 were also HIV positive. Of the 852 HCV antibody-negative samples, 626 were HIV positive and 226 were HIV negative. Two hundred and ten samples had indeterminate HCV status due to discrepancies between EIA and LIA results or indeterminate LIA results and were excluded from further analyses as per the composite reference standard algorithm (Figure 1). Although the sample size was not as large as was estimated to be required based on the previously stated test performance assumptions, based on the average test performance observed in this study, the sample size allowed for ≥ 80% power with a precision of ± 5 in all subgroups (Supplementary Table 2).
Figure 1.
Number of samples by HCV and HIV status. Abbreviations: EIA, enzyme immunoassay; HCV, hepatitis C virus; HIV, human immunodeficiency virus; LIA, line immunoassay.
The numbers of samples with genotype, CD4, count and HCV VL/cAg availability, and the country of sample origin for each sample type, are shown in Table 2. The majority of genotyped samples were of HCV genotype 1, 1a, or 1b (63.2% of HIV-uninfected and 54.2% of HIV-infected samples), followed by genotype 3 in HIV-uninfected samples (31.6%) and genotype 6 in HIV-infected samples (22.9%). The majority of HIV-positive samples had CD4 counts greater than 200 cells/mm3 (>93%). The majority (89%) of HCV-infected and HIV-infected samples were from patients receiving treatment for HIV at the time of sample collection (HIV treatment status was known for 256 of 264 HCV-positive and HIV-positive samples). None of the samples from Nigeria, Cambodia, or Georgia were from people receiving treatment for HCV; of the Belgian samples, 107 were from people who had never received treatment for HCV, 10 were from people who were on active interferon treatment, 7 were from people who had previously received interferon treatment, and 2 had no treatment information available.
Table 2.
Number of Samples With Genotype, CD4 Count, and HCV VL/cAg Information, and Country of Sample Origin
| HCV Positive/ HIV Negative (n = 384) | HCV Positive/ HIV Positive (n = 264) | HCV Negative/HIV Positive (n = 626) | HCV Negative/HIV Negative (n = 226) | |
|---|---|---|---|---|
| Country of sample origin, n (%) | ||||
| Nigeria | 70 (18.2) | 20 (7.6) | 292 (46.6) | 186 (82.3) |
| Georgia | 314 (81.8) | 0 | 0 | 40 (17.7) |
| Cambodia | 0 | 126 (47.7) | 332 (53.0) | 0 |
| Belgium | 0 | 118 (44.7) | 2 (0.3) | 0 |
| Genotype available, n (%) | 114 (29.7) | 179 (67.8) | … | … |
| Genotype 1, 1a or 1ba | 72 (63.2) | 97 (54.2) | … | … |
| Genotype 2a | 5 (4.4) | 5 (2.8) | … | … |
| Genotype 3a | 36 (31.6) | 10 (5.6) | … | … |
| Genotype 4a | 1 (0.9) | 26 (14.5) | … | … |
| Genotype 6a | 0 | 41 (22.9) | … | … |
| CD4 count available, n (%) | … | 261 (98.9) | 622 (99.4) | … |
| <200 cells/mm3a | … | 18 (6.9) | 41 (6.6) | … |
| 200–<500 cells/mm3a | … | 117 (44.8) | 266 (42.8) | … |
| ≥500 cells/mm3a | … | 126 (48.3) | 315 (50.6) | … |
| HCV VL/cAg available, n (%) | 350 (91.1) | 234 (88.7) | … | … |
| HCV VL/cAg detectablea | 262 (74.9) | 181 (77.4) | … | … |
| Mean HCV VL, cp/mL (SD) | 1.9E + 06 (2.55E + 06) n = 144 | 4.27E + 06 (6.97E + 06) n = 181 | … | … |
| Mean HCV cAg, fmol/L (SD) | 3.93E + 03 (4.74E + 03) n = 118 | … | … | … |
| HbsAg status positive, n/N (%) | 11/266 (4.1) | 11/223 (5.0) | 44/626 (7.0) | 3/226 (1.3) |
Abbreviations: cp, copies; fmol, femto molecules; HBsAg, hepatitis B surface antigen; HCV, hepatitis C virus; HIV, human immunodeficiency virus; SD, standard deviation; VL/cAg, viral load/core antigen
aExpressed as percentage of samples with available information.
Sensitivity and Specificity
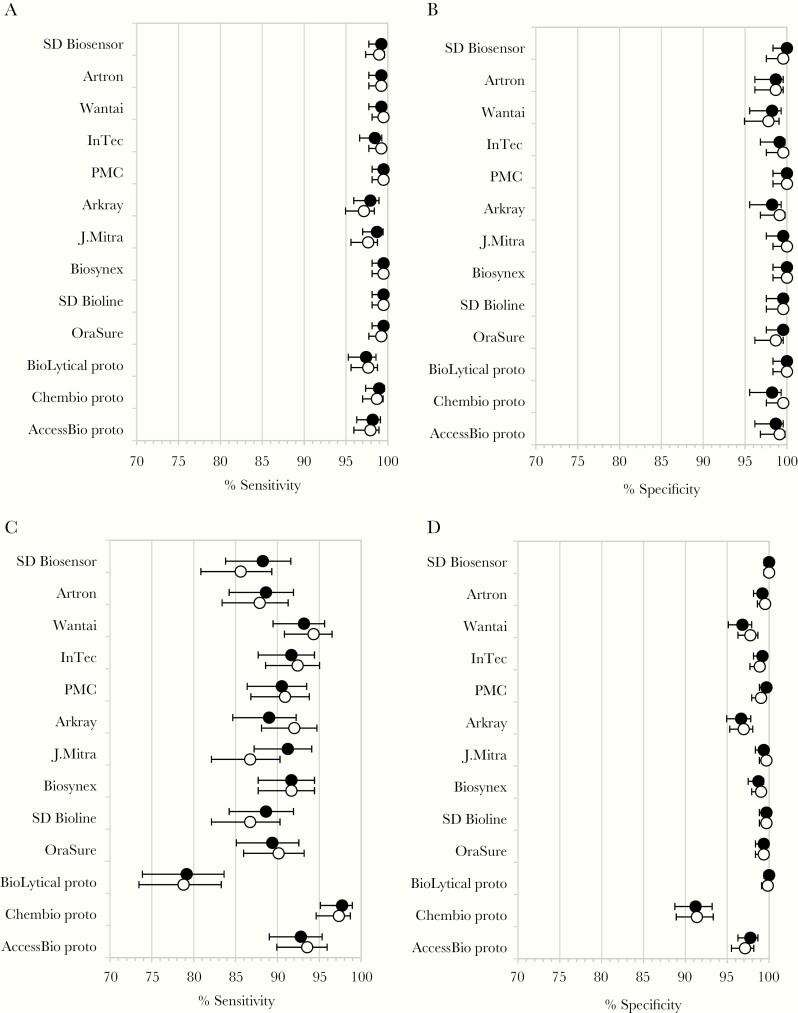
In the samples from HIV-uninfected patients, most RDTs showed high sensitivity, with the majority reaching ≥98% in 1 or both lots (Figure 2A and Table 3). The large majority of tests showed a specificity of ≥99% and several reached 100% (Figure 2B and Table 3). In HIV-infected samples, sensitivity was markedly lower than in HIV-uninfected samples, with only 1 RDT reaching >95% (Prototype DPP HCV; Chembio Diagnostic Systems) (Figure 2C and Table 3). For the large majority of RDTs, confidence intervals between HIV-uninfected and -infected samples did not overlap. Specificity was comparatively high in HIV-infected samples, with only 4 RDTs showing a specificity of <97% in at least 1 lot (Figure 2D and Table 3). In the combined sample set of HIV-uninfected and HIV-infected samples, results reflected the lower sensitivity for most RDTs, and lower specificity for some RDTs, observed in the HIV-infected samples (Table 3).
Figure 2.
Sensitivity and specificity of 13 HCV rapid diagnostic tests in human immunodeficiency virus (HIV)-uninfected and -infected samples (circles, % sensitivity or specificity; closed circles, lot 1; open circles, lot 2; error bars, upper and lower 95% confidence intervals): (A) sensitivity in HIV-uninfected samples; (B) specificity in HIV-uninfected samples; (C) sensitivity in HIV-infected samples; and (D) specificity in HIV-infected samples.
Table 3.
Summary of Sensitivity and Specificity of RDTs in All Study Populations Based on the Main Composite Reference Standard
| Overall Population | HIV Uninfected | HIV Infected | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Sensitivity | Specificity | Sensitivity | Specificity | ||||||||||||||
| RDT | Lot | TP | FN | % (LCI, UCI) | TN | FP | % (LCI, UCI) | TP | FN | % (LCI, UCI) | TN | FP | % (LCI, UCI) | TP | FN | % (LCI, UCI) | TN | FP | % (LCI, UCI) |
| SD Biosensor Standard Q HCV Ab | Lot 1 | 614 | 34 | 94.8 (92.8, 96.2) | 852 | 0 | 100 (99.6, 100) | 381 | 3 | 99.2 (97.7, 99.7) | 226 | 0 | 100 (98.3, 100) | 233 | 31 | 88.3 (83.8, 91.6) | 626 | 0 | 100 (99.4, 100) |
| Lot 2 | 606 | 42 | 93.5 (91.4, 95.2) | 851 | 1 | 99.9 (99.3, 100) | 380 | 4 | 99.0 (97.4, 99.6) | 225 | 1 | 99.6 (97.5, 99.9) | 226 | 38 | 85.6 (80.9, 89.3) | 626 | 0 | 100 (99.4, 100) | |
| Artron HCV Antibody Test | Lot 1 | 615 | 33 | 94.9 (92.9, 96.4) | 844 | 8 | 99.1 (98.2, 99.5) | 381 | 3 | 99.2 (97.7, 99.7) | 223 | 3 | 98.7 (96.2, 99.6) | 234 | 30 | 88.6 (84.2, 91.9) | 621 | 5 | 99.2 (98.1, 99.7) |
| Lot 2 | 613 | 35 | 94.6 (92.6, 96.1) | 846 | 6 | 99.3 (98.5, 99.7) | 381 | 3 | 99.2 (97.7, 99.7) | 223 | 3 | 98.7 (96.2, 99.6) | 232 | 32 | 87.9 (83.4, 91.3) | 623 | 3 | 99.5 (98.6, 99.8) | |
| Wantai HCV Rapid Test | Lot 1 | 627 | 32 | 96.8 (95.1, 97.9) | 828 | 24 | 97.2 (95.8, 98.1) | 381 | 3 | 99.2 (97.7, 99.7) | 222 | 4 | 98.2 (95.5, 99.3) | 246 | 18 | 93.2 (89.5, 95.6) | 606 | 20 | 96.8 (95.1, 97.9) |
| Lot 2 | 631 | 17 | 97.4 (95.8, 98.4) | 833 | 19 | 97.8 (96.5, 98.6) | 382 | 2 | 99.5 (98.1, 99.9) | 221 | 5 | 97.8 (94.9, 99.1) | 249 | 15 | 94.3 (90.8, 96.5) | 612 | 14 | 97.8 (96.3, 98.7) | |
| InTec Rapid Anti- HCV Test | Lot 1 | 620 | 28 | 95.7 (93.8, 97.0) | 845 | 7 | 99.2 (98.3, 99.6) | 378 | 6 | 98.4 (96.6, 99.3) | 224 | 2 | 99.1 (96.8, 99.8) | 242 | 22 | 91.7 (87.7, 94.4) | 621 | 5 | 99.2 (98.1, 99.7) |
| Lot 2 | 625 | 23 | 96.5 (94.7, 97.6) | 844 | 8 | 99.1 (98.2, 99.5) | 381 | 3 | 99.2 (97.7, 99.7) | 225 | 1 | 99.6 (97.5, 99.9) | 244 | 20 | 92.4 (88.6, 95.0) | 619 | 7 | 98.9 (97.7, 99.5) | |
| PMC First Response HCV Card Test | Lot 1 | 621 | 27 | 95.8 (94.0, 97.1) | 850 | 2 | 99.8 (99.2, 99.9) | 382 | 2 | 99.5 (98.1, 99.9) | 226 | 0 | 100 (98.3, 100) | 239 | 25 | 90.5 (86.4, 93.5) | 624 | 2 | 99.7 (98.8, 99.9) |
| Lot 2 | 622 | 26 | 96.0 (94.2, 97.3) | 846 | 6 | 99.3 (98.5, 99.7) | 382 | 2 | 99.5 (98.1, 99.9) | 226 | 0 | 100 (98.3, 100) | 240 | 24 | 90.9 (86.8, 93.8) | 620 | 6 | 99.0 (97.9, 99.6) | |
| Arkray Signal HCV Version 3.0 | Lot 1 | 610 | 37 | 94.3 (92.2, 95.8) | 827 | 25 | 97.1 (95.7, 98.0) | 375 | 8 | 97.9 (95.9, 98.9) | 222 | 4 | 98.2 (95.5, 99.3) | 235 | 29 | 89.0 (84.7, 92.2) | 605 | 21 | 96.7 (94.9, 97.8) |
| Lot 2 | 615 | 32 | 95.1 (93.1, 96.5) | 830 | 21 | 97.5 (96.3, 98.4) | 373 | 11 | 97.1 (94.9, 98.4) | 224 | 2 | 99.1 (96.8, 99.8) | 242 | 21 | 92.0 (88.1, 94.7) | 606 | 19 | 97.0 (95.3, 98.1) | |
| J. Mitra TRI DOT HCV | Lot 1 | 619 | 28 | 95.7 (93.8, 97.0) | 842 | 5 | 99.4 (98.6, 99.8) | 379 | 5 | 98.7 (97.0, 99.4) | 225 | 1 | 99.6 (97.5, 99.9) | 240 | 23 | 91.3 (87.2, 94.1) | 617 | 4 | 99.4 (98.4, 99.8) |
| Lot 2 | 603 | 44 | 93.2 (91.0, 94.9) | 847 | 2 | 99.8 (99.2, 99.9) | 374 | 9 | 97.7 (95.6, 98.8) | 226 | 0 | 100 (98.3, 100) | 229 | 35 | 86.7 (82.1, 90.3) | 621 | 2 | 99.7 (98.8, 99.9) | |
| Biosynex HCV-only | Lot 1 | 624 | 24 | 96.3 (94.6, 97.5) | 844 | 8 | 99.1 (98.2, 99.5) | 382 | 2 | 99.5 (98.1, 99.9) | 226 | 0 | 100 (98.3, 100) | 242 | 22 | 91.7 (87.7, 94.4) | 618 | 8 | 98.7 (97.5, 99.4) |
| Lot 2 | 624 | 24 | 96.3 (94.6, 97.5) | 846 | 6 | 99.3 (98.5, 99.7) | 382 | 2 | 99.5 (98.1, 99.9) | 226 | 0 | 100 (98.3, 100) | 242 | 22 | 91.7 (87.7, 94.4) | 620 | 6 | 99.0 (97.9, 99.6) | |
| Abbott SD Bioline HCV | Lot 1 | 616 | 32 | 95.1 (93.1, 96.5) | 849 | 3 | 99.7 (99.0, 99.9) | 382 | 2 | 99.5 (98.1, 99.9) | 225 | 1 | 99.6 (97.5, 99.9) | 234 | 30 | 88.6 (84.2, 91.9) | 624 | 2 | 99.7 (98.8, 99.9) |
| Lot 2 | 611 | 37 | 94.3 (92.2, 95.8) | 849 | 3 | 99.7 (99.0, 99.9) | 382 | 2 | 99.5 (98.1, 99.9) | 225 | 1 | 99.6 (97.5, 99.9) | 229 | 35 | 86.7 (82.1, 90.3) | 624 | 2 | 99.7 (98.8, 99.9) | |
| OraSure OraQuick HCV | Lot 1 | 618 | 30 | 95.4 (93.5, 96.7) | 847 | 5 | 99.4 (98.6, 99.8) | 382 | 2 | 99.5 (98.1, 99.9) | 225 | 1 | 99.6 (97.5, 99.9) | 236 | 28 | 89.4 (85.1, 92.6) | 622 | 4 | 99.4 (98.4, 99.8) |
| Lot 2 | 619 | 29 | 95.5 (93.7, 96.9) | 845 | 7 | 99.2 (98.3, 99.6) | 381 | 3 | 99.2 (97.7, 99.7) | 223 | 3 | 98.7 (96.2, 99.6) | 238 | 26 | 90.2 (86.0, 93.2) | 622 | 4 | 99.4 (98.4, 99.8) | |
| BioLytical prototype HCV Ab Test | Lot 1 | 583 | 65 | 90.0 (87.4, 92.1) | 852 | 0 | 100 (99.6, 100) | 374 | 10 | 97.4 (95.3, 98.6) | 226 | 0 | 100 (98.3, 100) | 209 | 55 | 79.2 (73.9, 83.6) | 626 | 0 | 100 (99.4, 100) |
| Lot 2 | 583 | 65 | 90.0 (87.4, 92.1) | 851 | 1 | 99.9 (99.3, 100) | 375 | 9 | 97.7 (95.6, 98.8) | 226 | 0 | 100 (98.3, 100) | 208 | 56 | 78.8 (73.5, 83.3) | 625 | 1 | 99.8 (99.1, 100) | |
| Chembio prototype DPP HCV | Lot 1 | 638 | 10 | 98.5 (97.2, 99.2) | 793 | 59 | 93.1 (91.2, 94.6) | 380 | 4 | 99.0 (97.4, 99.6) | 222 | 4 | 98.2 (95.5, 99.3) | 258 | 6 | 97.7 (95.1, 99.0) | 571 | 55 | 91.2 (88.7, 93.2) |
| Lot 2 | 636 | 12 | 98.2 (96.8, 98.9) | 797 | 55 | 93.5 (91.7, 95.0) | 379 | 5 | 98.7 (97.0, 99.4) | 225 | 1 | 99.6 (97.5, 99.9) | 257 | 7 | 97.4 (94.6, 98.7) | 572 | 54 | 91.4 (88.9, 93.3) | |
| AccessBio prototype CareStart HCV | Lot 1 | 622 | 26 | 96.0 (94.2, 97.3) | 835 | 17 | 98.0 (96.8, 98.8) | 377 | 7 | 98.2 (96.3, 99.1) | 223 | 3 | 98.7 (96.2, 99.6) | 245 | 19 | 92.8 (89.0, 95.3) | 612 | 14 | 97.8 (96.3, 98.7) |
| Lot 2 | 623 | 25 | 96.1 (94.4, 97.4) | 832 | 20 | 97.7 (96.4, 98.5) | 376 | 8 | 97.9 (95.9, 98.9) | 224 | 2 | 99.1 (96.8, 99.8) | 247 | 17 | 93.6 (89.9, 95.9) | 608 | 18 | 97.1 (95.5, 98.2) |
Abbreviations: Ab, antibody; FN, false negative, FP, false positive; LCI, lower 95% confidence interval; HCV, hepatitis C virus; HIV, human immunodeficiency virus; RDT, rapid diagnostic test; TN, true negative; TP, true positive; UCI, upper 95% confidence interval.
False negatives were distributed across 86 different samples. Of these, only 26 (30.2%) had genotype information available, with the most common genotype being genotype 6 (n = 9, 34.6%). The distribution of false negatives per CD4 count range (<200, 200–500, and >500 cells/mm3) in HIV-infected samples showed that false negatives occurred at a similar frequency in all CD4 count ranges (Supplementary Table 3).
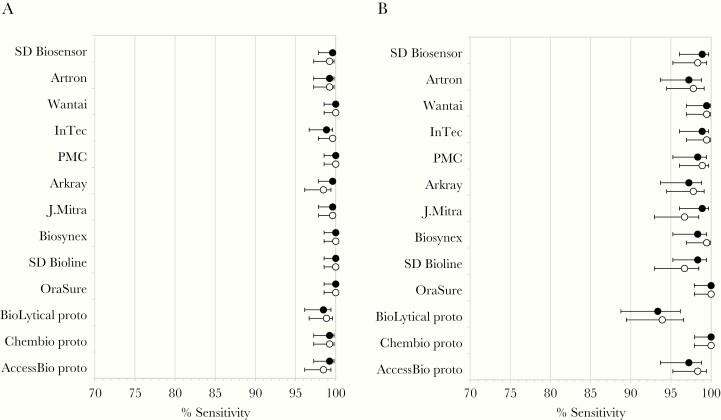
To evaluate whether RDT sensitivity was associated with detectable HCV VL/cAg, point estimates of sensitivity were calculated for HIV-uninfected and HIV-infected samples only for samples with detectable HCV VL/cAg. In HIV-uninfected samples with detectable HCV VL/cAg, sensitivity increased moderately compared with the overall sample set, while in HIV-infected samples with detectable HCV VL/cAg, sensitivity increased markedly compared with the overall sample set (Figure 3 and Supplementary Table 4). The majority of confidence intervals did not overlap between HIV-infected samples with detectable HCV VL/cAg and all HIV-infected samples (Figure 2A and 2C and Figure 3A and 3B).
Figure 3.
Sensitivity of 13 HCV rapid diagnostic tests in samples with detectable HCV VL/cAg (circles, % sensitivity or specificity; closed circles, lot 1; open circles, lot 2; error bars, upper and lower 95% confidence intervals): (A) HIV-uninfected samples; and (B) HIV-infected samples. Abbreviations: cAg, core antigen; HCV, hepatitis C; HIV, human immunodeficiency virus; VL, viral load.
Operational Characteristics
There was a very high concordance among readers in terms of interreader variability, with a coefficient of agreement ≥95% for all RDTs and lots. Furthermore, there was a high percentage of agreement between lots per RDT, with 10/13 RDTs achieving an agreement of >98% between both tested lots. Invalid runs were uncommon; 11/13 RDTs generated no or only very few invalid results during the first run and none during the repeat (Table 4).
Table 4.
Operational Characteristics of HCV RDTs
| RDT | Lot | Interreader Variability, κ | Lot-to-Lot Variability, % Agreement | Invalid Test Runs, % | |
|---|---|---|---|---|---|
| First Run | Repeat Run | ||||
| SD Biosensor Standard Q HCV Ab | Lot 1 | 0.99 | 99.42 | 0 | 0 |
| Lot 2 | 0.98 | 0 | 0 | ||
| Artron HCV Antibody Test | Lot 1 | 0.98 | 98.65 | 0 | 0 |
| Lot 2 | 0.98 | 0 | 0 | ||
| Wantai HCV Rapid Test | Lot 1 | 0.97 | 99.01 | 0 | 0 |
| Lot 2 | 0.97 | 0 | 0 | ||
| InTec Rapid Anti-HCV Test | Lot 1 | 0.98 | 99.18 | 0 | 0 |
| Lot 2 | 0.99 | 0 | 0 | ||
| PMC First Response HCV Card Test | Lot 1 | 0.98 | 99.12 | 0 | 0 |
| Lot 2 | 0.98 | 0.06 | 0 | ||
| Arkray Signal HCV Version 3.0 | Lot 1 | 0.95 | 96.19 | 1.05 | 0.12 |
| Lot 2 | 0.96 | 1.35 | 0.18 | ||
| J. Mitra TRI DOT HCV | Lot 1 | 0.95 | 97.88 | 1.93 | 0.41 |
| Lot 2 | 0.97 | 1.70 | 0.35 | ||
| Biosynex HCV-only | Lot 1 | 0.98 | 99.12 | 0 | 0 |
| Lot 2 | 0.98 | 0 | 0 | ||
| Abbott SD Bioline HCV | Lot 1 | 0.98 | 99.42 | 0 | 0 |
| Lot 2 | 0.98 | 0 | 0 | ||
| OraSure OraQuick HCV | Lot 1 | 0.98 | 99.36 | 0.12 | 0 |
| Lot 2 | 0.98 | 0.06 | 0 | ||
| BioLytical prototype HCV Ab Test | Lot 1 | 0.97 | 98.83 | 0.06 | 0 |
| Lot 2 | 0.97 | 0.12 | 0 | ||
| Chembio prototype DPP HCV | Lot 1 | 0.95 | 98.13 | 0.35 | 0 |
| Lot 2 | 0.96 | 0.06 | 0 | ||
| AccessBio prototype CareStart HCV | Lot 1 | 0.97 | 97.84 | 0.06 | 0 |
| Lot 2 | 0.95 | 0 | 0 |
Abbreviations: Ab, antibody; HCV, hepatitis C virus; RDT, rapid diagnostic test.
DISCUSSION
To our knowledge, this is the first study to evaluate the performance of HCV RDTs using a large number of samples representing different geographical regions and with a substantial proportion of HIV coinfected samples. As such, our findings provide valuable insights into HCV RDT performance on archived plasma samples and highlight a number of areas for future study.
WHO guidance on performance criteria for in vitro diagnostics for HCV recommends a sensitivity of ≥98% and a specificity of ≥97% for HCV serology RDTs [8]. In HIV-uninfected plasma samples in this study, the performance of the 13 RDTs was high; all tests met the WHO specificity criteria and 11 of 13 met the sensitivity criteria for 1 or both lots. This is consistent with previous studies demonstrating high sensitivity and specificity of the SD Bioline [10], First Response HCV Card Test [14], and OraQuick HCV [9, 10, 15] in plasma samples, and high performance of a number of the RDTs in other sample types including serum and oral fluid [9, 10, 16–19]. In 2 systematic reviews of HCV RDTs that included studies with varying designs, references and sample types, overall pooled sensitivity was 98%–99% [17, 19]. These findings suggest that a number of the RDTs tested may be suitable for in-country use in the HIV-uninfected population.
In HIV-infected samples, however, while specificity remained high (12 of 13 RDTs met the WHO specificity criteria for 1 or both lots), none of the tests evaluated met the WHO sensitivity criteria. The fact that sensitivity improved in the subset of HIV-infected samples with detectable HCV VL/cAg suggests that the reduced sensitivity in HIV-infected samples overall may have been due to low HCV antibody titers. However, the reasons for low HCV antibody titers in HIV-infected samples are unclear, as CD4 counts were generally high, suggesting that the sample donors were not severely immunosuppressed. Other studies have noted declines in HCV antibody levels following treatment-induced or spontaneous HCV clearance in HIV-infected men [20, 21]. In general, our observation of lower HCV RDT performance in HIV-infected individuals is consistent with observations made in other studies, in which 1 or more of the evaluated RDTs showed poor sensitivity in samples from HIV-positive individuals [9–11]. The reasons for lower sensitivity in HIV-positive samples in these studies also remains unclear. More detailed information on HCV VL/cAg, time of coinfection, and initiation of and adherence to HIV treatment should be collected in future studies, in order to further assess the impact of HIV status on RDT performance.
Of the 71 million people worldwide with chronic HCV infection, 2.3 million are also infected with HIV [1]. As such, good RDT performance in HCV and HIV coinfected people is essential, particularly in LMICs where the burden of both diseases is high [3, 22]. However, while the false negatives observed in HIV samples in this study are technically concerning, from a clinical perspective, it is reassuring that the diagnostic performance of the evaluated RDTs improved in HIV-infected samples with detectable HCV VL/cAg. HCV VL or cAg testing is used to confirm viremic infection in people who test positive for HCV antibodies [5], thus these samples represent patients who had active HCV infection and are ultimately in need of treatment. As RDT performance was high regardless of HIV status in these samples, the impact of HIV infection on test performance may not be that dramatic.
The majority (69.7%) of samples that were false negative in at least 1 lot of any RDT in this study did not have genotype information available, making it difficult to associate the occurrence of false negatives with any particular HCV genotype. Notably, 34.6% of all samples giving at least 1 false negative were of HCV genotype 6. Given the relatively low total number of genotype 6 samples (41 out of a total of 293 samples with genotype information available), this could potentially have been a contributing factor to the high number of false-negative samples. However, verification of this by statistical analysis was not possible due to the aforementioned low number of samples with genotype information available.
The WHO guidance on performance criteria for HCV serology RDTs recommends an interreader variability and device failure rate of ≤5% [8]. All of the RDTs evaluated in this study met both criteria. Additionally, the performance of all of the RDTs was in high agreement between the 2 lots evaluated, demonstrating a low technical lot-to-lot variability. These data show that the consistency of HCV serology RDTs is high, providing confidence in the operational quality of the tests across different lots and devices.
The data from this study contribute to the growing evidence on the use of HCV RDTs for HCV screening in LMICs, providing that they are first evaluated in the intended target population to determine whether sensitivity is impacted by population factors. A number of populations are commonly targeted for HCV screening, including sex workers, men who have sex with men, people who inject drugs, and people living with HIV [1, 23]. Procurement of high-performance RDTs will be key to the improvement of HCV testing services for these key populations. Although we did not collect data on sample donor characteristics, it is likely that samples from these target groups were tested in our study, given the countries included. For example, Georgia has one of the highest prevalences of injection drug use globally, with up to 40% of HCV infections attributable to injection drug use [24]. Additionally, in some countries HCV screening is indicated for the general population, as a result of historical unsafe medical practices [25], as is the case in Nigeria [26] and Cambodia [27].
Limitations of this study include the uneven geographical distribution of sample types. Sensitivity in the HIV-uninfected population was primarily assessed in samples originating from Georgia and Nigeria, while sensitivity in the HIV-infected population was assessed almost exclusively in samples from Cambodia and Belgium. This makes comparisons between sensitivity in the HIV-uninfected and -infected populations challenging. We cannot exclude the possibility that differences in population characteristics, such as different types of HIV/HCV risk groups, impacted the results. Notably, Belgium (from which 120 [14.7%] samples were obtained) is a high-income country, thus population characteristics such as HIV prevalence or HCV cohort may not be comparable to those of LMICs.
While we cannot exclude the possibility that differences in storage conditions between countries had an effect on sample quality, evidence suggests that antibodies remain stable in frozen samples for several years and after multiple freeze-thaw cycles [28–31]; furthermore, we minimized any potential impact by only including samples that appeared nonhemolytic upon visual inspection. A further limitation is the low number of HCV-negative and HIV-negative samples compared with HCV-negative HIV-positive samples, which may have influenced specificity in the overall population. The impact of this was likely minor, however, as most of the RDTs performed well in both study populations.
The design of the composite reference standard led to 210 samples being excluded from the study. It is possible that inclusion of these samples would have affected the sensitivity and specificity estimates. This study did not take into account the impact of treatment for HCV, although only a small number of samples (n = 17) were from people who were receiving or who had previously received interferon treatment. Additionally, the HCV-negative samples used may not have precisely represented the target populations for HCV serology testing, leading to patient bias. Finally, tests were performed by well-trained laboratory personnel using archived samples, thus this study does not represent a real-world setting. The performance of the RDTs in primary or community care settings using prospectively collected fresh samples is yet to be established.
In conclusion, the findings from this study show that a number of available HCV RDTs may be suitable for WHO prequalification and use in HCV screening programs in LMICs. However, HCV RDTs should always be evaluated in the intended target population, as sensitivity can be impacted by population factors such as HIV status. Any evaluation panels used for assessment of HCV RDTs should contain HIV-positive samples. These findings serve as a valuable baseline to investigate RDT performance in prospectively collected whole blood samples in the intended use settings. This will yield further insights into the robustness of the RDTs when used in primary health care settings by local health workers and tested on the most common sample type used for RDTs.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments . The authors express their gratitude to all those involved in the execution of the project. Medical writing assistance and editorial support, under the direction of the authors, was provided by Rachel Wright, PhD, funded by Foundation for Innovative New Diagnostics, according to Good Publication Practice guidelines.
With the exception of the Abbott SD Bioline HCV RDT, all HCV RDTs under evaluation have been provided free of charge or at reduced cost by the respective manufacturers.
Financial support . This work was supported by Unitaid (grant number UA_HCV01).
Potential conflicts of interest . All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: HIV Diagnostics Conference, 25–28 March 2019, Atlanta, Georgia; International Liver Congress, 10–14 April 2019, Vienna, Austria; International AIDS Society 10th Conference on HIV Science, 21–24 July 2019, Mexico City, Mexico; 6th Nigerian Institute of Medical Research International Scientific Conference, 12–14 November 2019, Lagos, Nigeria.
Contributor Information
Beatrice N Vetter, Foundation for Innovative New Diagnostics, Geneva, Switzerland.
Elena Ivanova Reipold, Foundation for Innovative New Diagnostics, Geneva, Switzerland.
Stefano Ongarello, Foundation for Innovative New Diagnostics, Geneva, Switzerland.
Rosemary Audu, Nigerian Institute of Medical Research, Lagos, Nigeria.
Fehintola A Ige, Nigerian Institute of Medical Research, Lagos, Nigeria.
Maia Alkhazashvili, National Center for Disease Control and Public Health/R. Lugar Center for Public Health Research, Tbilisi, Georgia.
Nazibrola Chitadze, National Center for Disease Control and Public Health/R. Lugar Center for Public Health Research, Tbilisi, Georgia.
Fien Vanroye, Institute of Tropical Medicine HIV/STD Reference Laboratory, Antwerp, Belgium.
Anja De Weggheleire, Institute of Tropical Medicine HIV/STD Reference Laboratory, Antwerp, Belgium.
Sokkab An, Sihanouk Hospital Center of Hope, Phnom Penh, Cambodia.
Katrien Fransen, Institute of Tropical Medicine HIV/STD Reference Laboratory, Antwerp, Belgium.
References
- 1. World Health Organization. Global hepatitis report, 2017. https://www.who.int/hepatitis/publications/global-hepatitis-report2017/en/. Accessed 9 July 2020.
- 2. Reipold EI, Trianni A, Krakower D, et al. Values, preferences and current hepatitis B and C testing practices in low- and middle-income countries: results of a survey of end users and implementers. BMC Infect Dis 2017; 17(suppl 1):702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tordrup D, Hutin Y, Stenberg K, et al. Additional resource needs for viral hepatitis elimination through universal health coverage: projections in 67 low-income and middle-income countries, 2016-30. Lancet Glob Health 2019; 7:e1180–8. [DOI] [PubMed] [Google Scholar]
- 4. World Health Organization. Global health sector strategy on viral hepatitis 2016–2021. https://www.who.int/hepatitis/strategy2016-2021/ghss-hep/en/. Accessed 9 July 2020.
- 5. World Health Organization. Guidelines on hepatitis B and C testing. https://www.who.int/hepatitis/publications/guidelines-hepatitis-c-b-testing/en/. Accessed 9 July 2020.
- 6. World Health Organization. Guidelines on HIV self-testing and partner notification. https://www.who.int/hiv/pub/vct/hiv-self-testing-guidelines/en/. Accessed 9 July 2020.
- 7. World Health Organization. WHO list of prequalified in vitro diagnostic products. https://www.who.int/diagnostics_laboratory/evaluations/190918_prequalified_product_list.pdf?ua=1. Accessed 9 July 2020.
- 8. World Health Organization. Selecting and purchasing HIV, HBsAg and HCV in vitro diagnostics. https://www.who.int/diagnostics_laboratory/procurement/purchase/en/. Accessed 9 July 2020.
- 9. Kosack CS, Nick S. Evaluation of two rapid screening assays for detecting hepatitis C antibodies in resource-constrained settings. Trop Med Int Health 2016; 21:603–9. [DOI] [PubMed] [Google Scholar]
- 10. Mane A, Sacks J, Sharma S, et al. Evaluation of five rapid diagnostic tests for detection of antibodies to hepatitis C virus (HCV): a step towards scale-up of HCV screening efforts in India. PLoS One 2019; 14:e0210556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Smith BD, Teshale E, Jewett A, et al. Performance of premarket rapid hepatitis C virus antibody assays in 4 national human immunodeficiency virus behavioral surveillance system sites. Clin Infect Dis 2011; 53: 780–6. [DOI] [PubMed] [Google Scholar]
- 12. World Health Organization. WHO protocol for performance laboratory evaluation of HCV serology assays. https://www.who.int/diagnostics_laboratory/evaluations/alter/171219_protocol_pqdx_040_v6_hcv.pdf?ua=1. Accessed 9 July 2020.
- 13. Zhou X-H, Obuchowski NA, McClish DK.. Statistical methods in diagnostic medicine. 2nd ed. Hoboken, NJ: Wiley, 2011. [Google Scholar]
- 14. Fondjo CLK, Ngoupo PAT, Ngono L, Plantier JC, Njouom R. Performance evaluation of three rapid screening assays for detection of antibodies to hepatitis C virus in Cameroon. BMC Res Notes 2018; 11:352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. O’Connell RJ, Gates RG, Bautista CT, et al. Laboratory evaluation of rapid test kits to detect hepatitis C antibody for use in predonation screening in emergency settings. Transfusion 2013; 53:505–17. [DOI] [PubMed] [Google Scholar]
- 16. Chevaliez S, Poiteau L, Rosa I, et al. Prospective assessment of rapid diagnostic tests for the detection of antibodies to hepatitis C virus, a tool for improving access to care. Clin Microbiol Infect 2016; 22:459.e1–6. [DOI] [PubMed] [Google Scholar]
- 17. Tang W, Chen W, Amini A, et al. Diagnostic accuracy of tests to detect hepatitis C antibody: a meta-analysis and review of the literature. BMC Infect Dis 2017; 17:695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Poiteau L, Soulier A, Lemoine M, et al. Performance of a new rapid diagnostic test for the detection of antibodies to hepatitis C virus. J Virol Methods 2018; 261:153–5. [DOI] [PubMed] [Google Scholar]
- 19. Shivkumar S, Peeling R, Jafari Y, Joseph L, Pant Pai N. Accuracy of rapid and point-of-care screening tests for hepatitis C: a systematic review and meta-analysis. Ann Intern Med 2012; 157:558–66. [DOI] [PubMed] [Google Scholar]
- 20. Aebi-Popp K, Wandeler G, Salazar-Vizcaya L, et al. ; the Swiss HIV Cohort Study . Rapid decline of anti-hepatitis C virus (HCV) antibodies following early treatment of incident HCV infections in HIV-infected men who have sex with men. HIV Med 2018; 19:420–5. [DOI] [PubMed] [Google Scholar]
- 21. Vanhommerig JW, Thomas XV, van der Meer JT, et al. ; MOSAIC (MSM Observational Study for Acute Infection with Hepatitis C) Study Group . Hepatitis C virus (HCV) antibody dynamics following acute HCV infection and reinfection among HIV-infected men who have sex with men. Clin Infect Dis 2014; 59:1678–85. [DOI] [PubMed] [Google Scholar]
- 22. World Health Organization. Number of people (all ages) living with HIV: estimates by country. http://apps.who.int/gho/data/view.main.22100?lang=en. Accessed 9 July 2020.
- 23. Applegate TL, Fajardo E, Sacks JA. Hepatitis C virus diagnosis and the holy grail. Infect Dis Clin North Am 2018; 32:425–45. [DOI] [PubMed] [Google Scholar]
- 24. Stvilia K, Spradling PR, Asatiani A, et al. Progress in testing for and treatment of hepatitis C virus infection among persons who inject drugs—Georgia, 2018. MMWR Morb Mortal Wkly Rep 2019; 68:637–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Umutesi J, Liu CY, Penkunas MJ, et al. Screening a nation for hepatitis C virus elimination: a cross-sectional study on prevalence of hepatitis C and associated risk factors in the Rwandan general population. BMJ Open 2019; 9:e029743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ejiofor OS, Emechebe GO, Igwe WC, Ifeadike CO, Ubajaka CF. Hepatitis C virus infection in Nigerians. Nigerian Med J 2010; 51:173–6. [Google Scholar]
- 27. Nouhin J, Iwamoto M, Prak S, et al. Molecular epidemiology of hepatitis C virus in Cambodia during 2016–2017. Sci Rep 2019; 9:7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Allison KM, Faddy HM, Margaritis A, Ismay S, Marks DC. The impact on blood donor screening for human immunodeficiency virus, hepatitis C virus, and hepatitis B virus using plasma from frozen-thawed plasma preparation tubes. Transfusion 2016; 56:449–56. [DOI] [PubMed] [Google Scholar]
- 29. Rastawicki W, Smietańska K, Rokosz N, Jagielski M. Effect of multiple freeze-thaw cycles on detection of IgA, IgG and IgM antibodies to selected bacterial antigens [in Polish]. Med Dosw Mikrobiol 2012; 64:79–85. [PubMed] [Google Scholar]
- 30. Pinsky NA, Huddleston JM, Jacobson RM, Wollan PC, Poland GA. Effect of multiple freeze-thaw cycles on detection of measles, mumps, and rubella virus antibodies. Clin Diagn Lab Immunol 2003; 10:19–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Castejon MJ, Yamashiro R, Cardosa de Oliveira C, Aparecida de Freitas Oliveira C, Ueda M. Stability of anti-HIV antibodies in serum samples stored for two to eighteen years periods. J Bras Patol Med Lab 2014; 50:272–7. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.