Abstract
Bacterial protein translocation is mediated by translocase, a multisubunit membrane protein complex that consists of a peripheral ATPase SecA and a preprotein-conducting channel with SecY, SecE, and SecG as subunits. Like Escherichia coli SecG, the Bacillus subtilis homologue, YvaL, dramatically stimulated the ATP-dependent translocation of precursor PhoB (prePhoB) by the B. subtilis SecA-SecYE complex. To systematically determine the functional exchangeability of translocase subunits, all of the relevant combinations of the E. coli and B. subtilis secY, secE, and secG genes were expressed in E. coli. Hybrid SecYEG complexes were overexpressed at high levels. Since SecY could not be overproduced without SecE, these data indicate a stable interaction between the heterologous SecY and SecE subunits. E. coli SecA, but not B. subtilis SecA, supported efficient ATP-dependent translocation of the E. coli precursor OmpA (proOmpA) into inner membrane vesicles containing the hybrid SecYEG complexes, if E. coli SecY and either E. coli SecE or E. coli SecG were present. Translocation of B. subtilis prePhoB, on the other hand, showed a strict dependence on the translocase subunit composition and occurred efficiently only with the homologous translocase. In contrast to E. coli SecA, B. subtilis SecA binds the SecYEG complexes only with low affinity. These results suggest that each translocase subunit contributes in an exclusive manner to the specificity and functionality of the complex.
In the gram-negative bacterium Escherichia coli, secretory proteins are transported from the cytosol to the periplasm by the translocase (13, 16, 53). The translocase consists of the membrane-peripheral ATPase SecA, which is bound with high affinity to a heterotrimeric integral membrane protein complex composed of the SecY, SecE, and SecG subunits. The SecYEG complex is thought to function as a preprotein-conducting channel in the inner membrane (36). In E. coli, some preproteins associate first with the export-dedicated chaperone SecB, which stabilizes the preprotein in the cytosol and targets it to the membrane-bound SecA (18). SecA drives the stepwise translocation of the preprotein across the membrane by nucleotide-modulated cycles of SecA membrane insertion and deinsertion (17, 44, 49). SecA, SecY, and SecE are the essential components of the translocase and are needed for the viability of E. coli. SecG is dispensable in vivo but stimulates translocation in vitro (39, 40). SecG can be isolated as part of a stable complex together with SecY and SecE from the E. coli inner membrane (7, 8, 21). In addition, translocation involves the products of the secD and secF genes, both of which code for integral membrane proteins with large periplasmic domains (20). SecD and SecF are also not essential for the viability of E. coli, but they can associate with the SecYEG complex and functionally substitute for the SecG protein (14).
The translocase complex of gram-positive bacteria like Bacillus subtilis is homologous to the system found in E. coli. In recent years, the components of the B. subtilis translocase, i.e., SecA (divA) (41, 42), SecY (45), SecE (25), and SecG (52), have been identified genetically. The B. subtilis SecD and SecF subunits (6) form a single polypeptide in the membrane. SecB appears to be absent in gram-positive bacteria.
Except for the results of some in vivo studies using conditional lethal sec mutants, few data on the functional interaction between translocase subunits in a heterologous complex are available. SecA of the gram-positive bacterium Staphylococcus carnosus complements the temperature-sensitive B. subtilis divA (secA) mutant, but it cannot functionally replace E. coli SecA (28). E. coli SecA fails to complement the B. subtilis divA mutant (46). Under conditions of low expression, B. subtilis SecA can complement SecA mutants in E. coli K-12 strains (29) but not in an E. coli B strain (34). This finding shows that the complementation by B. subtilis SecA can be very critical. Chimeras of the E. coli and B. subtilis SecA proteins have been reported, and one of these is able to effectively complement the E. coli secA mutants. This chimera consists of the first 242 amino acids of B. subtilis SecA, including the ATP-binding site and the carboxy-terminal part of E. coli SecA (34). B. subtilis SecY is unable to restore the growth defect of E. coli secY24 at the nonpermissive temperature, but it does support translocation of the precursor OmpA (proOmpA) (38). Likewise, B. subtilis SecE was shown to complement a cold-sensitive E. coli secE strain (25). The B. subtilis SecG and SecDF proteins are unable to complement the cold-sensitive growth phenotypes of the corresponding E. coli mutant proteins (6, 52). In consideration of these data, it appears that one or more Sec proteins function in a host-specific manner.
The complementation experiments described herein mainly score for restoration of growth and do not address the catalytic activities of the heterologous complexes formed. To study the host specificities of translocase subunits in a more systematic manner, we have expressed all relevant combinations of the three major integral membrane subunits, SecY, SecE, and SecG, of the E. coli and B. subtilis translocases in E. coli. Membranes harboring these hybrid translocase complexes were analyzed for their preprotein translocation activities in the presence of E. coli or B. subtilis SecA. The results suggest that each subunit of the translocase is directly involved in defining the host specificity of the complex.
MATERIALS AND METHODS
Materials.
E. coli SecA (10, 12), B. subtilis SecA (48), proOmpA (11), and His-precursor PhoB (prePhoB) (51) were purified as described previously. E. coli SecA, B. subtilis SecA, proOmpA, and His-prePhoB were labeled with carrier-free 125I (Amersham), Little Chalfont, Buckinghamshire, United Kingdom) with Iodo-Beads (Pierce Rockford) (51).
Strains and construction of plasmids.
Strains and plasmids used are shown in Table 1. Overproduction of the SecY, SecE, and SecG proteins was performed with E. coli SF100 as described previously (47). The synthetic operon containing B. subtilis and E. coli SecY, SecE, and SecG under the control of an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible trc promoter was constructed basically as described by van der Does et al. (47). Individual genes were amplified from the chromosomes of E. coli DH5α or B. subtilis DB104 (54) by PCR with primers containing the appropriate restriction sites and ribosome-binding sites. Nucleotide sequences of the cloned genes were checked on a Vistra DNA sequencer 725 (Amersham) with an automated sequencing kit from Amersham.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Relevant propertiesa | Reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | recA Δlac | 22 |
| E. coli SF100 | recA Δlac ΔompT | 3 |
| B. subtilis DB104 | NprE18 aprEΔ3 | 54 |
| Plasmids | ||
| pET324 | Ampr expression vector | 47 |
| pET340 | Ec-secY Ec-secE Ec-secG | 47 |
| pET811 | Ec-secY Bs-secE Ec-secG | This work |
| pET812 | Bs-secY Bs-secE Ec-secG | This work |
| pET813 | Bs-secY Ec-secE Ec-secG | This work |
| pET819 | Bs-secY Bs-secE | This work |
| pET821 | Ec-secY Bs-secE Bs-secG | This work |
| pET822 | Bs-secY Bs-secE Bs-secG | This work |
| pET823 | Bs-secY Ec-secE Bs-secG | This work |
| pET825 | Ec-secY Ec-secE Bs-secG | This work |
| pET865 | Bs-secY | This work |
Ec, E. coli, Bs, B. subtilis.
Isolation of IMVs.
Cells overexpressing the various combinations of SecY, SecE, and SecG were harvested by centrifugation, washed, and resuspended in 50 mM Tris-HCl, pH 8.0. The cell suspension was passed three times through a French press at 16,000 lb/in2 to obtain inside-out inner membrane vesicles (IMVs), and the cell debris was removed by low-spin centrifugation (10,000 × g for 5 min). Membranes were collected by high-spin centrifugation (200,000 × g, 1 h) and resuspended in buffer containing 1 mM dithiothreitol (DTT), and subsequently the inner membranes were separated from the outer membranes by sucrose density gradient centrifugation (47). IMVs were frozen in liquid N2 and stored at −80°C. Protein content was determined by the DC protein assay (Bio-Rad).
For translocation and SecA-binding reactions, IMVs were treated with a polyclonal antibody (PAb) raised against E. coli SecA. One hundred microliters of IMVs (10 mg/ml) was incubated and mixed continuously for 1 h with 20 μl of PAb (47) and subsequently spun through a sucrose cushion (25% sucrose, 50 mM Tris [pH 8.0], 1 mM DTT) (44).
In vitro translocation assay.
In vitro translocation of 125I-proOmpA or 125I-prePhoB into E. coli IMVs was assayed by the accessibility of the precursors to added proteinase K (12, 47). Reactions were performed in 50 μl of a solution containing 50 mM HEPES–KOH (pH 7.6), 30 mM KCl, 0.5 mg of bovine serum albumin per ml, 10 mM DTT, 2 mM Mg–acetate, 2 mM ATP, 10 mM phosphocreatine–phosphate, 50 μg of creatine kinase per ml, IMVs (10 μg of membrane protein), and, where indicated in the figures, purified E. coli or B. subtilis SecA (2 μg, unless indicated otherwise). Translocation was initiated by the addition of 1 μl of 125I-labeled proOmpA or 125I-labeled prePhoB (in 6 M urea–50 mM Tris, pH 7.2), which corresponds to about 0.2 μg of protein. After 30 min of incubation at 37°C, the mixture was chilled on ice and treated with proteinase K (1 mg/ml) for 15 min. Samples were precipitated with 7.5% trichloroacetic acid, acetone washed, and resuspended in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) sample buffer and separated on SDS–10% (prePhoB) or –12% (proOmpA) polyacrylamide gel (31), followed by autoradiography by exposure to Kodak Biomax MR film.
SecA binding.
Binding of SecA to IMVs was assayed essentially as described previously (24). IMVs (10 μg of membrane protein) were suspended in 100 μl of translocation buffer (50 mM HEPES–KOH [pH 7.6], 30 mM KCl, 0.5 mg of bovine serum albumin per ml, 10 mM DTT, 2 mM Mg-acetate) and incubated for 15 min on ice with 1 nM 125I-labeled E. coli or B. subtilis SecA and the concentration of nonlabeled SecA indicated in the figures. Samples were subsequently loaded on a sucrose cushion (0.25 mM sucrose in translocation buffer) and fractionated by centrifugation (10 min, 30 lb/in2 in a Beckman Airfuge, 4°C). The amounts of 125I-labeled SecA in the supernatant and the pellet were quantitated with a gamma counter.
Miscellaneous methods.
To measure precursor-stimulated SecA ATPase activity, IMVs were treated with 4 M urea as described before (19, 24). IMV bearing overexpressed SecY, SecE, and SecG proteins were analyzed by SDS–15% PAGE (31), stained with Coomassie brilliant blue stain, or blotted on polyvinylide difluoride membranes (Millipore) with a semidry blotter (Bio-Rad). Immunodetection was carried out with PAb raised against SecY, SecE, or SecG of E. coli (47) or SecE (a PAb raised against a synthetic peptide of B. subtilis SecE [KDVGKEMKKV] by Research Genetics) or SecG of B. subtilis (51, 52). The PAb raised against B. subtilis SecY was a generous gift of R. Freudl (Forschungs Institute, Jülich, Germany).
RESULTS
B. subtilis SecG stimulates in vitro prePhoB translocation by SecYE.
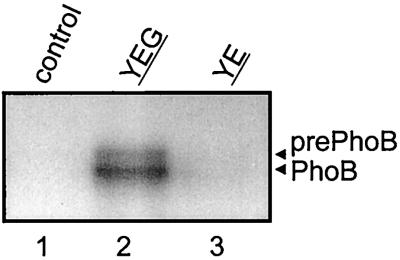
Recently, we have reported the in vivo identification of the product of the yvaL gene as the B. subtilis homologue of SecG (52). For E. coli, SecG has been found to dramatically stimulate the in vitro SecYE-mediated translocation of various precursor proteins (7, 8, 21). To further substantiate that the B. subtilis YvaL protein is a SecG homologue, its ability to stimulate preprotein translocation into IMVs bearing the B. subtilis SecYE complexes was determined. To circumvent the instability of SecY in B. subtilis due to proteolytic degradation (51), the B. subtilis SecY and SecE proteins were overproduced in E. coli SF100, the host, and coexpressed with (pET822) or without (pET819) YvaL. SDS-PAGE and Coomassie brilliant blue staining of the IMVs isolated from SF100 cells transformed with pET819 showed high-level expression of the B. subtilis SecY and SecE proteins (Fig. 1A, lane 9) compared to the levels of expression in IMVs of the host strain transformed with the empty vector (pET324) (lane 1). Identical results were obtained with IMVs derived from cells harboring pET822 that expressed only SecY and SecE (data not shown). Immunoblotting was employed to demonstrate the expression of the B. subtilis YvaL protein (Fig. 1C, where YvaL is indicated as SecG). Next, the IMVs were analyzed for the translocation of the urea-denatured 125I-labeled precursor of the B. subtilis alkaline phosphatase (prePhoB) (51). IMVs were treated with a PAb against SecA to inactivate endogenous membrane-bound E. coli SecA. Even when supplemented with B. subtilis SecA and ATP, IMVs derived from the host strain E. coli SF100 transformed with the empty vector were inactive for 125I-prePhoB translocation (Fig. 2, lane 1). The presence of YvaL (lane 2) dramatically enhanced the 125I-prePhoB translocation activity of B. subtilis SecYE (lane 3). No translocation was observed in the absence of B. subtilis SecA (see Fig. 4A, lane 9). These results further demonstrate that B. subtilis YvaL is functionally homologous to E. coli SecG.
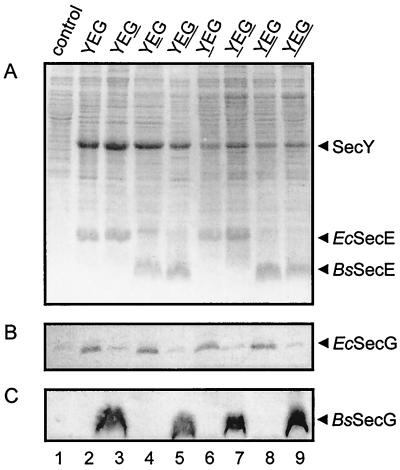
FIG. 1.
Overproduction of E. coli and B. subtilis SecY, SecE, and SecG proteins in E. coli SF100 cells. (A) Coomassie brilliant blue-stained SDS–15% polyacrylamide gel of membranes derived from SF100 cells bearing a control plasmid (wild type) and the hybrid SecYEG complex; the loci of B. subtilis Sec proteins are underlined. (B and C) Immunoblots of E. coli SF100 membranes developed with PAbs raised against synthetic polypeptides corresponding to SecG of E. coli (B) and B. subtilis (C).
FIG. 2.
In vitro translocation of 125I-prePhoB into SecA-depleted IMVs of E. coli SF100 cells transformed with the control vector, pET324 (lane 1); with pET822, which allows overproduction of B. subtilis SecYEG (lane 2); or with pET819, which directs overproduction of B. subtilis SecYE (lane 3). Translocation activity was measured in the presence of ATP and B. subtilis SecA as described in Materials and Methods.
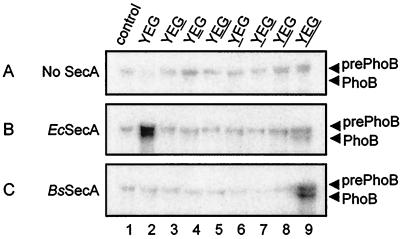
FIG. 4.
In vitro translocation of 125I-labeled prePhoB into SecA-depleted IMVs bearing SecYEG complexes. Experimental conditions were as described in the legend of Fig. 2 and Materials and Methods. The loci of B. subtilis SecA proteins are underlined. Ec, E. coli; Bs, B. subtilis.
Overproduction of hybrid E. coli and B. subtilis SecYEG complexes.
Plasmid pET340 harbors a synthetic operon of the E. coli secY, secE, and secG genes under the control of an IPTG-inducible trc promoter and allows high-level functional overproduction of the SecYEG complex in E. coli (47). To analyze the functional interchangeability of the E. coli and B. subtilis translocase subunits, pET340 was used to construct hybrid SecYEG complexes. The E. coli secY, secE, and secG genes were systematically replaced by their B. subtilis counterparts (Table 1), which yielded a total of eight different SecYEG complexes, including the two homologous systems. SDS-PAGE and immunoblotting of IMVs were used to analyze the IPTG-induced overproduction of the subunits. Coomassie brilliant blue staining of the SDS-polyacrylamide gel revealed that, irrespective of the construct used, a high level of overproduction of the E. coli and B. subtilis SecY and SecE proteins could be achieved (Fig. 1). These protein bands were not visible in the control transformed with the expression vector only (Fig. 1, lane 1). The identities of the proteins were further verified by means of immunoblotting with peptide-specific antibodies (data not shown), permitting also the detection of overexpressed E. coli SecG (Fig. 1B) (40) and B. subtilis SecG (Fig. 1C). Based on the Coomassie brilliant blue staining, the expression level of the B. subtilis SecY protein appeared to be about 25% of that of the E. coli SecY protein. Consistent with its smaller molecular mass, i.e., 6.9 versus 13.6 kDa, B. subtilis SecE was found to migrate much faster by SDS-PAGE than E. coli SecE (Fig. 1A).
E. coli SecY can be stably produced only when it is cooverexpressed with SecE (33). FtsH, a membrane-bound protease (1), degrades the uncomplexed form of SecY. By analogy, B. subtilis SecY could be overproduced in E. coli only when it was coexpressed with SecE (i.e., with pET819) and not in its absence (i.e., with pET865) (data not shown). Since all constructs showed a high level of overproduction of hybrid SecYEG complexes, we conclude that E. coli and B. subtilis SecY and SecE proteins form stable complexes.
Translocation activity of hybrid translocase.
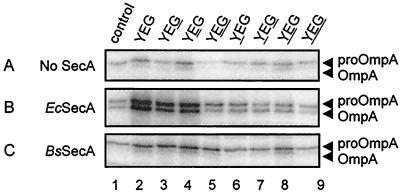
To establish whether the hybrid SecYEG complexes were functional, the SecA- and ATP-dependent translocation of 125I-labeled E. coli proOmpA and B. subtilis prePhoB was analyzed. Since the IMVs bearing overexpressed SecYEG contained substantial amounts of tightly bound endogenous E. coli SecA, membranes were first incubated with PAbs raised against E. coli SecA to reduce the background translocation activity in the absence of added SecA (44). The efficiency of this treatment varied with the hybrid SecYEG complex, but in all cases the endogenous translocation activity could be reduced to a low level (Fig. 3A). When IMVs were supplemented with a saturating concentration of E. coli SecA, a substantial amount of 125I-proOmpA (20 to 25% of total input) was translocated by the IMVs bearing overexpressed E. coli SecYEG (Fig. 3B, lane 2) and by the hybrid complexes that contained E. coli SecY with either the SecG (lane 3) or SecE (lane 4) subunit substituted for its B. subtilis counterpart. With all of the other hybrid SecYEG complexes, only a low translocation activity of proOmpA was observed (lanes 5 to 9). When instead of E. coli SecA, B. subtilis SecA was used, proOmpA translocation was inefficient with each of the hybrids (Fig. 3C). The small amount of translocated proOmpA in the presence of B. subtilis SecA was not processed by signal peptidase.
FIG. 3.
In vitro translocation of 125I-labeled proOmpA into SecA-depleted IMVs bearing SecYEG complexes. E. coli (Ec) or B. subtilis (Bs) SecA was added as indicated. The loci of B. subtilis Sec proteins are underlined. Translocation in the absence of added SecA reflects the activity of the remaining endogenous SecA. No translocation activity was observed in the absence of ATP. Experimental conditions were as described in Materials and Methods.
In contrast to that of proOmpA, translocation of B. subtilis prePhoB depends much more critically on the origin of the translocase components (Fig. 4). E. coli SecA supported efficient translocation of prePhoB (15 to 20% of total input) when it was combined with the homologous SecYEG complex (Fig. 4B, lane 2), while it supported a low level of translocation (about 5%) when it was combined with the B. subtilis SecYEG complex (lane 9). On the other hand, B. subtilis SecA promoted translocation of prePhoB (20 to 25%) only when it was assayed together with the B. subtilis SecYEG complex (Fig. 4C, lane 9). Replacement of only one of the integral subunits of the translocase for its heterologous counterpart resulted in a nearly complete loss of prePhoB translocation activity. These data demonstrate that prePhoB translocation exhibits a very narrow requirement for the translocase subunits.
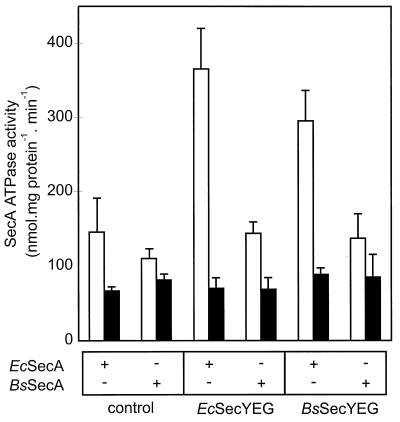
SecA translocation ATPase activity of hybrid translocase.
The ATPase activity of SecYEG-bound SecA stimulated by a translocation-competent preprotein is termed “translocation ATPase” (32) because with the wild-type translocase it correlates with translocation activity. Previous studies have demonstrated that proOmpA is an exceptionally good substrate for SecA ATPase activity in comparison to many other E. coli preproteins (4). IMVs were treated with urea to reduce background ATPase activity, and the proOmpA-stimulated SecA ATPase was measured with wild-type E. coli IMVs or IMVs bearing the overexpressed E. coli or B. subtilis SecYEG complex. E. coli SecA supported a substantial translocation ATPase both with overexpressed E. coli SecYEG and with overexpressed B. subtilis SecYEG (Fig. 5). On the other hand, with B. subtilis SecA, only a low level of proOmpA-stimulated ATPase activity was observed (Fig. 5), consistent with its poor ability to translocate proOmpA. The prePhoB-stimulated ATPase activity of the E. coli or B. subtilis SecA was very low, irrespective the nature of the SecYEG complex (data not shown), even though prePhoB was translocated efficiently by the IMVs containing the homologous systems (Fig. 4).
FIG. 5.
SecA ATPase activity of IMVs derived from control cells and cells that overexpress the E. coli and B. subtilis SecYEG complexes. The ATPase activities of E. coli (Ec) and B. subtilis (Bs) SecA proteins were measured in the presence (open bars) and absence (filled bars) of proOmpA. IMVs were treated with 4 M urea to inactivate endogenous SecA and other ATPases.
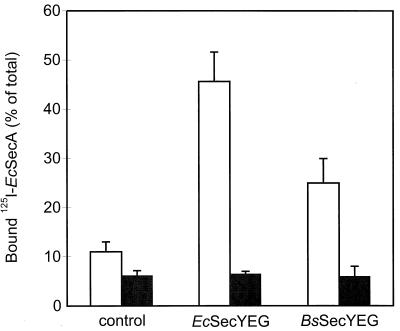
Bacillus subtilis SecA binds SecYEG with low affinity.
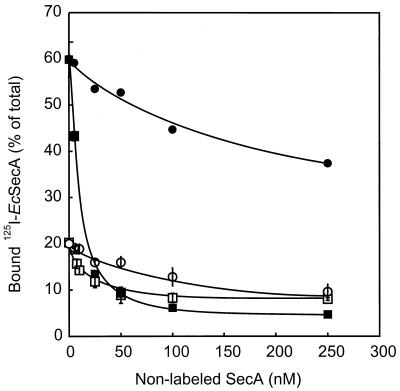
In E. coli, SecYEG functions as a high-affinity membrane-binding site for SecA (24). The ability of the E. coli and B. subtilis SecYEG complexes to bind 125I-labeled SecA (at a 1 nM concentration) was determined. IMVs bearing overexpressed E. coli or B. subtilis SecYEG supported high levels of binding of E. coli 125I-SecA compared to that of the wild-type control (Fig. 6). 125I-SecA binding was effectively reduced to the background level by the addition of a 500-fold excess of nonlabeled SecA. In contrast, it was not possible to discern specific binding of 125I-labeled B. subtilis SecA to any of the SecYEG complexes (data not shown). This result suggests a much lower affinity for the SecYEG complex than that of E. coli SecA, even though the translocation assays demonstrated that the B. subtilis SecA is active (Fig. 4). To investigate this phenomenon further, we determined the ability of B. subtilis SecA to compete with E. coli 125I-SecA for binding to the E. coli and B. subtilis SecYEG complexes. Increasing amounts of nonlabeled E. coli SecA efficiently chased 125I-SecA bound to E. coli SecYEG (Fig. 7). However, B. subtilis SecA appeared far less efficient in this chase. The IMVs retained, even at a 250-fold excess, up to 60% of specifically bound E. coli 125I-SecA. Also the E. coli 125I-SecA that bound to B. subtilis SecYEG was more efficiently chased by nonlabeled E. coli SecA than by B. subtilis SecA. The lower level of specific binding of E. coli 125I-SecA observed with IMVs bearing B. subtilis SecYEG is consistent with the lower level of expression of B. subtilis SecY than that of E. coli SecY (Fig. 1). It is concluded that E. coli SecA binds the SecYEG complex with a much higher affinity than B. subtilis SecA.
FIG. 6.
Binding of E. coli 125I-SecA to IMVs derived from control cells and cells that overproduce the E. coli (Ec) and B. subtilis (Bs) SecYEG complexes in the absence (open bars) or presence (filled bars) of a 500-fold excess of nonlabeled SecA. 125I-SecA was used at a final concentration of 1 nM. Endogenous levels of SecA were removed by treatment of the IMVs with a PAb directed against SecA as described in Materials and Methods.
FIG. 7.
Binding of E. coli 125I-SecA to IMVs bearing the overexpressed E. coli (filled symbols) and B. subtilis SecYEG (open symbols) complexes in the presence of various concentrations of non-labeled E. coli (■ and □) or B. subtilis (● and ○) SecA. 125I-SecA was used at a final concentration of 1 nM. Concentrations of SecA indicated are for the monomer. Ec, E. coli.
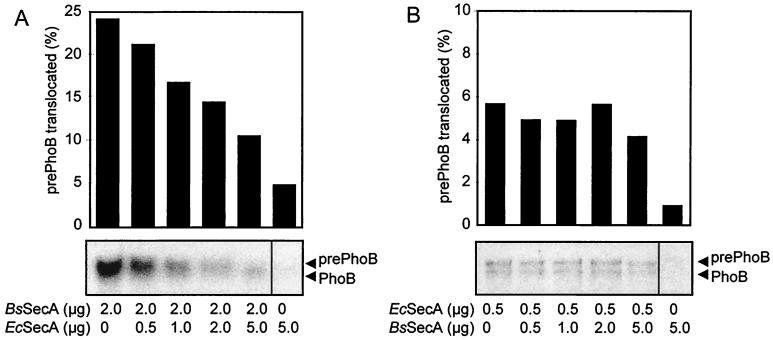
The functional consequence of the low-affinity binding of B. subtilis SecA to the SecYEG complex was further assessed in a competition experiment between E. coli SecA and B. subtilis SecA for the translocation of prePhoB. Since efficient translocation of prePhoB is observed only with a homologous translocase (Fig. 4), addition of a heterologous SecA may prevent translocation by the formation of a nonfunctional complex. To avoid depletion of the preprotein substrate, a large amount of prePhoB was added to the translocation reaction (about 0.7 μM, which equals the highest concentration of the competing SecA dimer used). The B. subtilis SecA-dependent translocation of prePhoB into IMVs bearing B. subtilis SecYEG was progressively inhibited by increasing amounts of E. coli SecA (Fig. 8A). In the reverse experiment, B. subtilis SecA was hardly capable of inhibiting the E. coli SecA-dependent translocation of prePhoB in E. coli SecYEG IMVs (Fig. 8B), even though E. coli SecA was present at a subsaturating amount (0.5 μg) while B. subtilis SecA was added at a 10-fold excess. These results are consistent with the notion that E. coli SecA binds SecYEG with a higher affinity than does B. subtilis SecA.
FIG. 8.
Translocation of prePhoB into IMVs bearing the overexpressed B. subtilis (A) or E. coli (B) SecYEG complex in the presence of various amounts of E. coli (Ec) and B. subtilis (Bs) SecA. PrePhoB was used at 2 μg per translocation reaction mixture.
DISCUSSION
Most of our knowledge on the catalysis of bacterial preprotein translocation is based on studies of E. coli. Here we report on the formation and activities of hybrid translocase complexes composed of subunits originating from E. coli and B. subtilis. Our data demonstrate that the translocase subunits of these bacteria cannot be unconditionally exchanged and provide evidence for host-specific functions. Each translocase subunit seems to contribute in an exclusive manner to the specificity and functionality of the complex.
A previous study of E. coli has shown that SecY can be stably overexpressed only together with SecE (33). The formation of a complex of SecE with SecY prevents the degradation of SecY by FtsH (27). FtsH is a membrane-integrated ATP-dependent metalloprotease that degrades incorrectly folded or assembled cytosolic and inner membrane proteins (1). Likewise, stable overproduction of B. subtilis SecY in E. coli was possible only when the protein was coexpressed with either B. subtilis or E. coli SecE. Therefore, the proper interaction between the SecE and SecY subunits is a prerequisite for the overproduction of hybrid SecYEG complexes that are composed of B. subtilis and E. coli subunits. Both the E. coli and B. subtilis SecE proteins stabilize SecY, irrespective of the origin of SecY. Even though a stable SecY-SecE interaction was apparent for each of the heterologous pairs, none of the hybrid complexes supported prePhoB translocation while the homologous translocase complexes were highly active. These data indicate that SecE not only functions in the stabilization of SecY but also fulfills a catalytic function. Efficient translocation of proOmpA strictly required E. coli SecA, but with the integral subunits a great flexibility was apparent. Apparently, precursor substrates differ in their levels of dependency on the translocase subunits, while each of the subunits may critically contribute to the specificity of the complex.
The subunit swapping experiments with the E. coli and B. subtilis translocases demonstrate that structural and functional aspects of the translocase subunit interactions can be separated. With SecY and SecE being essential subunits of the translocase, the SecY-SecE interaction has been studied in great detail. The SecE proteins of E. coli and B. subtilis are rather distinct. E. coli SecE is a 13.6-kDa integral membrane protein with three transmembrane segments (TMS) (43), whereas B. subtilis SecE is a small membrane protein of 6.9 kDa with only one TMS (25). B. subtilis SecE is homologous to the carboxy-terminal portion of E. coli SecE, which corresponds to the minimal functional size of this protein. The first two TMS of E. coli SecE can be deleted without loss of function (37, 43), whereas the third TMS can be replaced by a related sequence of the B. subtilis SecE (30) or by an unrelated TMS (37). Residues essential for the function of SecE are located in the highly conserved cytoplasmic region (30, 37). This domain is thought to interact with the fourth cytosolic loop of SecY (2). The second periplasmic loop of SecE interacts with the first periplasmic loop of SecY (23), allowing the proximity of TMS 2 of SecY to TMS 3 of SecE (26). It remains to be determined which of these interacting sites is involved in the catalytic function of SecE. In this respect, it is of interest that the reduced specificities observed for prlA mutants of SecY (5) correlate with a loosened interaction between the SecY and SecE subunits (15) and a tighter binding of SecA (50).
The low translocation activity of B. subtilis SecA with proOmpA may relate to poor recognition of this preprotein substrate. B. subtilis SecA can interact with the signal sequence of proOmpA (9), but its ATPase activity is only poorly stimulated by complete proOmpA (this study). proOmpA may thus not be properly recognized by SecYEG-bound B. subtilis SecA, and this in turn may prevent SecA from functionally associating with SecYEG. In vivo experiments indicated that proOmpA can be translocated by B. subtilis (35), but the conditions used differed from those used in the in vitro experiments, as SecDF and the proton motive force may add to the efficiency of translocation. In the in vitro system, no processing of proOmpA was observed. One may speculate that at the initiation of translocation, B. subtilis SecA exposes the signal sequence cleavage site less efficiently to the E. coli signal peptidase than does E. coli SecA.
B. subtilis SecA binds the E. coli SecYEG only with very poor affinity. Although this was noticed before (48), we can now relate this poor binding affinity to the functionality of the complex in an in vitro translocation assay. The observation that E. coli SecA efficiently competes with B. subtilis SecA for binding to the B. subtilis SecYEG complex is remarkable. In the in vitro translocation reaction, this competition resulted in the formation of an inactive complex of E. coli SecA and B. subtilis SecYEG. Although inhibition may also relate to competition for the available preprotein, this possibility seems less likely, as in our experiments the precursor was added in excess relative to the level of SecA. The remarkable difference between the SecYEG binding affinities of E. coli and B. subtilis SecA also makes in vivo complementation studies of temperature-sensitive E. coli SecA mutants more difficult to interpret (28, 29, 34, 41, 46). The presence of residual E. coli SecA, either active or nonactive, may prevent heterologous SecA from interacting efficiently with SecYEG. Future studies should reveal whether the dramatic difference in binding affinities for SecYEG reflects a mechanistic difference in the way E. coli and B. subtilis SecA support translocation.
ACKNOWLEDGMENTS
We thank Chris van der Does, Erik Manting, Andreas Kaufmann, and Martin van der Laan for valuable suggestions.
These investigations were supported by CEC Biotech grants BIO2 CT 930254 and BIO4 CT 960097.
REFERENCES
- 1.Akiyama Y, Kihara A, Tokuda H, Ito K. FtsH (HflB) is an ATP-dependent protease selectively acting on SecY and some other membrane proteins. J Biol Chem. 1996;271:31196–31201. doi: 10.1074/jbc.271.49.31196. [DOI] [PubMed] [Google Scholar]
- 2.Baba T, Taura T, Shimoike T, Akiyama Y, Yoshihisa T, Ito K. A cytoplasmic domain is important for the formation of a SecY-SecE translocator complex. Proc Natl Acad Sci USA. 1994;91:4539–4543. doi: 10.1073/pnas.91.10.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baneyx F, Georgiou G. In vivo degradation of secreted fusion proteins by the Escherichia coli outer membrane protease OmpT. J Bacteriol. 1990;172:491–494. doi: 10.1128/jb.172.1.491-494.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassilana M, Arkowitz R A, Wickner W. The role of the mature domain of proOmpA in the translocation ATPase reaction. J Biol Chem. 1992;267:25246–25250. [PubMed] [Google Scholar]
- 5.Bieker K L, Silhavy T J. PrlA (SecY) and PrlG (SecE) interact directly and function sequentially during protein translocation in E. coli. Cell. 1990;61:833–842. doi: 10.1016/0092-8674(90)90193-i. [DOI] [PubMed] [Google Scholar]
- 6.Bolhuis A, Broekhuizen C P, Sorokin A, van Roosmalen M L, Venema G, Bron S, Quax W J, van Dijl J M. SecDF of Bacillus subtilis, a molecular Siamese twin required for the efficient secretion of proteins. J Biol Chem. 1998;273:21217–21224. doi: 10.1074/jbc.273.33.21217. [DOI] [PubMed] [Google Scholar]
- 7.Brundage L, Fimmel C J, Mizushima S, Wickner W. SecY, SecE, and band 1 form the membrane-embedded domain of Escherichia coli preprotein translocase. J Biol Chem. 1992;267:4166–4170. [PubMed] [Google Scholar]
- 8.Brundage L, Hendrick J P, Schiebel E, Driessen A J M, Wickner W. The purified E. coli integral membrane protein SecY/E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell. 1990;62:649–657. doi: 10.1016/0092-8674(90)90111-q. [DOI] [PubMed] [Google Scholar]
- 9.Bunai K, Yamada K, Hayashi K, Nakamura K, Yamane K. Enhancing effect of Bacillus subtilis Ffh, a homologue of the SRP54 subunit of the mammalian signal recognition particle, on the binding of SecA to precursors of secretory proteins in vitro. J Biochem (Tokyo) 1999;125:151–159. doi: 10.1093/oxfordjournals.jbchem.a022252. [DOI] [PubMed] [Google Scholar]
- 10.Cabelli R J, Chen L, Tai P C, Oliver D B. SecA protein is required for secretory protein translocation into E. coli membrane vesicles. Cell. 1988;55:683–692. doi: 10.1016/0092-8674(88)90227-9. [DOI] [PubMed] [Google Scholar]
- 11.Crooke E, Brundage L, Rice M, Wickner W. ProOmpA spontaneously folds in a membrane assembly competent state which trigger factor stabilizes. EMBO J. 1988;7:1831–1835. doi: 10.1002/j.1460-2075.1988.tb03015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cunningham K, Lill R, Crooke E, Rice M, Moore K, Wickner W, Oliver D. SecA protein, a peripheral protein of the Escherichia coli plasma membrane, is essential for the functional binding and translocation of proOmpA. EMBO J. 1989;8:955–959. doi: 10.1002/j.1460-2075.1989.tb03457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Driessen A J, Fekkes P, van der Wolk J P W. The Sec system. Curr Opin Microbiol. 1998;1:216–222. doi: 10.1016/s1369-5274(98)80014-3. [DOI] [PubMed] [Google Scholar]
- 14.Duong F, Wickner W. Distinct catalytic roles of the SecYE, SecG and SecDFyajC subunits of preprotein translocase holoenzyme. EMBO J. 1997;16:2756–2768. doi: 10.1093/emboj/16.10.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duong F, Wickner W. The PrlA and PrlG phenotypes are caused by a loosened association among the translocase SecYEG subunits. EMBO J. 1999;18:3263–3270. doi: 10.1093/emboj/18.12.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Economou A. Bacterial preprotein translocase: mechanism and conformational dynamics of a processive enzyme. Mol Microbiol. 1998;27:511–518. doi: 10.1046/j.1365-2958.1998.00713.x. [DOI] [PubMed] [Google Scholar]
- 17.Economou A, Wickner W. SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell. 1994;78:835–843. doi: 10.1016/s0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 18.Fekkes P, Driessen A J M. Protein targeting to the bacterial cytoplasmic membrane. Microbiol Mol Biol Rev. 1999;63:161–173. doi: 10.1128/mmbr.63.1.161-173.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fekkes P, van der Does C, Driessen A J M. The molecular chaperone SecB is released from the carboxy-terminus of SecA during initiation of precursor protein translocation. EMBO J. 1997;16:6105–6113. doi: 10.1093/emboj/16.20.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gardel C, Johnson K, Jacq A, Beckwith J. The secD locus of E. coli codes for two membrane proteins required for protein export. EMBO J. 1990;9:3209–3216. doi: 10.1002/j.1460-2075.1990.tb07519.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanada M, Nishiyama K I, Mizushima S, Tokuda H. Reconstitution of an efficient protein translocation machinery comprising SecA and the three membrane proteins, SecY, SecE, and SecG (p12) J Biol Chem. 1994;269:23625–23631. [PubMed] [Google Scholar]
- 22.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 23.Harris C R, Silhavy T J. Mapping an interface of SecY (PrlA) and SecE (PrlG) by using synthetic phenotypes and in vivo cross-linking. J Bacteriol. 1999;181:3438–3444. doi: 10.1128/jb.181.11.3438-3444.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartl F-U, Lecker S, Schiebel E, Hendrick J P, Wickner W. The binding cascade of SecB to SecA to SecY/E mediates preprotein targeting to the E. coli plasma membrane. Cell. 1990;63:269–279. doi: 10.1016/0092-8674(90)90160-g. [DOI] [PubMed] [Google Scholar]
- 25.Jeong S M, Yoshikawa H, Takahashi H. Isolation and characterization of the secE homologue gene of Bacillus subtilis. Mol Microbiol. 1993;10:133–142. doi: 10.1111/j.1365-2958.1993.tb00910.x. [DOI] [PubMed] [Google Scholar]
- 26.Kaufmann A, Manting E H, Veenendaal A K J, Driessen A J M, van der Does C. Cysteine-directed cross-linking demonstrates that helix 3 of SecE is close to helix 2 of SecY and helix 3 of a neighbouring SecE. Biochemistry. 1999;38:9115–9125. doi: 10.1021/bi990539d. [DOI] [PubMed] [Google Scholar]
- 27.Kihara A, Akiyama Y, Ito K. FtsH is required for proteolytic elimination of uncomplexed forms of SecY, an essential protein translocase subunit. Proc Natl Acad Sci USA. 1995;92:4532–4536. doi: 10.1073/pnas.92.10.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klein M, Meens J, Freudl R. Functional characterization of the Staphylococcus carnosus SecA protein in Escherichia coli and Bacillus subtilis secA mutant strains. FEMS Microbiol Lett. 1995;131:271–277. doi: 10.1016/0378-1097(95)00267-9. [DOI] [PubMed] [Google Scholar]
- 29.Klose M, Schimz K L, van der Wolk J, Driessen A J M, Freudl R. Lysine 106 of the putative catalytic ATP-binding site of the Bacillus subtilis SecA protein is required for functional complementation of Escherichia coli secA mutants in vivo. J Biol Chem. 1993;268:4504–4510. [PubMed] [Google Scholar]
- 30.Kontinen V P, Yamanaka M, Nishiyama K, Tokuda H. Roles of the conserved cytoplasmic region and non-conserved carboxy-terminal region of SecE in Escherichia coli protein translocase. J Biochem (Tokyo) 1996;119:1124–1130. doi: 10.1093/oxfordjournals.jbchem.a021358. [DOI] [PubMed] [Google Scholar]
- 31.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 32.Lill R, Cunningham K, Brundage L A, Ito K, Oliver D, Wickner W. SecA protein hydrolyzes ATP and is an essential component of the protein translocation ATPase of Escherichia coli. EMBO J. 1989;8:961–966. doi: 10.1002/j.1460-2075.1989.tb03458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuyama S, Akimaru J, Mizushima S. SecE-dependent overproduction of SecY in Escherichia coli. Evidence for interaction between two components of the secretory machinery. FEBS Lett. 1990;269:96–100. doi: 10.1016/0014-5793(90)81128-b. [DOI] [PubMed] [Google Scholar]
- 34.McNicholas P, Rajapandi T, Oliver D. SecA proteins of Bacillus subtilis and Escherichia coli possess homologous amino-terminal ATP-binding domains regulating integration into the plasma membrane. J Bacteriol. 1995;177:7231–7237. doi: 10.1128/jb.177.24.7231-7237.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meens J, Frings E, Klose M, Freudl R. An outer membrane protein (OmpA) of Escherichia coli can be translocated across the cytoplasmic membrane of Bacillus subtilis. Mol Microbiol. 1993;9:847–855. doi: 10.1111/j.1365-2958.1993.tb01743.x. [DOI] [PubMed] [Google Scholar]
- 36.Meyer T H, Ménétret J F, Breitling R, Miller K R, Akey C W, Rapoport T A. The bacterial SecY/E translocation complex forms channel-like structures similar to those of the eukaryotic Sec61p complex. J Mol Biol. 1999;285:1789–1800. doi: 10.1006/jmbi.1998.2413. [DOI] [PubMed] [Google Scholar]
- 37.Murphy C K, Beckwith J. Residues essential for the function of SecE, a membrane component of the Escherichia coli secretion apparatus, are located in a conserved cytoplasmic region. Proc Natl Acad Sci USA. 1994;91:2557–2561. doi: 10.1073/pnas.91.7.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakamura K, Takamatsu H, Akiyama Y, Ito K, Yamane K. Complementation of the protein transport defect of an Escherichia coli secY mutant (secY24) by Bacillus subtilis secY homologue. FEBS Lett. 1990;273:75–78. doi: 10.1016/0014-5793(90)81054-r. [DOI] [PubMed] [Google Scholar]
- 39.Nishiyama K, Hanada M, Tokuda H. Disruption of the gene encoding p12 (SecG) reveals the direct involvement and important function of SecG in the protein translocation of Escherichia coli at low temperature. EMBO J. 1994;13:3272–3277. doi: 10.1002/j.1460-2075.1994.tb06628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishiyama K, Mizushima S, Tokuda H. A novel membrane protein involved in protein translocation across the cytoplasmic membrane of Escherichia coli. EMBO J. 1993;12:3409–3415. doi: 10.1002/j.1460-2075.1993.tb06015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Overhoff B, Klein M, Spies M, Freudl R. Identification of a gene fragment which codes for the 364 amino-terminal amino acid residues of a SecA homologue from Bacillus subtilis: further evidence for the conservation of the protein export apparatus in gram-positive and gram-negative bacteria. Mol Gen Genet. 1991;228:417–423. doi: 10.1007/BF00260635. [DOI] [PubMed] [Google Scholar]
- 42.Sadaie Y, Takamatsu H, Nakamura K, Yamane K. Sequencing reveals similarity of the wild-type div+ gene of Bacillus subtilis to the Escherichia coli secA gene. Gene. 1991;98:101–105. doi: 10.1016/0378-1119(91)90110-w. [DOI] [PubMed] [Google Scholar]
- 43.Schatz P J, Bieker K L, Ottemann K M, Silhavy T J, Beckwith J. One of three transmembrane stretches is sufficient for the functioning of the SecE protein, a membrane component of the E. coli secretion machinery. EMBO J. 1991;10:1749–1757. doi: 10.1002/j.1460-2075.1991.tb07699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schiebel E, Driessen A J M, Hartl F U, Wickner W. ΔμH+ and ATP function at different steps of the catalytic cycle of preprotein translocase. Cell. 1991;64:927–939. doi: 10.1016/0092-8674(91)90317-r. [DOI] [PubMed] [Google Scholar]
- 45.Suh J W, Boylan S A, Thomas S M, Dolan K M, Oliver D B, Price C W. Isolation of a secY homologue from Bacillus subtilis: evidence for a common protein export pathway in eubacteria. Mol Microbiol. 1990;4:305–314. doi: 10.1111/j.1365-2958.1990.tb00597.x. [DOI] [PubMed] [Google Scholar]
- 46.Takamatsu H, Fuma S, Nakamura K, Sadaie Y, Shinkai A, Matsuyama S, Mizushima S, Yamane K. In vivo and in vitro characterization of the secA gene product of Bacillus subtilis. J Bacteriol. 1992;174:4308–4316. doi: 10.1128/jb.174.13.4308-4316.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van der Does C, den Blaauwen T, de Wit J G, Manting E H, Groot N A, Fekkes P, Driessen A J M. SecA is an intrinsic subunit of the Escherichia coli preprotein translocase and exposes its carboxyl terminus to the periplasm. Mol Microbiol. 1996;22:619–629. doi: 10.1046/j.1365-2958.1996.d01-1712.x. [DOI] [PubMed] [Google Scholar]
- 48.van der Wolk J P, Klose M, Breukink E, Demel R A, de Kruijff B, Freudl R, Driessen A J M. Characterization of a Bacillus subtilis SecA mutant protein deficient in translocation ATPase and release from the membrane. Mol Microbiol. 1993;8:31–42. doi: 10.1111/j.1365-2958.1993.tb01200.x. [DOI] [PubMed] [Google Scholar]
- 49.van der Wolk J P, de Wit J G, Driessen A J M. The catalytic cycle of the Escherichia coli SecA ATPase comprises two distinct preprotein translocation events. EMBO J. 1997;16:7297–7304. doi: 10.1093/emboj/16.24.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Wolk J P, Fekkes P, Boorsma A, Huie J L, Silhavy T J, Driessen A J M. PrlA4 prevents the rejection of signal sequence defective preproteins by stabilizing the SecA-SecY interaction during the initiation of translocation. EMBO J. 1998;17:3631–3639. doi: 10.1093/emboj/17.13.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Wely K H M, Swaving J, Driessen A J M. Translocation of the precursor of α-amylase into Bacillus subtilis membrane vesicles. Eur J Biochem. 1998;255:690–697. doi: 10.1046/j.1432-1327.1998.2550690.x. [DOI] [PubMed] [Google Scholar]
- 52.van Wely K H M, Swaving J, Broekhuizen C P, Rose M, Quax W J, Driessen A J M. Functional identification of the product of the Bacillus subtilis yvaL gene as a SecG homologue. J Bacteriol. 1999;181:1786–1792. doi: 10.1128/jb.181.6.1786-1792.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wickner W, Leonard M R. Escherichia coli preprotein translocase. J Biol Chem. 1996;271:29514–29516. doi: 10.1074/jbc.271.47.29514. [DOI] [PubMed] [Google Scholar]
- 54.Yang M Y, Ferrari E, Henner D J. Cloning of the neutral protease gene of Bacillus subtilis and the use of the cloned gene to create an in vitro-derived deletion mutation. J Bacteriol. 1984;160:15–21. doi: 10.1128/jb.160.1.15-21.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]