Abstract
The Bacillus subtilis des gene encodes the cold-inducible Δ5 lipid desaturase involved in the formation of unsaturated fatty acids from saturated phospholipid precursors. Here, we describe the expression pattern of the des gene in response to a temperature downshift from 37 to 20°C. We found that the synthesis of des mRNA is undetectable at 37°C but dramatically induced upon the temperature downshift. Decay characteristics of the des transcript as well as the in vivo decay of B. subtilis bulk mRNA were investigated. The results showed that the stability of the des transcript as well as of bulk mRNA lasted substantially longer at 20°C than at 37°C. Functional expression of des at 37°C was achieved by exchanging its promoter with the non-cold shock spac promoter. These data provide the first direct evidence that temperature-mediated control of transcription is the major mechanism regulating the mRNA levels of the B. subtilis desaturase. The present results also demonstrate that the only component of the desaturation system regulated by temperature is the desaturase enzyme.
Unsaturated fatty acids (UFAs) are essential structural components of the cell membrane (3). They are also sophisticated signaling molecules that can mediate a myriad of processes involved in DNA replication (30), cellular differentiation (25), and cell death (7). It is for these reasons that all organisms require tight regulation of the lipid composition of the cell (3, 5). Perturbations in the levels of different types of lipids may be deleterious due to disruption of membrane structures and metabolic or regulatory processes (5). A universally conserved adaptation response observed among bacteria and most (if not all) poikilothermic organisms is the adjustment of membrane lipid composition at low temperatures (3, 4). As the growth temperature is lowered, the proportion of UFAs in the membrane lipids increases (for a review, see reference 3). This regulatory mechanism system, called thermal control of fatty acid synthesis, is thought to be designed to ameliorate the effects of temperature changes on the physical state of the membrane phospholipids (3). Thus, the membrane lipid composition can be altered to yield optimal membrane function at the new growth temperature. There are a variety of mechanisms that can alter the membrane phospholipid composition in response to a temperature change. Bacillus cells respond to a decrease in ambient growth temperature by desaturating the fatty acids of their membrane lipids (1, 8, 9). The introduction of an unsaturated bond into fatty acids that are esterified to the glycerol moiety of the glycerolipids is catalyzed by acyl-lipid desaturases (19). Bacillus subtilis contains a single acyl-lipid desaturase that inserts a double bond at the Δ5 position into the acyl chains of membrane phospholipids (1).
The des gene encoding the Δ5 desaturase of B. subtilis was recently isolated by members of our group (1). This gene encodes a polypeptide of 352 amino acid residues containing the three conserved histidine cluster motifs and two putative membrane-spanning domains characteristic of the membrane-bound desaturases of plants and cyanobacteria (1). Analysis of operon fusions in which the des promoter directed the synthesis of a lacZ reporter gene showed that des expression is repressed at 37°C, but shift of cultures from 37 to 20°C resulted in a 10- to 15-fold increase in transcription. This analysis shows that at least at some level control of des expression is transcriptional (1).
In contrast with B. subtilis, which possesses a single desaturase, cyanobacterial cells contain four distinct desaturases (18, 19, 21). It was recently reported that in Synechococcus sp. strain PCC 7000s the stability of the transcript from the genes that encode the Δ12 and ω3 desaturases is significantly increased at low temperatures (21). A similar temperature-mediated mRNA stabilization was also reported for the genes encoding the Δ6, Δ9, and ω3 desaturases from Synechocystis sp. strain PCC 6803 (18). Although it is not known whether the mRNA stabilization is specific to the desaturase mRNAs or is an effect that is common to all cyanobacterial mRNAs, it has been suggested that desaturase genes are controlled by a combination of mRNA synthesis and stabilization (18, 21).
In the work presented here, we describe the expression pattern of the B. subtilis des gene in response to changes in ambient temperature. We also show that the stability of des RNA at 37°C or upon a shift from 37 to 20°C is similar to the stability of bulk mRNA from B. subtilis cells subjected to the same conditions. This was further substantiated by an experiment in which we demonstrated that the des gene could be functionally expressed at 37°C by the exchange of the des promoter with the spac promoter, which is a non-cold shock promoter. The present results demonstrate that cold shock induction of des is almost exclusively controlled at the level of transcription and that the des gene product is the only component of the B. subtilis desaturation system regulated by growth temperature.
MATERIALS AND METHODS
Bacterial strains and plasmids.
B. subtilis was propagated in Spizizen minimal salts medium (24) supplemented with glucose (0.5%), vitamin-free casein hydrolysate (0.1%), tryptophan (50 μg/ml), and phenylalanine (50 μg/ml).
The parental bacterial strain was JH642 (trpC2 pheA1). Strain AKP5 carrying the integrated plasmid that introduces des under the control of the spac promoter was obtained by transformation as described previously (24). This strain was grown in media containing 5 μg of chloramphenicol ml−1 (final concentration). Plasmid pAG58, suitable for placing genes under the control of the inducible spac promoter in B. subtilis, was previously described (15).
Construction of plasmid pPA13.
Plasmid pPA13 (see Fig. 5) was constructed as follows. The oligomers 5′-TTAGCGTCGACTGAACCGAGACACACAATG-3′ and 5′-TCTGTATGCATGCTTTGTTCGTCTGGATGC-3′ (restriction sites are underlined) were synthesized so that the PCR technique could be used to amplify a chromosomal segment of DNA containing a promoterless fragment of des. This DNA fragment contains the 28 nucleotides preceding the initiation ATG codon and extends through the first 147 of the 352 codons of the des coding sequence. The PCR product was cloned into the region between the SalI and SphI sites of pAG58, giving rise to pPA13 (see Fig. 5).
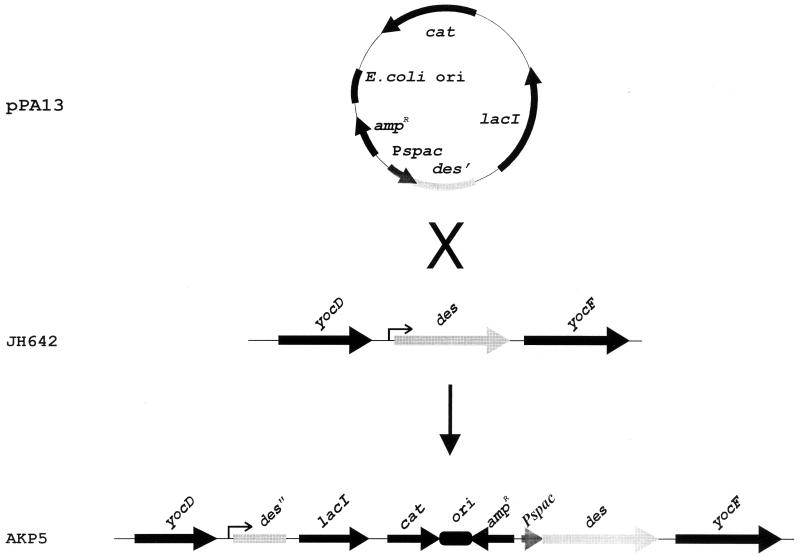
FIG. 5.
Construction of AKP5 strain with des under spac promoter control. Plasmid pPA13, which contains the 28-nucleotide-long 5′ UTR and the first 147 codons of the des gene downstream of the spac promoter, was used to transform wild-type strain JH642 to Cmr, yielding strain AKP5. Campbell insertion of this plasmid places the des gene under the control of the spac promoter.
RNA analysis.
B. subtilis strains were grown in supplemented Spizizen minimal salts medium, and the RNA was isolated as described previously (20). Northern blot analysis was performed with formaldehyde–0.8% agarose gels (17) or 5% denaturing polyacrylamide gels (6). The size of the des transcript was determined by comparison with RNA molecular weight standards (Promega) by Northern analysis of RNA fractionated on 5% denaturing polyacrylamide gels. Dot blot analysis was performed as previously described (22), with 4, 8, and 16 μg of total RNA. In all cases hybridization was performed with a single-strand des DNA probe synthesized with T4 DNA polymerase (Pharmacia), [α-32P]dATP, and the antisense oligonucleotide 5′-TCTTCAAGCTTATAGTTAGGCACCTTTGGACTC-3′ as the primer of a DNA fragment obtained by PCR amplification of the chromosome of strain JH642 with the oligonucleotides 5′-ACACGAATTCTTATCATCTTCCATGACTGCTGC-3′ and 5′-TCTTCAAGCTTATAGTTAGGCACCTTTGGACTC-3′. The 23S rRNA DNA probe was synthesized by random priming (22) on a DNA fragment obtained by PCR amplification of the chromosome of strain JH642 with the oligonucleotides 5′-TTAACGGGTGATGGCGTGCCTTTTG-3′ and 5′-ACTAACCCTGAGCGGACGAGCCTTC-3′. For analysis of des mRNA stability, radioactivity in the bands on Northern blots and in the spots of dot blots was directly quantified by using a phosphorimager instrument (ImageQuant).
Primer extension analysis and DNA sequencing.
The transcriptional mapping of the des gene was carried out by primer extension with Moloney murine leukemia virus reverse transcriptase (Promega) with the 32P-labeled (5′ end) oligonucleotide 5′-TCGAGGCTGAGATAAGCAAGAAACCATAGGC-3′ and 5 μg of total RNA extracted from cells cultures shifted to 20°C for 1 h. DNA sequencing was determined by the dideoxy chain terminator method (23) by using a T7 polymerase sequencing kit (Promega) and 32P-labeled dCTP. The double-stranded plasmid pDM10 (1), which harbors the des gene, was used as the template.
Measurement of bulk mRNA decay.
Bulk mRNA decay was measured exactly as described by Wang and Bechhofer (28). B. subtilis RNA was pulse-labeled in vivo by the addition of 30 μCi of [3H]uridine (35 to 50 Ci/mmol; New England Nuclear) to 30 ml of a mid-exponential phase culture for 2 min at 37°C and 12 min at 20°C, and then the transcription was stopped by the addition of actinomycin D (final concentration, 4 μg/ml; Sigma), nalidixic acid (final concentration, 20 μg/ml; Sigma), and unlabeled uridine (final concentration, 200 μg/ml; Sigma). At the times indicated below (see Fig. 4), duplicate 1-ml samples were removed, nucleic acids were precipitated with cold 20% trichloroacetic acid, and the precipitable counts were collected and quantified as described previously (28).
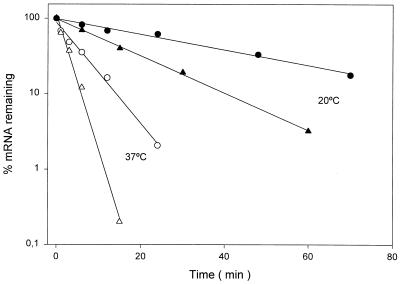
FIG. 4.
Stabilities of des and bulk mRNA. The des mRNA levels at 37°C (open triangles) or 20°C (filled triangles) were determined by quantifying the radioactivity of the bands of the Northern blot shown in Fig. 3 and from dot blots of RNA hybridized with a radiolabeled probe specific for des, as described in Materials and Methods. The samples for dot blot analysis were taken at the same times as those for the Northern analysis, as indicated in the legend of Fig. 3. Each value is the average of the results of two independent experiments. The bulk RNA decay was measured as described in Materials and Methods. Samples at 37°C (open circles) were taken at 0, 1, 3, 6, 12, and 24 min after inhibition of RNA synthesis. Samples at 20°C (filled circles) were taken at 0, 6, 12, 24, 48, and 70 min after inhibition of RNA synthesis.
Fatty acid analysis.
For measurements of fatty acid synthesis, B. subtilis cells were grown to exponential phase at 37°C. Aliquots (2 ml) of these cells were exposed to 10 μCi of sodium [14C]acetate for 12 h at the temperatures described in the legend of Fig. 7. Following incubation, the labeled lipids were extracted from whole cells and the fatty acids of the glycerolipids were transesterified, separated by argentation chromatography, and visualized as previously described (1).
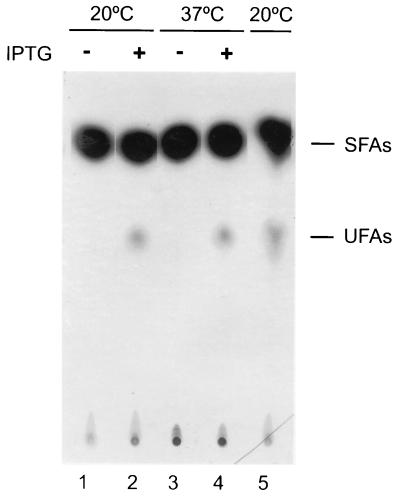
FIG. 7.
Fatty acids synthesized by strain AKP5 at 37 and 20°C. Cultures of strain AKP5 (lanes 1 to 4) were grown to mid-exponential phase at 37°C. One half of this culture was supplemented with 1 mM of IPTG. Two milliliters of treated (+) and untreated (−) cultures was challenged with 10 μCi of [14C]acetate and further shifted to 20°C (lanes 1 and 2) or maintained at 37°C (lanes 3 and 4) for 12 h. The lipids were then extracted and transesterified, and the resulting methyl esters were separated into saturated fatty acid (SFA) and UFA fractions by chromatography on 20% silver nitrate impregnated silica gel thin-layer plates. The plates were developed at −17°C and autoradiographed for 5 days. The UFA synthesized by strain JH642 grown at 37°C and then shifted to 20°C in the presence of 10 μCi of [14C]acetate for 12 h is shown in lane 5. The samples in lanes 1 and 3 contained 11,000 cpm in the SFA fractions, while the UFA fractions contained only background levels of radioactivity. The samples in lanes 2 and 4 contained 10,000 and 1,300 cpm of radioactivity in the SFA and UFA fractions, respectively. The sample in lane 5 contained 9,000 and 1,350 cpm of radioactivity in the SFA and UFA fractions, respectively.
RESULTS
Analysis of des mRNA levels following cold shock.
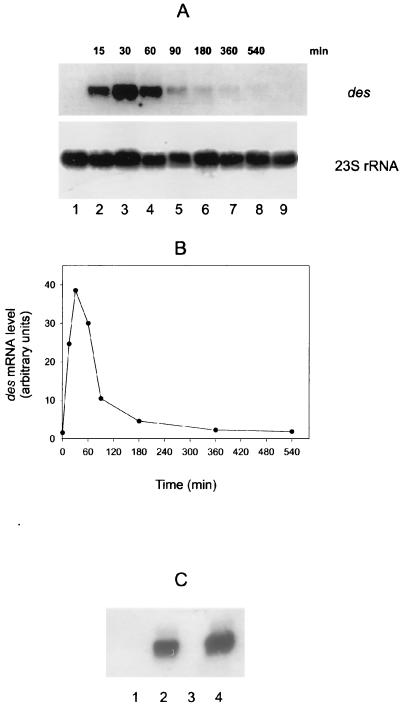
To examine the level and size of the des transcript, total cellular RNA was isolated from cultures of strain JH642 grown at 37°C and then shifted to 20°C for various times. Northern blot analysis indicated that the size of the des transcript is approximately 1.1 kb (Fig. 1A, lanes 2 to 9) and that this mRNA was not detected when cells were maintained at 37°C (Fig. 1A, lane 1). These results show that the des gene is transcribed as a monocistronic mRNA and that the des transcript only accumulates in cells downshifted to 20°C. The des mRNA was detected only 15 min after the downward shift in temperature (Fig. 1A, lane 2). The levels increased when the time of treatment was expanded to 30 min at 20°C (Fig. 1A, lane 3). However, the levels of the des transcript decreased after continuous growth at 20°C, being very low after 360 min at this temperature (Fig. 1A, lane 7). These data, plotted in Fig. 1B, indicated that cold shock results in a transient increase in the des mRNA levels.
FIG. 1.
des mRNA production before and after cold shock at 20°C and in the absence or presence of 0.2 mg of chloramphenicol per ml. (A) Northern blot analysis with formaldehyde-agarose gels was carried out as described in Materials and Methods. Total RNA was isolated from strain JH642 grown until mid-exponential phase at 37°C (lane 1), from cells shifted from 37 to 20°C at different times (lanes 2 to 8) and from cells grown continuously at 20°C (lane 9). Each lane contains 8 μg of total RNA. RNA blots were probed with a des-specific probe. After autoradiography, blots were stripped and reprobed with a B. subtilis 23S rRNA-specific probe. Autoradiographs of the resulting blots are shown. (B) Graph of the results shown in panel A. The densities of the bands corresponding to des mRNA and 23S rRNA for each lane were measured by a densitometer. The ratio of the intensities of 23S and des was used for the plot. (C) Northern blot analysis was performed as described for panel A. Cells were grown at 37°C until mid-exponential phase (lane 1) and then treated with chloramphenicol (200 μg/ml) for 15 min (lane 3) or then shifted to 20°C for 45 min in the absence (lane 2) or presence (lane 4) of chloramphenicol (200 μg/ml).
It has been demonstrated that a cold shock response (27) as well as transcription of the cold shock cspA gene (16), is specifically induced by chloramphenicol at 37°C. When this antibiotic was added at 37°C we did not observe synthesis of des mRNA (Fig. 1C, lane 3), showing that unlike cspA, the desaturase gene is not induced by inhibition of protein synthesis. However, the addition of chloramphenicol did not suppress the synthesis of the des transcript after a temperature downshift (Fig. 1C, lane 4), demonstrating that the induction of the desaturase gene at low temperature does not require the synthesis of any new protein(s).
Determination of the tsp of des.
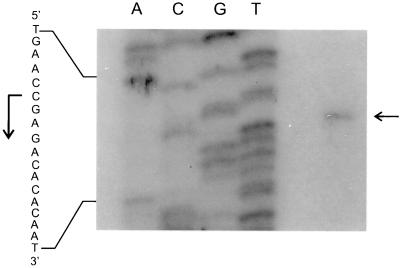
A conserved feature which is thought to contribute to the regulation by temperature of cold shock-induced genes (csp genes) from both Escherichia coli and B. subtilis is an unusually long 5′ untranslated leader region (5′UTR) contained in csp gene mRNAs (for recent reviews, see references 12, 26, and 29). To identify the promoter region responsible for cold induction of des transcription and to examine whether a 5′UTR-like region is present in des mRNA, the transcription start point (tsp) of the desaturase gene transcript was determined by primer extension analysis (Fig. 2). A single extension product was detected in extension experiments using total RNA from cells shifted from 37 to 20°C (Fig. 2). The tsp mapped to nucleotide −29 with respect to the first translated nucleotide, indicating that unlike csp gene mRNAs, the mRNA from des does not contain a long leader region. A sequence with five out of six nucleotides matching the −10 ςA-dependent B. subtilis-like promoter consensus sequence occurred upstream of the des tsp. However, no obvious −35 sequence was observed at the appropriate distance, as expected for a gene which is not expressed under normal growth conditions.
FIG. 2.
Primer extension analysis of the transcription start site. The autoradiogram shows a primer extension experiment performed with RNA extracted from strain JH642 incubated at 20°C for 45 min after shifting from 37°C. Lanes A, C, G, and T show sequencing reactions performed on des with the primer extension oligonucleotide. The horizontal arrow indicates the hybridizing extension product corresponding to the C residue taken to be the start of transcription. The vertical arrow indicates the direction of transcription.
Analysis of des mRNA stability.
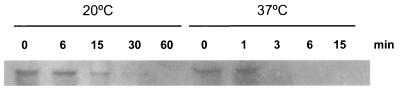
It has been reported that the increases in the levels of desaturase transcripts from cyanobacteria were caused both by enhanced transcription and by increased stability of the mRNAs at low temperatures (18, 21). To examine whether the accumulation of the transcript of the B. subtilis des gene at low temperature was also caused by increased stability of the transcript, we studied the kinetics of disappearance of the des mRNA after the addition of an inhibitor of transcription, rifampin. For this purpose, transcription of des mRNA was induced at 20°C for 1 h, rifampin was added, and the cells were transferred to 37°C or kept at 20°C. After various times of incubation at these temperatures, the total RNA was extracted and subjected to electrophoresis in 5% denaturing polyacrylamide gels, Northern blotting, hybridization with a radioactive des probe, and quantification by phosphorimaging. As seen from Fig. 3 and 4, the half-life of the transcript of the des gene increased from 1.7 min at 37°C to 11.3 min at 20°C. This result shows that the stability of the des transcript increased about sixfold at the low temperature. Because stabilization of the des mRNA at the lower temperature may simply reflect an effect that is common to all cellular mRNAs and therefore be nonspecific, the rate of des mRNA decay at 37 and 20°C was compared with that of bulk mRNA. The decay rate of bulk mRNA in strain JH642 was measured by pulse labeling RNA with [3H]uridine, after which transcription was blocked (see Materials and Methods) and trichloroacetic acid-precipitable counts were quantified. When this experiment was performed the overall mRNA half-life increased from 4.5 min at 37°C to 29.2 min at 20°C (Fig. 4). This result clearly demonstrates that similarly to the des transcript, the overall mRNA half-life is increased sixfold when cells are shifted to 20°C (Fig. 4). The results illustrated in Fig. 3 and 4 allow us to conclude that although the stability of des mRNA is increased at 20°C, the increased half-life of B. subtilis transcripts at low temperatures is a property inherent to cells that are subjected to cold shock.
FIG. 3.
Northern blot analysis of des mRNA decay after rifampin addition. Total RNA (5 μg per lane) from strain JH642 was separated on a denaturing 5% polyacrylamide gel. Cells were grown at 37°C until mid-exponential phase, des mRNA synthesis was induced by transferring the culture to 20°C for 45 min, rifampin (final concentration, 500 μg/ml) and nalidixic acid (final concentration, 20 μg/ml) were added, and the cells were kept at 20°C or transferred to 37°C. Total RNA from aliquots of 25 ml was extracted after the addition of the antibiotics (times shown indicate minutes elapsed following addition of antibiotics).
Promoter-dependent cold shock induction of des.
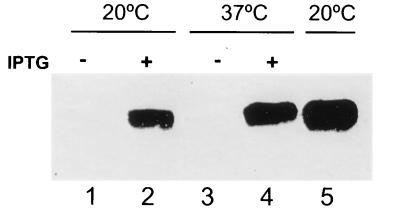
Results described above suggested that cold shock induction of des expression is regulated at the level of des transcription rather than at the level of its RNA stability. These conclusions were reached by inducing the transcription of des by cold shock and then studying the stability of the transcript at 37°C (Fig. 3). Since the mechanism(s) by which mRNA stability is controlled in bacteria by temperature is not well understood (26), this experimental approach did not rule out the possibility that incubation of the cell cultures at 20°C in some way increases the stability of the des transcript at 37°C. Thus, to uncouple the potential effect of temperature-mediated mRNA stabilization from cold shock transcriptional regulation, we exchanged the des promoter with the spac promoter. This hybrid promoter contains the RNA polymerase recognition site from an early promoter of the B. subtilis SPO1 phage and the lac operator (13). The des promoter was replaced with the spac promoter in such a way that the transcript from the new construct, spac-des, is identical to that of des mRNA. In order to eliminate des induction from the chromosomal des gene, the single chromosomal copy of this gene was placed under the control of spac as outlined in Fig. 5. To do this a fragment containing the 5′ end of des was cloned along with the spac promoter and the lacI gene into an integrational plasmid vector to generate plasmid pPA13 (Fig. 5). Integration of this plasmid by Campbell insertion into B. subtilis JH642 placed the single copy of des downstream from the spac promoter. The resulting strain was designated AKP5. This strain was grown at 37°C to exponential phase, at which time portions of the culture were supplemented with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and shifted to 20°C or maintained at 37°C. Northern blot analysis of total RNA of cells supplemented with IPTG or not clearly show that transcription of des was dependent on the spac inductor at both 37 and 20°C (Fig. 6). Moreover, the levels of des transcript obtained at either 37 or 20°C in the presence of IPTG were approximately the same (Fig. 6). In addition, we determined that the mRNA half-life of the des transcript produced in AKP5 at 37°C was about 2 min (data not shown), indicating that the des mRNA stability at this temperature is approximately the same whether the transcript is synthesized with the des wild-type promoter or the spac promoter. Therefore, we conclude that cold shock induction of des transcription is strictly dependent on the des promoter and that mRNA stability is not involved in the regulation by temperature of the B. subtilis desaturase gene.
FIG. 6.
des mRNA production under the control of the spac promoter. Total RNA (10 μg/ml) from strain AKP5 (lanes 1 to 4) or JH642 (lane 5) was separated on formaldehyde-agarose gels. Strain AKP5 was grown at 37°C to mid-exponential phase, and half of this culture was supplemented with 1 mM of IPTG. Treated (+) and untreated (−) cultures were further shifted to 20°C for 45 min (lanes 1 and 2) or maintained at 37°C for 15 min (lanes 3 and 4). Strain JH642 was grown at 37°C to mid-exponential phase and then shifted to 20°C for 45 min (lane 5).
Temperature-independent synthesis of UFAs.
The presence of des transcript in AKP5 cells at 37°C does not ensure per se that the desaturase enzyme is also present, since this mRNA could be functionally inactive or the translational apparatus of these cells could be incapable of using it. Thus, the fatty acid profile of strain AKP5 was studied. The fatty acids were labeled by growth of the strain in [14C]acetate, followed by argentation chromatography of the radioactive fatty acids. Strain AKP5 synthesized UFAs at both 20 and 37°C only when IPTG was added to the culture medium (Fig. 7). These experiments demonstrated that the Δ5 desaturase is active at 37°C and that at this temperature B. subtilis synthesizes all the cofactors necessary for desaturation of membrane lipids. Therefore, the des gene product is the only component of the B. subtilis desaturation system which is regulated by growth temperature.
DISCUSSION
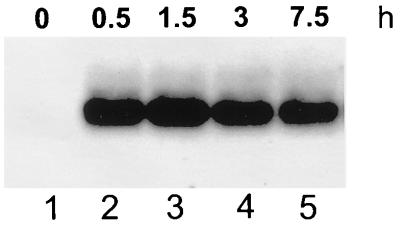
The studies of des gene expression reported in this paper were designed to dissect the mechanism of regulation of UFA synthesis in B. subtilis. Our results show that des is tightly regulated during cold shock. While the des transcript is barely detectable at 37°C, the production of des mRNA is transiently induced in the absence of protein synthesis upon temperature downshift. Derepression of des occurs at the level of transcription in a promoter-dependent fashion but was not caused by stabilization of des mRNA, which was reported to be a major cause of desaturase induction in cyanobacteria after cold shock (18, 21). In addition, we report here that the factor limiting the introduction of a double bond into fatty acids at 37°C is the desaturase enzyme, while other desaturation cofactors such as the electron donor or components of the electron transport, appear to be present at the restrictive temperature. It is worth noting that the amount of UFA synthesized by strains JH642 and AKP5 is approximately the same after 12 h of growth at 20°C (Fig. 7). However, although the efficiencies of the spac and des promoters are similar (data not shown), the des mRNA levels in strain AKP5 are 10- to 15-fold higher than the levels found in strain JH642 grown overnight at 20°C (Fig. 8 and data not shown). This suggests that either the synthesis or the activity of the desaturase in strain AKP5 is decreased after continuous growth at 20°C.
FIG. 8.
Pattern of spac-des expression at 20°C. Total RNA (10 μg/ml) from strain AKP5 was separated on formaldehyde-agarose gels. Cells were grown at 37°C to mid-exponential phase, supplemented with 1 mM IPTG, and then shifted to 20°C. Total RNA was extracted after the addition of IPTG (times shown indicate hours elapsed following addition of IPTG).
How could the transcription of des be regulated by cold shock? Our results show that the desaturase transcript can be produced upon cold shock by using resources already existing in the cell at the time of temperature downshift (Fig. 1C). Thus, if a transcriptional regulatory protein (activator or repressor) participates in des transcription, a temperature-dependent change in protein conformation could take place. This mode of regulation has been described for the Salmonella typhimurium virulence factor TlpA (14). This protein is an autoregulatory dimeric repressor which uses its folding equilibrium to regulate the DNA binding activity. Temperature upshifts lead to a shift in the equilibrium that favors the nonfunctional and unfolded monomeric form of TlpA. Thus, a temperature upshift could favor the formation of an unfolded des transcriptional activator. Alternatively, a cold-sensitive repressor could be responsible for the repression of des at higher temperatures.
We have recently hypothesized that DNA gyrase activity is required for transcriptional induction of UFA synthesis at low growth temperatures (10). Moreover, we have determined that the increase in negative supercoiling of plasmid DNA extracted from cells shifted from 37 to 20°C does not require de novo protein synthesis (2). Therefore, another possibility is that increased superhelicity of the DNA template could be important for expression of the desaturase gene. It has been suggested that local melting of DNA by RNA polymerase could be a considerable problem at low growth temperatures (11); thus, an increase in bacterial DNA negative supercoiling at these temperatures (10) should facilitate melting of the DNA strands and hence RNA polymerase action.
How could the transcription of des be downregulated after cold shock induction? We show here that the levels of the des transcript produced in strain AKP5, in which the des wild-type promoter was exchanged with the spac promoter, are not decreased after continuous growth at 20°C (Fig. 8). This finding indicates that the transient induction of the wild-type des gene at low growth temperatures seems to be due to shutoff of transcription rather than to the instability of the des mRNA. Our results agree with those of Fujii and Fulco (8). They provided in vivo evidence that within 10 min after a culture of Bacillus megaterium was shifted from 35 to 20°C, Δ5 desaturase synthesis began and continued at a very high rate for about 1 h. This phase was termed hyperinduction phase because the levels of desaturase synthesized during this interval far exceeded the levels found in cells growing at 20°C (8). Fujii and Fulco suggested that the shutdown of hyperinduction at 20°C resulted from the action of a repressor, which would inhibit the synthesis of the mRNA coding for the desaturase (8). Notably, the time course of accumulation of B. subtilis des mRNA showed in this study (Fig. 1A and B) is like the hyperinduction and modulation of desaturase synthesis observed by Fujii and Fulco for B. megaterium (8). It should be noted that, similar to B. megaterium, the amount of UFA observed to be synthesized by B. subtilis during the first growth division cycle was much higher than that observed for cultures grown for several generations at 20°C (9). Therefore, the phenomena of hyperinduction of desaturase reported by Fujii and Fulco (8) and the analysis of expression of the des gene shown here are consistent with the hypothesis that UFA synthesis in bacilli is downregulated after cold shock by shutoff of des transcription. We are currently attempting to identify the elements involved in the control of the des gene at low growth temperatures by the isolation of regulation-defective mutants.
ACKNOWLEDGMENTS
We thank R. Raya for generously providing Macaloid clay for RNA extraction.
This work was supported by the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Agencia de Promoción Científica y Tecnológica (FONCYT), Fundación Antorchas, and the exchange program Consejo Superior de Investigaciones Científicas (CSIC)/CONICET. P. S. Aguilar is a fellow from CONICET, and D. de Mendoza is a Career Investigator of the same institution.
REFERENCES
- 1.Aguilar P S, Cronan J E, Jr, de Mendoza D. A Bacillus subtilis gene induced by cold shock encodes a membrane phospholipid desaturase. J Bacteriol. 1998;180:2194–2200. doi: 10.1128/jb.180.8.2194-2200.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguilar, P. S., and D. de Mendoza. 1998. Unpublished results.
- 3.Cronan J E, Jr, Rock C O. Biosynthesis of membrane lipids. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: American Society for Microbiology; 1996. pp. 612–636. [Google Scholar]
- 4.de Mendoza D, Grau R, Cronan J E., Jr . Biosynthesis and function of membrane lipids. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 411–421. [Google Scholar]
- 5.Dowhan W. Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu Rev Biochem. 1997;66:199–232. doi: 10.1146/annurev.biochem.66.1.199. [DOI] [PubMed] [Google Scholar]
- 6.Drider D, Santos J M, Arraiano C M, López P. RNA processing is involved in the post-transcriptional control of the citQRP operon from Lactococcus lactis biovar diacetylactis. Mol Gen Genet. 1998;258:9–15. doi: 10.1007/s004380050701. [DOI] [PubMed] [Google Scholar]
- 7.English N, Palmer C N A, Alworth W L, Kang L, Hughes V, Wolf C R. Fatty acid signals in Bacillus megaterium are attenuated by cytochrome P-450-mediated hydroxylation. Biochem J. 1997;327:363–368. doi: 10.1042/bj3270363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujii D K, Fulco A J. Biosynthesis of unsaturated fatty acid in bacilli: hyperinduction and modulation of desaturase synthesis. J Biol Chem. 1977;252:3660–3670. [PubMed] [Google Scholar]
- 9.Grau R, de Mendoza D. Regulation of the synthesis of unsaturated fatty acids in Bacillus subtilis. Mol Microbiol. 1993;8:535–542. doi: 10.1111/j.1365-2958.1993.tb01598.x. [DOI] [PubMed] [Google Scholar]
- 10.Grau R, Gardiol D, Glikin G, de Mendoza D. DNA supercoiling and thermal regulation of unsaturated fatty acid synthesis in Bacillus subtilis. Mol Microbiol. 1994;11:933–941. doi: 10.1111/j.1365-2958.1994.tb00372.x. [DOI] [PubMed] [Google Scholar]
- 11.Graumann P L, Marahiel M A. Some like it cold: response of microorganisms to cold shock. Arch Microbiol. 1996;166:293–300. doi: 10.1007/s002030050386. [DOI] [PubMed] [Google Scholar]
- 12.Graumann P L, Marahiel M A. A superfamily of proteins that contain the cold-shock domain. Trends Biochem Sci. 1998;23:286–290. doi: 10.1016/s0968-0004(98)01255-9. [DOI] [PubMed] [Google Scholar]
- 13.Henner D J. Inducible expression of regulatory genes in Bacillus subtilis. Methods Enzymol. 1990;185:223–228. doi: 10.1016/0076-6879(90)85022-g. [DOI] [PubMed] [Google Scholar]
- 14.Hurme R, Berndt K D, Normark S J, Rhen M. A proteinaceous gene regulatory thermometer in Salmonella. Cell. 1997;90:55–64. doi: 10.1016/s0092-8674(00)80313-x. [DOI] [PubMed] [Google Scholar]
- 15.Jaacks K J, Healy J, Losick R, Grossman A D. Identification and characterization of genes controlled by the sporulation-regulatory gene spo0H in Bacillus subtilis. J Bacteriol. 1989;171:4121–4129. doi: 10.1128/jb.171.8.4121-4129.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang W, Jones P, Inouye M. Chloramphenicol induces the transcription of the major cold shock gene of Escherichia coli, cspA. J Bacteriol. 1993;175:5824–5828. doi: 10.1128/jb.175.18.5824-5828.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.López de Felipe F, Magni C, de Mendoza D, López P. Citrate utilization gene cluster of the Lactococcus lactis biovar diacetylactis: organization and regulation of expression. Mol Gen Genet. 1995;246:590–599. doi: 10.1007/BF00298965. [DOI] [PubMed] [Google Scholar]
- 18.Los D A, Ray M K, Murata N. Differences in the control of the temperature-dependent expression of four genes for desaturases in Synechocystis sp. PCC 6803. Mol Microbiol. 1997;25:1167–1175. doi: 10.1046/j.1365-2958.1997.5641912.x. [DOI] [PubMed] [Google Scholar]
- 19.Murata N, Wada H. Acyl-lipid desaturases and their importance in the tolerance and acclimation to cold of cyanobacteria. Biochem J. 1995;308:1–8. doi: 10.1042/bj3080001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raya R, Bardowski J, Andersen P S, Ehrlich S D, Chopin A J. Multiple transcriptional control of the Lactococcus lactis trp operon. J Bacteriol. 1998;180:3174–3180. doi: 10.1128/jb.180.12.3174-3180.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakamoto, T., and D. A. Bryant. Temperature-regulated mRNA accumulation and stabilization for fatty acid desaturase genes in the cyanobacterium Synechococcus sp. strain PCC 7002. Mol. Microbiol. 23:1281–1292. [DOI] [PubMed]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 23.Sanger F, Nicklen S, Coulson R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spizizen J. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc Natl Acad Sci USA. 1958;44:1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strauch M A, de Mendoza D, Hoch J A. cis-Unsaturated fatty acids specifically inhibit a signal-transducing protein kinase required for initiation of sporulation in Bacillus subtilis. Mol Microbiol. 1992;6:2909–2917. doi: 10.1111/j.1365-2958.1992.tb01750.x. [DOI] [PubMed] [Google Scholar]
- 26.Thieringer H A, Jones P G, Inouye M. Cold shock and adaptation. Bioessays. 1998;20:49–57. doi: 10.1002/(SICI)1521-1878(199801)20:1<49::AID-BIES8>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 27.VanBogelen R A, Neidhardt F C. Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:5589–5593. doi: 10.1073/pnas.87.15.5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang W, Bechhofer D H. Properties of a Bacillus subtilis polynucleotide phosphorylase deletion strain. J Bacteriol. 1996;178:2375–2382. doi: 10.1128/jb.178.8.2375-2382.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamanaka K, Fang L, Inouye M. The CspA family in Escherichia coli: multiple gene duplication for stress adaptation. Mol Microbiol. 1998;27:247–255. doi: 10.1046/j.1365-2958.1998.00683.x. [DOI] [PubMed] [Google Scholar]
- 30.Yung B Y, Kornberg A. Membrane attachment activates DnaA protein, the initiation protein of chromosome replication in Escherichia coli. Proc Natl Acad Sci USA. 1988;19:7202–7205. doi: 10.1073/pnas.85.19.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]