Abstract
The Bacillus subtilis spore is encased in a resilient, multilayered proteinaceous shell, called the coat, that protects it from the environment. A 181-amino-acid coat protein called CotE assembles into the coat early in spore formation and plays a morphogenetic role in the assembly of the coat’s outer layer. We have used a series of mutant alleles of cotE to identify regions involved in outer coat protein assembly. We found that the insertion of a 10-amino-acid epitope, between amino acids 178 and 179 of CotE, reduced or prevented the assembly of several spore coat proteins, including, most likely, CotG and CotB. The removal of 9 or 23 of the C-terminal-most amino acids resulted in an unusually thin outer coat from which a larger set of spore proteins was missing. In contrast, the removal of 37 amino acids from the C terminus, as well as other alterations between amino acids 4 and 160, resulted in the absence of a detectable outer coat but did not prevent localization of CotE to the forespore. These results indicate that changes in the C-terminal 23 amino acids of CotE and in the remainder of the protein have different consequences for outer coat protein assembly.
In response to nutrient limitation, the gram-positive soil bacterium Bacillus subtilis forms an alternative cell type, called the spore, during a process known as sporulation. A multilayered proteinaceous shell, called the coat, surrounds the spore and provides an extraordinarily high degree of resistance to a variety of environmental assaults, including mechanical stress, and degradative proteins, such as lysozyme (2). In the electron microscope, two major coat layers are visible: a lightly staining lamellar inner coat and a darkly staining, coarsely layered outer coat (Fig. 1A) (2, 31). Coat formation involves the ordered assembly and cross-linking of upwards of 20 coat protein species (6); elucidating this assembly process can give insight into the general cell biological problem of how complex macromolecular structures are formed as well as help to explain how the coat protects the spore.
FIG. 1.
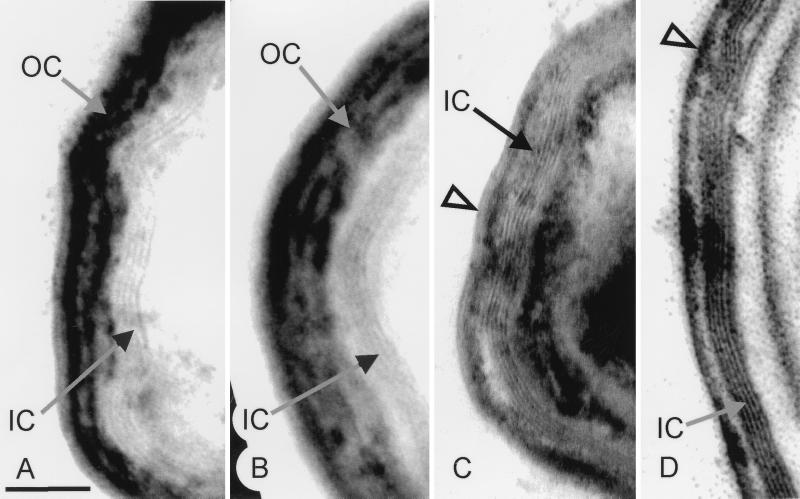
Electron microscopic analysis of wild-type and mutant spore coats. Arcs of wild-type (A), AD408 (B), TB50 (C), and TB70 (D) spore coats. The open arrowheads indicate thin remnants of the outer coat. Bar, 100 nm. IC, inner coat; OC, outer coat.
Spores are built in an 8-h process comprised of several morphologically distinct stages (28). Soon after the commitment to sporulation, an asymmetrically positioned septum that divides the cell into two compartments is formed. The smaller compartment, known as the forespore, ultimately becomes the spore, while the larger chamber, called the mother cell, will surround and nurture the developing spore. As sporulation proceeds, the septum engulfs the smaller cell, creating a membrane-bound compartment. Soon after engulfment, the coat becomes visible as a darkly staining shell encircling the forespore. After assembly of the coat and other spore-associated structures is complete, the mother cell lyses and releases the spore into the environment.
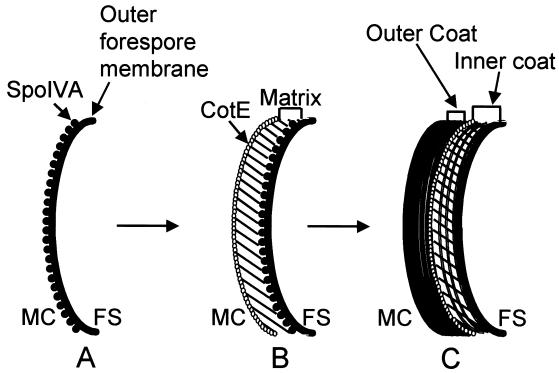
In the current model of coat assembly (6, 7), the formation of the coat begins just after the formation of the sporulation septum. At this time, a protein called SpoIVA binds at or very close to the mother cell-adjacent forespore membrane (Fig. 2A) (7, 17–19, 26). SpoIVA serves to connect a preliminary coat structure, called the precoat, to the mother cell side of the forespore (Fig. 2B). The only known component of the precoat is a 181-amino-acid protein called CotE (7, 34) that does not resemble other proteins in the databases but possesses one notable sequence feature, a stretch of acidic residues between amino acids 150 and 169. CotE forms a layer standing a short distance away from the septum (7, 17). A hypothetical precoat component, called the matrix, fills the gap between the forespore membrane and CotE, connecting the two. After the precoat has assembled, the inner coat proteins infiltrate into the skeleton formed by the precoat and the outer coat proteins assemble around the shell of CotE (Fig. 2C).
FIG. 2.
Model of the stages of coat formation. (A) SpoIVA localizes to the mother cell side of the sporulation septum, of which only an arc is shown. (B) The precoat, consisting of the matrix and the layer of CotE, assembles at the forespore surface, under the direction of SpoIVA. (C) The inner coat proteins assemble into the matrix, and the outer coat proteins bind around the shell of CotE. FS, forespore; MC, mother cell.
Spores from a cotE null mutant strain are missing the outer coat (34). However, CotE is not required for the synthesis of at least two of the outer coat proteins, CotA and CotC. It is likely, therefore, that CotE does not play a major role in outer coat protein synthesis but rather is involved at the level of assembly of the outer coat proteins. To understand how CotE participates in coat assembly, we have generated a set of point and deletion mutant versions of cotE and determined their effects on outer coat formation and the ability of CotE to localize to the forespore.
MATERIALS AND METHODS
Bacterial strains and recombinant DNA methods.
The strains and plasmids used in this study are described in Table 1, and the primers are described in Table 2. We carried out genetic manipulations in B. subtilis as described previously (3). To build the cotE deletion strains, we carried out PCR, using pAD105 (harboring epitope-tagged CotE [7]) as the template. For the C-terminal deletion mutants TB50, TB70, and TB97, the 5′ primer was OL3, permitting amplification of all known 5′ regulatory sequences of cotE (35). For the 3′ primer, we used OL37, OL36, and OL35R, respectively (Fig. 3). Each 3′ primer contained 18 to 21 nucleotides of cotE, the sequence for nine amino acids of the HA1 epitope (8) (in AD408, these nine amino acids are followed by a serine), and a stop codon (TAA). To generate internal deletions in cotE (in strains TB51, TB53, TB71, TB83, and TB95), we used PCR, with pAD105 as the template, to generate two pieces of DNA flanking the region to be deleted. The 5′ piece was generated by using OL3 and a primer immediately upstream of the region to be deleted (Fig. 3). The 3′ piece was generated with OL4 and a primer immediately downstream of the region to be deleted. The primers flanking the regions to be deleted had the general structure 5′ AAAACTCTTCA 3′, followed by a glutamic acid-encoding codon (GAA), and ended with a cotE sequence that varied depending on the region to be deleted. Nucleotides 5 through 10 of the primer were the recognition sequence for the type IIS restriction endonuclease Eam1104I, which cleaves outside its recognition sequence, generating a 3-nucleotide 5′ overhang (14). Because the primers flanking the regions to be deleted contained a glutamic acid codon within the cleavage site of the enzyme, after digestion we were able to ligate the two PCR fragments without the addition of any extraneous sequence. After digestion with Eam1104I and ligation, we carried out PCR amplification with the primers OL3 and OL4 to generate a contiguous internally deleted version of cotE. To insert these cotE alleles into the B. subtilis genome, we digested the PCR products with EcoRI and BamHI and cloned them into similarly digested pDG364 (10). We linearized these plasmids with BstEII and used them to transform TB1, generated by transformation of AD28 with linearized pJL62 (12). To sequence the DNA constructs used in this study, we carried out cycle sequencing with the Perkin-Elmer direct cycle sequencing kit, following the manufacturer’s instructions, or we used the Iowa State University DNA Sequencing and Synthesis Facility. All other recombinant DNA methods are described in reference 21. We used Western blot analysis to demonstrate that lysates of the mutant strains produced versions of CotE of the expected sizes (data not shown).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| B. subtilis | ||
| PY79 | Wild type | 33 |
| AD18 | spoIVAΔ::neo | 7 |
| AD28 | cotEΔ::cat | 7 |
| AD408 | cotEΔ::cat amyE::cotEepi1 (Knr) | 7 |
| TB1 | cotEΔ::cat::spec | This study |
| TB50 | cotEΔ::cat::spec amyE::cotEΔ173-181 | This study |
| TB70 | cotEΔ::cat::spec amyE::cotEΔ159-181 | This study |
| TB97 | cotEΔ::cat::spec amyE::cotEΔ145-181 | This study |
| TB51 | cotEΔ::cat::spec amyE::cotEΔ147-160 | This study |
| TB71 | cotEΔ::cat::spec amyE::cotEΔ80-102 | This study |
| TB95 | cotEΔ::cat::spec amyE::cotEΔ58-75 | This study |
| TB83 | cotEΔ::cat::spec amyE::cotEΔ30-55 | This study |
| TB53 | cotEΔ::cat::spec amyE::cotEΔ4-29 | This study |
| AD648 | cotEΔ::cat amyE::cotEV64G (Knr) | This study |
| AD650 | cotEΔ::cat amyE::cotEY66N (Knr) | This study |
| AD903 | cotEΔ::cat::spec amyE::cotEΔ173-181 spoIVAΔ::neo | This study |
| AD904 | cotEΔ::cat::spec amyE::cotEΔ147-160 spoIVAΔ::neo | This study |
| AD905 | cotEΔ::cat::spec amyE::cotEΔ4-29 spoIVAΔ::neo | This study |
| AD906 | cotEΔ::cat::spec amyE::cotEΔ159-181 spoIVAΔ::neo | This study |
| AD907 | cotEΔ::cat::spec amyE::cotEΔ80-102 spoIVAΔ::neo | This study |
| AD908 | cotEΔ::cat::spec amyE::cotEΔ30-55 spoIVAΔ::neo | This study |
| AD909 | cotEΔ::cat::spec amyE::cotEΔ58-75 spoIVAΔ::neo | This study |
| AD911 | cotEΔ::cat::spec amyE::cotEΔ145-181 spoIVAΔ::neo | This study |
| AD912 | cotEΔ::cat::spec amyE::cotEepi1 spoIVAΔ::neo | This study |
| AD913 | cotEΔ::cat::spec amyE::cotEV64G spoIVAΔ::neo | This study |
| AD914 | cotEΔ::cat::spec amyE::cotEY66N spoIVAΔ::neo | This study |
| ER203 | cotG::erm trpC2 | 20 |
| SL111 | cotEΔ::cat amyE::cotEepi1 ΔcotA::cat::spec | This study |
| E. coli | ||
| NR9232 | ara thi zaf13::Tn10 mutD5 Δprolac F′ prolac | 24 |
| BL21(DE3) | Overexpression host | Laboratory stock |
| Plasmids | ||
| pAD105 | pER82::cotEepi1 | 7 |
| pJL62 | Converts cat strains to cat::spec | 12 |
| pDG364 | Permits recombination into the chromosome at amyE | 10 |
TABLE 2.
Primers
| Oligonucleotide | Sequencea | Enzyme | Nucleotidesb |
|---|---|---|---|
| OL3 | 5′ AAAAAGAATTCAGCTCGTTGCACACACC 3′ | EcoRI | −386 to −370 |
| OL4 | 5′ AAAAAGGATCCTACTGTCTCCCCTAGTCCC 3′ | BamHI | +551 to +569 |
| OL22R | 5′ AAAAACTCGAGTTCTTCAGGGCTGGCGTAGT 3′ | XhoI | +536 to +544 |
| OL23 | 5′ AAAACTCTTCAGAAAAAAAACCGAGCAGC 3′ | Eam1104I | +85 to +102 |
| OL24 | 5′ AAAACTCTTCAGAAATTGAAGGGTATTAT 3′ | Eam1104I | +163 to +180 |
| OL25 | 5′ AAAACTCTTCATTCAGACATTCCGGCATG 3′ | Eam1104I | −9 to +9 |
| OL26R | 5′ AAAACTCTTCAGAAGATGAGCTTGATGAA 3′ | Eam1104I | +478 to +495 |
| OL27 | 5′ AAAACTCTTCATTCAACAACTACCTTTGTTTCCCC 3′ | Eam1104I | +415 to +438 |
| OL31 | 5′ AAAAAAAACATATGTCTGAATACAGGGAAATTAT 3′ | NdeI | +1 to +23 |
| OL35R | 5′ AAAAAGGATCCTTAGGCGTAGTCGGGGACGTCGTAGGGGTAAACTACCTTTGTTTCCCC 3′ | BamHI | +415 to +432 |
| OL36 | 5′ AAAAAGGATCCTTAGGCGTAGGACGTCGTAGGGGTAATCTTCCTCGTCATCCTC 3′ | BamHI | +457 to +474 |
| OL37 | 5′ AAAAAGGATCCTTAGGCGTAGGACGTCGTAGGGGTACGGGTTGATGTCTTCAAGCTC 3′ | BamHI | +496 to +516 |
| OL41 | 5′ AAAACTCTTCATTCTGTGACAACCTCTGT 3′ | Eam1104 | +220 to +237 |
| OL42 | 5′ AAAACTCTTCAGAAGTGATTGCCAAAGTG 3′ | Eam1104I | +304 to +321 |
| OL49 | 5′ AAAACTCTTCATTCAGGCGAGATGGT 3′ | Eam1104I | +73 to +87 |
| OL50 | 5′ AAAACTCTTCATTCAATTTCTACCGTTTTTCC 3′ | Eam1104I | +151 to +171 |
| OL51R | 5′ AAAACTCTTCAGAAGTTGTCACAGAACGG 3′ | Eam1104I | +223 to +240 |
FIG. 3.
Deletion constructs of CotE and resulting phenotypes. The diagram in the upper left of the figure indicates the cotE open reading frame (box) and its upstream sequences (dotted line). cotE mutant constructs used in this study are indicated below the diagram. The triangles indicate oligonucleotides used to generate the deletion mutant versions of cotE. Asterisks indicate the positions of point mutations in AD648 and AD650. To the right of each construct is the strain name. The third column indicates which amino acids have been deleted or altered in each construct. The fourth column indicates the coat layers detected by electron microscopy (EM). OC, outer coat; IC, inner coat. The rightmost column gives the degree of lysozyme resistance, relative to that of the wild type. cotE null mutant spores have 6% resistance compared to the wild type. The numbers in parentheses are standard errors of the means.
To build double mutant strains harboring spoIVAΔ::Neor and each one of the mutant alleles of cotE, we transformed competent cells of strains TB50, TB51, TB53, TB70, TB71, TB83, TB95, TB97, AD408, AD648, and AD650 with DNA from the strain AD18 (7), resulting in strains AD903 to AD909 and AD911 to AD914, respectively. To create SL111, we transformed a cotA::cat strain (4) with linearized plasmid pJL62 to inactivate cat and introduce a spectinomycin resistance gene, made chromosomal DNA from the resulting strain, and used it to transform competent AD408.
Creation of point mutations in cotE.
To generate point mutations in cotE, we transformed the Escherichia coli mutator strain NR9232 (24) with pAD105. We plated transformants on 2× YT plates (21), grew them at 42°C for 48 h, prepared plasmid DNA from the resulting lawn of E. coli, and used this DNA to transform a fresh culture of NR9232. We repeated the growth step on 2× YT plates, again prepared plasmid DNA, and used it to transform AD28 to kanamycin resistance. We plated about 500 colonies on Difco sporulation medium (DSM) and identified spores with altered coats using the tetrazolium overlay germination assay (3) (this is possible because germination mutants are good candidates for coat assembly mutants [2]) and by examining the spores for the loss of the CotA-associated brown color (22).
Preparation of anti-CotE antibodies.
To produce a histidine-tagged, HA1 epitope-tagged version of CotE in E. coli, we first carried out a PCR with pAD105 as the template and the primers OL22R and OL31, subcloned the resulting DNA fragment into the overproduction vector pAGS05 (27), and used the resulting plasmid to transform BL21(DE3). We purified CotE from these cells by nickel chromatography according to the manufacturer’s directions (Novagen) and injected approximately 200 μg of this protein into rabbits to generate antibodies.
Preparation of spores and spore protein extractions.
To generate spores for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), we cultured cells in DSM for 24 to 48 h (22) and purified phase-bright spores by centrifugation in Renografin gradients (3). To prepare protein extracts for SDS-PAGE analysis of coat proteins, we added SDS-PAGE loading buffer (21) to spores prepared from 2.5 ml of culture, boiled the sample for 5 min, mixed it vigorously for 1 min, and boiled it again for 5 min. After mixing the sample again, we centrifuged it and electrophoresed the supernatant on a 15% acrylamide gel.
Western blot assays.
We prepared spores as described previously for SDS-PAGE analysis and loaded 0.15 optical density unit (at 600 nm) in each lane of a 15% acrylamide gel. We carried out electrotransfer and detection of bound protein as described previously (21), except that we used polyvinylidene difluoride membranes instead of nitrocellulose. We used anti-CotE antibodies at a dilution of 1:10,000.
Phase-contrast and electron microscopy and lysozyme assays.
We judged spore coats to be wild type in appearance if they showed a well-demarcated, contiguous dark layer encircling the spore core under phase-contrast microscopy. We judged the coat as mutant if the dark layer showed regions of discontinuity, as is seen in the case of a cotE null mutant (5). For an example of this appearance, see reference 32, in which the authors analyze a strain bearing a CotE-green fluorescent protein fusion that causes a similar phenotype. Electron microscopy was performed as described previously (13). We measured resistance to lysozyme according to the method described in reference 3. The percentage of survivors after lysozyme assay was normalized to the wild type, and the data presented are an average of the results from either three or four experiments.
Immunofluorescence microscopy.
Immunofluorescence microscopy and deconvolution image processing were carried out in the laboratory of Kit Pogliano at the University of California at San Diego, according to the methods described in references 16 and 17. Fixation was carried out with paraformaldehyde alone, and Cy-5-conjugated anti-immunoglobulin G antibodies (Jackson Laboratories) were used as the secondary reagent. Cells were sporulated by resuspension (3), and aliquots were collected at the fourth hour of sporulation.
RESULTS
Insertion of an epitope tag alters the binding of CotG.
To assess the effects of the addition of the HA1 epitope tag on CotE, we examined spores from strain AD408, in which the epitope tag is inserted near the C terminus of CotE (7). AD408 spores appeared to be wild type by light microscopy. Electron microscopic examination revealed that the coats of these spores were similar in morphology to those of wild-type spores (Fig. 1B), although they often failed to fully encircle the spore, as previously reported (7).
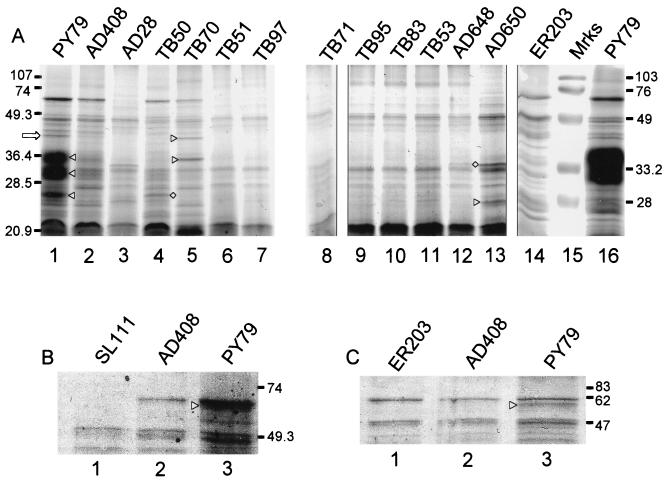
The protein composition of AD408 spores differed from that of wild-type spores, as determined by SDS-PAGE analysis of extractable spore proteins (Fig. 4A). Several species were absent or their amounts were significantly reduced, including species migrating as bands or tight clusters of bands of approximately 43, 36, 30, and 26 kDa (Fig. 4A, lane 2). To determine the identity of the 43-kDa band, we isolated protein from this band, after fractionation of wild-type spore extracts by SDS-PAGE, and carried out matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry on a peptide fragment generated after trypsin digestion. This analysis indicated that the 43-kDa band represents the GerE-controlled, CotE-dependent inner coat protein CotS (1, 29). The 36- and 30-kDa bands are likely to represent CotG (20), based on the apparent molecular masses and because bands of identical mobility were absent in an extract from cotG mutant spores (Fig. 4A, lanes 14 and 16). CotG is a 24-kDa protein, but it typically migrates as a broad band that is difficult to resolve, centered around 36 kDa. To further address the possibility that this protein is CotG, we asked whether CotB was also absent, as CotB relies on CotG for its assembly into the coat (20). To distinguish CotB from CotA, which usually migrated as a single band of about 60 kDa under these conditions of electrophoresis, we generated a strain that does not produce CotA, SL111, carrying cotEΔ::cat, amyE::cotEepi1, and ΔcotA::cat::spec. SDS-PAGE analysis of spore proteins from this strain indicated that CotB was reduced in amount or absent (Fig. 4B, lane 1), consistent with the view that the 36-kDa band is CotG. On rare occasions, CotA and CotB resolved as distinct bands. In these instances, we did not observe CotB in spores from a cotG null mutant strain (ER203) or from AD408 (Fig. 4C, lanes 1 and 2, respectively). The identity of the 26-kDa band is unknown, but it might be YrbB (see below). These results indicate that the addition of the epitope tag does not grossly impair the outer coat but does interfere with the assembly of at least three outer coat proteins: CotS and, most likely, CotB and CotG.
FIG. 4.
SDS-PAGE analysis of spore coat extracts. Coat protein extracts were electrophoresed on 15% polyacrylamide gels. Extracts in each lane were prepared from spores of the indicated strain. (A) Coomassie blue-stained acrylamide gel. Lane 15 contains markers. The open arrow indicates CotS and the triangles between lanes 1 and 2 indicate the positions of bands of about 36, 30, and 26 kDa, discussed in the text. The triangles between lanes 4 and 5 indicate the positions of YvdO (upper) and YveN (lower) and the diamond indicates YrbB. The diamond between lanes 12 and 13 indicates the 31-kDa band, and the triangle indicates the 25-kDa band. Molecular masses indicated on the left are for lanes 1 to 13, and those on the right are for lanes 14 to 16. (B) Upper region of an SDS-polyacrylamide gel showing the consequence of deletion of cotA in AD408. (C) An experiment in which CotA and CotB were successfully resolved. In panels B and C, triangles indicate the positions of CotB. Molecular masses are indicated in kilodaltons.
Mutations that result in a thin outer coat.
We further investigated the role of the C terminus of CotE in coat assembly with a series of deletion constructs. We found that spores of strains TB50 and TB70, in which the C-terminal 9 or 23 amino acids, respectively, were missing from CotE, were indistinguishable from wild-type spores by light microscopy (data not shown). By electron microscopic analysis, however, the coats of TB50 and TB70 spores were similar to each other but different from the coats of wild-type, AD408, or cotE null mutant spores. Unlike wild-type and AD408 spores, in which the outer coat is usually thick (Fig. 1A and B), TB50 and TB70 outer coats were usually thin (Fig. 1C and D), although not entirely absent, as they were from cotE null mutant spores (34).
We found significant differences in the SDS-PAGE profiles of extractable spore proteins from TB50 and TB70 compared to those of the wild type and AD408 (Fig. 4A, lanes 1, 2, 4, and 5). Notably, a species of close to 43 kDa was present in TB70 spores but was reduced or absent in TB50 and AD408 spores. We used MALDI-TOF mass spectrometry analysis to demonstrate that the protein in this band is encoded by yveN, an open reading frame identified during sequencing of the B. subtilis genome (11) and potentially encoding a protein with a predicted mass of 43.4 kDa. A band of about 35 kDa was present in TB70 spores but was undetectable in spores from TB50. MALDI-TOF mass spectrometry analysis indicated that this band represents the product of yvdO, encoding a protein with a predicted mass of 35.2 kDa. We also found that a 26-kDa species was readily detected only in spores from TB50 and the wild type. This band corresponds to the spore protein YrbB (30), as determined by MALDI-TOF mass spectrometry analysis. Immunoelectron microscopy analysis indicates that YrbB is associated with the cortex and the inner coat. Spores from TB50, and especially TB70, had reduced amounts of CotE relative to the wild type and AD408, as detected by Western blot analysis (Fig. 5). Lower levels of CotE did not prevent outer coat assembly, as a partial outer coat was assembled in the majority of these spores. Consistent with the structural and biochemical changes in the coats of TB50 and TB70, we found that lysozyme resistance, which relies on the integrity of the coat (2), was reduced to 80 and 37% of that of the wild type, respectively (Fig. 3). Therefore, the deletion of 9 or 23 amino acids from the C terminus of CotE significantly altered coat structure, biochemical composition, and function.
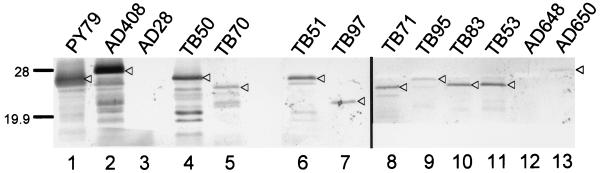
FIG. 5.
Western blot analysis of spore coat extracts. Coat protein extracts were electrophoresed on 15% polyacrylamide gels and transferred to polyvinylidene difluoride membranes. CotE was detected with anti-CotE antibodies. Extracts in each lane were prepared from spores from the indicated strain. The positions of CotE are indicated by triangles. Bands below CotE are likely to be degradation products. The difference in migration between CotE in spores from PY79 and AD408 and the similarity in mobility of PY79 and TB50 are due to the additional mass of the HA1 epitope. Molecular masses are indicated in kilodaltons.
Mutations that largely prevent outer coat assembly.
To further delineate functional regions of CotE, we constructed a large set of mutants in which various regions between amino acids 4 and 160 were deleted or altered by point mutation. This set included a C-terminal truncation mutant missing 37 amino acids (TB97), as well as five internal in-frame deletions, removing amino acids 4 to 29, 30 to 55, 58 to 75, 80 to 102, or 147 to 160 (in strain TB53, TB83, TB95, TB71, or TB51, respectively). We also identified two point mutants, from a random mutagenesis experiment, in which we screened for an apparent cotE null phenotype (see Materials and Methods). We found two such mutants, both of which had single-amino-acid alterations in the resulting versions of CotE: in one, the alteration of a valine to a glycine at amino acid 64 (AD648), and in the other, a change of a tyrosine to an asparagine at amino acid 66 (AD650). These amino acid changes fell within the region deleted in TB95 (amino acids 80 to 102).
These mutants were similar to each other and to a cotE null mutant by light microscopic examination (data not shown), suggesting a defect in coat formation. Consistent with this, we did not detect an outer coat in spores of any of these strains, similar to results obtained with cotE null mutant spores (data not shown). By SDS-PAGE analysis, the complements of spore proteins in these mutants were very similar to that of a cotE null mutant (Fig. 4A, lanes 3 and 6 to 13). One difference was a 31-kDa protein of unknown identity that was present in spores from the cotE null mutant, AD648, and AD650 (Fig. 4A, lanes 3, 12, and 13) but not the other mutants. Additionally, a band of about 25 kDa was prominent in AD650 spores but reduced in intensity or absent in spores from the other mutants in this set. As expected from spores missing a significant amount of the outer coat, the levels of lysozyme resistance varied from that of a cotE null mutant spore, 6% of the wild-type level, to 25% of the wild-type level (Fig. 3).
We had found that the deletion of the C-terminal 23 amino acids of CotE (in TB70) resulted in an outer coat with altered structure and composition but that the removal of 37 C-terminal amino acids (in TB97) largely eliminated the outer coat. This difference in phenotypes raised the question of whether residues between 146 and 158 were required for outer coat assembly. To test this possibility, we built TB51, bearing an in-frame deletion of amino acids 147 through 160. Our inability to detect an outer coat by electron microscopy (data not shown) or by examination of extracted spore proteins (Fig. 4A, lane 6) indicated that the region of CotE containing a stretch of largely acidic residues between amino acids 150 and 169 plays a necessary role in outer coat formation.
The levels of CotE in spores from this set of mutants varied considerably, as detected by Western blot analysis, and were significantly less than levels in wild-type or AD408 spores (Fig. 5). However, with the exception of AD648, for which we did not detect CotE in spores (Fig. 5, lane 12), it is unlikely that the low amounts of CotE in the mutant spores are solely responsible for the observed defects in outer coat assembly. For example, the levels of CotE in spores from TB95, TB97, and TB70 are similar, but the degree of coat assembly is very different; spores from TB95 and TB97 do not possess any detectable outer coat, but TB70 spores have a partial outer coat.
We also used Western blot analysis to measure the steady-state levels of CotE in extracts of sporangia of these strains, harvested at the fourth hour of sporulation (data not shown). As expected from the analysis of CotE in spores, we found relatively less CotE in the sporangia of mutants than in that of the wild type. For most of the mutants, the degree to which CotE steady-state levels were reduced in the sporangia of the mutants relative to the wild type was similar to the decrease of CotE in mutant spores, relative to wild-type spores. The exceptions were TB95 and AD650, in which the amounts of CotE in sporangial extracts were approximately 20-fold greater than the amounts in spores.
Localization of CotE to the spore.
The failure of most of the altered versions of CotE to foster outer coat protein deposition could be due to impairments in localization to the forespore. To address this, we used immunofluorescence microscopy to directly determine the location of CotE. We fixed aliquots of cells at the fourth hour of sporulation, after the majority of cells had undergone engulfment. Cells of each strain were treated with anti-CotE antibodies and examined by immunofluorescence deconvolution microscopy (9, 16, 23, 25). As a positive control, we treated separate aliquots of cells with anti-SpoIVA antibodies. SpoIVA localization should not be disturbed by mutations in cotE (7, 17). Consistent with the results of previous studies (17), we found that SpoIVA formed a ring around the forespore in all the cotE mutant strains (Fig. 6K and data not shown). spoIVA null mutant cells (AD18) showed a low level of fluorescence distributed throughout the cell (data not shown).
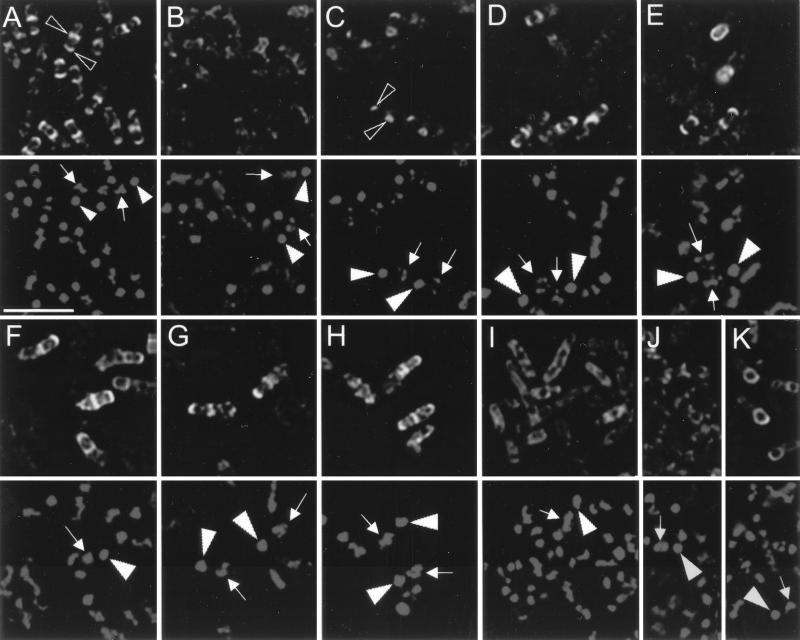
FIG. 6.
Immunofluorescence localization of CotE. At hour 4 of sporulation, cells were fixed for immunofluorescence microscopy, treated with either anti-CotE antibodies (panels A to J) or anti-SpoIVA antibodies (panel K), followed by a secondary Cy-5-conjugated antibody and then with the DNA stain DAPI (4′,6′-diamidino-2-phenylindole). Cells were examined for antibody staining (upper portion of each panel) or DNA staining (lower portion of each panel). Solid arrowheads indicate the more brightly fluorescing forespore chromosomes and the arrows indicate the diffuse mother cell chromosomes (17). Open arrowheads indicate positions of CotE decoration. (A) PY79; (B) AD28; (C) AD18; (D) AD408; (E) TB50; (F) TB51; (G) TB53; (H) TB83; (I) AD907; (J and K) AD648. Bar, 3 μm.
When we applied the anti-CotE antibody to wild-type cells, we saw caps and, very occasionally, rings of fluorescence surrounding the forespore in the majority of cells (Fig. 6A). We also saw clumps of fluorescence not associated with the forespore, often at the mother cell poles. It appeared that this additional fluorescence was largely restricted to nucleoid-free regions of the cytoplasm. This cap-like pattern of decoration is consistent with previous results achieved by using immunoelectron microscopy (7), fluorescence localization of a CotE-green fluorescent protein fusion (32), and immunofluorescence microscopy with a strain bearing a cotE-lacZ fusion and an anti-β-galactosidase antibody (17). When we examined cotE null mutant spores (AD28), we detected either clumps of faint fluorescence that occupied positions in the cytoplasm varying randomly from cell to cell (Fig. 6B) or no fluorescence at all. Very occasionally, we saw faint fluorescence around the forespore as well. As a further control, we examined the localization of CotE in a spoIVA null mutant strain (spoIVAΔ::neo). This mutation liberates CotE and the coat from the forespore, causing them to form a clump or a swirl in the mother cell cytoplasm (7, 15, 32). In this strain, we found that CotE no longer formed forespore-associated caps or rings, but rather formed detached clumps that sat between the nucleoids (Fig. 6C), consistent with results obtained in previous studies (7).
Localization of altered versions of CotE.
When we analyzed the location of CotE in the epitope-tagged version of CotE (AD408) we found caps of fluorescence around the forespore in the majority of cells, similar to those found in the wild type (Fig. 6D). We also found rings or caps of fluorescence encircling the forespores in all strains bearing altered versions of CotE, except for TB70 and AD648 (discussed below) (representative examples are shown in Fig. 6E to H). The number of cells that showed labeling with antibody varied from experiment to experiment, ranging from 10 to 50% of the number of wild-type cells that were decorated. Where binding occurred, it was similar in intensity and pattern to localization in wild-type cells. This lower percentage of binding does not necessarily indicate that CotE is inactive, as we saw this lower degree of binding in TB50 (Fig. 6E), in which some outer coat assembly still occurred (and, therefore, in which CotE was active) (Fig. 1C and 4A, lane 4). The failure of CotE to show localization in TB70 (data not shown) is surprising, in light of the presence of a partial outer coat in TB70 spores (Fig. 1D) and the accumulation of CotE in spores (Fig. 5). Possibly, this version of CotE adopts a conformation that prevents reaction with the antiserum under the relatively mild fixation conditions used for immunofluorescence microscopy.
We had demonstrated that a spoIVA mutation disrupted the cap-like pattern of CotE localization (Fig. 6C), as predicted by previous work (7). To show that spoIVA would likewise disrupt the caps formed by the altered versions of CotE, we generated strains harboring spoIVAΔ::neo in combination with each of the cotE alleles (AD903 to AD909 and AD911 to AD914; Table 1). We saw no caps or rings in these strains, confirming that the antibody staining was due to the presence of a coat-related structure (a representative case, AD907, is shown in Fig. 6I). Unlike the pattern of decoration seen in the spoIVA null mutant strain (Fig. 6C), we did not see bright foci of fluorescence, but rather faint fluorescence in nucleoid-free regions of the cells. Possibly, these versions of CotE are sufficiently impaired such that, unlike wild-type CotE, they are unable to aggregate in the absence of SpoIVA (7, 15). The dependency of CotE localization on SpoIVA supports the view that the caps of fluorescence accurately represent the position of CotE.
In contrast to the other mutants (except TB70, see above), we found that the majority of AD648 cells did not possess caps of fluorescence at the forespore after treatment with the anti-CotE antibody. Rather, the pattern of fluorescence resembled that of the cotE null mutant (Fig. 6, compare panel B with panel J). Staining with the anti-SpoIVA antibody showed rings around the forespores in these strains (Fig. 6K), confirming that the forespores in these cells were intact and could be immunodecorated. These data are consistent with our failure to detect CotE by Western blot in spores of this mutant (Fig. 5, lane 12).
DISCUSSION
The primary finding from this work is the effect of alterations in the C terminus of CotE on outer coat deposition. The addition of an epitope tag at the C terminus of CotE (as in AD408) reduced or eliminated the presence of several spore coat proteins. Among them are the CotE-dependent inner coat protein CotS and two proteins likely to be the known outer coat components CotG and CotB. Clearly, the region between amino acids 158 and 181 plays a role in outer coat assembly, as the deletion of 9 or 23 amino acids of the C terminus of CotE (in TB50 and TB70) resulted in a partially formed outer coat with a significantly altered polypeptide profile. In contrast to changes within the 23 C-terminal amino acids, mutations between amino acids 4 and 160 appeared to largely prevent outer coat assembly without preventing localization of CotE to the forespore.
Although the version of CotE in AD648 fails to localize, it is unlikely that the mutation in this allele defines a region of specific contact with the matrix. This is because the version of CotE produced in TB95 localizes to the forespore but is missing the amino acid changed in AD648. We speculate that the valine-to-glycine change at residue 64 in AD648 alters the conformation of CotE such that binding to the matrix is prevented.
It is possible that the phenotypes we observed were, in part, a result of the lower levels of the altered forms of CotE. However, the lower amounts of CotE are unlikely to be the predominant cause of these phenotypes since, as noted above, the apparent functionality of CotE did not correlate with the steady-state level of CotE in either spores or sporangia. For example, spores of TB53, which do not assemble any detectable outer coat, have more CotE than those of TB70, which do assemble a partial outer coat. Likewise, as noted above, the ability of CotE to localize is not strictly related to the level of CotE on the spore. AD650, whose spores have the lowest level of CotE of any of the mutants, is able to assemble CotE at the forespore, as judged by immunofluorescence microscopy. Therefore, the phenotypes of the mutant strains are, to a significant degree, the result of alterations in the ability of CotE to bind outer coats and to associate with the spore.
A direct interaction between the CotE shell and every outer coat protein seems unlikely. Rather, it would appear more plausible that CotE directly contacts a relatively small number of morphogenetic proteins that help link the outer coat proteins together. CotG, which requires CotE for its assembly and directs the assembly of the outer coat protein CotB, is a reasonable candidate for such a factor (20). In support of this idea, we found that the incorporation of a 36- and a 30-kDa protein, likely to be CotG, was reduced in AD408 as well as in the other mutants. However, cotG spores are deficient in only a small subset of outer coat proteins. Therefore, it is likely that additional morphogens participate in outer coat protein deposition.
ACKNOWLEDGMENTS
Immunofluorescence deconvolution microscopy was carried out at the laboratory of Kit Pogliano by Kit Pogliano and Marc Sharpe. We are deeply indebted to them and to Joseph Pogliano for their critical input and generosity in making their facilities available to us. MALDI-TOF mass spectrometry analysis was performed by the PAN facility (Stanford University) under the direction of Alan J. Smith. We thank Jean Greenberg for critically reading the manuscript; Patrick Stragier, Alan Grossman and Ezio Ricca for gifts of strains; John Foster for alerting us to seamless cloning; Fran Catalano for strain construction; Margarita Correa for preparing the anti-SpoIVA antibody; and Simon Foster and Anne Moir for very helpful discussions.
This work was supported by Public Health Service grant GM539898 from the National Institutes of Health and by the Schweppe Foundation. A.G.S. was funded, in part, by a Schmitt dissertation fellowship.
REFERENCES
- 1.Abe A, Koide H, Kohno T, Watabe K. A Bacillus subtilis spore coat polypeptide gene, cotS. Microbiology. 1995;141:1433–1442. doi: 10.1099/13500872-141-6-1433. [DOI] [PubMed] [Google Scholar]
- 2.Aronson A I, Fitz-James P. Structure and morphogenesis of the bacterial spore coat. Bacteriol Rev. 1976;40:360–402. doi: 10.1128/br.40.2.360-402.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cutting S M, Vander Horn P B. Molecular biological methods for Bacillus. Chichester, United Kingdom: John Wiley & Sons Ltd.; 1990. [Google Scholar]
- 4.Donovan W, Zheng L, Sandman K, Losick R. Genes encoding spore coat polypeptides from Bacillus subtilis. J Mol Biol. 1987;196:1–10. doi: 10.1016/0022-2836(87)90506-7. [DOI] [PubMed] [Google Scholar]
- 5.Driks, A. Unpublished observations.
- 6.Driks A. The Bacillus subtilis spore coat. Microbiol Mol Biol Rev. 1999;63:1–20. doi: 10.1128/mmbr.63.1.1-20.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Driks A, Roels S, Beall B, Moran C P, Jr, Losick R. Subcellular localization of proteins involved in the assembly of the spore coat of Bacillus subtilis. Genes Dev. 1994;8:234–244. doi: 10.1101/gad.8.2.234. [DOI] [PubMed] [Google Scholar]
- 8.Field J, Nikawa J, Broek D, MacDonald B, Rodgers L, Wilson I A, Lerner R A, Wigler M. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol Cell Biol. 1988;8:2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiraoka Y, Sedat J W, Agard D A. Determination of three-dimensional imaging properties of a light microscope system. Partial confocal behavior in epifluorescence microscopy. Biophys J. 1990;57:325–333. doi: 10.1016/S0006-3495(90)82534-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karmazyn-Campelli C, Fluss L, Leighton T, Stragier P. The spoIIN279(ts) mutation affects the FtsA protein of Bacillus subtilis. Biochimie. 1992;74:689–694. doi: 10.1016/0300-9084(92)90141-z. [DOI] [PubMed] [Google Scholar]
- 11.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Danchin A, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 12.LeDeaux J R, Grossman A D. Isolation and characterization of kinC, a gene that encodes a sensor kinase homologous to the sporulation sensor kinases KinA and KinB in Bacillus subtilis. J Bacteriol. 1995;177:166–175. doi: 10.1128/jb.177.1.166-175.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Margolis P S, Driks A, Losick R. Sporulation gene spoIIB from Bacillus subtilis. J Bacteriol. 1993;175:528–540. doi: 10.1128/jb.175.2.528-540.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Padgett K A, Sorge J A. Creating seamless junctions independent of restriction sites in PCR cloning. Gene. 1996;168:31–35. doi: 10.1016/0378-1119(95)00731-8. [DOI] [PubMed] [Google Scholar]
- 15.Piggot P J, Coote J G. Genetic aspects of bacterial endospore formation. Bacteriol Rev. 1976;40:908–962. doi: 10.1128/br.40.4.908-962.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pogliano J, Osborne N, Sharp M D, Abanes-De Mello A, Perez A, Sun Y L, Pogliano K. A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol Microbiol. 1999;31:1149–1159. doi: 10.1046/j.1365-2958.1999.01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pogliano K, Harry L, Losick R. Visualization of the subcellular localization of sporulation proteins in Bacillus subtilis using immunofluorescence microscopy. Mol Microbiol. 1995;18:459–470. doi: 10.1111/j.1365-2958.1995.mmi_18030459.x. [DOI] [PubMed] [Google Scholar]
- 18.Price K D, Losick R. A four-dimensional view of assembly of a morphogenetic protein during sporulation in Bacillus subtilis. J Bacteriol. 1999;181:781–790. doi: 10.1128/jb.181.3.781-790.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roels S, Driks A, Losick R. Characterization of spoIVA, a sporulation gene involved in coat morphogenesis in Bacillus subtilis. J Bacteriol. 1992;174:575–585. doi: 10.1128/jb.174.2.575-585.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sacco M, Ricca E, Losick R, Cutting S. An additional GerE-controlled gene encoding an abundant spore coat protein from Bacillus subtilis. J Bacteriol. 1995;177:372–377. doi: 10.1128/jb.177.2.372-377.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 22.Sandman K, Kroos L, Cutting S, Youngman P, Losick R. Identification of the promoter for a spore coat protein gene in Bacillus subtilis and studies on the regulation of its induction at a late stage of sporulation. J Mol Biol. 1988;200:461–473. doi: 10.1016/0022-2836(88)90536-0. [DOI] [PubMed] [Google Scholar]
- 23.Scalettar B A, Swedlow J R, Sedat J W, Agard D A. Dispersion, aberration and deconvolution in multi-wavelength fluorescence images. J Microsc. 1996;182:50–60. doi: 10.1046/j.1365-2818.1996.122402.x. [DOI] [PubMed] [Google Scholar]
- 24.Schaaper R M. Mechanisms of mutagenesis in the Escherichia coli mutator mutD5: role of DNA mismatch repair. Proc Natl Acad Sci USA. 1988;85:8126–8130. doi: 10.1073/pnas.85.21.8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaw P. Deconvolution in 3-D optical microscopy. Histochem J. 1994;26:687–694. doi: 10.1007/BF00158201. [DOI] [PubMed] [Google Scholar]
- 26.Stevens C M, Daniel R, Illing N, Errington J. Characterization of a sporulation gene spoIVA involved in spore coat morphogenesis in Bacillus subtilis. J Bacteriol. 1992;174:586–594. doi: 10.1128/jb.174.2.586-594.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stöver A G, Driks A. Secretion, localization, and antibacterial activity of TasA, a Bacillus subtilis spore-associated protein. J Bacteriol. 1999;181:1664–1672. doi: 10.1128/jb.181.5.1664-1672.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 29.Takamatsu H, Chikahiro Y, Kodama T, Koide H, Kozuka S, Tochikubo K, Watabe K. A spore coat protein, CotS, of Bacillus subtilis is synthesized under the regulation of ςK and GerE during development and is located in the inner coat layer of spores. J Bacteriol. 1998;180:2968–2974. doi: 10.1128/jb.180.11.2968-2974.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takamatsu H, Hiraoka T, Kodama T, Koide H, Kozuka S, Tochikubo K, Watabe K. Cloning of a novel gene yrbB, encoding a protein located in the spore integument of Bacillus subtilis. FEMS Microbiol Lett. 1998;166:361–367. doi: 10.1111/j.1574-6968.1998.tb13913.x. [DOI] [PubMed] [Google Scholar]
- 31.Warth A D, Ohye D F, Murrell W G. The composition and structure of bacterial spores. J Cell Biol. 1963;16:579–592. doi: 10.1083/jcb.16.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Webb C D, Decatur A, Teleman A, Losick R. Use of green fluorescent protein for visualization of cell-specific gene expression and subcellular protein localization during sporulation in Bacillus subtilis. J Bacteriol. 1995;177:5906–5911. doi: 10.1128/jb.177.20.5906-5911.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Youngman P, Perkins J B, Losick R. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid. 1984;12:1–9. doi: 10.1016/0147-619x(84)90061-1. [DOI] [PubMed] [Google Scholar]
- 34.Zheng L, Donovan W P, Fitz-James P C, Losick R. Gene encoding a morphogenic protein required in the assembly of the outer coat of the Bacillus subtilis endospore. Genes Dev. 1988;2:1047–1054. doi: 10.1101/gad.2.8.1047. [DOI] [PubMed] [Google Scholar]
- 35.Zheng L, Losick R. Cascade regulation of spore coat gene expression in Bacillus subtilis. J Mol Biol. 1990;212:645–660. doi: 10.1016/0022-2836(90)90227-d. [DOI] [PubMed] [Google Scholar]