Abstract
The current model for autodisplay suggests a mechanism that allows a passenger protein to be translocated across the outer membrane by coordinate action of a C-terminal β-barrel and its preceding linking region. The passenger protein, linker, and β-barrel are together termed the autotransporter, while the linker and β-barrel are here referred to as the translocation unit (TU). We characterized the minimal TU necessary for autodisplay with the adhesin-involved-in-diffuse-adherence (AIDA-I) autotransporter. The assumed β-barrel structure at the C terminus of the AIDA-I autotransporter was studied by constructing a set of seven AIDA-I–cholera toxin B subunit fusion proteins containing various portions of AIDA-I. Surface exposure of the cholera toxin B moiety was assessed by dot blot experiments and trypsin accessibility of the chimeric proteins expressed in Escherichia coli JK321 or UT5600. Export of cholera toxin B strictly depended on a complete predicted β-barrel region. The absolute necessity for export of a linking region and its influence on expression as an integral part of the TU was also demonstrated. The different electrophoretic mobilities of native and denatured chimeras indicated that the proposed β-barrel resides within the C-terminal 312 amino acids of AIDA-I. Together these data provide evidence for the predicted β-barrel structure and support our formerly proposed model of membrane topology of the AIDA-I autotransporter.
The cell envelope of gram-negative bacteria consists of two membranes, the cytoplasmic or inner membrane and the outer membrane. Transport of proteins across the inner membrane in most cases follows the general secretory pathway (GSP) (36, 40). Therefore, in gram-negative bacteria, proteins end up in the periplasm. To translocate proteins to the outer surface or into the supernatant, gram-negative bacteria have developed several distinct mechanisms. Type I secretion systems usually consist of three components: an ATPase located in the inner membrane, a membrane fusion protein which connects the inner and outer membranes and thereby possibly excludes the periplasm, and an outer membrane component, all three of which are necessary to export a passenger protein into the supernatant. For efficient transport of the export proteins, a specific C-terminal recognition sequence is required (10, 43, 53). The best known type II system, mediating pullulanase export from Klebsiella oxytoca, requires 16 different gene products, including some which have been localized to the periplasmic space and inner and outer membranes (41). The exported pullulanase itself and most of the accessory components employ the GSP for translocation to the periplasm or insertion into the membranes. Similarly, the type III or contact-dependent systems consist of at least 20 gene products (17, 50). Some of these proteins are thought to build platforms in the inner and outer membranes, while others form a channel which excludes the periplasmic space between these platforms and which protrudes above the cell surface (29). The channel seems to be regulated by a molecular stop valve protein that opens upon contact with a specific receptor (12, 42). A variety of passengers which contain an mRNA-dependent export signal are then released from the cells (1).
In contrast to the secretory systems that require a variety of specialized accessory proteins that, often in combination with the GSP, are responsible for the export of one or several passenger proteins into the supernatant (reviewed in reference 16), the autotransporter protein family members carry the export signal and machinery within a single polypeptide chain. A mechanism for autotransporters has been previously suggested (39) and supported by other reports (25, 28, 33, 37, 49). The N-terminally located signal peptide directs export of the precursor molecule by action of the GSP into the periplasm, where it is cleaved by the signal peptidase; this action sets free the passenger domain and the C-terminal translocation unit (TU), consisting of the linking region and the β-barrel (at the extreme C terminus), which are together required for autodisplay. The β-barrel portion of the autotransporter presumably folds from a periplasmic intermediate conformation to build an aqueous pore channel across the membrane. The linker attached to the periplasmic side of the β-barrel presumably folds through the aqueous pore, pulling the as-yet-unfolded translocation-competent passenger domain through the pore, beginning with its C-terminal part (26). Autodisplayed passenger proteins are often cleaved from the autotransporter and set free into the supernatant (6, 20, 22, 27, 34, 45, 54); however, others instead remain covalently connected or attached to the cell surface (8, 9, 19) and their outer membrane localization has been shown (11, 46, 48). Several approaches to identify the minimal TU of autotransporters have been employed (25, 35, 49), but so far no attempt to distinguish between the linker and assumed β-barrel has been made. β-Barrel topologies have thus been postulated to exist in the TU of autotransporters but have not been demonstrated (46, 48), and the contribution of the linking region to the translocation still remains to be shown.
The adhesin-involved-in-diffuse-adherence (AIDA-I) autotransporter has been identified as a virulence factor of the enteropathogenic Escherichia coli (EPEC) strain 2787 (7) and predicted to be a member of the autotransporter protein family (23). Previously, we fused the cholera toxin B subunit (CTB) to the C-terminal part of AIDA-I (33) that remains associated with the outer membrane after natural processing (48). This fusion protein is located in the outer membrane, and surface exposure of the CTB moiety and protease resistance of the membrane-embedded part have been demonstrated. On the basis of this, a model for the structure of the membrane-embedded part of AIDA-I was proposed (33). In this paper, we define the TU of the AIDA-I autotransporter. We generated a series of CTB–AIDA-I fusion proteins which contain different portions of AIDA-I. Expression of the various fusion proteins was monitored, and surface exposure of the CTB moiety was examined. We here present data demonstrating (i) the influence of the length of the linker on expression, (ii) the linker region’s influence on and essential role for export, (iii) evidence for the predicted β-barrel structure of the C-terminal domain of AIDA-I autotransporter and its necessity for export, and (iv) the conformity of the new data with our former structural prediction (33).
MATERIALS AND METHODS
Bacterial strains.
E. coli UT5600 (F− ara-14 leuB6 azi-6 lacY1 proC14 tsx-67 entA403 trpE38 rfbD1 rpsL109 xyl-5 mtl-1 thi1 ΔompT-fepC266) (15), E. coli JK321 (UT5600 zih-12::Tn10 dsbA::kan) (24), and E. coli EPEC 2787 harboring the plasmid containing AIDA-I (7) were described previously. For all purposes, bacteria were grown routinely at 37°C on Luria-Bertani agar plates containing 50 mg of ampicillin liter−1.
Recombinant DNA techniques.
The aida 3′ regions used to generate constructs employed in this study were amplified from a plasmid preparation of pJM7 by PCR with the primers JM1 (5′-GGAAGATCTG CCTCAGAAAT GAGGGCC-3′), JM2 (5′-ATTCCGCGGG CACTGGCTAA CTCACTGT-3′), JM3 (5′-ACTCCGCGGC TTCCCACATC TGATACCC-3′), JM4 (5′-GGTCCGCGGG TTGCCGGAGC TTATGATTAC-3′), JM7 (5′-CGGGGTACCC TTAATCCTAC AAAAGAAAGT-3′), JM11 (5′-AACGGTACCC TTAATGACGG GCAAAATAA-3′), and JM12 (5′-AGAGGTACCA GAAACACACT GGATGGT-3′). JM2, JM3, and JM4 contain SacII sites, while JM11 and JM12 contain Asp718I sites. The PCR products from JM1 and JM11 or JM12 were cleaved with Asp718I and BamHI, while PCR products from JM1 and JM2, JM3, or JM4 were cloned blunt-ended into pBluescript KS+. Cloning from pBluescript into Asp718I-BamHI-digested pJM7 allowed the generation of all plasmids used in this study.
Standard molecular techniques were performed in accordance with common procedures (44). Ligation products were used to transform competent E. coli JK321 or UT5600. The enzymes used in this study were obtained from Applied Gene Technology Systems, New England Biolabs, Boehringer Mannheim, and Pharmacia.
In vivo techniques.
For whole-cell protease treatment, E. coli cells were collected from agar plates, suspended in phosphate-buffered saline (PBS), and adjusted to an optical density at 578 nm OD578 of 10.0. To 0.2 ml of cell suspension was added 2 μl of protease stock solution to yield a final concentration of 50 mg of trypsin liter−1. Suspensions were incubated for 10 min at 37°C, and digestion was stopped by washing the cells three times with 750 μl of PBS containing 10% fetal calf serum. Cells were sedimented by a brief centrifugation, suspended in sample buffer (30), and then immediately heat treated by incubation at the indicated temperatures for 10 min for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting.
Outer membrane preparation of E. coli.
E. coli cells grown overnight at 37°C were collected from agar plates and suspended in PBS to yield an OD578 of 5.0. The suspension was passed through a French pressure cell once at 20,000 lb/in2 to lyse the bacteria. The lysate was centrifuged at 5,000 × g for 10 min to remove intact bacteria and large debris. The clarified bacterial lysate was centrifuged for 30 min at 100,000 × g, and the resulting supernatant was completely aspirated. The pellet was suspended with PBS plus 1% Sarkosyl (N-lauroyl sarcosinate, sodium salt) and centrifuged for 30 min at 100,000 × g. The pelleted proteins were derived from the outer membranes and used for heat modification experiments.
Cell envelope preparation of E. coli.
Cells were harvested and suspended in PBS. Cell lysis resulted from passing the bacterial suspensions through a French pressure cell once at 20,000 lb/in2. Raw envelopes were pelleted by centrifugation at 14,000 × g for 20 min. Envelopes were washed with PBS and suspended in SDS-PAGE sample buffer.
Protein preparation and Western blotting.
Fragments derived from whole-cell digestion of fusion proteins with trypsin were copurified with outer membranes and subjected to SDS-PAGE. The proteins were electrotransferred to a polyvinylidene difluoride membrane (Trans-Blot; Bio-Rad). Protein bands were stained with Coomassie brilliant blue R250 or incubated after blocking with one of the different antisera at the following dilutions: 1:1,000 for anti-CTB AK55, 1:500 for anti-OmpA AK57, and 1:300 for the anti-PEYFK hybridoma supernatant Dü142. After incubation with goat anti-mouse–peroxidase or goat anti-rabbit–peroxidase conjugates, the blots were developed by enhanced chemiluminescence.
RESULTS AND DISCUSSION
Construction of aida-ctxB gene fusions.
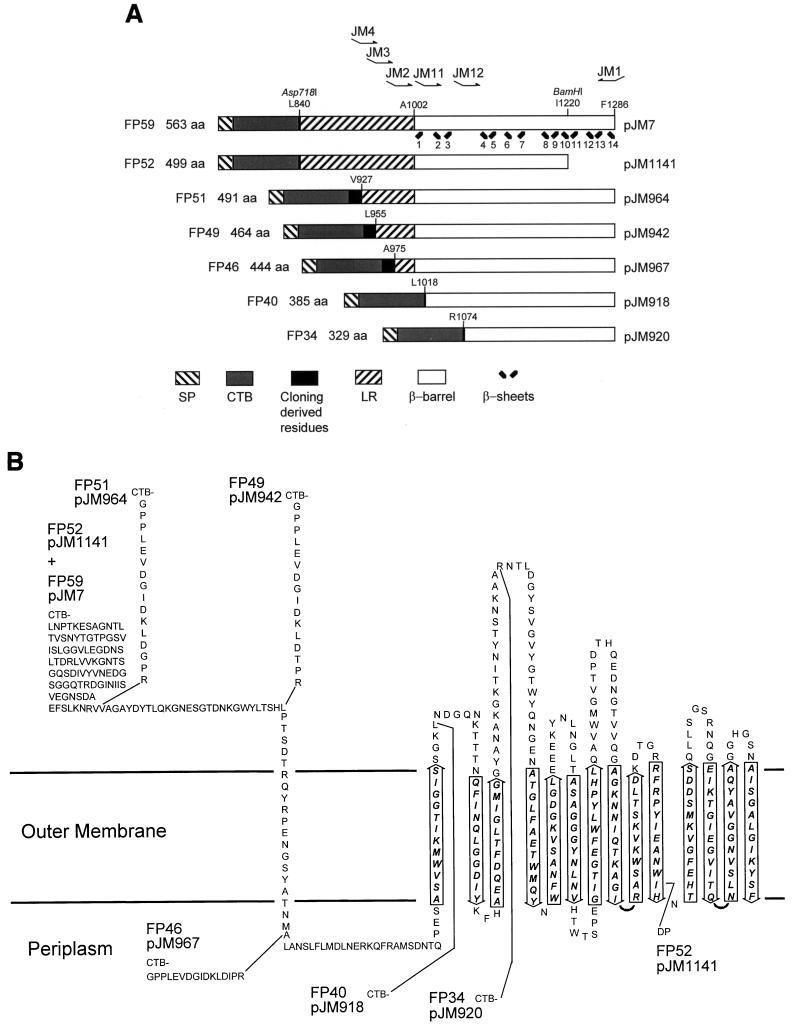
Different portions of the 3′ region of aida were amplified from the pJM7 template by PCR with oligonucleotide primers JM1 and either JM2, JM3, JM4, JM11, or JM12 (Fig. 1A). pJM7 contains a sequence, coding for a CTB–AIDA-I fusion protein, in which the ctxB sequences are located in-frame before the linker region of the EPEC strain 2787 aida-I gene. pJM7 contains an Asp718I cleavage site between ctxB and aida and a BamHI cleavage site in the domain coding for the predicted β-barrel of AIDA-I (33). The PCR products from JM1 and JM11 or JM12 were cleaved with Asp718I and BamHI and cloned in pJM7.
FIG. 1.
(A) Scheme of the composition of the various CTB–AIDA-I fusion proteins. The orientation and location of primers are indicated, as are the sites used for cloning and amino acid positions according to the native AIDA-I sequence. Total lengths of the various fusion proteins and the respective plasmids are shown. The arrangement of β-sheets in the proposed β-barrel and their numbering are given. LR, linking region; SP, signal peptide. (B) Topological model of the organization of the AIDA-I TU and the CTB fusion proteins. β-Barrel sequences (in italics) and linker sequences are shown relative to the lipid/LPS bilayer of the outer membrane (depicted by the horizontal lines). The fusion points of CTB (and any cloning-derived residues) in the respective CTB–AIDA-I fusion proteins are indicated by connecting lines. Arrows represent the predicted amphipathic β-sheets and their assumed directions relative to the (horizontal) plane of the outer membrane.
The PCR products from JM1 (upstream) and JM2, JM3, or JM4 did not contain Asp718I cleavage sites and were therefore cloned into pBluescript KS+ hydrolyzed with EcoRV. The resulting plasmids were cleaved with Asp718I and BamHI, and the fragments were cloned into pJM7 also digested with Asp718I and BamHI. Due to this procedure, a fragment coding for 16 amino acids derived from the pBluescript KS+ multiple-cloning site was included in these gene fusions between the ctxB portion and aida (Fig. 1).
The ctxB used for this experiment was a mutated form that lacks cysteines and therefore was not expected to pentamerize (32). All fusion proteins were expressed under control of the strong constitutive promoter PTK (26). The resulting proteins were named according to their deduced molecular weights after processing by signal peptidase, as indicated in Fig. 1. The fusion points were designed to give rise to four different lengths of the proposed linking region that is thought to be necessary for export by the AIDA-I autotransporter. Two additional N-terminal deletions were set within the predicted β-barrel of the AIDA-I autotransporter. These six CTB–AIDA-I fusion proteins were stepwise N-terminally deleted variants of the AIDA-I autotransporter. The largest one started at AIDA-I’s natural processing site 447 amino acids (in FP59) before the C terminus with L840 (with respect to AIDA’s complete sequence of 1,286 amino acids) and thereby defined the beginning of the proposed linking region. The next fusions were positioned 359 (FP51; V927), 332 (FP49; L955), 312 (FP46; A975), 269 (FP40; L1018), and 213 (FP34; R1074) amino acids in front of the C terminus, respectively. The predicted β-barrel started at 284 amino acids upstream of the C terminus with A1002, while FP34 ended within the second predicted extracellular loop of the predicted β-barrel. One more ctxB-aida fusion was produced by cleaving pJM7 with BamHI, and subsequent fill-in and self-ligation resulted in a frameshift mutation. The respective plasmid was named pJM1141, and the corresponding fusion protein FP52 comprised the complete AIDA-I linking region from FP59 while missing the C-terminal four transmembrane regions contained in the deleted C-terminal 66 amino acids of the proposed β-barrel of AIDA-I (Fig. 1B), thereby ending with I1220.
Expression of aida-ctxB gene fusions.
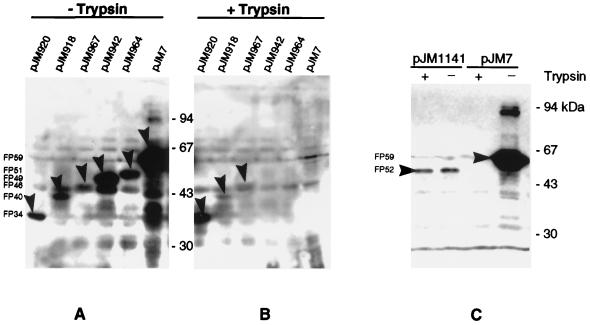
This set of seven different CTB–AIDA-I fusion proteins was expressed by using E. coli JK321 (dsbA ompT) (24) or its parent strain UT5600 (ompT) (15) as the host. Fusion protein expression levels varied significantly despite the identical backgrounds used (Fig. 2A and C). In a Western blot experiment, whole-cell lysates of E. coli JK321 or UT5600 containing one of the different plasmids were separated by SDS-PAGE. The presence of the CTB moiety of the respective fusion proteins was detected with the anti-CTB serum AK55. The expression level in UT5600 was generally higher than that in JK321, a strain that grows slightly slower. FP59 was expressed at the highest level, followed by FP51 and FP49, comprising smaller portions of AIDA-I linking region. Very poor expression was observed for FP46 and FP52, while the two shortest fusion proteins, FP34 and FP40, were expressed at moderate levels.
FIG. 2.
Expression and trypsin sensitivity of CTB–AIDA-I fusion proteins. (A) Equal amounts of whole-cell protein from E. coli JK321 harboring the indicated plasmids were separated by SDS-PAGE and Western blotted with anti-CTB serum AK55. Specific CTB-derived bands are indicated with arrowheads, and the migration of molecular size markers is indicated in kilodaltons. (B) Cells of E. coli JK321 were subjected to trypsin treatment, and subsequently treated as described for panel A. (C) E. coli UT5600 harboring pJM1141 (or pJM7 as a control) was treated as described for panels A and B. Panels A to C employed equivalent cell numbers.
Mapping the minimal translocation unit of the AIDA-I autotransporter.
The surface exposure of the passenger protein domain CTB was judged by two means, the accessibility of the CTB moiety in whole cells to anti-CTB serum AK55 in a dot blot experiment, and the sensitivity of CTB to trypsin treatment of whole cells. The dot blot experiment (data not shown) clearly showed surface exposure of CTB in cells expressing the CTB–AIDA-I fusions FP59, FP51, and FP49, each of which, based on our previous model, is expected to contain a complete β-barrel. However, FP46, which is predicted to have a complete β-barrel but lacks most of the adjacent linking region, was not labeled by AK55. This suggests that the linker domain is essential for membrane translocation of the passenger domain. The three clones expressing fusion proteins predicted to have incomplete β-barrels, FP52 (C-terminal deletion), FP40, and FP34 (both N-terminal deletions), were tested negative in the dot blot experiment, showing that the CTB moiety was not surface exposed.
In the second approach, the seven different CTB–AIDA-I-expressing clones were assayed for the accessibility of the CTB moiety to treatment of whole cells with trypsin. After digestion, whole-cell protein was separated by SDS-PAGE, and CTB was detected in a Western blot with undigested cells used as controls (Fig. 2). CTB was detected in all seven clones. After digestion of whole cells with trypsin, the signals of FP59, FP51, and FP49 disappeared completely. Thus, the CTB moieties of these fusion proteins were accessible to trypsin externally added to whole cells, thereby indicating translocation of CTB through the outer membrane. The four remaining fusion proteins, FP52, FP46, FP40, and FP34, were resistant to trypsin digestion, indicating the absence of CTB on the bacterial surface. According to our model, three of the four trypsin-resistant AIDA-I–CTB fusion proteins, FP52, FP40, and FP34, are not expected to possess a complete β-barrel. The finding that these proteins were resistant to trypsin indicates a blockade in either membrane insertion or translocation of the passenger across the outer membrane. Interestingly, despite the fact that FP46 covered both the complete predicted β-barrel and a short portion of the preceding linking region, the CTB moiety was not exported. This indicates that the linker region preceding the assumed β-barrel may be essential for autotransporter function. Together these data indicate that the minimal TU of AIDA-I extends to within the 20 amino acids of the linking region between the fusion points of FP46 (A975) and FP49 (L955).
Several points regarding the extreme differences in steady-state levels of the various fusion proteins have to be considered. The variable amounts of detected proteins probably were caused by their different levels of accessibility and sensitivity to cellular proteases. The well-expressed fusion proteins are likely able to completely export the CTB passenger domain to the bacterial surface. Here, the CTB and linking region would no longer be prone to degradation by proteases of the inner membrane and the periplasmic space (2, 4), while the assumed β-barrel portion of the autotransporter is embedded in the membrane. The poorly expressed fusion proteins probably do not form the predicted C-terminal β-barrel. Consequently, they might be unable to insert into, or to translocate the passenger across, the outer membrane, possibly floating freely in the periplasm or being attached to the outer membrane. In both cases, and due to the lack of stable secondary structures, these fusion proteins are likely exposed to rapid degradation by envelope proteases (3, 5). Although FP49 and FP51 were both shown to possess functional TUs as did FP59, the former two proteins exhibited lower steady-state levels than FP59. This difference might be due to the truncated linkers in FP49 and FP51 and reflect the elongated times necessary for the β-barrel folding and for the passenger translocation.
Evidence for a β-barrel structure at the C terminus of the AIDA-I TU.
Recently, we proposed a topological model of the membrane-embedded β-barrel portion of the AIDA-I autotransporter (33). To strengthen that prediction, we used the criterion of heat modification. β-Barrels are known to form very stable secondary or tertiary structures which are stable in SDS-containing sample buffer at room temperature and only unfold upon heating to high temperatures (14, 47). This feature can be easily detected because outer membrane proteins with intact β-barrels, in comparison to the completely denatured, random-coiled polypeptide chain that exists after boiling in sample buffer, show an altered electrophoretic mobility in SDS-PAGE. This property can be explained with the differing dodecyl sulfate binding capacity of the two protein forms: due to their compact secondary structure, intact β-barrels bind relatively few dodecyl sulfate molecules while denatured proteins bind more, thereby having an increased charge/molecular weight ratio. Proteins with high charge/weight ratios migrate faster in SDS-PAGE; those with low ratios migrate more slowly. Proteins such as OmpA and AIDA-I autotransporter possess, in addition to their β-barrels, additional domains. These domains likely unfold in SDS sample buffer at room temperature, binding normal amounts of dodecyl sulfate. This leads to different outcomes: intact β-barrels alone (not linked to other domains) tend to migrate more slowly in SDS-PAGE than do their completely denatured variants, while intact β-barrels linked to another domain that is completely denatured at room temperature in SDS sample buffer tend to migrate faster in SDS-PAGE because the compact, low-charged β-barrels are “dragged” through the gel matrix by the highly-charged second domain (47).
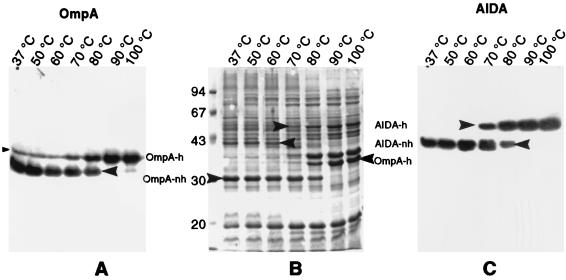
To show the existence of heat modification for the predicted β-barrel of the AIDA-I autotransporter, a pJM7-derived plasmid (pJM1013) in which ctxB was almost entirely deleted was constructed. The portion coding for the signal peptide was left, while the region coding for the mature CTB was replaced by an oligonucleotide linker coding for a reporter epitope. Hence, the resulting fusion protein was expressed and targeted in the same manner as FP59 but the CTB was replaced by the reporter epitope PEYFK. Outer membranes of E. coli UT5600(pJM1013) were prepared and resuspended in SDS sample buffer. Aliquots heated to different temperatures were separated by SDS-PAGE, and proteins were transferred to an Immobilon P membrane. The membranes were probed either with anti-OmpA serum AK57 (as a control) or anti-PEYFK monoclonal antibody Dü142. The different electrophoretic mobilities of OmpA and PEYFK–AIDA-I were also visualized by staining with Coomassie briliant blue R250 (Fig. 3). OmpA and PEYFK–AIDA-I both showed shifts in electrophoretic mobility depending on the sample temperature preceding electrophoresis. Also, the temperatures at which the conformational changes took place were similar for both proteins. Below 70°C, no slowly migrating form of PEYFK–AIDA-I and very little slowly migrating OmpA was observed, while above 80°C, no fast-migrating forms could be detected. These observations match observations of other groups for OmpA (18, 47), clearly supporting the existence of a β-barrel in the AIDA-I TU.
FIG. 3.
Heat modification of AIDA-I. An outer membrane preparation of E. coli UT5600(pJM1013) was prepared by using the Sarkosyl method as described in Materials and Methods. Aliquots were incubated for 10 min at the indicated temperature. (A and C) Western blots probed with anti-OmpA serum AK57 (A) or anti-CTB-serum AK55 (C). (B) SDS-PAGE gel stained with Coomassie brilliant blue R250. Arrowheads pointing to the right indicate heat-modified (h) forms of the respective proteins, while arrowheads pointing to the left indicate not-heat-modified (nh) forms of the proteins.
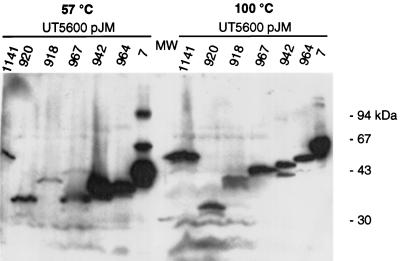
To assess the extent of protein sequence which is contained within the assumed β-barrel, we subjected the seven CTB–AIDA-I fusions to a similar experiment. After preincubation at 57 or 100°C, cell envelope proteins from a crude preparation were separated by SDS-PAGE, and a Western blot analysis employing anti-CTB serum AK55 was performed (Fig. 4). All seven CTB–AIDA-I fusion proteins were found exclusively in the crude envelope preparations (data not shown), thus indicating their membrane insertion or, at least, membrane association. Three fusion proteins, FP52 (pJM1141) with a C-terminal deletion and FP40 (pJM918) and FP34 (pJM920), the latter two proteins N-terminally deleted, did not undergo heat modification, while the remaining four fusion proteins, FP59 (pJM7), FP51 (pJM964), FP49 (pJM942), and FP46 (pJM967), did. Interestingly, FP46, which was found not to support export of the CTB moiety, clearly showed an altered electrophoretic mobility, thus emphasizing the essential role of the linker for translocation but not for the assumed β-barrel conformation. The N terminus of the hypothetical β-barrel can therefore be located to the region between the fusion points of FP46 (A975) and FP40 (L1018).
FIG. 4.
Heat modification of CTB–AIDA-I fusion proteins. Cell envelopes from recombinant E. coli UT5600 were prepared. After incubation at the indicated temperatures, aliquots corresponding to 3.0 ml of culture with an OD578 of 1.0 for E. coli UT5600 containing either plasmid pJM1141, pJM918, or pJM920, 3.5 ml for pJM967, 0.7 ml for pJM942 and pJM964, and, finally, 0.2 ml for pJM7 were Western blotted with anti-CTB serum AK55.
Conclusions.
This report supports the notion that the AIDA-I autotransporter protein is a member of the family of β-barrel-forming outer membrane proteins as predicted previously (23). Heat modifiability of AIDA-I was shown to begin between amino acids A975 and L1018 and to extend beyond amino acid I1220, supporting the structural prediction according to which the β-barrel spans the amino acids A1002 to F1286. Considering our data and the recent reports on sequence homologies in predicted β-sheets of autotransporters (21, 31), it is likely that all autotransporters possess membrane-embedded β-barrels like monomeric OmpA (38) and the trimeric porins (13, 51, 52).
Export by the autotransporter requires a TU comprising a hypothetical C-terminal β-barrel and a preceding linking region (39). The assumed C-terminal β-barrel, according to the commonly accepted model, makes up an aqueous pore in the outer membrane through which the unfolded N-terminal passenger domain is thought to be exported via a hairpin-like structure guided by the linking region. After export, the C-terminal portion of the linking region is assumed to remain within the β-barrel pore, possibly thereby inhibiting leakage through the pore, while the N-terminal portion of the linker anchors the passenger domain to the cell surface.
Here we could confirm the existence and requirement of such a linking region for the function of autotransporters. For AIDA-I, the minimal N-terminal extension of the linker necessary for export could be restricted to an area between amino acids A975 in FP46 and L955 in FP49. Since the predicted β-barrel of AIDA-I starts at A1002, the minimal essential linking region spans 28 to 48 amino acids, although the contribution of the 16 amino acids introduced by cloning to the TU cannot be excluded from our data. Further studies may elucidate the role of the composition of the linking region necessary for export.
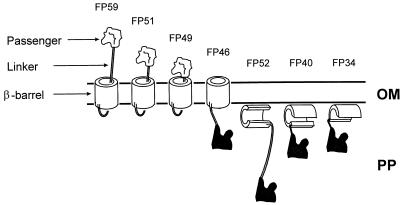
Strength of expression as well as surface exposure of the passenger domain were found to depend both on the length of the linking region and the integrity of the assumed β-barrel. One explanation is that incomplete fusion proteins with impaired targeting are likely to become rapidly degraded by envelope proteases. Alternatively, it is possible that only stable protein forms can be targeted to the outer membrane. Our findings are illustrated in Fig. 5 and summarized in Table 1 and provide strong evidence supporting the accuracy of our model for the β-barrel of AIDA-I autotransporter.
FIG. 5.
Schematic model and features of CTB–AIDA-I fusion proteins. The assumed positions, export, and folding behavior of the β-barrel of the various CTB–AIDA-I fusion proteins are visualized. OM, outer membrane; PP, periplasm.
TABLE 1.
Features of CTB–AIDA-I fusion proteins
| Fusion protein | Plasmid | Deletiona | Trypsin sensitivity | Heat modifiability | Expression |
|---|---|---|---|---|---|
| FP59 | pJM7 | None (full size; 447 aa) | + | + | ++++ |
| FP51 | pJM964 | ΔL (359 aa) | + | + | +++ |
| FP49 | pJM942 | ΔL (332 aa) | + | + | +++ |
| FP46 | pJM967 | ΔL (312 aa) | − | + | + |
| FP52 | pJM1141 | ΔTM11-14 (66 aa)b | − | − | + |
| FP40 | pJM918 | ΔTM1 (269 aa) | − | − | ++ |
| FP34 | pJM920 | ΔTM1-3 (213 aa) | − | − | ++ |
The listed amino acids correspond to the fusion points in the respective CTB–AIDA-I fusion proteins beginning with the C terminus. ΔL, deletion within the linking region; ΔTM, deletion of transmembrane regions.
In FP52, the N-terminal fusion point is identical to the fusion point in FP59 but the C-terminal 66 amino acids are deleted.
ACKNOWLEDGMENTS
We thank S. G. Gray-Owen and C. P. Gibbs for comments on the manuscript and helpful discussions.
REFERENCES
- 1.Anderson D M, Schneewind O. A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science. 1997;278:1140–1143. doi: 10.1126/science.278.5340.1140. [DOI] [PubMed] [Google Scholar]
- 2.Baneyx F, Ayling A, Palumbo T, Thomas D, Georgiou G. Optimization of growth conditions for the production of proteolytically-sensitive proteins in the periplasmic space of Escherichia coli. Appl Microbiol Biotechnol. 1991;36:14–20. doi: 10.1007/BF00164691. [DOI] [PubMed] [Google Scholar]
- 3.Baneyx F, Georgiou G. In vivo degradation of secreted fusion proteins by the Escherichia coli outer membrane protease OmpT. J Bacteriol. 1990;172:491–494. doi: 10.1128/jb.172.1.491-494.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baneyx F, Georgiou G. Construction and characterization of Escherichia coli strains deficient in multiple secreted proteases: protease III degrades high-molecular-weight substrates in vivo. J Bacteriol. 1991;173:2696–2703. doi: 10.1128/jb.173.8.2696-2703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baneyx F, Georgiou G. Degradation of secreted proteins in Escherichia coli. Ann NY Acad Sci. 1992;665:301–308. doi: 10.1111/j.1749-6632.1992.tb42593.x. [DOI] [PubMed] [Google Scholar]
- 6.Benjelloun-Touimi Z, Sansonetti P J, Parsot C. SepA, the major extracellular protein of Shigella flexneri: autonomous secretion and involvement in tissue invasion. Mol Microbiol. 1995;17:123–135. doi: 10.1111/j.1365-2958.1995.mmi_17010123.x. [DOI] [PubMed] [Google Scholar]
- 7.Benz I, Schmidt M A. Cloning and expression of an adhesin (AIDA-I) involved in diffuse adherence of enteropathogenic Escherichia coli. Infect Immun. 1989;57:1506–1511. doi: 10.1128/iai.57.5.1506-1511.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benz I, Schmidt M A. AIDA-I, the adhesin involved in diffuse adherence of the diarrhoeagenic Escherichia coli strain 2787 (O126:H27) is synthesized via a precursor molecule. Mol Microbiol. 1992;6:1539–1546. doi: 10.1111/j.1365-2958.1992.tb00875.x. [DOI] [PubMed] [Google Scholar]
- 9.Benz I, Schmidt M A. Isolation and serologic characterization of AIDA-I, the adhesin mediating the diffuse adherence phenotype of the diarrhea-associated Escherichia coli strain 2787 (O126:H27) Infect Immun. 1992;60:13–18. doi: 10.1128/iai.60.1.13-18.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Binet R, Letoffe S, Ghigo J M, Delepelaire P, Wandersman C. Protein secretion by gram-negative bacterial ABC exporters—a review. Gene. 1997;192:7–11. doi: 10.1016/s0378-1119(96)00829-3. [DOI] [PubMed] [Google Scholar]
- 11.Charles I, Fairweather N, Pickard D, Beesley J, Anderson R, Dougan G, Roberts M. Expression of the Bordetella pertussis P.69 pertactin adhesin in Escherichia coli: fate of the carboxy-terminal domain. Microbiology. 1994;140:3301–3308. doi: 10.1099/13500872-140-12-3301. [DOI] [PubMed] [Google Scholar]
- 12.Cornelis G R, Wolf-Watz H. The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Mol Microbiol. 1997;23:861–867. doi: 10.1046/j.1365-2958.1997.2731623.x. [DOI] [PubMed] [Google Scholar]
- 13.Cowan S W, Schirmer T, Rummel G, Steiert M, Ghosh R, Pauptit R A, Jansonius J N, Rosenbusch J P. Crystal structures explain functional properties of two E. coli porins. Nature. 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 14.de Cock H, van Blokland S, Tommassen J. In vitro insertion and assembly of outer membrane protein PhoE of Escherichia coli K-12 into the outer membrane. Role of Triton X-100. J Biol Chem. 1996;271:12885–12890. doi: 10.1074/jbc.271.22.12885. [DOI] [PubMed] [Google Scholar]
- 15.Elish M E, Pierce J R, Earhart C F. Biochemical analysis of spontaneous fepA mutants of Escherichia coli. J Gen Microbiol. 1988;134:1355–1364. doi: 10.1099/00221287-134-5-1355. [DOI] [PubMed] [Google Scholar]
- 16.Finlay B B, Falkow S. Common themes in microbial pathogenicity revisited. Microbiol Mol Biol Rev. 1997;61:136–169. doi: 10.1128/mmbr.61.2.136-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galan J E. Molecular genetic bases of Salmonella entry into host cells. Mol Microbiol. 1996;20:263–271. doi: 10.1111/j.1365-2958.1996.tb02615.x. [DOI] [PubMed] [Google Scholar]
- 18.Garten W, Hindennach I, Henning U. The major proteins of the Escherichia coli outer cell envelope membrane. Characterization of proteins II* and III, comparison of all proteins. Eur J Biochem. 1975;59:215–221. doi: 10.1111/j.1432-1033.1975.tb02444.x. [DOI] [PubMed] [Google Scholar]
- 19.Goldberg M B, Bârzu O, Parsot C, Sansonetti P J. Unipolar localization and ATPase activity of IcsA, a Shigella flexneri protein involved in intracellular movement. J Bacteriol. 1993;175:2189–2196. doi: 10.1128/jb.175.8.2189-2196.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halter R, Pohlner J, Meyer T F. IgA protease of Neisseria gonorrhoeae: isolation and characterization of the gene and its extracellular product. EMBO J. 1984;3:1595–1601. doi: 10.1002/j.1460-2075.1984.tb02016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henderson I R, Navarrogarcia F, Nataro J P. The great escape—structure and function of the autotransporter proteins. Trends Microbiol. 1998;6:370–378. doi: 10.1016/s0966-842x(98)01318-3. [DOI] [PubMed] [Google Scholar]
- 22.Hendrixson D R, de la Morena M L, Stathopoulos C, St. Geme J W. Structural determinants of processing and secretion of the Haemophilus influenzae hap protein. Mol Microbiol. 1997;26:505–518. doi: 10.1046/j.1365-2958.1997.5921965.x. [DOI] [PubMed] [Google Scholar]
- 23.Jose J, Jahnig F, Meyer T F. Common structural features of IgA1 protease-like outer membrane protein autotransporters. Mol Microbiol. 1995;18:378–380. doi: 10.1111/j.1365-2958.1995.mmi_18020378.x. [DOI] [PubMed] [Google Scholar]
- 24.Jose J, Kramer J, Klauser T, Pohlner J, Meyer T F. Absence of periplasmic DsbA oxidoreductase facilitates export of cysteine-containing passenger proteins to the Escherichia coli cell surface via the Iga beta autotransporter pathway. Gene. 1996;178:107–110. doi: 10.1016/0378-1119(96)00343-5. [DOI] [PubMed] [Google Scholar]
- 25.Klauser T, Kramer J, Otzelberger K, Pohlner J, Meyer T F. Characterization of the Neisseria Iga beta-core. The essential unit for outer membrane targeting and extracellular protein secretion. J Mol Biol. 1993;234:579–593. doi: 10.1006/jmbi.1993.1613. [DOI] [PubMed] [Google Scholar]
- 26.Klauser T, Pohlner J, Meyer T F. Extracellular transport of cholera toxin B subunit using Neisseria IgA protease beta-domain: conformation-dependent outer membrane translocation. EMBO J. 1990;9:1991–1999. doi: 10.1002/j.1460-2075.1990.tb08327.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klauser T, Pohlner J, Meyer T F. Selective extracellular release of cholera toxin B subunit by Escherichia coli: dissection of Neisseria Iga beta-mediated outer membrane transport. EMBO J. 1992;11:2327–2335. doi: 10.1002/j.1460-2075.1992.tb05292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klauser T, Pohlner J, Meyer T F. The secretion pathway of IgA protease-type proteins in gram-negative bacteria. Bioessays. 1993;15:799–805. doi: 10.1002/bies.950151205. [DOI] [PubMed] [Google Scholar]
- 29.Kubori T, Matsushima Y, Nakamura D, Uralil J, Laratejero M, Sukhan A, Galan J E, Aizawa S. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 30.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 31.Loveless B J, Saier M H., Jr A novel family of channel-forming, autotransporting, bacterial virulence factors. Mol Membr Biol. 1997;14:113–123. doi: 10.3109/09687689709048171. [DOI] [PubMed] [Google Scholar]
- 32.Ludwig D S, Holmes R K, Schoolnik G K. Chemical and immunochemical studies on the receptor binding domain of cholera toxin B subunit. J Biol Chem. 1985;260:12528–12534. [PubMed] [Google Scholar]
- 33.Maurer J, Jose J, Meyer T F. Autodisplay: one-component system for efficient surface display and release of soluble recombinant proteins from Escherichia coli. J Bacteriol. 1997;179:794–804. doi: 10.1128/jb.179.3.794-804.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyer T F, Halter R, Pohlner J. Mechanism of extracellular secretion of an IgA protease by gram-negative host cells. Adv Exp Med Biol. 1987;216B:1271–1281. [PubMed] [Google Scholar]
- 35.Miyazaki H, Yanagida N, Horinouchi S, Beppu T. Characterization of the precursor of Serratia marcescens serine protease and COOH-terminal processing of the precursor during its excretion through the outer membrane of Escherichia coli. J Bacteriol. 1989;171:6566–6572. doi: 10.1128/jb.171.12.6566-6572.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murphy C, Prinz W, Pohlschroder M, Derman A, Beckwith J. Essential features of the pathway for protein translocation across the Escherichia coli cytoplasmic membrane. Cold Spring Harbor Symp Quant Biol. 1995;60:277–283. doi: 10.1101/sqb.1995.060.01.032. [DOI] [PubMed] [Google Scholar]
- 37.Ohnishi Y, Nishiyama M, Horinouchi S, Beppu T. Involvement of the COOH-terminal pro-sequence of Serratia marcescens serine protease in the folding of the mature enzyme. J Biol Chem. 1994;269:32800–32806. [PubMed] [Google Scholar]
- 38.Pautsch A, Schulz G E. Structure of the outer membrane protein A transmembrane domain. Nat Struct Biol. 1998;5:1013–1017. doi: 10.1038/2983. [DOI] [PubMed] [Google Scholar]
- 39.Pohlner J, Halter R, Beyreuther K, Meyer T F. Gene structure and extracellular secretion of Neisseria gonorrhoeae IgA protease. Nature. 1987;325:458–462. doi: 10.1038/325458a0. [DOI] [PubMed] [Google Scholar]
- 40.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pugsley A P, Francetic O, Possot O M, Sauvonnet N, Hardie K R. Recent progress and future directions in studies of the main terminal branch of the general secretory pathway in gram-negative bacteria—a review. Gene. 1997;192:13–19. doi: 10.1016/s0378-1119(96)00803-7. [DOI] [PubMed] [Google Scholar]
- 42.Rosqvist R, Hakansson S, Forsberg A, Wolf-Watz H. Functional conservation of the secretion and translocation machinery for virulence proteins of Yersiniae, Salmonellae and Shigellae. EMBO J. 1995;14:4187–4195. doi: 10.1002/j.1460-2075.1995.tb00092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salmond G P, Reeves P J. Membrane traffic wardens and protein secretion in gram-negative bacteria. Trends Biochem Sci. 1993;18:7–12. doi: 10.1016/0968-0004(93)90080-7. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 45.Schmitt W, Haas R. Genetic analysis of the Helicobacter pylori vacuolating cytotoxin: structural similarities with the IgA protease type of exported protein. Mol Microbiol. 1994;12:307–319. doi: 10.1111/j.1365-2958.1994.tb01019.x. [DOI] [PubMed] [Google Scholar]
- 46.Shikata S, Shimada K, Ohnishi Y, Horinouchi S, Beppu T. Characterization of secretory intermediates of Serratia marcescens serine-protease produced during its extracellular secretion from Escherichia-coli-cells. J Biochem. 1993;114:723–731. doi: 10.1093/oxfordjournals.jbchem.a124244. [DOI] [PubMed] [Google Scholar]
- 47.Sugawara E, Steiert M, Rouhani S, Nikaido H. Secondary structure of the outer membrane proteins OmpA of Escherichia coli and OprF of Pseudomonas aeruginosa. J Bacteriol. 1996;178:6067–6069. doi: 10.1128/jb.178.20.6067-6069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suhr M, Benz I, Schmidt M A. Processing of the AIDA-I precursor: removal of AIDAc and evidence for the outer membrane anchoring as a beta-barrel structure. Mol Microbiol. 1996;22:31–42. doi: 10.1111/j.1365-2958.1996.tb02653.x. [DOI] [PubMed] [Google Scholar]
- 49.Suzuki T, Lett M C, Sasakawa C. Extracellular transport of VirG protein in Shigella. J Biol Chem. 1995;270:30874–30880. doi: 10.1074/jbc.270.52.30874. [DOI] [PubMed] [Google Scholar]
- 50.Van Gijsegem F, Genin S, Boucher C. Conservation of secretion pathways for pathogenicity determinants of plant and animal bacteria. Trends Microbiol. 1993;1:175–180. doi: 10.1016/0966-842x(93)90087-8. [DOI] [PubMed] [Google Scholar]
- 51.Weiss M S, Abele U, Weckesser J, Welte W, Schiltz E, Schulz G E. Molecular architecture and electrostatic properties of a bacterial porin. Science. 1991;254:1627–1630. doi: 10.1126/science.1721242. [DOI] [PubMed] [Google Scholar]
- 52.Weiss M S, Kreusch A, Schiltz E, Nestel U, Welte W, Weckesser J, Schulz G E. The structure of porin from Rhodobacter capsulatus at 1.8 Å resolution. FEBS Lett. 1991;280:379–382. doi: 10.1016/0014-5793(91)80336-2. [DOI] [PubMed] [Google Scholar]
- 53.Welch R A. Pore-forming cytolysins of gram-negative bacteria. Mol Microbiol. 1991;5:521–528. doi: 10.1111/j.1365-2958.1991.tb00723.x. [DOI] [PubMed] [Google Scholar]
- 54.Yanagida N, Uozumi T, Beppu T. Specific excretion of Serratia marcescens protease through the outer membrane of Escherichia coli. J Bacteriol. 1986;166:937–944. doi: 10.1128/jb.166.3.937-944.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]