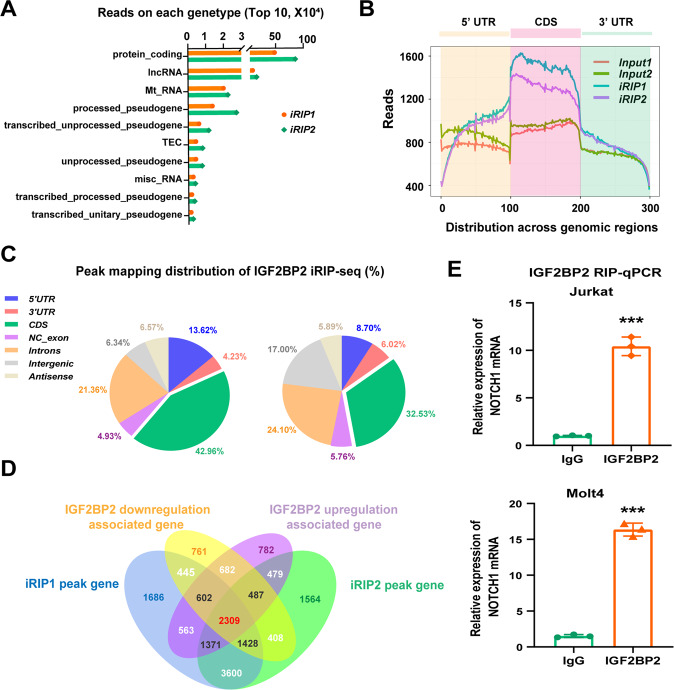
Fig. 3. Transcriptome-wide iRIP-sequence assays identifies potential targets of IGF2BP2 in T-ALL.
A The Top 10 RNA reads within different gene types bound by IGF2BP2 from two replicate IGF2BP2 binding iRIP-sequence data. Protein_coding, contains an open reading frame (ORF); for detailed explanations of other gene types, refer to following website, https://www.gencodegenes.org/pages/biotypes.html. B Metagene profiles of enrichment of IGF2BP2-binding sites and their Input control analyzed by two replicate iRIP-sequence. CDS, coding sequence. C The distribution of IGF2BP2-binding peaks within different gene regions from two replicate IGF2BP2 binding iRIP-sequence data. D Venn diagram was plotted to show the intersected genes from IGF2BP2 binding iRIP-sequence, RNA-sequence of IGF2BP2 downregulation associated gene and IGF2BP2 upregulation associated gene. 2309 common genes were screened out. E The enrichment of NOTCH1 was analyzed by IGF2BP2 RIP-qPCR in Jurkat and Molt4 cells. Data are mean ± SD values. *P < 0.05; **P < 0.01; ***P < 0.001.