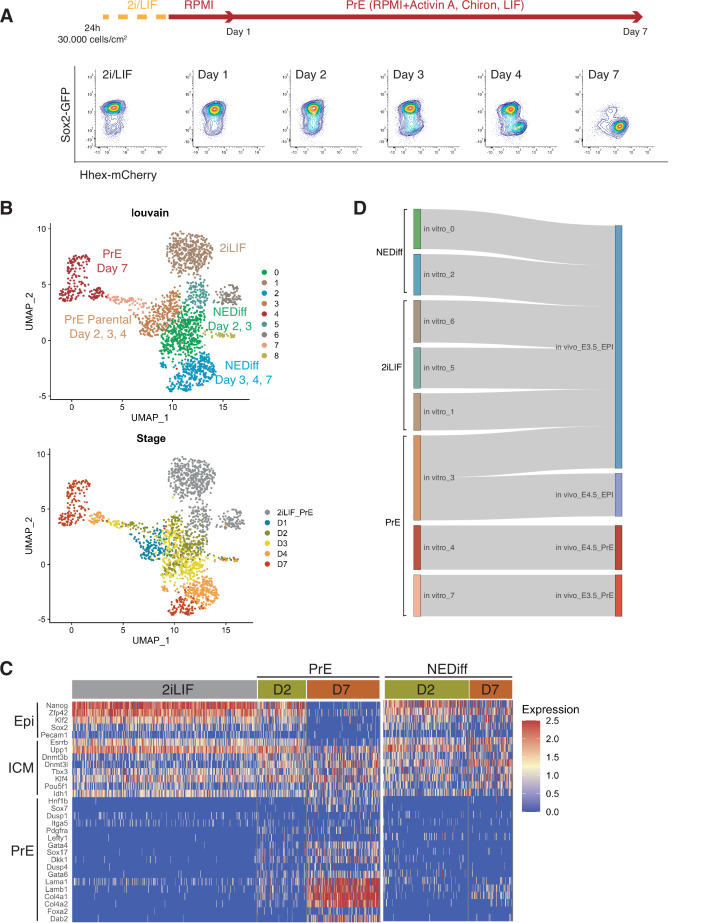
Figure 1. Transcriptome profiling of Primitive Endoderm (PrE) in vitro differentiation.
(A) Schematic of the experiment. Cells were passaged twice in 2i/LIF and then plated in RPMI base media 24 hr before starting the experiment. Bottom panel: Flow cytometry plots showing the time points selected for single-cell RNA-seq. The fluorescent information of Sox2 and Hhex was recorded prior to sequencing. Cells from all the populations shown in the plots were collected for sequencing. (B) UMAP projection of the in vitro experiment showing nine identified clusters using Louvain (upper panel) and stages of differentiation (bottom panel). (C) Heatmap showing expression of selected Epi, Inner Cell Mass (ICM), and PrE markers in 2i/LIF, days 2 and 7 of differentiation. Left panel: PrE Diff branch. Right panel: NEDiff branch. Cells at day 2 in the PrE branch already are upregulating endoderm genes while the NEDiff cells are not. (D) Sankey plot visualizing cluster similarity comparison between identified in vitro clusters and in vivo (Nowotschin et al., 2019) experiment using the Cluster Alignment Tool (CAT).