Abstract
The deep biosphere is the largest microbial habitat on Earth and features abundant bacterial endospores. Whereas dormancy and survival at theoretical energy minima are hallmarks of microbial physiology in the subsurface, ecological processes such as dispersal and selection in the deep biosphere remain poorly understood. We investigated the biogeography of dispersing bacteria in the deep sea where upward hydrocarbon seepage was confirmed by acoustic imagery and geochemistry. Thermophilic endospores in the permanently cold seabed correlated with underlying seep conduits reveal geofluid-facilitated cell migration pathways originating in deep petroleum-bearing sediments. Endospore genomes highlight adaptations to life in anoxic petroleum systems and bear close resemblance to oil reservoir microbiomes globally. Upon transport out of the subsurface, viable thermophilic endospores reenter the geosphere by sediment burial, enabling germination and environmental selection at depth where new petroleum systems establish. This microbial dispersal loop circulates living biomass in and out of the deep biosphere.
Subsurface microbial dispersal facilitated by geological processes circulates living biomass in and out of the deep biosphere.
INTRODUCTION
Identifying dispersal vectors that distribute microorganisms throughout the biosphere is critical to understanding biogeography and Earth system functioning. Whereas distributions of animals and plants have been studied since the time of Darwin (1), related ecological processes are harder to elucidate in the microbial realm where the effects of dispersal and environmental selection must be disentangled (2–4). Dormant populations of microbes retain viability while enduring inhospitable conditions in relation to growth requirements, allowing dispersal to be studied directly without the influence of conflating factors such as selection, drift, or mutation (3). Bacterial endospores are equipped to survive dispersal over long distances and time scales (5), with reports of viable spores ~2.5 km beneath the seafloor (6) suggesting journeys lasting millions of years. This points to a genetically and functionally diverse seed bank of microbes that can be revived if subsurface environmental conditions select for their traits (7, 8).
The marine subsurface biosphere contains an estimated 1029 microbial cells contributing up to 2% of the total living biomass on Earth (9). Whereas deep biosphere populations exhibit exponentially decreasing numbers with depth (10), endospores experience less pronounced declines during burial and are estimated to outnumber vegetative cells in deeper marine sediments (11, 12). Measurements of the endospore-specific biomarker dipicolinic acid (DPA) indicate remarkably high numbers of endospores buried in warm sediments hundreds to thousands of meters deep, with depth profiles revealing that temperature influences sporulation and germination (10). This is consistent with the prevalence of endospore-forming Firmicutes in microbiome surveys of hot oil reservoirs from around the world (13), where they actively contribute to biogeochemical cycling. In the deep biosphere, these petroleum systems represent energy rich oases (14, 15) that select for thermophilic organotrophs, including anaerobic hydrocarbon degraders, sulfate reducers, and other groups. Accordingly, cell densities in oil reservoirs can be an order of magnitude higher than those in surrounding sediments (16).
Hydrocarbon seepage up and out of deep petroleum systems is a widespread process in the world’s oceans (17). Studies of thermophilic spores in cold surface sediments globally (18, 19) have invoked warm-to-cold dispersal routes such as hydrocarbon seeps to explain these observations (20). In the Gulf of Mexico where cold seeps are common (21), spore-forming thermophiles in surface sediments are correlated with the presence of migrated liquid hydrocarbons (22). While buoyant gas migration mediates upward dispersal of cells in the top few centimeters (23), whether viable cells from deeper and hotter subsurface layers can be similarly circulated over greater depths and time scales remains hypothetical. Here, we compare deep-sea sediments from the northwest (NW) Atlantic Ocean (fig. S1) using geophysics, hydrocarbon geochemistry, spore germination dynamics, and genomics to demonstrate dispersal of viable cells throughout the marine subsurface. This geologically mediated microbial dispersal loop transports living biomass via upward seepage and downward burial and represents a previously overlooked mechanism for ecological maintenance and preservation of life in the subsurface biosphere.
RESULTS
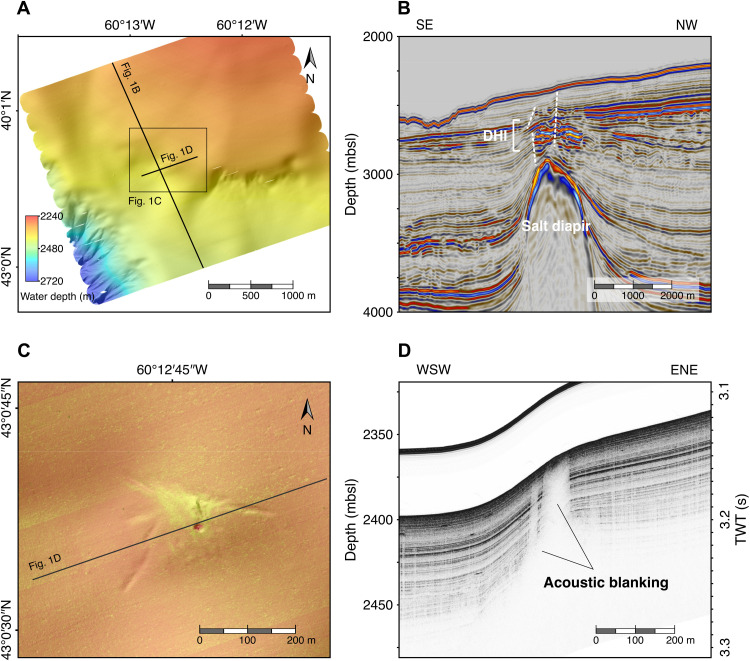
Structural geology indicative of deep subsurface to surface geofluid conduits with the potential to transport microbial cells was determined by multichannel two-dimensional (2D) and 3D seismic reflection surveys along the NW Atlantic Scotian Slope. Geophysical surveys covered ~70,000 km2 and obtained ~10,000 m of subsurface stratigraphic imagery in up to 3400 m of water depth (Fig. 1A). Global estimates of the extent of the deep biosphere predict that marine sediments in this study area harbor at least 1025 microbial cells (24) and even higher numbers of bacterial endospores (11). Detecting seabed hydrocarbon seepage in deep-sea settings similar to this is very challenging (25); thus, a multidisciplinary strategy was used. Colocation at the seabed of the updip limit of deep-seated faults and seismic reflection anomalies considered to be direct hydrocarbon indicators were used to identify potential subsurface seep networks (Fig. 1B) (26). These large-scale geophysical survey results were refined through high-resolution seismic reflection, side scan sonar, and multibeam bathymetry. Morphological features included a mounded structure with high backscatter intensity intersected by an elongated fracture-like depression (Fig. 1C) and a circular pockmark, suggesting the presence of a seep-like structure. Immediately beneath these features, high-resolution subsurface seismic profiling revealed a localized acoustic blanking zone (Fig. 1D), suggesting the presence of gas (27) and a hydrocarbon migration pathway through subsurface sediments. On this basis, sites were prioritized for sediment coring to enable further geochemistry, microbiology, and genomics.
Fig. 1. Deep subsurface to surface geofluid migration.
(A) Seafloor surface map derived from autonomous underwater vehicle (AUV) multibeam bathymetric sonar data. (B) Seismic cross section through the Tangier 3D extending to 4000 m below sea level (mbsl) and >1500 m below sea floor (mbsf), showing a buried salt diapir and the location and direction of crestal faults (white dashed lines), including an interval with direct hydrocarbon indicators (DHI). SE, southeast. (C) Combined mosaic of side scan sonar data and shaded relief bathymetry of the area surrounding a seep structure, indicating a pockmark feature and a small mound morphology. High backscatter intensity, related to distinctive properties of near-surface sediment, is shown in light-yellowish tones. (D) AUV-based sub-bottom profiling showing localized acoustic blanking under the seep structure, indicative of upward migration of fluid originating deeper in the sediment. WSW, west-southwest; ENE, east-northeast; TWT, two-way travel time.
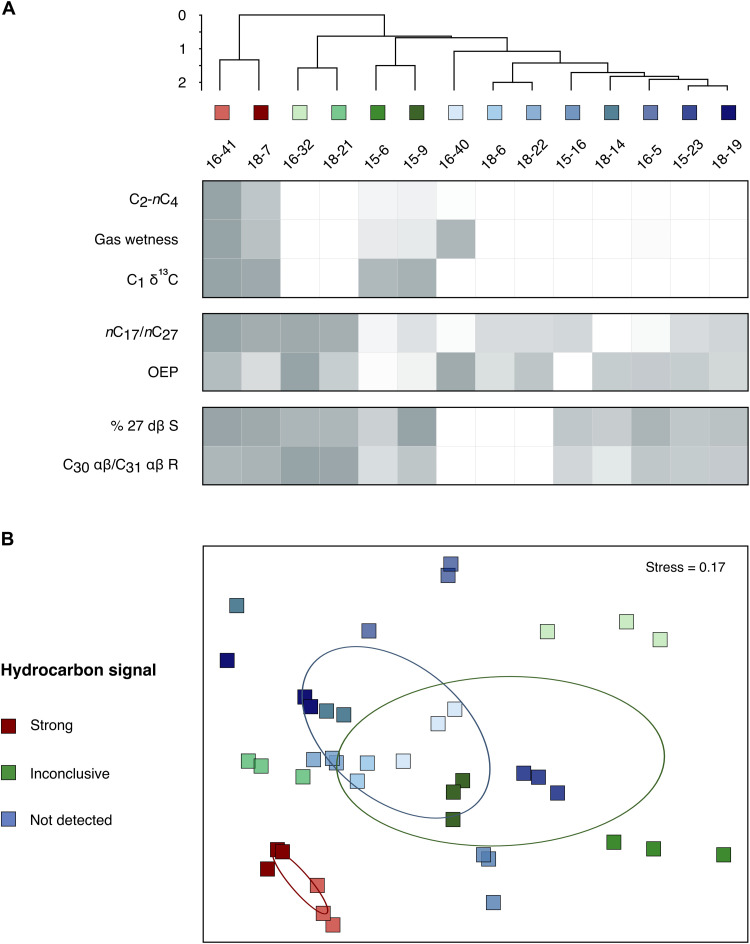
Locations showing seismic evidence of migrated hydrocarbons originating from a deep subsurface source (table S1) were examined in greater detail by comparing chemical differences in hydrocarbon signals from 14 different sites (Fig. 2A and table S2). Higher concentrations of thermogenic C2-nC4 compounds, elevated wet gas ratios and heavy δ13C values for methane (−42 to −52 per mil) in interstitial gas, coupled with liquid hydrocarbon extracts featuring elevated nC17/nC27 ratios and a lack of odd-over-even alkane distributions in the nC23–33 range, provided clear evidence of thermogenic hydrocarbons at two sites. Thermogenic hydrocarbons are formed at depth, making these sites candidates for geofluid dispersal of deep biosphere microbes. A deep origin of these hydrocarbons is further supported by higher proportions of thermally derived diasteranes relative to regular steranes (% 27 dβ S) and more thermally mature terpane distributions (C30 αβ relative to C31 αβ 22R hopane) in these two cores. These signatures of seepage are indicative of an underlying petroleum system (17, 27, 28), consistent with the geophysical evidence shown in Fig. 1. At the 12 other sites, thermogenic hydrocarbon signals were either inconclusive (n = 4) or not detected (n = 8).
Fig. 2. Hydrocarbon geochemistry and microbial community variance between seabed sampling sites.
(A) Gas (∑C2-nC4, gas wetness, and C1 δ13C), liquid hydrocarbon extract [nC17/nC27 and odd-over-even predominance (OEP)], and biomarker (% 27 dβ S and C30 αβ/C31 αβ 22R) measurements to assess the presence of thermogenic hydrocarbons. Each parameter is scaled between 0 and 1, as shown in the heatmap. Cores are represented by maximum values in instances where multiple depths from a core were tested (see table S2). Hierarchical clustering of the Euclidean distance between scaled values highlights groups of sites where the evidence for the presence of thermogenic hydrocarbons is strong (red), inconclusive (green), or not detected (blue). (B) The same three groupings are reflected in Bray-Curtis dissimilarity in microbial community composition after sediment incubation at 50°C (ellipses indicate SDs of weighted averaged means of within-group distances for each of the three groups). Results from 40° and 60°C incubations showed similar clustering (see fig. S2). This demonstrates that sites with strong thermogenic hydrocarbon signals have distinct populations of seep-associated thermophilic spores (see also table S4), relative to the sites without thermogenic hydrocarbon signals.
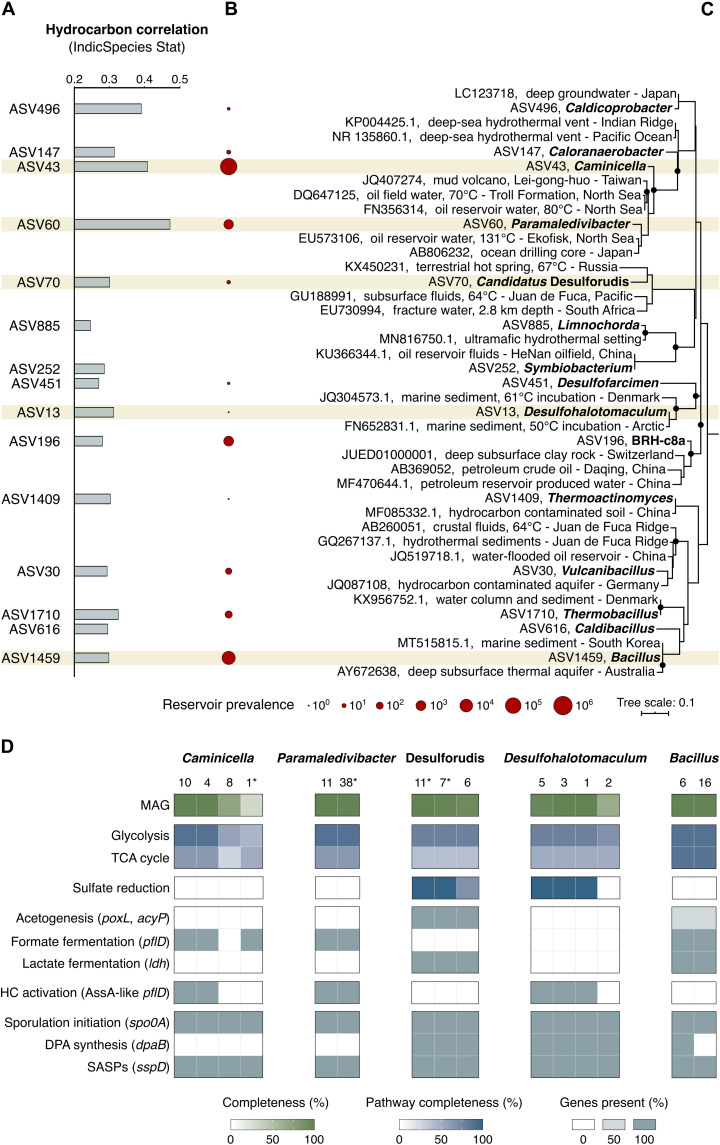
To compare thermophilic spore-forming bacterial populations in cores with and without evidence of thermogenic hydrocarbons, endospore germination and thermophile enrichment were stimulated in high-temperature anoxic incubations (40° to 60°C following pasteurization at 80°C; fig. S3). Assessing microbial community composition by 16S ribosomal RNA (rRNA) gene profiling of incubated surface sediments from all 14 locations showed divergent profiles of enriched communities of thermophiles in hydrocarbon-positive locations (Fig. 2B and table S3). These differences were further assessed by statistical comparisons that revealed 42 unique amplicon sequence variants (ASVs), all belonging to spore-forming Firmicutes (29), correlated with upward seepage of thermogenic hydrocarbons (IndicSpecies, P < 0.05; table S5). Putative fermentative organotrophs such as Paramaledivibacter and Caminicella, as well as sulfate-reducing Desulfotomaculales and Candidatus Desulforudis, showed strong hydrocarbon seep associations (Fig. 3A). None of these groups were detected by applying the same DNA sequencing method to the unincubated sediment (fig. S3 and table S3), likely owing both to the corresponding endospores in situ having low relative abundance (30) and the multilayered endospore coat not yielding to the standard cell lysis protocol used here for DNA extraction from vegetative cells (31).
Fig. 3. Subsurface oil reservoir origins of seep-associated thermophiles.
(A) Correlation of thermophilic spore-forming bacterial ASVs with thermogenic hydrocarbons (IndicSpecies, P < 0.05). Highest-ranking ASVs from each of 15 different genera are shown (representing 42 hydrocarbon-correlated ASVs in total). (B) Prevalence of these genera in 59 oil reservoir microbiome assessments (11 million 16S rRNA gene sequences in total). (C) Maximum likelihood phylogeny showing the 15 representative hydrocarbon-correlated ASVs and close relatives (see fig. S4 for all 42 indicator ASVs). Black circles at the branch nodes indicate >80% bootstrap support (1000 resamplings), and the scale bar indicates 10% sequence divergence as inferred from PhyML. (D) Metagenome-assembled genomes (MAGs) matching ASVs of interest [corresponding to brown bands in (A) to (C)] were assessed for anaerobic alkane degradation, sporulation, and other metabolic features characteristic of deep biosphere populations living in oil reservoirs. Metagenome bin numbers are indicated below the corresponding taxonomy (see table S8 for pathway definitions and additional gene annotations). Asterisks (*) denote bins with exact 16S rRNA gene sequence matches to ASVs. TCA, tricarboxylic acid; HC, hydrocarbon; SASPs, small acid-soluble proteins.
To assess the prevalence of these bacteria in deep petroleum systems, we curated a dataset of bacterial and archaeal 16S rRNA gene sequences from 59 different oil reservoir microbiomes from around the world (table S6). Seep-associated thermophilic endospore lineages identified in the cold deep-sea sediments analyzed here are found in high proportions in subsurface petroleum systems. For example, Caminicellaceae and Desulfotomaculales each make up 2 to 3% of the global oil reservoir microbiome (Fig. 3B) and are commonly encountered in high-temperature oil reservoirs deeper than 1500 m below sea floor (mbsf) (15, 32, 33). ASV assessment at finer taxonomic resolution confirms close genetic relatedness between thermophilic endospores in Scotian Slope sediments and bacteria found in different subsurface oil reservoirs (Fig. 3C and fig. S4). Metagenomic analysis of heated sediments revealed that these dormant spores encode the potential for anaerobic hydrocarbon biodegradation, favoring their selection and growth in deep petroleum-bearing sediments (Fig. 3D). Metagenome-assembled genomes (MAGs) of Caminicella, Paramaledivibacter, and Desulfohalotomaculum included contigs with rRNA sequences matching the indicator ASVs (Fig. 3 and table S7) and encoded glycyl radical enzymes related to alkylsuccinate synthases proposed to mediate anaerobic alkane biodegradation via addition to fumarate (34, 35). Using newly developed hidden Markov models for annotating alkylsuccinate synthases (36), putative assA gene sequences in thermophilic spores are shown here to diverge from canonical assA found in mesophilic Proteobacteria (fig. S5). This divergent clade includes thermophiles from hot oil reservoirs such as UPetromonas tenebris (33), Archaeoglobus fulgidus (37), and Thermococcus sibiricus (38). Sediment incubation MAGs also contain sporulation genes (Fig. 3D) including the spo0A master transcriptional response regulator (39) and genes for synthesizing α/β-type small acid-soluble proteins (e.g., sspD) and DPA (e.g., dpaB) involved in DNA protection (for a full list of sporulation genes, see table S9) (5). Other genomic features relevant to life in deep hot oil reservoirs are summarized in Fig. 3D.
DISCUSSION
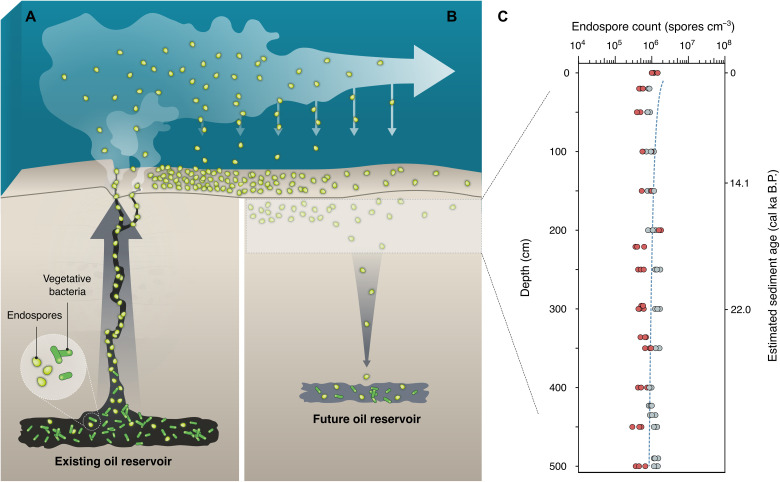
Maintenance of dormancy has been proposed as a necessary prerequisite for microbial taxa to exhibit biogeographic patterns over large distances and time scales (8). The thermophilic endospores revealed here as originating from deep petroleum-bearing sediments exemplify large-scale biogeography by connecting anaerobic hydrocarbon biodegradation and other microbial activities in these subsurface habitats with intervening periods of migration in a dormant, sporulated state. Recurrent cyclical dispersal facilitates this scenario, consistent with a “microbial conveyor belt” framework for understanding microbial biogeography where the same environment features as both the origin and eventual destination for migrating populations (8). Upward dispersal of spores originating in ~2000 mbsf petroleum reservoirs in this region (40) would require tens to hundreds of years, based on seepage rates on the order of 10 cm day−1 for hydrocarbons (41, 42). Spores dispersed by geofluids exiting the subsurface (see movie S1) and entering the marine environment (Fig. 4A) then experience relatively rapid lateral dispersal via bottom water currents on the order of 10 cm s−1 (43), preceding their eventual reentry into the seabed (Fig. 4B). In the cold surface sediment of the Scotian Slope, thermophilic spores were detected in all sampling locations, including those lacking geochemical evidence of hydrocarbon seepage (table S3). DPA concentrations within the top few meters in nonseep sites demonstrate constant deposition and burial of endospores (Fig. 4C), with numbers in this study area similar to or exceeding the seabed global average (11). High-temperature anoxic incubation of sediments from the deepest cores (down to 9 mbsf) causes thermophilic spores to germinate, demonstrating that viability is maintained during burial (fig. S6). Survival during sedimentation (44) enables dispersal of dormant populations down into deep, hot environments, which may require on the order of 10 million years based on a sediment deposition rate of 0.15 mm year−1 (45, 46), a geothermal gradient of 3°C per 100 mbsf (47), and spore germination at >40°C (fig. S2). These temperatures can support high rates of metabolism by thermophiles (48), thus completing a cyclical subsurface migration via passive dispersal of viable cells looping out of and back into the deep biosphere (Fig. 4). In buried sediments that get filled with petroleum from deeper organic-rich source rocks to create oil reservoirs (28), dispersing populations are further replenished.
Fig. 4. Subsurface microbial dispersal loop mediated by seepage, sedimentation, dormancy, and environmental selection.
(A) Endospore-forming microbial populations that are active and abundant in deep petroleum systems get dispersed upward as dormant spores by hydrocarbon seepage along geological conduits. Endospores entering the deep sea are dispersed laterally by bottom water currents as part of the marine rare biosphere. (B) Endospores get deposited on the seabed and undergo burial. (C) DPA was quantified in the upper meters of the seabed in two Scotian Slope sediment cores where hydrocarbons were not detected. On the basis of the average sediment accumulation rate in this region, this corresponds to a burial rate on the order of 108 spores m−2 year−1. The dashed regression line reflects the global average estimated for endospores in the marine subseafloor biosphere (11). Survival of individual endospores over long burial time scales (6, 44, 49) enables environmental selection (i.e., germination, activity, and population replenishment) by suitable substrates and favorable conditions that exist in petroleum-bearing sediments such as oil reservoirs (28). The sequence shown in (A) and (B) completes a subsurface “microbial dispersal loop” that incorporates cell dispersal and biogeochemical cycling in Earth’s deep biosphere.
Marine sediments contain 12 to 45% of Earth’s microbial biomass and are central to the planet’s biogeochemical cycling (49). This is especially true in subsurface petroleum systems that control and are controlled by deep biosphere communities (34, 50). Despite the importance of these processes, research on the subsurface microbiome rarely focuses on ecological factors such as dispersal and selection, preventing a more complete understanding of this vast ecosystem (51). The results presented here demonstrate that geological processes of geofluid flow and sedimentation connect deep petroleum systems with the ocean, where thermophiles contribute to the marine rare biosphere seed bank (52) and can travel great distances (18). While some dispersing thermophiles may encounter and colonize suitable new environments, e.g., at warm mid-ocean ridges (see fig. S1), many of the endospores ejected from the deep biosphere will reenter the seabed and begin a long downward journey with other sedimentary particles. These processes thus mediate extensive microbial dispersal throughout the subsurface. Circulation of living biomass characterized by intermittent episodes of microbial activity in petroleum-bearing sediments interspersed by intervals of passive dispersal is an ecological sequence that is difficult to delineate in other environmental settings (51). By connecting the physical and physiological factors that govern survival and evolution in the deep biosphere, this subsurface microbial dispersal loop connecting major Earth compartments showcases the geosphere as a model system for understanding the interplay between dispersal and selection in the biosphere at large spatial and temporal scales.
MATERIALS AND METHODS
Subsurface imaging via seismic data acquisition and processing
Multiple 2D and 3D multichannel seismic surveys interpreted here for the identification of seafloor hydrocarbon seeps relied on an earlier regional 28,000-km 2D seismic survey. The earlier survey was shot in a 6-km grid, acquiring 14 s of data with 80- to 106-fold and a 2-ms sampling interval. The 1998 vintage used was processed to prestack time migrated data. 2D seismic survey interpretations were refined using surveys such as the Shelburne 3D Wide Azimuth Seismic survey. This survey was acquired over 12,000 km2 in the deep-water Shelburne sub-basin at 6.25-m by 50-m bin spacing with a fold of 100, as documented in the acquisition report. This vintage of data used both full 3D anisotropic Kirchhoff prestack time migration (PSTM) and full-volume anisotropic Kirchhoff prestack depth migration (PSDM) with vertical transverse isotropy. PSTM had a processed bin size of 12.5 m by 25 m, while PSDM had an output bin size of 25 m by 25 m. Data were interpreted using the Petrel E&P Software Platform (Schlumberger Limited).
High-resolution seismic reflection profiles were used to investigate the subsurface stratigraphy in the vicinity of seabed fluid flow prospects to inform autonomous underwater vehicle (AUV) survey and piston coring locations. Profiles were collected during three expeditions between 2015 and 2018 onboard the CCGS Hudson (53–55) using a Huntec single-channel deep-tow seismic (DTS) sparker system. Tow depth was ~100 m beneath the sea surface with the source fired at a moving time interval between 1 and 3 s. The peak frequency for the Huntec DTS sparker is approximately 1500 Hz and spans from 500 to 2500 Hz. Raw sparker data were processed using the VISTA Desktop Seismic Data Processing software (Schlumberger Limited) and included Ormsby band-pass filtering, scaling correction, automatic gain control, and trace mixing.
An AUV was deployed from the vessel Pacific Constructor to collect high-resolution geophysical data over a 2.5-km by 2.5-km area at the location of cores 16-41 and 18-7 in August 2020. The HUGIN 6000 AUV (Kongsberg Maritime) was steered approximately 40 m above the seafloor for multibeam bathymetric, side scan sonar, and sub-bottom profiling data collection using a Kongsberg EM 2040 multibeam echosounder and an EdgeTech 2205 sonar system, respectively. EM 2040 multibeam bathymetric data were acquired at a frequency of 400 kHz, with a continuous waveform pulse and synchronized with Doppler velocity log. Multibeam bathymetric data were processed using the CARIS and EIVA suite. Side scan sonar data were acquired at a frequency of 230 kHz, and postprocessing of the data was completed in SonarWiz. The sub-bottom profiler was operated over the frequency range of 1 to 9 kHz with a 20-ms pulse. The high-resolution seismic data were integrated and analyzed using IHS Kingdom Suite (IHS Markit Ltd.). Acoustic travel times for high-resolution sub-bottom profiler lines were converted into depths using an average seismic velocity of 1500 m s−1.
Imagery of seabed hydrocarbon seepage was collected using a Triton XLX (Helix Robotics Solutions Ltd.) remotely operated underwater vehicle deployed from the vessel Atlantic Condor (56). High-definition video (movie S1) was recorded using a Simrad OE 1366 SD color zoom camera (Kongsberg, Norway).
Marine sediment sampling
Seabed surface sediments in 2000 to 3400 m of water depth were collected by piston and gravity coring from different locations on the Scotian Slope, offshore Nova Scotia, Canada (Fig. 1 and table S1) during May to June expeditions aboard the CCGS Hudson in 2015, 2016, and 2018 (53–55). Piston cores, trigger weight cores (i.e., smaller cores that release the head weight of the piston core as they enter the seabed), and gravity cores ranged from 0.18 to 8.34 m in length. Upon recovery, cores were split longitudinally onboard the ship. Sediment intervals from the base of the core (5 to 10 cm) were transferred to gas-tight IsoJars (Isotech Laboratories Inc., USA), immediately flushed with nitrogen, and stored at −20°C before interstitial gas analysis. Similar intervals from variable depths along the cores, selected on the basis of indications of visible hydrocarbon staining or odor, fluorescence, or sandy lithology, were stored in aluminum foil at −20°C for eventual hydrocarbon analysis and in sterile Whirl-Pak bags or glass jars at 4°C for eventual high-temperature endospore germination experiments. Sediment intervals at the top of the core (either 0 to 10 or 0 to 20 cm below seabed) were similarly transferred to sterile Whirl-Pak bags or sterile glass jars. Sedimentation rates of 0.1 and 0.2 mm year−1 were determined in core 18-19 for sections between 0 and 141 cm and 141 and 302 cm, respectively, using a chronostratigraphic framework based on the correlation with sediment cores from the Scotian Slope (45) and the identification at 141 cm of the brick red mud regional marker bed “d” dated at 14 thousand years before the present (ka B.P.) (46).
Hydrocarbon geochemical analysis
Interstitial gas analysis was performed on aliquots of IsoJar (Isotech Laboratories Inc., USA) headspace transferred into Exetainers (Labco Limited, UK). Sample volumes of 1 ml were injected into an Agilent 7890 RGA gas chromatograph (Agilent Technologies, USA). A flame ionization detector determined C1-C5 hydrocarbon gas concentrations that were used to calculate gas wetness [(∑C2-nC4)/(∑C1-nC4) × 100]. Carbon isotopic composition (δ13C) of hydrocarbon gas components was determined by gas chromatography combustion isotope ratio mass spectrometry; headspace aliquots were analyzed on a Trace 1310 gas chromatograph (Thermo Fisher Scientific, USA) interfaced to a Delta V isotope ratio mass spectrometer (Thermo Fisher Scientific, USA).
Sediments were analyzed for hydrocarbon biomarkers in subsamples where sufficient extract yields were recovered. Accordingly, no extract yield, or insufficient yields to determine biomarker concentrations, was considered, indicative of the absence of hydrocarbon seepage. Organic matter was extracted from sediment by adding dichloromethane with 7% (v/v) methanol, mixing the solution in an ultrasonic bath for 15 min, and then leaving at room temperature for 24 hours. Extractable organic matter (EOM) was evaporated to dryness and weighed. Asphaltenes were removed by pentane addition in excess (40 times the volume of EOM), storage for 12 hours, and centrifugation. Gas chromatography analysis of the EOM was performed on an Agilent 7890A gas chromatograph (Agilent Technologies, USA). Saturate and aromatic hydrocarbon fractions showing possible evidence of deeply sourced thermogenic hydrocarbons were analyzed further using a Micromass ProSpec gas chromatography–mass spectrometer (Waters Corporation, USA). Geochemical analyses were performed by Applied Petroleum Technology, Norway, to the standards used in industrial hydrocarbon assessments. Geochemistry data were collectively interpreted for evidence of thermogenic hydrocarbons likely derived from subsurface hydrocarbon seeps. Hierarchical clustering (complete linkage clustering based on Euclidean distance) of geochemical measurements scaled between 0 and 1 over the range of values (0 representing the weakest thermogenic signal and 1 representing the strongest thermogenic signal) was used to further assess and visualize groups of sites with similar geochemical signatures (Fig. 2A).
Sediment incubation at elevated temperatures
Sediments were investigated for the germination and growth of dormant bacterial endospores. Following homogenizing by stirring within the sample container, up to 100 g of sediment was transferred into separate 250-ml serum bottles that were sealed with butyl rubber stoppers (Chemglass Life Sciences, USA) and the headspace exchanged with N2:CO2 (90:10%). Sediment slurries were prepared in a 1:2 (w/w) ratio with sterile, anoxic, synthetic seawater medium (57) containing 20 mM sulfate and amended with acetate, butyrate, formate, lactate, propionate, and succinate (5 mM each for surface sediments and either 5 or 1 mM each for deeper sediments). The same medium was used with all sediment samples. Master slurries for a given sediment sample were subdivided into replicate, sterile, anoxic 50-ml serum bottles sealed with butyl rubber stoppers. Slurries were pasteurized at 80°C for 1.5 hours to kill vegetative cells and select for heat-resistant endospores. Triplicate pasteurized slurries were immediately incubated at 40°, 50°, or 60°C for up to 56 days to promote germination and growth of thermophilic endospore-forming bacteria. Subsamples (2 ml) were periodically removed using sterile N2:CO2-flushed syringes and stored at −20°C for molecular analysis.
16S rRNA gene amplicon sequencing
Genomic DNA was extracted from triplicate slurries subsampled immediately before incubation (i.e., after pasteurization), and periodically during the incubation, using the DNeasy PowerLyzer PowerSoil Kit (QIAGEN, USA). Extractions were performed on 300 μl of slurry according to the manufacturer’s protocol, except for inclusion of a 10-min incubation at 70°C immediately after the addition of solution C1 to enhance cell lysis. Extraction blanks (Milli-Q water) were processed in parallel. DNA was quantified using the Qubit dsDNA High Sensitivity Assay Kit on a Qubit 2.0 fluorometer (Thermo Fisher Scientific, Canada). The V3 and V4 hypervariable regions of the 16S rRNA gene were amplified in triplicate polymerase chain reaction (PCR) reactions per extraction using the primer pair SD-Bact-341-bS17/SD-Bact-785-aA21 (58) modified with Illumina MiSeq overhang adapters. This primer pair has been shown to preferentially amplify bacterial 16S rRNA genes and provides excellent overall coverage in detecting bacterial diversity (58). PCR reactions included 30 cycles and were performed in triplicate. All DNA extraction blanks and PCR reagent blanks were confirmed for negative amplification using agarose gel electrophoresis. Triplicate PCR products were pooled, purified using a NucleoMag NGS clean-up and size select kit (Macherey-Nagel Inc., USA), and indexed. Sizes of indexed amplicons were verified using the high-sensitivity DNA kit on an Agilent 2100 Bioanalyzer system (Agilent Technologies, Canada). Indexed amplicons were pooled in equimolar amounts and sequenced on an in-house Illumina MiSeq benchtop sequencer (Illumina Inc., USA) using Illumina’s v3 600-cycle reagent kit to obtain 300–base pair (bp) paired-end reads.
16S rRNA gene amplicon sequence processing
A total of 20,589,990 raw paired-end reads were generated across six separate runs on an Illumina MiSeq sequencer. Primers were trimmed using Cutadapt version 2.7 (59) before ASV inference using DADA2 version 1.16 (60) in base R version 3.6.1 (61). Forward and reverse read pairs were trimmed to a run-specific length defined by a minimum quality score of 25. Read pairs were filtered, allowing no ambiguous bases and requiring each read to have less than two expected errors, and PhiX sequences were removed. Reads were dereplicated, providing unique sequences with their corresponding abundance. Error rates were estimated from sequence composition and quality by applying a core denoising algorithm for each sequencing run to account for run-to-run variability. Unique ASVs were inferred independently from the forward and reverse reads of each sample, using the run-specific error rates, and then pairs were merged if they overlapped with no mismatches. Chimeras were identified and removed, and an additional length trimming step removed sequence variants shorter than 400 nucleotides and larger than 435 nucleotides. A total of 32,018 ASVs were resolved from 11,355,683 quality-controlled reads. Taxonomy was assigned using the Ribosomal Database Project’s k-mer–based naïve Bayesian classifier with the DADA2-formatted Silva database version 138 (62). Reads were randomly subsampled without replacement to the smallest library size (n = 4635) using the phyloseq R package (63) before comparative analysis.
Metagenome sequencing
Genomic DNA extracted from four separate sediment slurries after 56 days of incubation was used for metagenomic sequencing. Library preparation and sequencing were conducted at the Center for Health Genomics and Informatics in the Cumming School of Medicine, University of Calgary. DNA was sheared using a Covaris S2 ultrasonicator (Covaris, USA), and fragment libraries prepared using the NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs, USA). Metagenomic libraries were sequenced on the Illumina NovaSeq platform (Illumina Inc., USA) using an S4 flow cell with Illumina 300 cycle (2 × 150 bp) V1.5 sequencing kit.
Metagenome sequence processing
A total of 65,786,766 raw reads from four metagenomic libraries were quality-controlled by trimming technical sequences (primers and adapters) and low-quality additional bases, as well as filtering artifacts (phiX), low-quality reads, and contaminated reads using BBDuk (BBTools suite; http://jgi.doe.gov/data-and-tools/bbtools). Trimmed and filtered reads from each metagenome were assembled separately, as well as coassembled, using MEGAHIT version 1.2.2 (64) with default parameters and with <500-bp contigs removed. Binning of the four assemblies and one coassembly was performed using MetaBAT 2 version 2.12.1 (65). Contamination and completeness of the resulting MAGs were estimated using CheckM version 1.0.11 (66) with the lineage-specific workflow. rRNA genes were identified in unbinned reads using phyloFlash (67) and in binned reads using rRNAFinder implemented in MetaErg version 1.2.0 (68). Protein coding genes were predicted and annotated against curated protein sequence databases (Pfam, TIGRFAM, and Swiss-Prot) using MetaErg version 1.2.0 (68). Metabolic pathways were identified using KEGG Decoder (69) to parse genes annotated with KEGG Orthology using BlastKOALA (70). Hydrocarbon degradation genes were additionally annotated using CANT-HYD (36) following gene predictions made using Prodigal version 2.6.3 (71). MAGs were classified with GTDB-Tk version 1.3.0 (72) and by alignment with Silva database version 138 (62) using mothur version 1.39.5 (73) in instances where 16S rRNA gene was recovered by rRNAFinder (68).
MAGs for the seep-associated taxa were identified by alignment of predicted 16S rRNA gene sequences recovered from bins with the seep indicator ASV sequences highlighted by IndicSpecies (see below). An alignment identity of 100% across the full length of the amplicon was required to confirm association. In instances where a V3-V4 overlapping 16S rRNA gene sequence was not recovered in the MAG, taxonomic classification of the partial 16S rRNA gene, the MAG (GTDB-Tk version 1.3.0) (72), or the sample by phyloFlash (67) was used to identify possible associations to seep-associated taxa. If the most abundant ASV in an unrarefied 16S rRNA gene amplicon library, with the same taxonomic classification as the recovered 16S rRNA gene or MAG, corresponded to the most abundant ASV in that sample, then a probabilistic association was assumed and the MAG was retained for further analysis. Replicate MAGs were identified from cluster groups based on metagenome distance estimation using a rapid primary algorithm (Mash) and average nucleotide identity using dRep version 2.3.2 (74) and included in the analysis.
Analysis of global oil reservoir microbiome sequences
Raw high-throughput sequence data, totaling 53,019,792 reads from 10 separate studies, was obtained from the National Center for Biotechnology Information’s (NCBI) Sequence Read Archive (SRA) (75) by compiling sequence accession lists and using the SRA Toolkit. Initial sequence data processing was performed using VSEARCH version 2.11.1 (76). If necessary, then paired-end sequence files were merged on the basis of a minimum overlap length of 10 bp and a maximum permitted mismatch of 20% of the length of the overlap. Merged reads were filtered with a maximum expected error of 0.5 for all bases in the read and minimum and maximum read lengths of 150 and 500 bp, respectively. Identical reads were dereplicated and annotated with their associated total abundance for each sample, before de novo chimera detection. Rereplication resulted in 10,857,433 quality-controlled reads. In addition to these amplicons generated by high-throughput sequencing platforms, 2850 near full-length amplicon sequences from 49 separate clone library and/or cultivation-based studies were downloaded from the NCBI’s GenBank database using published accession numbers. Taxonomy was assigned to the combined 10,860,283 sequences using the Ribosomal Database Project’s k-mer–based naïve Bayesian classifier with the Silva database version 138 (62). Genus-level taxonomic comparisons were made to ASVs derived from marine sediment incubation experiments, because not all of the compiled oil reservoir studies sequenced the same V3-V4 hypervariable region of the 16S rRNA gene that heated sediment amplicon libraries used.
Statistical analysis and data visualization
Statistical analyses and visualization were performed using base R version 3.6.1 (61) or the specific R packages described below. Nonmetric multidimensional scaling of Bray-Curtis dissimilarity was calculated using the metaMDS function of the vegan package (77) in R and visualized using the ggplot2 package (78). Analysis of similarity (ANOSIM) tests measured significant differences between sediment communities and were performed using the anosim function of the vegan package (77).
Microbial indicator sequence analysis, designed to test the association of a single ASV with an environment through multilevel pattern analysis, was used to identify sequences that best represent specific sediments or groups of sediments under variable test conditions based on both ASV presence/absence and relative abundance patterns. Indicator ASVs were calculated using the multipatt function of the indicspecies package in R, using a point-biserial correlation index (79). Tests were performed on amplicon libraries constructed after 28 and 56 days of high-temperature incubation, omitting preincubation (day 0) libraries as representing samples before endospore enrichment. Among the 32,018 ASVs, only those present in >1% relative abundance in at least one sample across the entire dataset were included in the analysis. The strength of the association is represented by the IndicSpecies Stat value (plotted in Fig. 3A). Only observations with P < 0.05 were considered statistically significant and reported.
Phylogenetic analysis
ASVs associated with thermogenic hydrocarbons, together with their five most closely related sequences from GenBank [determined by BLAST (basic local alignment search tool) searches], were aligned using the web-based multiple sequence aligner SINA (80). Aligned sequences were imported into the ARB-SILVA 138 SSU Ref NR 99 database (62) and visualized using the open-source ARB software package (81). A maximum likelihood (PhyML) tree was calculated with near full-length (>1300 bases) bacterial 16S rRNA gene reference sequences and those from closest cultured isolates. In total, 172 sequences were used to calculate phylogeny (bootstrapped with 1000 resamplings), accounting for 1006 alignment positions specified based on positional variability and termini filters for bacteria. Using the ARB Parsimony tool, ASV and GenBank sequences were added to the newly calculated tree using positional variability filters, covering the length of the representative sequences for each sequence without changing the overall tree topology (fig. S4). Trees were annotated using iTOL version 5.5 (82).
DPA measurement
Sediment samples from cores 18-14 and 18-19 that showed no geochemical evidence of thermogenic hydrocarbons were prepared in triplicate using the methods described by Lomstein and Jørgensen (83) and Rattray et al. (84). To extract DPA, 0.1 g of freeze-dried sediment was hydrolyzed by addition of 6 M HCl and heating at 95°C for 4 hours, before quenching on ice to stop hydrolysis. The hydrolysate was freeze-dried, reconstituted in Milli-Q water, frozen, and freeze-dried again. Samples were then dissolved in 1 M sodium acetate, and aluminum chloride was added. Sediment extracts were filtered (0.2 μm) and mixed with terbium (Tb3+) prepared in 1 M sodium acetate. DPA was separated and eluted using gradient chromatography over a Kinetex 2.6-μm EVO C18 100-Å liquid chromatography column (150 mm by 4.5 mm; Phenomenex, USA) fitted with a guard column. Solvent A was 1 M sodium acetate amended with 1 M acetic acid to pH 5.6, and solvent B was 80% methanol:20% water pumped with a Thermo RS3000 pump (Dionex, Thermo Fisher Scientific, USA). The sample injection volume was 50 μl, and the total run time was 10 min (including flushing). Detection was performed using a Thermo FLD-3000RS fluorescence detector (Dionex, Thermo Fisher Scientific, USA) set at an excitation wavelength of 270 nm and emission of 545 nm. To determine DPA concentrations under the limit of detection, samples were analyzed using standard addition (83). For this, a known concentration of DPA standard/Tb3+ sodium acetate was sequentially added to the sediment exact and analyzed. Concentrations were calculated using methods described by Lomstein and Jørgensen (83). DPA concentrations were converted into endospore abundances by assuming 2.24 fmol of DPA per endospore as determined previously (85) and used in other studies (10, 11, 84).
Acknowledgments
We thank the crew of CCGS Hudson and Natural Resources Canada for collection of piston cores and onboard core processing. Ship time funding and support were provided by the Department of Natural Resources and Renewables, the Nova Scotia Offshore Energy Research Association, and Natural Resources Canada. We thank S. Larter, L. Gieg, B. B. Jørgensen, W. Richardson, K. Jenner, R. Clark, M. Strous, S. Bryant, and C. Ryan for helpful discussions and suggestions and research support.
Funding: This work was supported by Mitacs Accelerate Fellowship (to D.A.G.), Genome Canada Genomics Applications Partnership Program grant facilitated by Genome Atlantic and Genome Alberta (to C.R.J.H. and A.M.), Canada Foundation for Innovation grant (CFI-JELF 33752) for instrumentation (to C.R.J.H.), and Campus Alberta Innovates Program Chair (to C.R.J.H.).
Author contributions: C.R.J.H. and A.M. secured research funding. P.-A.D., N.M., D.C.C., and A.M. acquired, processed, and analyzed geophysical data. D.A.G., A.C., C.L., M.A.C., R.B., J.W., A.M., D.C.C., and C.R.J.H. collected and processed marine sediment samples for testing. M.F., J.W., and D.A.G. generated and analyzed hydrocarbon geochemical data. D.A.G., A.C., C.L., O.H., M.A.C., F.B., and C.R.J.H. generated, processed, and interpreted microbial community data. D.A.G. and S.B. curated and analyzed reservoir microbiome data. D.A.G. and J.Z. processed and analyzed metagenomic data. J.E.R. processed and interpreted DPA signals. D.A.G. and C.R.J.H. drafted the manuscript with feedback from all authors during refinement and finalization.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Amplicon and metagenome sequences generated in this study are available through the NCBI SRA (www.ncbi.nlm.nih.gov; BioProject accession number PRJNA604781).
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
References
Other Supplementary Material for this manuscript includes the following:
Movie S1
Tables S1 to S9
REFERENCES AND NOTES
- 1.C. Darwin, On the Origin of Species by Means of Natural Selection, or Preservation of Favoured Races in the Struggle for Life (J. Murray, 1859). [PMC free article] [PubMed] [Google Scholar]
- 2.L. G. M. Baas Becking, Geobiologie of Inleiding Tot de Milieukunde (W. P. Van Stockum & Zoon, 1934). [Google Scholar]
- 3.Hanson C. A., Fuhrman J. A., Horner-Devine M. C., Martiny J. B. H., Beyond biogeographic patterns: Processes shaping the microbial landscape. Nat. Rev. Microbiol. 10, 497–506 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Ward B. A., Cael B. B., Collins S., Young C. R., Selective constraints on global plankton dispersal. Proc. Natl. Acad. Sci. U.S.A. 118, e2007388118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Setlow B., Atluri S., Kitchel R., Koziol-Dube K., Setlow P., Role of dipicolinic acid in resistance and stability of spores of Bacillus subtilis with or without DNA-protective α/β-type small acid-soluble proteins. J. Bacteriol. 188, 3740–3747 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang J. S., Kato C., Runko G. M., Nogi Y., Hori T., Li J. T., Morono Y., Inagaki F., Predominance of viable spore-forming piezophilic bacteria in high-pressure enrichment cultures from ~1.5 to 2.4 km-deep coal-bearing sediments below the ocean floor. Front. Microbiol. 8, 137 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lennon J. T., Jones S. E., Microbial seed banks: The ecological and evolutionary implications of dormancy. Nat. Rev. Microbiol. 9, 119–130 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Mestre M., Höfer J., The microbial conveyor belt: Connecting the globe through dispersion and dormancy. Trends Microbiol. 29, 482–492 (2021). [DOI] [PubMed] [Google Scholar]
- 9.Bar-On Y. M., Phillips R., Milo R., The biomass distribution on Earth. Proc. Natl. Acad. Sci. U.S.A. 115, 6506–6511 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heuer V. B., Inagaki F., Morono Y., Kubo Y., Spivack A. J., Viehweger B., Treude T., Beulig F., Schubotz F., Tonai S., Bowden S. A., Cramm M., Henkel S., Hirose T., Homola K., Hoshino T., Ijiri A., Imachi H., Kamiya N., Kaneko M., Lagostina L., Manners H., McClelland H., Metcalfe K., Okutsu N., Pan D., Raudsepp M. J., Sauvage J., Tsang M., Wang D. T., Whitaker E., Yamamoto Y., Yang K., Maeda L., Adhikari R. R., Glombitza C., Hamada Y., Kallmeyer J., Wendt J., Wörmer L., Yamada Y., Kinoshita M., Hinrichs K., Temperature limits to deep subseafloor life in the Nankai Trough subduction zone. Science 370, 1230–1234 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Wörmer L., Hoshino T., Bowles M. W., Viehweger B., Adhikari R. R., Xiao N., Uramoto G., Könneke M., Lazar C. S., Morono Y., Inagaki F., Hinrichs K., Microbial dormancy in the marine subsurface: Global endospore abundance and response to burial. Sci. Adv. 5, eaav1024 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lomstein B. A., Langerhuus A. T., D’Hondt S., Jørgensen B. B., Spivack A. J., Endospore abundance, microbial growth and necromass turnover in deep sub-seafloor sediment. Nature 484, 101–104 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Hubert C. R., Oldenburg T. B., Fustic M., Gray N. D., Larter S. R., Penn K., Rowan A. K., Seshadri R., Sherry A., Swainsbury R., Massive dominance of Epsilonproteobacteria in formation waters from a Canadian oil sands reservoir containing severely biodegraded oil. Environ. Microbiol. 14, 387–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orphan V. J., Taylor L. T., Hafenbradl D., Delong E. F., Culture-dependent and culture independent characterization of microbial assemblages associated with high-temperature petroleum reservoirs. Appl. Environ. Microbiol. 66, 700–711 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vigneron E., Alsop B., Lomans B. P., Kyrpides N. C., Head I. M., Tsesmetzis N., Succession in the petroleum reservoir microbiome through an oil field production lifecycle. ISME J. 11, 2141–2154 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bennett B., Adams J. J., Gray N. D., Sherry A., Oldenburg T. B. P., Huang H., Larter S. R., Head I. M., The controls on the composition of biodegraded oils in the deep subsurface—Part 3. The impact of microorganism distribution on petroleum geochemical gradients in biodegraded petroleum reservoirs. Org. Geochem. 56, 94–105 (2013). [Google Scholar]
- 17.Judd A. G., The global importance and context of methane escape from the seabed. Geo-Mar. Lett. 23, 147–154 (2003). [Google Scholar]
- 18.Müller A. L., de Rezende J. R., Hubert C. R. J., Kjeldsen K. U., Lagkouvardos I., Berry D., Jørgensen B. B., Loy A., Endospores of thermophilic bacteria as tracers of microbial dispersal by ocean currents. ISME J. 8, 1153–1165 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanson C. A., Müller A. L., Loy A., Dona C., Appel R., Jørgensen B. B., Hubert C. R. J., Historical factors associated with past environments influence the biogeography of thermophilic endospores in Arctic marine sediments. Front. Microbiol. 10, 245 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hubert C., Loy A., Nickel M., Arnosti C., Baranyi C., Bruchert V., Ferdelman T., Finster K., Christensen F. M., de Rezende J. R., Vandieken V., Jørgensen B. B., A constant flux of diverse thermophilic bacteria into the cold Arctic seabed. Science 325, 1541–1544 (2009). [DOI] [PubMed] [Google Scholar]
- 21.MacDonald I. R., Natural and unnatural oil slicks in the Gulf of Mexico. J. Geophys. Res. Oceans 120, 8364–8380 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chakraborty A., Ellefson E., Li C., Gittins D., Brooks J. M., Bernard B. B., Hubert C. R. J., Thermophilic endospores associated with migrated thermogenic hydrocarbons in deep Gulf of Mexico marine sediments. ISME J. 12, 1895–1906 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chakraborty A., Ruff S. E., Dong X., Ellefson E. D., Li C., Brooks J. M., McBee J., Bernard B. B., Hubert C. R. J., Hydrocarbon seepage in the deep seabed links subsurface and seafloor biospheres. Proc. Natl. Acad. Sci. U.S.A. 117, 11029–11037 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kallmeyer J., Pockalny R., Adhikari R. R., Smith D. C., D’Hondt S., Global distribution of microbial abundance and biomass in subseafloor sediment. Proc. Natl. Acad. Sci. U.S.A. 109, 16213–16216 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abrams M. A., Marine seepage variability and its impact on evaluating the surface migrated hydrocarbon seep signal. Mar. Pet. Geol. 121, 104600 (2020). [Google Scholar]
- 26.N. C. Nanda, Direct hydrocarbon indicators (DHI), in Seismic Data Interpretation and Evaluation for Hydrocarbon Exploration and Production, N. C. Nanda, Ed. (Springer, 2016), pp. 103–113. [Google Scholar]
- 27.A. G. Judd, M. Hovland, in Seabed Fluid Flow: The Impact on Geology, Biology and the Marine Environment (Cambridge Univ. Press, 2007), pp. 163–178. [Google Scholar]
- 28.L. B. Magoon, W. G. Dow, The Petroleum System—From Source to Trap (American Association of Petroleum Geologists, 1994). [Google Scholar]
- 29.Galperin M. Y., Genome diversity of spore-forming Firmicutes. Microbiol. Spectr. 1, 10.1128/microbiolspectrum.TBS-0015-2012 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Rezende J. R., Hubert C. R. J., Røy H., Kjeldsen K. U., Jørgensen B. B., Estimating the abundance of endospores of sulfate-reducing bacteria in environmental samples by inducing germination and exponential growth. Geomicrobiol. J. 34, 338–345 (2017). [Google Scholar]
- 31.Wunderlin T., Junier T., Roussel-Delif L., Jeanneret N., Junier P., Endospore-enriched sequencing approach reveals unprecedented diversity of Firmicutes in sediments. Environ. Microbiol. Rep. 6, 631–639 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Dahle H., Garshol F., Madsen M., Birkeland N. L., Microbial community structure analysis of produced water from a high-temperature North Sea oil-field. Antonie Van Leeuwenhoek 93, 37–49 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Christman G. D., León-Zayas R. I., Zhao R., Summers Z. M., Biddle J. F., Novel clostridial lineages recovered from metagenomes of a hot oil reservoir. Sci. Rep. 10, 8048 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones D. M., Head I. M., Gray N. D., Adams J. J., Rowan A. K., Aitken C. M., Bennett B., Huang H., Brown A., Bowler B. F. J., Oldenburg T., Erdmann M., Larter S. R., Crude-oil biodegradation via methanogenesis in subsurface petroleum reservoirs. Nature 451, 176–180 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Gray N. D., Sherry A., Larter S. R., Erdmann M., Leyris J., Liengen T., Beeder J., Head I. M., Biogenic methane production in formation waters from a large gas field in the North Sea. Extremophiles 13, 511–519 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Khot V., Zorz J., Gittins D. A., Chakraborty A., Bell E., Bautista M. A., Paquette A. J., Hawley A. K., Novotnik B., Hubert C. R. J., Strous M., Bhatnagar S., CANT-HYD: A curated database of phylogeny-derived Hidden Markov Models for annotation of marker genes involved in hydrocarbon degradation. Front. Microbiol. 12, 764058 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Birkeland N. K., Schönheit P., Poghosyan L., Fiebig A., Klenk H. P., Complete genome sequence analysis of Archaeoglobus fulgidus strain 7324 (DSM 8774), a hyperthermophilic archaeal sulfate reducer from a North Sea oil field. Stand Genomic Sci. 12, 79 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mardanov A. V., Ravin N. V., Svetlitchnyi V. A., Beletsky A. V., Miroshnichenko M. L., Bonch-Osmolovskaya E. A., Skryabin K. G., Metabolic versatility and indigenous origin of the archaeon Thermococcus sibiricus, isolated from a Siberian oil reservoir, as revealed by genome analysis. Appl. Environ. Microbiol. 75, 4580–4588 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoch J. A., Regulation of the phosphorelay and the initiation of sporulation in Bacillus subtilis. Annu. Rev. Microbiol. 47, 441–465 (1993). [DOI] [PubMed] [Google Scholar]
- 40.M. E. Deptuck, K. L. Kendell, “Atlas of 3D seismic surfaces and thickness maps, central and southwestern Scotian Slope” (Geoscience Open File Report 2020-002MF and Geoscience Open File Report 2020-001MF to 006MF, Canada-Nova Scotia Offshore Petroleum Board, 2020). [Google Scholar]
- 41.Arp G. K., Effusive microseepage: A first approximation model for light hydrocarbon movement in the subsurface. Assoc. Pet. Geol. Bull. 8, 1–17 (1992). [Google Scholar]
- 42.Rice G. K., Vertical migration in theory and in practice. Interpretation 10, SB17–SB26 (2022). [Google Scholar]
- 43.Hannah C. G., Shore J. A., Loder J. W., Naimie C. E., Seasonal circulation on the western and central Scotian Shelf. J. Phys. Oceanogr. 31, 591–615 (2001). [Google Scholar]
- 44.de Rezende J. R., Kjeldsen K. U., Hubert C. R. J., Finster K., Loy A., Jørgensen B. B., Dispersal of thermophilic Desulfotomaculum endospores into Baltic Sea sediments over thousands of years. ISME J. 7, 72–84 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.K. A. Jenner, D. C. Campbell, J. M. Barnett, J. Higgins, A. Normandeau, Piston cores and supporting high-resolution seismic data, CCGS Hudson Expedition 2015018, Scotian Slope, Canada (Open File 8637, Geological Survey of Canada, 2022). [Google Scholar]
- 46.Piper D. J. W., Skene K. I., Latest Pleistocene ice-rafting events on the Scotian Margin (eastern Canada) and their relationship to Heinrich events. Paleoceanography 13, 205–214 (1998). [Google Scholar]
- 47.Lund J. W., Bjelm L., Bloomquist G., Mortensen A. K., Characteristics, development and utilization of geothermal resources—A Nordic perspective. Episodes 31, 140–147 (2008). [Google Scholar]
- 48.Beulig F., Schubert F., Adhikari R. R., Glombitza C., Heuer V. B., Hinrichs K. U., Homola K. L., Inagaki F., Jørgensen B. B., Kallmeyer J., Krause S. J., Rapid metabolism fosters microbial survival in the deep, hot subseafloor biosphere. Nat. Commun. 13, 312 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morono Y., Ito M., Hoshino T., Terada T., Hori T., Ikehara M., D’Hondt S., Inagaki F., Aerobic microbial life persists in oxic marine sediment as old as 101.5 million years. Nat. Commun. 11, 3626 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilhelms A., Larter S. R., Head I., Farrimond P., di-Primio R., Zwach C., Biodegradation of oil in uplifted basins prevented by deep-burial sterilization. Nature 411, 1034–1037 (2001). [DOI] [PubMed] [Google Scholar]
- 51.Biddle J. F., Sylvan J. B., Brazelton W. J., Tully B. J., Edwards K. J., Moyer C. L., Heidelberg J. F., Nelson W. C., Prospects for the study of evolution in the deep biosphere. Front. Microbiol. 2, 285 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lynch M. D., Neufeld J. D., Ecology and exploration of the rare biosphere. Nat. Rev. Microbiol. 13, 217–229 (2015). [DOI] [PubMed] [Google Scholar]
- 53.D. C. Campbell, A. W. A. MacDonald, CCGS Hudson Expedition 2015–018 Geological investigation of potential seabed seeps along the Scotian Slope, June 25–July 9, 2015 (Open File 8116, Geological Survey of Canada, 2016). [Google Scholar]
- 54.D. C. Campbell, CCGS Hudson Expedition 2016–011, phase 2. Cold seep investigations on the Scotian Slope, offshore Nova Scotia, June 15–July 6, 2016 (Open File 8525, Geological Survey of Canada, 2019). [Google Scholar]
- 55.D. C. Campbell, A. Normandeau, CCGS Hudson Expedition 2018–041: High-resolution investigation of deep-water seabed seeps and landslides along the Scotian Slope, offshore Nova Scotia, May 26–June 15, 2018 (Open File 8567, Geological Survey of Canada, 2019). [Google Scholar]
- 56.R. Bennett, P.-A. Desiage, Expedition report 21CONDOR: Scotian Slope, August 14–29, 2021 (Open File 8889, Geological Survey of Canada, 2022). [Google Scholar]
- 57.Isaksen M. F., Bak F., Jørgensen B. B., Thermophilic sulfate-reducing bacteria in cold marine sediment. FEMS Microbiol. Ecol. 14, 1–8 (1994). [Google Scholar]
- 58.Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M., Glöckner F. O., Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41, e1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011). [Google Scholar]
- 60.Callahan J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P., DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.R Core Team, R: A language and environment for statistical computing (R Foundation for Statistical Computing, 2014);www.R-project.org/.
- 62.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F. O., The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, 590–596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McMurdie P. J., Holmes S., Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLOS ONE 8, e61217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li D., Liu C.-M., Luo R., Sadakane K., Lam T.-W., MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676 (2015). [DOI] [PubMed] [Google Scholar]
- 65.Kang D. D., Li F., Kirton E., Thomas A., Egan R., An H., Wang Z., MetaBAT 2: An adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ. 7, e7359 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parks D. H., Imelfort M., Skennerton C. T., Hugenholtz P., Tyson G. W., CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gruber-Vodicka H. R., Seah B. K. B., Pruesse E., phyloFlash: Rapid small-subunit rRNA profiling and targeted assembly from metagenomes. mSystems 5, e00920-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dong X., Strous M., An integrated pipeline for annotation and visualization of metagenomic contigs. Front. Genet. 10, 999 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Graham E. D., Heidelberg J. F., Tully B. J., Potential for primary productivity in a globally-distributed bacterial phototroph. ISME J. 350, 1–6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kanehisa M., Sato Y., Morishima K., BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428, 726–731 (2016). [DOI] [PubMed] [Google Scholar]
- 71.Hyatt D., Chen G.-L., LoCascio P. F., Land M. L., Larimer F. W., Hauser L. J., Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chaumeil P. A., Mussig A. J., Hugenholtz P., Parks D. H., GTDB-Tk: A toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36, 1925–1927 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schloss P. D., Westcott S. L., Ryabin T., Hall J. R., Hartmann M., Hollister E. B., Lesniewski R. A., Oakley B. B., Parks D. H., Robinson C. J., Sahl J. W., Stres B., Thallinger G. G., Van Horn D. J., Weber C. F., Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Olm M. R., Brown C. T., Brooks B., Banfield J. F., dRep: A tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 11, 2864–2868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leinonen R., Sugawara H., Shumway M., The sequence read archive. Nucleic Acids Res. 39, D19–D21 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rognes T., Flouri T., Nichols B., Quince C., Mahé F., VSEARCH: A versatile open source tool for metagenomics. PeerJ 4, e2584 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.J. Oksanen, F. G. Blanchet, M. Friendly, R. Kindt, P. Legendre, D. McGlinn, P. R. Minchin, R. B. O’Hara, G. L. Simpson, P. Solymos, M. H. H. Stevens, E. Szoecs, H. Wagner, Package “vegan.” R Package version 2.4–1 (R Foundation for Statistical Computing, 2016).
- 78.H. Wickham, ggplot2: Elegant Graphics for Data Analysis (Springer, ed. 3, 2009). [Google Scholar]
- 79.De Cáceres M., Legendre P., Moretti M., Improving indicator species analysis by combining groups of sites. Oikos 119, 1674–1684 (2010). [Google Scholar]
- 80.Pruesse E., Peplies J., Glöckner F. O., SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28, 1823–1829 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ludwig W., Strunk O., Westram R., Richter L., Meier H., Yadhukumar, Buchner A., Lai T., Steppi S., Jobb G., Förster W., Brettske I., Gerber S., Ginhart A. W., Gross O., Grumann S., Hermann S., Jost R., König A., Liss T., Lüssmann R., May M., Nonhoff B., Reichel B., Strehlow R., Stamatakis A., Stuckmann N., Vilbig A., Lenke M., Ludwig T., Bode A., Schleifer K., ARB: A software environment for sequence data. Nucleic Acids Res. 32, 1363–1371 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Letunic I., Bork P., Interactive tree of life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lomstein B. A., Jørgensen B. B., Pre-column liquid chromatographic determination of dipicolinic acid from bacterial endospores. Limnol. Oceanogr. Methods 10, 227–233 (2012). [Google Scholar]
- 84.Rattray J. E., Chakraborty A., Li C., Elizondo G., John N., Wong M., Radović J. R., Oldenburg T. B. P., Hubert C. R. J., Sensitive quantification of dipicolinic acid from bacterial endospores in soils and sediments. Environ. Microbiol. 23, 1397–1406 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fichtel J., Köster J., Rullkötter J., Sass H., Spore dipicolinic acid contents used for estimating the number of endospores in sediments. FEMS Microbiol. Ecol. 61, 522–532 (2007). [DOI] [PubMed] [Google Scholar]
- 86.Khelifi N., Ali O. A., Roche P., Grossi V., Brochier-Armanet C., Valette O., Ollivier B., Dolla A., Hirschler-Réa A., Anaerobic oxidation of long-chain n-alkanes by the hyperthermophilic sulfate-reducing archaeon, Archaeoglobus fulgidus. ISME J. 8, 2153–2166 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
References
Movie S1
Tables S1 to S9