Abstract
A lipase-negative deletion mutant of Pseudomonas aeruginosa PAO1 still showed extracellular lipolytic activity toward short-chain p-nitrophenylesters. By screening a genomic DNA library of P. aeruginosa PAO1, an esterase gene, estA, was identified, cloned, and sequenced, revealing an open reading frame of 1,941 bp. The product of estA is a 69.5-kDa protein, which is probably processed by removal of an N-terminal signal peptide to yield a 67-kDa mature protein. A molecular mass of 66 kDa was determined for 35S-labeled EstA by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and autoradiography. The amino acid sequence of EstA indicated that the esterase is a member of a novel GDSL family of lipolytic enzymes. The estA gene showed high similarity to an open reading frame of unknown function located in the trpE-trpG region of P. putida and to a gene encoding an outer membrane esterase of Salmonella typhimurium. Amino acid sequence alignments led us to predict that this esterase is an autotransporter protein which possesses a carboxy-terminal β-barrel domain, allowing the secretion of the amino-terminal passenger domain harboring the catalytic activity. Expression of estA in P. aeruginosa and Escherichia coli and subsequent cell fractionation revealed that the enzyme was associated with the cellular membranes. Trypsin treatment of whole cells released a significant amount of esterase, indicating that the enzyme was located in the outer membrane with the catalytic domain exposed to the surface. To our knowledge, this esterase is unique in that it exemplifies in P. aeruginosa (i) the first enzyme identified in the outer membrane and (ii) the first example of a type IV secretion mechanism.
Pseudomonas aeruginosa is a gram-negative soil bacterium that is also well known to be an important opportunistic human pathogen, which secretes a variety of proteins into the extracellular medium. Three of these are lipolytic enzymes: two extracellular phospholipases C (PLC) and a lipase (20, 53). Apart from the phospholipases (EC 3.1.4.3), the term “lipolytic enzymes” comprises lipases (EC 3.1.1.3) and esterases (EC 3.1.1.1), which hydrolyze glycerol esters of both short- and long-chain fatty acids. Lipases are, by definition, carboxylesterases that have the ability to hydrolyze long-chain acylglycerols (≥C10), whereas esterases hydrolyze ester substrates with short-chain fatty acids (≤C10) (57). However, it should be emphasized that lipases are perfectly capable of hydrolyzing esterase substrates. In P. aeruginosa, the phospholipases PLC-H (heat-labile hemolysin) and PLC-N (nonhemolytic) have molecular masses of 78,000 and 73,000, respectively (56), and hydrolyze a variety phosphoric monoester substrates. The lipase LipA has a molecular mass of 29,000 and hydrolyzes water-insoluble carboxylic esters of long-chain fatty acids, e.g., trioleoyl glycerol and p-nitrophenylpalmitate (53). In addition to these secreted enzymes, an esterase tightly bound to the outer membrane of P. aeruginosa which has a molecular mass of 55,000 and preferentially hydrolyzes long-chain acyl thio- or oxyesters has been described (37).
There are two main reasons to study lipolytic enzymes of P. aeruginosa: (i) their important role as virulence factors and (ii) their biotechnological potential.
Clinical P. aeruginosa strains isolated from cystic fibrosis patients produce both lipase and PLC (21). A synergistic effect of PLC-H and LipA which led to the complete hydrolysis in vitro of the major lung surfactant lipid dipalmitoylphosphatidyl-choline has been demonstrated (20). Furthermore, these enzymes induce the release of the inflammatory mediator 12-hydroxyeicosatetraenoic acid from human platelets (27). These findings suggest that the lipolytic enzymes of P. aeruginosa act as virulence factors. The outer membrane-bound esterase may enable P. aeruginosa to utilize a variety of acyl esters as carbon sources; however, its role in pathogenicity has not been studied (37).
Lipases also play an important role in a variety of biotechnological applications (23). This potential is based on their ability to catalyze not only the hydrolysis of triglycerides but also their synthesis from glycerol and fatty acids, which may proceed with high specificity and enantioselectivity (24). In particular, P. aeruginosa lipase catalyzes the stereoselective conversion of a variety of amines as well as primary and secondary alcohols (25). Recently, this lipase was used to demonstrate the principle of creating a biocatalyst with high enantioselectivity toward a given substrate by applying the technique of directed evolution (41).
In the culture supernatant of the lipase-negative deletion mutant P. aeruginosa PABS1, we detected residual lipolytic activity, which led us to identify a novel esterase. The corresponding gene was cloned and expressed, and the encoded protein was analyzed with respect to its cellular location.
MATERIALS AND METHODS
Strains and plasmids.
The strains and plasmids used in this study are listed in Table 1. P. aeruginosa PAO1 and PABS1 were used throughout this study. Escherichia coli JM109 was used as a host for cloning, E. coli S17-1 was used for conjugational transfer of mobilizable plasmids, and E. coli BL21(DE3)(pLysS) (Novagene) was used for selective expression of plasmid-encoded esterase.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| P. aeruginosa PAO1 | wild-type | 19 |
| P. aeruginosa PABS1 | ΔlipA ΔlipH | 45 |
| P. aeruginosa 2B18 | pilD(xcpA)::Tn5 | 49 |
| P. aeruginosa PUS13 | ΔxcpQ | 1 |
| E. coli JM109 | F′ traD36 lacIq Δ(lacZ)M15 proA+B+/el4(McrA−) Δ(lac-proAB) thi gyrA96 (Nalr) endA1 hsdR17 (rK− mK+) relA1 supE44 recA1 | 65 |
| E. coli S17-1 | thi pro hsdRM+, chromosomally integrated [RP4-2-Tc::Mu:Kmr::Tn7, Tra+ Trir Strr] | 46 |
| E. coli BL21(DE3)(pLysS) | F−ompT hsdSB(rB− mB−) gal dcm (λcIts857 ind1 Sam7 nin5 lacUV5-T7 gene1) [pLysS Cmr T7-Lysozyme] | 51, 52 |
| Plasmids | ||
| pLAFR3 | Cos sites Tcr Plac mob | 47 |
| pUCPKS/SK | AmprlacZα Plac PT7 | 60 |
| pBBR1MCS | Cmr mob lacZα Plac | 29 |
| pLAFR3-21.P | pLAFR3 containing a 22.1-kb fragment of chromosomal DNA of P. aeruginosa PAO1, including estA | This study |
| pSKX+ | pUCPSK containing a 3.3-kb XhoI DNA fragment of P. aeruginosa, including estA, under control of Plac | This study |
| pSKX− | As pSKX+, but estA in the opposite orientation under control of PT7 | This study |
| pBBX+ | pBBR1MCS containing a 3.3-kb XhoI DNA fragment of P. aeruginosa, including estA, under control of Plac | This study |
| pLip1 | pUCPSK containing a 2.85-kb fragment carrying the lipA-lipH lipase operon | Unpublished data |
Media and growth conditions.
Bacteria were grown in glass tubes overnight at 37°C, used to inoculate 5 ml of fresh medium to an optical density at 580 nm (OD580) of 0.05, and grown for 24 h under aeration. P. aeruginosa was grown in nutrient broth (Oxoid), supplemented when necessary with 100 μg of tetracycline per ml, 300 μg of chloramphenicol per ml, or 500 μg of carbenicillin per ml. E. coli was grown in Luria broth (LB) medium or M9 minimal medium (42), supplemented when necessary with 25 μg of tetracycline per ml, 100 μg of ampicillin per ml, or 50 μg of chloramphenicol per ml.
General DNA manipulations.
Plasmid DNA was prepared as described by Birnboim and Doly (5) and purified by anion-exchange chromatography on Qia-tips (Qiagen). Chromosomal DNA was prepared as described by Gamper et al. (15). Recombinant DNA techniques were performed essentially as described by Sambrook et al. (42). Restriction endonuclease reactions and bacteriophage T4 DNA ligase treatments were done as recommended by the manufacturers. DNA fragments were analyzed on 0.4 to 1% (wt/vol) agarose gels.
Construction of a genomic library.
A genomic library of P. aeruginosa PAO1 chromosomal DNA in cosmid pLAFR3 was constructed as described by Visca et al. (58). Chromosomal DNA was partially digested with Sau3A to obtain fragments of >15 kb. Cosmid DNA was linearized with either EcoRI or HindIII, dephosphorylated, mixed, and digested with BamHI. Chromosomal DNA fragments were added and ligated. The λ-DNA in vitro packaging module (Amersham) was used for DNA packaging and infection of E. coli S17-1. The genomic library consisted of 15,000 individual clones each carrying an insert of 21 to 28 kb (average size), statistically representing 99.9% of the P. aeruginosa genome.
Screening of the genomic library.
The individual clones were screened in P. aeruginosa PABS1 on esterase indicator plates. The library clones were conjugated from E. coli S17-1 into P. aeruginosa PABS1 and plated on esterase indicator plates to detect the formation of halos.
Cell fractionation.
Cultures (volume, 100 ml) of P. aeruginosa and E. coli grown in LB medium for 24 h were separated by centrifugation (8,000 × g for 5 min) into cells and culture supernatant, which was used to determine the extracellular enzyme activity. The cells were resuspended in 20 ml of 100 mM Tris-HCl (pH 8.0) and disrupted by two passages through a French press. Cell debris was removed by centrifugation (5,000 × g for 15 min), and the supernatant was subjected to ultracentrifugation at 100,000 × g for 2 h to collect the crude membrane fraction, which was resuspended in 5 ml of 100 mM potassium phosphate buffer (pH 7.2) (membrane fraction). The resulting supernatant was used as the cytoplasmic/periplasmic (cp/pp) fraction.
Enzyme assays. (i) Plate assay.
Esterase indicator plates (28) were prepared by addition of 15 ml of an emulsion of 50% (vol/vol) tributyrin and 5% (wt/vol) gum arabic to 500 ml of molten nutrient broth agar medium. Tributyrin was emulsified by sonication for 3 min at 75 W (duty cycle 100%) in a Branson 250 sonifier. Esterase and lipase activity is indicated by the formation of clear halos around the colonies.
(ii) Liquid assays.
For esterase assay 1, 23.7 mg of p-nitrophenyl caproate (pNPC; Sigma) was dissolved in 10 ml of 2-propanol and added to 90 ml of Sørensen phosphate buffer (pH 8.0) supplemented with sodium deoxycholate (207 mg) and gum arabic (100 mg), yielding a final pNPC concentration of 1 mM. Culture supernatant (5 to 50 μl) was added to the substrate solution to give a final volume of 2.5 ml, the solution was incubated for 15 min at 30°C, and the OD410 was recorded with a Zeiss PMQ II spectrophotometer. The OD410/OD580 ratio was used to estimate clone-specific extracellular enzyme activities during library screening. For esterase assay 2, 23.7 mg of pNPC was dissolved in ethanol (5 ml) and added to 95 ml of 100 mM potassium phosphate buffer (pH 7.0) containing 10 mM MgSO4 to yield a final concentration of 1 mM pNPC. Samples (5 to 50 μl) were added to the substrate solution to give a final volume of 1 ml, and the ΔOD410/min was recorded for 5 to 10 min at room temperature. The molar absorption coefficient of pNP at pH 7.0 was determined as 10,400 M−1. One unit of enzyme activity is defined as the amount of enzyme forming 1 μmol of pNP per min. For the lipase assay 30 mg of p-nitrophenyl palmitate (pNPP; Sigma) was dissolved in 10 ml of 2-propanol at 60°C. The test was done as described above for esterase assay 1, except that the reaction was carried out at 37°C (63).
(iii) Glucose-6-phosphate dehydrogenase assay.
Glucose-6-phosphate dehydrogenase was used as a cytoplasmic marker enzyme (10). A stock solution of NADP (45 mM) and a stock solution of glucose-6-phosphate (110 mM) were diluted 1:100 in a buffer containing 55 mM Tris-HCl (pH 7.5) and 11 mM MgCl2. A 950-μl volume of this test solution was mixed with 50 μl of samples, and the decrease in optical density (ΔOD340/min) was monitored spectrophotometrically at 30°C for 5 min.
Trypsin treatment of whole cells.
P. aeruginosa and E. coli were grown overnight in LB medium. The cells were collected by centrifugation and resuspended to an OD580 of 5 in 100 mM Tris-HCl (pH 8.0). Trypsin (10 μl of a stock solution containing 2 mg of trypsin per ml in 100 mM Tris-HCl [pH 8.0]) was added to 100 μl of the cell suspension. After 1 h of incubation at 37°C, the protease was inhibited by the addition of 1 mM phenylmethylsulfonyl fluoride. Cells were collected by centrifugation and resuspended in 100 μl of 100 mM Tris-HCl (pH 8.0). Esterase activity was determined in supernatants and whole cells obtained after trypsin treatment. As a control, cells were treated in the same way except that no trypsin was added.
Selective labeling of plasmid-encoded proteins.
The selective labeling of esterase encoded by pSKX−/+ was performed as described previously (54). Bacteria precultured in LB/M9 medium were used to inoculated 5 ml of fresh medium to an OD580 of 0.1. At an OD580 of 0.6, the cells were harvested and added to 12 ml of M9 medium supplemented with 0.2% glucose and 0.2% methionine assay mix (Difco). E. coli BL21(DE3) harboring pUCPSK was used as a control. The expression of T7-RNA polymerase in E. coli BL21(DE3)(pLysS) was induced by the addition at t = 0 of 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After 30 min of incubation (t = 30), a sample was taken and E. coli RNA polymerase was inhibited by the addition of 200 μg of rifampin per ml. Additional samples were taken after 30 (t = 60) and 60 (t = 90) min of incubation with rifampin. These samples were labeled in vivo by supplementing 1 ml of culture with 1 μl of l-[35S]methionine–l-[35S]cysteine (10 μCi/ml; Amersham) and incubating the mixture for 10 min at 37°C. The cells were harvested, lysed, and denatured, and the proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by autoradiographic detection on hyperfilm (Amersham) for 48 h.
SDS-PAGE.
SDS-PAGE was performed as described by Laemmli (30) with a 5% stacking gel and a 15% separating gel.
DNA sequencing.
DNA sequencing was kindly performed by Genencor International, Delft, The Netherlands, using the method described by Sanger et al. (43).
Software.
The program BLAST X (2, 16) was used for protein homology searching, PSORT was used for prediction of protein localization (34), and DNAstar (Lasergene) was used for sequence alignments.
Nucleotide sequence accession number.
The DNA sequence of estA from P. aeruginosa has been deposited in GenBank (accession no. AF005091).
RESULTS AND DISCUSSION
Screening of a genomic library.
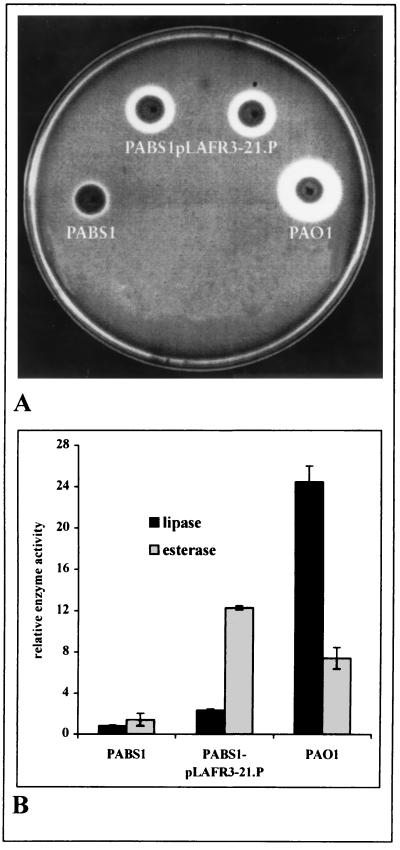
A genomic library containing P. aeruginosa DNA cloned in pLAFR3 was transferred from E. coli S17-1 into the lipase-negative mutant P. aeruginosa PABS1 by conjugation, and bacteria were plated on esterase indicator plates containing tributyrin. Although this triglyceride is a classical lipase substrate, it is partially soluble in water and is therefore also hydrolyzed by esterases (7). Colonies with extracellular esterase or lipase activity formed clear halos on these plates. As shown in Fig. 1A, lipase production in the wild-type strain P. aeruginosa PAO1 caused the formation of a large halo, which appeared after overnight incubation, whereas the halo formed by the lipase-negative mutant PABS1 was much smaller and appeared only after incubation of the plates for at least 48 h. Our screening was based on the assumption that such a halo would appear earlier and with increased diameter if several copies of the esterase gene cloned in pLAFR3 were expressed in the lipase-negative mutant. After a library consisting of 15,000 clones was screened, 11 positive clones with halos of intermediate size, i.e., smaller than that formed by the wild type but larger than that of the lipase-negative mutant, were identified. Several clones identified on esterase indicator plates were further assayed for lipase and esterase activity in the culture supernatant (Fig. 1B). The wild-type strain, PAO1, showed high lipase activity and significantly lower esterase activity. However, its esterase activity was still higher than that of the lipase-negative mutant, PABS1, presumably because of the ability of lipase to cleave the esterase substrate. Introduction of pLAFR3-21.P into P. aeruginosa PABS1 yielded a 10-fold increase in esterase activity (Fig. 1B). The slightly increased lipase activity was presumably caused by the ability of esterase to hydrolyze the lipase substrate p-nitrophenylpalmitate.
FIG. 1.
Lipolytic activity of P. aeruginosa strains, wild-type PAO1, lipase-negative mutant PABS1, and PABS1pLAFR3-21.P containing a cosmid with a 22.1-kb insert of chromosomal DNA from P. aeruginosa PAO1. (A) Halo formation of bacterial colonies on esterase indicator plates after incubation for 3 days at 30°C. (B) Lipolytic activities of culture supernatants in liquid assays, assayed with p-nitrophenylpalmitate (C16) for lipase activity and p-nitrophenylcaproate (C6) for esterase activity. Relative enzyme activities were determined as the ratio of OD410 (enzyme activity) to OD580 (cell density) per milliliter of culture supernatant.
Identification of the esterase.
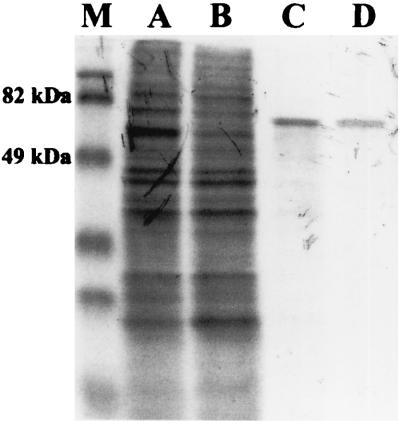
The cosmid pLAFR3-21.P was digested with various restriction endonucleases, and fragments were subcloned into plasmid pUCPSK, transferred into the lipase-negative mutant, PABS1, and assayed for esterase activity. After several subcloning steps, one clone containing plasmid pUCPSK carrying a 3.3-kb XhoI fragment was isolated. This fragment was cloned in both orientations, and the corresponding plasmids were named pSKX+ and pSKX−, respectively. P. aeruginosa PABS1(pSKX+) showed high esterase activity (data not shown), whereas no increase in esterase activity was observed for P. aeruginosa PABS1(pSKX−). The DNA fragment cloned in plasmid pUCPSK in the positive orientation is expressed from the lac promoter, which is constitutively expressed in P. aeruginosa, thereby explaining the observed esterase activity, whereas in the opposite (negative) orientation it is under control of the T7 promoter. To identify a putative protein encoded by this fragment, we used an E. coli T7-RNA polymerase expression system. Samples were taken at several time intervals, and each of them was immediately labeled with l-[35S]methionine–l-[35S]cysteine in vivo. Expression from pSKX− yielded a single 66-kDa protein band visible upon autoradiography, which appeared when cellular protein synthesis was inhibited by addition of rifampin (Fig. 2). Since no protein bands were detected in cells harboring either the control plasmid pUCPSK or pSKX+, we concluded that the 3.3-kb XhoI fragment of P. aeruginosa DNA cloned in pSKX− contained a single open reading frame (ORF) expressed from PT7 which encoded a protein with Mr = 66,000.
FIG. 2.
Autoradiography of l-[35S]methionine–l-[35S]cysteine-labeled proteins from cell lysates of E. coli BL21(DE3)(pLysS) containing pSKX− separated by SDS-PAGE. Samples were labeled prior to induction of T7 RNA polymerase expression (t = 0; lane A), 30 min after induction but prior to inhibition of E. coli RNA polymerase with rifampin (t = 30; lane B), and 30 min (t = 60; lane C) and 60 min (t = 90; lane D) after addition of rifampin. Lane M contains prestained molecular mass markers (Bio-Rad).
Nucleotide sequence analysis.
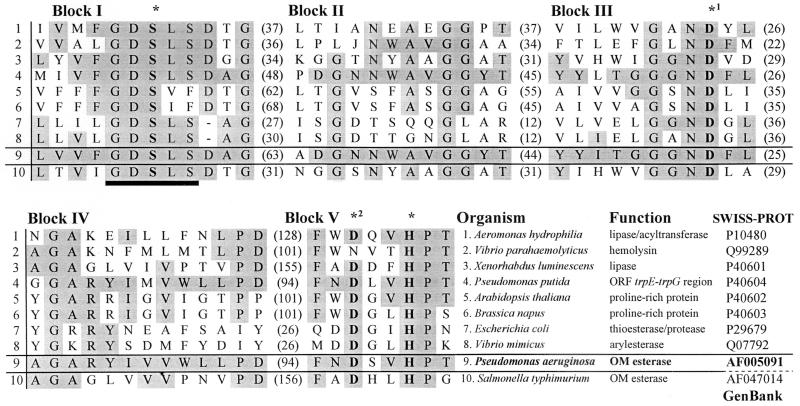
Determination of the nucleotide sequence of this DNA fragment (Fig. 3) revealed an ORF of 1,941 bp, which was designated estA. A putative Shine-Dalgarno sequence was located 7 bp upstream of the ATG start codon, and a consensus sequence typical for an RpoN-dependent promoter [TGGCACN5TTGC(a/t)] (33) was identified 135 bp upstream of the translational start codon. The G+C content was 66.9% and the frequency of C or G at the third codon position was 89.2%, indicating a typical P. aeruginosa codon usage (62). The esterase encoded by estA is synthesized as a 646-amino-acid precursor with a calculated Mr of 69,526, including a predicted 24-amino-acid signal sequence (35). The mature esterase has a calculated molecular weight of 67,000, which is consistent with the Mr determined by SDS-PAGE and autoradiography. A BLAST X (2, 16) analysis of estA revealed homologies to an ORF located in the trpE-trpG region of P. putida (11), to apeE encoding an outer membrane esterase from Salmonella typhimurium (9), to lip-1 encoding a lipase from Xenorhabdus (Photorhabdus) luminescens (59), and to the GCAT gene encoding a acyltransferase of Aeromonas hydrophila (32). These proteins and the esterase of P. aeruginosa belong to a novel family of lipolytic enzymes identified on the basis of sequence homology (55). Many of the prokaryotic proteins of this new family exhibited lipolytic activity; eukaryotic members were found in higher plants (6), and their physiological functions are as yet unknown. Five conserved blocks of high amino acid homology were identified (Fig. 4). The enzymes belonging to this novel family differ from other lipases in the location and structure of the active-site consensus motif G-X-S-X-G (22). In this family, this motif is located close to the N terminus and consists of G-D-S-X-S, with the terminal glycine of the characteristic lipase consensus motif replaced in most cases by serine. The active site of these enzymes consists of a catalytic triad formed by the amino acids serine, histidine, and aspartate. For A. hydrophilia lipase/acyltransferase, serine 16, histidine 291, and aspartate 116, located in blocks I, V, and III, respectively (Fig. 4), were shown by site-directed mutagenesis to form the catalytic triad (8). Aspartate 116 could not unequivocally be assigned to the active site, and a second aspartate residue at position 288 was identified as another likely candidate (8). Recently, the three-dimensional structure of an esterase from Streptomyces scabies was determined by X-ray crystallography (61). This enzyme is unique among all known lipolytic enzymes in that instead of a triad, it contains an active-site dyad consisting of a serine and a histidine residue. The correct orientation of the histidine imidazole ring is normally ensured by hydrogen bonding to the carboxyl group of a third residue (aspartate or glutamate), which is replaced here by the backbone carbonyl of a tryptophan. The three-dimensional structure of an esterase isolated from bovine brain has a similar active-site architecture. In this case, however, the tryptophan is indeed replaced by aspartate (18). A comparison of the amino acid sequences of S. scabies and bovine brain esterases with the sequences of A. hydrophila lipase/acyltransferase and of P. aeruginosa EstA (3) indicated that in these enzymes, aspartate residues at positions 288 and 286, respectively, belong to the catalytic triad, as indicated in Fig. 4. Therefore, we predict that the catalytic triad of the P. aeruginosa esterase is formed by serine 14, histidine 289, and aspartate 286.
FIG. 3.
Nucleotide sequence and derived amino acid sequence of the esterase gene estA. The putative Shine-Dalgarno sequence is underlined, the consensus sequence for the RpoN (ς54)-dependent promoter is marked by dashed lines, and the putative signal sequence is indicated by an arrow.
FIG. 4.
Sequence comparison between P. aeruginosa esterase (EstA) and members of a novel family of lipolytic enzymes (55). Identical amino acids are shaded in grey; numbers in parentheses refer to the number of amino acid residues between the conserved blocks. The putative catalytic triad residues (∗) are printed in bold, and the G-D-S-L-S consensus motif is underlined.
Secretion.
Esterase activity could be detected in the bacterial culture supernatant (Fig. 1B). Furthermore, a putative signal sequence precedes the mature esterase protein, leading us to assume that this enzyme could be secreted by the general secretory pathway (GSP) (12). This pathway consists of two steps: the secreted protein is translocated through the inner membrane via the Sec-dependent mechanism and is subsequently translocated through the outer membrane by the Xcp machinery (12). The lipase-negative mutant P. aeruginosa PABS1 and two different xcp mutants were transformed with plasmids encoding either lipase (pLip1) or esterase (pSKX+), and the extracellular enzyme activities were determined. In mutant 2B18, the xcpA gene, encoding the prepilin-peptidase, which is required for the processing of several Xcp components, is disrupted (36, 49), and in mutant PUS13, the outer membrane secretin XcpQ is absent. Since the lipase is known to be secreted via the GSP (24), it served as a control. As shown in Table 2, xcp mutants did not show extracellular lipase activity. However, the extracellular esterase activity remained unaffected, demonstrating that the esterase is not secreted via the GSP.
TABLE 2.
Extracellular enzyme activities of a lipase-negative P. aeruginosa strain and mutants defective in the general secretory pathway
| Mutant | Genotype | Gene expressed from plasmid | Relative enzyme activity (%)a
|
|
|---|---|---|---|---|
| Esterase | Lipase | |||
| PABS1 | lipA | estA | 100 | 17 |
| 2B18 | xcpA | estA | 97 | 0 |
| PUS13 | xcpQ | estA | 98 | 0 |
| PABS1 | lipA | lipA | 15 | 100 |
| 2B18 | xcpA | lipA | 0 | 0 |
| PUS13 | xcpQ | lipA | 0.1 | 0 |
Extracellular enzyme activities of P. aeruginosa PABS1 harboring either the cloned esterase gene on pSKX+ or the lipase gene on pLip1 were arbitrarily set at 100%. Relative enzyme activities were determined as the ratio of OD410 (enzyme activity) to OD580 (cell density) per milliliter of culture supernatant with the substrates p-NPC for esterase and p-NPP for lipase.
Cellular localization.
EstA was predicted to be anchored to the outer membrane by the computer program PSORT (34). Additional observations suggested an outer membrane location of the 28-kDa C-terminal fragment of EstA (amino acids 374 to 622): (i) it contains many putative amphipathic β-strands, and (ii) as in the vast majority of bacterial outer membrane proteins (50), the C-terminal amino acid residue is a phenylalanine. These observations led us to compare the C-terminal fragment of EstA with other outer membrane proteins (Fig. 5), revealing a significant similarity to a family of proteins which is secreted by gram-negative bacteria and has been designated the autotransporter family (31). These proteins, which are believed to be virulence factors, are secreted by the so-called type IV secretion mechanism (13), which has so far not been found to operate in P. aeruginosa (17). Autotransporter proteins are translocated through the inner membrane via a Sec-dependent mechanism and cross the periplasm, and their β-domain inserts into the outer membrane. The catalytically active α-domain (i) remains attached to the outer membrane, (ii) is autoproteolytically cleaved off, or (iii) is cleaved off by another protease (17). Significant amino acid homologies to recognized members of the autotransporter family, such as Ssp (22.6%), a serine protease from Serratia marcescens (64), and VacA (23.3%), a vacuolating cytotoxin from Helicobacter pylori (44), were detected. However, the highest homology scores were found to an outer membrane-located esterase from S. typhimurium (24.6%) (9) and to a secreted lipase from X. luminescens (29.3%) (59). Interestingly, this lipase inserts into the outer membrane upon expression in E. coli (59). So far, these lipolytic enzymes have not been found to possess a C-terminal autotransporter domain. Autotransporters share a number of characteristic features, which also exist in the P. aeruginosa esterase. (i) They possess an N-terminal leader peptide. (ii) The mature protein consists of a surface-exposed N-terminal passenger domain (or α-domain), which harbors the catalytic site, and a C-terminal β-domain located in the outer membrane (17). The α-domain usually contains the N-terminally located active-site motifs followed by a stretch of amino acids with few cysteine residues (Cys258 and Cys264 in EstA). The C-terminal β-domains of autotransporters are predicted to contain 14 amphipathic β-strands, which may form a β-barrel pore, allowing the translocation of the α-domain to the bacterial cell surface (31). (iii) The terminal amino acid residue is always a phenylalanine or tryptophan, which is preceded by four alternating charged/polar and hydrophobic/aromatic residues (31, 50). In EstA the terminal amino acid residue is F622 and is preceded by D-L-S-L-A-L-S-V. (iv) Proline 171 and glycine 275 have been identified as two fully conserved residues by the aligning autotransporter domains of several proteins (31). These residues are also present in EstA (Fig. 5).
FIG. 5.
Multiple alignment of C-terminal domains. EstA, outer membrane proteins, and secreted proteins belonging to the autotransporter family (31) were compared. Boxes indicate putative amphiphathic β-barrels, and the two fully conserved amino acids from the ATF are marked with an asterisk.
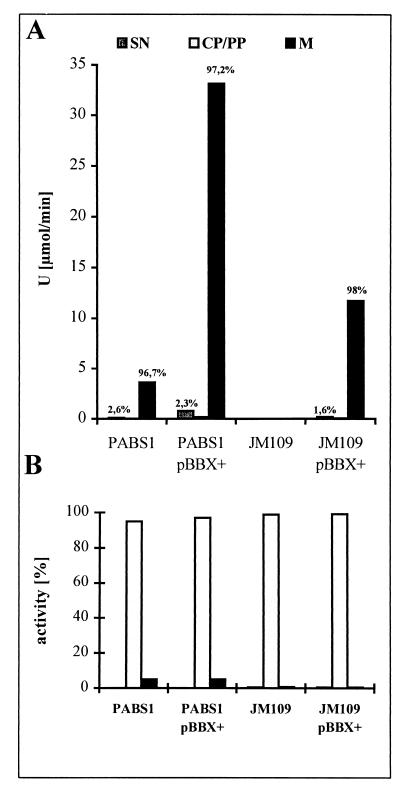
Based on these predictions, we investigated experimentally the localization of EstA in the lipase-negative mutant P. aeruginosa PABS1 (wild type for esterase) and the esterase-overexpressing strain P. aeruginosa PABS1 containing estA on pBBX+. Cellular compartments were fractionated into culture supernatant, cytoplasmic-periplasmic fraction, and a crude membrane fraction. The absolute esterase activity was about 10-fold higher in the overexpressing strain, but in both strains more than 95% of the esterase activity was detected in the crude membrane fraction (Fig. 6A). Only 2 to 3% of the enzyme activity was released into the culture supernatant, which did not contain glucose-6-phosphate dehydrogenase activity (Fig. 6B), indicating that no significant cell lysis had occurred. These results showed that EstA is tightly membrane bound in P. aeruginosa. A comparable result was obtained when estA was expressed in the heterologous host E. coli, although the absolute amounts of esterase activity were smaller. The small amount of extracellular esterase may have two possible explanations: (i) the enzyme was cleaved by a protease and remained bound to the outer membrane, as found for the Ag43 protein from E. coli (39), with only a small amount released into the extracellular medium; or (ii) the enzyme remained bound to the outer membrane, as shown for Hsr from Helicobacter mustelae (38). A small part of the esterase might have been released either in a free form or in the form of membrane vesicles which contain in P. aeruginosa periplasmic, membrane-bound, and extracellular proteins (4).
FIG. 6.
Distribution of esterase (A) and glucose-6-phosphate dehydrogenase (B) activity in cellular compartments of P. aeruginosa PABS1 and E. coli JM109 containing estA on pBBX+. The percentages of total enzyme activities present in culture supernatants (SN), cytoplasmic-periplasmic fractions (CP/PP), and membranes (M) are given.
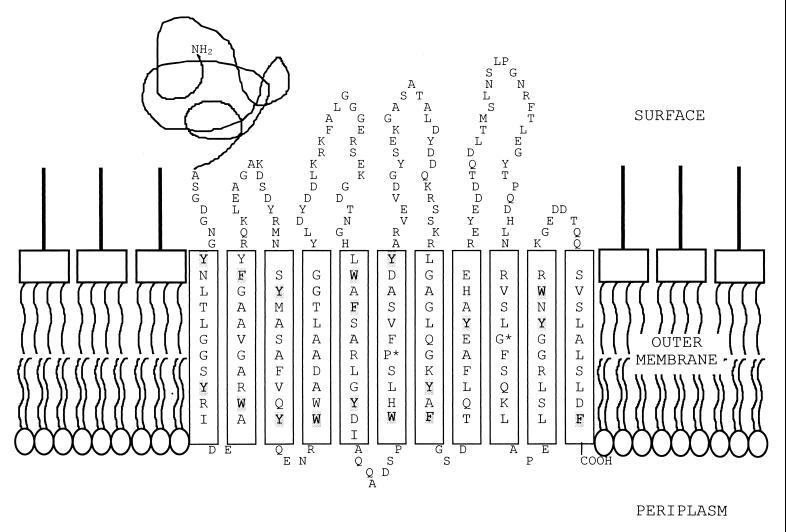
Inspection of the C-terminal domain of EstA revealed several segments that could form amphipathic β-strands. Assuming that the β-domains of all autotransporters have similar tertiary structures, we expect that the amphipathic character should be conserved in all β-strands that are actually part of the β-barrel. When the new members of the autotransporter family identified in this study are aligned with the recognized members of the family (examples are given in Fig. 5), 11 segments that could form amphipathic β-strands in all sequences could be distinguished. These segments are almost devoid of β-turn predictions, when the criteria described by Paul and Rosenbusch (40) are applied, whereas turn predictions are generally found in the intervening segments (data not shown). Since a closed β-barrel should consist of an even number of β-strands, the β-barrel may consist of as few as 10 β-strands, with the first segment reaching through the interior of the barrel, thereby exposing the α-domain to the cell surface. Alternatively, the barrel formed may not be closed, creating an instability that may result in the destruction of the β-domain after the translocation of the α-domain. In this respect, it should be noted that the fate of the β-domain after having performed its job has hardly been investigated so far. A 10- or 11-stranded β-barrel could enclose only a small pore, which indicates that the passenger domain should be transported in a delineated fashion. In agreement, the periplasmic folding of an artificial passenger domain has been demonstrated to prevent outer membrane translocation (26). However, it should be noted that the β-domain does not necessarily have to form a β-barrel with an enclosed pore to exert its function. The molecular mechanism of transport may be entirely different. For example, it has been reported that a lipid-modified domain of OmpA, encompassing only five β-strands, can transport a periplasmic passenger protein to the cell surface (14). Clearly, these five β-strands cannot form a pore through which the passenger is transported. A tentative topology model of the β-domain of EstA is depicted in Fig. 7. As in the porins, the β-strands are connected by short periplasmic turns and longer extracellular loops. Furthermore, two girdles of aromatic residues that may surround the putative β-barrel at the height of the polar head groups of the lipids in the membrane can readily be discerned. Further experiments are required to investigate the structure and function of the autotransporter domain of P. aeruginosa EstA as well as of other members of the family.
FIG. 7.
One possible model of the C-terminal domain of P. aeruginosa esterase (EstA) located in the outer membrane. The C-terminal domain (∼G 374 to F 622) is predicted to consist of 11 amphipathic β-strands with 10 to 12 amino acid residues per strand, sufficient to traverse the hydrophobic core of the membrane. The two amino acids fully conserved in proteins belonging to the autotransporter family (31) are marked with asterisks; amino acids printed in bold indicate the hydrophobic side of the β-strands; and amino acids shaded in grey represent two girdles of aromatic residues, which seem to be present in outer membrane proteins of known structure.
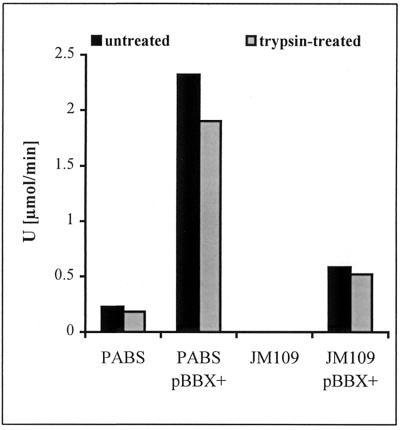
Our model predicts that the catalytically active N-terminal domain of EstA is exposed to the surface and should at least partly be accessible to proteolytic cleavage. Therefore, whole cells of P. aeruginosa and E. coli expressing estA were treated with trypsin and the residual cell-bound esterase activity was determined. As shown in Fig. 8 a significant amount of esterase activity was removed (20% for P. aeruginosa and 10% for E. coli). Increasing the amount of trypsin did not result in an increased amount of esterase removed from cells. Increasing the concentration of MgCl2 during trypsin treatment led to a decrease in the amount of esterase activity removed, whereas increasing the concentration of EDTA led to a increase in the amount of esterase activity removed (data not shown). These results indicate the following. (i) The esterase expressed in P. aeruginosa and in E. coli is partly accessible to trypsin treatment of whole cells. Since the localization and the protease accessibility in E. coli and in P. aeruginosa are very similar, the information allowing the enzyme to reach the cell surface must be independent of specific factors present only in the homologous host and should therefore reside in EstA itself. (ii) Removal of a significant part of the esterase activity from whole cells of P. aeruginosa and E. coli by trypsin treatment strongly suggests that EstA is attached to the bacterial outer membrane, as predicted by our model (Fig. 7). (iii) The trypsin-resistant portion of the esterase may be shielded by association with lipopolysaccharide and/or exopolysaccharide as described for outer membrane proteins (48) and also for extracellular lipase from P. aeruginosa (53). Further experiments are required to decide whether the accessibility of esterase to trypsin is different in appropriate mutants altered in lipopolysaccharide or exopolysaccharide (e.g., alginate) composition.
FIG. 8.
Effect of trypsin treatment on esterase activities of whole cells of P. aeruginosa and E. coli containing estA on pBBX+.
In summary, a novel esterase was identified which is located in the outer membrane of P. aeruginosa. This enzyme presumably belongs to a family of putative virulence factors which are self-secreted via a C-terminally located autotransporter domain. Determination of esterase activity in a cell-free culture supernatant led us to assume that the catalytically active N-terminal esterase domain may be released into the external medium by a so far unknown mechanism. At present, we are trying to identify the enzyme responsible for proteolytic cleavage of surface-exposed EstA and are investigating the physiological significance of this novel lipolytic enzyme.
ACKNOWLEDGMENTS
We thank Alain Filloux, IBSM-CNRS, Marseille, France, and Steve Lory, University of Washington, Seattle, Wash., for providing P. aeruginosa mutant strains.
This work was supported by grant BIO4-CT96-0119 from the European Commission in the framework of the Biotechnology Program. S.W. is a recipient of a graduate fellowship from the Deutsche Forschungsgemeinschaft (Graduiertenkolleg: “Biogenese und Mechanismen komplexer Zellfunktionen”).
REFERENCES
- 1.Akrim M, Bally M, Ball G, Tommassen J, Teerink H, Filloux A, Lazdunski A. Xcp-mediated protein secretion in Pseudomonas aeruginosa: identification of two additional genes and evidence for regulation of xcp gene expression. Mol Microbiol. 1993;10:431–443. doi: 10.1111/j.1365-2958.1993.tb02674.x. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Arpigny J L, Jaeger K-E. Bacterial lipolytic enzymes: classification and properties. Biochem J. 1999;343:177–189. [PMC free article] [PubMed] [Google Scholar]
- 4.Beveridge T J. Structures of gram-negative cell walls and their derived membrane vesicles. J Bacteriol. 1999;181:4725–4733. doi: 10.1128/jb.181.16.4725-4733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birnboim H C, Doly J. A rapid alkaline extraktion procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brick D J, Brumlik M J, Buckley J T, Cao J-X, Davies P C, Misra S, Tranbarger T J, Upton C. A new family of lipolytic plant enzymes with members in rice, arabidopsis and maize. FEBS Lett. 1995;377:475–480. doi: 10.1016/0014-5793(95)01405-5. [DOI] [PubMed] [Google Scholar]
- 7.Brockerhoff H. Action of pancreatic lipase on emulsions of water-soluble esters. Arch Biochem Biophys. 1969;134:366–371. doi: 10.1016/0003-9861(69)90295-1. [DOI] [PubMed] [Google Scholar]
- 8.Brumlik M J, Buckley J T. Identification of the catalytic triad of the lipase/acyltransferase from Aeromonas hydrophilia. J Bacteriol. 1996;178:2060–2064. doi: 10.1128/jb.178.7.2060-2064.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carinato M E, Collin-Osdoby P, Yang X, Knox T M, Conlin C, Miller C G. The apeE gene of Salmonella typhimurium encodes an outer membrane esterase not present in Escherichia coli. J Bacteriol. 1998;180:3517–3521. doi: 10.1128/jb.180.14.3517-3521.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Smet M J, Kingma J, Witholt B. The effect of toluene on the structure and permeability of the outer and cytoplasmic membranes of Escherichia coli. Biochim Biophys Acta. 1978;506:64–80. doi: 10.1016/0005-2736(78)90435-2. [DOI] [PubMed] [Google Scholar]
- 11.Essar D W, Eberly L, Crawford I P. Evolutionary differences in chromosomal locations of four early genes of the tryptophan pathway in fluorescent Pseudomonads: DNA sequence and characterization of Pseudomonas putida trpE and trpGDC. J Bacteriol. 1990;172:867–883. doi: 10.1128/jb.172.2.867-883.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filloux A, Michel G, Bally M. GSP-dependent protein secretion in Gram-negative bacteria: the Xcp system of Pseudomonas aeruginosa. FEMS Microbiol Rev. 1998;22:177–198. doi: 10.1111/j.1574-6976.1998.tb00366.x. [DOI] [PubMed] [Google Scholar]
- 13.Finlay B B, Falkow S. Common themes in microbial pathogenicity revisited. Microbiol Mol Biol Rev. 1997;61:136–169. doi: 10.1128/mmbr.61.2.136-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Francisco J A, Earhart C F, Georgiou G. Transport and anchoring of beta-lactamase to the external surface of Escherichia coli. Proc Natl Acad Sci USA. 1992;89:2713–2717. doi: 10.1073/pnas.89.7.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gamper M, Ganter B, Polito M R, Haas D. RNA processing modulates the expression of the arcDABC operon in Pseudomonas aeruginosa. J Mol Biol. 1992;226:943–957. doi: 10.1016/0022-2836(92)91044-p. [DOI] [PubMed] [Google Scholar]
- 16.Gish W, States D J. Identification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 17.Henderson I R, Navarro-Garcia F, Nataro J P. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 1998;6:370–378. doi: 10.1016/s0966-842x(98)01318-3. [DOI] [PubMed] [Google Scholar]
- 18.Ho Y S, Swenson L, Derewenda U, Serre L, Wei Y, Dauter Z, Hattori M, Adachi T, Aoki J, Arai H, Inoue K, Derewenda Z S. Brain acetylhydrolase that inactivates platelet-activating factor is a G-protein-like trimer. Nature. 1997;385:89–92. doi: 10.1038/385089a0. [DOI] [PubMed] [Google Scholar]
- 19.Holloway B W, Krishnapillai V, Morgan A F. Chromosomal genetics of Pseudomonas. Microbiol Rev. 1979;43:73–102. doi: 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaeger K-E. Extrazelluläre Enzyme von Pseudomonas aeruginosa als Virulenzfaktoren. Immun Infekt. 1994;22:177–180. [PubMed] [Google Scholar]
- 21.Jaeger K E, Kinscher D A, Koenig B, Koenig W. Extracellular lipase of Pseudomonas aeruginosa: biochemistry and potential role as a virulence factor. In: Hoiby N, Pedersen S S, editors. Cystic fibrosis, basic and clinical research. Amsterdam, The Netherlands: Elsevier; 1992. pp. 113–119. [Google Scholar]
- 22.Jaeger K-E, Ransac S, Dijkstra B W, Colson C, van Heuvel M, Misset O. Bacterial lipases. FEMS Microbiol Rev. 1994;15:29–63. doi: 10.1111/j.1574-6976.1994.tb00121.x. [DOI] [PubMed] [Google Scholar]
- 23.Jaeger K-E, Reetz M T. Microbial lipases form versatile tools for biotechnology. Trends Biotechnol. 1998;16:396–403. doi: 10.1016/s0167-7799(98)01195-0. [DOI] [PubMed] [Google Scholar]
- 24.Jaeger K-E, Schneidinger B, Liebeton K, Haas D, Reetz M T, Philippou S, Gerritse G, Ransac S, Dijkstra B W. Lipase of Pseudomonas aeruginosa: Molecular biology and biotechnological application. In: Nakazawa T, et al., editors. Molecular biology of pseudomonads. Washington, D.C.: American Society for Microbiology; 1996. pp. 319–330. [Google Scholar]
- 25.Jaeger K-E, Schneidinger B, Rosenau F, Werner M, Lang D, Dijkstra B W, Zonta A, Reetz M T. Bacterial lipases for biotechnological applications. J Mol Catal Ser B. 1997;3:3–12. [Google Scholar]
- 26.Klauser T, Pohlner J, Meyer T F. Extracellular transport of cholera toxin B subunit using Neisseria IgA protease β-domain: conformation-dependent outer membrane translocation. EMBO J. 1990;9:1991–1999. doi: 10.1002/j.1460-2075.1990.tb08327.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koenig B, Jaeger K-E, Koenig W. Induction of inflammatory mediator release (12-hydroxyeicosatetraenoic acid) from human platelets by Pseudomonas aeruginosa. Int Arch Allergy Immunol. 1994;104:33–41. doi: 10.1159/000236706. [DOI] [PubMed] [Google Scholar]
- 28.Kok R G, Christoffels V M, Vosman B, Hellingwerf K J. Growth-phase-dependent expression of the lipolytic system of Acinetobacter calcoaceticus BD413: cloning of a gene, encoding one of the esterases. J Gen Microbiol. 1993;139:2329–2342. doi: 10.1099/00221287-139-10-2329. [DOI] [PubMed] [Google Scholar]
- 29.Kovach M E, Phillips R W, Elzer P H, Roop II R M, Peterson K M. pBBR1MCS: a broad-host-range cloning vector. BioTechniques. 1994;16:800–802. [PubMed] [Google Scholar]
- 30.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 31.Loveless B J, Saier M H., Jr A novel family of channel-forming, autotransporting, bacterial virulence factors. Mol Membr Biol. 1997;14:113–123. doi: 10.3109/09687689709048171. [DOI] [PubMed] [Google Scholar]
- 32.MacIntyre S, Trust T J, Buckley J T. Distribution of glycerophospholipid-cholesterol acyltransferase in selected bacterial species. J Bacteriol. 1979;139:132–136. doi: 10.1128/jb.139.1.132-136.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merrick M J. In a class of its own—the RNA polymerase sigma factor ς54 (ςN) Mol Microbiol. 1993;10:903–909. doi: 10.1111/j.1365-2958.1993.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 34.Nakai K, Kanehisa M. Expert system for predicting protein localization sites in Gram-negative bacteria. Protein Struct Funct Genet. 1991;11:95–110. doi: 10.1002/prot.340110203. [DOI] [PubMed] [Google Scholar]
- 35.Nielsen H, Engelbrecht J, Bunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 36.Nunn D, Bergman S, Lory S. Products of three accessory genes, pilB, pilC, and pilD, are required for biogenesis of Pseudomonas aeruginosa pili. J Bacteriol. 1990;172:2911–2919. doi: 10.1128/jb.172.6.2911-2919.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohkawa I, Shiga S, Kageyama M. An esterase on the outer membrane of Pseudomonas aeruginosa for the hydrolysis of long chain acyl esters. J Biochem. 1979;86:643–656. doi: 10.1093/oxfordjournals.jbchem.a132568. [DOI] [PubMed] [Google Scholar]
- 38.O’Toole P W, Austin J W, Trust T J. Identification and molecular characterization of a major ring-forming surface protein from the gastric pathogen Helicobacter mustelae. Mol Microbiol. 1994;11:349–361. doi: 10.1111/j.1365-2958.1994.tb00315.x. [DOI] [PubMed] [Google Scholar]
- 39.Owen P, Meehan M, de Loughry-Doherty H, Henderson I. Phase-variable outer membrane proteins in Escherichia coli. FEMS Immunol Med Microbiol. 1996;16:63–76. doi: 10.1111/j.1574-695X.1996.tb00124.x. [DOI] [PubMed] [Google Scholar]
- 40.Paul C, Rosenbusch J P. Folding patterns of porin and bacteriorhodopsin. EMBO J. 1985;4:1593–1597. doi: 10.1002/j.1460-2075.1985.tb03822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reetz M T, Zonta A, Schimossek K, Liebeton K, Jaeger K-E. Creation of enatioselective biocatalysts for organic chemistry by in vitro evolution. Angew Chem Int Ed English. 1997;36:2830–2832. [Google Scholar]
- 42.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 43.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmitt W, Haas R. Genetic analysis of the Helicobacter pylori vacuolating cytotoxin: structural similarities with the IgA protease type of exported protein. Mol Microbiol. 1994;12:307–319. doi: 10.1111/j.1365-2958.1994.tb01019.x. [DOI] [PubMed] [Google Scholar]
- 45.Schneidinger B. Überexpression und transkriptionelle Regulation des Lipaseoperons von Pseudomonas aeruginosa und funktionelle Charakterisierung der Lipase-spezifischen Foldase LipH. Ph.D. thesis. Bochum, Germany: Ruhr-Universität; 1997. [Google Scholar]
- 46.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vitro genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 47.Staskawicz B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stinnett J D, Gilleland H E, Jr, Eagon R G. Proteins released from cell envelopes of Pseudomonas aeruginosa on exposure to ethylenediaminetetraacetate: comparison with dimethylformamide-extractable proteins. J Bacteriol. 1973;114:399–407. doi: 10.1128/jb.114.1.399-407.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strom M S, Nunn D, Lory S. Multiple roles of the pilus biogenesis protein PilD: involvement of PilD in excretion of enzymes from Pseudomonas aeruginosa. J Bacteriol. 1991;173:1175–1180. doi: 10.1128/jb.173.3.1175-1180.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Struyvé M, Moons M, Tommassen J. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J Mol Biol. 1990;218:141–148. doi: 10.1016/0022-2836(91)90880-f. [DOI] [PubMed] [Google Scholar]
- 51.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 52.Studier F W. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J Mol Biol. 1991;219:37–44. doi: 10.1016/0022-2836(91)90855-z. [DOI] [PubMed] [Google Scholar]
- 53.Stuer W, Jaeger K-E, Winkler U K. Purification of extracellular lipase from Pseudomonas aeruginosa. J Bacteriol. 1986;168:1070–1074. doi: 10.1128/jb.168.3.1070-1074.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tabor S. Expression using the T7 RNA polymerase/promoter system. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: John Wiley & Sons, Inc.; 1990. pp. 16.2.1–16.2.11. [Google Scholar]
- 55.Upton C, Buckley J T. A new family of lipolytic enzymes? Trends Biochem Sci. 1995;20:178–179. doi: 10.1016/s0968-0004(00)89002-7. [DOI] [PubMed] [Google Scholar]
- 56.Vasil M L, Prince R W, Shortridge V D. Exoproducts: Pseudomonas exotoxin A and phospholipase C. In: Fick R B Jr, editor. Pseudomonas aeruginosa. The opportunist. Pathogenesis and disease. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 59–77. [Google Scholar]
- 57.Verger R. Interfacial activation of lipases: facts and artifacts. Trends Biotechnol. 1997;15:32–38. [Google Scholar]
- 58.Visca P, Ciervo A, Orsi N. Cloning and nucleotide sequence of the pvdA gene encoding the pyoverdine biosynthetic enzyme l-ornithine-N5-oxygenase in Pseudomonas aeruginosa. J Bacteriol. 1994;176:1128–1140. doi: 10.1128/jb.176.4.1128-1140.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang H, Dowds B C A. Phase variation in Xenorhabdus luminescens: Cloning and sequencing of the lipase gene and analysis of its expression in primary and secondary phases of the bacterium. J Bacteriol. 1993;175:1665–1673. doi: 10.1128/jb.175.6.1665-1673.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Watson A A, Alm R A, Mattick J S. Construction of improved vectors for protein production in Pseudomonas aeruginosa. Gene. 1996;172:163–164. doi: 10.1016/0378-1119(96)00026-1. [DOI] [PubMed] [Google Scholar]
- 61.Wei Y, Schottel J L, Derewenda U, Swenson L, Patkar S, Derewenda Z S. A novel variant of the catalytic triad in the Streptomyces scabies esterase. Struct Biol. 1995;3:218–223. doi: 10.1038/nsb0395-218. [DOI] [PubMed] [Google Scholar]
- 62.West S E H, Iglewski B H. Codon usage in Pseudomonas aeruginosa. Nucleic Acids Res. 1988;16:9323–9335. doi: 10.1093/nar/16.19.9323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Winkler U K, Stuckmann M. Glycogen, hyaluronate, and some other polysaccharides greatly enhance the formation of exolipase by Serratia marcescens. J Bacteriol. 1979;138:663–670. doi: 10.1128/jb.138.3.663-670.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yanagida N, Uozumi T, Beppu T. Specific excretion of Serratia marcescens protease through the outer membrane of Escherichia coli. J Bacteriol. 1986;166:937–944. doi: 10.1128/jb.166.3.937-944.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yanisch-Peron C, Vieria J, Messing J. Improved M13 cloning vectors and host strains: nucleotide sequences of M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]