Abstract
Cholesterol, an essential molecule for cell structure, function, and viability, plays crucial roles in the development, progression, and survival of cancer cells. Earlier studies have shown that cholesterol-lowering drugs can inhibit the high expression of programmed-death ligand 1 (PD-L1) that contributes to immunoevasion in cancer cells. However, the regulatory mechanism of cell surface PD-L1 abundance by cholesterol is still controversial. Here, using nuclear magnetic resonance and biochemical techniques, we demonstrated that cholesterol can directly bind to the transmembrane domain of PD-L1 through two cholesterol-recognition amino acid consensus (CRAC) motifs, forming a sandwich-like architecture and stabilizing PD-L1 to prevent downstream degradation. Mutations at key binding residues prohibit PD-L1–cholesterol interactions, decreasing the cellular abundance of PD-L1. Our results reveal a unique regulatory mechanism that controls the stability of PD-L1 in cancer cells, providing an alternative method to overcome PD-L1–mediated immunoevasion in cancers.
Cholesterol plays a crucial role in PD-L1 stability by direct binding to two CRAC motifs in the transmembrane domain of PD-L1.
INTRODUCTION
Cholesterol, a major constituent of plasma membranes, affects structural integrity, thickness, permeability, and fluidity of cell membranes (1, 2). As a precursor of several hormones, cholesterol is a key factor in modulating cell function and signaling (3). Cell functions can be regulated by cholesterol through effects on the structure, function, and dynamics of numerous transmembrane proteins (4, 5). For example, cholesterol enhances the structural integrity and activation of voltage-dependent anion channels (VDACs) and affects the interactions of VDACs with other proteins (6). In addition, cholesterol can maintain the stability or alter the function of G protein–coupled receptors (GPCRs) (7–10), e.g., cholesterol depletion abolishes signaling of the chemokine receptor CXCR4 by impeding ligand binding (11, 12). Cholesterol can modulate the function of such membrane-embedded proteins directly (through binding of cholesterol to the protein), indirectly (through changes in membrane properties), or through a combination of direct and indirect mechanisms (5, 9). Three major cholesterol-binding motifs, which facilitate cholesterol-sensitive functions, have been identified in transmembrane proteins, including a cholesterol recognition amino acid consensus motif (CRAC, L/V–X1–5–Y/F–X1–5–K/R) (13, 14), an inverted cholesterol recognition motif (CARC, K/R–X1–5–Y/F–X1–5–L/V) (15), and a cholesterol consensus motif (16).
Recently, emerging evidence has demonstrated that cholesterol plays essential roles in cancer cells (17). A high level of cholesterol is required for rapid proliferation and invasion of cancer cells (18). Several clinical and experimental studies have revealed that cholesterol accumulation in cancer cells increases the incidence rate for some cancers (19), whereas cholesterol reduction by statins, one of the major cholesterol-lowering medications, has shown beneficial effects in many different types of cancers (20). Moreover, high cholesterol levels in the tumor microenvironment affect the phenotype and activity of tumor-infiltrating immune cells (21, 22), although the reported effects of cholesterol on immune cells remain controversial. Therefore, modulating cholesterol levels has been highlighted as a promising strategy to treat cancers and improve immunotherapy effectiveness.
Programmed death ligand-1 (PD-L1), a highly expressed protein on the plasma membrane of cancer cells (23), showed substantially lower expression in response to statin treatment (20), suggesting a strong relationship between PD-L1 and cholesterol. Two typical CRAC motifs (fig. S1) exist in the transmembrane domain (TMD) of PD-L1, indicating a strong potential for direct binding of cholesterol. However, whether interactions with cholesterol can influence PD-L1 stability or function has not been established. To experimentally test this possibility, we investigated the effects of cholesterol on PD-L1. First, biochemical assays showed that cholesterol enhances the stability of PD-L1. Using versatile nuclear magnetic resonance (NMR) spectroscopy, we found that cholesterol can directly bind to the two CRAC motifs in the TMD of PD-L1. Molecular dynamics (MD) simulations, a powerful complementary approach, revealed detailed structural and energetic information about the interactions between PD-L1 and cholesterol. Functional mutagenesis further confirmed that the CRAC motifs are critical in mediating the cellular abundance of PD-L1. Our results provide molecular-level insight into the PD-L1–cholesterol interaction and its effects on tumor therapy.
RESULTS
Cholesterol maintains the stability of PD-L1
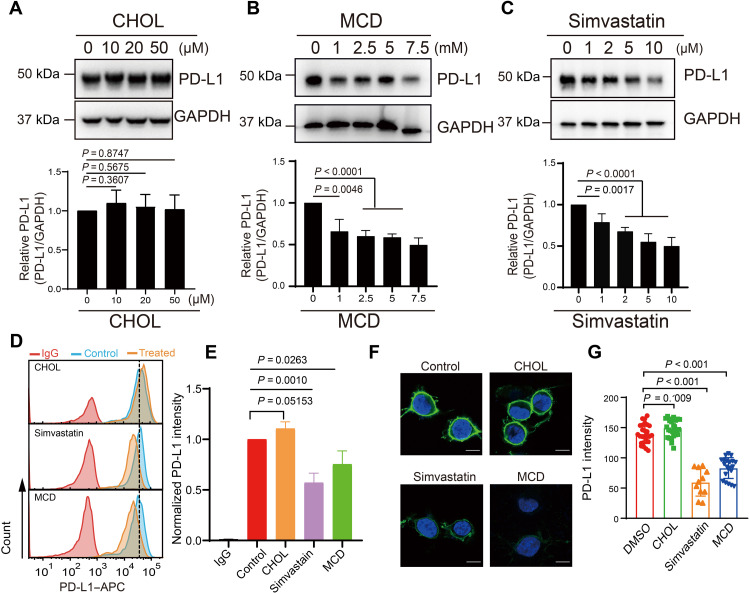
To investigate the relationship between cholesterol and PD-L1, we first explored the spatial relationship between PD-L1 and cholesterol in RKO cells (a human colon cancer cell line) by immunofluorescence staining and colocalization analysis and found that PD-L1 and cholesterol have a strong correlation with a Pearson’s correlation coefficient of 0.83 determined by ImageJ (fig. S2, A and B) (24). When RKO cells were treated with the protein synthesis inhibitor cycloheximide (CHX) (25), PD-L1 exhibited a slower turnover rate when cholesterol was added (fig. S2, C and D). We next examined endogenous PD-L1 expression in RKO cells treated with cholesterol, methyl-β-cyclodextrin (MCD), or simvastatin. MCD is a water-soluble polymer that depletes cholesterol from the cells (26), and simvastatin is a U.S. Food and Drug Administration–approved drug used to lower total cholesterol (27). Western blot analyses showed that the addition of cholesterol to RKO cells, which increases the amount of cholesterol in the plasma membrane, up-regulated PD-L1 level (Fig. 1A), whereas the addition of MCD or simvastatin substantially down-regulated the cell abundance of PD-L1 (Fig. 1, B and C). The effects of cholesterol, MCD, and simvastatin on cell surface expression of PD-L1 were further confirmed by flow cytometry (Fig. 1D) and confocal imaging (Fig. 1E). Consistently, the cell surface levels of PD-L1 slightly increased after cholesterol treatment but substantially decreased after removal of cholesterol by MCD or simvastatin treatment. Furthermore, the ubiquitination of endogenous PD-L1 was substantially increased after addition of MCD or simvastatin (fig. S2, E and F). Together, these results suggest that cellular cholesterol plays a critical role in PD-L1 localization and stability.
Fig. 1. Cholesterol maintains the stability of PD-L1.
(A to C) RKO cells were treated with several concentrations of cholesterol (CHOL, 0 to 50 μM for 12 hours), MCD (0 to 7.5 mM for 6 hours), or simvastatin (0 to 10 μM for 12 hours). Western blot was performed to measure PD-L1 expression in response to each treatment (top). The intensity of PD-L1 expression (relative to GAPDH) was quantified with ImageJ (bottom). Data shown are the means ± SD (n = 3). P values were calculated using unpaired two-sided Student’s t test. (D and E) Flow cytometric analysis of PD-L1 expressed on the cell surface after treatment with CHOL (25 μM for 12 hours), MCD (5 mM for 6 hours), or simvastatin (10 μM for 12 hours) (D). Levels of PD-L1 were quantified with FlowJo (E). Expression data are shown as fold change values relative to the control and indicate the means ± SD (n = 3). P values were calculated with one-way ANOVA. (F and G) Immunofluorescence staining shows PD-L1 in RKO cells treated with CHOL (25 μM for 12 hours), MCD (5 mM for 6 hours), or simvastatin (10 μM for 12 hours). PD-L1 was labeled with anti–PD-L1 antibody (green), and the cell nucleus was stained with DAPI (blue) (F). Scale bars, 10 μm. Fluorescence intensities were then quantified (G); n = 26, 27, 11, and 21 for control, cholesterol, simvastatin, and MCD treatments, respectively. Statistical differences were determined with one-way ANOVA. All results are representative of three independent experiments.
We further verified the effect of cholesterol on PD-L1 expression using K562 cells (a human immortalized myelogenous leukemia cell line). Up-regulation of PD-L1 in K562 cells has been observed after addition of the cytokine interferon-γ (IFN-γ), which is a key stimulator of PD-L1 expression in several types of tumor cells (28). Decreasing cholesterol levels in the K562 cells by MCD or simvastatin treatment down-regulated PD-L1 in the presence of IFN-γ (fig. S3A). Treating K562 cells with natural killer (NK) cell–derived supernatants showed similar results; PD-L1 levels decreased by ~70% in response to MCD or simvastatin treatment in the presence of NK cell supernatants (fig. S3B). In another experiment, we pretreated K562 cells with IFN-γ and used anti–PD-L1 antibodies to label cell surface PD-L1. PD-L1 levels decreased by nearly 50% within 6 hours after the removal of IFN-γ, but high PD-L1 expression was maintained in the presence of cholesterol (fig. S3, C to E). Together, the in vitro results showed that changing cholesterol content specifically modulated cellular abundance of PD-L1.
NMR characterization of PD-L1-TC
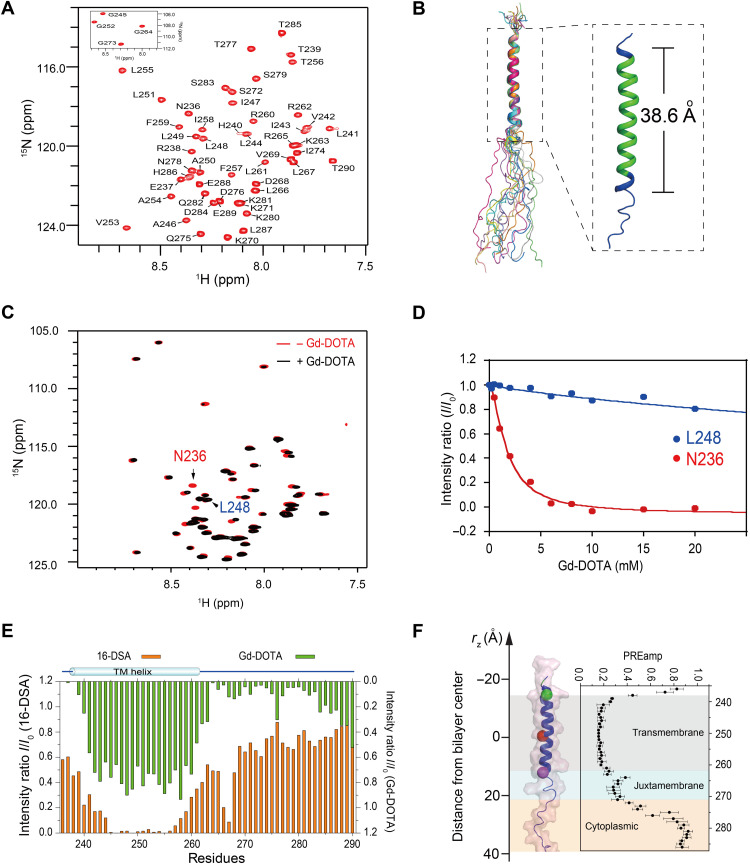
The above results demonstrated that cholesterol plays an essential role in controlling PD-L1 levels. The presence of the cholesterol recognition motifs (L255)-X-F257-XX-R260 (CRAC1) and (L255)-XXX-F259-XX-R262 (CRAC2) (fig. S1) in the TMD of PD-L1 suggests that cholesterol has the ability to directly interact with PD-L1 and enhance protein stability, as has been reported for other receptors. To explore this possibility, we first characterized the interactions between the human PD-L1 and cholesterol using solution NMR spectroscopy. The transmembrane/cytoplasmic domain of PD-L1 (PD-L1-TC; amino acid residues 232 to 290) was expressed and purified following a previously published protocol (fig. S4, A and B) (29). Purified PD-L1-TC was reconstituted in zwitterionic 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC)/1,2-dihexanoyl-sn-glycero-3-phosphocholine (DH6PC) bicelles (molar ratio of DMPC:DH6PC, q = 0.5) (fig. S4, C and D). The backbone (Fig. 2A) and side-chain (fig. S5A) resonance assignments of PD-L1-TC were determined through a standard set of transverse relaxation optimized spectroscopy (TROSY)–based triple-resonance experiments (30). The secondary structures of PD-L1-TC were derived from analysis of the backbone chemical shifts using the TALOS+ program (fig. S5B) (31). The final ensemble (Fig. 2B) of the 15 lowest-energy structures [Protein Data Bank (PDB): 7DCV] was calculated using 240 protein nuclear Overhauser effect (NOE)–derived distance restraints (fig. S5C) and 38 hydrogen bond restraints (table S1) and converged to a root mean square deviation (RMSD) of 0.619 and 1.194 Å for backbone and heavy atoms, respectively. Residues 238 to 261 formed the helical region of PD-L1-TC, which had a length of 38.6 Å (Fig. 2B). The cytoplasmic domain of PD-L1 was largely dynamic because there were no NOE signals characteristic of secondary structures (fig. S5C).
Fig. 2. NMR characterizations of PD-L1-TC in bicelles.
(A) The 2D 1H, 15N-TROSY-HSQC spectrum of PD-L1-TC samples in DMPC/DH6PC bicelles recorded at 310 K on a Bruker 600-MHz spectrometer. ppm, parts per million. (B) Cartoon representation of the 15 lowest-energy structures of PD-L1-TC in DMPC/DH6PC bicelles. The calculated helical region contains amino acids 238 to 261 (blue) with a length of 38.6 Å, covered by the predicted transmembrane region (amino acids 239 to 259, green) from UniProt (right). (C and D) Measurement of membrane immersion depth of PD-L1-TC in DMPC/DH6PC bicelles using the water-soluble probe Gd-DOTA. 1H, 15N-TROSY NMR spectra of PD-L1-TC samples in the absence (red) and presence (black) of 15 mM Gd-DOTA; both spectra were acquired at 310 K on a Bruker 600-MHz spectrometer (C). PRE effects (I/I0) versus [Gd-DOTA] for N236 and L248 were plotted at different concentrations of Gd-DOTA (D). I, peak intensity in the presence of Gd-DOTA; I0, peak intensity in the absence of Gd-DOTA. (E) Assessment of PD-L1-TC in DMPC/DH6PC bicelles from the PRE effects of hydrophilic Gd-DOTA and lipophilic16-DSA paramagnetic probes. Peak intensity reductions induced by 16-DSA (2 mM) and Gd-DOTA (15 mM) relative to a reference spectrum are shown as bar graphs in orange and green, respectively. Measurements were carried out for PD-L1-TC in DMPC/DH6PC bicelles at an 1H frequency of 700 MHz (pH 6.5) and 310 K. (F) Membrane partitioning of PD-L1-TC as determined by PRE effects. The position of the PD-L1-TC structure is shown along the membrane axis of the lipid bilayer. The transmembrane region (amino acids 239 to 261) is highlighted in gray, the juxtamembrane region (amino acids 262 to 272) is highlighted in light blue, and the cytoplasmic region (amino acids 273 to 290) is highlighted in pink, with residues 236, 251, and 261 colored green, red, and purple, respectively. The complete PREamp plot from Gd-DOTA titrations is shown on the right.
Paramagnetic relaxation enhancement (PRE) titrations were further performed to evaluate the conformation of membrane-embedded PD-L1-TC (32). The water-soluble paramagnetic probe gadolinium(III)1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetate (Gd-DOTA) and the lipophilic paramagnetic probe 16-doxyl stearic acid (16-DSA) were each titrated to PD-L1-TC in DMPC/DH6PC bicelles. A series of two-dimensional (2D) TROSY–heteronuclear single quantum coherence (HSQC) spectra were recorded to determine the PRE effects (Fig. 2C). The intensities of residues (e.g., N236) in the aqueous phase obviously decreased as the concentration of Gd-DOTA increased, whereas the intensities of residues buried in the lipid bilayer (e.g., L248) decreased at a much slower rate (Fig. 2D). These results were consistent with complementary PRE titrations of lipophilic 16-DSA (Fig. 2E), demonstrating that the membrane-embedded residues included T239-L261, with the membrane center located near L251 (Fig. 2F).
Cholesterol binds to CRAC motifs in PD-L1-TC
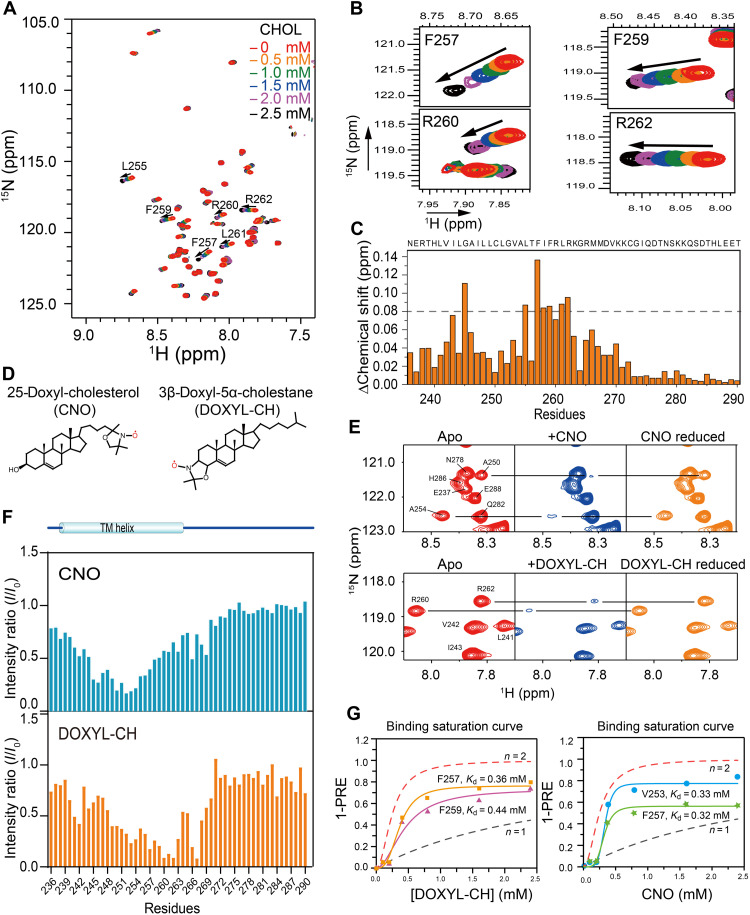
Next, we titrated cholesterol into PD-L1-TC in DMPC/DH6PC bicelles to monitor cholesterol-induced chemical shift changes via 2D TROSY-HSQC experiments. NMR spectra were recorded at cholesterol concentrations ranging from 0 to 2.5 mM. Cholesterol titration of PD-L1-TC resulted in substantial perturbations in NMR resonance positions for a subset of peaks (Fig. 3A). The PD-L1-TC resonances that exhibited the largest changes in response to cholesterol were mostly located at the CRAC motif region, namely, F257, F259, R260, and R262 (Fig. 3, B and C). To more precisely identify the cholesterol binding site(s), two spin-labeled cholesterol derivatives, 3β-DOXYL-5α-cholestane (DOXYL-CH) and 25-doxyl-cholesterol (CNO) (33), were used to measure PRE effects on PD-L1-TC (Fig. 3D). DOXYL-CH has a nitroxide moiety at the 3β position, and CNO has a nitroxide at position 25 near the terminal methyl group. Spin-labeled DOXYL-CH and CNO were each titrated separately into a PD-L1-TC sample, and the intensity changes of individual residues were detected with a series of 2D TROSY-HSQC spectra (Fig. 3E). DOXYL-CH induced strong PREs for residues R260 to R262, indicating that the cholesterol head group was located in the C-terminal juxtamembrane region, whereas CNO strongly decreased the intensities of residues near G252-A254 (Fig. 3F), indicating that the cholesterol tail bound to the center of the membrane. The apparent dissociation constant (Kd) values further determined using the PRE titration data are at a range of 0.32 to 0.44 mM, and the experimental binding curves agree better with the simulation curve of n = 2 (Fig. 3G) (34), indicating positive cooperativity in cholesterol binding between the two PD-L1 binding sites. Collectively, the PRE data suggested that cholesterol binds at the CRAC motif region from G252 to R262, oriented so the head group is associated with cytoplasmic compartments and the acyl tail is near the membrane center.
Fig. 3. Characterization of cholesterol binding sites in PD-L1-TC.
(A) Superimposed 2D 1H, 15N-TROSY-HSQC spectra of 0.2 mM 15N-labeled PD-L1-TC were acquired at 700 MHz (1H frequency) and 310 K in the presence of cholesterol at 0 mM (red), 0.5 mM (orange), 1.0 mM (green), 1.5 mM (blue), 2.0 mM (purple), or 2.5 mM (black). (B) Enlarged view of four regions in the plot displayed in (A), showing specific chemical shift perturbations with cholesterol titration of the two CRAC motifs (CRAC1: F257 and R260; CRAC2: F259 and R262) (right). (C) NMR chemical shift changes of PD-L1-TC after the addition of 1.5 mM cholesterol in DMPC/DH6PC bicelles. The plot shows chemical shift changes between the spectra of PD-L1-TC with or without cholesterol. (D) Chemical structures of CNO (left) and DOXYL-CH (right) are shown on in the topmost panel. (E) PRE (I/I0) effects of different nitroxide-labeled cholesterols (DOXYL-CH and CNO) on PD-L1-TC in DMPC/DH6PC bicelles. Specific broadening of 1H-15N correlation peaks by 2.4 mM CNO and DOXYL-CH and intensity recovery of the broadened peaks with the addition of ascorbic acid are shown in the middle and bottom panels, respectively. (F) PRE (I/I0) analysis of CNO and DOXYL-CH. The addition of 2.4 mM CNO showed obvious PRE effects at L252 to A254, whereas addition of 2.4 mM DOXYL-CH caused the most notable PRE effects at R260 to R262. I, peak intensity in the presence of the paramagnetic agent; I0, peak intensity in the absence of the paramagnetic agent. (G) Comparison of (1-PRE) caused by DOXYL-CH (left) and CNO (right) with simulated binding curves. For the PRE data, PD-L1-TC NMR samples were titrated with 0, 0.1, 0.2, 0.4, 0.8, 1.6, and 2.4 mM DOXYL-CH and CNO, respectively. The simulated binding curves with n = 1 (no cooperativity, dotted black line) and n = 2 (positive cooperativity, dotted red line) are shown in each diagram, respectively.
Evaluation of cholesterol binding sites with MD simulation
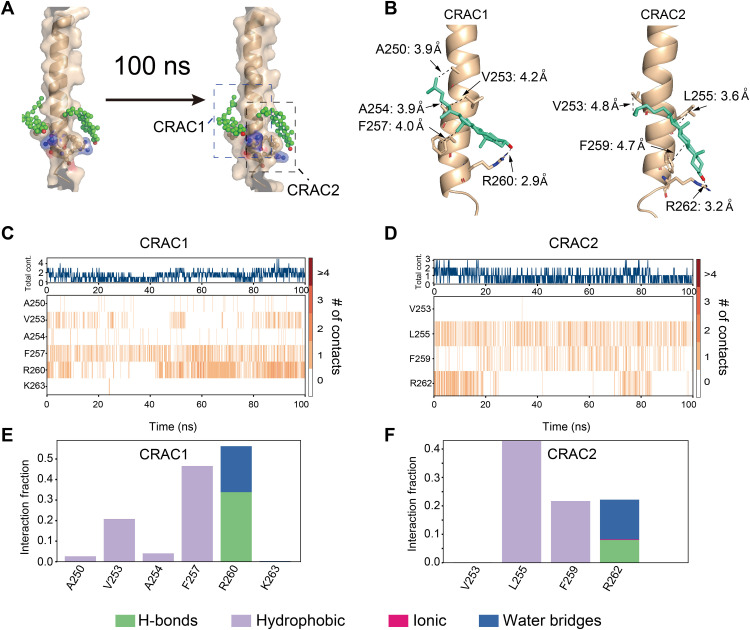
We further evaluated cholesterol binding to PD-L1-TC using MD simulation, a valuable tool to study protein-ligand interactions. We first docked cholesterol in vacuo onto PD-L1-TC using AutoDock Vina 1.1.2 (35) with the NMR constraints described above. The NMR structure (PDB: 7DCV) of PD-L1-TC was used as a target protein for molecular docking. Cholesterol was docked to PD-L1-TC in a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) lipid bilayer environment built with Desmond 2020 software (https://schrodinger.com/desmond, accessed on 17 January 2022). Cholesterol was initially positioned at the sites identified from our PRE analysis; the hydroxyl head group pointed to F257 and R260, F259 and R262, respectively, and the acyl chain pointed to the transmembrane center of PD-L1-TC (Fig. 4A). The PD-L1-TC side chains within 5 Å of cholesterol were allowed to move freely during docking. The docking with two molecules of cholesterol on one PD-L1-TC showed no steric clash with the binding free energy −4.6 kcal/mol, indicating that the interaction of PD-L1 with cholesterol is thermodynamically stable. The docked conformation that most closely matched our NMR data was selected as the initial model for further MD simulations to evaluate the stability of cholesterol binding to PD-L1-TC.
Fig. 4. Evaluation of the NMR-derived cholesterol binding site with MD simulation.
(A) Models showing the positions of cholesterol and PD-L1-TC over the MD simulation trajectory (0 ns, left to 100 ns, right). CRAC1 and CRAC2 are the key motifs in PD-L1 that interact with cholesterol (green). (B) Detailed description of the interactions between CRAC1 and CRAC2 on PD-L1-TC and cholesterol (green) from the final snapshot of MD simulations. (C and D) The binding heatmap shows that cholesterol mainly interacts with the CRAC1 (C) and CRAC2 (D) motifs in PD-L1-TC through the whole trajectory. The orange lines show the number of contacts between cholesterol and PD-L1-TC. (E and F) Quantification of heatmap intensity showing the number of contacts for CRAC1 and CRAC2 motifs. Cholesterol interacts with the CRAC1 motif mainly through hydrophobic interactions with V253 and F257 and through hydrogen bond and water bridges with R260 (E). Cholesterol interacts with the CRAC2 motif mainly through hydrophobic interactions with L255 and F259 and through hydrogen bonds and water bridges with R262 (F).
Three-stage MD simulations were carried out using the docked model. The first two stages were restrained minimization and equilibration runs using the default membrane relax protocol in Desmond. Upon reaching equilibrium, the production was run for a third stage. After a 100-ns simulation, RMSD plots of the starting and final positions for a selection of atoms in the PD-L1-TC–cholesterol complex indicated thermal convergence (fig. S6), and root mean square fluctuation plots showed that the protein backbones and ligand atoms remained locally steady over the entire trajectory (fig. S7). The simulation results showed that F257 and R260 in the CRAC1 motif and F259 and R262 in the CRAC2 motif could bind to cholesterol (Fig. 4, B to E). The interaction analysis showed that F257 and F259 interacted with cholesterol via hydrophobic forces, whereas R260 and R262 interacted with cholesterol through hydrogen bonds and water bridges (Fig. 4, D and E). The relatively flexible acyl chain of cholesterol leaned toward V253 (the CRAC1 motif) or L255 (the CRAC2 motif) through hydrophobic interactions. The binding energy between cholesterol and the CARC1 and CRAC2 motifs is −20 and −18 kcal/mol, respectively, indicating tight binding affinity between PD-L1 and cholesterol for both motifs.
CRAC motifs are critical for the interaction between PD-L1-TC and cholesterol
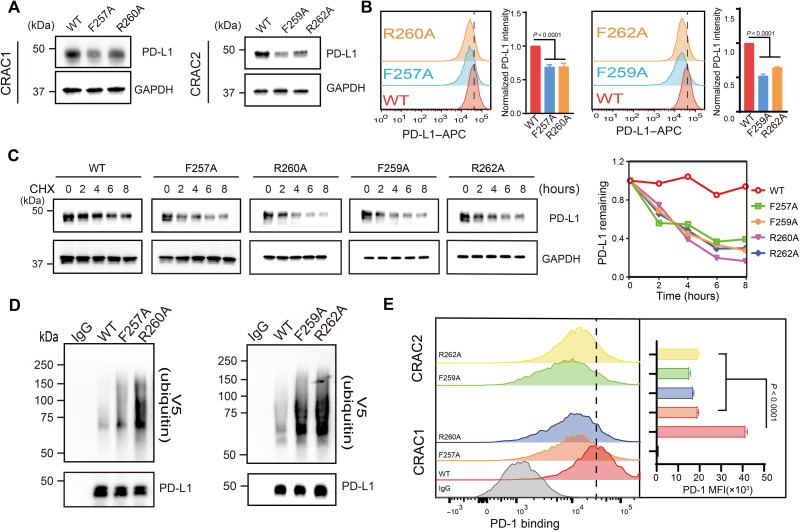
To verify the specific residues identified by NMR and MD simulations as critical to cholesterol binding, we used alanine scanning mutagenesis to replace each residue in the two CRAC motifs. Four single mutants (PD-L1-TCF257A, PD-L1-TCF259A, PD-L1-TCR260A, and PD-L1-TCR262A) were examined with cholesterol titrations. 2D TROSY spectra showed that the addition of cholesterol to PD-L1-TCF259A and PD-L1-TCR262A mutants induced few chemical shift changes, and only small chemical shift perturbations were observed in the NMR spectra of PD-L1-TCF257A and PD-L1-TCR260A mutants with the addition of cholesterol (fig. S8). These results indicated that the mutations disrupted cholesterol binding to PD-L1-TC. Additional mutants were assayed to examine PD-L1 localization: human embryonic kidney (HEK) 293FT stable cells expressing enhanced green fluorescent protein fused to PD-L1-TCWT, CRAC1 motif double mutants (PD-L1-TCF257A/R260A), CRAC2 motif double mutants (PD-L1-TCF259A/R262A), and a combined double motif (CRAC1 and CRAC2) quadruple mutant (PD-L1-TC4M). In CRAC1 and CRAC2 single motif mutants (PD-L1-TCF257A/R260A and PD-L1-TCF259A/R262A), cell surface levels of PD-L1-TC were comparable to those of cells expressing the wild-type PD-L1-TCWT; double motif mutant PD-L1-TC4M was not observed on the membrane (fig. S9). This indicated that PD-L1-TC could maintain normal surface levels when one cholesterol binding site was disrupted but that structural integrity on the membrane was lost when both cholesterol binding sites were destroyed.
The relationship between the CRAC motifs and the stability of PD-L1 was further validated with cell assays. The full-length wild-type PD-L1 (PD-L1WT) and PD-L1F257A, PD-L1F259A, PD-L1R260A, and PD-L1R262A mutants were individually expressed in endogenous PD-L1–knockdown RKO cells (fig. S10A), and PD-L1 protein expression was analyzed using Western blot. All of the mutants showed an obvious reduction in protein expression compared to PD-L1WT (Fig. 5A), whereas PD-L1 mRNA levels were similar in the mutants and the wild type (fig. S10B). Cell surface expression of PD-L1 (monitored by flow cytometry) also showed a more substantial decrease in the four single mutants compared to the wild type (Fig. 5B). A CHX-chase experiment demonstrated faster degradation of PD-L1 in all four mutants (Fig. 5C). Furthermore, higher ubiquitination levels in the presence of MG132 were detected for PD-L1 mutants compared to those expressing the wild-type protein (Fig. 5D). PD-L1 suppression resulting from CRAC motif mutations affected binding of PD-L1 to programmed cell death protein 1 (PD-1). When a fusion protein containing human PD-1 and the Fc fragment of immunoglobulin G (IgG) (25) was incubated with RKO cells expressing PD-L1WT or one of the four single mutants, flow cytometry revealed notably decreased mean fluorescence intensity (MFI) of the mutants. This indicated that the reduction in PD-L1 membrane expression of CRAC mutants led to a substantial decrease in PD-1 binding (Fig. 5E). Collectively, these results suggested that CRAC motifs are critical for the interaction between PD-L1 and cholesterol and, consequently, for PD-1 binding to PD-L1.
Fig. 5. CRAC motifs are critical for the interaction between PD-L1 and cholesterol.
(A) Cellular levels of exogenous PD-L1WT, PD-L1F257A, PD-L1F259A, PD-L1R260A, and PD-L1R262A in endogenous PD-L1–knockdown RKO stable cells were analyzed using Western blot. (B) Flow cytometric analysis of membrane PD-L1WT, PD-L1F257A, PD-L1F259A, PD-L1R260A, and PD-L1R262A in endogenous PD-L1–knockdown RKO stable cells. Bar graphs depict the fold change relative to the control; values shown are the means ± SD from four independent experiments (n = 4). Statistical differences were determined with one-way ANOVA. (C) CHX-chase assay showing the degradation of PD-L1WT and mutants. RKO stable cells expressing PD-L1WT, PD-L1F257A, PD-L1F259A, PD-L1R260A, and PD-L1R262A were treated with 50 μM CHX for 2, 4, 6, or 8 hours, and then, PD-L1 was detected by Western blot (shown on the left). The intensity of relative PD-L1 protein (PD-L1 remaining) was quantified using ImageJ analysis (shown on the right). Two independent experiments were performed with similar results. (D) Ubiquitination levels of wild-type PD-L1 (PD-L1WT) and the mutant proteins PD-L1F257A, PD-L1F259A, PD-L1R260A, and PD-L1R262A in HEK293FT cells. Ubiquitination levels were measured with V5 immunoblotting after immunoprecipitation with anti–PD-L1 antibody. Two independent experiments were performed with similar results. (E) Flow cytometry detection of PD-1 binding in RKO stable cells expressing PD-L1WT, PD-L1F257A, PD-L1F259A, PD-L1R260A, or PD-L1R262A. The y axis indicates PD-1 mean fluorescence intensity (MFI). Data are shown as the means ± SD from three independent experiments (n = 3). Statistical differences were determined with one-way ANOVA.
DISCUSSION
Cholesterol appears to play a critical role in the prevention and treatment of cancer and has therefore attracted increasing attention in cancer research. Accumulated evidence indicates that cholesterol affects structure, dynamics, and functions of cell membrane proteins (36, 37). Despite numerous examples of cholesterol-mediated regulation, the underlying mechanisms have long remained enigmatic because of the small size, complex dynamics, and poor solubility of cholesterol. NMR spectroscopy is a versatile tool commonly used to fill gaps in the understanding of interactions between lipids and membrane proteins. We previously applied NMR techniques to demonstrate that acidic phospholipids regulate PD-L1 stability by attracting basic residues in the juxtamembrane region and inhibiting downstream degradation (38). Here, we have provided visual and biochemical evidence that cholesterol itself stabilizes PD-L1 on the cell membrane surface. We further identified direct binding of cholesterol to PD-L1 transmembrane region, which plays a crucial role in PD-L1 stability. Two CRAC motifs in human PD-L1 were revealed to bind with cholesterol; mutating CRAC1 or CRAC2 motif disrupted the binding of cholesterol in PD-L1 and enhanced PD-L1 degradation. Sequence alignment analysis showed that the CRAC1 motif in human is not conserved among homologs; F257 is replaced by valine or alanine, and R260 is replaced by cysteine or tyrosine in other species (fig. S1). In contrast, the CRAC2 motif is highly conserved with the phenylalanine residues binding cholesterol rings through π-π interactions and arginine or lysine residues binding the –OH group of cholesterol through hydrogen bonding. This indicates that binding one cholesterol molecule is enough to stabilize PD-L1 on the plasma membranes in most species. The CRAC2 motif may be the critical cholesterol binding motif in regulating PD-L1 stability across species, whereas the extra CRAC1 cholesterol binding site facilitates stabilization of human PD-L1 specifically. The stoichiometry of cholesterol binding to human PD-L1 could be different from that of other homologs.
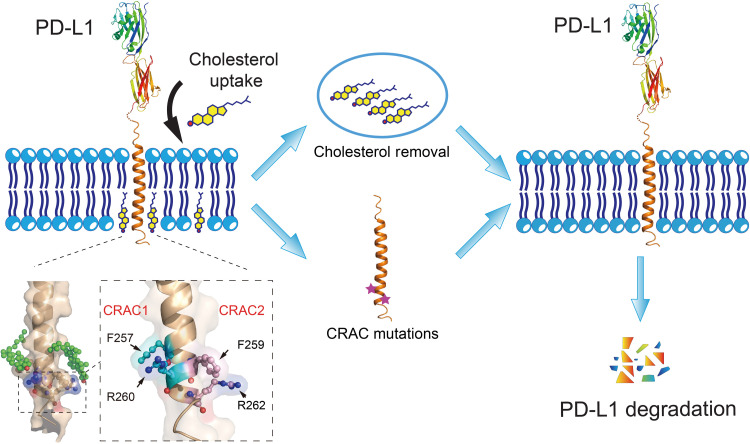
Typically, it is very difficult to determine the stoichiometry of cholesterol to membrane proteins because of the strong partition of cholesterol in the membrane system of the protein samples. The small size and high dynamics of PD-L1 further impede the determination of how many cholesterol molecules bind to this single-transmembrane protein using x-ray crystallography or cryo–electron microscopy. Nevertheless, our NMR titration data indicated positively cooperative binding between PD-L1 and cholesterol that PD-L1-TC can bind more than one molecule of cholesterol (Fig. 3G). MD simulations of PD-L1 with two cholesterol molecules also showed a good stability, and the interactions between CARC1 and CRAC2 on PD-L1 and cholesterol could coexist. These results suggest that human PD-L1 has the possibility to bind two cholesterol molecules at the two opposite CRAC motifs simultaneously, forming a sandwich-like architecture (Fig. 6). Such a unique disposition of two cholesterol molecules in PD-L1 plays an important role in enhancing PD-L1 stability, and thus facilitating cancer cells to evade immune surveillance.
Fig. 6. Schematic diagram depicting the effect of cholesterol on PD-L1 stability.
Cholesterol can directly bind to the transmembrane domain of PD-L1 at two CRAC motifs, stabilizing PD-L1 on the cell membrane. Reducing cholesterol levels with a 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase inhibitor (simvastatin) or cholesterol depletion reagent (MCD) decreases levels of membrane-bound PD-L1 by promoting PD-L1 ubiquitination and degradation. Mutations in the CRAC motifs have similar effects, reducing levels of PD-L1 on the cell surface. The magnified region shows key residues (CRAC1: F257/R260; CRAC2: F259/R262) in the two cholesterol-binding sites of PD-L1-TC.
Our results further demonstrated that disruption of the PD-L1–cholesterol interaction by mutating the CRAC motifs decreased cellular levels of PD-L1. Multiple pathways are involved in regulating PD-L1 levels, including ubiquitination-dependent proteosomal (39) or lysosomal degradation (25) and palmitoylation by palmitoyltransferase DHHC3 (40), but it is not clear which pathway is affected by cholesterol. Our data show that the PD-L1 mutants have increased levels of ubiquitination; it is possible that cholesterol stabilizes PD-L1 in the membrane, inhibiting downstream ubiquitination-dependent degradation. However, we cannot exclude the possibility that cholesterol may enhance the palmitoylation-mediated stabilization of membrane-bound PD-L1. The reduction of PD-L1 cellular levels may also be a combined outcome of disrupting PD-L1–cholesterol interactions in multiple pathways, including enhancing PD-L1 degradation and impairing DHHC3 palmitoylation. Further experiments are needed to investigate whether or how each of these pathways is affected by cholesterol.
Lim et al. recently reported that statins decrease PD-L1 expression in cancer cells by inhibiting protein kinase B (AKT) and β-catenin signaling (41). Another study revealed that simvastatin inhibits PD-L1 expression by suppressing expression of a long noncoding RNA, SNHG29, thus promoting antitumor immunity (42). Both studies showed indirect effects of statins that are more complex than simply reducing cholesterol in cancer cells; instead, they cause changes in the activities of other PD-L1–related factors, which in turn down-regulate PD-L1 expression. In contrast, the present study unveiled a direct effect of cholesterol on PD-L1. In addition to using simvastatin as a cholesterol-lowering drug, we treated cells with MCD to reduce the cholesterol levels in the membrane; in response, the cellular level of PD-L1 decreased (Fig. 1, B and C). Moreover, PD-L1 levels slightly increased when cholesterol was added to RKO cells (Fig. 1A) and remained steady in the presence of cholesterol when a stimulator of PD-L1 expression (IFN-γ) was removed (fig. S3D) in K562 cells. Together, our results shed light on a unique role of cholesterol in stabilizing PD-L1 on cancer cell membranes.
Anti–PD-L1 drugs have achieved great clinical success in treating a wide range of cancer patients by recovering the tumor-killing activity of T cells (43, 44). However, some issues have emerged with the current PD-L1–based cancer treatments, especially increasing drug resistance in patients (45), highlighting the need for alternative antitumor strategies. Small-molecule inhibitors have been reported to sensitize patients to achieve therapeutic efficacy (46). Our observations that cholesterol can directly bind to PD-L1 and enhance PD-L1 expression in cancer cells provide a unique strategy to develop small-molecule inhibitors for removal or replacement of cholesterol, reducing PD-L1 levels and inhibiting tumor growth. Designing or screening competitive chemical analogs of cholesterol may be a complementary strategy to increase patient responses to immunotherapy. The direct involvement of cholesterol in the modulation of PD-L1 demonstrated here at the molecular level is expected to aid in development of anticancer therapies.
MATERIALS AND METHODS
Reagents and cells
Lipids and detergents (DMPC, DH6PC, and 25-DOXYL-cholesterol) were obtained from Avanti Polar Lipids (Alabaster, AL, USA). The free radical 3β-DOXYL-5α-cholestane was purchased from Sigma-Aldrich (St. Louis, MO, USA). Stable isotopes for NMR spectroscopy experiments were obtained from Cambridge Isotope Laboratories (Tewksbury, MA, USA). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) polyclonal antibody (10494-1-AP) and horseradish peroxidase (HRP)–conjugated goat anti-mouse IgG (SA00001-1) were obtained from Proteintech (Rosemont, IL, USA). Anti-human PD-L1 antibodies (ab213524), rabbit monoclonal IgG (isotype control) (ab172730), and goat anti-rabbit IgG H&L (HRP) (ab97051) were purchased from Abcam (Cambridge, UK). Anti-V5 antibodies (96025) and Alexa Fluor 647–conjugated goat anti-human IgG (A21445) were obtained from Thermo Fisher Scientific (Waltham, MA, USA). Allophycocyamin (APC) anti-human PD-L1 antibodies (329708) and APC mouse IgG2b (Isotype control) (400322) were obtained from BioLegend (San Diego, CA, USA). Mouse anti-rabbit IgG (Conformation Specific) (5127S), PD-L1 extracellular domain-specific rabbit monoclonal antibody (86744S), Alexa Fluor 488–conjugated goat anti-rabbit IgG (4412S), and Alexa Fluor 647–conjugated goat anti-rabbit IgG (4414S) were obtained from Cell Signaling Technology (Danvers, MA, USA). Recombinant human PD-1 FC chimera protein (1086-PD050) was purchased from R&D Systems (Minneapolis, MN, USA). MG132, MCD, CHX, and simvastatin were obtained from MedChem Express (Monmouth Junction, NJ, USA). Cholesterol and other biochemical reagents were purchased from Sigma-Aldrich. Escherichia coli DH5α (C2987I) and BL21 (DE3) (C2527I) were purchased from New England BioLabs (Ipswich, MA, USA). The HEK293FT cell line was a gift from L. Sun [Chinese Academy of Sciences Center for Excellence in Molecular Cell Science (CEMCS)]. The RKO cell line was a gift from J. Xu (Fudan University). All other cell lines were obtained from the Cell Bank of the Chinese Academy of Sciences.
Expression and purification of wild-type and mutant PD-L1-TC
The TC domain of human PD-L1 (residues 232 to 290), designated as PD-L1-TC, was synthesized by GenScript (Piscataway, NJ, USA). Expression constructs were created by fusing the PD-L1-TC fragment to the C terminus of the His9-TrpLE expression sequence in the pMM-LR6 vector (a gift from S. C. Blacklow, Harvard Medical School), with an added methionine between PC-L1-TC and His9-TrpLE for cleavage by cyanogen bromide (CNBr). Mutant constructs were generated using standard polymerase chain reaction (PCR) protocols and confirmed with DNA sequencing. For NMR sample preparation, transformed E. coli strain BL21 (DE3) cells were grown in M9 minimal media supplemented with Centrum adult multivitamins and stable isotopes. Cultures were grown at 37°C to an optical density at 600 nm (OD600) of 0.6 to 0.8 and then cooled to 18°C before induction with 250 μM isopropyl β-d-thiogalatopyranoside (IPTG) at 18°C overnight. For fully deuterated proteins, bacterial cultures were grown in 99.8% D2O (Sigma-Aldrich) with deuterated glucose (Cambridge Isotope Laboratories). The expressed fusion proteins were extracted and purified with nickel affinity resin (Thermo Fisher Scientific), followed by CNBr digestion in 80% formic acid. The digest was dialyzed against water to remove the majority of formic acid, lyophilized, and redissolved in 80% formic acid. The PD-L1-TC protein was further purified by reverse-phase high-performance liquid chromatography with a Zorbax 300SB-C3 PrepHT column (Agilent Technologies, Santa Clara, CA, USA) using an elution gradient from 40% (v/v) acetonitrile (ACN) with 0.1% (v/v) trifluoroacetic acid (TFA) to 100% (v/v) ACN and 0.1% (v/v) TFA. The fractions corresponding to pure PD-L1-TC peptide were identified by matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry and SDS–polyacrylamide gel electrophoresis (SDS-PAGE) analysis. All mutant proteins were expressed and purified following the same protocol.
Reconstitution of PD-L1-TC into bicelles
To reconstitute wild-type and mutant PD-L1-TC proteins in bicelles, 1 to 2 mg of purified and lyophilized protein was mixed with 10 mg of protonated or deuterated DMPC (Avanti Polar Lipids) and dissolved in hexafluoro-isopropanol (HFIP). The mixture was slowly dried to a thin film under a nitrogen stream and then lyophilized overnight. The dried film was redissolved in 0.5 ml of 25 mM MES buffer (pH 6.5) containing 60 mM protonated or deuterated DH6PC (Avanti Polar Lipids). The DMPC:DH6PC ratio was monitored with 1D NMR to verify the q value. The final NMR samples contained 0.4 to 0.5 mM PD-L1-TC (wild type or mutant), ~30 mM DMPC, ~60 mM DH6PC, 25 mM MES (pH 6.5), and 10% D2O. For all NOE experiments, proteins were reconstituted using DMPC and DH6PC with deuterated acyl chains (Avanti Polar Lipids).
Assignment of NMR resonances in bicelles
All NMR spectra were acquired at 310 K on Bruker Avance III spectrometers (600 or 900 MHz) equipped with cryogenic probes and Agilent DD2 spectrometers (800 or 700 MHz) equipped with triple-resonance cold probes. NMR data were processed using NMRPipe (47) and analyzed with XEASY (48). Sequence-specific assignments of backbone chemical shifts were performed with a suite of standard triple resonance experiments, including the TROSY version of HNCA, HN(CO)CA, HN(CA)CO, HNCO, and HNCACB on an (15N, 13C, 85% 2H)–labeled sample at an 1H frequency of 600 MHz. Side-chain assignments were completed with 15N NOESY-HSQC (49), NOESY-13C HSQC, and 13C (CT)–HSQC experiments, which also provided information for backbone assignments. 15N-edited and 13C-edited NOESY data were both acquired with 120-ms mixing times.
NMR structure calculation of PD-L1-TC into bicelles
PD-L1-TC structures were calculated using the program XPLOR-NIH (50). The structures were derived using intramonomer, backbone dihedral, and hydrogen bond restraints. The backbone dihedral restraints were determined from chemical shifts using the TALOS+ program (31). The dihedral restraints and uncertainties from the TALOS+ program were used as restraints in XPLOR calculations; the statistics for N, Cα, and C′ chemical shift–derived ϕ and ψ values were considered “good” by the TALOS+ program. Hydrogen bond restraints were also implemented for residues in the transmembrane segment. Two distance restraints were defined for each hydrogen bond: 1.8 to 2.0 Å for the HN-O distance and 2.7 to 3.0 Å for the N-O distance. A total of 200 structures were calculated, and the 15 lowest-energy structures were selected for the final structural ensemble. The assignment completeness for different nuclei was as follows: N, 92.5%; H, 94.87%; CO, 97.50%; CA, 97.50%; HA, 95.35%; methyl C, 97.30%; methyl H, 97.30%; and all other H, 71.95%.
Cholesterol titration assay
Because of the low solubility of cholesterol, different amounts of cholesterol were first dissolved in trichloromethane. These solutions were then added to 0.4 mM PD-L1-TC peptides and 30 mM DMPC dissolved in HFIP. Each mixture was dried to a thin film under a nitrogen stream, followed by overnight lyophilization. The dried thin film was dissolved in 500 μl of NMR buffer (25 mM MES, pH 6.5) containing 60 mM DH6PC. The final cholesterol concentrations of the NMR samples were 0, 0.5, 1.0, 1.5, 2.0, and 2.5 mM. For each titration experiment, a 2D TROSY-HSQC spectrum was recorded to monitor the chemical shift changes. The combined chemical shift (δcomb) difference was calculated for each residue using the following formula (51)
| (1) |
where ΔδH and ΔδN are chemical shift changes (in parts per million) in the 1H and 15N dimensions, respectively, and ωH and ωN are normalization factors (ωH = 1.00 and ωN = 0.15).
All of the cholesterol titrations for the single mutants were prepared as described above with 0.4 mM peptides and 30 mM DMPC. 2D 1H-15N TROSY-HSQC spectra were recorded in the absence or presence of 1.5 mM cholesterol. All NMR titration experiments were performed at 37°C on a 700-MHz Agilent spectrometer.
PRE titrations
DOXYL-CH and CNO titrations were performed as previously described (33). Briefly, we mixed PD-L1-TC and DMPC with spin-labeled cholesterol derivatives in each of the organic solvents, dried the solution under a nitrogen stream, lyophilized overnight to remove the organic solvent, and then dissolved the mixture in NMR buffer (25 mM MES, pH 6.5) with DH6PC. At each titration point, a 2D TROSY-HSQC spectrum was recorded to measure the residue-specific PRE, which is here defined as I/I0; I and I0 are the intensities of a peak in the presence of the paramagnetic agent DOXYL-CH or CNO at 0, 0.1, 0.2, 0.4, 0.8, 1.6, and 2.4 mM, respectively. Peak intensities were analyzed using CcpNmr (52). After the final titration point, a freshly prepared ascorbic acid stock (500 mM) in NMR buffer was added to reach a final concentration of 10 mM to recover the broadened peaks. To interpret the PRE titration data more quantitatively, we estimated the apparent dissociation constant (Kd) using Eq. 2. The PRE readouts were plotted with the experimental (1 − PRE) versus [DOXYL-CH] or [CNO] to estimate the apparent Kd
| (2) |
where PRE is the ratio of the intensity over the initial intensity (I/I0); [PCD] is the concentration of the paramagnetic cholesterol derivatives (PCD), DOXYL, and CNO; Bmax is the maximum value when the binding is saturated by DOXYL-CH and CNO; n represents Hill coefficient; and Kd is the dissociation constant.
For Gd-DOTA and 16-DSA titrations, we prepared 0.4 mM 15N-labeled PD-L1-TC reconstituted in bicelles. The water-soluble paramagnetic agent Gd-DOTA was titrated into the 500-μl bicelle sample at 0, 0.5, 1, 2, 4, 8, 10, 15, and 20 mM, respectively. The lipophilic paramagnetic agent 16-DSA was titrated into the 500-μl bicelle sample at 2 mM. At each titration point, a 2D 15N TROSY-HSQC spectrum was acquired using a 700-MHz Agilent spectrometer. Peak intensities in the presence (I) and absence (I0) of the paramagnetic agent from the NMR spectra were analyzed using CcpNmr (52). The residue-specific PREamp was derived by fitting the peak intensity decay as a function of the paramagnetic agent. For individual peaks, Origin was used to fit I/I0 versus paramagnetic agent concentrations to the following equation (Eq. 3)
| (3) |
where [PA] is the concentration of the paramagnetic agent (Gd-DOTA), and τ is the decay constant.
Generation of PD-L1–knockdown RKO stable cells
Using pLKD-CMV-G&PR-U6-shPD-L1 and pLVX-hPD-L1-IRES expression constructs, we established an RKO stable cell line with endogenous PD-L1 knocked down in which to express PD-L1WT, PD-L1F257A, PD-L1F259A, PD-L1R260A, and PD-L1R262A. To package lentivirus, the pLKD-CMV-G&PR-U6-shPD-L1 and pLVX-hPD-L1-IRES constructs were transfected into HEK293FT cells using two packaging plasmids (psPAX2 and pMD2.G) with Lipofectamine 2000 reagent (Invitrogen, Waltham, MA, USA). The medium was changed at 8 hours after transfection, and virus-containing supernatants were collected at 24 and 48 hours after transfection. The centrifuged supernatant was filtered using a 0.45-μm filter. RKO cells at 50% confluency were incubated in lentivirus-containing medium with polybrene. After infection, double-positive cells were collected with the BD Influx cell sorter. The sequences of short hairpin RNAs used to knock down PD-L1 were as follows: nontargeting (NC), 5′-CCTAAGGTTAAGTCGCCCTCG-3′; sh1, 5′-GCATTTGCTGAACGCATTT-3′; sh2, 5′-CCGCTGCATGATCAGCTAT-3′; and sh3, 5′-CCAGCACACTGAGAATCAA-3′. Cells expressing sh3 showed the best knockdown efficiency and were therefore selected for all experiments using PD-L1–knockdown RKO stable cells.
Quantitative real-time PCR analysis
Total RNA was extracted using TRIzol reagent (Invitrogen); 1 μg of total RNA was reverse transcribed using the Evo M-MLV RT Kit with genomic DNA Clean (AG11705, Accurate Biotechnology, Changsha, Hunan, CN). Reverse transcription quantitative (RT-q) PCR was performed using a ViiA 7 Real-Time PCR System (Applied Biosystems, Waltham, MA, USA) with SYBR Green Premix Pro Taq HS qPCR Kit (AG11701, Accurate Biotechnology). Gene expression was calculated using the 2−∆∆Ct method (53) and presented as fold change. ∆∆Ct is the difference between the ∆Ct values of the experimental sample and the control sample; ∆Ct is the difference in Ct values for the gene of interest and the internal reference gene for a given sample. Human PD-L1 mRNA was normalized to expression of the GAPDH gene. Primer sequences used for RT-qPCR were as follows: human PD-L1,,5′-TGGCATTTGCTGAACGCATTT-3′ (forward) and 5′-TGCAGCCAGGTCTAATTGTTTT-3′ (reverse); and human GAPDH, 5′-AAGGTGAAGGTCGGAGTCAA-3′ (forward) and 5′-AATGAAGGGGTCATTGATGG-3′ (reverse).
Confocal imaging analysis
For immunofluorescence, RKO cells were seeded at approximately 50% confluency in a glass-bottomed dish (801001, NEST, Wuxi, Jiangsu, CN). After removal of culture medium, the dish was washed twice with phosphate-buffered saline (PBS). Cells were then fixed with 4% paraformaldehyde (PFA) (P0099, Beyotime Biotechnology, Shanghai, CN) for 15 min and then washed three times with PBS. After blocking with 5% normal goat serum (36119ES03, Yeasen Biotechnology, Shanghai, CN) in PBS for 30 min at room temperature, cells were incubated with anti–PD-L1 primary antibodies overnight at 4°C. After rinsing with PBS three times, cells were stained with Alexa Fluor 488–conjugated goat anti-rabbit IgG for 2 hours at room temperature and then washed three times with PBS for 10 min. Before imaging, cells were treated with SlowFade Diamond Antifade reagent with 4′,6-diamidino-2-phenylindole (DAPI) (S36968, Thermo Fisher Scientific) and sealed with a coverslip. Cells were observed on a Leica SP8 confocal microscope fitted with a 100× oil immersion objective.
Western blot
For Western blot analysis, cells were washed with PBS and then lysed in radioimmunoprecipitation assay (RIPA) buffer [150 mM NaCl, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM EDTA, and 50 mM tris-HCl (pH 7.4)] supplemented with 1 mM phenylmethanesulfonylfluoride (PMSF) and protease inhibitor cocktail (P8340, Sigma-Aldrich). Protein concentrations were measured with a bicinchoninic acid assay. Equal amounts of protein were resolved by SDS-PAGE, and protein transfer was performed for 30 min at 120 mA to polyvinylidene difluoride membranes (IPVH00010, Merck Millipore, Darmstadt, Germany). After blocking with 5% bovine serum albumin in TBS-T buffer (50 mM tris, 1.37 mM NaCl, and 2.7 mM KCl at pH 8.0 with 0.05% Tween 20) for 1 hour at room temperature, the membrane was incubated with antibodies (PD-L1 antibody, 1:5000; GAPDH antibody, 1:5000) overnight at 4°C. The membrane was washed three times with TBS-T buffer and then incubated with HRP-conjugated goat anti-rabbit IgG antibodies (1:5000) for 1 hour at room temperature. After washing three times with TBS-T buffer, protein bands were detected using an ECL Western blotting substrate (T7101A, Takara, San Jose, CA, USA) and analyzed using ImageJ software (54).
Flow cytometry for detection of cell surface PD-L1
For PD-L1 detection on cell surfaces, cells were collected and washed three times with PBS. Cells were then resuspended in 200 μl of PBS and incubated with APC-conjugated anti-human PD-L1 antibody (1:50) at room temperature for 30 min. After washing three times with PBS, stained cells were analyzed using flow cytometry (BD, LSRFortessa). Data were analyzed with FlowJo software.
Filipin III staining
Filipin III was dissolved in ethanol to reach a final concentration of 5 mg/ml. Cells were fixed with 4% PFA and stained with Filipin III (50 μg/ml) for 30 min. After imaging, cells were treated with SlowFade Diamond Antifade reagent. Images were collected using a Leica SP8 confocal microscope and analyzed using ImageJ software (54).
PD-L1 ubiquitination assay
To detect PD-L1 ubiquitination in HEK293FT cells, cells were transfected with PD-L1 and V5-ubiquitin constructs for 48 hours and then incubated with 20 μM MG132 for 4 hours before analysis. Cells were then lysed in RIPA lysis buffer containing 1 mM PMSF and protease inhibitor cocktail (Sigma-Aldrich). Lysates (200 μl per sample) were incubated with anti–PD-L1 antibody (1:50) overnight at 4°C and then pulled down with protein A/G mix magnetic beads (88802, Thermo Fisher Scientific) at room temperature for 1 hour. Beads were washed three times with TBS-T buffer and eluted with 2× SDS loading buffer at 95°C for 10 min. Ubiquitination of PD-L1 was measured by immunoblotting with anti-V5 antibody (R96025, Thermo Fisher Scientific).
Analysis of PD-L1 protein stability
A 30 mM cholesterol stock solution was prepared in ethanol and then added to RKO cells, which was plated in six-well plates and allowed to reach 80 to 90% confluency, to reach final cholesterol concentrations of 10, 20, and 50 μM, respectively. Pure ethanol with each corresponding volume was used as a control. Cells were collected with the trypsin-digestion method and transferred to separate tubes at 0, 2, and 4 hours. Collected cells were washed twice with PBS and then lysed by incubation with 100 μl of RIPA buffer (containing 1 mM PMSF and protease inhibitor cocktail) for 30 min on ice. Lysates were collected and centrifuged for 15 min at 18,000 rpm and 4°C. The supernatant was collected and analyzed via Western blot using anti–PD-L1 antibody. The above experimental procedure was also performed in the presence of CHX in which 50 mM CHX stock solution in dimethyl sulfoxide (DMSO) was added to RKO cells to reach a 50 μM CHX final concentration, and DMSO only was used as a control.
Analysis of membrane PD-L1 protein stability
K562 cells were pretreated with IFN-γ (50 U/ml) for 12 hours, and cell surface PD-L1 was labeled with APC–PD-L1 antibodies for 1 hour at 4°C. Cells were washed three times to remove unbound antibody and IFN-γ and then incubated at 37°C to allow PD-L1 recycling in the absence and presence of cholesterol. Samples were collected at 0, 2, 4, and 6 hours and immediately diluted in ice-cold PBS to halt further recycling. Cells were washed twice, resuspended in PBS, and then analyzed with flow cytometry in the same protocol as described above.
PD-L1/PD-1–binding assay
To measure protein-protein interactions between PD-1 and PD-L1, cells were seeded into six-well plates and allowed to reach 90 to 100% confluency. Cells were collected and transferred into centrifuge tubes, and the PD-L1WT, PD-L1F257A, PD-L1F259A, PD-L1R260A, and PD-L1R262A RKO stable cells were resuspended in PBS with recombinant human PD-1 FC chimera protein (5 μg/ml) (1:80) at room temperature for 30 min. Cells were pelleted at 800g for 3 min, and then, the supernatant was removed. Cells were washed three times with PBS for 10 min each. Then, cells were incubated with anti-human Alexa Fluor 647 dye–conjugated antibody (1:400) for 30 min at room temperature. Treated cells were gently washed three times with PBS and then analyzed with flow cytometry. Cells were protected from exposure to light. Flow cytometry data were analyzed using FlowJo software.
MD simulation
The TMD of PD-L1 with docked cholesterol was inserted into a POPC membrane and solvated with explicit solvent simple point charge water under an OPLS2005 force field (55). The periodic boundary of the water box was set to X: 20 Å, Y: 20 Å, and Z: 30 Å as an orthorhombic shape to enclose the whole complex system, which was neutralized by adding counter ions and 0.15 M NaCl. The simulation implemented Nose-Hoover temperature coupling (56) and the Martina-Tobias-Klein method (57) with isotropic scaling to control the temperature (300 K) and atmospheric pressure (1 atm) under the normal pressure and temperature ensemble class; this represents an isothermal-isobaric ensemble with constant normal pressure and lateral surface tension, especially for membrane-containing systems. The particle-mesh Ewald method (58) was used to calculate long-range electrostatic interactions with a grid spacing of 0.8 Å. Van der Waals and short-range electrostatic interactions were smoothly truncated at 9.0 Å. The trajectory frame was set to 10 ps, and the energy frequency was set to 2 ps to record dynamic changes. Before running each MD production simulation, the system was minimized and equilibrated using the default membrane relax protocol enabled in Desmond, which comprised a series of restrained minimizations and a heating process to slowly reach equilibration. After 2-ns relaxation, each system (either CRAC1 or CRAC2) was analyzed with a 100-ns production simulation with three repeats in which cholesterol and side chains of PD-L1-TC were unrestrained while the backbone atoms of the protein were under restraining force (0.1 kcal/mol) to maintain the protein conformation in correspondence to our NMR structure.
Statistical analysis
GraphPad PRISM v8 was used for statistical analyses. Unpaired two-sided Student’s t test or one-way analysis of variance (ANOVA) was performed to compare different groups of independent samples as indicated for each experiment. Immunofluorescence results were based on three independent cell culture experiments and analyzed with ImageJ v1.53n. Flow cytometry results were based on three independent experiments and analyzed with FlowJo v10. P < 0.05 was considered statistically significant. Additional information can be found in the figure legends.
Acknowledgments
We thank the staff at the Large-scale Protein Preparation System/Nuclear Magnetic Resonance System/Mass Spectrometry System/Integrated Laser Microscopy System/Molecular Imaging System at the National Facility for Protein Science in Shanghai, Zhangjiang Laboratory (NFPS, ZJLab), China, for providing technical support and assistance in data collection and analysis.
Funding: This work was supported by grants from the National Key R&D Program of China (2017YFA0504804), the National Natural Science Foundation of China (31872722), the Key Research Program of Frontier Sciences, CAS (QYZDB-SSW-SMC043), and CAS Major Science and Technology Infrastructure Open Research Projects to B.O.
Author contributions: B.O. conceived of the study. Y.C., Q.W., and L.D. prepared NMR samples. Y.C., Q.W., H.W., and M.W. performed NMR experiments and data collection. M.W., B.W., Y.C., and Q.W. analyzed the NMR data. Q.W., Y.C., L.S., T.X., R.C., and M.T. performed the biochemical and imaging experiments with the help of Y.Y. S.Wa. and S.We. performed the molecular dynamics simulations. B.O. and M.W. wrote the paper. All authors contributed to editing the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. 1H, 13C, and 15N chemical shifts have been deposited in the Biological Magnetic Resonance Bank under accession number BMRB50525, related to the PDB code 7DCV (Structure of the transmembrane domain of human PD-L1).
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
Table S1
References
REFERENCES AND NOTES
- 1.Subczynski W. K., Pasenkiewicz-Gierula M., Widomska J., Mainali L., Raguz M., High cholesterol/low cholesterol: Effects in biological membranes: A review. Cell Biochem. Biophys. 75, 369–385 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yeagle P. L., Non-covalent binding of membrane lipids to membrane proteins. Biochim. Biophys. Acta 1838, 1548–1559 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Hu J., Zhang Z., Shen W. J., Azhar S., Cellular cholesterol delivery, intracellular processing and utilization for biosynthesis of steroid hormones. Nutr. Metab. 7, 47 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grouleff J., Irudayam S. J., Skeby K. K., Schiott B., The influence of cholesterol on membrane protein structure, function, and dynamics studied by molecular dynamics simulations. Biochim. Biophys. Acta 1848, 1783–1795 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Song Y., Kenworthy A. K., Sanders C. R., Cholesterol as a co-solvent and a ligand for membrane proteins. Protein Sci. 23, 1–22 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiser B. P., Salari R., Eckenhoff R. G., Brannigan G., Computational investigation of cholesterol binding sites on mitochondrial VDAC. J. Phys. Chem. B 118, 9852–9860 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherezov V., Rosenbaum D. M., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Kuhn P., Weis W. I., Kobilka B. K., Stevens R. C., High-resolution crystal structure of an engineered human β2-adrenergic G protein–coupled receptor. Science 318, 1258–1265 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu W., Chun E., Thompson A. A., Chubukov P., Xu F., Katritch V., Han G. W., Roth C. B., Heitman L. H., IJzerman A. P., Cherezov V., Stevens R. C., Structural basis for allosteric regulation of GPCRs by sodium ions. Science 337, 232–236 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zocher M., Zhang C., Rasmussen S. G., Kobilka B. K., Muller D. J., Cholesterol increases kinetic, energetic, and mechanical stability of the human β2-adrenergic receptor. Proc. Natl. Acad. Sci. U.S.A. 109, E3463–E3472 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sejdiu B. I., Tieleman D. P., Lipid-protein interactions are a unique property and defining feature of G protein-coupled receptors. Biophys. J. 118, 1887–1900 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gahbauer S., Bockmann R. A., Membrane-mediated oligomerization of G protein coupled receptors and its implications for GPCR function. Front. Physiol. 7, 494 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen D. H., Taub D., CXCR4 function requires membrane cholesterol: Implications for HIV infection. J. Immunol. 168, 4121–4126 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Jamin N., Neumann J. M., Ostuni M. A., Vu T. K., Yao Z.-X., Murail S., Robert J.-C., Giatzakis C., Papadopoulos V., Lacapère J.-J., Characterization of the cholesterol recognition amino acid consensus sequence of the peripheral-type benzodiazepine receptor. Mol. Endocrinol. 19, 588–594 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Li H., Papadopoulos V., Peripheral-type benzodiazepine receptor function in cholesterol transport. Identification of a putative cholesterol recognition/interaction amino acid sequence and consensus pattern. Endocrinology 139, 4991–4997 (1998). [DOI] [PubMed] [Google Scholar]
- 15.Baier C. J., Fantini J., Barrantes F. J., Disclosure of cholesterol recognition motifs in transmembrane domains of the human nicotinic acetylcholine receptor. Sci. Rep. 1, 69 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanson M. A., Cherezov V., Griffith M. T., Roth C. B., Jaakola V. P., Chien E. Y., Velasquez J., Kuhn P., Stevens R. C., A specific cholesterol binding site is established by the 2.8 A structure of the human β2-adrenergic receptor. Structure 16, 897–905 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang B., Song B. L., Xu C., Cholesterol metabolism in cancer: Mechanisms and therapeutic opportunities. Nat. Metab. 2, 132–141 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Xu H., Zhou S., Tang Q., Xia H., Bi F., Cholesterol metabolism: New functions and therapeutic approaches in cancer. Biochim. Biophys. Acta Rev. Cancer 1874, 188394 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Liu W., Chakraborty B., Safi R., Kazmin D., Chang C. Y., McDonnell D. P., Dysregulated cholesterol homeostasis results in resistance to ferroptosis increasing tumorigenicity and metastasis in cancer. Nat. Commun. 12, 5103 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Bello E., Zwergel C., Mai A., Valente S., The innovative potential of statins in cancer: New targets for new therapies. Front. Chem. 8, 516 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma X., Bi E., Lu Y., Su P., Huang C., Liu L., Wang Q., Yang M., Kalady M. F., Qian J., Zhang A., Gupte A. A., Hamilton D. J., Zheng C., Yi Q., Cholesterol induces CD8+ T cell exhaustion in the tumor microenvironment. Cell Metab. 30, 143–156.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang W., Bai Y., Xiong Y., Zhang J., Chen S., Zheng X., Meng X., Li L., Wang J., Xu C., Yan C., Wang L., Chang C. C., Chang T.-Y., Zhang T., Zhou P., Song B.-L., Liu W., Sun S.-c., Liu X., Li B.-l., Xu C., Potentiating the antitumour response of CD8+ T cells by modulating cholesterol metabolism. Nature 531, 651–655 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi Y., Regulatory mechanisms of PD-L1 expression in cancer cells. Cancer Immunol. Immunother. 67, 1481–1489 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zinchuk V., Zinchuk O., Quantitative colocalization analysis of confocal fluorescence microscopy images. Curr. Protoc. Cell Biol. Chapter 4, Unit 4.19 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Wang H., Yao H., Li C., Shi H., Lan J., Li Z., Zhang Y., Liang L., Fang J. Y., Xu J., HIP1R targets PD-L1 to lysosomal degradation to alter T cell-mediated cytotoxicity. Nat. Chem. Biol. 15, 42–50 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Mohammad N., Malvi P., Meena A. S., Singh S. V., Chaube B., Vannuruswamy G., Kulkarni M. J., Bhat M. K., Cholesterol depletion by methyl-β-cyclodextrin augments tamoxifen induced cell death by enhancing its uptake in melanoma. Mol. Cancer 13, 204 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Branchi A., Fiorenza A. M., Torri A., Muzio F., Berra C., Colombo E., Dalla Valle E., Rovellini A., Sommariva D., Effects of low doses of simvastatin and atorvastatin on high-density lipoprotein cholesterol levels in patients with hypercholesterolemia. Clin. Ther. 23, 851–857 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Bellucci R., Martin A., Bommarito D., Wang K., Hansen S. H., Freeman G. J., Ritz J., Interferon-γ-induced activation of JAK1 and JAK2 suppresses tumor cell susceptibility to NK cells through upregulation of PD-L1 expression. Onco. Targets. Ther. 4, e1008824 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu Q., Piai A., Chen W., Xia K., Chou J. J., Structure determination protocol for transmembrane domain oligomers. Nat. Protoc. 14, 2483–2520 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salzmann M., Pervushin K., Wider G., Senn H., Wuthrich K., TROSY in triple-resonance experiments: New perspectives for sequential NMR assignment of large proteins. Proc. Natl. Acad. Sci. U.S.A. 95, 13585–13590 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen Y., Delaglio F., Cornilescu G., Bax A., TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piai A., Fu Q., Dev J., Chou J. J., Optimal bicelle size q for solution NMR studies of the protein transmembrane partition. Chemistry 23, 1361–1367 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu H., Cao R., Wei S., Pathan-Chhatbar S., Wen M., Wu B., Schamel W. W., Wang S., OuYang B., Cholesterol binds in a reversed orientation to TCRβ-TM in which its OH group is localized to the center of the lipid bilayer. J. Mol. Biol. 433, 167328 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Weiss J. N., The Hill equation revisited: Uses and misuses. FASEB J. 11, 835–841 (1997). [PubMed] [Google Scholar]
- 35.Eberhardt J., Santos-Martins D., Tillack A. F., Forli S., AutoDock vina 1.2.0: New docking methods, expanded force field, and Python bindings. J. Chem. Inf. Model 61, 3891–3898 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jaipuria G., Leonov A., Giller K., Vasa S. K., Jaremko L., Jaremko M., Linser R., Becker S., Zweckstetter M., Cholesterol-mediated allosteric regulation of the mitochondrial translocator protein structure. Nat. Commun. 8, 14893 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elkins M. R., Williams J. K., Gelenter M. D., Dai P., Kwon B., Sergeyev I. V., Pentelute B. L., Hong M., Cholesterol-binding site of the influenza M2 protein in lipid bilayers from solid-state NMR. Proc. Natl. Acad. Sci. U.S.A. 114, 12946–12951 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wen M., Cao Y., Wu B., Xiao T., Cao R., Wang Q., Liu X., Xue H., Yu Y., Lin J., Xu C., Xu J., OuYang B., PD-L1 degradation is regulated by electrostatic membrane association of its cytoplasmic domain. Nat. Commun. 12, 5106 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J., Bu X., Wang H., Zhu Y., Geng Y., Nihira N. T., Tan Y., Ci Y., Wu F., Dai X., Guo J., Huang Y. H., Fan C., Ren S., Sun Y., Freeman G. J., Sicinski P., Wei W., Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature 553, 91–95 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yao H., Lan J., Li C., Shi H., Brosseau J. P., Wang H., Lu H., Fang C., Zhang Y., Liang L., Zhou X., Wang C., Xue Y., Cui Y., Xu J., Inhibiting PD-L1 palmitoylation enhances T-cell immune responses against tumours. Nat. Biomed. Eng. 3, 306–317 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Lim W. J., Lee M., Oh Y., Fang X. Q., Lee S., Lim C. H., Park J., Lim J. H., Statins decrease programmed death-ligand 1 (PD-L1) by inhibiting AKT and beta-catenin signaling. Cells 10, 2488 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ni W., Mo H., Liu Y., Xu Y., Qin C., Zhou Y., Li Y., Li Y., Zhou A., Yao S., Zhou R., Huo J., Che L., Li J., Targeting cholesterol biosynthesis promotes anti-tumor immunity by inhibiting long noncoding RNA SNHG29-mediated YAP activation. Mol. Ther. 29, 2995–3010 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garon E. B., Rizvi N. A., Hui R., Leighl N., Balmanoukian A. S., Eder J. P., Patnaik A., Aggarwal C., Gubens M., Horn L., Carcereny E., Ahn M. J., Felip E., Lee J. S., Hellmann M. D., Hamid O., Goldman J. W., Soria J. C., Dolled-Filhart M., Rutledge R. Z., Zhang J., Lunceford J. K., Rangwala R., Lubiniecki G. M., Roach C., Emancipator K., Gandhi L.; KEYNOTE-001 Investigators , Pembrolizumab for the treatment of non-small-cell lung cancer. N. Engl. J. Med. 372, 2018–2028 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Brahmer J., Reckamp K. L., Baas P., Crino L., Eberhardt W. E., Poddubskaya E., Antonia S., Pluzanski A., Vokes E. E., Holgado E., Waterhouse D., Ready N., Gainor J., Aren Frontera O., Havel L., Steins M., Garassino M. C., Aerts J. G., Domine M., Paz-Ares L., Reck M., Baudelet C., Harbison C. T., Lestini B., Spigel D. R., Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N. Engl. J. Med. 373, 123–135 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nowicki T. S., Hu-Lieskovan S., Ribas A., Mechanisms of resistance to PD-1 and PD-L1 blockade. Cancer J. 24, 47–53 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu Q., Jiang L., Li S. C., He Q. J., Yang B., Cao J., Small molecule inhibitors targeting the PD-1/PD-L1 signaling pathway. Acta Pharmacol. Sin. 42, 1–9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A., NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995). [DOI] [PubMed] [Google Scholar]
- 48.Bartels C., Xia T. H., Billeter M., Güntert P., Wüthrich K., The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR 6, 1–10 (1995). [DOI] [PubMed] [Google Scholar]
- 49.Xia Y., Sze K., Zhu G., Transverse relaxation optimized 3D and 4D 15n/15N separated NOESY experiments of 15N labeled proteins. J. Biomol. NMR 18, 261–268 (2000). [DOI] [PubMed] [Google Scholar]
- 50.Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M., The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160, 65–73 (2003). [DOI] [PubMed] [Google Scholar]
- 51.OuYang B., Xie S., Berardi M. J., Zhao X., Dev J., Yu W., Sun B., Chou J. J., Unusual architecture of the p7 channel from hepatitis C virus. Nature 498, 521–525 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vranken W. F., Boucher W., Stevens T. J., Fogh R. H., Pajon A., Llinas M., Ulrich E. L., Markley J. L., Ionides J., Laue E. D., The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins 59, 687–696 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Livak K. J., Schmittgen T. D., Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 54.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jorgensen W. L., Tirado-Rives J., The OPLS [optimized potentials for liquid simulations] potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. J. Am. Chem. Soc. 110, 1657–1666 (1988). [DOI] [PubMed] [Google Scholar]
- 56.Hoover W. G., Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A Gen. Phys. 31, 1695–1697 (1985). [DOI] [PubMed] [Google Scholar]
- 57.Deng Z., Martyna G. J., Klein M. L., Structure and dynamics of bipolarons in liquid ammonia. Phys. Rev. Lett. 68, 2496–2499 (1992). [DOI] [PubMed] [Google Scholar]
- 58.Petersen H. G., Accuracy and efficiency of the particle mesh Ewald method. J. Chem. Phys. 103, 3668–3679 (1995). [Google Scholar]
- 59.Fantini J., Epand R. M., Barrantes F. J., Cholesterol-recognition motifs in membrane proteins. Adv. Exp. Med. Biol. 1135, 3–25 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Crooks G. E., Hon G., Chandonia J. M., Brenner S. E., WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10
Table S1
References