Abstract
How neurons can maintain cellular identity over an entire life span remains largely unknown. Here, we show that maintenance of identity in differentiated dopaminergic and serotonergic neurons is critically reliant on the Polycomb repressive complex 2 (PRC2). Deletion of the obligate PRC2 component, Eed, in these neurons resulted in global loss of H3K27me3, followed by a gradual activation of genes harboring both H3K27me3 and H3K9me3 modifications. Notably, H3K9me3 was lost at these PRC2 targets before gene activation. Neuronal survival was not compromised; instead, there was a reduction in subtype-specific gene expression and a progressive impairment of dopaminergic and serotonergic neuronal function, leading to behavioral deficits characteristic of Parkinson’s disease and anxiety. Single-cell analysis revealed subtype-specific vulnerability to loss of PRC2 repression in dopamine neurons of the substantia nigra. Our study reveals that a PRC2-dependent nonpermissive chromatin state is essential to maintain the subtype identity and function of dopaminergic and serotonergic neurons.
Epigenetic control of transcriptional stability maintains identity and function in dopaminergic and serotonergic neurons.
INTRODUCTION
The brain contains a large number of different neuronal subtypes that maintain their distinct cellular identities over several decades despite continuous environmental fluctuation. Instructive information, provided by transcription factors controlling cell type–specific gene programs, is crucial for maintenance of neuronal identity and function. Many of these factors, denominated terminal selectors, are also involved in subtype specification during development and continue to be expressed in the differentiated neurons where they govern the expression of effector genes encoding factors necessary for neuronal function (e.g., receptors, transporters, ion channels, and neurotransmitter-synthesizing enzymes) (1). Autoregulation by terminal selectors provides further stability, enabling maintenance of identity despite a fluctuating environment. Aberrant expression of genes governing alternative identities or of regulators of progenitor features potentially threatens the transcriptional integrity of terminal selector–dependent endogenous gene programs. Hence, besides instructive regulation, which arguably defines neuronal identities, there is also a need to maintain silencing of transcriptional programs promoting other cellular fates or progenitor properties (1, 2). However, the mechanisms governing permanent repression of aberrant transcription in mature neurons are not well understood.
Within this context, it is key to understand the mechanisms regulating chromatin structure, e.g., dynamic modification of histones and how this is coupled to changes in gene expression. A prominent example of how chromatin-associated gene silencing contributes to maintained cellular identity is the sustained repression of Hox genes during segmentation of the Drosophila melanogaster embryo, which is dependent on Polycomb group proteins (3). The Polycomb repressive complex 2 (PRC2) maintains established cell type–specific gene repression through the deposition of the repressive histone modification H3K27me3 in promoter regions of silenced genes, thus facilitating chromatin compaction (4–6). PRC2 is required for proper differentiation during the development of the vertebrate central nervous system (CNS) (7, 8), but except for medium spiny neurons (MSNs), whether PRC2 is an essential component for maintaining differentiated neuronal identity remains unclear. H3K27me3 is also found in domains that harbor the H3K4me3 modification associated with active transcription, and these bivalent domains have been proposed to be silent but “poised” for rapid activation at a later developmental stage (9, 10). Thus, H3K4me3/H3K27me3 bivalency potentially represents a more relaxed chromatin state, amenable to rapid activation.
Efforts have been directed to understand how deviant gene regulation is involved in neurodegenerative and psychiatric disorders (11). The complex etiology, often lacking a distinct and identifiable genetic component, of many such pathological conditions suggests that alterations of the epigenome contribute to the disease (11). Changes in PRC2 activity and in H3K27me3 levels and distribution have been associated with neurodegenerative disease (12–14) and mood disorders (15); however, whether or how these processes contribute to disease remains poorly understood.
To understand the fundamental molecular mechanisms underlying the role of gene repression for maintenance of neuronal identity and function, we have focused on the well-defined midbrain dopaminergic (mDA) and serotonergic (5HT) neuronal subpopulations as model systems. These monoaminergic neurons are involved in several psychiatric disorders and drug addiction. In addition, degeneration of mDA neurons in the substantia nigra pars compacta (SNpc) is a hallmark of Parkinson’s disease (PD), and dysregulated 5HT function is causing depression and anxiety and contributes to l-dopa dyskinesia in patients with PD (16). Hence, it is also relevant from a clinical perspective to understand the basal mechanisms important for maintaining intact mDA and 5HT neuronal identity and function. Notably, in a preclinical model of PD, mDA neurons treated with the neurotoxin 6-hydroxydopamine (6-OHDA) exhibited substantial reduction in H3K27me3 levels (17). Besides the well-known toxic effects of 6-OHDA on mitochondria resulting in increased free radicals, this also couples exposure to PD-associated cellular stressors to the induction of a more relaxed chromatin state and potential derepression of aberrant non-mDA genes. Furthermore, exposure to l-dopa in a mouse model of PD leads to loss of Polycomb-mediated repression (12). In differentiated MSN, it has been shown that loss of PRC2 activity induced expression of cell death–promoting genes, which resulted in neuronal loss, leading to neurodegeneration (6). In these neurons, derepression primarily affected bivalent poised H3K4me3/H3K27me3 genes.
H3K9me3 is a histone modification involved in the establishment and maintenance of heterochromatin (18). Primarily associated with constitutive heterochromatin at centromeres and telomeres, H3K9me3 has also been shown to regulate facultative heterochromatin, provide an obstacle for cell reprogramming, and be required for the establishment and maintenance of cellular identity (18, 19). Furthermore, H3K9me3 has been shown to be disrupted along with H3K27me3 upon 6-OHDA treatment (17). In a recent study, we generated global integrated maps of transcription and histone modifications (H3K4me3, H3K27me3, and H3K9me3) of transitory and stable cellular states in mDA and 5HT neurons as well as their progenitors (20). This study also showed that in a mouse model of PD, there was a substantial enrichment of H3K27me3 targets among the up-regulated genes, implying a role for PRC2 in the transcriptional response to PD-associated cellular stressors.
To address the functional role of Polycomb-mediated gene silencing for postmitotic mDA and 5HT neuronal identity, we have conditionally deleted the obligate PRC2 member Embyonic ectoderm development (Eed), which is necessary for PRC2 binding to H3K27me3 (21) and subsequent propagation of the modification, in both these neuronal subtypes. Together, our study reveals a common logic in mDA and 5HT neurons wherein PRC2 activity is required for the maintenance of subtype-specific gene patterns and neuronal function; consequently, loss of PRC2 function generates phenotypes that mirror key aspects of PD and mood disorders, without compromising neuronal survival. In addition, our single-cell analysis reveals a specific vulnerability in mDA neurons of the SNpc to reduced H3K27me3 levels.
RESULTS
Conditional deletion of Eed in differentiated mDA neurons
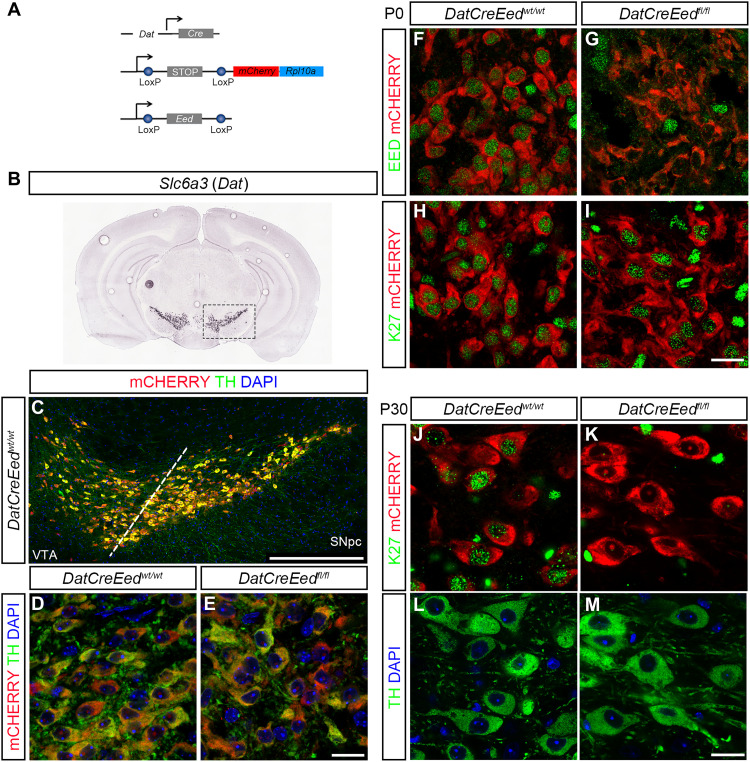
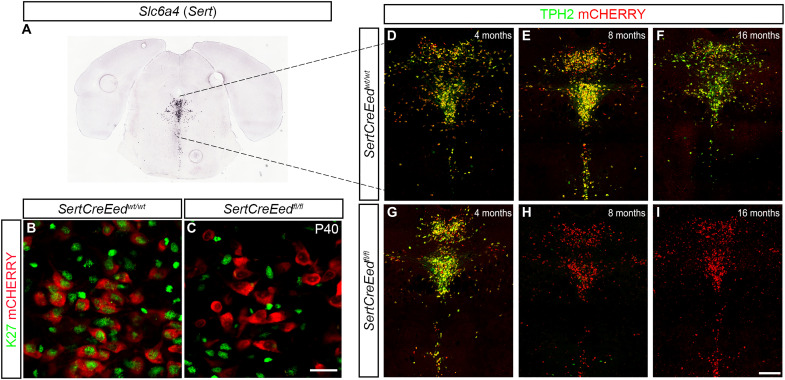
To address the role of PRC2-mediated repression in differentiated mDA neurons, we generated a compound mouse mutant by crossing mice carrying floxed alleles for a ribosomal protein fused to mCherry (RPL10a-mCherry). The Rpl10a-mCherryflox/flox mouse line was crossed with mice carrying a floxed Eed allele (22) and lastly with DatCre (23) (Fig. 1A). This enabled the deletion of Eed and the expression of mCherry in postmitotic mDA neurons under the control of Dat (Slc6a3) expression, which is first detected in the midbrain at around embryonic day 13.5 (E13.5). In this study, the full compound mutants will be denominated DatCreEedfl/fl. Pups from the mutants were born at expected Mendelian ratios and were indistinguishable from wild-type (WT) littermates. Immunostaining of sections showed that the mCHERRY reporter colocalized with the rate-limiting enzyme for dopamine synthesis, tyrosine hydroxylase (TH), in both the ventral tegmental area (VTA) and SNpc of both WT and mutant mice (Fig. 1, B to E). At E13.5, EED protein is readily detectable in TH+ cells in both WTs and mutants (fig. S1, A and B).
Fig. 1. Deletion of Eed leads to progressive loss of H3K27me3 in differentiated mDA neurons.
(A) Schematic representation of the three constructs used to generate the mutant mice. (B) In situ hybridization for Slc6a3 (Dat) taken from the Allen Brain Atlas (mouse.brain-map.org, image credit: Allen Institute). (C) Immunostaining of the ventral midbrain in DatCreEedwt/wt as indicated by the box in (B), showing overlap between mCHERRY and TH. DAPI, 4′,6-diamidino-2-phenylindole. (D and E) Immunostaining at ×63 magnification showing overlap of TH and mCHERRY in the SNpc of DatCreEedwt/wt mice (D) and DatCreEedfl/f mice (E). (F) mDA neurons double positive for EED and mCHERRY immunostaining at P0 in DatCreEedwt/wt SNpc. (G) Loss of EED in mCHERRY+ cells at P0 in DatCreEedft/fl SNpc. (H) mDA neurons double positive for H3K27me3 and mCHERRY immunostaining at P0 in DatCreEedwt/wt SNpc. (I) mDA neurons double positive for H3K27me3 and mCHERRY immunostaining at P0 in DatCreEedfl/fl SNpc. (J) mDA neurons double positive for H3K27me3 and mCHERRY immunostaining at P30 in DatCreEedwt/wt SNpc. (K) Loss of H3K27me3 in mCHERRY+ cells at P30 in DatCreEedfl/fl SNpc. (L) Staining with TH and DAPI of the DatCreEedwt/wt cells from (J). (M) Staining with TH and DAPI of the DatCreEedfl/fl cells from (K). Scale bars, 20 μm (E, I, and M).
Intact PRC2 function is required for long-term maintenance of H3K27me3
To understand whether induction of Cre resulted in deletion of EED protein at birth, we performed immunostaining of midbrains from newborn pups at postnatal day 0 (P0) with an antibody specific for EED. In DatCreEedwt/wt mice, there was clear nuclear EED immunoreactivity in mCHERRY+ mDA neurons (Fig. 1F), whereas mCHERRY+ cells in the DatCreEedfl/fl mutants exhibited a lack of nuclear EED (Fig. 1G). To investigate whether the absence of EED protein in the mutants also resulted in loss of H3K27me3, we performed immunostaining with an H3K27me3-specific antibody. Although EED was lost in DatCreEedfl/fl, H3K27me3 was retained (Fig. 1, H and I), showing that the H3K27me3 modification is remarkably stable in postmitotic mDA neurons. To further gauge whether long-term H3K27me3 stability depends on intact PRC2 function, we stained the midbrain sections of juvenile mice, at P30, which revealed a virtually complete lack of H3K27me3 immunoreactivity in DatCreEedfl/fl mice (Fig. 1, J and K). Notably, immunoreactivity levels of TH were retained in both genotypes (Fig. 1, L and M). We could not detect any differences in the nuclear localization of mCHERRY between WT and mutant mDA neurons (fig. S1C and movies S1 and S2).
Progressive activation of silent non-mDA PRC2 targets and repression of mDA identity genes
To investigate the effects on global distribution of H3K27me3 and the possible consequences of altered H3K27me3 levels for gene expression, we dissected out midbrains from which we dissociated nuclei and used fluorescence-activated cell sorting (FACS) to isolate mCHERRY+ nuclei from DatCreEedwt/wt and DatCreEedfl/fl mice at 4 and 8 months of age. Sorted nuclei were collected in batches of 1000. Batches were used to generate chromatin immunoprecipitation (ChIP) libraries not only for H3K27me3 (K27) but also for the permissive modification H3K4me3 (K4) and the facultative heterochromatin–associated modification H3K9me3 (K9). In addition, one batch per mouse brain was used to generate libraries for bulk RNA sequencing (RNA-seq).
Initially, we examined the presence of K27 in the promoter region ±10 kb around transcription start sites (TSSs) together with expression levels in WT cells, which revealed an inverse correlation (Fig. 2A). We then determined how distinct chromatin states in the same promoter region correlated with expression levels in WT cells at 4 months as described in our previous study (20). The results show that among the eight possible different states, the genes containing K4 exhibit the highest expression levels. Conversely, chromatin states harboring K27 and/or K9 without K4 exhibit the lowest levels of expression (fig. S2A).
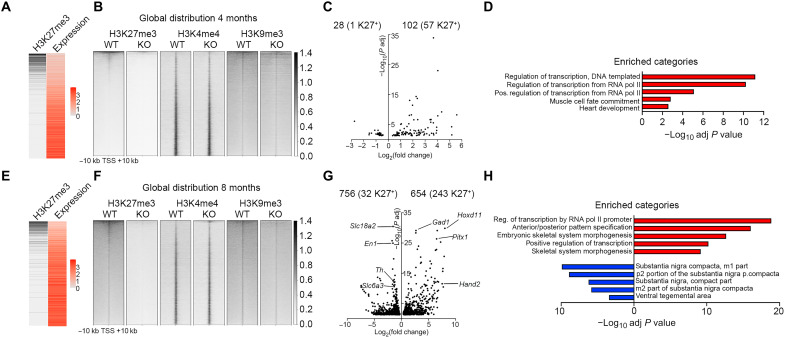
Fig. 2. Loss of Eed results in progressive up-regulation of PRC2 targets and reduced expression of mDA neuronal genes.
(A) Heatmaps showing inverse correlation between H3K27me3 and expression levels at 4 months in DatCreEedwt/wt. (B) Genome-wide abundance of histone modifications at 4 months in DatCreEedwt/wt (WT) and DatCreEedfl/fl [knockout (KO)] in 20-kb regions centered on TSS of individual genes ranked by H3K27me3 abundance in WT. (C) Volcano plot showing DEGs at 4 months in isolated mCHERRY+ nuclei from KO ventral midbrain. Numbers of up- and down-regulated genes are indicated above the plot, with the number of H3K27me3+ genes in WT at 4 months within brackets. (D) Enriched categories (GO biological process) for up-regulated genes are characterized by activation of transcription and early developmental non-neuronal processes. (E) Heatmaps showing inverse correlation between H3K27me3 and expression levels at 8 months in WT. Heatmap for H3K27me3 as in (A), with promoter chromatin states at 8 months. (F) Genome-wide abundance of histone modifications at 8 months in WT and KO as in (B). (G) Volcano plot showing DEGs at 8 months in isolated mCHERRY+ nuclei from KO ventral midbrain. Numbers of up- and down-regulated genes are indicated above the plot, with the number of H3K27me3+ genes in WT at 8 months within brackets. Examples of up-regulated genes that are PRC2 targets in WT mDA neurons at 8 months are labeled on the right side of the plot. Examples of down-regulated mDA identity genes are labeled on the left side of the plot. (H) Enriched categories for up-regulated genes (red) (GO biological process) and down-regulated genes (blue) (Up in Allen Brain Atlas, as calculated by Enrichr).
We then investigated the consequences of Eed deletion for the distribution of K27, K4, and K9 at 4 and 8 months. In 4-month mutant mDA neurons, there was a substantial loss of K27, reducing the number of detected K27+ genes by 75% (2973 in WT compared to 751 in mutants) (Fig. 2B and table S1). To understand the consequence of such a major loss of K27 for gene expression, we integrated the expression data with the chromatin analysis. Despite the substantial global loss of K27, only 102 genes were significantly up-regulated (adjusted P < 0.05) (Fig. 2C and table S2), and of those, 57 genes were K27 targets (Fig. 2C and table S2), a 4.7-fold enrichment (Fisher’s exact test, P = 5 × 10−27) over the expected ratio. Of the 57 up-regulated K27 targets, 22 were determined as lacking K27 at ±10 kb of TSS in mutant nuclei, which represents a 2.4-fold enrichment among the up-regulated genes (Fisher’s exact test, P = 9 × 10−5). The 35 up-regulated genes that were still determined as K27+ in mutants do retain K27 around the TSS, albeit at low levels (e.g., Foxg1, Phox2b, and Hand2). Several quintessential PRC2 targets such as the Hoxd cluster did not exhibit increased expression in the mutants. Despite a reduction in K27 enrichment in such genes, a significant proportion of the modification remained (fig. S2B), underscoring the resilience of this modification in mDA neurons lacking PRC2 activity. The up-regulated genes exhibited strong enrichment of gene ontology (GO) categories associated with regulation of transcription and early developmental processes [GO Biological Process 2021, as calculated by Enrichr (24, 25)] (Fig. 2D). Concomitantly, loss of K27 also resulted in significant (adjusted P < 0.05) down-regulation of 28 genes, with 1 of them (Myo7a) being a K27 target (Fig. 2C).
ChIP sequencing (ChIP-seq) and RNA-seq analysis in nuclei sorted from 8-month-old animals also showed an inverse correlation between K27 and expression level in WT nuclei (Fig. 2E). Furthermore, despite a slightly higher background in the K27 ChIP-seq than at 4 months (Fig. 2, B and F), it revealed a complete loss of genes determined as K27+ in the DatCreEedfl/fl mutants (Fig. 2F and table S1). Inspection of differentially expressed genes (DEGs) showed that 654 genes were significantly up-regulated (Fig. 2G), with 55 of them up-regulated at 4 months and 243 of 654 carrying the K27 modification (Fig. 2G and table S2), representing a 4.3-fold enrichment over expected (Fisher’s exact test, P = 5 × 10−95). Up-regulated PRC2 targets included several members of the Hox family; transcription factors involved in determining other cell fates during development, e.g., Pitx1, Gata2, and Foxd3; stem cell factors such as Pax6; genes typically expressed in other neuronal types, e.g., Gad1 and Gad2; and cell cycle regulators including Ccnd1, Ccnd2, and Cdnkn2a (table S2). In contrast to the 4-month-old mutant mDA neurons, several members of the Hoxd clusters were up-regulated at 8 months. This was reflected by the acquisition of H3K4me3 combined with the loss of K27 and K9 (fig. S2C). Moreover, in the 8-month-old mutant mDA neurons, there were 756 significantly down-regulated genes, with 32 of them carrying the K27 modification (Fig. 2G and table S2), representing a twofold reduction compared to what would be expected by chance (Fisher’s exact test, P = 7 × 10−5). Among the down-regulated genes, several transcription factors critical for mDA neuronal function were present, e.g., En1/2, Nr4a2 (Nurr1), Lmo3, Pitx3, and Pou3f2 (table S2). Up-regulated genes exhibited strong enrichment of GO categories associated with regulation of transcription and early developmental processes, whereas down-regulated genes were enriched for ventral midbrain categories (GO Biological Process 2021 and Allen Brain Atlas Up, as calculated by Enrichr) (Fig. 2H).
Combined H3K9me3/H3K27me3 is associated with higher probability of derepression upon loss of PRC2 activity
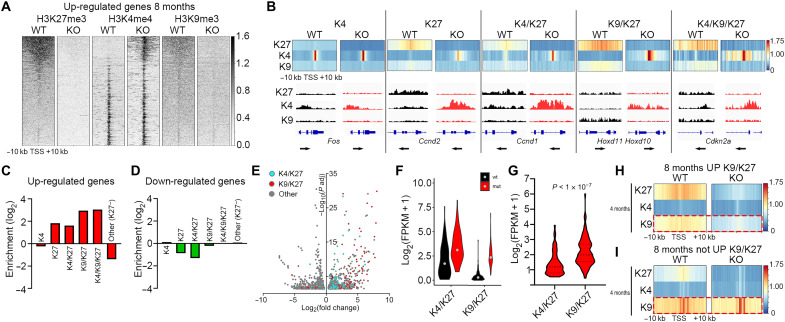
Inspection of the H3K27me3, H3K4me3, and H3K9me3 modifications ±10 kb around the TSSs of the genes up-regulated in the mutants revealed a pronounced increase in K4 and loss of K9 (Fig. 3A). To further understand whether any specific chromatin state in WT mDA neurons would predispose for derepression, we inspected the distribution of K4, K27, K4/K27, K9/K27, and K4/K9/K27 in WT mDA nuclei and correlated them with the DEGs. Among the up-regulated genes, there was a significant enrichment in all states containing K27 but not in the K4 only, where there rather was a depletion (Fig. 3, B and C). Closer inspection revealed that the most enriched chromatin states of up-regulated genes in the mutants were those that contained both HK27me3 and HK9me3 (K4/K9/K27 and K9/K27) in WT cells. Compared to genes with a poised, bivalent K4/K27 state in the WT, which had a 3.1-fold enrichment, the K9/K27 state exhibited 7.8-fold and the K4/K9/K27 state 8.3-fold enrichment (Fig. 3C). Notably, for down-regulated genes, there was no enrichment but rather a depletion of chromatin states containing K27 (Fig. 3D). In addition, when we compared the relative fold change and statistical significance of the DEGs in the 8-month mutants, there was a clear difference between K4/K27 and K9/K27 genes, with K9/K27 genes generally exhibiting higher fold increase and lower adjusted P values (Fig. 3E). This was also true for the 4-month mutants (fig. S2D). To understand whether this difference was a consequence of K9/K27 genes being derepressed from absolute expression levels close or equal to zero, we investigated the absolute expression levels of K4/K27 and K9/K27 genes in 8-month-old WT and mutants. This revealed that the WT expression levels of K9/K27 genes were substantially lower than those of K4/K27 genes (Fig. 3F). However, the difference in absolute increase in the mutants was significantly larger in the K9/K27 genes (Fig. 3G). Thus, this analysis revealed that the presence of the additional heterochromatin modification K9 actually increases both the probability to activate repressed K27 genes and the magnitude of increased expression. Notably, GO analysis showed that K9/K27 genes are enriched for categories typically including regulation of transcription and early developmental events such as embryonic organ morphogenesis, whereas K4/K27 genes include GO categories associated with regulation of extracellular matrix, neuronal differentiation, and proliferation (fig. S2E). The stronger enrichment of K9/K27 genes over K4/K27 genes prompted us to inspect the chromatin states at 4 months for genes that were up-regulated at 8 months but not at 4 months. This analysis revealed that reduction of K9 occurs before derepression of expression (Fig. 3H). Hence, the early loss of K9 is not a mere consequence of activated transcription. Loss of K27 could potentially influence stability of the K9 modification. However, when inspecting the 4-month mutant chromatin states for WT K9/K27 genes that were not up-regulated at 8 months, no loss of K9 was detected despite loss of K27 (Fig. 3I). Hence, loss of K27 could be required but is not sufficient to induce loss of K9 at these loci. GO analysis of the K9/K27 genes at 4 months that were up-regulated at 8 months showed a strong enrichment for positive regulation of transcription and early developmental processes (fig. S2F). Notably, the GO term “negative regulation of transcription, DNA templated” was one of the top enriched terms, potentially reflecting the down-regulation of several mDA neuronal identity genes (Fig. 2, G and H). We have previously shown that early developmental regulators already silent and harboring K27 in neural stem cells gain K9 in differentiated mDA neurons (20), suggesting that both the acquisition and maintenance of K9 at these TSSs are closely connected and potentially dependent on PRC2 activity.
Fig. 3. Combined H3K9me3/H3K27me3 is associated with higher probability of derepression upon loss of PRC2 activity.
(A) Heatmaps showing abundance of H3K27me3, H3K4me3, and H3K9me3 at genes up-regulated in 8-month KO in 20-kb regions centered on the TSS of individual genes ranked by H3K27me3 abundance in WT. (B) Heatmap profiles of average H3K27me3, H3K4me3, and H3K9me3 RPKMs ±10 kb around the TSS of genes up-regulated in 8-month KO per defined chromatin state (denoted as K4, K27, K4/K27, K9/K27, and K4/K9/K27) in 8-month WT and how these states are resolved in 8-month KO. Below each chromatin state, Integrative Genomics Viewer tracks exemplify how the histone marker abundances compare at representative genes between WT and KO. (C) Enrichment/depletion of chromatin states for up-regulated genes in KO mDA cells at 8 months. (D) Enrichment/depletion of chromatin states for down-regulated genes in KO mDA cells at 8 months. (E) Volcano plot as in Fig. 2G showing DEGs belonging to H3K4me3/H3K27me3 (cyan) or H3K9me3/H3K27me3 (red) chromatin states in 8-month WT mDA cells. (F) Violin plot showing the absolute expression level [log2(FPKM + 1)] of up-regulated genes in KO mDA cells belonging to K4/K27 or K9/K27 chromatin states in 8-month WT mDA cells. (G) Difference (Δ) in gene expression between WT and KO of genes belonging to the K4/K27 or K9/K27 states at 8 months. Student’s t test. (H) Genes belonging to the K9/K27 chromatin state in 4-month WT mDA cells and up-regulated in KO mDA cells at 8 months but not at 4 months have reduced H3K9me3 surrounding their TSSs in KO mDA cells already at 4 months. (I) Genes belonging to the K9/K27 chromatin state in 4-month WT mDA cells and not up-regulated in KO mDA cells at 8 months do not have reduced K9 surrounding their TSSs at 4 months in 4-month KO mDA cells.
Progressive loss of TH, dopamine, and dopamine-associated metabolites upon loss of PRC2 activity
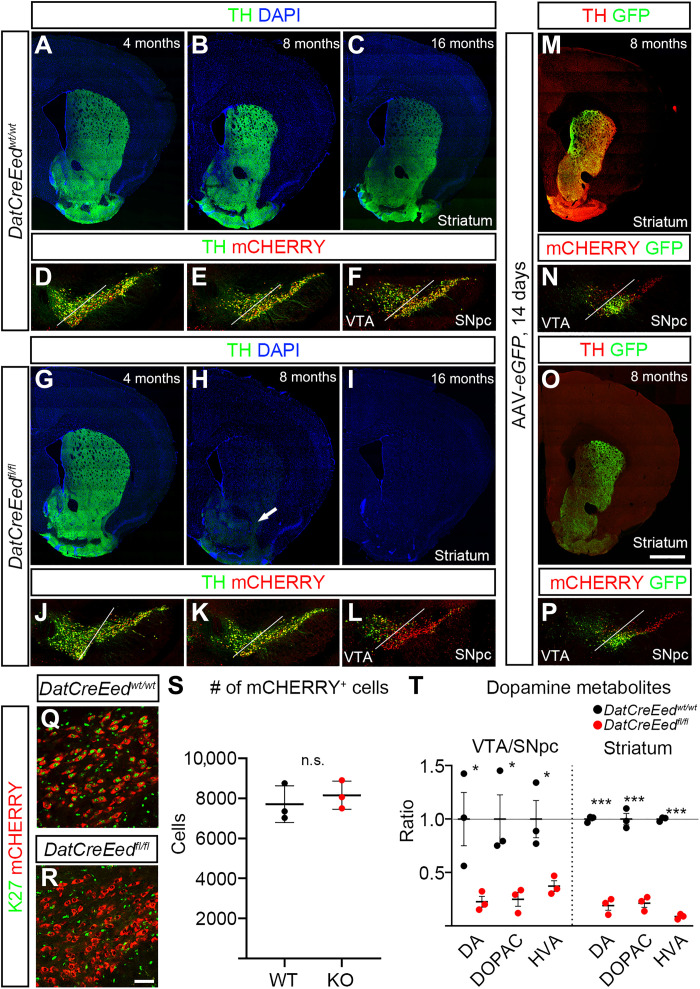
To understand the phenotypic consequences of the progressive loss of K27, we stained for TH at 4 and 8 months, both in the midbrain VTA/SNpc and in the striatal target region. At 4 months, there was no apparent difference between WT and mutant mice, neither in the VTA/SNpc nor in the striatum (Fig. 4, A, D, G, and J), suggesting that in the mutants, the establishment of this circuitry during development is not perturbed. Reflecting the reduction of Th expression in 8-month mutants (Fig. 2G), there was a slight reduction of TH immunoreactivity in mutant midbrain at 8 months (Fig. 4, E and K). However, in the dorsal striatum of DatCreEedfl/fl mutants, TH was completely lost, while low levels of TH could still be detected in the nucleus accumbens (Fig. 4, B and H). To investigate whether this loss of TH progressed over time, we performed staining of 16-month-old mutants. At this time point also, mCHERRY+ cells in the SNpc displayed an almost complete loss of TH, whereas cells in the mutant VTA still exhibited substantial TH immunoreactivity (Fig. 4, C, F, I, and L). The loss of TH immunostaining in mutant mice at 8 months could be the result of a progressive loss of established projections from the SN/VTA to their striatal targets. To investigate this possibility, we performed intracranial injections targeting the SN/VTA with adeno-associated virus (AAV) vectors [pCAG-FLEX-EGFP-WPRE virus (26)] to anterograde trace the projections from the midbrain to the striatum at 8 months. Three weeks after injection, we euthanized the animals for analysis. Green fluorescent protein (GFP) was exclusively expressed from the mDA neurons expressing the recombinant protein CRE at the site of injection (Fig. 4, N and P), as well as extending until their target area in the striatum (Fig. 4, M and O). Despite an almost total absence of TH immunoreactivity in mutant striatum, there was a strong GFP signal in all mutants analyzed (Fig. 4, M to P). Hence, the projections from the midbrain to the striatal target area are largely intact in the DatCreEedfl/fl mutants. Because it has been reported that Ezh1/2 deletion in MSNs caused severe neuronal loss through cell death (6), we analyzed and counted the mCHERRY+ cells in representative areas of 8-month-old midbrains but failed to detect any significant difference in the number of cells between DatCreEedwt/wt control mice and DatCreEedfl/fl mutants (Fig. 4, Q to S). To further validate that there was no increase in cell death at different time points, we inspected mCHERRY+ nuclei in the midbrains of both DatCreEedwt/wt control mice and DatCreEedfl/fl mutants for any signs of fragmented nuclei through 4′,6-diamidino-2-phenylindole (DAPI) staining. However, we could not detect any fragmented nuclei in this population, neither at P0, at 4 months, at 8 months, nor at 16 months (fig. S3, A to P). We next wanted to understand whether the reduced levels of TH affected levels of dopamine and associated metabolites. To investigate this, we dissected out the midbrain and striatum and performed high-performance liquid chromatography (HPLC) to measure the metabolites of the dopamine synthesis pathway. In DatCreEedfl/fl mutants, both regions displayed a significant reduction of dopamine, 3,4-dihydroxyphenylacetic acid (DOPAC), and homovanillic acid (HVA) (Fig. 4T). The decrease was most prominent in the striatum, where mutant animals exhibited a more than fivefold reduction of all three metabolites compared with control animals (Fig. 4T).
Fig. 4. Reduced levels of TH and dopamine metabolites in the striatum and midbrain upon inactivation of PRC2.
(A to C) TH immunostaining in the striatum of DatCreEedwt/wt mice at 4 months (A), 8 months (B), and 16 months (C). (D to F) TH immunostaining and mCHERRY fluorescence in the ventral midbrain of DatCreEedwt/wt mice at 4 months (D), 8 months (E), and 16 months (F). (G to I) TH immunostaining in the striatum of DatCreEedfl/fl mice at 4 months (G), 8 months (H), and 16 months (I); arrow in (H) indicates reduced albeit detectable TH immunostaining in the nucleus accumbens. (J to L) TH immunostaining and mCHERRY fluorescence in the ventral midbrain of DatCreEedfl/fl mice at 4 months (J), 8 months (K), and 16 months (L). (M) Expression of eGFP in the striatum of DatCreEedwt/wt mice 21 days after injection of AAV-eGFP in the ventral midbrain. (N) Injection site of AAV-eGFP in DatCreEedwt/wt mice 21 days after injection. (O) eGFP expression in the striatum of DatCreEedfl/fl mice 21 days after injection of AAV-eGFP in the ventral midbrain. (P) Injection site of AAV-eGFP in DatCreEedfl/fl mice 21 days after injection. (Q) H3K27me3 immunostaining in mCHERRY+ cells in the SN of 8-month DatCreEedwt/wt mice. (R) No H3K27me3 immunostaining in mCHERRY+ cells in the SN of 8-month DatCreEedwt/wt mice. (S) Quantification of mCHERRY+ cells in the ventral midbrain of DatCreEedwt/wt and DatCreEedfl/fl mice at 8 months shows no loss of mCHERRY+ cells in mutant midbrains. nWT = 3 and nMUT = 3. (T) Reduced levels of dopamine metabolites in the ventral midbrain and striatum of DatCreEedfl/fl mice. nWT = 3 and nMUT = 3. In (T), *P < 0.05 and ***P < 0.001, unpaired t test with Welch’s correction. Scale bars, 1000 μm (O) and 50 μm (R). n.s., not significant.
Basic electrophysiological properties of mDA neurons are perturbed upon loss of PRC2 activity
The reduction in dopamine and associated metabolites combined with the loss of TH immunoreactivity in the striatal target area suggest that, besides loss of identity, a severe perturbation of mDA neuron function also occurred in DatCreEedfl/fl mutants. To address this, we analyzed basal physiological properties through whole-cell patch-clamp recordings in slice preparations from the 8-month mutant and WT SNpc. Several of the measured parameters were significantly perturbed in the mutant midbrains (Fig. 5, A to M). Cell capacitance was reduced (Fig. 5A), whereas membrane resistance was slightly increased (Fig. 5B). This implies that mutant mDA neurons exhibit smaller surface area and reduced open channel activity, generating a higher input resistance. Spontaneous pacemaker spiking typical of mDA neurons exhibited no difference in frequency (Fig. 5C), but the pacemaker pattern was disturbed with significant loss of consistency of interspike intervals (Fig. 5, D and E). The hyperpolarization-activated current Ih, mediated by hyperpolarization-activated cyclic nucleotide–gated channels, which contributes to mDA neuronal pacemaker firing integrity, was reduced (Fig. 5, F and G). Also, slow afterhyperpolarization (AHP) current (AHC) generated by small-conductance Ca2+-sensitive K+ channels was reduced (Fig. 5, H and I). Although the action potential amplitude was unchanged, the threshold was decreased, whereas AHP amplitude was decreased (Fig. 5, J to M). These anomalies of spontaneous firing in the mDA neuron population combined with the low levels of dopamine in the striatum indeed suggest severe loss of mDA neuronal function.
Fig. 5. Altered electrophysiological properties and animal behavior upon inactivation of PRC2.
(A) Reduced capacitance in 8-month DatCreEedfl/fl mDA neurons. (B) Increased membrane resistance in 8-month DatCreEedfl/fl mDA neurons. (C to E) Increased coefficient of variation (CV) of interspike intervals of autonomous pacemaker currents in 8-month DatCreEedfl/fl mDA neurons. (F and G) Decreased hyperpolarization current (Ih) in 8-month DatCreEedfl/fl mDA neurons. (H and I) Decreased slow AHC in 8-month DatCreEedfl/fl mDA neurons. (J and K) Action potential (AP) threshold is reduced (J), whereas AHP is decreased (K) in DatCreEedfl/fl mDA neurons. (L) Action potential amplitude was not significantly reduced in DatCreEedfl/fl mDA neurons. (M) Phase plot (dV/dt versus Vm) of action potential in DatCreEedwt/wt and DatCreEedfl/fl mDA neurons of the SNpc. (N to P) Open-field test at 4 months (N) (nWT = 10 and nMUT = 12), 8 months (O) (nWT = 19 and nMUT = 16), and 16 months (P) (nWT = 36 and nMUT = 14) shows a progressive decrease in distance moved by DatCreEedfl/fl mice. (Q) Progressive decrease in the number of rearings for DatCreEedfl/fl mice. nWT4 = 10, nMUT4 = 12, nWT8 = 18, nMUT8 = 16, nWT16 = 15, and nMUT = 12. (R) Progressive increase in total time needed for DatCreEedfl/fl mice to complete the pole test. nWT4 = 8, nMUT4 = 12, nWT8 = 17, nMUT8 = 16, nWT16 = 17, and nMUT = 15, average from five trials per mouse. (S) CPP score before and after exposure to cocaine in WT and mutant mice. nWT = 11 and nMUT = 10. In (A) to (M), the datasets were checked for normality with Shapiro-Wilk test. For normally distributed datasets, unpaired t test was used (*P < 0.05 and ****P < 0.0005). If the datasets did not pass the normality test, then Mann-Whitney test was applied. In (N) to (P), P values were calculated by two-way repeated-measures analysis of variance (ANOVA). In (Q) and (R), *P < 0.05 and ***P < 0.001 calculated by one-way ANOVA with Tukey’s multiple comparisons test. In (S), P values were calculated with paired t test.
Progressive impairment of overall locomotor activity and fine motor movement in Eed mutants
To test whether reduced levels of TH and dopamine metabolites as well as altered electrophysiological properties would generate any behavioral consequences, we performed several behavioral tests to detect potential motor skill impairments in the mutant mice (27). Coordination, endurance, and muscle strength were not affected in the mutants as shown by the rotarod and grip strength tests, respectively (fig. S4, A to C). However, when analyzed in the open-field test, their overall locomotor activity exhibited progressive reduction with age as recorded from the total moved distance (Fig. 5, N to P). In addition, the number of rearings was significantly reduced in the mutants (Fig. 5Q). When challenged by the pole test, assessing fine locomotor function, mutant mice exhibited a significant delay in initiation of descent and frequent failure to climb down the pole by falling sideways (Fig. 5R and movies S3 to S5). Hence, the mutants exhibited deficits primarily in the initiation of voluntary movement as seen in the open-field test, in the number of rearings, and in the pole test, whereas, when exposed to a forced movement paradigm testing coordination and balance, such as the rotarod, the mutant mice performed on par with the WT mice. Motor function has mainly been coupled to the SNpc, whereas reward and motivation processes, to a larger degree, involve the VTA (28). Therefore, we challenged the mice by conditioning them to cocaine and examined whether perturbation in the reward system of 8-month-old mutant mice occurred. We conducted conditional place preference (CPP) for cocaine, where a weak CPP occurred in the WT mice (Fig. 5S). The CPP in mutant mice was more distinct (Fig. 5S); however, the difference in posttest CPP between WT and mutant mice was not significant, indicating that response to cocaine conditioning was unaffected by genotype. To investigate whether the motor response to cocaine was affected in the mutants, we also performed an open-field test after injection of cocaine (10 mg/kg). In both WT and mutant mice, there was a substantial increase in movement compared to nontreated animals (fig. S4D). However, there was no significant difference between cocaine-treated animals with different genotypes (fig. S4D). Thus, the capacity to respond to cocaine in the mutant is not impaired, implicating that this aspect of SNpc/VTA function is intact.
Together, the loss of Eed results in progressive loss of H3K27me3, leading to up-regulation of Polycomb target genes and reduced expression of mDA neuronal genes. This loss of cellular identity severely disrupts mDA neuronal function at the cellular level, ultimately altering the behavior of DatCreEedfl/fl mutants.
Loss of PRC2 function in 5HT neurons results in loss of cellular identity and function
An earlier study reported that cell-specific loss of PRC2 function in striatal MSNs caused substantial cell death (6). As mentioned, a corresponding cell death was not noted in the mDA neurons of DatCreEedfl/fl mutants. To understand whether deletion of Eed can cause loss of neuronal identity without cellular loss also in a different neuronal population, we crossed the Rpl10a-mCherryflox/flox/Eedflox/flox mice with SertCre mice (29). In this SertCreEedfl/fl mutant, Eed is selectively deleted in hindbrain 5HT neurons expressing the serotonin [serotonin hydrochloride (5-HT)] transporter Slc6a4 (a.k.a. Sert) (Fig. 6A). Expression of Slc6a4 first occurs in the hindbrain at around E12.5 of the mouse embryo. At this time point, tryptophan hydroxylase–positive (TPH+) cells in both WT and mutant hindbrain expressed EED (fig. S5, A and B). As in mDA neurons, there was, in P0 mCHERRY+ 5HT neurons, a substantial reduction in EED immunoreactivity in the SertCreEedfl/fl mutants, whereas levels of H3K27me3 remained as in SertCreEedwt/wt mice (fig. S5, C to F). At P40, the levels of H3K27me3 were not detectable in SertCreEedfl/fl mutant cells (Fig. 6, B and C). To investigate the possible consequences of Eed loss in 5HT neurons, we stained the hindbrains of WT and mutants with an antibody specific for the rate-limiting enzyme in serotonin synthesis, TPH2. At 4 months of age, the number of mCHERRY+ cells in the SertCreEedfl/fl mutants was unaltered compared to the WT mice, with a distinct overlap of TPH2 immunoreactivity and mCHERRY+ fluorescence in both WT and mutant animals (Fig. 6, D and G). In contrast, at 8 and 16 months, immunostaining revealed a major loss of TPH2 in SertCreEedfl/fl mutants but no loss of mCHERRY+ cells (Fig. 6, E to I).
Fig. 6. Eed deficiency in 5HT neurons results in loss of H3K27me3 and progressive decrease in TPH2 expression.
(A) In situ hybridization for Slc6a4 (Sert) taken from the Allen Brain Atlas (mouse.brain-map.org, image credit: Allen Institute). (B) Immunostaining of H3K27me3 in mCHERRY+ 5HT neurons in the dorsal raphe of SertCreEedwt/wt mice. (C) Lack of H3K27me3 immunostaining in mCHERRY+ 5HT neurons in the dorsal raphe of SertCreEedfl/fl mice. (D to F) TPH2 immunostaining localized in mCHERRY+ 5HT neurons in the dorsal raphe of SertCreEedwt/wt mice aged 4 months (D), 8 months (E), and 16 months (F). (G to I) Progressive loss of TPH2 in 5HT neurons in the dorsal raphe of SertCreEedfl/fl mice aged 4 months (G), 8 months (H), and 16 months (I). Scale bars, 20 μm (C) and 200 μm (I).
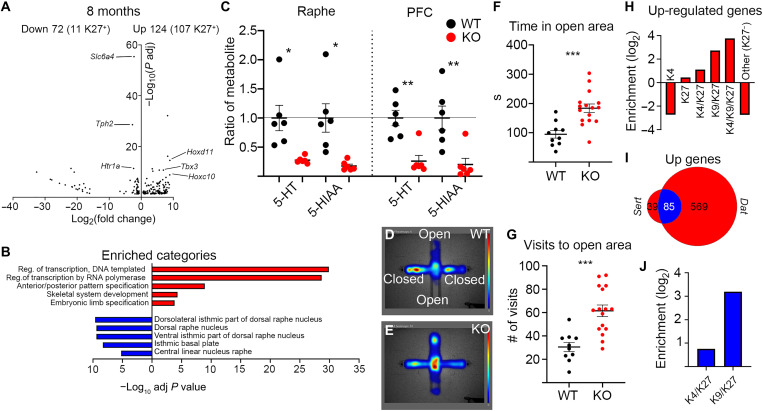
We proceeded to perform RNA-seq analysis of sorted 5HT nuclei at 4 months, which revealed up-regulation of 84 transcripts, of which 36 were K27+, as previously determined (20). Twenty-six transcripts were down-regulated, of which 2 were K27+ (fig. S5G). Similar analysis of sorted 5HT nuclei at 8 months revealed 124 up-regulated genes, of which 107 were K27+ (Fig. 7A). As in the mutant mDA nuclei, the up-regulated genes were enriched for several members of the Hox family; transcription factors involved in determining other cell fates during development, e.g., Gata6, Foxg1, and Dlx1; stem cell factors such as Pax6; genes typically expressed in other neuronal types, e.g., Gad1; and cell cycle regulators including Ccnd2 and Cdkn2a (Fig. 7A and table S3). Down-regulated genes numbered 72 and included 5HT-specific genes such as Slc6a4, Tph2, and Htr1a (Fig. 7A and table S3). Of these 72 genes, 11 were H3K27me3+, which does not constitute an enrichment. GO analysis of the DEGs showed that among up-regulated genes, there was a strong enrichment of categories related to transcriptional activation and early developmental processes. For the down-regulated genes, there was an enrichment of dorsal raphe nucleus–associated categories (GO Biological Process 2021 and Allen Brain Atlas Up, as calculated by Enrichr) (Fig. 7B). The substantial reduction of TPH2 in 8-month mutants caused a decrease in the levels of serotonin (5-HT) and the serotonin-associated metabolite [5-hydroxyindole-3-acetic acid (5-HIAA)] both in the hindbrain and in the prefrontal cortex target area as measured by HPLC (Fig. 7C).
Fig. 7. Eed deficiency in 5HT neurons results in impaired 5HT-specific gene expression and function.
(A) Volcano plot showing DEGs in mCHERRY+ nuclei from SertCreEedfl/fl mutants at 8 months. Numbers of up- and down-regulated genes are indicated above the plot, with the number of H3K27me3+ genes in 8-month SertCreEedwt/wt 5HT neurons within brackets. Examples of up-regulated genes that are PRC2 targets in SertCreEedwt/wt 5HT neurons are labeled on right side of the plot. Examples of down-regulated 5HT identity genes are labeled on left side of the plot. (B) Enriched categories for up-regulated genes (red) (GO biological process) are characterized by activation of transcription and early developmental non-neuronal processes, whereas down-regulated genes (blue) show enrichment for raphe categories (Up in Allen Brain Atlas, as calculated in Enrichr). (C) Reduced levels of serotonin metabolites in the raphe and prefrontal cortex (PFC) of SertCreEedfl/fl mice. nWT = 6 and nMUT = 6. (D and E) Heatmap of time spent in indicated areas in EPM for SertCreEedwt/wt mice (D) and SertCreEedfl/fl mice (E). (F) Increased time spent in open area for SertCreEedfl/fl mice in EPM. nWT = 10 and nMUT = 16. (G) Increased number of visits to open area for SertCreEedfl/fl mice in EPM. nWT = 10 and nMUT = 16. (H) Enrichment/depletion of chromatin states (in 8-month WT 5HT neurons) of genes up-regulated in SertCreEedfl/fl mCHERRY+ nuclei at 8 months. (I) Overlap of up-regulated genes between 8-month DatCreEedfl/fl and SertCreEedfl/fl mice. (J) Commonly up-regulated genes in 8-month DatCreEedfl/fl and SertCreEedfl/fl mice are more enriched for the K9K/K27 chromatin state than the K4/K27 state. In (C), (F), and (G), *P < 0.05, **P < 0.01, and ***P < 0.001, unpaired t test with Welch’s correction.
To understand whether loss of 5HT identity was followed by altered behavior, as seen in the DatCreEedfl/fl mutants, we investigated whether aspects of behavior that depend on an intact 5HT function were perturbed in the 5HT mutants. To examine whether loss of Eed evoked depressive behavior, we subjected 8-month SertCreEedwt/wt and SertCreEedfl/fl mice to the forced swim test. Notably, there was no significant difference between WT and mutant mice for the time they spent swimming (fig. S5H). However, when subjected to the elevated plus maze (EPM) (30), the mutants spent significantly more time in the open arms and visited them more frequently (Fig. 7, D to G). This change in behavior could be a consequence of hyperactivity or less anxiety, behavioral phenotypes that have previously been associated with deficient serotonin neurotransmission (31, 32).
Activated PRC2 targets are enriched for H3K9me3 in 5HT nuclei
The strong enrichment of K27 targets among the up-regulated genes is reminiscent of the effects of Eed deletion in mDA neurons. Because the presence of the heterochromatin modification H3K9me3 was associated with a higher probability of derepression and activated transcription in mutant mDA nuclei, we assigned up-regulated genes in the SertCreEedfl/fl nuclei to the same set of chromatin states by using the dataset we generated for WT 5HT neurons in our previous study (20). This analysis showed that also in mutant 5HT nuclei, the presence of K9 in a chromatin state was associated with a higher probability to activate transcription than the presence of K4 (Fig. 7H). In contrast, down-regulated genes exhibited no such enrichment (fig. S5J).
Common enrichment of up-regulated H3K9me3/H3K27me3 targets in mutant mDA and 5HT neurons
Because we noted that transcriptional response to loss of Eed included similar PRC2 targets in mDA and 5HT neurons, we compared the overlap of up-regulated genes at 8 months. There was a large overlap of up-regulated genes with 85 common transcripts of 124 (5HT nuclei) and 654 (mDA nuclei) (Fig. 7I). This represents a more than 23-fold higher number than expected by chance (P = 1 × 10−100, Fisher’s exact test). Of these 85 transcripts, 83 were K27+ PRC2 targets in 5HT neurons and 77 in mDA neurons, which represents a substantial enrichment in both cell types. In addition, the 85 commonly up-regulated targets were also strongly enriched for the K9/K27 state when compared to the poised K4/K27 state (Fig. 7J). The number of commonly down-regulated genes also exhibited a fivefold significant enrichment but not to the same extent as for the up-regulated genes (P = 3 × 10−5, Fisher’s exact test) (fig. S5K).
When comparing Enrichr (24, 25) analysis between the up-regulated genes in mDA and 5HT neurons, the similarity is clear. In both neuronal cell types, there was a strong enrichment of early developmental regulators, e.g., Hox genes (Figs. 2G and 7A). Furthermore, another similarity is that both types of neurons exhibit reduced expression of transcripts specific to their identity, e.g., Th, Slc6a3, En1, Nr4a2, and Pitx3 in the mDA neurons and Tph2, Htr1a, Slc6a4, and Htr5b in 5HT neurons (Figs. 2G and 7A). Notably, in both mutants, substantially increased expression of genes previously described as “death promoting” (e.g., Cdkn2a, Hoxa5, and Wt1) was evident (tables S2 and S3) but without inducing any cell death, which clearly distinguishes these neuronal subtypes from MSNs (6).
Single-nucleus expression analysis reveals SNpc-specific vulnerability to loss of PRC2 activity
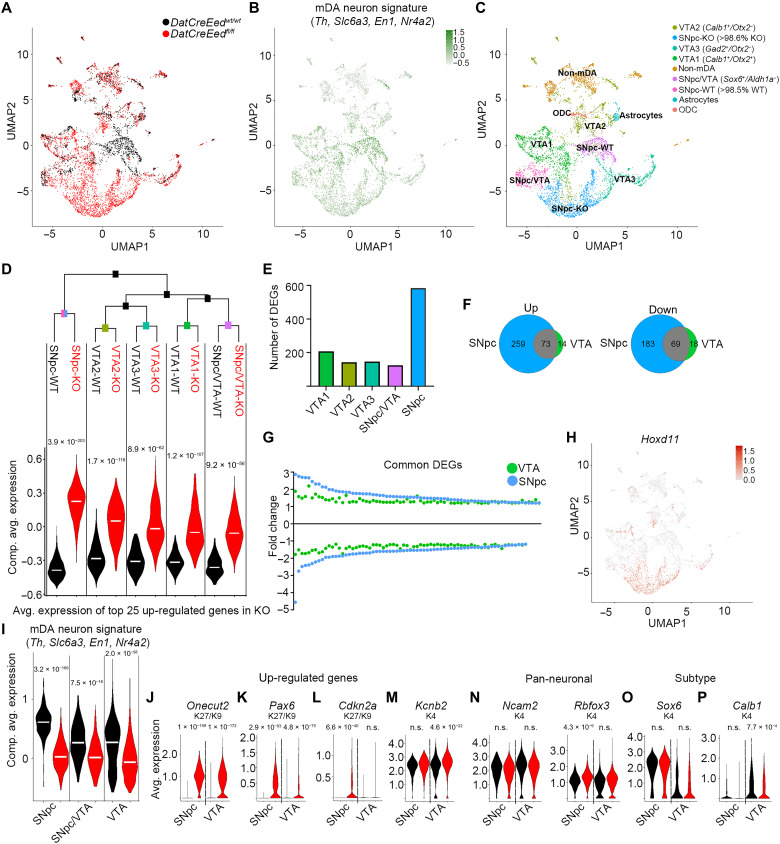
The selective and progressive loss of TH in the SNpc at 8 and 16 months (Fig. 4) indicates that different mDA neuron subtypes may respond differently to PRC2 deficiency. To explore whether the changes in gene expression upon loss of H3K27me3 are distinct between mDA neuron subtypes, we analyzed gene expression with single-nucleus RNA-seq (snRNA-seq) of sorted mCHERRY+ nuclei from the midbrain of 8-month-old WT and mutant mice. Following quality control, sequencing data from 1772 nuclei from WT brains and 3968 nuclei from mutant brains were obtained. Uniform manifold approximation and projection (UMAP) plots of these nuclei revealed a considerable diversity of WT and mutant nuclei (Fig. 8, A and C). Composite average expression of an mDA neuron signature (Th, Slc6a3, Nr4a2, and En1) was evident in a substantial proportion of the nuclei (Fig. 8B). To further characterize the snRNA-seq data, we assigned distinct identities to different UMAP clusters on the basis of expression of markers previously described in the literature (Fig. 8C, fig. S6A, and table S7) (33). Nuclei lacking robust expression of the mDA neuron signature, potentially isolated along with mCHERRY+ mDA neuron nuclei when sorting, were defined as astrocytes, oligodendrocytes, and non-mDA neurons on the basis of typical neural cell markers (table S6). On the basis of the classification reported in (33), the six clusters that robustly expressed the mDA neuron signature were divided into VTA1 (Calb1+/Otx2+), VTA2 (Calb1+/Otx2−), VTA3 (Gad2+/Otx2−), SNpc/VTA (Sox6+/Aldh1a−), SNpc-WT (>98.5% WT), and SNpc-KO (knockout) (>98.6% KO) of both groups of Sox6+/Aldh1a+ (Fig. 8C). One mDA neuron group (SNpc-WT) was almost exclusively enriched for WT nuclei (>98.5%), while another group (SNpc-KO) was almost exclusively enriched (>98.6%) for nuclei from mutant mice. In contrast, WT and mutant nuclei were distributed in roughly equal proportions in all other groups (Fig. 8A and table S8). SNpc-KO nuclei had diminished mDA neuron marker gene expression but did express Sox6 and Aldh1a1, consistent with a relationship to SNpc mDA neurons. The expression of the SNpc markers Sox6 and Aldh1a1 and of the VTA marker Calb1 was distributed as shown in fig. S6 (B and C). Thus, these observations indicate a rather drastic influence on gene expression in SNpc neurons as compared to the other mDA neuron groups as a consequence of disrupted PRC2 function.
Fig. 8. mDA neurons of the SNpc exhibit selective increased vulnerability to loss of PRC2 activity.
(A) UMAP showing distribution of WT (black) and mutant (red) nuclei. (B) Composite average expression levels of mDA identity signature genes. (C) Classification of defined subgroups in UMAP space. ODC, oligodendrocyte. (D) Top: Hierarchical clustering of mDA neuron subgroups with identity classes defined as subgroup genotype. Bottom: Violin plots highlighting subtype-specific differences between genotypes based on the composite expression of the 25 most up-regulated genes in the mutant nuclei. (E) Number of DEGs in mutant nuclei of different mDA neuron groups according to (C). (F) Venn diagram showing overlap of up- and down-regulated genes in mutant nuclei from the combined VTA and SNpc. (G) Fold change of gene expression for the mutant versus WT DEGs, which are common between the VTA and SNpc. To normalize scales, the inverted fold change (−1/FC) is plotted for down-regulated genes. (H) UMAP visualization of Hoxd11 expression. (I) Composite average expression of mDA neuronal signature in WT (black) and mutant (red) nuclei for mDA neuron subgroups (VTA = VTA1 + VTA2 + VTA3). (J to M) Violin plots exemplifying genes up-regulated in both mutant SNpc and VTA (J), of genes more up-regulated in mutant SNpc than in mutant VTA (K), of genes only up-regulated in mutant SNpc and not in the VTA (L), and of genes only up-regulated in mutant VTA and not in SNpc (M). (N to P) Violin plots of pan-neuronal markers (N), of the SNpc marker Sox6 (O), and of the VTA marker Calb1 in WT (black) and mutant (red) SNpc and VTA nuclei (P). Wilcoxon rank sum test with Bonferroni corrections for adjusted P values. Adjusted P values are included in (D) and (I) to (P).
Hierarchical clustering based on the 2000 most variable genes in all nuclei from the mDA neuron groups revealed that the SNpc-WT and SNpc-KO clusters grouped separately from the VTA1 to VTA3 and SNpc/VTA groups (Fig. 8D and fig. S6D). To understand whether loss of PRC2-mediated repression had selective effects in different mDA neuron subgroups, the signature expression of the 25 most up-regulated genes in mutant nuclei was visualized in violin plots. This analysis revealed that the most profound increase occurred in the SNpc-KO versus SNpc-WT groups (Fig. 8D and fig. S6E). We then generated a heatmap of all mDA neuron groups based on the 205 DEGs between all mutant and WT nuclei, of which 93 exhibited increased expression and 112 decreased expression. This revealed a block of genes that are strongly up-regulated in most of the SNpc-KO nuclei but only in subsets of nuclei in the VTA1-KO, VTA2-KO, VTA3-KO, and SNpc/VTA-KO nuclei (fig. S6F). This group of genes was strongly enriched for PRC2 targets and for genes also up-regulated in the bulk RNA-seq at 8 months.
To further understand the differential effects resulting from loss of PRC2 function, we analyzed genes differentially expressed between WT and mutant nuclei for each group separately. As expected, major effects on gene expression were only seen in mDA neuron clusters derived from cells in which PRC2 had been disrupted by targeted KO of Eed, while marginal effects were seen in non-mDA neuron groups. Notably, the largest number of DEGs was seen in the SNpc group (Fig. 8E and table S4), in which 584 genes were differentially expressed when comparing SNpc-KO versus SNpc-WT. In contrast, only 174 genes were differentially expressed when comparing WT and mutant nuclei in the three VTA groups combined into one VTA group (Fig. 8F). The more substantial response to loss of PRC2 activity in the SNpc group was also reflected in the magnitude of increase or decrease in expression in common up- or down-regulated genes between SNpc and VTA (Fig. 8G) and is also clearly illustrated by plotting highly up-regulated genes, such as Hoxd11, in the UMAP plot (Fig. 8H). Moreover, composite average expression of the mDA neuron signature was more significantly reduced in the mutant nuclei of the SNpc group (Fig. 8I). Together, the greatest impact of Eed deletion occurred in the SNpc population, which was also reflected by how TH immunoreactivity was substantially reduced in the SNpc at 8 and 16 months but was largely intact in the VTA (Fig. 4, C to L).
Comparison of DEGs between mutant and WT nuclei in the combined VTA and the SNpc showed equal (Fig. 8J and fig. S6G), stronger (Fig. 8K and fig. S6H), or exclusive (Fig. 8L and fig. S6I) up-regulation of PRC2 targets in the SNpc. The few genes that were more robustly induced in the VTA are not PRC2 targets. Instead, they are typically expressed at robust levels in WT cells and harbor the H3K4me3 modification (Fig. 8M and fig. S6J). Expression of pan-neuronal genes was not decreased in any of the groups, and Rbfox3 was actually slightly increased in the SNpc-KO nuclei (Fig. 8N). Notably, expression of the SNpc subtype–specific gene Sox6 was not decreased in the SNpc-WT versus SNpc-KO subgroups, whereas expression of the VTA-specific Calb1 gene was reduced in the mutant VTA cells (Fig. 8, O and P). We also plotted the individual expression of the four genes that constituted the mDA neuron signature used in Fig. 8 (B and I) (fig. S6K).
To compare the results of the single-cell nuclei analysis with the results we previously obtained with the bulk RNA-seq, we examined common or unique DEGs. This revealed that genes identified as up- or down-regulated in both the single-nucleus and bulk analysis exhibited significantly larger differences in gene expression than genes only identified as differentially regulated in the single-nucleus analysis (fig. S6L). Notably, this trend was most pronounced in the SNpc group (fig. S6L and table S5). To ensure that non-mDA populations do not add noise to the bulk RNA-seq and ChIP-seq analysis, we performed DE analysis between WT non-mDA nuclei and WT mDA neuronal nuclei. A total of 252 genes were significantly up-regulated in the non-mDA nuclei group. When compared to genes up-regulated in mutants in bulk sequencing at 4 months, there was an overlap of 4 genes, which represents ~3× enrichment (Fisher’s exact test, P = 0.04) and, at 8 months, an overlap of 18 genes (~2× enrichment, Fisher’s exact test, P = 0.004). Although these overlaps represent a significant enrichment, the number of genes is low and represents a minute fraction of the genes up-regulated in each dataset. Furthermore, a total of 30 genes are uniquely up-regulated in the mutant non-mDA group; of these, only 2 are up-regulated in the bulk sequencing at 8 months, and none are up-regulated at 4 months. We have now also investigated the expression of mCherry in the bulk RNA-seq data. It is expressed both in mDA (baseMean = 44.9) and in 5HT (baseMean = 66.0). It is not differentially expressed between WT and mutant samples (8-month mDA: log2FoldChange = 0.45, unadjusted P = 0.26; 8-month 5HT: log2FoldChange = −0.49, unadjusted P = 0.44), which further supports that contaminant non-mDA/non-5HT cells are not biasing WT versus mutant comparisons. Hence, potential noise added by contaminant populations is not likely to affect the analysis or the conclusions in any fundamental aspect.
To understand whether genes with specific chromatin states were regulated equally between mutant and WT nuclei in the VTA and SNpc, we used the chromatin states generated from the bulk ChIP-seq. As in the bulk RNA-seq, up-regulated genes both in the VTA group and the SNpc group were strongly enriched for the K9/K27 chromatin state (VTA: 8.3× increase over expected, P = 4.2 × 10−12; SNpc: 4.3× increase over expected, P = 1.4 × 10−13, Fisher’s exact test). In contrast, in the SNpc group, there was no significant enrichment of K4/K27 genes, and in the VTA group, the enrichment was less (2.9× increase over expected, P = 0.0015, Fisher’s exact test) than for the K9/K27 state (fig. S6M).
DISCUSSION
How long-term maintenance of cellular identity is coupled to permanent silencing of alternative lineages is largely unknown. This question is of particular interest for CNS neurons because their functional integrity and identity need to be maintained for several decades in the human brain. Our study reveals that in two well-characterized neuronal populations of high clinical relevance, intact PRC2 function is essential for the repression of aberrant gene expression and for the maintenance of cell type–specific gene expression but not for neuronal survival. Although PRC2-mediated gene silencing has previously been shown to be required for proper neurogenesis in the neocortex (7, 34), the role of PRC2 in differentiated postmitotic cells, such as neurons, is not understood. A previous report showed that in MSNs and Purkinje cells, PRC2 is required to maintain silencing of death-promoting genes (6). Similar “death-promoting” genes were up-regulated in mutant mDA and 5HT neurons upon loss of PRC2 activity. However, there was no reduction in cell numbers, neither in mDA nor in 5HT neurons, even across an extended time span (up to 16 months). Instead, there was a profound reduction in the expression of subclass-specific genes in both types of neurons. This is in contrast to a study wherein the methyltransferase Ezh2, another member of the PRC2 complex, was deleted in postmitotic mDA neurons (35). Notably, deletion of Ezh2 resulted in a selective and progressive loss of VTA neurons. Because the levels of K27 were not changed in the mutants, it is possible that this effect was uncoupled from the canonical methyltransferase function of Ezh2. In addition, in differentiated mDA neurons, the expression level of Ezh2 is lower than that of Ezh1, which also harbors methyltransferase capacity and thus could act as a redundant factor.
Because most down-regulated genes are not H3K27me3+ in the WT cells and the main function of PRC2 is to maintain repression, the reduced expression is most likely an indirect effect of Eed deletion. A similar effect was reported in MSNs and in differentiated β cells wherein cell type–specific genes were down-regulated upon loss of PRC2 activity (6, 36). Using the single-nucleus dataset, we have tried to identify crucial up-regulated factors that repeatedly correspond with a decrease in mDA neuronal identity genes. However, we could not couple any single up-regulated factor with the decrease in mDA identity genes. This would argue for a combined effect of several up-regulated factors, which, during development, harbor the capacity to induce other cell lineages and to silence the mDA or 5HT neuronal lineages. This protracted process is in stark contrast with the phenotype upon combined deletion of the transcriptional coactivators adenosine 3′,5′-monophosphate response element–binding protein–binding protein and p300 (i.e., KAT3) in the hippocampus. Upon deletion of these coactivators, there was a rapid loss of expression of neuronal genes followed by loss of neuronal identity and function (37). Not unexpectedly, inactivation of a repressor does not itself induce expression of its targets. Potential additional repressive mechanisms might interfere with recruitment of transcriptional activators, further extending the time required for target gene activation. Reduced expression of identity genes is, with high probability, an indirect consequence of up-regulation of PRC2 targets, further extending the time between deletion of Eed and effects on gene expression. Notably, despite other major differences, neither inactivation of PRC2 nor KAT3 activity induced any detectable cell death.
Because the emergence of phenotypes in both mDA and 5HT neurons coincides with significantly reduced expression of genes necessary for neuronal subtype-specific function (e.g., Th, Slc6a3, and En1 for mDA neurons and Tph2, Slc6a4, and Htr1a for 5HT neurons), this down-regulation is the most probable cause of the observed phenotypes. However, it cannot be ruled out that up-regulated PRC2 targets themselves directly target aspects of neuronal function and identity, thus contributing to the phenotypes. Notably, several of the up-regulated PRC2 target genes at 8 months, in both mDA and 5HT neurons, are transcription factors endowed with autoregulatory and gene-activating capacity. Thus, the emergence of gene regulatory networks capable of disrupting the integrity of endogenous transcriptional programs required for the maintenance of identity is a probable major contributor to the phenotypes observed. A similar process was described in a recent study wherein inactivation of the PRC1 in developing spinal motor neurons resulted in the derepression and up-regulation of transcription factors harboring cell fate–determinant capacity, among them the caudal Hox paralogs Hox10 to Hox13. This coincided with the down-regulation of genes associated with motor neuron subtype identity (38). Furthermore, the authors showed that up-regulation of Hox13 paralogs had the capacity to directly repress other Hox genes, e.g., Hox4 to Hox8, necessary for the correct specification of the anterior-posterior identity of spinal motor neurons. In these cell types at this developmental window, loss of PRC2 function had no major effect on subtype-specific identity. However, analysis was not extended beyond embryogenesis, which limits conclusions concerning any long-term effects on the maintenance of differentiated motor neurons.
Because neurons are postmitotic, the progressive reduction of K27 in mutant neurons is not the result of failure to establish novel K27 marks during cell division. Previous reports that histone turnover in the rodent brain occurs at an extended time scale (~220 days) have recently been contrasted by studies showing that the histone variant H3.3 exhibits rapid and continuous turnover in differentiated CNS neurons (39). Thus, the loss of K27 evident in the mDA and 5HT neurons likely is a consequence of a combination of histone turnover and demethylase activity by Kdm6a and Kdm6b, both of which are expressed in mDA and 5HT neurons.
It has previously been proposed that the primary role for PRC2 in differentiated cells is to suppress transcription of bivalent K4/K27 genes (6, 40). In both mDA and 5HT neurons, there was an enrichment of K4/K27 bivalent genes among up-regulated transcripts. However, the presence of K9/K27 was associated with a higher probability of derepression and a higher increase in both relative and absolute expression levels. This preference for activating K9/K27 genes over K4/K27 genes poses several questions. Is this a cell-specific feature? What does it reveal about potential interactions between K9- and K27-associated factors, as well as how these modifications are interpreted by the transcriptional machinery? GO analysis of K9/K27 genes in 8-month DatCreEedwt/wt mCHERRY+ nuclei clearly shows strong enrichment of categories such as regulation of transcription and early developmental categories, for example, anterior/posterior pattern specification. K4/K27 genes, on the other hand, are more enriched for categories such as extracellular matrix organization, proliferation control, and neuronal differentiation (fig. S2E). Notably, the categories enriched for the K4/K27 state reflect transitions from K4 state in neural progenitor cells (NPCs) to K4/K27 state in mDA neurons as determined in our previous study (20). Similarly, the categories enriched for the K9/K27 state are reminiscent of silent genes carrying K27 already in NPCs but that gain K9 to become K9/K27 in mDA neurons. Thus, after terminal differentiation, K9 appears to be gained as an additional layer of repression; hence, the higher probability of derepression of K9/K27 genes is an unexpected result. In particular, in differentiated MSNs, loss of PRC2 activity led to the activation of predominantly poised K4/K27 genes (6). However, in that study, no analysis of K9 was performed, inactivation of PRC2 was achieved by targeting other components of the complex, and a different Cre promotor was used. Hence, differences could be due to different cell types or the system used to delete PRC2 activity, which makes any direct comparison difficult. A recent study questions the whole concept of bivalency, implicating that the combined presence of K4 and K27 does not represent a poised state (41). Our data showing that the K9/K27 state is a better predictor of derepression than K4/K27 would reflect that K4/K27 does not represent a poised state wherein activation of transcription has a high probability to occur after loss of K27. However, further studies are required to solve this question.
Derepression of K9/K27 genes results in a more substantial absolute increase in expression when compared to K4/K27 genes. Given that the K9/K27 genes have nearly undetectable expression levels and the K4/K27 genes have substantially higher expression levels in the WT cells, the identified K9/K27 likely represents true K9/K27 promoters. The loss of K9 precedes the up-regulation of expression, which implies that the loss of K9 is not a mere consequence of increased expression upon loss of K27. Rather, this suggests that the presence of K9 at K27+ promoters is coupled to intact PRC2 function, but not simply a function of K27 loss, because several genes lose K27 without losing K9. How the loss of PRC2 activity and/or K27 promotes loss of K9 is not clear. It has previously been shown that PRC2 and K27 cooperate with K9 to maintain the K9-associated heterochromatin protein 1α (42). A more direct link between K27 and K9 has been reported for telomeric heterochromatin assembly, wherein PRC2 and K27 are essential for K9 as well (43). In differentiated mouse embryonic stem cells, K9 is dependent on intact SUZ12 function (44).
The selective vulnerability to loss of PRC2 activity in mDA neurons of the SNpc is reminiscent of how the same cells are hypersensitive in the response to cellular stressors such as 6-OHDA. Thus, loss of identity upon deletion of Eed is mirrored by cell death upon increased cellular stress. Both vulnerability to cellular stressors and mouse genetic models of PD have been coupled to diverse processes such as mitochondrial dysfunction, inflammation, and protein misfolding, whereas the phenotype we report here is the consequence of dysregulation of transcriptional processes. Notably, the electrophysiological alterations of the DatCreEedfl/fl mutants are reminiscent of the age-dependent decline of similar parameters in the MitoPark mouse (45), whereas progressive loss of TH in the SNpc and progressive development of motor deficits mirror key aspects of PD. Thus, the more significant impact of the loss of PRC2 function in SNpc mDA neurons shows that these cells harbor an additional selective vulnerability in addition to the death-promoting effects of cellular stressors, inflammation, and α-synuclein overexpression previously described. Whether this dual vulnerability is mechanistically coupled remains to be addressed. To reveal early divergence in the transcriptional response to PRC2 inactivation between the SNpc and VTA subpopulations, single-cell analysis at earlier time points (e.g., P30 or 4 months) would be an option. Such analysis could, for example, identify potential factors uniquely up-regulated in the VTA with the capacity to protect these cells from the detrimental effects of Eed deletion.
Expression of the homeobox gene Engrailed1 (En1), which is a key survival factor for mDA neurons (46), is reduced in both the mutant SNpc and VTA (fig. S6K). It has previously been shown that there is a pronounced reduction of K27 in mDA neurons of En+/− mice (17). Furthermore, in the same study, En1+/− mutant exhibited heightened sensitivity to 6-OHDA treatment and reduced expression of both Ezh1 and Ezh2. Hence, it appears that there is a link between En1 and levels of K27, wherein En1 facilitates the expression of Ezh1 and Ezh2, thus helping to maintain K27 levels. Reciprocally, inhibition of PRC2 function, in turn, potentially leads to the up-regulation of factors capable of repressing the expression of En1. Given the fundamental role for En1 in the maintenance of mDA neurons, it is possible that the loss of mDA neuronal traits is closely coupled to the reduced levels of En1 in the mutants.
Together, our study elucidates how an epigenetic mechanism controls permanent gene silencing and maintenance of 5HT and dopaminergic identity. It also reveals how loss of such epigenetic control does not compromise neuronal survival but leads to loss of subtype-specific function and to phenotypes that recapitulate symptoms characteristic of PD and mood disorders, providing a deeper understanding of how epigenetic mechanisms could contribute to the etiology of these multifactorial diseases.
MATERIALS AND METHODS
Ethical considerations
All animal experiments were performed according to Swedish guidelines and regulations, and the ethical permits N189/15 and 6259-2020 were granted by “Stockholms Norra djurförsöksetiska nämnd, Sweden.”
Mice
The generation of DatCre, SertCre (23, 29), Rpl10a-mCherry, and Eedflo (22) mice has been previously described. Mouse lines were crossed and generated the DatCreEedfl/flRpl10a-mCherry and the SertCreEedfl/flRpl10a-mCherry lines used in our study. Mice were kept in ventilated cages with controlled 12-hour light/dark cycles, temperature, and humidity, with water and food provided ad libitum. Mice were housed at a maximum number of four males or six females per cage. Both genders were represented in similar numbers for different type of experiments.
Histological analyses
Animals were deeply anesthetized with Avertin intraperitoneal sodium pentobarbital (Apoteksbolaget AB) and perfused with room temperature phosphate buffer (PB) saline (PBS) through the ascending aorta, followed by ice-cold 4% paraformaldehyde. The brains were subsequently removed, postfixed in the same fixative for 16 to 18 hours, and cryoprotected for 24 to 48 hours in 30% sucrose at 4°C, before being cut on a Leica microtome at a thickness of 30 μm. Sections were permeabilized in 5% bovine serum albumin (BSA) in PBS-Tx100 (PBS with 0.5% Triton X-100), followed by primary antibody incubation at 4°C for 16 to 18 hours using sheep anti-TH (1:1000; catalog no. P60101-150, Pel-Freeze), anti-TPH2 (1:500; catalog no. T0678, Sigma-Aldrich), anti-H3K27me3 (1:500; catalog no. 9733, Cell Signaling Technology), and anti-EED (1:500; catalog no. 85322, Cell Signaling Technology). Fluorescent detection was done with an Alexa-tagged secondary antibody from Molecular Probes, donkey anti-sheep (1:500; catalog no. A21448), goat anti-mouse (1:500; catalog no. A21151), and donkey anti-rabbit (1:500; catalog no. A21206). Section images were obtained in an LSM-700 confocal microscope from Zeiss.
Tissue processing for imaging and cell counting
Mouse brains were cleared using the advanced clear, unobstructed brain imaging cocktails and computational analysis (CUBIC) protocol (47) with minor modifications (48). In brief, mice were perfused with 4% paraformaldehyde, and after postfixation, brains were washed in phosphate buffer [PB; 0.1 M (pH 7.6 to 7.8)] at 4°C for 24 hours. Brains were cut in 1-mm slices using a brain matrix. Slices (1 mm) were incubated in CUBIC reagent 1 [25% urea, 25% N,N,N′,N′-tetrakis-(2-hydroxypropyl)ethylenediamine, and 15% Triton X-100] at 37°C for 2 days. Slices were transferred to fresh CUBIC reagent 1 and incubated for a further 24 hours at 37°C, before washing in PB (0.1 M) for 8 hours at room temperature (8 × 1 hour of shaking). Tissue was incubated in blocking solution (5% BSA) for 24 hours at 37°C and switched to anti-MCHERRY (1:5000; catalog no. AB0040-200, SICGEN) for 2 days at 37°C. After washing in PB (0.1 M) for 8 hours at room temperature (8 × 1 hour of shaking), slices were incubated in Alexa 555 and donkey anti-goat (1:500; catalog no. A21432, Invitrogen) for 24 hours at 37°C and washed for 8 hours (8 × 1 hour of shaking) with PB (0.1 M) at room temperature. Slices were afterward incubated in CUBIC reagent 2 (50% sucrose, 25% urea, 10% 2,2′,2″-nitrilotriethanol, and 0.1% Triton X-100) while shaking for 16 to 18 hours at 37°C. Tissue slices were placed in 1-mm-height chambers on a glass slide and imaged in CUBIC reagent 2 in an LSM-700 confocal microscope from Zeiss. Acquired images were analyzed with Imaris Cell Imaging software, and cell bodies of mCHERRY-positive cells were counted.
FACS of cell type–specific nuclei
DatCreEedfl/fl-Rpl10a-mCherry and SertCreEedfl/fl-Rpl10a-mCherry mice were euthanized with CO2, and brains were rapidly removed and transferred into cold PBS. The midbrain and hindbrain, respectively, were dissected under a fluorescent stereoscope and snap-frozen in dry ice. Tissue was thawed and dissociated using a 1-ml Dounce homogenizer (Wheaton) in ice-cold lysis buffer [0.32 M sucrose, 5 nM CaCl2, 3 mM MgAc, 0.1 mM Na2EDTA, 10 mM tris-HCl (pH 8.0), 1 mM dithiothreitol, and 1× complete proteinase inhibitor, EDTA-free (Roche)]. The homogenate was centrifuged for 5 min at 500g, and the pelleted nuclei were resuspended in a nuclear storage buffer [15% sucrose, 10 mM tris-HCl (pH 7.2), 70 mM KCl, and 2 mM MgCl2] supplemented with a ribonuclease (RNase) inhibitor (RNaseOUT, Invitrogen) and a proteinase inhibitor (cOmplete, Roche). When nuclei were used for 10x Genomics single-cell RNA-seq, pelleted nuclei were instead resuspended in 2% BSA with an RNase inhibitor. Resuspended nuclei were filtered through a 30-μm cup (Filcon; BD Biosciences, catalog no. 340625) into a BSA-coated tube for sorting. Nuclei were stained with DAPI for 30 min before sorting.
FACS was performed using a FACSAria Fusion cell sorter and the FACSDiva software (BD Biosciences). The nuclei were identified by forward- and side-scatter gating, a 561-nm laser with a 610/20 filter, and a 405-nm laser with a 450/50 filter, quantifying DNA content per event to assure that only singlets were collected. A 100-mm nozzle, a sheath pressure of 138 to 172 kPa, and an acquisition rate of up to 1000 events per second were used. Cell type–specific nuclei were collected in batches of 1000, supplemented with a nuclear pellet buffer [10 mM tris (pH 8.0), 100 mM NaCl, 2 mM MgCl2, 0.3 M sucrose, and 0.25% IGEPAL CA-630] to a volume of 10 μl when used for bulk RNA-seq or ChIP-seq, whereas for 10x Genomics single-cell RNA-seq, nuclei were preserved in 2% BSA until downstream procedure.
Cell type–specific nuclear RNA-seq and analysis
Sequencing libraries were generated from total RNA of 1000 FACS-sorted cell type–specific nuclei as previously described (20). In brief, total RNA was extracted from nuclei, and the Smartseq2 protocol (49) was implemented to generate libraries. Sequencing was performed on an Illumina NovaSeq 6000 within the National Genomics Infrastructure in SciLifeLab, Stockholm, Sweden. Raw 51–base pair (bp) paired-end reads were aligned to the mouse genome (mm10 assembly) using STAR v2.7.0a with default settings (50). Gene expression was quantified as both counts and fragments per kilobase transcript per million mapped reads (FPKM) using rpkmforgenes (51). Differential gene expression analysis was performed on counts data for genes using DESeq2 (52) with two separate DESeqDataSets, one for all mDA samples and one for all 5HT samples. A total of 35 mDA samples (9 WTs and 10 mutants at 4 months and 8 WTs and 8 mutants at 8 months) and a total of 16 5HT samples (4 WTs and 4 mutants at both 4 and 8 months) were used. Each dataset was filtered for genes with at least a total count of 10 summed across all samples. DEGs were identified by requiring adjusted P < 0.05 and using design formulas controlling for sex and contrasting WT with mutant samples separately for 4 and 8 months. GO terms were obtained from Enrichr (24, 25).
ChIP-seq: ChIP, sequencing, and mapping
ChIP and library preparation for sequencing were performed as previously described in the ULI-NChIP protocol with previously described minor modifications (20). Chromatin from one batch of 1000 nuclei per animal and antibody was immunoprecipitated for 16 to 18 hours at 4°C with 0.25 mg of anti-H3K27me3 (Cell Signaling Technology, 9733), anti-H3K9me3 (Active Motif, 39161), or anti-H3K4me3 (Cell Signaling Technology, 9751). Libraries were sequenced on an Illumina NovaSeq 6000 with 51-bp paired-end reads within the National Genomics Infrastructure in SciLifeLab, Stockholm, Sweden.
A total of 64 ChIP-seq libraries were sequenced (two genotypes: WT and mutants; two time points: 4 and 8 months; four IPs: input, H3K4me3, H3K27me3, and H3K9me3; and lastly, four biological replicates for each combination). Nuclei (1000) were used for each library, and biological replicates correspond to different animals. Reads were mapped to the mm10 mouse genome using bowtie2 v2.3.5.1 with default settings (53). Duplicate reads were marked using Picard v2.10.3. Coverage of mapped ChIP-seq libraries was generated using the tool bamCoverage in deepTools v3.1 with parameters ignoreDuplicates, binSize 50, and normalizeUsing RPKM (54). Signal-to-background relationships were investigated using the plotFingerprint tool in deepTools. On the basis of manual inspection of fingerprint and coverage plots, we decided to use all 64 samples in further analyses.
The median fraction of duplicated reads for all samples was 34% (range, 17 to 52). The median fraction of unmapped reads was 18% (range, 7 to 60). The median total number of million mapped unique reads (after removal of duplicates) was 70 for inputs (range, 38 to 78), 31 for H3K4me3 (range, 17 to 40), 24 for H3K27me3 (range, 8 to 63), and 60 for H3K9me3 (range, 41 to 86). It is important to note that these numbers are per library. In our analyses, we used four biological replicates (four libraries) for each experiment. This means that for H3K27me3, the median number of reads used in total to identify marked regions was approximately 96 million (48 million read pairs). The median number of reads was similar for WT and mutant libraries for input, H3K4me3, and H3K9me3 but not for H3K27me3. For WT libraries, the median number of reads for H3K27me3 was 40 million, and therefore, approximately 160 million reads (80 million read pairs) were used to identify H3K27me3 in WT experiments. For mutants, the number of reads for each biological replicate dropped for H3K27me3 but not for the other markers, as expected. For mutant libraries, the median number of reads for H3K27me3 was 16 million, and therefore, approximately 64 million reads (32 million read pairs) were used to identify H3K27me3 in mutant experiments. On the basis of our previous analysis of data obtained with this ChIP-seq protocol starting with only 1000 nuclei per ChIP (20), we conclude that we have obtained data at a sequencing depth that is more than enough for our analysis strategy.
ChIP-seq: Identifying marked genes and TSS chromatin states
For each histone modification, genotype, and time point, we identified marked genes by comparing the ChIP experiments to the input experiments using the csaw package version 1.22.1 in R as previously described (20). Reads were counted into sliding windows along the genome and duplicate reads, reads with a mapping quality smaller than five, and reads mapping to regions in the curated blacklist of problematic regions available as the bed-file ENCFF547MET from the ENCODE project were removed. Window widths were set to reflect a broad or more narrow distribution of the investigated histone modifications: A width of 1000 bp and a spacing interval of 100 bp were used for H3K27me3 and H3K9me3, whereas a width of 150 bp and a spacing of 50 bp were used for H3K4me3. Read counting was restricted to chromosomes 1 to 19, X and Y. Low-abundance windows were filtered out in two steps: first, by only keeping windows with at least 20 reads across all the eight experiments (i.e., all input and ChIP experiments). Second, only windows displaying an enrichment of reads above the global background were kept by requiring a minimal log2 fold change (lfc) of two for all experiments except lfc = 3 for H3K4me3 at 4 months and lfc = 1.5 for H3K9me3 at 8 months. Composition biases across libraries were normalized using the trimmed mean of M-values method on binned counts.
Counts for windows that passed filtering were tested for differential binding between the four replicated ChIP experiments and the four replicated input experiments as previously described (20). We defined TSS regions as all regions of a size of 20,000 bp centered on a TSS in the org.Mm.eg.db package. We calculated a combined P value for differential binding in each TSS region based on all windows in the region and corrected for multiple testing to obtain a false discovery rate (FDR) using the Benjamini-Hochberg method. We defined a gene as marked by a histone modification in a cell type if it had a TSS region that was differentially bound by the marker compared to the inputs with FDR < 0.05. Last, we assigned each gene a promoter chromatin state in a cell type by combining this binary binding status of H3K27me3, H3K9me3, and H3K4me3 into the eight possible states. In each comparison, four biological replicates were used both for the ChIP and the input experiments.
ChIP-seq: Visualizations
For visualizations, biological replicates were pooled. ChIP-seq coverage files of pooled samples were generated in deepTools using the bamCoverage function as for individual replicates. From the pooled coverage files, ChIP-seq heatmaps and average profile heatmaps were generated with deepTools using the plotHeatmap function and the plotProfile function, respectively. Coverages were plotted using the Integrative Genomics Viewer version 2.8.3.
Enrichment analyses
Genes were mapped between ChIP-seq and gene expression data based on gene symbols. Associations between two gene sets were investigated using 2 × 2 contingency tables and Fisher’s exact tests as in the following example. For mDA, more than 21,000 genes were mapped between the ChIP-seq and cell type–specific nuclear RNA-seq datasets, and each of these genes can be placed uniquely in the 2 × 2 contingency table defined as marked by H3K27me27 in mDA WT at 8 months or not versus up-regulated in WT compared to mutated mDA at 8 months or not. To test whether there was a significant association (enrichment or depletion) between H3K27me3 and up-regulation, two-sided Fisher’s exact test was calculated for this 2 × 2 contingency table. Enrichment of H3K27me3 for the up-regulated genes was calculated as the ratio between the frequency of H3K27me3 marked genes among the up-regulated genes and the frequency of H3K27me3 among all genes in the 2 × 2 contingency table.
Analysis of neurotransmitters by HPLC
HPLC with electrochemical detection (ECD) was based on previously published protocols (55, 56). Briefly, ice-cold 0.1 M perchloric acid was added to the tissue sample, with 50 μl of perchloric acid per 10 mg of tissue. Samples were incubated on ice for 10 min, vortexed, and centrifuged at 16,000g for 10 min at 4°C. Resulting supernatants were filtered through 0.2-μm nylon membrane inserts and centrifuged at 4000g for 5 min. Eluents were immediately stored at −80°C and subjected to HPLC-ECD analysis within 1 week. Standard solutions of l-norepinephrine hydrochloride, (+/−)-epinephrine hydrochloride, DOPAC, 3,4-dihydroxy-l-phenylalanine (dopa), dopamine hydrochloride (DA), 5-HIAA, HVA, 5-HT, 4-hydroxy-3-methoxyphenylglycol hemipiperazinium salt, dl-4-hydroxy-3-methoxymandelic acid, and 3-methoxytyramine hydrochloride were prepared in 0.1 M perchloric acid to obtain final standard concentrations of 200, 100, 50, 10, 5, 2, and 1 ng/ml. Calibration curves were obtained with the Chromeleon software through linear regression of peak area versus concentration. The HPLC-ECD system used was a Dionex Ultimate 3000 series (Dionex, Thermo Fisher Scientific, USA). Analyte separation was performed on a Dionex C18 reversed-phase MD-150 3.2 mm by 250 mm column (3 μm, particle size). Column and analytical cell were kept at 30°C. The mobile phase, which was pumped at a flow rate of 0.4 ml/min, consisted of 75 mM monobasic sodium phosphate, 2.2 mM 1-octanesulfonic acid sodium salt, triethylamine (100 μl/liter), 25 μM EDTA disodium salt, and 10% acetonitrile (v/v) (pH 3.0) adjusted with 85% phosphoric acid. For detection of neurotransmitters and metabolites, the first and second analytical cells were set to −100 and +300 mV, respectively. Processed tissue samples were thawed on ice in the dark for about 1 hour before analysis, placed in the autosampler, and kept at 5°C before injection. Chromatograms were acquired with Dionex Chromeleon 7 software over an acquisition time of 55 min. Analyte concentrations in tissue samples were expressed as nanograms per milligram of tissue.
Electrophysiology of mDA neurons
Mice were perfused with artificial cerebrospinal fluid (aCSF) containing NaCl (126 mM), KCl (2.5 mM), sodium phosphate buffer (1.2 mM), MgCl2 (1.3 mM), CaCl2 (2.4 mM), glucose (10 mM), and NaHCO3 (26 mM). Their brains were rapidly removed, and coronal brain slices containing the SN, 200 μm thick, were prepared with a microslicer (VT 1000S, Leica Microsystem, Heppenheim, Germany) in oxygenated (95% O2 + 5% CO2) ice-cold modified aCSF containing NaCl (15.9 mM), KCl (2 mM), sodium phosphate buffer (1 mM), sucrose (219.7 mM), MgCl2 (5.2 mM), CaCl2 (1.1 mM), glucose (10 mM), and NaHCO3 (26 mM). Slices were incubated, for 1 hour at 32°C and thereafter at 28°C, in oxygenated modified aCSF containing NaCl (126 mM), KCl (2.5 mM), sodium phosphate buffer (1.2 mM), MgCl2 (4.7 mM), CaCl2 (1 mM), glucose (9 mM), and NaHCO3 (23.4 mM). Slices were transferred to a recording chamber and were continuously perfused with oxygenated aCSF at 32° to 34°C.
Whole-cell patch-clamp recordings of visually identified DA neurons in the SN were made as described previously by Yao and co-workers (57). Borosilicate patch electrodes (3 to 5 megohms) were filled with a solution containing 120 mM d-gluconic acid potassium salt, 20 mM KCl, 2 mM MgCl2, 1 mM CaCl2, 10 mM Hepes, 10 mM EGTA, 2 mM MgATP, and 0.3 mM Na3GTP, pH adjusted to 7.3 with KOH. Whole-cell membrane currents and potentials were recorded with a MultiClamp 700B (Axon Instruments, Foster City, CA, USA), acquired at 10 kHz, and filtered at 2 kHz. Data were acquired and analyzed with the pClamp 11 software (Axon Instruments, Foster City, CA, USA).
Viral tracing injections
Mice were anesthetized with isoflurane (4 mg/ml) supplemented in the air, while placed in a separate cage, and afterward, they were mounted in a stereotaxic frame (David Kopf Instruments, Tujunga, CA) equipped with a mouse adapter. Mice were kept anesthetized throughout the procedure by inhaling isoflurane at a concentration of 2 mg/ml. Anterograde tracing of the mDA neurons was achieved by injecting a Cre-dependent AAV expressing EGFP (AAV2/2.pCAG.FLEX.EGFP.WPRE.bGH) (26) in the midbrain of 8-month-old mice. Using bregma as a reference point, 1 μl of the AAV virus was injected unilaterally reaching the SN [anteroposterior (AP): −2.9, mediolateral (ML): −1.25, and dorsoventral (DV): −4.5] and 1 μl reaching the VTA (AP: −3.1, ML: −0.5, and DV: −4.2). All coordinates are in millimeters relative to bregma according to The Mouse Brain in Stereotaxic Coordinates (Academic Press, San Diego, CA, 2012). Virus injection was executed at a dispense rate of 0.2 μl/min. Animals were euthanized 3 weeks after surgery by CO2.
Open-field test
For assessment of general ambulatory ability, mice were placed in 45 cm by 45 cm by 28 cm plastic, nontransparent boxes, and activity was recorded for 60 min using the Ethovision XT 15 Noldus software. Activity is measured as the mean of total distance covered in 1 hour with 5-min time bins.
Pole test
Motor coordination and movement initiation were assessed by the pole test. To perform this task, mice were placed on the top of a wooden pole (50 cm in height and 1 cm in diameter) that is fixed to a wooden stable base. Each animal was placed with all four limbs grasping the pole and facing the tip of it. The total time they spent to turn themselves downward and climb down the pole was calculated.
The day before the experiment, mice were trained to orient themselves and descent the pole 5 to 10 times. On the test day, each animal was recorded performing the test five times, and the average time for every mouse is presented. To avoid exhaustion of the mice, a maximum time for every trial was defined at 120 s, a time score that was also given to objects that failed the experiment, by either not turning themselves downward at all or by descending the pole by rolling down.
Elevated plus maze
The EPM test evaluates anxiety-like behavior in mice. The setup consisted of a cross with two open arms and two enclosed ones, elevated ~1 m from the ground. Mice were placed in the center of the cross, facing toward the open compartment. Mobility and preference over the two types of arms were recorded for 5 min using Ethovision XT Noldus software.
Rearing
Locomotion activity and exploratory behavior were evaluated by the rearing events. Mice were placed in a plexiglass 15-cm-diameter cylinder and videotaped for 10 min. Vertical rearing events supported on the cylinder wall or unsupported were manually counted. Mice were tested individually, without any visual interaction with each other.
Forced swim test
The forced swim test or Porsolt test was used to evaluate depression-like traits in rodents. More specifically, mice were forced to swim in a plexiglass cylinder (15 cm in diameter and 30 cm in height) halfway filled with water at ±25°C for 6 min. During the session, mice were recorded, and the time of immobility was assessed by a trained observer. The first 2 min were counted as habituation time, and the remaining 4 min served as the actual experiment. During those minutes, immobility time was counted as the time mice spent floating in the water without any effort to move but only the necessary moves that would let them keep the head above the surface. Video recordings were scored twice from the same observer, and the average of those scores was calculated.
Rotarod
Motor coordination was evaluated by using the rotarod test with increasing speed. Mice were familiarized with the apparatus for three trials at a fixed speed of 4 rpm. After a resting period of 2 hours, mice performed the test with increasing speed over time. The acceleration protocol spans from 4 to 38 rpm within 5 min, where the latency to fall was measured. Every mouse participated three times, and the average time was calculated for every individual.
Grip strength
Mice were placed horizontally on a metal net-shaped frame with all four limbs, and instantly, the apparatus was twisted 180°. With that setup, the back of the mice was facing the ground, and the time they managed to hold gripped on the frame was calculated. Each individual was monitored for three trials, and the average time was noted.
Conditioned place preference
Experiment was performed in a rectangular apparatus with three chambers measuring 15 cm by 25 cm each. One compartment was colored gray, the middle one was white, and the last one was colored with black and white stripes. One the first day, mice were allowed to explore the whole apparatus for 20 min. On days 2 and 3, mice were confined to one compartment and injected with cocaine (20 mg/kg) or saline. After injection, they remained in the compartment for 30 min. On days 4 and 5, the same pairings were repeated. Pairing of drug and compartment was counterbalanced across animals. On day 6, mice were placed in the middle compartment and were freely allowed to move between all three of them. While there was no injection on that day, mice were recorded, and their preference for every compartment was calculated. The following day, all mice were injected with cocaine (20 mg/kg), and their motor skills and mobility were examined in the open-field test, where they were recorded for 60 min.
snRNA-seq: Sequencing, mapping, and expression levels
Tissues from mouse ventral midbrain (3 controls and 3 KOs) were used to obtain single nuclei (1883 controls and 4103 KOs). Single-nucleus libraries were made using Single Cell 3′ v3 on Chromium platform (10x Genomics and SciLifeLab, Stockholm) in accordance with the manufacturer’s protocols. Libraries were sequenced on a NovaSeq 6000 system (NGI, SciLifeLab, Stockholm). Sequenced reads were demultiplexed and aligned to transcriptome: mm10-820 3.0.0_premrna using CellRanger Pipeline version 3.1.0 (10x Genomics). Quality control and filtering of data were performed in multiple steps. First, percent_mito% and percent_ribo% were computed on the basis of the percentage of transcripts that map to mitochondrial genes and ribosomal genes, respectively. Then, CellCycleScoring (score cell cycle phases), S.Score, G2M.Score, and phase columns were calculated on the basis of the expression of G2-M and S phase canonical markers. Next, the average relative expression of each gene per cell, i.e., the ratio of a gene’s unique molecular identifier (UMI) counts to the sum of all UMIs per library (nucleus), was calculated. Malat1 was, on average, the most abundant gene per library (nucleus), with a mean fraction of total UMI counts per library of about 2% and being expressed in all nuclei. Malat1 is frequently detected in polyadenylate captured RNA-seq libraries, independent of the used protocol. However, compared to other methods, it is even more abundant in snRNA-seq and therefore used to estimate the nuclear proportion of total mRNA (57). Malat1 was filtered out for downstream analysis. Genes detected in fewer than three libraries were removed. Libraries with less than 500 detected genes, with more than 10,000 detected genes (doublets), and with percent mito > 0.9 were also filtered out. After the filtering steps, 1772 control and 3968 KO nuclei remained. The remaining nuclei have UMIs per library of more than 620, genes per library of more than 500, with average UMIs per library of 23,474, and average genes per library of 4965. Then, a Seurat object was made with these filtered data (Seurat version “4.0.1”) (58). Data were log-normalized with Seurat NormalizeData function and a scale.factor of 10 × 104.
snRNA-seq: UMAP clustering and identification of marker genes
The 2000 most highly variable genes (HVGs) were identified using the FindVariableFeatures function [selection.method = “vst,” with clip.max = (n.Cells)2]. The log-normalized data were then scaled and centered using the ScaleData function (model.use = “linear”). Principal components analysis was done using the 2000 HVGs, which reduced the dimensionality of the data into the calculated components while maintaining the most important gene expression differences across libraries. We identified the most significant principal components (PCs) based on the JackStraw procedure using the JackStraw (num.replicate = 100) and ScoreJackStraw functions. After plotting the JackStraw scores, noticeable gaps in P values were observed at PCs 18, 22, and 32. We also used a heuristic method called ElbowPlot to visualize the SDs of the PCs, and at PCs 18, 22, and 32, there were noticeable inflection points (“elbows”). We chose to continue with the first 32 PCs, which we think contain most of the variance without losing any true signal that reflects biological heterogeneity. Next, we used PCs 1 to 32 as input to the Seurat FindNeighbors function (k.param = 20, dims = 1:32), as well as in the FindClusters function with the resolution parameter between 0.3 and 2.0 in 0.1 intervals, using the Louvain algorithm. We found the most optimal resolution for res = 0.3 with 18 clusters (a purely heuristic approach based on known markers for cell types, neuroanatomical regions, and hierarchical dendrograms). The final nine subgroups were created by combining related clusters together, based on cluster-specific markers in UMAP visualization and hierarchical dendrograms. The same PCs 1 to 32 were used in the Seurat function RunUMAP. Subgroup-enriched markers (up- and down-regulated) were identified using Seurat FindAllMarkers function with Wilcoxon rank sum test (logfc.threshold = 0.25, min.pct = 0.1, min.diff.pct = -Inf, and only.pos = FALSE). In this function, P values were adjusted for multiple comparisons with the Bonferroni correction. Differential gene expression analysis was also performed between all WT nuclei and all KO nuclei, irrespective of clusters/subgroups, as well as per each subgroup between KO and WT, using Seurat FindMarkers function with the same setting and parameters as above.
snRNA-seq: Hierarchical clustering and heatmaps
Hierarchical clustering was done for all nuclei with subgroups defined in UMAP visualization as the identity classes (fig. S6A). Hierarchical clustering was done for mDA neuron nuclei with identity classes defined as subgroup_genotype, subgroups (SNpc, VTA1 to VTA3, and SNpc/VTA), and by genotype (WT and KO), using either the 2000 most HVGs (Fig. 8D and fig. S6D) or the 205 DEGs between KO and WT nuclei (fig. S6F). Seurat BuildClusterTree function was used to make the dendrograms (Fig. 8D and fig. S6A). This clustering results in a phylogenetic tree based on calculating the “average” cell from each “identity class.” The distance matrix for this tree was calculated using normalized gene expression. Heatmaps with clustering dendrograms were made for the subgroup genotype identity classes in mDA neurons, using either the 2000 HVGs (fig. S6D) or the 205 DEGs between KO and WT nuclei (fig. S6F). Heatmaps were generated using ComplexHeatmap version2.6.261.
snRNA-seq: Composite average expression of gene sets
The Seurat AddModuleScore function was used to calculate the average expression levels of gene sets (modules) on a single-nucleus level. An mDA neuron composite average expression (mDA module) was calculated using the (Th, Slc6a3, En1, Nr4a2) gene set. Another composite average expression (module) was calculated on the basis of the top 25 most highly up-regulated genes in the KO nuclei when compared to the WT nuclei, irrespective of their clusters.
snRNA-seq: DESeq2 analysis
To enable a more thorough comparison between KO and WT DEGs from bulk RNA-seq (8 months old) and snRNA-seq data, DESeq2 (DESeq2_1.34.0) package was used to perform KO versus WT DEG analysis in snRNA-seq data. Next, the overlap of DEGs from both datasets was compared. Comparison of means of lfc values of the DEGs that are either common between bulk and snRNA-seq datasets or unique in snRNA-seq dataset was done for all cells, in the SNpc and VTA subgroups, using Wilcoxon rank sum test with continuity correction and 95% confidence interval.
Acknowledgments
Funding: Support was provided by the Knut and Alice Wallenberg Foundation (grant 2013.0075 to J.H., T.P., P.S., and K.C.), The Swedish Research Council (VR 2016-02536 to J.H.; VR 2016-02506 and VR 2020-00884 to T.P.), The Swedish Brain Foundation (to J.H. and T.P.), and Torsten Söderbergs Stiftelse (to T.P.). M.R. was financially supported by the Knut and Alice Wallenberg Foundation as part of the National Bioinformatics Infrastructure Sweden at SciLifeLab. The computations were enabled by resources in project SNIC 2021/23-184 provided by the Swedish National Infrastructure for Computing (SNIC) at UPPMAX, partially funded by the Swedish Research Council through grant agreement no. 2018-05973.
Author contributions: Conceptualization: J.H., K.T., and E.S. Methodology: K.T., E.S., M.R., and B.Y.-S. Formal analysis: J.H., K.T., B.Y.-S., M.R., and O.S. Investigation: K.T., B.Y.-S., O.S., W.P., L.G., V.S., and I.A. Data curation: K.T., B.Y.-S., and M.R. Visualization: J.H., K.T., B.Y.-S., M.R., and T.P. Supervision: J.H., T.P., P.S., and K.C. Writing—original draft: J.H. and K.T. Writing—review and editing: J.H., T.P., O.S., K.C., K.T., B.Y.-S., and M.R.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The datasets generated and analyzed during the current study are available in the GEO repository with accession number GSE189018. All data needed to evaluate the conclusions in the paper are present in the paper, the Supplementary Materials, and the GEO repository.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S8
Movies S1 to S5
REFERENCES AND NOTES
- 1.Deneris E. S., Hobert O., Maintenance of postmitotic neuronal cell identity. Nat. Neurosci. 17, 899–907 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holmberg J., Perlmann T., Maintaining differentiated cellular identity. Nat. Rev. Genet. 13, 429–439 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Bantignies F., Cavalli G., Cellular memory and dynamic regulation of polycomb group proteins. Curr. Opin. Cell Biol. 18, 275–283 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Di Croce L., Helin K., Transcriptional regulation by Polycomb group proteins. Nat. Struct. Mol. Biol. 20, 1147–1155 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Corley M., Kroll K. L., The roles and regulation of Polycomb complexes in neural development. Cell Tissue Res. 359, 65–85 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.von Schimmelmann M., Feinberg P. A., Sullivan J. M., Ku S. M., Badimon A., Duff M. K., Wang Z., Lachmann A., Dewell S., Ma’ayan A., Han M. H., Tarakhovsky A., Schaefer A., Polycomb repressive complex 2 (PRC2) silences genes responsible for neurodegeneration. Nat. Neurosci. 19, 1321–1330 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsuboi M., Hirabayashi Y., Gotoh Y., Diverse gene regulatory mechanisms mediated by Polycomb group proteins during neural development. Curr. Opin. Neurobiol. 59, 164–173 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Wever I., Wagemans C., Smidt M. P., EZH2 is essential for fate determination in the mammalian isthmic area. Front. Mol. Neurosci. 12, 76 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohn F., Weber M., Rebhan M., Roloff T. C., Richter J., Stadler M. B., Bibel M., Schubeler D., Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol. Cell 30, 755–766 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Jaenisch R., Wagschal A., Feil R., Schreiber S. L., Lander E. S., A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Nestler E. J., Pena C. J., Kundakovic M., Mitchell A., Akbarian S., Epigenetic basis of mental illness. Neuroscientist 22, 447–463 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sodersten E., Feyder M., Lerdrup M., Gomes A. L., Kryh H., Spigolon G., Caboche J., Fisone G., Hansen K., Dopamine signaling leads to loss of Polycomb repression and aberrant gene activation in experimental parkinsonism. PLOS Genet. 10, e1004574 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J., Hart R. P., Mallimo E. M., Swerdel M. R., Kusnecov A. W., Herrup K., EZH2-mediated H3K27 trimethylation mediates neurodegeneration in ataxia-telangiectasia. Nat. Neurosci. 16, 1745–1753 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seong I. S., Woda J. M., Song J. J., Lloret A., Abeyrathne P. D., Woo C. J., Gregory G., Lee J. M., Wheeler V. C., Walz T., Kingston R. E., Gusella J. F., Conlon R. A., MacDonald M. E., Huntingtin facilitates polycomb repressive complex 2. Hum. Mol. Genet. 19, 573–583 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vialou V., Feng J., Robison A. J., Nestler E. J., Epigenetic mechanisms of depression and antidepressant action. Annu. Rev. Pharmacol. Toxicol. 53, 59–87 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Munoz A., Lopez-Lopez A., Labandeira C. M., Labandeira-Garcia J. L., Interactions between the serotonergic and other neurotransmitter systems in the basal ganglia: Role in parkinson’s disease and adverse effects of L-DOPA. Front. Neuroanat. 14, 26 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rekaik H., Blaudin de The F. X., Fuchs J., Massiani-Beaudoin O., Prochiantz A., Joshi R. L., Engrailed homeoprotein protects mesencephalic dopaminergic neurons from oxidative stress. Cell Rep. 13, 242–250 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicetto D., Zaret K. S., Role of H3K9me3 heterochromatin in cell identity establishment and maintenance. Curr. Opin. Genet. Dev. 55, 1–10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaefer A., Sampath S. C., Intrator A., Min A., Gertler T. S., Surmeier D. J., Tarakhovsky A., Greengard P., Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron 64, 678–691 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sodersten E., Toskas K., Rraklli V., Tiklova K., Bjorklund A. K., Ringner M., Perlmann T., Holmberg J., A comprehensive map coupling histone modifications with gene regulation in adult dopaminergic and serotonergic neurons. Nat. Commun. 9, 1226 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Margueron R., Justin N., Ohno K., Sharpe M. L., Son J., Drury W. J. III, Voigt P., Martin S. R., Taylor W. R., De Marco V., Pirrotta V., Reinberg D., Gamblin S. J., Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461, 762–767 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie H., Xu J., Hsu J. H., Nguyen M., Fujiwara Y., Peng C., Orkin S. H., Polycomb repressive complex 2 regulates normal hematopoietic stem cell function in a developmental-stage-specific manner. Cell Stem Cell 14, 68–80 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ekstrand M. I., Terzioglu M., Galter D., Zhu S., Hofstetter C., Lindqvist E., Thams S., Bergstrand A., Hansson F. S., Trifunovic A., Hoffer B., Cullheim S., Mohammed A. H., Olson L., Larsson N. G., Progressive parkinsonism in mice with respiratory-chain-deficient dopamine neurons. Proc. Natl. Acad. Sci. U.S.A. 104, 1325–1330 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen E. Y., Tan C. M., Kou Y., Duan Q., Wang Z., Meirelles G. V., Clark N. R., Ma’ayan A., Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuleshov M. V., Jones M. R., Rouillard A. D., Fernandez N. F., Duan Q., Wang Z., Koplev S., Jenkins S. L., Jagodnik K. M., Lachmann A., McDermott M. G., Monteiro C. D., Gundersen G. W., Ma’ayan A., Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oh S. W., Harris J. A., Ng L., Winslow B., Cain N., Mihalas S., Wang Q., Lau C., Kuan L., Henry A. M., Mortrud M. T., Ouellette B., Nguyen T. N., Sorensen S. A., Slaughterbeck C. R., Wakeman W., Li Y., Feng D., Ho A., Nicholas E., Hirokawa K. E., Bohn P., Joines K. M., Peng H., Hawrylycz M. J., Phillips J. W., Hohmann J. G., Wohnoutka P., Gerfen C. R., Koch C., Bernard A., Dang C., Jones A. R., Zeng H., A mesoscale connectome of the mouse brain. Nature 508, 207–214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brooks S. P., Dunnett S. B., Tests to assess motor phenotype in mice: A user’s guide. Nat. Rev. Neurosci. 10, 519–529 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Collins A. L., Saunders B. T., Heterogeneity in striatal dopamine circuits: Form and function in dynamic reward seeking. J. Neurosci. Res. 98, 1046–1069 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhuang X., Masson J., Gingrich J. A., Rayport S., Hen R., Targeted gene expression in dopamine and serotonin neurons of the mouse brain. J. Neurosci. Methods 143, 27–32 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Pellow S., Chopin P., File S. E., Briley M., Validation of open:closed arm entries in an elevated plus-maze as a measure of anxiety in the rat. J. Neurosci. Methods 14, 149–167 (1985). [DOI] [PubMed] [Google Scholar]
- 31.Mosienko V., Bert B., Beis D., Matthes S., Fink H., Bader M., Alenina N., Exaggerated aggression and decreased anxiety in mice deficient in brain serotonin. Transl. Psychiatry 2, e122 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitney M. S., Shemery A. M., Yaw A. M., Donovan L. J., Glass J. D., Deneris E. S., Adult brain serotonin deficiency causes hyperactivity, circadian disruption, and elimination of siestas. J. Neurosci. 36, 9828–9842 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poulin J. F., Gaertner Z., Moreno-Ramos O. A., Awatramani R., Classification of midbrain dopamine neurons using single-cell gene expression profiling approaches. Trends Neurosci. 43, 155–169 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirabayashi Y., Suzki N., Tsuboi M., Endo T. A., Toyoda T., Shinga J., Koseki H., Vidal M., Gotoh Y., Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron 63, 600–613 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Wever I., von Oerthel L., Wagemans C., Smidt M. P., EZH2 influences mdDA neuronal differentiation, maintenance and survival. Front. Mol. Neurosci. 11, 491 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu T. T., Heyne S., Dror E., Casas E., Leonhardt L., Boenke T., Yang C. H., Sagar, Arrigoni L., Dalgaard K., Teperino R., Enders L., Selvaraj M., Ruf M., Raja S. J., Xie H., Boenisch U., Orkin S. H., Lynn F. C., Hoffman B. G., Grun D., Vavouri T., Lempradl A. M., Pospisilik J. A., The polycomb-dependent epigenome controls β cell dysfunction, dedifferentiation, and diabetes. Cell Metab. 27, 1294–1308.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lipinski M., Munoz-Viana R., Del Blanco B., Marquez-Galera A., Medrano-Relinque J., Carames J. M., Szczepankiewicz A. A., Fernandez-Albert J., Navarron C. M., Olivares R., Wilczynski G. M., Canals S., Lopez-Atalaya J. P., Barco A., KAT3-dependent acetylation of cell type-specific genes maintains neuronal identity in the adult mouse brain. Nat. Commun. 11, 2588 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sawai A., Pfennig S., Bulajic M., Miller A., Khodadadi-Jamayran A., Mazzoni E. O., Dasen J. S., PRC1 sustains the integrity of neural fate in the absence of PRC2 function. eLife 11, e72769 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maze I., Wenderski W., Noh K. M., Bagot R. C., Tzavaras N., Purushothaman I., Elsasser S. J., Guo Y., Ionete C., Hurd Y. L., Tamminga C. A., Halene T., Farrelly L., Soshnev A. A., Wen D., Rafii S., Birtwistle M. R., Akbarian S., Buchholz B. A., Blitzer R. D., Nestler E. J., Yuan Z. F., Garcia B. A., Shen L., Molina H., Allis C. D., Critical role of histone turnover in neuronal transcription and plasticity. Neuron 87, 77–94 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jadhav U., Nalapareddy K., Saxena M., O’Neill N. K., Pinello L., Yuan G. C., Orkin S. H., Shivdasani R. A., Acquired tissue-specific promoter bivalency is a basis for PRC2 necessity in adult cells. Cell 165, 1389–1400 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.R. N. Shah, A. T. Grzybowski, J. Elias, Z. Chen, T. Hattori, C. C. Lechner, P. W. Lewis, S. Koide, B. Fierz, A. J. Ruthenburg, Re-evaluating the role of nucleosomal bivalency in early development. bioRxiv 2021.09.09.458948 [Preprint]. 10 September 2021. 10.1101/2021.09.09.458948. [DOI]
- 42.Boros J., Arnoult N., Stroobant V., Collet J. F., Decottignies A., Polycomb repressive complex 2 and H3K27me3 cooperate with H3K9 methylation to maintain heterochromatin protein 1α at chromatin. Mol. Cell. Biol. 34, 3662–3674 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Montero J. J., Lopez-Silanes I., Megias D., Fraga M. F., Castells-García Á., Blasco M. A., TERRA recruitment of polycomb to telomeres is essential for histone trymethylation marks at telomeric heterochromatin. Nat. Commun. 9, 1548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de la Cruz C. C., Kirmizis A., Simon M. D., Isono K., Koseki H., Panning B., The polycomb group protein SUZ12 regulates histone H3 lysine 9 methylation and HP1 alpha distribution. Chromosome Res. 15, 299–314 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Branch S. Y., Chen C., Sharma R., Lechleiter J. D., Li S., Beckstead M. J., Dopaminergic neurons exhibit an age-dependent decline in electrophysiological parameters in the mitopark mouse model of parkinson’s disease. J. Neurosci. 36, 4026–4037 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sonnier L., Le Pen G., Hartmann A., Bizot J. C., Trovero F., Krebs M. O., Prochiantz A., Progressive loss of dopaminergic neurons in the ventral midbrain of adult mice heterozygote for Engrailed1. J. Neurosci. 27, 1063–1071 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Susaki E. A., Tainaka K., Perrin D., Yukinaga H., Kuno A., Ueda H. R., Advanced CUBIC protocols for whole-brain and whole-body clearing and imaging. Nat. Protoc. 10, 1709–1727 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Calvigioni D., Máté Z., Fuzik J., Girach F., Zhang M. D., Varro A., Beiersdorf J., Schwindling C., Yanagawa Y., Dockray G. J., McBain C. J., Hökfelt T., Szabó G., Keimpema E., Harkany T., Functional differentiation of cholecystokinin-containing interneurons destined for the cerebral cortex. Cereb. Cortex 27, 2453–2468 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Picelli S., Faridani O. R., Bjorklund A. K., Winberg G., Sagasser S., Sandberg R., Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramskold D., Wang E. T., Burge C. B., Sandberg R., An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput. Biol. 5, e1000598 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramirez F., Ryan D. P., Gruning B., Bhardwaj V., Kilpert F., Richter A. S., Heyne S., Dundar F., Manke T., deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hubbard K. E., Wells A., Owens T. S., Tagen M., Fraga C. H., Stewart C. F., Determination of dopamine, serotonin, and their metabolites in pediatric cerebrospinal fluid by isocratic high performance liquid chromatography coupled with electrochemical detection. Biomed. Chromatogr. 24, 626–631 (2010). [DOI] [PubMed] [Google Scholar]
- 56.Yang L., Beal M. F., Determination of neurotransmitter levels in models of Parkinson’s disease by HPLC-ECD. Methods Mol. Biol. 793, 401–415 (2011). [DOI] [PubMed] [Google Scholar]
- 57.Bakken T. E., Hodge R. D., Miller J. A., Yao Z., Nguyen T. N., Aevermann B., Barkan E., Bertagnolli D., Casper T., Dee N., Garren E., Goldy J., Graybuck L. T., Kroll M., Lasken R. S., Lathia K., Parry S., Rimorin C., Scheuermann R. H., Schork N. J., Shehata S. I., Tieu M., Phillips J. W., Bernard A., Smith K. A., Zeng H., Lein E. S., Tasic B., Single-nucleus and single-cell transcriptomes compared in matched cortical cell types. PLOS ONE 13, e0209648 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hao Y., Hao S., Andersen-Nissen E., Mauck W. M. III, Zheng S., Butler A., Lee M. J., Wilk A. J., Darby C., Zager M., Hoffman P., Stoeckius M., Papalexi E., Mimitou E. P., Jain J., Srivastava A., Stuart T., Fleming L. M., Yeung B., Rogers A. J., McElrath J. M., Blish C. A., Gottardo R., Smibert P., Satija R., Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Tables S1 to S8
Movies S1 to S5