Abstract
The Bacillus subtilis two-dimensional (2D) protein index contains almost all glycolytic and tricarboxylic acid (TCA) cycle enzymes, among them the most abundant housekeeping proteins of growing cells. Therefore, a comprehensive study on the regulation of glycolysis and the TCA cycle was initiated. Whereas expression of genes encoding the upper and lower parts of glycolysis (pgi, pfk, fbaA, and pykA) is not affected by the glucose supply, there is an activation of the glycolytic gap gene and the pgk operon by glucose. This activation seems to be dependent on the global regulator CcpA, as shown by 2D polyacrylamide gel electrophoresis analysis as well as by transcriptional analysis. Furthermore, a high glucose concentration stimulates production and excretion of organic acids (overflow metabolism) in the wild type but not in the ccpA mutant. Finally, CcpA is involved in strong glucose repression of almost all TCA cycle genes. In addition to TCA cycle and glycolytic enzymes, the levels of many other proteins are affected by the ccpA mutation. Our data suggest (i) that ccpA mutants are unable to activate glycolysis or carbon overflow metabolism and (ii) that CcpA might be a key regulator molecule, controlling a superregulon of glucose catabolism.
The sequencing of entire genomes opens new perspectives for the comprehensive description and understanding of living cells. To obtain new and complete information on the regulation of global gene expression is one of the most exciting prospects of the postgenome era. The highly sensitive two-dimensional (2D) gel electrophoresis technique, introduced more than 20 years ago (32), has received renewed interest by recent genome sequencing programs. By 2D gel electrophoresis, more than 1,500 bacterial proteins can be separated on a gel of 20 by 20 cm. If alkaline or extracellular proteins are also included in those studies, the majority of proteins synthesized within a bacterial cell can be visualized. The identification of large numbers of protein spots has been facilitated by recent developments in mass spectrometric techniques (e.g., matrix-assisted laser desorption ionization–time of flight and electrospray ionization) that rely on genome sequence data. When proteins collected in a 2D protein index are allocated to physiological function groups to establish a protein function map, two main groups can be distinguished: proteins synthesized during growth, which mainly fulfill housekeeping functions (vegetative proteins), and proteins produced especially in starved or stressed cells, which may have protective functions against stress or starvation in the slow-growing or nongrowing cell.
Vegetative or housekeeping proteins can be allocated to the basic domains of metabolism, such as glycolysis, the tricarboxylic acid (TCA) cycle, and amino acid or pyrimidine/purine biosynthesis, etc. Proteins produced in nongrowing cells can be allocated to starvation- or stress-specific responses with special adaptive functions against one single stimulus or to more general stress responses that protect the nongrowing cell against “future stress” (17, 19).
In many cases, the expression of genes whose products belong to specific function groups is coordinately regulated by environmental stimuli that control the expression of entire regulons. It is quite usual that regulation and function are unified. If the global regulators that control the regulons are known, proteomics is a useful approach for defining the structure and function of individual regulons simply by comparing the proteins produced under appropriate physiological circumstances in the wild-type strain and in the corresponding mutant. The allocation of proteins to stimulons and regulons is an essential step toward understanding of global regulation of the expression of entire genomes (“response regulation map” [42]).
The genome sequence of Bacillus subtilis, a soil-living bacterium, became available 2 years ago (27). Using this sequence information we were able to establish a 2D protein database comprising almost 200 entries (3a). The proteomic approach has been used to characterize and define specific stimulons and regulons of B. subtilis, such as the heat stress stimulon, which was dissected into three main regulons (16, 17). Furthermore, the 2D protein gel electrophoresis technique was also used for defining a network of interacting regulons or modulons (1, 3, 6).
We are interested in the regulation of carbon catabolism in B. subtilis. B. subtilis cells are able to use a variety carbohydrates as sources of carbon and energy. The genes and operons required for the utilization of specific carbon sources are in most cases expressed only if (i) the carbon source is present in the medium and if (ii) preferred carbon sources, such as glucose and other glycolytically metabolizable sugars, are absent from the growth medium. These two processes are referred to as substrate induction and carbon catabolite repression, respectively (for a review, see reference 22). Central to the regulation of carbon catabolism is catabolite control protein A (CcpA) (20, 22). In the presence of glucose, the CcpA protein is a repressor of several catabolic operons involved in the degradation of secondary carbon sources (22, 40). Moreover, CcpA activates the expression of some genes whose products are involved in excretion of excess carbon, such as the ackA gene encoding acetate kinase (14). The main route of entry of many sugars into the central metabolism is glycolysis (9). Despite its great importance for cellular physiology, only limited information about the regulation of the enzymes of glycolysis in B. subtilis is available.
The B. subtilis 2D protein index contains many vegetative proteins involved in glycolysis, the TCA cycle, the pentose phosphate cycle, amino acid or nucleotide biosynthesis, or translation (1, 35, 43). Among the most abundant vegetative proteins of growing cells, almost all glycolytic and TCA cycle enzymes were identified. The knowledge of this “glycolytic and TCA cycle proteome” stimulated the idea of analyzing the regulation of glucose catabolism in B. subtilis by this comprehensive proteomic approach. The data indicate that glycolysis and the TCA cycle are regulated by glucose availability and that CcpA might be involved in this global regulation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
B. subtilis IS58 (trpC2 lys-3) (36) and BGW2 (trpC2 lys-3 ccpA::Tn917) (25) were grown under vigorous agitation at 37°C in a synthetic minimal medium (ASM) with citrate and glutamate as basic sources of carbon and nitrogen, as described previously (38). Glucose was added to final concentrations of 0.025, 0.1, and 1.0%. Ribose was added to a final concentration of 0.1%.
Genetic techniques.
Chromosomal DNA of B. subtilis IS58 was isolated as described previously (29).
PCR products were obtained under the following conditions: denaturation for 1 min at 94°C, annealing for 1.5 min at 55°C, and extension for 1.5 min at 72°C (30 cycles). DNA fragments were amplified by using chromosomal DNA (100 ng) of B. subtilis IS58 with 1 U of Taq polymerase in the appropriate buffer by adding 200 μmol of each deoxynucleoside triphosphate and 100 pmol of each primer in a final volume of 100 μl. The PCR products were analyzed by electrophoresis and purified with the QIAquick purification kit (Qiagen).
Northern blot and slot blot analyses.
Digoxigenin-labeled RNA probes were obtained by in vitro transcription with T7 RNA polymerase by using PCR-generated fragments as templates. Primers used for PCR are listed in Table 1. Reverse primers contain the T7 RNA polymerase recognition sequence (except reverse primers for citH and sucD). Probes for citH and sucD were generated with the Dig Chem Link labeling and detection set (Boehringer Mannheim). Total RNA from exponentially growing B. subtilis cells (optical density at 500 nm of about 0.4) was isolated with the High Pure RNA isolation kit (Boehringer Mannheim). Cultures were grown in ASM supplemented with the appropriate carbon sources. Northern blot analysis was carried out as described previously with equal amounts of RNA as determined spectrophotometrically (26). For quantification of specific mRNA (slot blot), serial dilutions of total RNA in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) were transferred to a positively charged nylon membrane and hybridized with a gene-specific probe. Hybridization signals were detected according to the manufacturer’s instructions (Boehringer Mannheim). Chemiluminographs were analyzed with a Lumi-Imager and the Lumi-Analyze software package (Boehringer Mannheim). Signals corresponding to mRNA of ribose-grown cells served as a standard (set to 1).
TABLE 1.
Primers used in this study
| Designation | Sequence (5′→3′)a |
|---|---|
| gapfor | GTAACGTATTCCGCGCAG |
| gaprev | CTAATACGACTCACTATAGGGAGAGCTGCAAGGTCAACAACG |
| fbaAfor | GCGATTCACTTAGACCACG |
| fbaArev | CTAATACGACTCACTATAGGGAGACGTGGATCGTACTCGTCAG |
| pgkfor | CGATACACGTATCCGTGC |
| pgkrev | CTAATACGACTCACTATAGGGAGACGTACCAATGTCGATTGC |
| enofor | GCACGCGAAGTATTAGACTC |
| enorev | CTAATACGACTCACTATAGGGAGAGGTCGTCACCAACAAGC |
| yvbQfor | CGAAGAAGCCTGTCTGC |
| yvbQrev | CTAATACGACTCACTATAGGGAGACGGTAAGTGCCTGAAGC |
| ackAfor | CGGTATCGCCGACAGCGTATTC |
| ackArev | CTAATACGACTCACTATAGGGAGAAACCGCGAAGAACGCGTTCTC |
| acsAfor | GAAGCGATTGACCGCCATGC |
| acsArev | CTAATACGACTCACTATAGGGAGATTCAATCGTTCCATACCAGC |
| alsSfor | CGTGATCCGTGCATATCG |
| alsSrev | CTAATACGACTCACTATAGGGAGAGGAGTGCGAACCGATATCGC |
| acuBfor | CGAAGACGGATACGCTTG |
| acuBrev | CTAATACGACTCACTATAGGGAGAGACGTGGTGTCCATTTC |
| citHfor | GACAGAGCTTTAGGATAATACTTTCATGACATTT |
| citHrev | TCACAGAGCAGAGAGACATGGGAAATACTC |
| sucDfor | GACAGAGCTTATTAATGCGTTTTACAAGTTTC |
| sucDrev | TCACAGAGCATGAGTGTTTTCATTAATAAAGATA |
T7 recognition sequences are underlined.
pH measurements.
Cells were grown as described above. Probes (2 ml) of each culture were harvested by centrifugation (5 min at 10,000 × g), and the supernatant was used for measurement of pH with a pH checker (Hanna Instruments).
Analytical 2D gel electrophoresis.
Pulse labeling of the bacterial culture for 5 min with 10 μCi of l-[35S]methionine ml−1 during exponential growth was carried out as described previously (35). After sonication of the harvested cells, the protein amount was determined (4). Crude protein extracts (60 μg of protein) were loaded onto IPG strips for the first dimension of 2D gel electrophoresis, as recommended by Völker et al. (43). The second dimension was carried out as described previously (3). After fixation and silver staining, the wet gels were scanned with a Hewlett-Packard ScanJet 6000 in transmission mode at a resolution of 300 dpi and a color depth of 8-bit–256 gray level (10-bit–1,024 gray level internal). For autoradiography of the radiolabeled protein pattern (after separation of 2 mio cpm), the dried gels were exposed to storage phosphor screens (Molecular Dynamics Storage Phosphor Screen, 20 by 25 cm) for 24 hours and scanned with a STORM 840 PhosphorImager (Molecular Dynamics) at a resolution of 200 μm and a color depth of 16-bit (65,536 gray level).
For computer-aided analysis of the gels, the MELANIE II package (Bio-Rad) was used. The protein spots were reallocated from the Sub2D 2D database. The data were incorporated into Sub2D, the 2D protein index of Bacillus subtilis, which is available via the World Wide Web (3a).
RESULTS
Regulation of glycolysis and the TCA cycle by glucose.
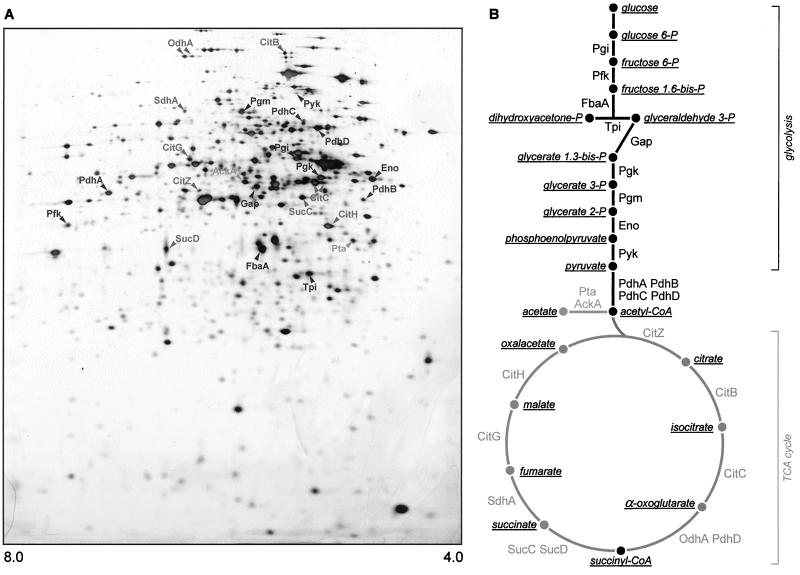
Almost all glycolytic, as well as TCA cycle, enzymes were identified on 2D gels (Fig. 1). Furthermore, acetate kinase AckA and phosphotransacetylase Pta, enzymes involved in carbon source overflow metabolism, were also localized.
FIG. 1.
Main pathways of carbohydrate metabolism. (A) Silver-stained 2D gel showing identified proteins which are involved in glycolysis (Pgi, Pfk, FbaA, Tpi, Gap, Pgk, Pgm, Eno), pyruvate dehydrogenesis (PdhA, PdhB, PdhC, PdhD), the TCA cycle (CitZ, CitB, CitC, OdhA, PdhD, SucC, SucD, SdhA, CitG, CitH), and overflow metabolism (Pta, AckA). (B) Schematic representation of glycolysis and the TCA cycle. Enzymes and metabolites are indicated. Note that not all the proteins of these pathways are identified on 2D gels.
In order to get information on the regulation of glycolysis, cells of the B. subtilis wild-type strain IS58 were cultivated on 1% glucose and on a nonglycolytic carbon source (pure ASM). The protein patterns were analyzed by 2D gel electrophoresis and visualized by silver staining (Fig. 2). To study the rate of synthesis of the proteins of interest, radioactively labeled proteins were also separated (data not shown). In the presence of glucose, there was strong synthesis of the glycolytic enzymes encoded by the gap gene and the pgk operon (Fig. 2), whereas a much lower synthesis rate of these enzymes was found in cells grown without glucose. By contrast, expression of genes encoding enzymes of the upper part (pgi, pfk, fbaA) and lower part (pykA, pdh operon) of glycolysis does not seem to be controlled by glucose. The increased synthesis of glycolytic enzymes was already observed at low glucose concentrations (0.025%) compared to cells grown in the absence of glucose. In glucose-starved stationary-phase cells, the synthesis of glycolytic enzymes was repressed (data not shown).
FIG. 2.
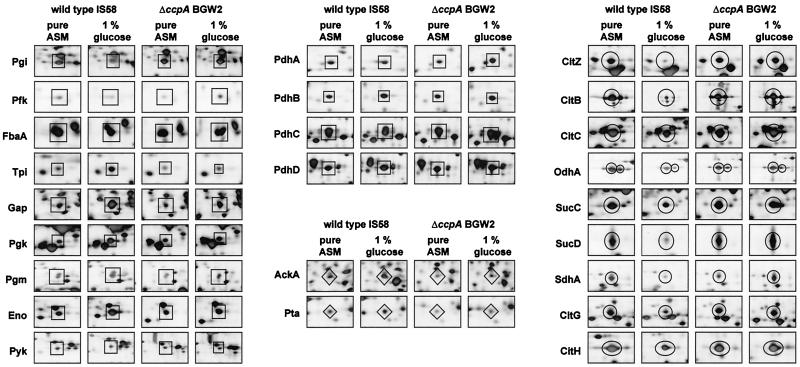
Comparison of CcpA-dependent expression of proteins involved in glycolysis (□), the TCA cycle (○), and overflow metabolism (◊) in B. subtilis. The spots correspond to enzymes of central carbon metabolism. They are derived from silver-stained 2D electrophoretograms of B. subtilis IS58 and the ccpA mutant strain BGW2, which were grown in pure ASM without any additional carbon source or with 1% glucose.
In contrast to glycolytic enzymes, the synthesis of TCA cycle enzymes is controlled in the opposite way: glucose seems to repress expression of the genes citH, citC, citB, odhA, sucCD, and citG, which are derepressed in glutamate–citrate (pure ASM)- or ribose-grown cells (Fig. 2) (see reference 37 for a review).
CcpA is necessary for activation of glycolysis.
The coordinate regulation of genes whose products are required for glycolysis and the TCA cycle implied a regulon structure. Therefore, the involvement of global regulators that can measure the availability of glucose as a carbon source was considered. In B. subtilis and other gram-positive bacteria, CcpA controls carbon catabolite repression. This protein can receive information on the intracellular glucose concentration via interaction with different effector molecules (see Discussion and reference 40). Therefore, the involvement of CcpA in global regulation of glycolysis and the TCA cycle was analyzed. As shown in Fig. 2, stimulation of the synthesis of glycolytic enzymes by glucose in the ccpA mutant strain BGW 2 was weaker than in the wild type.
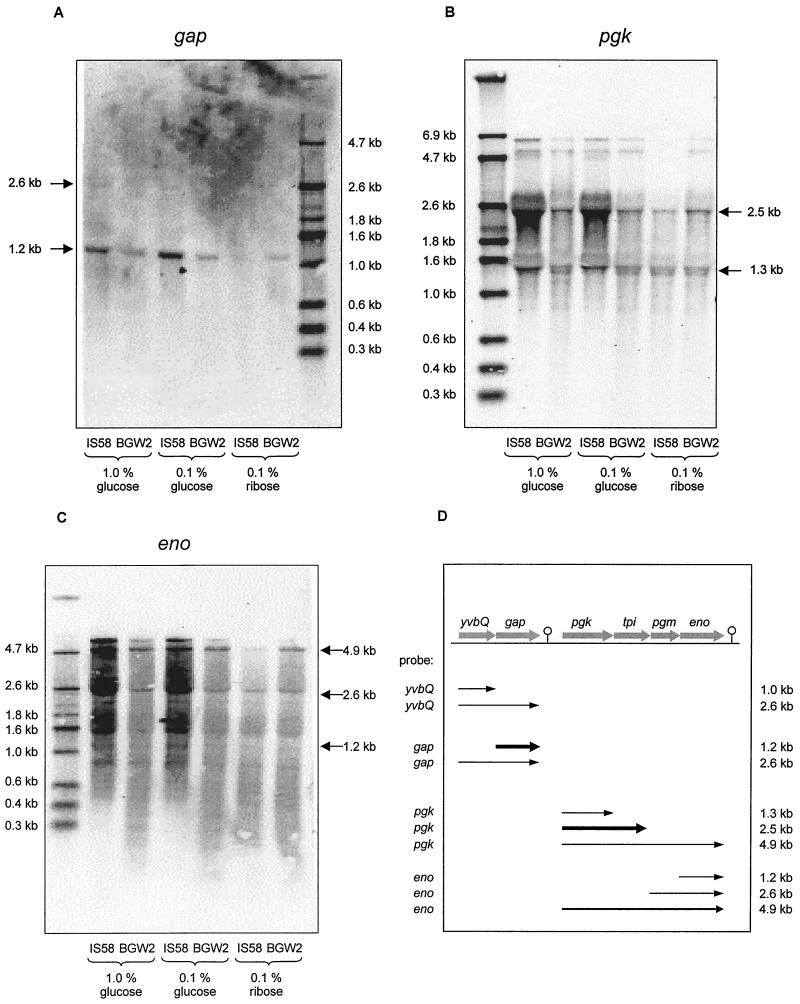
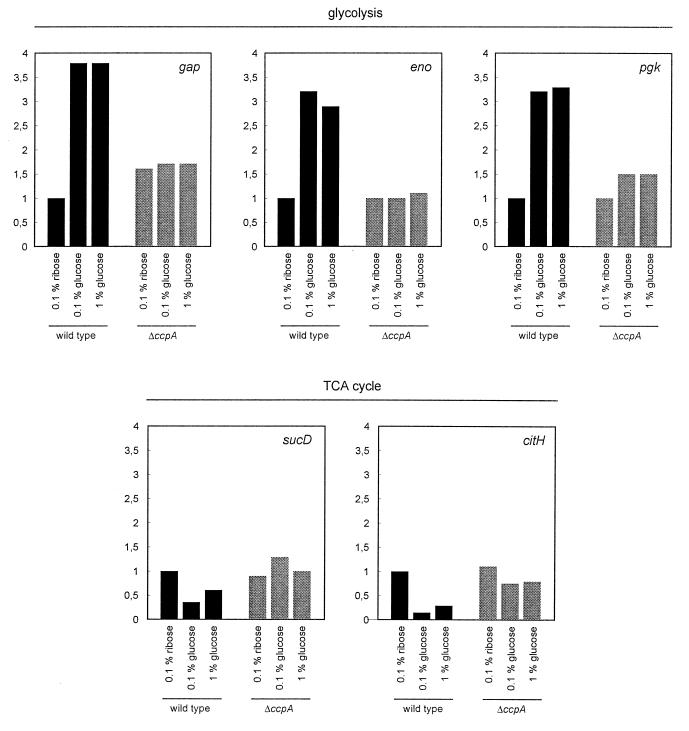
As a complementary approach to obtain evidence for the hypothesis that CcpA acts as global regulator of glycolytic gene expression, transcriptional studies were carried out. The structure of the gap pgk tpi pgm eno glycolytic gene cluster is shown in Fig. 3D. The Northern blotting data indicate that gap, encoding glyceraldehyde-3-phosphate dehydrogenase, is transcribed monocistronically (or forms an operon with the upstream gene yvbQ), while the remaining four genes, encoding phosphoglycerate kinase (pgk), triose phosphate isomerase (tpi), phosphoglycerate mutase (pgm), and enolase (eno), form an operon (Fig. 3). Moreover, we performed a slot blot analysis of the amounts of gap, eno, and pgk mRNAs. The results of the quantification are shown in Fig. 4. This transcriptional analysis confirmed the hypothesis that glucose strongly stimulates the transcription of pgk, eno, and gap in the wild-type strain. In the ccpA mutant strain, however, there was only marginal (if any) stimulation of expression of these genes by glucose. These results strongly support the possibility that CcpA might act as a global activator of transcription.
FIG. 3.
Northern blot analysis of genes encoding glycolytic enzymes. RNA of exponentially growing B. subtilis cells was hybridized with probes specific for gap (A), pgk (B), and eno (C). Sizes of transcripts are indicated, and possible interpretations are illustrated (D); thicknesses of arrows correspond with the amount of transcript.
FIG. 4.
Quantitative analysis of mRNAs of glycolytic and TCA cycle enzymes. Different concentrations of RNA (1 and 0.5 μg) from B. subtilis IS58 and BGW2 were blotted onto nylon membranes and hybridized with probes specific for the genes indicated. The mRNA amount obtained for IS58 in the presence of 0.1% ribose was set at 1.
Glucose stimulates overflow metabolism in a ccpA-dependent manner.
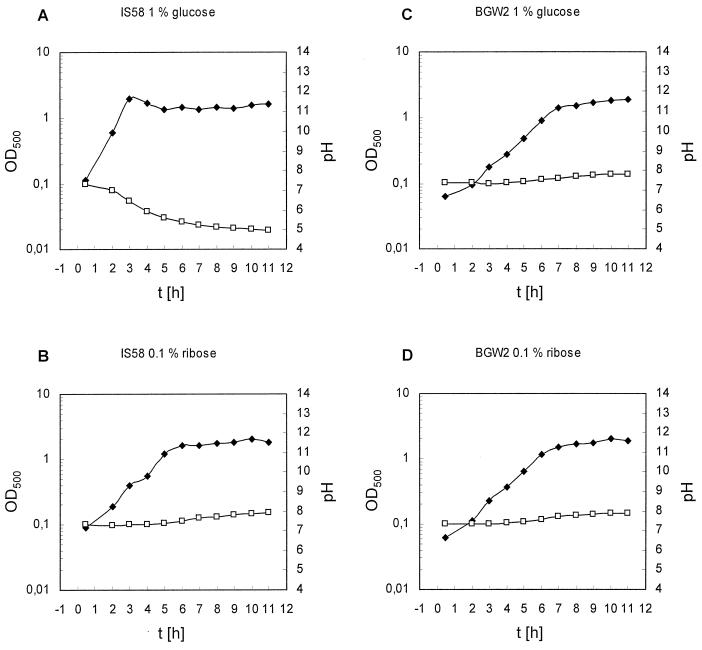
B. subtilis cells grown with high concentrations of glucose produce acetate, lactate, and other organic acids as products of overflow metabolism (30). In the wild-type strain IS58 grown in ASM minimal medium containing 1% glucose, the pH value of the medium declined from pH 7 to pH 5 during the growth and stationary phases. This pH shift did not occur in cells grown without extra glucose, on ribose (Fig. 5), or on complex medium (LB) (data not shown). Only a weak pH shift was observed in cells grown on 0.1% glucose. However, the weak shift to pH 6 was readjusted to pH 7 in later stages of the stationary growth phase (data not shown).
FIG. 5.
CcpA- and carbon source-dependent acidification of growth medium. Measurement of pH (□) during growth (⧫) was carried out as described in Materials and Methods for the B. subtilis IS58 wild-type strain in ASM containing 1% glucose (A) or 0.1% ribose (B), as well as for the B. subtilis BGW2 ccpA mutant strain in ASM containing 1% glucose (C) or 0.1% ribose (D).
Surprisingly, the pH shift did not occur in growing or stationary-phase cells of the ccpA mutant strain BGW2 grown at high glucose concentrations (Fig. 5C). These results indicate that the wild type, but not the ccpA mutant, is able to induce an overflow metabolism at high glucose concentrations to excrete excess carbon sources in the form of acetate, lactate or acetoin, which are responsible for the acidification of the growth media. The expression of genes involved in this overflow metabolism, such as ackA, pta, and alsS, encoding acetate kinase, phosphotransacetylase, and acetolactate synthase, respectively, is also activated by high glucose concentrations in a CcpA-dependent manner, as demonstrated by slot blot quantification of mRNA levels (data not shown; see also Fig. 2). The observation that activation of ackA or alsS expression by high levels of glucose is dependent on a functional ccpA gene is in agreement with previous reports (14, 21).
Involvement of CcpA in regulation of TCA cycle enzymes.
A comparison of the protein synthesis patterns of TCA cycle enzymes revealed that synthesis of these enzymes was subject to carbon catabolite repression in the wild-type strain IS58 (see above) (Fig. 2). In contrast, there was a high enzyme level even in the presence of glucose in the ccpA mutant strain BGW2 (Fig. 2). It was therefore considered conceivable that CcpA mediates carbon catabolite repression of TCA cycle enzymes. This idea was tested by analyzing the amounts of mRNAs of the citH and sucD genes, encoding malate dehydrogenase and the alpha subunit of succinyl coenzyme A (CoA) synthetase, respectively, in the wild type and a ccpA mutant strain (Fig. 4). As observed for protein synthesis, the levels of citH and sucD mRNAs were reduced in the presence of glucose in the wild-type strain. This repression was partially relieved in the ccpA mutant strain, suggesting that the regulation acts at the transcriptional level.
CcpA has a pleiotropic effect on gene expression.
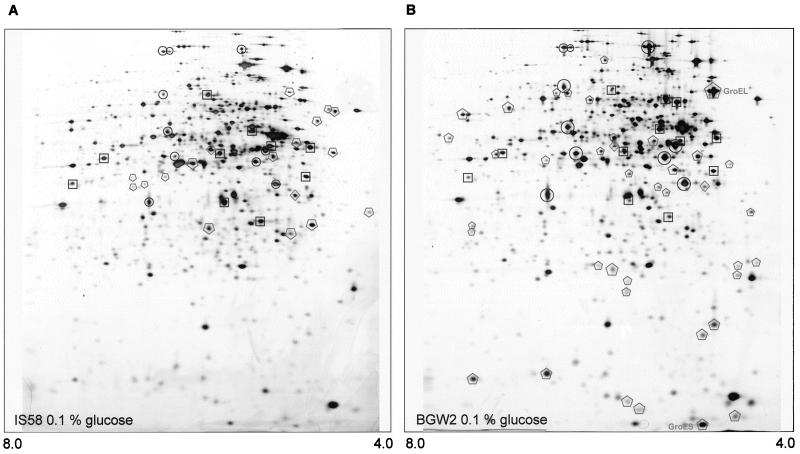
CcpA is known to be a pleiotropic regulator, controlling gene expression in B. subtilis in response to the availability of glucose and other glycolytically metabolized carbon sources. In addition to genes and operons required for the catabolism of secondary sources of carbon and energy, CcpA is involved in the regulation of ammonium assimilation, glycolysis, and the TCA cycle (references 8 and 21 and this work). We asked therefore whether CcpA would affect also the synthesis of other B. subtilis proteins. A comparison of the protein synthesis pattern of the wild-type and ccpA strains revealed that several proteins are affected by the ccpA mutation (Fig. 6). Among these are many proteins that have not yet been identified. Catabolic enzymes that are repressed by CcpA were probably not found in this study, because the corresponding carbon sources as inducers were not supplied. Two prominent proteins, the levels of which were strongly enhanced in the ccpA mutant, are the chaperones GroEL and GroES. These proteins are encoded by the groESL operon. It is interesting to note that a cre box potentially involved in CcpA-dependent control of the operon is located in front of groESL.
FIG. 6.
Silver-stained 2D gels of B. subtilis wild-type strain IS58 (A) and isogenic ccpA mutant strain BGW2 (B) protein extracts. Glycolytic enzymes (□), enzymes of the TCA cycle (○), and enzymes of overflow metabolism (◊) are indicated, as are proteins which are repressed ( ) or induced (
) or induced ( ) in the absence of CcpA.
) in the absence of CcpA.
DISCUSSION
Glycolysis and the TCA cycle are the major catabolic pathways of B. subtilis for generation of carbon backbones and energy (9, 18). The results presented in this work suggest that these pathways are coordinately controlled to achieve a balance between the need for energy production and the generation of precursors for anabolism.
Based on the regulation of the pathway, glycolysis can be divided into four parts: sugar transport and phosphorylation, as the first part, are inducible by the specific sugars present in the medium. The enzymes required for the interconversion of hexose phosphates and the subsequent generation of triose phosphate (glucose phosphate isomerase, phosphofructokinase, and fructose-1,6-bisphosphate aldolase) are synthesized constitutively. The results presented in this paper clearly show that the gap gene and the pgk operon, encoding enzymes catalyzing the conversions from triose phosphate to phosphoenolpyruvate, are not expressed in a constitutive manner, as commonly suggested, but are activated by the addition of glucose. This conclusion was drawn from proteomic studies that indicated an up-regulation of these glycolytic enzymes by glucose and reinforced by transcript analyses which also demonstrated a transcriptional activation of the corresponding genes in the presence of glucose. An increased expression of gap in glucose medium was recently also observed by Aymerich and coworkers (1a). The enzyme performing the final reaction of glycolysis, the conversion of PEP to pyruvate, is again synthesized constitutively. Interestingly, the pyk gene, encoding pyruvate kinase, seems to be the distal gene of an operon with pfk, which encodes phosphofructokinase.
In contrast to glycolytic enzymes, TCA cycle enzymes are repressed at a high glucose concentration in the presence of glutamate. The proteomic approach provided an overall view on the synthesis of almost all TCA cycle enzymes and was in good agreement with previous reports on enzyme activities and transcription of TCA cycle genes (15, 23, 34). Differential repression efficiency, suggesting a fine-tuning of expression, was described for citZ and citB, which are more strongly repressed than citC (23) (Fig. 2). An excess of ATP might be produced at high glucose concentrations mainly by glycolysis. Therefore, the capacity for a complete oxidation of the excess glucose via the TCA cycle is no longer available (and necessary), demanding an overflow metabolism and an excretion of incompletely oxidized intermediates. The pyruvate and acetyl-CoA that are generated but not fed into the TCA cycle in the presence of high glucose concentrations are subsequently converted to acetoin and acids such as acetate or lactate. This suggestion is in agreement with the observation that the medium is acidified after growth in the presence of glucose and with the stimulation of synthesis of enzymes of overflow metabolism in the presence of glucose (references 14 and 21 and this work).
Three major mechanisms of gene regulation by glucose in B. subtilis have been documented. Expression of the ptsG gene, encoding the glucose-specific component of the PEP-dependent phosphotransferase system, is inducible by glucose. This induction occurs at the level of transcription elongation by an antiterminator protein, GlcT (39). The pleiotropic regulatory protein CcpA is known to repress catabolic genes and operons in the presence of glucose and to activate some genes under the same conditions (20–22). Induction of several catabolic operons is negatively controlled by glucose. In the absence of glucose, the HPr protein of the phosphotransferase system phosphorylates operon-specific positive regulators or enzymes that generate the inducers, while no phosphorylation, and consequently no induction, occurs in the presence of glucose (40).
Our data suggest that the global regulator that activates glycolysis and overflow metabolism and represses the TCA cycle in response to the availability of glucose is CcpA. The mechanism of this predicted regulation is still a matter for speculation. No obvious cre boxes are present in front of the genes shown here to be controlled by CcpA. Therefore, an indirect effect of CcpA on controlling another regulator(s) should be taken into consideration. Recently, it was reported that CcpA and the CcpB repressor are not involved in anaerobic regulation of TCA cycle genes (31). Thus, CcpA might control expression of Krebs cycle genes only under aerobic conditions. CcpA activity is modulated by the binding of cofactors, either the HPr protein of the phosphotransferase system or the Crh protein, if these proteins are phosphorylated at a regulatory residue, Ser-46 (5, 11). A fructose-1,6-bisphosphate-activated kinase has been shown to phosphorylate HPr and Crh on Ser-46, thus providing a link between glycolytic activity and the CcpA protein (12, 33). In addition, CcpA can interact with glucose-6-phosphate and NADP (13, 24). Binding of any of the cofactors of CcpA enhances the binding of CcpA to its target cre sites (10, 13, 24). Very recently, Luesink et al. (28) also found a transcriptional activation of the glycolytic las operon mediated by CcpA in Lactococcus lactis and proposed a similar role for CcpA in this organism. Concerning the overflow metabolism in B. subtilis at glucose excess, the situation seems to be more clear: alsS and ackA, encoding α-acetolactate synthase and acetate kinase and containing cre boxes upstream from the promoter regions, are activated by CcpA (14, 41).
Disruption of ccpA genes in gram-positive bacteria generally decreases the growth rate on glucose (2, 7, 8, 22) (Fig. 5). The low glycolytic capacity of ccpA mutants due to the lack of activation of glycolysis in the presence of glucose might be one of several factors to explain the growth deficiency. Among these are the unbalanced expression of catabolic enzymes that might be a burden to the cell, lack of ammonium assimilation, or an accumulation of glycolytic intermediates that cannot be excreted (8, 21, 40). The finding that addition of glutamate to the medium restores growth of ccpA mutants suggests that the defect in induction of glycolytic enzymes is only one of many factors that ultimately result in the growth defect of the ccpA mutant (8, 44). All in all, CcpA seems to be responsible for intracellular glucose sensing following fast glycolytic reactions at high glucose concentrations and ending in overflow metabolism, acidification of the medium, and finally a high growth rate. Kinetic analyses of glucose intermediate concentrations as well as fluxes in the wild type and ccpA mutant could strengthen such a suggestion.
Taken together, the data presented in this work clearly show that the global regulator CcpA not only is involved in carbon catabolite repression but is a key regulatory molecule obviously controlling wide branches of carbon and nitrogen metabolism (8). The latter conclusion is also underlined by our finding that, in addition to glycolytic or TCA cycle enzymes, the level of many other proteins is significantly modified in a ccpA mutant (Fig. 6). To identify these proteins, which may belong to the CcpA regulon, is another of the experimental strategies relying on the proteomic approach. The aim of these studies is to get more comprehensive information on the CcpA regulon in B. subtilis, and presumably in other gram-positive bacteria, and on the global control of carbon metabolism by this regulatory protein.
ACKNOWLEDGMENTS
We are grateful to K. Binder, A. Harang, and A. Tschirner for excellent technical assistance. We are grateful to Uwe Völker and Stephane Aymerich for sharing results prior to publication.
This work was supported by the Deutsch Forschungsgemeinschaft, the Fonds der Chemischen Industrie (to M.H. and J.S), and the EU Biotechnology Programme (BIO-4CT95-0278).
REFERENCES
- 1.Antelmann H, Bernhardt J, Schmid R, Mach H, Völker U, Hecker M. First steps from a two-dimensional protein index towards a response-regulation map for Bacillus subtilis. Electrophoresis. 1997;18:1451–1463. doi: 10.1002/elps.1150180820. [DOI] [PubMed] [Google Scholar]
- 1a.Aymerich, S. Personal communication.
- 2.Behari J, Youngman P. A homolog of CcpA mediates catabolite control in Listeria monocytogenes but not carbon source regulation of virulence genes. J Bacteriol. 1998;180:6316–6324. doi: 10.1128/jb.180.23.6316-6324.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernhardt J, Völker U, Völker A, Antelmann H, Schmid R, Mach H, Hecker M. Specific and general stress proteins in Bacillus subtilis—a two dimensional protein electrophoresis study. Microbiology. 1997;143:999–1017. doi: 10.1099/00221287-143-3-999. [DOI] [PubMed] [Google Scholar]
- 3a.Berhardt J, Werner H. revision date. The 2D protein index of Bacillus subtilis—Sub2D. 19 August 1999. http://microbiol.biologie.uni-greifswald.de::88801 [Online.] http://microbiol.biologie.uni-greifswald.de::88801. Ernst-Moritz-Arndt-Universität Greifswald, Griefswald, Germany. . Ernst-Moritz-Arndt-Universitt Greifswald, Griefswald, Germany. [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Deutscher J, Küster E, Bergstedt U, Charrier V, Hillen W. Protein kinase-dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in Gram-positive bacteria. Mol Microbiol. 1995;15:1049–1053. doi: 10.1111/j.1365-2958.1995.tb02280.x. [DOI] [PubMed] [Google Scholar]
- 6.Drzewiecki K, Eymann C, Mittenhuber G, Hecker M. The yvyD gene of Bacillus subtilis is under dual control of ςB and ςH. J Bacteriol. 1998;180:6674–6680. doi: 10.1128/jb.180.24.6674-6680.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egeter O, Brückner R. Catabolite repression mediated by the catabolite control protein CcpA in Staphylococcus xylosus. Mol Microbiol. 1996;21:739–749. doi: 10.1046/j.1365-2958.1996.301398.x. [DOI] [PubMed] [Google Scholar]
- 8.Faires N, Tobisch S, Bachem S, Martin-Verstraete I, Hecker M, Stülke J. The catabolite control protein CcpA controls ammonium assimilation in Bacillus subtilis. J Mol Microbiol Biotechnol. 1999;1:141–148. [PubMed] [Google Scholar]
- 9.Fortnagel P. Glycolysis. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 171–180. [Google Scholar]
- 10.Fujita Y, Miwa Y, Galinier A, Deutscher J. Specific recognition of the Bacillus subtilis gnt cis-acting catabolite-responsive element by a protein complex formed between CcpA and seryl-phosphorylated HPr. Mol Microbiol. 1995;17:953–960. doi: 10.1111/j.1365-2958.1995.mmi_17050953.x. [DOI] [PubMed] [Google Scholar]
- 11.Galinier A, Haiech J, Kilhoffer M-C, Jaquinod M, Stülke J, Deutscher J, Martin-Verstraete I. The Bacillus subtilis crh gene encodes a HPr-like protein involved in catabolite repression. Proc Natl Acad Sci USA. 1997;94:8439–8444. doi: 10.1073/pnas.94.16.8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galinier A, Kravanja M, Engelmann R, Hengstenberg W, Kilhoffer M C, Deutscher J, Haiech J. New protein kinase and protein phosphatase families mediate signal transduction in bacterial catabolite repression. Proc Natl Acad Sci USA. 1998;95:1823–1828. doi: 10.1073/pnas.95.4.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gösseringer R, Küster E, Galinier A, Deutscher J, Hillen W. Cooperative and non-cooperative DNA binding modes of catabolite control protein CcpA from Bacillus megaterium result from sensing two different signals. J Mol Biol. 1997;266:665–676. doi: 10.1006/jmbi.1996.0820. [DOI] [PubMed] [Google Scholar]
- 14.Grundy F J, Waters D A, Allen S H, Henkin T M. Regulation of the Bacillus subtilis acetate kinase gene by CcpA. J Bacteriol. 1993;175:7348–7355. doi: 10.1128/jb.175.22.7348-7355.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanson R S, Cox D P. Effect of different nutritional conditions on the synthesis of tricarboxylic cycle enzymes. J Bacteriol. 1967;93:1777–1787. doi: 10.1128/jb.93.6.1777-1787.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hecker M, Schumann W, Völker U. Heat-shock and general stress response in Bacillus subtilis. Mol Microbiol. 1996;19:417–428. doi: 10.1046/j.1365-2958.1996.396932.x. [DOI] [PubMed] [Google Scholar]
- 17.Hecker M, Völker U. Non-specific, general and multiple stress resistance of growth restricted Bacillus subtilis cells by the expression of the ςB-regulon. Mol Microbiol. 1998;29:1129–1136. doi: 10.1046/j.1365-2958.1998.00977.x. [DOI] [PubMed] [Google Scholar]
- 18.Hederstedt L. The Krebs citric acid cycle. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 181–197. [Google Scholar]
- 19.Hengge-Aronis R. Interplay of global regulators and cell physiology in the general stress response of Escherichia coli. Curr Opin Microbiol. 1999;2:148–152. doi: 10.1016/S1369-5274(99)80026-5. [DOI] [PubMed] [Google Scholar]
- 20.Henkin T M, Grundy F J, Nicholson W L, Chambliss G H. Catabolite repression of alpha-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacI and galR repressors. Mol Microbiol. 1991;5:575–584. doi: 10.1111/j.1365-2958.1991.tb00728.x. [DOI] [PubMed] [Google Scholar]
- 21.Henkin T M. The role of the CcpA transcriptional regulator in carbon metabolism in Bacillus subtilis. FEMS Microbiol Lett. 1996;135:9–15. doi: 10.1111/j.1574-6968.1996.tb07959.x. [DOI] [PubMed] [Google Scholar]
- 22.Hueck C J, Hillen W. Catabolite repression in Bacillus subtilis: a global regulatory mechanism for Gram-positive bacteria? Mol Microbiol. 1995;15:395–401. doi: 10.1111/j.1365-2958.1995.tb02252.x. [DOI] [PubMed] [Google Scholar]
- 23.Jin S, Sonenshein A L. Transcriptional regulation of Bacillus subtilis citrate synthase genes. J Bacteriol. 1994;176:4680–4690. doi: 10.1128/jb.176.15.4680-4690.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J H, Voskuil M I, Chambliss G H. NADP, corepressor for the Bacillus subtilis catabolite control protein CcpA. Proc Natl Acad Sci USA. 1998;95:9590–9595. doi: 10.1073/pnas.95.16.9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krüger S, Stülke J, Hecker M. Catabolite repression of β-glucanase synthesis in Bacillus subtilis. J Gen Microbiol. 1993;139:2047–2054. doi: 10.1099/00221287-139-9-2047. [DOI] [PubMed] [Google Scholar]
- 26.Krüger S, Gertz S, Hecker M. Transcriptional analysis of bglPH expression in Bacillus subtilis: evidence for two distinct pathways mediating carbon catabolite repression. J Bacteriol. 1996;178:2637–2644. doi: 10.1128/jb.178.9.2637-2644.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 28.Luesink E, van Herpen R E M A, Grossiord B P, Kuipers O P, de Vos W M. Transcriptional activation of the glycolytic las operon and catabolite repression of the gal operon in Lactococcus lactis are mediated by the catabolite control protein CcpA. Mol Microbiol. 1998;30:789–798. doi: 10.1046/j.1365-2958.1998.01111.x. [DOI] [PubMed] [Google Scholar]
- 29.Maede H M, Long S R, Ruvkun G B, Brown S E, Ausubel F M. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J Bacteriol. 1982;149:114–122. doi: 10.1128/jb.149.1.114-122.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakano M N, Dailly Y P, Zuber P, Clark D P. Characterization of anaerobic fermentative growth of Bacillus subtilis: identification of fermentation end products and genes required for growth. J Bacteriol. 1997;179:6749–6755. doi: 10.1128/jb.179.21.6749-6755.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakano M N, Zuber P, Sonenshein A L. Anaerobic regulation of Bacillus subtilis Krebs cycle genes. J Bacteriol. 1998;180:3304–3311. doi: 10.1128/jb.180.13.3304-3311.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Farrell P H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 33.Reizer J, Hoischen C, Titgemeyer F, Rivolta C, Rabus R, Stülke J, Karamata D, Saier M H, Hillen W. A novel protein kinase that controls carbon catabolite repression in bacteria. Mol Microbiol. 1998;27:1157–1169. doi: 10.1046/j.1365-2958.1998.00747.x. [DOI] [PubMed] [Google Scholar]
- 34.Rosenkrantz M S, Dingman D W, Sonenshein A L. Bacillus subtilis citB gene is regulated synergistically by glucose and glutamine. J Bacteriol. 1985;164:155–164. doi: 10.1128/jb.164.1.155-164.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmid R, Bernhardt J, Antelmann H, Völker A, Mach H, Völker U, Hecker M. Identification of vegetative proteins for a two dimensional protein index of Bacillus subtilis. Microbiol. 1997;143:991–998. doi: 10.1099/00221287-143-3-991. [DOI] [PubMed] [Google Scholar]
- 36.Smith I, Paress P, Cabane K, Dubnau E. Genetics and physiology of the rel system of Bacillus subtilis. Mol Gen Genet. 1980;18:271–279. doi: 10.1007/BF00270472. [DOI] [PubMed] [Google Scholar]
- 37.Sonenshein A L. Metabolic regulation of sporulation and other stationary-phase phenomena. In: Smith I, Slepecky R A, Setlow P, editors. Regulation of prokaryotic development. Washington, D.C.: American Society for Microbiology; 1989. pp. 109–130. [Google Scholar]
- 38.Stülke J, Hanschke R, Hecker M. Temporal activation of β-glucanase synthesis in Bacillus subtilis is mediated by the GTP pool. J Gen Microbiol. 1993;139:2041–2045. doi: 10.1099/00221287-139-9-2041. [DOI] [PubMed] [Google Scholar]
- 39.Stülke J, Martin-Verstraete I, Zagorec M, Rose M, Klier A, Rapoport G. Induction of the Bacillus subtilis ptsGHI operon by glucose is controlled by a novel antiterminator, GlcT. Mol Microbiol. 1997;25:65–78. doi: 10.1046/j.1365-2958.1997.4351797.x. [DOI] [PubMed] [Google Scholar]
- 40.Stülke J, Hillen W. Carbon catabolite repression in bacteria. Curr Opin Microbiol. 1999;2:195–201. doi: 10.1016/S1369-5274(99)80034-4. [DOI] [PubMed] [Google Scholar]
- 41.Turinsky A J, Grundy F J, Kim J-H, Chambliss G H, Henkin T M. Transcriptional activation of the Bacillus subtilis ackA gene requires sequences upstream of the promoter. J Bacteriol. 1998;180:5961–5967. doi: 10.1128/jb.180.22.5961-5967.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Bogelen R A, Abshire K Z, Moldover B, Olson E R, Neidhardt F C. Escherichia coli proteome analysis using the gene-protein database. Electrophoresis. 1997;18:1243–1251. doi: 10.1002/elps.1150180805. [DOI] [PubMed] [Google Scholar]
- 43.Völker, A., M. Hecker, and U. Völker. Unpublished results.
- 44.Wray L W, Jr, Pettengill F K, Fisher S H. Catabolite repression of the Bacillus subtilis hut operon requires a cis-acting site located downstream of the transcription initiation site. J Bacteriol. 1994;176:1894–1902. doi: 10.1128/jb.176.7.1894-1902.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]